Abstract
Introduction
Lenz microphthalmia syndrome (LMS) is a genetically heterogeneous X-linked disorder characterised by microphthalmia/anophthalmia, skeletal abnormalities, genitourinary malformations, and anomalies of the digits, ears, and teeth. Intellectual disability and seizure disorders are seen in about 60% of affected males. To date, no gene has been identified for LMS in the microphthalmia syndrome 1 locus (MCOPS1). In this study, we aim to find the disease-causing gene for this condition.
Methods and results
Using exome sequencing in a family with three affected brothers, we identified a mutation in the intron 7 splice donor site (c.471+2T→A) of the N-acetyltransferase NAA10 gene. NAA10 has been previously shown to be mutated in patients with Ogden syndrome, which is clinically distinct from LMS. Linkage studies for this family mapped the disease locus to Xq27-Xq28, which was consistent with the locus of NAA10. The mutation co-segregated with the phenotype and cDNA analysis showed aberrant transcripts. Patient fibroblasts lacked expression of full length NAA10 protein and displayed cell proliferation defects. Expression array studies showed significant dysregulation of genes associated with genetic forms of anophthalmia such as BMP4, STRA6, and downstream targets of BCOR and the canonical WNT pathway. In particular, STRA6 is a retinol binding protein receptor that mediates cellular uptake of retinol/vitamin A and plays a major role in regulating the retinoic acid signalling pathway. A retinol uptake assay showed that retinol uptake was decreased in patient cells.
Conclusions
We conclude that the NAA10 mutation is the cause of LMS in this family, likely through the dysregulation of the retinoic acid signalling pathway.
INTRODUCTION
Lenz microphthalmia syndrome (LMS, also known as MCOPS1, MIM 309800) is a rare multisystem condition defined by the canonical features of unilateral or bilateral microphthalmia or anophthalmia and defects in the skeletal and genitourinary systems.1,2 Anomalies of the digits, teeth, and ears are also hallmarks of the disease. Intellectual disability ranges from mild to severe with self-mutilating behaviours and seizures in severely affected individuals. The multi-organ abnormalities of the disorder suggest that causative genes play a central role in biological pathways essential to human development.
LMS is an X-linked, genetically heterogeneous disorder. At least two disease loci have been described. The microphthalmia syndromic 1 locus (MCOPS1; OMIM 309800) was mapped in 1991 and again in 2001 to Xq27-q28,3,4 while the gene for the microphthalmia syndromic 2 locus (MCOPS2; OMIM 300166) showed linkage to Xp11.4–p21.2 in 2002.5,6 MCOPS1 has remained elusive since its initial mapping to Xq27-q28 more than 20 years ago. Using exome sequencing of three brothers with LMS we have re-evaluated this previously characterised family4 and now identify a mutation in the NAA10 gene as a disease-causing locus for MCOPS1.
NAA10 (ARD1) codes for the catalytic subunit (Naa10p) of the hNaTA complex. NAA10 transfers an acetyl group from acetyl-CoA to proteins with Ser, Ala, Thr, Gly, and Val N-termini.7 NAA10 is thought to acetylate about 8000 different proteins.8
Human NAA10 is ubiquitously expressed9 and absence of NAA10 expression is embryonic lethal. A single mutation in NAA10, p.Ser37Pro, resulting in reduced N-terminal acetyltransferase (NAT) function was reported to cause a perinatal lethal phenotype, known as Ogden syndrome, in affected males from two unrelated families.10 In addition to lethality, other clinical features included aged appearance, craniofacial abnormalities, hypotonia, global developmental delay, and cardiac arrhythmia. This study exemplifies NAA10′s importance in early development. Given the diversity of NAA10 function, it is clear that a mutation of this gene could result in multiple organ abnormalities observed in patients affected with LMS.
SUBJECTS AND METHODS
Affected individuals and controls
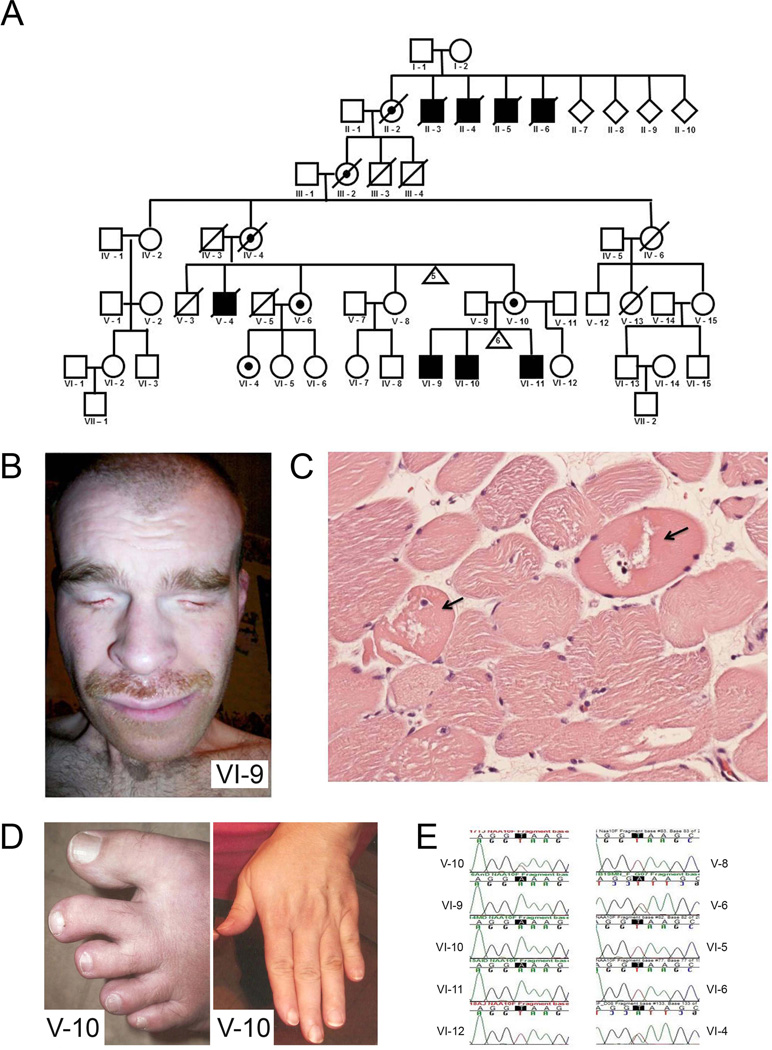
Written informed consent for genetic analysis was obtained from patients in accordance with the University of California, Irvine Institutional Review Board (IRB) (2005–4253) or from the National Human Genome Research Institute IRB. Clinical features of the family discussed in this study have been previously reported.4 Briefly, three brothers and a maternal uncle had congenital anophthalmia, delayed motor development, hypotonia, and mental retardation.4 They also have abnormal ears, high-arched palate, pectus excavatum, finger and toe syndactyly, clinodactyly, fetal pads, scoliosis, and cardiac and renal abnormalities.4 An obligate carrier has abnormal ears and syndactyly of the second to third toes bilaterally,4 probably due to the presence of X chromosome skewing. Linkage and haplotype analysis of this family suggested that the gene is located in a 17.65-cM region of Xq27.1-q28 (MCOPS1) with a logarithm of odds (LOD) score of 1.2. Based on the LOD score, MCOPS2 (Xpter-Xq24) was not likely linked.4 The updated pedigree and clinical information are shown in figure 1A.
Figure 1. Pedigree of family affected with Lenz microphthalmia syndrome (LMS), supporting clinical features, and confirmation by Sanger sequencing.
(A) Pedigree of LMS affected family. Further experimentation and analyses focused on the three living affected brothers (VI-9, VI-10, VI-11), one obligate heterozygote (V-10), one heterozygote identified previously by linkage (V-6) and her daughter (VI-4). Analysis of family pedigree was consistent with sex-linked inheritance. (B) Bilateral anophthalmia is an example of a dysmorphic feature present in patients affected by LMS (VI-9). (C) Haematoxylin and eosin staining of skeletal muscle showing neuropathic degeneration (arrow). (D) Dysmorphic features such as cutaneous syndactyly between the second and third toes and short terminal phalanges are present in a heterozygote (V-10). (E) Sanger sequencing of NAA10 gene (genomic DNA) shows the c.471+2T→A mutation present in all three affected males (VI-9, VI-10, VI-11), their obligate heterozygote mother (V-10), as well as an aunt who was previously suspected as a heterozygote by linkage (V-6) and one of her daughters (VI-4).
Whole exome sequencing
We pursued whole exome sequencing of three affected individuals (VI-9, VI-10, VI-11). Total DNA was obtained from peripheral blood of the patients and controls using standard protocols. Total DNA was sheared into approximately 300 bp fragments using a Covaris sonicator (Covaris). A paired-end exome library for Illumina sequencing was prepared using the Truseq DNA kit (Illumina) following the manufacturer’s instructions. Exonic regions covering most of the human genome were captured with the Illumina Exome Enrichment kit, following the manufacturer’s protocol. Massively parallel sequencing was performed using the Illumina HiSeq 2000 (Illumina) at the UCI High Throughput Facility. The data analysis was performed using the CLC Bio Genomics Workbench software.11 Reads were mapped to the human reference genome Hg19 to ensure optimal alignment conditions. We then extracted all X chromosomal reads from the 24 chromosome alignment and realigned only the paired-end reads to the X chromosome reference sequence (~3.9 M paired-end reads). Subsequently single nucleotide polymorphism (SNP) and deletion/insertion polymorphism (DIP) calling was performed only for the X chromosome alignment and parameters were set as follows: Due to the fact that all probands are male we chose a haploid algorithm. We increased the window length to 19 bp (determines the length of the initial segmentation of the genome, in base pairs, to analyse) and chose a minimum variant frequency of 75%. Although we had only aligned paired-end reads, the minimum paired coverage was set to 1, and maximum coverage was set to 500. SNP and DIP tables were filtered using CLC Bio discarding intergenic variants focusing on variants within known genes. We predicted the variant responsible for LMS to be rare, and therefore unlikely to be previously identified. Polymorphic variants from the 1000 Genome Project and variants from dbSNP were filtered out to distinguish potentially pathogenic mutations (rare) from other common variants using the ANNOVAR gene annotation and common variant filtering tool (http://www.openbioinformatics.org/annovar)12. We defined new variants as ones that did not exist in the dbSNP database (dbSNP132) or the 1000 genome project (1000g2010nov) and ultimately followed up on variants common to all three individuals found within an exon or a splice donor– acceptor site.
Mutation validation
Sanger sequencing of PCR amplicons from genomic DNA was used to confirm the candidate variants identified via exome sequencing. Genomic DNA was extracted from whole blood via the Gentra Puregene Blood Kit (Qiagen) following the manufacturer’s protocol. PCR of the region of interest was performed using the forward primer, GTGGAGGAGGGTGTCCCCGG, in intron 6 and the reverse primer, GGTCGTGACTCCTGGCAACGT, in intron 7, resulting in a 273 bp amplicon. PCR conditions were as follows: 95°C for 2 min, 95°C for 20 s, 63°C for 9 s, and 72°C for 20 s with 35 cycles and a final extension time of 20 s. Sanger sequencing was performed on the ABI 3130 Genetic Analyzer (Life Technologies) using primers from both directions.
RT-PCR analysis
Total RNA from control and patient fibroblasts and skeletal muscle was isolated using the RNeasy Plus Mini Kit (Qiagen) following the manufacturer’s instructions. Fifty nanograms of total RNA were used for cDNA synthesis and amplification using SuperScript One-Step RT-PCR with Platinum Taq (Invitrogen) with the following primers: NAA10 cDNA amplicon F1 (TGTGAAGCGTTCCCACCGGC), NAA10 cDNA F2 (GAAGAGTAACCGGGCCGCCC), NAA10 cDNA amplicon R1 (CCTCGCGACAGGCCTCTCCT), and NAA10 cDNA R2 (CCAGGCCCTTCTCCTCGCGA).
Quantitative real-time PCR analysis
RNA was isolated and purified using the RNeasy Plus Mini Kit (Qiagen), according to the manufacturer’s recommendations. SYBR Green quantitative real-time PCRs using the SuperScript One Step RT-qPCR kit (Invitrogen) were performed in triplicate for each primer set using the ABI 7900 HT Sequence Detection System (Life Technologies). Primers were designed to span introns or at exon junctions. To ensure specificity of PCR, melt curve analyses were performed at the end of all PCRs. Gene expression levels were normalised to GAPDH and then analysed using the 2ΔΔCt method.
Affymetrix gene expression array
For microarray analysis, RNA was extracted from control and patient fibroblasts using the RNeasy Kit (Qiagen). RNA quality was determined using the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, California, USA). Microarray analysis was performed using Human Gene 1.0 ST arrays (Affymetrix), according to the manufacturer’s guidelines. The data were then normalised and analysed using Cyber-T (http://cybert.ics.uci.edu/) and Partek (http://www.partek.com/microarray) software. Pathway and Bio-function analysis was performed using Ingenuity (Ingenuity Systems Inc) in order to put into context the differentially expressed probe sets. The following criteria were used: a value of p<0.001 and a minimum fold change > 1.5 (273 probe sets).
Western blot
Cultured fibroblast cells were washed with phosphate buffered saline (PBS) and lysed with lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA pH 8.0, 1% Triton X-100, and one protease inhibitor tablet). Protein concentrations were determined using a Bradford Assay (Bio-Rad). Forty micrograms of each lysate were resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) on a NuPAGE 4–12% gradient Bis-Tris gel (Invitrogen) and electroblotted onto a polyvinylidene difluoride membrane (Invitrogen). Membranes were incubated in 5% blocking buffer (non-fat dry milk in PBS containing 0.1% Tween) for 3 h at room temperature. Membranes were then blotted with either rabbit polyclonal anti-NAA10 (1:200; Santa Cruz Biotechnology) or rabbit polyclonal anti-tuberin (1:2000; Santa Cruz Biotechnology) antibody overnight at 4°C. Membranes were then washed three times with PBS and incubated with goat anti-rabbit horseradish peroxidase (HRP) conjugated secondary antibody (1:2000; Pierce) for 1.5 h at 4°C. HRP was detected using ECL Western Blotting Detection Reagents (Amersham Biosciences). Blots were stripped and re-probed with mouse anti-GAPDH antibody to verify equal loading.
Co-immunoprecipitation
293T cells were co-transfected with pcDNA3-TSC2 and pcDNA3-myc-NAA10 or its mutant, pcDNA-myc-NAA10-mutant, using Lipofectamine 2000 according to manufacturer’s instructions (Invitrogen). Forty-eight hours later, cells were harvested and lysed with 250 µL lysis buffer see above. Fifty microlitres of each lysate was placed aside as input. The cell lysates were pre-cleared by incubation with pre-washed protein G beads (Santa Cruz Biotechnology) for 2 h at 4°C. After centrifugation at 3000 rpm for 3 min, the supernatants were collected and incubated overnight at 4°C with 1 µg of anti-TSC2 (Santa Cruz Biotechnology) or anti-Myc antibody (Clonetech). The next day, samples were incubated with pre-washed protein G beads for 2 h at 4°C. Afterward, the beads were washed five times using lysis buffer and finally resuspended in SDS-PAGE loading buffer, heated to 100°C for 5 min, and analysed by western blotting using either anti-TSC2 or anti-Myc antibodies.
Immunocytochemistry
For immunofluorescence staining, fibroblast cells were fixed with 4% paraformaldehyde for 20 min at room temperature and washed three times in PBS. The fixed cells were blocked in 10% blocking buffer (10% goat serum, 0.1% Triton X-100 in PBS) for 1.5 h at room temperature before overnight incubation with rabbit anti-NAA10 antibody (1:50; Santa Cruz Biotechnology) at 4°C. The rabbit anti-NAA10 antibody was raised against amino acids 1–235, representing the full length NAA10 of human origin. After overnight incubation, cells were washed three times for 5 min in PBS and incubated with goat anti-rabbit Alexa fluor 488 (Molecular Probes, Invitrogen) in 10% blocking buffer for 1.5 h at 4°C. Cell nuclei were visualised by 4′-6-diamidino-2-phenylindole (DAPI, 1:400) staining. Immunofluorescence images were captured on a Nikon Eclipse Ti Inverted Microscope.
MTT assay
Cell proliferation was determined by the conventional 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) reduction assay using an MTT assay kit (Sigma) according to the manufacturer’s instructions. Briefly, patient fibroblast cells were plated at a density of 1×104 cells/well of a 12-well plate in triplicate. On days 1, 3, 5, and 7, the MTT solution was diluted in fresh medium and added to the cells. After the cells were incubated for 3.5 h in the dark, the medium was removed and the formazan dye crystals were solubilised in 1 mL of dimethyl sulfoxide (DMSO). Absorbance at 570 nm was measured with a microplate reader. The data were analysed with a Student t test. Significance was defined by a p value <0.05, n=3.
Cell growth competition assay
To compare the growth rate of patient cells, a competition assay was performed. Equal numbers of wild-type (control) fibroblast cells and fibroblast cells from patient VI-11 were mixed and a total of 100 000 cells were plated in one well of a 6-well plate. The cells were passaged several times when they reached confluence. Each time the cells were passaged, a portion was collected for mutant DNA analysis. Sanger sequencing was performed as described above. The ratio of wild type versus mutant cells was determined by chromatogram analysis; n=3.
Lentivirus production
Lentivirus was produced by transient co-transfection of three plasmids into 293T cells. Briefly, 293T cells were plated onto poly-D-lysine coated plates at approximately 80–90% confluence. 293T cells were transfected using Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer’s protocol. A total of 16 µg of plasmid DNA per 100 mm dish was used: 3 µg of the VSV-G envelope plasmid, pMD.G, 12 µg of the packaging plasmid, pCMVR8.91, and 9 µg of the lentiviral transducing vector. The medium was replaced 24 h after transfection with virus-collecting medium (Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS)). The viral supernatant was collected at 48 and 72 h after transfection and filtered through 0.45 µm filters. Virus was immediately added to fibroblast cells.
Generation of NAA10 knockdown fibroblast cell lines
Control and NAA10 shRNA plko. 1-lentiviral constructs were a generous gift from Dr Anand Ganesan. Twenty-four hours before transduction, 1×105 normal dermal human fibroblasts (NDHFs) were plated onto 6-well plates. The following day control and NAA10 shRNA lentivirus was supplemented with 6 µg/mL polybrene, and added to the cells. Forty-eight hours after transduction, puromycin (1 µg/mL) was used to select for transduced cells. Once the cell lines were expanded, NAA10 knockdown was verified by immunofluorescence and SYBR Green quantitative real-time PCR (qRT-PCR).
Assay of 3H-retinol uptake from 3H-retinol/RBP
Production of 3H-retinol/retinal binding protein (RBP ) and 3H-retinol uptake assay were performed as previously described.13 Briefly, wild-type (control) and patient fibroblast cells were washed with Hank’s balanced salt solution (HBSS) before incubation with 3H-retinol/RBP diluted in serum-free medium at 37°C. The reaction was stopped by removing the medium, washing the cells with HBSS, and solubilising the cells in 1% (w/v) Triton X-100 in PBS. Radioactivity was measured with a scintillation counter.
RESULTS
Pedigree analysis and clinical examination of family
The recently updated pedigree information included seven generations with a total of 10 participants in this family of mixed European descent, including German, Italian, and other northern European backgrounds (figure 1A). Analysis of the pedigree was consistent with X-linked inheritance. Only affected males without male-to-male transmission were observed. 4 Physical examinations were recently performed and clinical information was updated. At the most recent evaluation, there were three living affected brothers (VI-9, VI-10, VI-11), one obligate carrier (V-10), one carrier identified previously by linkage (V-6), and her daughter who was also a carrier (VI-4). All three affected individuals presented with anophthalmia or microphthalmia, prenatal onset of growth deficiency, and significant dysmorphic features (figure 1B). Although the affected male individuals presented with LMS phenotypes, there was also significant intra-familial variation. The affected males all developed scoliosis to varying degrees. Individuals VI-9 and VI-11, but not VI-10, had surgical procedures to correct scoliosis. Individuals VI-9 and VI-10, but not VI-11, developed body hair to an uncomfortable degree, requiring frequent shaving. In the past, individual VI-10 presented as energetic, outgoing, and physically strong, but at the time of the last evaluation displayed a loss of energy, increased muscle stiffness, loss of short term memory, and a worsened autism-like behaviour. A few months before to his evaluation, he had two strokes and an MRI showed arterial sclerosis with a 50% narrowing of the carotid artery. Individual VI-11 was the most severely affected and has a seizure disorder. He has not developed words, and relies completely on sign language. In addition, haematoxylin and eosin (H&E) staining of his skeletal muscle showed muscle degeneration (figure 1C). He also required surgical intervention to lengthen his heel cord.
Three heterozygotes (VI-4, V-6, V-10) had cutaneous syndactyly between the second and third toes and short terminal phalanges (figure 1D). These manifestations were not seen in the individual without the mutation. We concluded that the phenotypes of the three heterozygotes were causally related to the hand anomalies of the male individuals. Therefore, the condition could be considered as X-linked with reduced expressivity in the heterozygotes.
Exome sequencing identifies a novel splice site mutation in NAA10
To identify the gene mutated in this family, we first obtained informed consent from all patients and female carriers in accordance with the UC Irvine Human Subjects Committee. Although the locus in this family was linked to chromosome Xq27-q28 within an interval of 17.25 cM, the LOD score was only 1.2.4 The region was defined by one mildly affected individual.4 Since a small percentage of patients with LMS have a mutation in the BCOR gene on Xp11.4, patient VI-9 underwent BCOR sequencing, and no mutation was detected. An Affymetrix SNP6 chip did not identify any significant duplications or deletions. The 17.25 cM interval on Xq27-q28 where the MCOPS1 gene was originally mapped proved to be gene-rich, containing more than 100 genes. Many of these genes are expressed in the developing eye. Some of these genes were previously sequenced but no mutations were detected.4
We pursued exome sequencing of the three affected male individuals (VI-9, VI-10, VI-11) on the Illumina HiSeq 2000 sequencer. The resulting exome library resulted in 186 860 022 reads of which 179 177 640 mapped to the human reference sequence (95.9%). We extracted all X chromosomal reads from the 24 chromosome alignment and realigned only the paired-end reads to the X chromosome reference sequence (~3.9 M paired-end reads). The CLC Bio alignment and ANNOVAR filtering, as described in the methods section, resulted in 32/42 SNP/DIP variants in VI-9, 37/42 SNP/DIP variants in VI-10, and 37/36 SNP/DIP variants in VI-11. Five SNPs and 14 DIPs showed overlap among all three affected male individuals. Only one variant fulfilled the criteria to be either in an exon or a splice junction. This novel variant was in the NAA10 gene. This variant predicts a mutation at the intron 7 GT splice donor site (c.471+2T→A) of NAA10. The NAA10 splice mutation was confirmed via Sanger sequencing in all three affected males (VI-9, VI-10, VI-11), their obligate heterozygote mother (V-10), as well as an aunt who was previously suspected as a heterozygote by linkage (V-6) and one of her daughters (VI-4) (figure 1E). NAA10 is in Xq27, but was not inside the region previously defined by DXS1232 and DXS8043 markers. This may be the reason why no mutations were detected when sequencing the genes within these markers in the study by Forrester et al.4 The NAA10 c.471+2T> A mutation was not found in any of the four unaffected individuals in this family (V-8, VI-5, VI-6, VI-12) (figure 1E). Therefore, the mutation segregated with the disease in this pedigree.
NAA10 mutant splice site results in loss of wild-type NAA10 expression and multiple splice variants
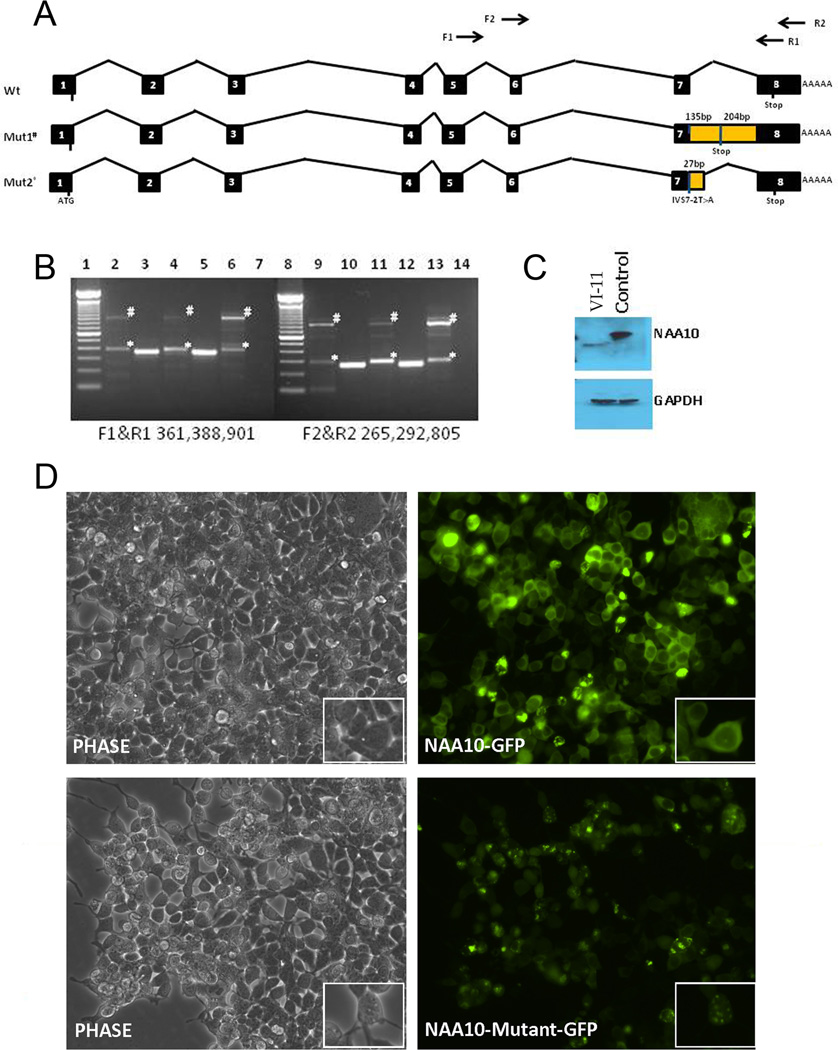
The +2T position of the GT splice donor site is highly conserved in the splice site consensus sequence, predicting that the c.471 +2T→A mutation severely alters exon 7 splicing. Although predicting the exact resulting splice mutation products would be difficult, we can anticipate that this mutation may lead to alternative splicing and aberrant transcripts. In mice, three isoforms of NAA10 have been identified consisting of 235, 225, and 190 amino acids.14 Generated by alternative splicing, these isoforms differ in the C-terminal region. In humans, however, alternative splicing events have not been reported for NAA10 and only a 235-full length amino acid form has been identified. To determine whether aberrant splicing is present in the affected males, RNA was isolated and RT-PCR was performed using primers that extended from exon 5 through exon 8 (figure 2A). While no NAA10 wild-type band was present in any of the affected male patient samples, we did observe at least five bands representing aberrant sizes of mRNA (figure 2B). Two primer sets were used in the RT-PCR reactions resulting in similar banding patterns with a 94 base pair size difference. We gel-purified the RT-PCR products for subsequent Sanger sequencing; sufficient quantities of cDNA could be isolated for only two out of the five bands (figure 2B; # and *). Sequencing of the 901/805 base pair band (#) showed it as the exon7-intron7-exon8 fusion transcript; mutant splice variant 1. Sequencing of the 388/292 base pair band (*) showed it as the mutant splice variant 2, having activated a cryptic splice site at position c.471+27 that resulted in a 27 nucleotide in-frame insertion derived from intron 7.
Figure 2. Transcript analysis of mutant NAA10 and implications of exon 8 truncation.
(A) Intron/exon structure of NAA10 and variant transcripts present in patient VI-11 with Lenz microphthalmia syndrome. (B) RT-PCR analysis of NAA10 transcripts in control and patients VI-9 and VI-11. Total RNA from patient VI-9 blood, patient VI-11 fibroblasts and skeletal muscle and control fibroblasts was isolated and used for cDNA synthesis and amplification of two NAA10 cDNA amplicons. No NAA10 wild-type band was present in any of the affected male patient samples. Mutant splice variant 1 and 2 are indicated by # and *, respectively. Lanes 2–6: Amplicon F1&R1 (exon 5 and exon 8); lane 1, 100 bp ladder; lane 2, VI-9 blood; lane 3, control blood; lane 4, patient VI-11 muscle; lane 5, control fibroblasts; lane 6, patient VI-11 fibroblasts; lane 7, negative control, no template; lane 8, 100 bp ladder. Lanes 9–14: Amplicon F2&R2 (exon 5/6 junction and exon 8); lane 9, VI-9 blood; lane 10, control blood; lane 11, patient VI-11 muscle; lane 12, control fibroblasts; lane 13, patient VI-11 fibroblasts; lane 14, negative control, no template. (C) Western blot analysis of NAA10 expression in normal and patient VI-11 fibroblast cells. Left lane, NAA10 from patient VI-11; right lane, control fibroblast. #, Splice mutant variant 1. Blots were stripped and re-probed with mouse anti-GAPDH antibody to verify equal loading as shown in the lower panel. (D) Immunofluorescence analysis of wild-type and mutant-NAA10 cellular localisation. NAA10-GFP and mutant-NAA10-GFP were over-expressed in 293T cells. Mutant NAA10-GFP aggregates in the cytoplasm. Top panel, left, phase contrast, right, NAA10-GFP; bottom panel, left, phase contrast, right, NAA10-mutant-GFP.
To test if any of these mutant isoforms produced protein, fibroblast cells derived from patient VI-11 were subjected to western blot analysis with a rabbit anti-human NAA10 antibody. As shown in figure 2C, a lower molecular weight band was visible in individual VI-11 derived fibroblast cells when compared to normal fibroblast cells. This lower molecular weight band may represent the read through mutant splice variant 1, resulting in the truncation of exon 8 and the addition of 45 amino acids derived from intron 7. The transcript with a 27 nucleotide insertion resulting in the addition of nine amino acids from intron 7, at the protein level (mutant splice variant 2), was not detected in fibroblast cells. It is important to note that there is no detection of wild-type NAA10 protein in the fibroblast cells from the affected male patient, coinciding with the RT-PCR data showing the lack of a wild-type band in his mRNA. Expression analysis using an Affymetrix array showed a reduction of NAA10 mRNA levels for all three affected male patient fibroblast samples, as compared to controls (data not shown).
To test if the c.471+2T→A mutation affected NAA10 cell localisation we performed immunofluorescence on control and affected individual, VI-11 derived, fibroblasts. NAA10 has been shown to be located within the cytoplasm and the nucleus.9 Results from immunofluorescence indicated that endogenous NAA10 protein is located within the cytoplasm and nucleus of control and patient VI-11 fibroblast cells, suggesting that this mutation does not affect cellular localisation (data not shown). However, the staining pattern of endogenous NAA10 in the fibroblast cells of patient VI-11 was suggestive of the formation of protein aggregates. To further investigate whether this punctate staining was a result of the NAA10-mutant, we created a mutant NAA10-GFP fusion protein containing the addition of 45 amino acids past exon 7, followed by the truncation of the original 79 amino acids from exon 8 (mutant splice variant 1). Over-expression of mutant NAA10-GFP fusion protein in 293T cells resulted in punctate staining suggestive of protein aggregation within the cytoplasm, as shown in figure 2D (insets; Mutant NAA10-GFP), further suggesting that the mutant NAA10 aggregated in the cytoplasm.
Patient fibroblasts with NAA10 splice site mutation are defective in cell proliferation
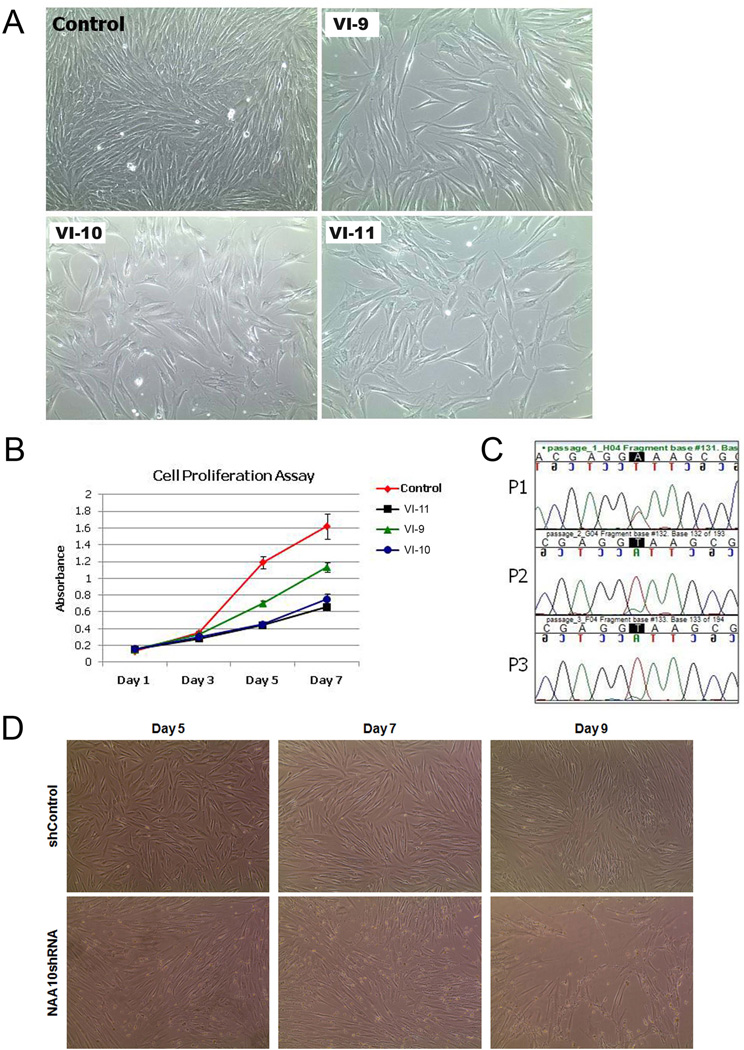
NAA10 has been shown to play a role in cell proliferation. In addition, all the affected male individuals in this pedigree had severe prenatal and postnatal growth deficiency, suggesting that cell proliferation may be affected by the c.471+2T→A mutation. To test if this mutation causes cellular growth deficiency, we plated an equal amount of control and patient fibroblast cells and observed whether they reached confluence at the same time. As shown in figure 3A, by the fifth day of culture, the growth rate of fibroblast cells from the affected male individuals appeared slower than in the normal control. To confirm and quantify this observation, we performed an MTT assay (figure 3B). Results from the MTT assay confirmed that the proliferation rate of fibroblast cells from the affected male individuals were dramatically reduced as compared to the normal control (p<0.01) (figure 3B), suggesting that the loss of wild-type NAA10 may inhibit cell proliferation. We also created a cell growth competition assay. To do this, we mixed an equal number of fibroblast cells, from control and affected patient VI-11, in one 6-well plate. Each time the cells reached confluence, they were passaged and a portion of the cells were harvested for DNA isolation. Since cell growth rate can be estimated by the sequencing chromatogram, the mutation was sequenced at every time point. As shown in figure 3C, after two passages in the mixed population, there was no detectable trace of the mutant in the sequence chromatogram, suggesting that the fibroblast cells with the NAA10 mutation grew more slowly than the wild-type control and were rapidly taken over during competition.
Figure 3. NAA10 mutation c.471+2T→A causes growth deficiency.
(A) Cell proliferation of patient VI-9, VI-10, and VI-11 fibroblasts as compared to control fibroblasts. An equal number of patient and fibroblast cells were plated and phase images were obtained on day 5 of culture; n=3. (B) MTT assay. Control and patient VI-9, VI-10, and VI-11 fibroblast cells were used for the MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay. 1×105 cells were plated for each cell type, in triplicate, and the assay was performed on days 1, 3, 5, and 7; n=3. (C) Cell growth competition assay. Equal number of control and patient fibroblast cells were mixed and plated in a single well of a 6-well plate. Cells were collected at every passage, for three passages, and DNA purified. The percentage of wild-type or patient cells present at each passage was estimated by chromatogram analysis at the c.471+2T→A mutation site. (D) NAA10 knockdown in normal human fibroblast cells affects cell proliferation. Normal human fibroblast cells were transduced with either shControl or NAA10 shRNA virus. Phase images were obtained on days 5, 7, and 9 after selection with puromycin. By day 9, NAA10 knockdown fibroblast cells became unstable and began to die while shControl fibroblast cells continued to proliferate.
To verify that the loss of NAA10 expression is directly responsible for the decrease in cell proliferation, NAA10 knockdown experiments were performed. NAA10 was knocked down in NDHF cells, using a shRNA lentivirus system. Initially, we started with five different NAA10 shRNA constructs. However, after transduction with the various NAA10 shRNA constructs, four out of five cell lines became apoptotic and proceeded to die within a week of transduction. Fibroblast cells transduced with control shRNA were normal (figure 3D). The only surviving NAA10 knockdown cell line was tested for knockdown by qPCR and showed over 90% reduction in NAA10 mRNA expression (data not shown). After an additional week, the only remaining NAA10 shRNA cell line also became unstable and began to die (figure 3D). Although we could not confirm the extent of NAA10 knockdown in the other cell lines, these results have been duplicated and may suggest that a high efficiency knockdown of NAA10 is lethal.
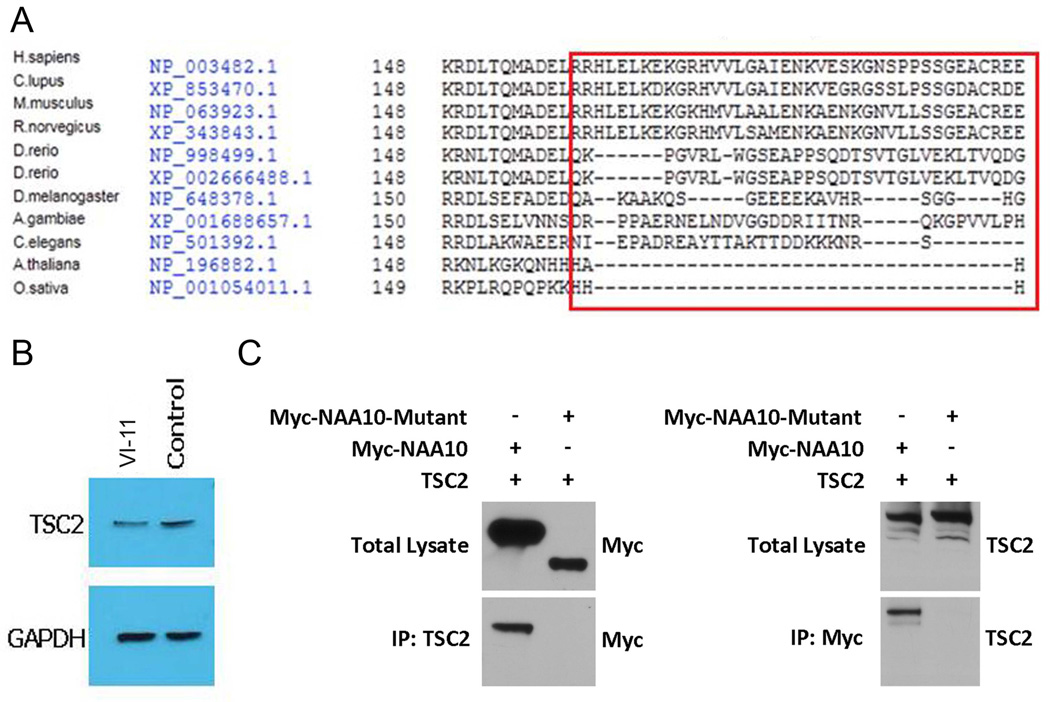
The function of NAA10 in cell proliferation is controversial. Several groups have found that NAA10 is essential for cell proliferation, particularly in several cancer cell lines. Therefore, NAA10 could be an oncogene.16–18 ~ Other groups have found the opposite, 19 suggesting that this function is cell line dependent. NAA10 has been identified as a substrate of IKK-β.20 IKK-β mediated phosphorylation of amino acid Ser209 on NAA10 destabilises NAA10 and induces its degradation 20. Therefore truncating the protein before this site will prevent IKK-β mediated degradation and may cause a concomitant up-regulation of NAA10 activity. In addition, the amino acid sequence encoded by exon 8 of NAA10 is conserved in mammals (figure 4A), suggesting that loss of exon 8, as in the mutant splice variant 1, could be detrimental to its function. It has been recently reported that the C-terminus of NAA10 is required for interaction with TSC2, an inhibitor of the mTOR (mammalian target of rapamycin) pathway 21 After forming a complex with TSC2, NAA10 acetylates TSC2, both stabilising TSC2 and increasing its cellular concentration.21 NAA10 may therefore play an important role in the regulation of the mTOR pathway and loss of exon 8 could disrupt this regulation. To test if the mutation c.471+2T→A in the NAA10 gene (truncating exon 8 and adding of 45 amino acids) affects the stability of TSC2, we examined TSC2 expression in fibroblast cells derived from patient VI-11 by western blot with a rabbit anti-human TSC2 antibody. TSC2 levels were decreased significantly compared to a normal control (figure 4B). The NAA10-TSC2 interaction was further tested by co-immunoprecipitation. Mutant NAA10 failed to precipitate TSC2 (figure 4C), suggesting that the interacting domain of NAA10 was truncated. These results suggested that the c.471 +2T→A mutation truncated exon 8 and leads to the loss of TSC2 binding and a reduction of TSC2 protein levels.
Figure 4. Mutation of NAA10 abolishes the interaction with TSC2.
(A). Conservation of amino acid sequence encoded by exon 8 of NAA10. Alignment was done using the Interactive Structure based Sequences Alignment Program Strap (http://3d-alignment.eu/). (B) Mutant NAA10 fails to precipitate TSC2. Western blot analysis of TSC2 expression in a male patient with Lenz microphthalmia syndrome. Left lane, TSC2 from patient VI-11; right lane, TSC2 from control fibroblast. Blots were stripped and re-probed with mouse anti-GAPDH antibody to verify equal loading as shown in the lower panel. (C) Co-immunoprecipitation of NAA10 and TSC2. Immunoprecipitation assays were performed using either anti-TSC2 or anti-Myc antibodies followed by western blot analysis. Mutant NAA10 failed to precipitate TSC2 and vice versa.
Mutant NAA10 expression array shows dysregulation of retinoic acid signalling pathways
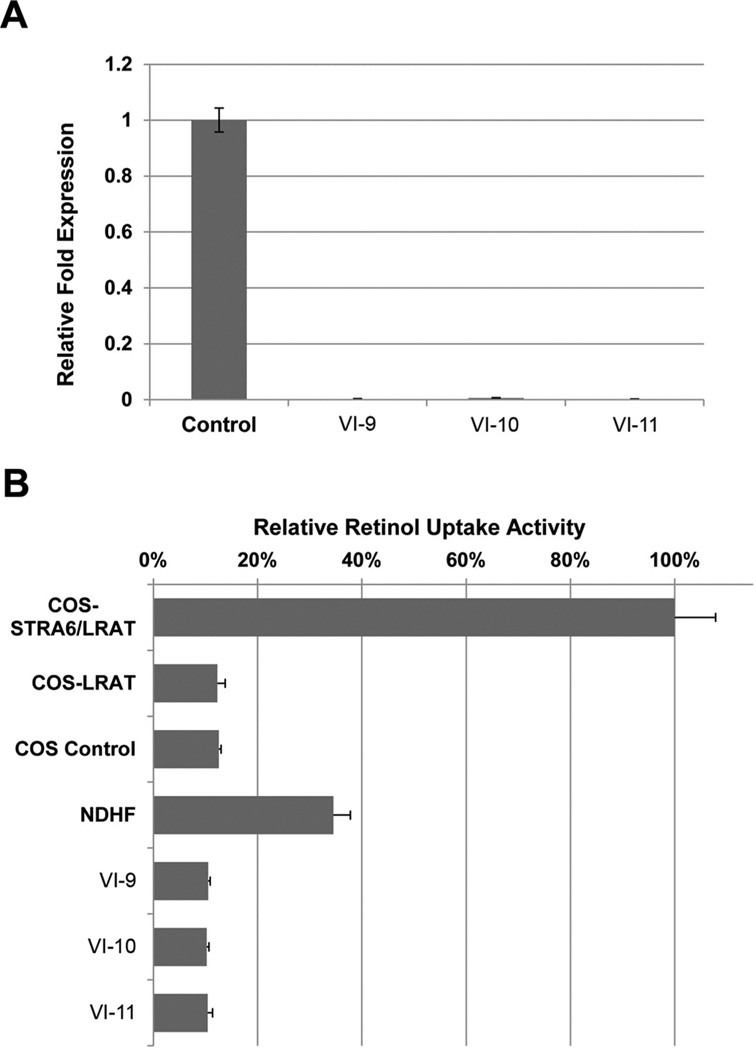
To identify the molecular pathways that may be disrupted by this mutation in NAA10 and further understand the pathogenesis of this disease, we performed gene expression profiles on fibroblasts from the three affected male patients and two normal controls using the human Affymetrix gene array and Ingenuity pathway and bio-function analysis. Since it has been suggested that NAA10 can affect up to 8000 genes, we were not surprised to see a large number of genes severely dysregulated; approximately 800 genes were identified exhibiting significant fold difference in expression levels compared to controls (p<0.002). NAA10 was included among these dysregulated genes. Genes involved in eye development are particularly interesting since one of the most striking features of LMS is microphthalmia/anophthalmia. Therefore, genes involved in eye development were our focus in the pathway analysis. It has been shown that the retinoic acid and canonical wnt signalling pathways play a critical role in eye development. Genes associated with these pathways, such as STRA6, FOXF1, HOXC8, and WNT2B were among the most highly dysregulated genes in the affected male patient samples. In addition, we performed analysis, using the Ingenuity software, to classify the differentially expressed genes into physiological functions. From our analysis, the most significant physiological functions are related to the following developmental categories: tissue development, embryonic development, organ development, skeletal and muscular system development. The genes associated with the retinoic acid and canonical wnt signalling pathways are listed within these highly significant biofunctions, (see online supplemental file 1). Mutations in the RBP4 (retinol binding protein) receptor, STRA6,22 cause the Matthew Wood syndrome (MCOPS9, OMIM 601186),23,24 a condition that overlaps with the LMS phenotype. STRA6 binds the RBP4-retinol (holo-RBP4) complex and mediates cellular uptake of vitamin A,22 which is the precursor to retinoic acid, a developmental morphogen. The RBP4 disease-associated mutations abolish STRA6’s vitamin A uptake activity.25 Data analysis from the human expression array showed that the STRA6 expression level was significantly reduced in all three of the affected male patients’ (VI-9, VI-10, VI-11) fibroblast cells, as compared to the controls (data not shown). This result was validated by SYBR Green qRT-PCR (figure 5A). To determine if the down-regulation of STRA6 affected the cellular uptake of retinol in the affected male patient samples, a retinol uptake assay was performed. Our results showed that retinol uptake from retinol/RBP4 complex was decreased significantly in patient VI-9, VI-10, or VI-11 fibroblast cells compared with normal controls (p<<0.01) (figure 5B). We conclude that the NAA10 c.471+2T→A mutation causes LMS through perturbation of the retinoic acid signalling pathway.
Figure 5. Retinol uptake is dramatically affected in patient fibroblast cells.
(A) Quantitative real-time PCR analysis of STRA6 expression. RNA was isolated from control and patient VI-9, VI-10, and VI-11 fibroblast cells. STRA6 is significantly down-regulated in patient fibroblast cells. (B) Retinol uptake in control and patient VI-9, VI-10, and VI-11 fibroblast cells. COS untransfected and COS eGFP/LRAT were used as negative controls. COS bSTRA6/LRAT was used as a positive control for the assay.
DISCUSSION
We have used exome sequencing to identify a novel NAA10 splice mutation, c.471+2T→A, in a family with LMS. This mutation resulted in no detectable normally spliced NAA10 at the RNA or protein level and produced a small amount of aberrant transcript and truncated protein. In addition, the NAA10 c.471+2T→A mutation caused fibroblast cell growth defects. This mutation disrupted association with TSC2, a protein known to interact with wild-type NAA10 through its C-terminal domain.21 The mutant protein had normal cellular localisation but tended to aggregate in the cytoplasm when over-expressed. Lastly, an Affymetrix expression array showed significant dysregulation of genes involved in the retinoic acid and WNT signalling pathways, both of which play important roles in neural and retinal development. Although exome sequencing covers only 95% of exons and the non-coding regions are generally not covered, the above studies provide convincing evidence that NAA10 mutation is disease-causing in this family.
NAA10 plays an important role in basic cell function and human development, and has been associated with Ogden syndrome.10 A c.109T>C (p.Ser37Pro) variant in NAA10 has been recently identified as a cause for Ogden syndrome, resulting in the lethality of male infants due to a deficiency in NAT activity. Data mining using in situ hybridisation and microarray gene expression databases,26,27 for human and mouse, showed that NAA10 is ubiquitously expressed in the eyes, the skeletal and genitourinary systems, the digits, teeth, ears, and central nervous system—tissues in which the development defects in our patients were observed (data not shown).
Due to the diverse functions of NAA10, it is no surprise that different mutations in this gene can have heterogeneous effects. A distinct difference between Ogden syndrome and LMS, besides lethality, is the effect of the NAA10 mutation on eye development. While individuals affected with Ogden syndrome had normal globe size eyes, patients with LMS had microphthalmia or anophthalmia. This major difference suggests that the NAA10 mutation identified here has important effects on pathways involved in eye development. Unlike the p.Ser37Pro mutation causing lethal Ogden syndrome, which directly affects the N-terminal of NAA10 and its NAT activity, the mutation, c.471 +2T>A, truncates the C-terminal end of NAA10, maintaining the N-acetyltransferase catalytic domain, resulting in a milder phenotype. The loss of exon 8 (C-terminal) affects NAA10’s ability to interact with other proteins, resulting in the dysregulation of important pathways. Thus, this study suggests that different mutations in NAA10 can cause different syndromes.
How the deletion of exon 8 affects NAA10 function is not completely understood. The serine residue at position 209 (Ser209), in NAA10, was shown to be a phosphorylation site of IKK-β.20 Phosphorylation by IKK-β destabilises NAA10 and induces its proteasome mediated degradation.20 Deletion of this IKK-β phosphorylation site is associated with NAA10 stability.20 In addition, immunofluorescence of mutant-NAA10 over-expression showed that the NAA10 variant aggregated within the nucleus and cytoplasm, suggesting that the loss of exon 8 causes conformational changes in the protein structure that result in aggregation. The c.471+2T>A splice site mutation, identified in this study, truncates the C-terminal end of NAA10 and keeps the N-acetyltransferase catalytic domain intact, suggesting that the NAT activity may be maintained. However, c.471+2T>A splice site mutation in affected patient cells causes no expression of wild-type NAA10 and minimal expression of the mutated NAA10. Even if the truncated mutant NAA10 protein maintains N-acetyltransferase activity, the overall activity would be significantly less than normal, considering its expression level is minimal compared to expression in wild-type cells (figure 2C). In addition, it has been reported that the C-terminus of NAA10 is required for interaction with TSC2, an inhibitor of the mTOR pathway. After forming a complex with TSC2, NAA10 acetylates TSC2. Data from our mutant-NAA10 and TSC-2 co-immunoprecipitation experiments support the proposal that the c.471+2T>A mutation truncates exon 8 and leads to the loss of TSC2 binding and a reduction of TSC2 protein levels (figure 4C). The ability of NAA10 to form a complex with TSC2 and acetylate it was impeded. This experiment suggests that the c.471+2T>A mutation not only exhibits diminished N-acetyltransferase activity, but also losses other functions, such as interacting with TSC2.
Although western blot and RT-PCR analyses showed that there is no detectable amount of normal NAA10 on the RNA or protein level in affected male patient fibroblasts, there is expression of the truncated NAA10 protein. However, expression of the truncated NAA10 protein is dramatically less in the affected individuals when compared to the expression of wild-type NAA10 in a normal control (figure 2). Previous studies showed that NAA10 knockout is embryonic lethal.28 We can therefore infer that this hypomorphic NAA10 mutation (c.471+2T→A) provides sufficient activity to allow viability of primary patient cells as well as human survival, although significant growth deficiencies are present. This is further suggested based on an attempt to create NAA10 knockdown primary human fibroblast cell lines. Upon knockdown with five different NAA10 shRNA constructs, all five cell lines became unstable and eventually died while our control cell line survived. This result is consistent with another study in which NAA10 knockdown led to apoptosis and cell cycle arrest.29 However, a previous study by Fisher et al15 performed NAA10 RNAi experiments, using human liver carcinoma cells, which resulted in 35% and 85% NAA10 knockdown. Although cell proliferation decreased, these cells were still viable. This could be explained by the difference in cell types used, their tissue specific expression patterns, and the level of NAA10 knockdown. Even though >80% knockdown of NAA10 decreased cell proliferation, in HepG2 cells, it did not induce cell death,15 suggesting that the 20% residual activity of NAA10 is sufficient to maintain cell survival or that these cancer cells may activate signalling pathways that complement NAA10 deficiency, preventing apoptosis or cell cycle arrest.
Studies regarding the role NAA10 plays in regulating cell proliferation have been controversial. While some studies show that expression of NAA10 promotes cell proliferation,30–32 another study shows that it inhibits cell proliferation, primarily through regulation of the mTOR pathway. NAA10 has been shown to form a complex with TSC2, an inhibitor of the mTOR pathway.21 NAA10 interacts with acetylates and stabilises TSC2, repressing mTOR activity and inhibiting cell proliferation.21 Although we showed that the interaction between mutant-NAA10 and TSC2 was lost, causing a significant decrease in the stability and expression of TSC2, we still observed cell proliferation defects in our patient cells and shRNA knockdown cells. Thus, our data suggest that NAA10 may act on pathways other than mTOR to regulate cell proliferation. Further study of this subject may elucidate additional functions of NAA10.
Gene expression analysis performed on fibroblasts from the three affected male patients showed many dysregulated genes. Genes involved in the retinoic acid signalling pathway and the WNT pathway were among the most highly dysregulated genes. Retinoic acid and canonical Wnt signalling is critical for normal eye development. Specifically, retinoic acid signalling is important for the embryonic development of a number of organs such as the central nervous system, heart, eyes, ears, and olfactory organs, all of which are affected in male patients with LMS. Various studies have shown that cellular uptake of vitamin A/retinol from its RBP4-bound form is dependent on the membrane receptor protein, STRA6.13,22,33 In humans, mutations of STRA6 results in Matthew-Wood syndrome characterised by defects in the eye similar to those seen in patients with LMS.23,24 In the fibroblasts of the affected male patient, STRA6 expression was significantly reduced, as compared to controls, suggesting that these patients may have retinol uptake deficiencies. Further experimentation confirmed that patients VI-9, VI-10, and VI-11 had deficiencies in retinol uptake. These results suggest that NAA10 may play a role in the retinoic acid signalling pathway and in normal eye development. Further studies to elucidate the exact mechanism by which NAA10 regulates STRA6 and the retinoic acid signalling pathway would facilitate our understanding of the pathogenesis of this disease.
A recent study showed that a p.Ser37Pro NAA10 mutation caused lethality in male infants in two unrelated families.10 The clinical features included lethality, aged appearance, craniofacial abnormalities, hypotonia, global developmental delay, and cardiac arrhythmia. Affected individuals had prominent eyes but no other eye abnormalities were reported in Ogden syndrome.10 The Ogden syndrome mutation is in the N-acetylation catalytic domain and resulted in reduced NAT activity. Using quantitative in vitro N-terminal acetylation assays employing four different substrates, it was shown that there was a 20% and 80% reduction of NAT activity, depending on protein class.10 The results of this and the present study show that Ogden syndrome and LMS are allelic disorders. However, minimum overlap in clinical features suggests the diverse function of NAA10 and demonstrates that the level of enzymatic activity is crucial. We also sequenced the NAA10 gene in 11 male patients with microphthalmia and atypical Lenz syndrome but no mutations were found. A separate group sequenced the NAA10 gene in five microphthalmic patients, and again no mutations were found.
We conclude that the NAA10 splice mutation identified in the present family can cause LMS and explains a component of the know locus heterogeneity. However, additional loci have yet to be identified. In a recent study by Piton et al, 34 the authors used the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project to reassess the implications of 106 genes proposed to be involved in monogenic forms of XLID. According to this paper, NAA10 was one of 15 genes characterised as ‘awaiting replication’ because this gene had been recently described but not yet replicated. In addition, no contradictory findings were found in Exome Variant Server (EVS). Nevertheless, in this study, the identified splice donor mutation in NAA10 demonstrates the importance of the C-terminus in maintaining the functionality of NAA10, and also emphasises the role of NAA10 in regulating cell proliferation and retinoic acid signalling, providing valuable insight into the pathology of LMS.
Supplementary Material
Acknowledgements
We thank the patients and their family for participating in our study. The authors thank the UC Irvine Genomic High-Throughput Facility where the exome seguencing was performed. The authors would also like to thank Joseph Yoo, from the Division of Human Genetics at the Cincinnati Children’s Hospital Medical Center, for his help in modifying the figures.
Funding This work is supported in part by UC Irvine Foundation and NEI 1R01EY018876 (TH).
Footnotes
Contributors TE, TH conceived idea; TE, HR JLL, SD, VH, SM, BMS, JJJ, RK performed experiments; AT collected data; TE, TH, AB, JW, RBC, AS, JJJ, HS analysed data; TE, TH, LGB, HS, VK wrote the manuscript. The project was designed and supervised by TH.
Competing interests None.
Patient consent Obtained.
Ethics approval UC Irvine IRB committee.
Provenance and peer review Not commissioned; externally peer reviewed.
REFERENCES
- 1.Traboulsi EI, Lenz W, Gonzales-Ramos M, Siegel J, Macrae WG, Maumenee IH. The Lenz microphthalmia syndrome. Am J Ophthalmol. 1988;105:40–45. doi: 10.1016/0002-9394(88)90119-5. [DOI] [PubMed] [Google Scholar]
- 2.Bardakjian TM, Schneider AS, Ng D, Johnston JJ, Biesecker LG. Association of a de novo 16q copy number variant with a phenotype that overlaps with Lenz microphthalmia and Townes-Brocks syndromes. BMC Med Genet. 2009;10:137. doi: 10.1186/1471-2350-10-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graham CA, Redmond RM, Nevin NC. X-linked clinical anophthalmos. Localization of the gene to Xq27-Xq28. Ophthalmic Paediatr Genet. 1991;12:43–48. doi: 10.3109/13816819109023084. [DOI] [PubMed] [Google Scholar]
- 4.Forrester S, Kovach MJ, Reynolds NM, Urban R, Kimonis V. Manifestations in four males with and an obligate carrier of the Lenz microphthalmia syndrome. Am J Med Genet. 2001;98:92–100. [PubMed] [Google Scholar]
- 5.Ng D, Hadley DW, Tifft CJ, Biesecker LG. Genetic heterogeneity of syndromic X-linked recessive microphthalmia-anophthalmia: is Lenz microphthalmia a single disorder? Am J Med Genet. 2002;110:308–314. doi: 10.1002/ajmg.10484. [DOI] [PubMed] [Google Scholar]
- 6.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 7.Arnesen T, Anderson D, Baldersheim C, Lanotte M, Varhaug JE, Lillehaug JR. Identification and characterization of the human ARD1-NATH protein acetyltransferase complex. Biochem J. 2005;386(Pt 3):433–443. doi: 10.1042/BJ20041071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Starheim KK, Gromyko D, Velde R, Varhaug JE, Arnesen T. Composition and biological significance of the human Nalpha-terminal acetyltransferases. BMC Proc. 2009;3(Suppl 6):S3. doi: 10.1186/1753-6561-3-S6-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugiura N, Adams SM, Corriveau RA. An evolutionarily conserved N-terminal acetyltransferase complex associated with neuronal development. J Biol Chem. 2003;278:40113–40120. doi: 10.1074/jbc.M301218200. [DOI] [PubMed] [Google Scholar]
- 10.Rope AF, Wang K, Evjenth R, Xing J, Johnston JJ, Swensen JJ, Johnson WE, Moore B, Huff CD, Bird LM, Carey JC, Opitz JM, Stevens CA, Jiang T, Schank C, Fain HD, Robison R, Dalley B, Chin S, South ST, Pysher TJ, Jorde LB, Hakonarson H, Lillehaug JR, Biesecker LG, Yandell M, Arnesen T, Lyon GJ. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am J Hum Genet. 2011;89:28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng SB, Buckingham KJ, Lee C, Bigham AW, Tabor HK, Dent KM, Huff CD, Shannon PT, Jabs EW, Nickerson DA, Shendure J, Bamshad MJ. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawaguchi R, Yu J, Ter-Stepanian M, Zhong M, Cheng G, Yuan Q, Jin M, Travis GH, Ong D, Sun H. Receptor-mediated cellular uptake mechanism that couples to intracellular storage. ACS Chem Biol. 2011;6:1041–1051. doi: 10.1021/cb200178w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SH, Park JA, Kim JH, Lee JW, Seo JH, Jung BK, Chun KH, Jeong JW, Bae MK, Kim KW. Characterization of ARD1 variants in mammalian cells. Biochem Biophys Res Commun. 2006;340:422–427. doi: 10.1016/j.bbrc.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 15.Fisher TS, Etages SD, Hayes L, Crimin K, Li B. Analysis of ARD1 function in hypoxia response using retroviral RNA interference. J Biol Chem. 2005;280:17749–17757. doi: 10.1074/jbc.M412055200. [DOI] [PubMed] [Google Scholar]
- 16.Arnesen T, Starheim KK, Van Damme P, Evjenth R, Dinh H, Betts MJ, Ryningen A, Vandekerckhove J, Gevaert K, Anderson D. The chaperone-like protein HYPK acts together with NatA in cotranslational N-terminal acetylation and prevention of Huntingtin aggregation. Mol Cell Biol. 2010;30:1898–1909. doi: 10.1128/MCB.01199-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu M, Huang C, Xiang M, Lai J, Yang H, Ma M, Tan D. [hARD1 antiserum preparation and primary immunohistochemical analysis of hARD1 in tumor tissues] Sheng Wu Gong Cheng Xue Bao. 2008;24:1155–1161. doi: 10.1016/s1872-2075(08)60057-6. [DOI] [PubMed] [Google Scholar]
- 18.Jiang B, Ren T, Dong B, Qu L, Jin G, Li J, Qu H, Meng L, Liu C, Wu J, Shou C. Peptide mimic isolated by autoantibody reveals human arrest defective 1 overexpression is associated with poor prognosis for colon cancer patients. Am J Pathol. 2010;177:1095–1103. doi: 10.2353/ajpath.2010.091178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hua KT, Tan CT, Johansson G, Lee JM, Yang PW, Lu HY, Chen CK, Su JL, Chen PB, Wu YL, Chi CC, Kao HJ, Shih HJ, Chen MW, Chien MH, Chen PS, Lee WJ, Cheng TY, Rosenberger G, Chai CY, Yang CJ, Huang MS, Lai TC, Chou TY, Hsiao M, Kuo ML. N-alpha-acetyltransferase 10 protein suppresses cancer cell metastasis by binding PIX proteins and inhibiting Cdc42/Rac1 activity. Cancer Cell. 2011;19:218–231. doi: 10.1016/j.ccr.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 20.Kuo HP, Lee DF, Xia W, Lai CC, Li LY, Hung MC. Phosphorylation of ARD1 by IKKbeta contributes to its destabilization and degradation. Biochem Biophys Res Commun. 2009;389:156–161. doi: 10.1016/j.bbrc.2009.08.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo HP, Lee DF, Chen CT, Liu M, Chou CK, Lee HJ, Du Y, Xie X, Wei Y, Xia W, Weihua Z, Yang JY, Yen CJ, Huang TH, Tan M, Xing G, Zhao Y, Lin CH, Tsai SF, Fidler IJ, Hung MC. ARD1 stabilization of TSC2 suppresses tumorigenesis through the mTOR signaling pathway. Sci Signal. 2009;3:ra9. doi: 10.1126/scisignal.2000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, Sun H. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- 23.Pasutto F, Sticht H, Hammersen G, Gillessen-Kaesbach G, Fitzpatrick DR, Nurnberg G, Brasch F, Schirmer-Zimmermann H, Tolmie JL, Chitayat D, Houge G, Fernandez-Martinez L, Keating S, Mortier G, Hennekam RC, von der Wense A, Slavotinek A, Meinecke P, Bitoun P, Becker C, Nurnberg P, Reis A, Rauch A. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am J Hum Genet. 2007;80:550–550. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Golzio C, Martinovic-Bouriel J, Thomas S, Mougou-Zrelli S, Grattagliano-Bessieres B, Bonniere M, Delahaye S, Munnich A, Encha-Razavi F, Lyonnet S, Vekemans M, Attie-Bitach T, Etchevers HC. Matthew-Wood syndrome is caused by truncating mutations in the retinol-binding protein receptor gene STRA6. Am J Hum Genet. 2007;80:1179–1187. doi: 10.1086/518177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaguchi R, Yu J, Wiita P, Honda J, Sun H. An essential ligand-binding domain in the membrane receptor for retinol-binding protein revealed by large-scale mutagenesis and a human polymorphism. J Biol Chem. 2008;283:15160–15168. doi: 10.1074/jbc.M801060200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Richardson L, Venkataraman S, Stevenson P, Yang Y, Burton N, Rao J, Fisher M, Baldock RA, Davidson DR, Christiansen JH. EMAGE mouse embryo spatial gene expression database: 2010 update. Nucleic Acids Res. 2010;38(Database issue):D703–D709. doi: 10.1093/nar/gkp763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu C, Macleod I, Su Al. BioGPS and MyGene.info: organizing online, gene-centric information. Nucleic Acids Res. 2013;41(Database issue):D561–D565. doi: 10.1093/nar/gks1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu M, Gong J, Ma M, Yang H, Lai J, Wu H, Li L, Li L, Tan D. Immunohistochemical analysis of human arrest-defective-1 expressed in cancers in vivo. Oncol Rep. 2009;21:909–915. doi: 10.3892/or_00000303. [DOI] [PubMed] [Google Scholar]
- 29.Arnesen T, Gromyko D, Pendino F, Ryningen A, Varhaug JE, Lillehaug JR. Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein N-alpha-acetyltransferase complex. Oncogene. 2006;25:4350–4360. doi: 10.1038/sj.onc.1209469. [DOI] [PubMed] [Google Scholar]
- 30.Lee CF, Ou DS, Lee SB, Chang LH, Lin RK, Li YS, Upadhyay AK, Cheng X, Wang YC, Hsu HS, Hsiao M, Wu CW, Juan LJ. hNaa10p contributes to tumorigenesis by facilitating DNMT1 -mediated tumor suppressor gene silencing. J Clin Invest. 2010;120:2920–2930. doi: 10.1172/JCI42275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo JH, Cha JH, Park JH, Jeong CH, Park ZY, Lee HS, Oh SH, Kang JH, Suh SW, Kim KH, Ha JY, Han SH, Kim SH, Lee JW, Park JA, Jeong JW, Lee KJ, Oh GT, Lee MN, Kwon SW, Lee SK, Chun KH, Lee SJ, Kim KW. Arrest defective 1 autoacetylation is a critical step in its ability to stimulate cancer cell proliferation. Cancer Res. 2010;70:4422–4432. doi: 10.1158/0008-5472.CAN-09-3258. [DOI] [PubMed] [Google Scholar]
- 32.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 33.Golczak M, Maeda A, Bereta G, Maeda T, Kiser PD, Hunzelmann S, von Lintig J, Blaner WS, Palczewski K. Metabolic basis of visual cycle inhibition by retinoid and nonretinoid compounds in the vertebrate retina. J Biol Chem. 2008;283:9543–9554. doi: 10.1074/jbc.M708982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piton A, Redin C, Mandel JL. XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am J Hum Genet. 2013;93:368–383. doi: 10.1016/j.ajhg.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.