Abstract
The cullin-4 (CUL4) complex DCDC (DIM-5/-7/-9/CUL4/DDB1 complex) is essential for DNA methylation and heterochromatin formation in Neurospora crassa. Cullins form the scaffold of cullin-RING E3 ubiquitin ligases (CRLs) and are modified by the covalent attachment of NEDD8, a ubiquitin-like protein that regulates the stability and activity of CRLs. We report that neddylation is not required for CUL4-dependent DNA methylation or heterochromatin formation but is required for the DNA repair functions. Moreover, the RING domain protein RBX1 and a segment of the CUL4 C terminus that normally interacts with RBX1, the E2 ligase, CAND1, and CSN are dispensable for DNA methylation and heterochromatin formation by DCDC. Our study provides evidence for the noncanonical functions of core CRL components.
INTRODUCTION
Ubiquitination, the addition of ubiquitin moieties to proteins, is a multistep process that regulates the intracellular stability, localization, and function of numerous proteins (1, 2). Ubiquitin is activated by an E1 enzyme and then transferred to a substrate by an E2 ubiquitin-conjugating enzyme under the direction of multisubunit cullin-RING E3 ubiquitin ligases (CRLs) (3, 4). Cullins form the scaffold of CRLs by bridging substrate adaptor proteins that interact with their N termini and catalytic proteins that interact with their C termini (4).
Cullin-4 (CUL4) complexes control cell cycle progression, DNA repair, and signal transduction (5, 6). The Neurospora crassa DCDC (DIM-5/-7/-9/CUL4/DDB1 complex) is essential for methylation of lysine 9 of histone H3 (H3K9) and DNA methylation (6). The DCDC resembles established CRLs, in which the substrate specificity adaptor protein DDB1/DIM-8 (DNA-damage-binding protein 1) serves as a bridge between CUL4 and the DCAF (DDB1 and CUL4-associated factor) DIM-9. These members of the DCDC, in turn, associate with the histone methyltransferase DIM-5 and its partner, DIM-7 (6). In addition, as in validated CRLs, CUL4 is modified by attachment of the ubiquitin-like protein NEDD8 in DCDC (6). Similarly, the Schizosaccharomyces pombe fission yeast CUL4 complex CLRC directs H3K9 methylation, although the potentially ubiquitinated substrate for this complex remains elusive (7–9), as with DCDC.
Fruitless efforts to find putative substrates for ubiquitination by DCDC led us to explore the possibility that this complex has an ubiquitination-independent function. The C terminus of cullins, which is essential for the catalytic activity of CRLs, directly interacts with the E2 ligase and RBX1/ROC1, a small RING domain catalytic protein that facilitates the attachment of ubiquitin moieties onto substrates (10). The flexible backbone of cullins undergoes conformational changes to bring RBX1 and the E2 ligase in close proximity to the substrates, which are recruited by the adaptor protein bound to the N terminus (11). Covalent attachment of NEDD8 to an invariant lysine in the C terminus regulates the stability and activity of cullins (12–15). To explore the contribution of neddylation in the regulation of N. crassa CUL4 functions, we mutated the CUL4 NEDD8 attachment site and found, surprisingly, that the neddylation-site mutants have normal histone H3K9 methylation and DNA methylation, although CUL4-mediated DNA repair is compromised. We also report that a deletion of the C terminus of CUL4, including the neddylation site and a region mediating the interaction with the E2 ligase and RBX1, does not affect DNA methylation or heterochromatin formation; again, only DNA repair is compromised. Furthermore, a deletion of the gene coding for RBX1 does not affect DNA methylation, although DNA repair is disrupted. Taken together, our findings imply that N. crassa DCDC directs H3K9 and DNA methylation through an ubiquitination-independent mechanism.
MATERIALS AND METHODS
N. crassa strains were maintained, grown, and crossed using previously described procedures (16). Construction of various plasmids and strains with cul4 neddylation-site substitutions, hH2A substitutions, and an rbx1 deletion is described in the supplemental material. All strains and primers used in this study are listed in Tables S2 and S3 in the supplemental material, respectively. N. crassa transformation (17), DNA isolation (18), Southern blotting (19), and protein isolation and Western blotting (20) were performed as previously described. For drug sensitivity assays, serial dilutions of conidia were spot tested on medium with or without methyl methanesulfonate (MMS; 0.015%), camptothecin (CPT; 0.3 μg/ml), or thiabendazole (TBZ; 0.5 μg/ml), obtained from Sigma-Aldrich (6).
RESULTS AND DISCUSSION
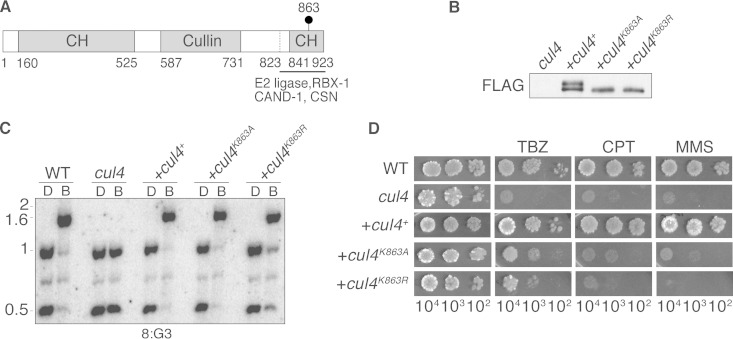
To test the possible role of CUL4 neddylation in DNA methylation and DNA repair, we replaced the predicted neddylated lysine (K863) with nonneddylatable residues and introduced the constructs, with or without a FLAG tag, at the his-3 locus of a cul4RIP1 null mutant (Fig. 1A; see also Fig. S1 in the supplemental material). The control strain, bearing FLAG-tagged wild-type cul4+, exhibited a protein doublet (Fig. 1B), presumably reflecting the neddylated and unneddylated forms of CUL4 (6, 21). Mutation of the predicted neddylation site eliminated the slower-migrating band (Fig. 1B). We tested DNA methylation in strains expressing mutant or wild-type forms of cul4. Strikingly, the DNA methylation defect of the cul4RIP1 null mutant was fully rescued by all of the constructs bearing mutations of K863 (Fig. 1C; see also Fig. S2 in the supplemental material). Neddylation-deficient mutants were also able to rescue the DNA methylation defect of a cul4 deletion mutant (see Fig. S3 in the supplemental material). Complementation of the DNA methylation defect implies that H3K9 trimethylation (H3K9me3) was also restored, as H3K9 methylation is required for DNA methylation in N. crassa (22, 23). After we obtained some of the findings reported here, Zhao and colleagues reported a neddylation site mutation of N. crassa CUL4 and claimed that it interfered with H3K9 trimethylation (24). We do not consider the reported chromatin immunoprecipitation assay results to be robust, however, and it appears that neither DNA methylation nor global H3K9 methylation was examined.
FIG 1.
Neddylation of CUL4 is dispensable for DNA methylation but not for DNA repair. (A) Schematic of CUL4 showing the NEDD8 attachment site (K863), cullin and cullin homology (CH) domains, and C-terminal region thought to interact with NEDD8 attachment machinery, E2 ligase, RBX1, CAND1, and CSN. (B) Western blots of extracts from an untransformed cul4RIP1 strain and cul4RIP1 strains expressing the indicated FLAG-CUL4 constructs. The strains are listed in Table S2 in the supplemental material. (C) DNA methylation analysis of wild-type (WT) and cul4RIP1 strains and cul4RIP1 strains expressing the indicated FLAG-tagged CUL4 constructs at the his-3 locus. DNA was digested with 5-methylcytosine-sensitive BfuCI (lanes B) or its 5-methylcytidine-insensitive isoschizomer, DpnII (lanes D), and the Southern blot was probed for methylated region 8:G3 (36); size standards (in kb) are indicated on the (left). Equivalent results were obtained for methylated regions 8:A6 and 5:B8 (36) (see Fig. S2 in the supplemental material). (D) Sensitivity of neddylation site mutants to DNA-damaging agents. Serial dilutions of conidia (indicated at the bottom) were tested on medium with or without MMS (0.015%), CPT (0.3 μg/ml), or TBZ (0.5 μg/ml).
CUL4 and DDB1, each of which is encoded by a single gene in N. crassa (25), interact with multiple substrate adaptors known as DCAFs (DDB1 and CUL4-associated factors) to carry out various functions (26). DIM-9/DCAF26 is required for the establishment of H3K9me3 by the DCDC (6, 21). To address the possibility that neddylation of N. crassa CUL4 might be required for another function, such as DNA repair, we tested the sensitivity of wild-type and K863 mutants to the alkylating agent methyl methanesulfonate (MMS) and the topoisomerase inhibitor camptothecin (CPT). The neddylation mutants, like the cul4RIP1 null mutant, were highly sensitive to both drugs (Fig. 1D), suggesting that neddylation of CUL4 is required for resistance to these drugs, presumably through DNA repair pathways. Interestingly, the neddylation-defective mutants showed less sensitivity to the microtubule inhibitor thiabendazole (TBZ) (6), consistent with heterochromatin formation being independent of CUL4 neddylation. To test if the absence of neddylation affects the integrity of DCDC, we performed a proteomic analysis of proteins immunoprecipitated with DIM-8–FLAG from the neddylation-site mutant (a cul4 mutant with K-to-A mutation at residue 863 [cul4K863A]; see Table S1 in the supplemental material). The recovery of all DCDC components implies that the neddylation of CUL4 is not required for DCDC formation.
The C termini of cullins are critical for their ubiquitin ligase activities (4, 13) and mediate interactions with the E2 ligases, a RING domain protein (ROC1/RBX1), CAND1, and the signalosome (4, 13) (Fig. 1A). Biochemical studies suggest that neddylation of cullins promotes polyubiquitination by increasing the affinity of cullins for the E2 ligase and RBX1 (14). The COP9 signalosome complex (CSN) removes NEDD8 from cullins to permit interaction of the cullin C terminus with CAND1 (13, 14, 27, 28), limiting ubiquitination and promoting the exchange of substrate receptors (28, 29). The absence of the COP9 signalosome causes the degradation of cullins by uncontrolled autoubiquitination (30). Interestingly, we found that DNA methylation is unaffected by the absence of an active signalosome (see the information and Fig. S4 in the supplemental material).
Recently, it was speculated that ubiquitination of histone H2A lysine 122 (H2AK122ub) by CRL4B may direct DNA methylation (31). We failed to detect ubiquitination of the corresponding lysine on N. crassa H2A (H2AK119) (see the supplemental material), consistent with observations in Saccharomyces cerevisiae (32), suggesting that H2A ubiquitination is absent, and we found that replacement of K119 with alanine, a nonmodifiable residue, does not affect DNA methylation (see Fig. S5 in the supplemental material). Clearly, DCDC does not operate by ubiquitination of H2AK119.
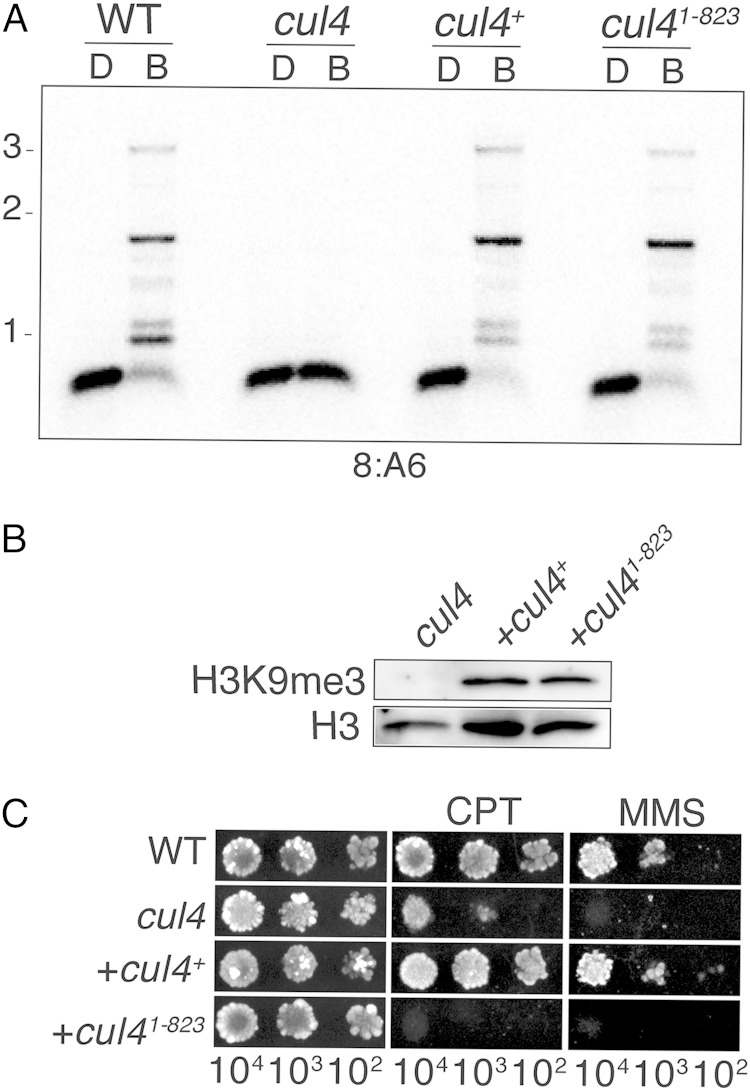
Our observation that neddlylation of CUL4 is not required for DNA methylation raised the possibility that N. crassa DCDC is not acting as a ubiquitin ligase for heterochromatin formation. To further explore this possibility, we generated and tested a deletion mutant that lacks the CUL4 C terminus (residues 824 to 923; Fig. 1A), including both the neddylation site and regions of interaction with CAND1 and RBX1. The deletion did not affect DNA methylation or H3K9me3 (Fig. 2A and B), suggesting that CUL4 is not involved in catalyzing ubiquitination in this context. In contrast, like the neddylation site mutant, the deletion mutant was sensitive to chemicals that cause DNA damage (Fig. 2C), consistent with a requirement for the ubiquitin ligase activity of a CUL4 complex for DNA repair.
FIG 2.
The CUL4 C terminus is not required for DCDC function. (A) Normal DNA methylation in a CUL4 C terminus deletion mutant. DNA methylation was tested for the wild-type (WT) strain, a cul4RIP1 strain, and cul4RIP1 strains bearing either the wild-type cul4+ allele or the C terminus truncation allele (the strain contained cul4 residues 1 to 823 [cul41-823] and a deletion of residues 824 to 923; Fig. 1A), as described in the legend to Fig. 1C. (B) H3K9me3 is unaffected by deletion of the C terminus of CUL4. Nuclear extracts from the indicated strains were probed by Western blotting to detect global trimethylated H3K9 or histone H3 levels. (C) The CUL4 C-terminal deletion abrogates DNA repair. Strains of the indicated genotypes were tested as described in the legend to Fig. 1D.
An expectation based on our findings is that the N. crassa RBX1 homolog is required for DNA repair but not DNA methylation. We found that the N. crassa rbx1 homologue is essential for the viability of the organism, but we were able to build a heterokaryotic strain in which the major nuclear component has a deletion of the gene, resulting in a strong reduction of the protein (see the information and Fig. S7 in the supplemental material). DNA repair was compromised in the heterokaryon, but DNA methylation was normal (Fig. 3A and B). Altogether, our findings render it highly unlikely that CUL4 is operating as a ubiquitin ligase for heterochromatin formation and DNA methylation. Perhaps CUL4 simply serves as a scaffold for assembly of the H3K9 methylation machinery.
FIG 3.
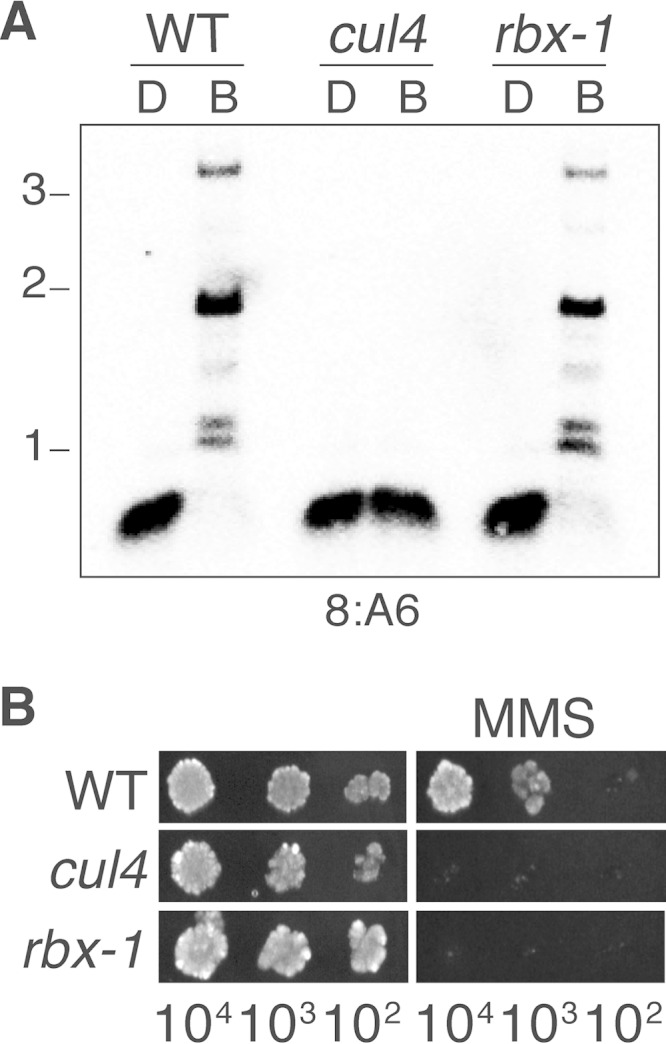
RBX1, the putative RING protein for CUL4 complexes, is not required for control of DNA methylation by DCDC. (A) Normal DNA methylation in an rbx1 deletion strain. DNA methylation was tested for the wild type (WT), a cul4RIP1 strain, and the rbx1 deletion mutant, as described in the legend to Fig. 1C. (B) Compromised DNA repair in the rbx1 mutant. Serial dilutions of conidia from strains of the indicated genotype were tested on medium with or without MMS (0.015%). The strains are listed in Table S2 in the supplemental material.
It is interesting to consider the possible generality of our findings. CUL4 is essential in Schizosaccharomyces pombe, complicating studies of its role in heterochromatin formation. Mutation of the S. pombe CUL4 neddylation site perturbs heterochromatic silencing (7, 8), consistent with the possibility that CLRC, the fission yeast counterpart of DCDC, is operating as a true E3 ubiquitin ligase, despite fruitless efforts to find a likely ubiquitination substrate for this apparent CRL (7–9). It is noteworthy, however, that S. pombe, unlike N. crassa, requires two CUL4-dependent complexes for heterochromatin formation: CLRC, which sports a DDB1-related protein (Rik1) in place of DDB1, methylates H3K9, whereas CUL4/DDB1CDT2 directs polyubiquitination and the subsequent degradation of the antisilencing protein Epe1. Defects in CUL4/DDB1CDT2 lead to the loss of gene silencing (33). Thus, it is possible that neddylation is required for the degradation of Epe1 by the CUL4/DDB1CDT2 complex but not for H3K9me3 by CLRC. There are other indications that cullin complexes do not always conform to accepted models. Inhibition of the NEDD8-activating enzyme (NAE) by MLN4924 causes rapid deneddylation of cullins without perturbing the global CRL network, and mutation of the neddylation site on CUL1 (CUL1K780R) does not prevent the assembly of CUL1 with SKP1 and the substrate adaptor F-box proteins (34). Knockdown experiments in HeLa cells have implicated CUL4 and DDB1 in histone methylation (35), raising the possibility that E3-like complexes similar to DCDC carry out ubiquitination-independent functions in a variety of organisms. Our finding that DCDC is not a true CRL raises the question of why and how H3K9 methylation requires such intricate multicomponent protein complexes.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by NIH grant GM035690. Jordan D. Gessaman was supported by an NIH genetics training grant (5T32GM007413-37).
Keyur K. Adhvaryu thanks Peter Cheung, Patricia Lakin-Thomas, and Roger Lew for helpful discussions.
We all contributed to designing and performing the experiments. K.K.A., J.D.G., and E.U.S. wrote the paper.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/EC.00212-14.
REFERENCES
- 1.Hershko A, Ciechanover A. 1998. The ubiquitin system. Annu Rev Biochem 67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Komander D, Rape M. 2012. The ubiquitin code. Annu Rev Biochem 81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM. 2001. Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 4.Petroski MD, Deshaies RJ. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 5.Jackson S, Xiong Y. 2009. CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem Sci 34:562–570. doi: 10.1016/j.tibs.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lewis ZA, Adhvaryu KK, Honda S, Shiver AL, Knip M, Sack R, Selker EU. 2010. DNA methylation and normal chromosome behavior in Neurospora depend on five components of a histone methyltransferase complex, DCDC. PLoS Genet 6:e1001196. doi: 10.1371/journal.pgen.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horn PJ, Bastie J-N, Peterson CL. 2005. A Rik1-associated, cullin-dependent E3 ubiquitin ligase is essential for heterochromatin formation. Genes Dev 19:1705–1714. doi: 10.1101/gad.1328005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jia S, Kobayashi R, Grewal SIS. 2005. Ubiquitin ligase component Cul4 associates with Clr4 histone methyltransferase to assemble heterochromatin. Nat Cell Biol 7:1007–1013. doi: 10.1038/ncb1300. [DOI] [PubMed] [Google Scholar]
- 9.Zhang K, Mosch K, Fischle W, Grewal SIS. 2008. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol 15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- 10.Zimmerman ES, Schulman BA, Zheng N. 2010. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol 20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Nussinov R. 2011. Flexible cullins in cullin-RING E3 ligases allosterically regulate ubiquitination. J Biol Chem 286:40934–40942. doi: 10.1074/jbc.M111.277236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duda D, Borg L, Scott D, Hunt H, Hammel M, Schulman B. 2008. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hotton SK, Callis J. 2008. Regulation of cullin RING ligases. Annu Rev Plant Biol 59:467–489. doi: 10.1146/annurev.arplant.58.032806.104011. [DOI] [PubMed] [Google Scholar]
- 14.Merlet J, Burger J, Gomes J-E, Pintard L. 2009. Regulation of cullin-RING E3 ubiquitin-ligases by neddylation and dimerization. Cell Mol Life Sci 66:1924–1938. doi: 10.1007/s00018-009-8712-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boh BK, Smith PG, Hagen T. 2011. Neddylation-induced conformational control regulates cullin RING ligase activity in vivo. J Mol Biol 409:136–145. doi: 10.1016/j.jmb.2011.03.023. [DOI] [PubMed] [Google Scholar]
- 16.Davis RH. 2000. Neurospora: contributions of a model organism. Oxford University Press, New York, NY. [Google Scholar]
- 17.Margolin BS, Freitag M, Selker EU. 1997. Improved plasmids for gene targeting at the his-3 locus of Neurospora crassa by electroporation. Fungal Genet Newsl 44:34–36. [Google Scholar]
- 18.Oakley CE, Weil CF, Kretz PL, Oakley BR. 1987. Cloning of the riboB locus of Aspergillus nidulans. Gene 53:293–298. [DOI] [PubMed] [Google Scholar]
- 19.Selker EU, Stevens JN. 1987. Signal for DNA methylation associated with tandem duplication in Neurospora crassa. Mol Cell Biol 7:1032–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honda S, Selker EU. 2008. Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora. Mol Cell Biol 28:6044–6055. doi: 10.1128/MCB.00823-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu H, Wang J, Hu Q, Quan Y, Chen H, Cao Y, Li C, Wang Y, He Q. 2010. DCAF26, an adaptor protein of Cul4-based E3, is essential for DNA methylation in Neurospora crassa. PLoS Genet 6:e1001132. doi: 10.1371/journal.pgen.1001132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamaru H, Selker EU. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 23.Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU. 2003. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet 34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- 24.Zhao Y, Shen Y, Yang S, Wang J, Hu Q, Wang Y, He Q. 2010. Ubiquitin ligase components Cullin4 and DDB1 are essential for DNA methylation in Neurospora crassa. J Biol Chem 285:4355–4365. doi: 10.1074/jbc.M109.034710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S, Rehman B, Elkins T, Engels R, Wang S, Nielsen CB, Butler J, Endrizzi M, Qui D, Ianakiev P, Bell-Pedersen D, Nelson MA, Werner-Washburne M, Selitrennikoff CP, Kinsey JA, Braun EL, Zelter A, Schulte U, Kothe GO, Jedd G, Mewes W, Staben C, Marcotte E, Greenberg D, Roy A, Foley K, Naylor J, Stange-Thomann N, Barrett R, Gnerre S, Kamal M, Kamvysselis M, Mauceli E, Bielke C, Rudd S, Frishman D, Krystofova S, Rasmussen C, Metzenberg RL, Perkins DD, Kroken S, et al. 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422:859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, Zhou P. 2007. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol Cell 26:775–780. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Wu J-T, Chan Y-R, Chien C-T. 2006. Protection of cullin-RING E3 ligases by CSN-UBP12. Trends Cell Biol 16:362–369. doi: 10.1016/j.tcb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Wu S, Zhu W, Nhan T, Toth JI, Petroski MD, Wolf DA. 2013. CAND1 controls in vivo dynamics of the cullin 1-RING ubiquitin ligase repertoire. Nat Commun 4:1642. doi: 10.1038/ncomms2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pierce NW, Lee JE, Liu X, Sweredoski MJ, Graham RL, Larimore EA, Rome M, Zheng N, Clurman BE, Hess S, Shan SO, Deshaies RJ. 2013. Cand1 promotes assembly of new SCF complexes through dynamic exchange of F box proteins. Cell 153:206–215. doi: 10.1016/j.cell.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Q. 2005. The COP9 signalosome regulates the Neurospora circadian clock by controlling the stability of the SCFFWD-1 complex. Genes Dev 19:1518–1531. doi: 10.1101/gad.1322205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, Liu R, Qiu R, Zheng Y, Huang W, Hu H, Ji Q, He H, Shang Y, Gong Y, Wang Y. 2013. CRL4B promotes tumorigenesis by coordinating with SUV39H1/HP1/DNMT3A in DNA methylation-based epigenetic silencing. Oncogene 2013:1–15. doi: 10.1038/onc.2013.522. [DOI] [PubMed] [Google Scholar]
- 32.Swerdlow PS, Schuster T, Finley D. 1990. A conserved sequence in histone H2A which is a ubiquitination site in higher eucaryotes is not required for growth in Saccharomyces cerevisiae. Mol Cell Biol 10:4905–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braun S, Garcia JF, Rowley M, Rougemaille M, Shankar S, Madhani HD. 2011. The cul4-ddb1(cdt2) ubiquitin ligase inhibits invasion of a boundary-associated antisilencing factor into heterochromatin. Cell 144:41–54. doi: 10.1016/j.cell.2010.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett EJ, Rush J, Gygi SP, Harper JW. 2010. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143:951–965. doi: 10.1016/j.cell.2010.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higa LA, Wu M, Ye T, Kobayashi R, Sun H, Zhang H. 2006. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol 8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 36.Selker EU, Tountas NA, Cross SH, Margolin BS, Murphy JG, Bird AP, Freitag M. 2003. The methylated component of the Neurospora crassa genome. Nature 422:893–897. doi: 10.1038/nature01564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.