Abstract
The ATP-binding cassette (ABC) family of transporters, including ABCC3, is a large family of efflux pumps that plays a pivotal role in the elimination of xenobiotics from the body. ABCC3 has been reported to be induced during hepatic stress conditions and through the progression of some forms of cancer. Several lines of evidence have implicated the transcription factor nuclear factor (erythroid-derived 2)–like 2 (Nrf2) in this induction. However, although rodent models have been investigated, a functional antioxidant response element (ARE) in the human ABCC3 gene has not been identified. The purpose of this study was to identify and characterize the ARE(s) responsible for mediating the Nrf2-dependent induction of the human ABCC3 gene. A high-throughput chromatin immunoprecipitation-sequencing analysis performed in A549 cells revealed a specific interaction between Nrf2 and the eighth intron of the human ABCC3 gene rather than the more prototypical flanking region of the gene. Subsequent in silico analysis of the intron identified two putative ARE elements that contained the core consensus ARE sequence commonly found in several Nrf2-responsive genes. Functional characterization of these two AREs using luciferase-reporter constructs with ARE mutant constructs revealed that one of these putative AREs is functionally active. Finally, DNA pull-down assays confirmed specific binding of these intronic AREs by Nrf2 in vitro. Our findings identify a functional Nrf2 response element within the eighth intron of the ABCC3 gene, which may provide mechanistic insight into the induction of ABCC3 during antioxidant response stimuli.
Introduction
Membrane transporters, such as ABCC3, serve important roles in mediating the absorption, distribution, and excretion of endobiotic and xenobiotic substrates. ABCC3 is an organic anion transporter belonging to the ATP-binding cassette (ABC) superfamily of transporters, and transports a variety of endogenous and exogenous compounds, such as bile acids, bilirubin, and chemotherapeutic agents (methotrexate and etoposide) as well as glutathione, sulfate, and glucuronide conjugates (Borst et al., 2007). ABCC3 is almost exclusively expressed on the basolateral membrane of polarized epithelial cells, and tissue distribution in humans is primarily confined to the adrenal glands, kidney, liver, small intestine, colon, pancreas, and bladder (Borst et al., 2007; Klaassen and Aleksunes, 2010). ABCC3 is also highly expressed in several forms of cancer, including non–small-cell lung carcinoma, gallbladder cancer, and hepatocellular carcinoma (Nies et al., 2001; Young et al., 2001; Wang et al., 2010).
In addition to being overexpressed in several malignant tumors, expression of ABCC3, particularly in the liver, is highly inducible in other nonmalignant conditions (Borst et al., 2007). Hepatic ABCC3 has been shown to be upregulated in primary biliary cirrhosis, hepatitis C viral infections, nonalcoholic steatohepatitis, and rodent models of cholestasis (Ros et al., 2003; Zollner et al., 2003; Hardwick et al., 2011). The mechanisms that mediate ABCC3 induction are hypothesized to be hepatoprotective in nature and primarily involve the activation of transcriptional regulators, such as nuclear receptors and transcription factors. It has been shown that nuclear receptors, such as the constitutive androstane receptor and pregnane X receptor as well as the transcription factor nuclear factor (erythroid 2)–like 2 (Nrf2), are important mediators of the xenobiotic-dependent induction of the Abcc3 gene (Klaassen and Aleksunes, 2010).
Nrf2 is a master regulator of the antioxidant response pathway and orchestrates the transcriptional activation of cellular defense mechanisms in combating oxidative stress. Nrf2 activation is highly regulated and controlled by the Kelch-like ECH-associated protein (Keap1), an adapter protein that belongs to an E3 ubiquitin ligase complex (Zhang, 2006). Under basal conditions, Nrf2 associates as an inactive complex with Keap1 and is targeted for ubiquitin-mediated proteasomal degradation. Under periods of oxidative/electrophilic stress, however, the Keap1/Nrf2 interaction is disrupted, leading to cytosolic accumulation and subsequent nuclear translocation of Nrf2, where it can drive the transcription of target genes (Jaiswal, 2004; Zhang, 2006).
Activation of Nrf2 is largely regarded as a cytoprotective mechanism, and target genes consist of a battery of phase I/II detoxifying enzymes and xenobiotic transporters that ultimately aid in defending the cell in response to oxidative stress (Shen and Kong, 2009). Multiple investigations using Nrf2 chemical activators have shown a clear role in the Nrf2-dependent induction of xenobiotic transporters (Maher et al., 2005, 2008; Aleksunes et al., 2008). Importantly, this has resulted in the identification of an ARE upstream of the promoter of the mouse Abcc3 gene (Maher et al., 2007); however, identification of a functional ARE in the human gene is lacking, despite clear evidence of Nrf2-dependent regulation of human ABCC3 (Mahaffey et al., 2009, 2012; Wang et al., 2010; Sasaki et al., 2012).
The purpose of the current study was to identify and characterize the functional ARE(s) responsible for the Nrf2-dependent induction of the human ABCC3 gene. Chromatin immunoprecipitation (ChIP) assays followed by sequencing analysis in Nrf2 overexpressing cell lines identified Nrf2 interaction within a region corresponding to the eighth intron of the human ABCC3 gene. Subsequent in silico analyses and reporter gene assays identify two functional AREs within the eighth intron, suggesting a novel mechanism of Nrf2-dependent regulation of ABCC3.
Materials and Methods
Unless otherwise specified, all other materials were obtained from Sigma Aldrich (St. Louis, MO).
Cell Culture
The HEK293 cells used were a gift from the Zhang Laboratory at the University of Arizona, Tucson, AZ. The cells were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, glutamine, and penicillin-streptomycin. Cultures were grown at 37°C in a 5% CO2 atmosphere.
Chromatin Immunoprecipitation (Sequencing)
Cell Culture.
A549 cells (ATCC CCL-185) were passaged in Dulbecco’s modified Eagle’s medium 5% fetal bovine serum with 1% penicillin-streptomycin. A 10-cm plate was grown to approximately 90% confluence, translating to roughly 1.5 × 106 cells.
Crosslinking and Sonication.
Formaldehyde (∼270 μl) was added to cell media to reach a final concentration of 1%. Cells were incubated at room temperature while shaking. Crosslinking was stopped by adding 1 ml of 1.25 M glycine, and cells were scraped and centrifuged at 800 rpm. Media was aspirated completely, rinsed with phosphate-buffered saline, and then resuspended in 1-ml cell lysis buffer (5 mM 1,4-piperazinediethanesulfonic acid, pH 8.0; 85 mM KCl; 0.5% NP40) for 10 minutes on ice. Samples were centrifuged at 2000 rpm, and the supernatant was removed and resuspended in 1-l nuclei lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.0) for 10 minutes on ice. Sonication of samples was conducted according to the following parameters: output 4, duty cycle 50%, 20 seconds, and 12 repetitions. Samples were centrifuged at full speed for 30 minutes at 4°C, and then diluted with dilution buffer (1.1% SDS, 10 mM EDTA, 50 mM Tris, pH 8.0)
Immunoprecipitation.
Preclearing was conducted for 12 hours at 4°C using 5 μg of rabbit IgG and 300 μl of preblocked protein A/G fast-flow sepharose beads. Samples were centrifuged at 3000 rpm for 5 minutes, and then transferred to a new tube. This process was repeated for a total of four times. Subsequently, into 1 ml of sample, the antibody of interest was added at the following amounts: IgG (2 μg), Nrf2 (2 μg), and dH3H4dime (3 μg). Samples were rotated for 24 hours at 4°C, and 60 μl of precleared beads was added. Samples were centrifuged at 3000 rpm, and the supernatant was removed. Beads were washed with a series of low salt, high salt, and LiCl and Tris EDTA buffers. To elute, 250 μl of elution buffer was added (1% SDS, 0.1 M NaHCO3, and 0.25 μM dithiothreitol) at 65°C overnight. Samples were treated with proteinase K (10 mg/ml) and then a phenol-chloroform extraction for final resuspension into 30 μl of polymerase chain reaction (PCR)–grade water.
Library Construction and Sequencing.
Library creation was accomplished using adapter ligation methods. Each unique library was sequenced along with an input control library using the Solexa Genome Analyzer (Illumina, San Diego, CA), and sequences were aligned to the human genome (http://www.genome.ucsc.edu) using ELAND software. Peaks were visualized using the Integrated Genome Browser (BioViz, University of North Carolina at Charlotte, Charlotte, NC).
In Silico Analysis
Identification of putative AREs within the eighth intron was performed using the CLC sequence viewer, version 6.4 (CLC bio, Aarhus, Denmark). Species alignments were performed using comparative genomics tools available at http://www.ensemble.org.
Molecular Cloning of ABCC3 Intron
A DNA construct containing the eighth intron [1.2 kilobase (kb)] of the ABCC3 gene was produced by PCR amplification of the RP11 BAC clone 121F10. Primers used included exogenous restriction sites (underlined) and are as follows: forward primer 5′-CGCCTCGAGTGGCTGGCTAGCCCAGAGGA-3′ (XhoI site), and reverse primer 5′-CGCAAGCTTGCACCACTGGCCCCACATGA-3′ (HindIII site). The resulting PCR product was cloned into a TOPO vector using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA) per the manufacture’s recommendations.
Subcloning of ABCC3 Intron in Reporter Vectors
The cloned ABCC3 intron (∼1.2 kb) was subcloned from the TOPO vector into the pGL3 basic reporter vector (Promega Corporation, Madison, WI) using the XhoI/HindIII restriction sites within the multiple cloning region upstream of the luciferase gene (1.2 kb). The intronic insert cloned into pGL3 basic was subsequently cloned into the pT81luc reporter vector containing a herpes simplex virus thymidine kinase promoter upstream of the promoter by PCR amplification of the insert using the following primers: forward primer 5′-TCGCGGATCCGAGGAAGGAACAACGTACAG-3′ and reverse primer 5′-ATAGCAAGCTTAGATCTTGAAGCCAGATG-3′. The underlined sequences represent exogenous restriction sites that were inserted for ligation of insert into the pT81luc vector (BamHI for forward and HindIII for reverse). The resulting PCR amplicon is ∼900 bp and was inserted into the pT81luc reporter vector using BamHI/HindIII restriction sites.
PCR Mutagenesis of ARE2 and ARE5 within Intron
Mutagenesis of ARE2 and ARE5 was carried out by PCR using a proofreading polymerase to reduce sequencing errors. Briefly, reverse-complementary primers (primer pairs) containing the mutated sequence of interest were used and were as follows: ARE2 mutant, 5′-CAGGAACTCAGAGAGGTGAAGCGGCTTGTTCAAGGGCACACAGCAAAGAAG-3′, along with reverse complement, and ARE5 mutant, 5′-AACAGGGGAAAGTTCAGCCTCAAATTAGCCGCAAAGCCCTGAAACAGCATGAC-3′, as well as its reverse complement. Underlined sequences represent the ARE sequence, whereas the bolded sequences are mutated (TGACTTGGC is wild-type (wt) sequence for ARE2 and AGCTTAGTCA is wt sequence for ARE5). The mutagenesis reaction was carried out using the cloned intron in pT81luc as a template (50 ng/reaction) and the PCR cycle was as follows: 1) 95°C for 1 minute, 2) 95°C for 30 seconds, 3) 55°C for 1 minute, and 4) 68°C for 10 minutes; steps 2–4 are repeated 24 more times. The reactions were cooled to room temperature followed by the addition of 1 μl DpnI restriction enzyme and incubated at 37°C for 1 hour to cleave parental plasmid DNA and leave the newly synthesized nascent strand intact. After incubation, 5 μl was transformed into competent bacteria cells and subsequently screened and sequenced for mutant sequence. The ARE2/5 double mutant construct was synthesized using the above mentioned methods using the ARE2 mutant as the starting template.
Transfection and Luciferase Reporter Gene Assay
HEK293 cells were transfected with pT81 constructs using Lipofectamine 3000 (Life Technologies, Carlsbad, CA) transfection reagent per the manufacturer’s protocol, including the use of pRL-tk transfection control (Promega Corporation). Twenty-four hours post-transfection, the cells were treated with either 5 μM sulforaphane in dimethylsulfoxide or dimethylsulfoxide control for 17 hours. After treatment, luciferase activity was determined using the Dual-Glo luciferase assay (Promega Corporation,) per the manufacturer’s instructions and normalized to transfection control. Each reporter construct was assayed in triplicate wells, and luciferase activity was measured using a GloMax 20/20 luminometer (Promega Corporation). In addition to chemical induction, a separate experiment was conducted using an Nrf2 overexpression platform by cotransfecting the pT81 constructs in HEK293 cells with an Nrf2 expression vector. The Nrf2 vector has been used and characterized previously (Zhang and Hannink, 2003), with slight modifications. Nrf2 induction was verified by using SDS-PAGE and Western blot analysis using primary antibodies against Nrf2 and glyceraldehyde 3-phosphate dehydrogenase (control) (Santa Cruz Biotechnology, Santa Cruz, CA).
Nrf2 DNA Pull-Down Assay
The pull-down assay was performed in vitro using recombinant glutathione S-transferase (GST)–tagged Nrf2 and His-tagged MafG protein, which were a kind gift from the laboratory of Dr. Donna Zhang at the Department of Pharmacology and Toxicology at the University of Arizona. Briefly, GST-Nrf2 and His-MafG (5:1) were incubated in microcentrifuge tubes with radioimmunoprecipitation assay buffer (RIPA) buffer (10-mM sodium phosphate pH 7.2, 150-mM NaCl, 1% sodium deoxycholate, 2-mM EDTA, 0.1% SDS, 1% NP-40) and double-stranded (hybridized), 5′ biotin-labeled oligonucleotide probes containing ARE2, ARE5, or NQO1 promoter as a positive control as well as their mutated counterparts. The sequences of the probes used were as follows (the lowercase are nucleotides that were mutated):
ARE2wt: 5′-CTCAGAGAGGTGAAGTGACTTGGCCAAGGGCACACAGCAAA-3′
AREmut: 5′-CTCAGAGAGGTGAAGcGgCTTGttCAAGGGCACACAGCAAA-3′
AREwt: 5′-GGAAAGTTCAGCCTCAGCTTAGTCACAAAGCCTGAAACAGC-3′
AREmut: 5′-GGAAAGTTCAGCCTCAaaTTAGcCgCAAAGCCCTGAAACAGC-3′
NQO1wt: 5′-AAATCGCAGTCACAGTGACTCAGCAGAATCTGAGCCTAGGG-3′
NQO1mut: 5′-AAATCGCAGTCACAGactCTCAcgAGAATCTGAGCCTAGGG-3′
Tubes were incubated at 4°C under constant rotation for 2 hours to allow for complexes to form. After incubation, 30 μl of streptavidin agarose resin (prewashed with RIPA buffer to remove alcohol) was added to each reaction vial and incubated overnight at 4°C under constant rotation to precipitate the bound complexes. After incubation, the beads were centrifuged, supernatant was removed, and beads were washed with RIPA buffer to remove any unbound protein. This previous step was performed three times to ensure removal of all unbound protein. After the final centrifugation, all the remaining supernatant was removed and the complexes were eluted by the addition of Laemmli sample buffer (20 μl) to each reaction tube and heated at 95°C for 5 minutes. The beads were then centrifuged and the supernatant was subjected to SDS-PAGE and Western blot analysis using a primary antibody directed against GST (Nrf2) and an anti-mouse secondary (Santa Cruz Biotechnology, Santa Cruz, CA). Densitometry analyses were performed using ImageJ software (National Institutes of Health, Bethesda, MD).
Statistical Analysis
Data were analyzed using one-way analysis of variance to determine significant findings in luciferase assay across reporter vectors with a Bonferroni posthoc analysis. A significance level of P ≤ 0.05 was used for all analyses. All analyses were carried out using GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA).
Results
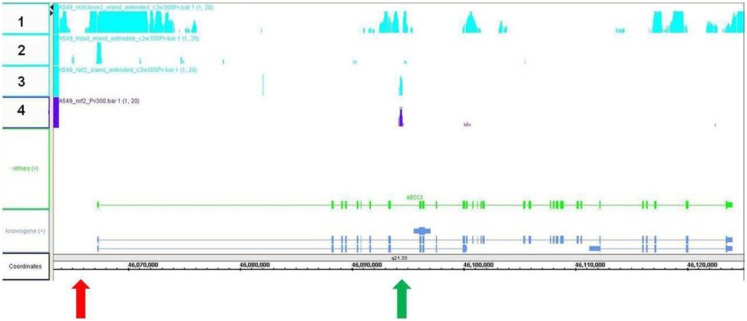
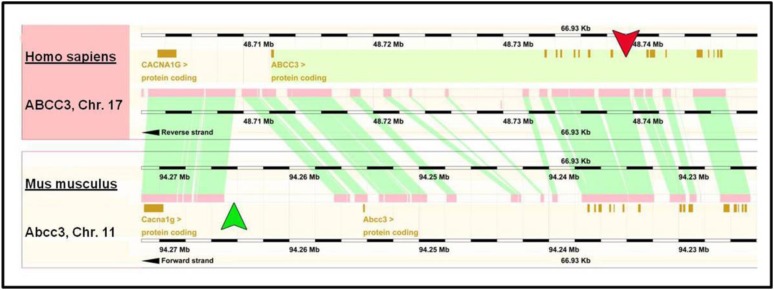
To determine the relative location of Nrf2 association within the ABCC3 gene, a ChIP assay followed by sequencing analysis was performed using A549 cells, a human lung carcinoma cell line that has constitutively active Nrf2. The results reveal a specific, robust interaction of Nrf2 within a genomic region located −26,258 to −28,823 upstream from the transcription start site, which corresponds to the eighth intronic region of the ABCC3 gene (green arrow), whereas no Nrf2 binding was detected within the promoter region (red arrow) (Fig. 1). Furthermore, histone 3, lysine 4 (H3K4) methylation status was assessed and was found to be hypermethylated within the intronic region corresponding to Nrf2-association as well as the promoter region of the ABCC3 gene. Interestingly, gene comparison analyses between human and mouse ABCC3 show that the previously characterized ARE within the mouse promoter region (Maher et al., 2007) (green arrow) is located within a nonconserved genomic region, suggesting that this ARE is possibly unique to the mouse species (Fig. 2). In contrast, the genomic region corresponding to Nrf2 association within the eighth intron of ABCC3 appears to be in a region that is conserved across human and mouse (Fig. 2, red arrow).
Fig. 1.
Chromatin immunoprecipitation (sequencing). Results obtained from a chromatin immunoprecipitation assay in A549 cells (Nrf2 constitutively active) followed by sequencing analysis. The DNA sequence of the ABCC3 gene is depicted as the green line, with exons shown as green bars. Positive results from the ChIP sequencing analysis are shown as binding intensity from two separate ChIP assays (teal, row 3; purple, row 4) and represents a strong Nrf2 interaction within the eighth intron of the ABCC3 gene (green arrow) (intron located −26,258 to −28,823 from TSS. H3K4 methylation status is highlighted in teal on the top row of the figure (color represents positive methylation status) (row 1). Input control is also shown as teal in row 2. No Nrf2 interaction was observed within the promoter region of the ABCC3 gene (red arrow).
Fig. 2.
ABCC3 gene comparison across human and mouse. Gene comparison of human (top) and mouse (bottom) ABCC3 is shown. Brown squares represent protein coding regions and pink represents conserved regions across both species. The previously identified mouse ARE is in a region shown by the green arrowhead, which is located in a nonconserved genomic region. Contrastingly, the intronic region corresponding with the Nrf2 association in human ABCC3 is located in a genetically conserved locus (red arrowhead).
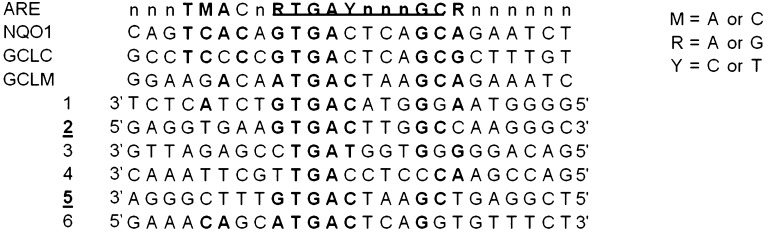
To identify potential ARE(s) within the eighth intronic region of the ABCC3 gene, in silico and sequencing analyses were performed using CLC sequence viewer software. A total of six putative ARE and ARE-like elements were identified within the eighth intronic region; however, only two (ARE2 and ARE5) matched the core ARE consensus sequence and are separated by ∼400 bp (Fig. 3). Furthermore, comparative analysis of the ARE2 and ARE5 across several eutherian mammal species showed that ARE5, and not ARE2, is evolutionarily conserved (data not shown).
Fig. 3.
Comparison of putative ABCC3 intronic AREs with consensus ARE. Comparison of the six ARE and ARE-like elements located within the eighth intron of the ABCC3 with the consensus ARE sequence is shown. For additional comparison purposes, the ARE of known Nrf2-target genes, NQO1, GLC, and GCLM, are also shown. The underlined bold sequences of the consensus sequence are required for activity. A total of two (ARE2 and ARE5) AREs from the six contain the required consensus sequence.
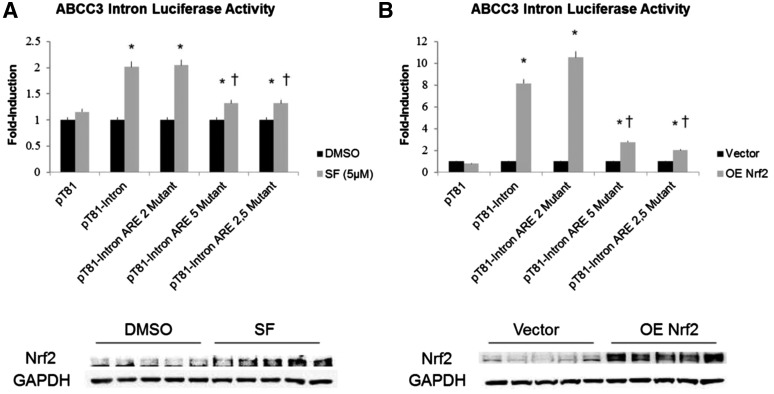
To confirm ARE functionality, luciferase reporter constructs containing a ∼900 bp region of the ABCC3 eighth intron were transfected into HEK293 cells and either treated with 10 μM of the Nrf2 activator, sulforaphane (SF) or cotransfected with a Nrf2 overexpressing vector (Fig. 4). SF treatment of pT81-intron (ABCC3 intron 8) transfectants resulted in a 2-fold induction of luciferase activity compared with the vehicle-treated transfectants, whereas overexpressing Nrf2 protein resulted in an 8-fold induction of luciferase acticity (Fig. 4, A and B, respectively). These results suggested the presence of at least one functional ARE within this genomic region that is regulated in an Nrf2-dependent manner.
Fig. 4.
Luciferase reporter activity of ABCC3 intron in HEK293 cells. Luciferase reporter activity of the eighth ABCC3 intron cloned into pT81 reporter vector and transfected into HEK293 cells. Nrf2 was subsequently activated by treatment of cells with 5 µM SF (A) or overexpressed by cotransfecting an Nrf2 expression vector (B) (Nrf2 OE). Constructs containing mutations of the two identified intronic AREs (ARE2 and ARE5) as well as an ARE2/5 double mutant were used to independently determine functional activity of each ARE. Data represent the results of three independent experiments done in triplicate for each group and are described as fold-change of control. *P ≤ 0.05 versus respective control; †P ≤ 0.05 versus pT81-intron.
To further characterize the ARE(s) mediating the Nrf2-dependent induction of luciferase activity, mutant ARE2 and ARE5 constructs as well as an ARE2/5 double mutant were generated (Fig. 4). Mutating ARE2 had no effect on luciferase activity in both experiments, whereas mutation of ARE5 caused a significant decrease and a near complete ablation of the Nrf2-dependent induction of luciferase activity in both SF-treated and Nrf2 overexpressing cells (Fig. 4). Interestingly, mutating both ARE2 and ARE5 did not result in further reduction of Nrf2-dependent luciferase activity in both treated and overexpression platforms, suggesting that ARE5 is the functionally dominant ARE.
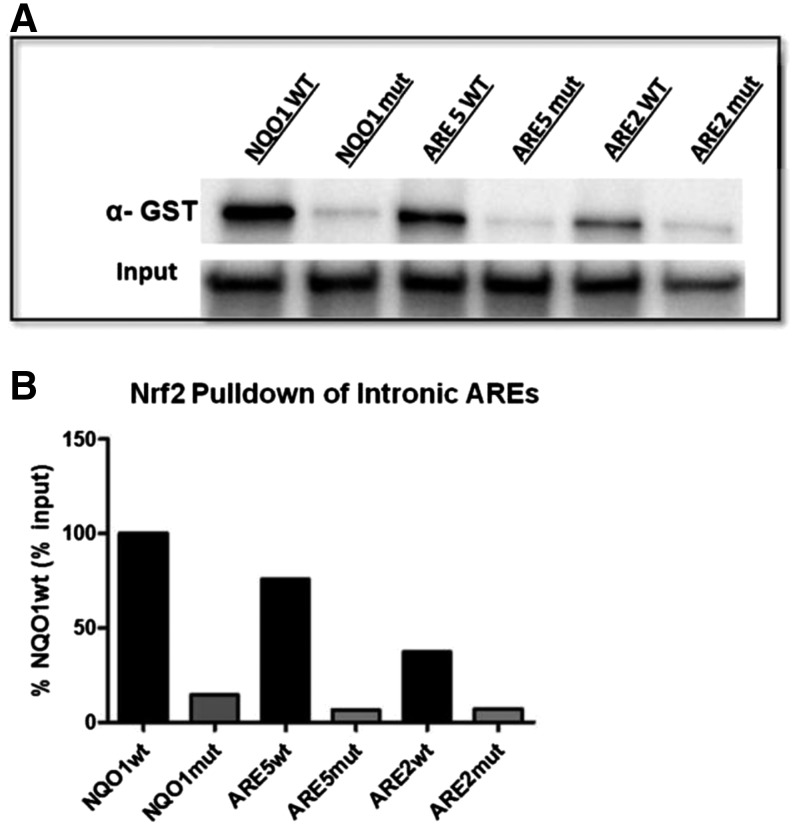
To further elucidate interaction of Nrf2 with the intronic DNA region, DNA pull-down assays were conducted using biotin-labeled DNA probes containing the ARE2 and ARE5 genomic sequences (Fig. 5). A strong, positive interaction of Nrf2 was observed with the ARE5 probe that was ∼75% of the positive control NQO1wt (Fig. 5B). Interestingly, Nrf2 interaction with the ARE2 probe was ∼50% of ARE5, and mutating both the ARE5 and ARE2 sequence significantly reduced the enrichment of Nrf2 (Fig. 5B).
Fig. 5.
DNA pull down of Nrf2. Recombinant Nrf2 (GST tagged) and its heterodimeric partner MafG were incubated with 5′ biotin-labeled oligonucleotide probes containing ARE2 or ARE5 along with their native flanking sequences. The complex was “pulled down” using streptavidin beads, and the precipitated complex was subjected to Western blot analysis using a commercially available antibody against GST (Nrf2). Probes containing the NQO1 wild-type and NQO1 mutant ARE were used as positive and negative controls, respectively. Input represents the amount of Nrf2-GST used in each incubation and directly in SDS-PAGE and Western blot analysis. Results of a single experiment are shown in (A), with respective densitometry analysis in (B).
Discussion
The purpose of the current study was to identify the ARE(s) responsible for the Nrf2-dependent induction of the human ABCC3 gene and our investigation herein describes a novel mechanism responsible for this induction. Using high-throughput ChIP sequencing followed by in silico analyses, we have successfully identified two putative AREs located within the eighth intron of the ABCC3 gene. Functional characterization using gene reporter assays confirms a functional ARE response element that is unresponsive to Nrf2 activity and binding upon mutation. Furthermore, comparative sequencing analyses identify one of these AREs, ARE5, as being evolutionarily conserved across several species, suggesting a potentially important role in mediating the Nrf2-dependent regulation of human ABCC3.
Xenobiotic membrane transporters are pivotal players in mediating the absorption, distribution, and excretion of both endobiotics and xenobiotics. The function of these transporters may directly impact the biologic effectiveness and toxicity of their chemical substrates, and understanding the multiple levels of membrane transporter regulation is crucial in determining functionality. There are multiple lines of evidence to support Nrf2-mediated transcriptional regulation of several ABCC gene family members, including Abcc3, in mice. Studies conducted in mice and rats treated with known Nrf2 activators, such as oltipraz, butylated hydroxyanisole, and ethoxyquin, have shown significant gene induction of Abcc3 along with other Abcc family members, such as Abcc2 and Abcc4 (Slitt et al., 2003; Maher et al., 2005; Merrell et al., 2008). Additionally, several investigators have reported a loss of Abcc3 induction in Nrf2-null mice after treatment with a variety of Nrf2 chemical activators (Maher et al., 2007, 2008; Aleksunes et al., 2008; Okada et al., 2008; Tanaka et al., 2009). Together, these findings suggest a clear role for the Nrf2-dependent regulation of Abcc3 in rodents; however, the mechanistic understanding of this regulation of human ABCC3 still remains unclear.
Investigation into the mechanism that drives the regulation of human ABCC3 by Nrf2 has been quite limited. However, several correlative reports strongly demonstrate the role of Nrf2-dependent induction of the ABCC3 gene. Previous observations in primary human hepatocytes show ABCC3 upregulation after treatment with the Nrf2 activator oltipraz (Jigorel et al., 2006). Similarly, human-derived HepG2 cells treated with tert-butylhydroquinone increased levels of ABCC3 mRNA (Adachi et al., 2007). Lastly, several malignancies that have been shown to have Nrf2 mutations are associated with high levels of ABCC3 expression (Wang et al., 2010; Sasaki et al., 2012). The first mechanistic investigation into Nrf2-dependent induction of ABCC3 was performed in human lung cancer cell lines in which the investigators describe the identification of four putative AREs upstream of the transcription start site (TSS), but did not attempt to functionally assess the contributions of these AREs (Mahaffey et al., 2009). Using ChIP PCR analyses, Mahaffey et al. later demonstrated that upon treatment of a lung carcinoma cell line with the Nrf2 activator, 4-hydroxynonenol, a specific interaction occurred between Nrf2 and a genomic region −805 bases from the transcription start site of the ABCC3 gene (Mahaffey et al., 2012). However, our results in A549 cells reveal no interaction between Nrf2 and the promoter region of the ABCC3 gene. This discrepancy may be due to the different approach used to investigate the interaction between Nrf2 and the ABCC3 gene. The investigators use a ChIP-PCR method and limited their genomic region of interest to just the promoter region. In contrast, using ChIP, followed by sequencing analyses, allows for a higher throughput method that does not limit the investigation to a particular chromosomal region.
In addition to Nrf2 association within the eighth intron, our analysis yields novel information regarding H3K4 methylation status of the ABCC3 gene. Our data suggest that the ABCC3 gene is hypermethylated within several regions, including the promoter and eighth intron, where we have identified Nrf2 association. Interestingly, unlike other methylation sites, methylation of H3K4 is almost exclusively found within regions of transcriptionally active genes (Shilatifard, 2008). The identification of H3K4 hypermethylation within the eighth intron further supports our findings that a functional ARE is present within this region. Recent studies have implicated an important role of the H3K4 methyltransferase, mixed lineage leukemia 3, in the transcriptional regulation of several clinically important membrane transporters, including MDR1 and the Farsenoid X receptor–dependent transcriptional regulation of hepatic ABCC2, ABCB11, and SLC10A1 (Huo et al., 2010; Ananthanarayanan et al., 2011). However, the role of H3K4 methylation and the Nrf2-dependent induction of ABCC3 have not been investigated, and further analyses are necessary to elucidate the importance of histone 3 methylation within the eighth intron of this gene.
A recent investigation by Malhotra et al. utilizes ChIP-seq investigation using Keap1-null mice (constitutive Nrf2 activation). Although the regulation of the mouse Abcc3 gene was not specifically examined, these studies did reveal interesting information concerning the ARE consensus sequence (Malhotra et al., 2010). Historically, the definition of the ARE is a core sequence of TGACnnnGC (see Fig. 3). Using a larger data set of 410 DNA sequences, these new studies reveal an expanded role of the intervening three base pairs (between TGAC and GC), with TCA being the most common sequence observed. Furthermore, previously published data on the topic clearly show that the TCA-intervening sequence is more sensitive to chemical induction compared with the wild-type AAA sequence, suggesting that this variation may be a more potent ARE sequence (Nguyen et al., 1994). Interestingly, our results demonstrate that the intervening bases (nnn) of this consensus sequence (TCA) are more homologous to the ARE5 core sequence (TAA) than the ARE2 (TTG). It is possible that the difference seen in the intervening bases between ARE2 and ARE5 may partially explain why Nrf2 binds more strongly to ARE5 as well as it being the functionally dominant ARE. It is interesting to note, however, that mutating ARE5 did not completely abolish reporter activity upon Nrf2 activation. This observation may be partially explained by the fact that mutant ARE5 still confers slight affinity for Nrf2, as seen by the DNA-binding assay.
The identification of putative AREs within the eighth intron of the ABCC3 gene occurred within a region that is conserved across mouse and human, suggesting that this region within the mouse genome may have ARE(s) as well. A previous study conducted in a mouse liver cell line has identified an ARE upstream of the promoter region using ChIP-PCR analysis (Maher et al., 2007). Interestingly, this region is not conserved in the human (see Fig. 2) despite a 3- to 4-fold induction in Abcc3 after oltipraz treatment in two independent studies in mice (Maher et al., 2007; Aleksunes and Klaassen, 2012). Using primary human hepatocytes, Jigorel et al. showed a 1.5-fold induction of ABCC3 upon oltipraz treatment, suggesting that mice may be more sensitive to chemical induction of ABCC3 by Nrf2 (Jigorel et al., 2006). Together, these findings support the need for further investigation into whether additional functional ARE(s) are present within the eighth intron of the mouse Abcc3 gene.
The identification of gene regulatory elements within regions downstream of the TSS is a rare occurrence but has been reported in various genes by various transcription factors. Many of these downstream elements are located in the first intron; however, there are several examples revealing distal elements that can regulate the transcription of genes. ChIP-seq experiments have reported that 38% of estrogen receptorα–binding elements were found within introns along with an additional 38% being further than 10 kb from the TSS (Levy et al., 2008). Other examples of nuclear receptors with identified response elements within an intronic region include the androgen receptor (Zheng et al., 2006), the vitamin D receptor (Zella et al., 2006), and peroxisome proliferator–activated receptorα (Sun et al., 2008). In addition, androgen regulation of the Nrf2-target gene GSTP1 appears to be dependent upon a 500-bp region of the fifth intron of the gene (Ikeda et al., 2002); however, to our knowledge, this is the first example of Nrf2 itself potentially regulating a target gene through an intronic response element.
In conclusion, we have identified two putative AREs in the eighth intron of the human ABCC3 gene. In silico analyses have shown that this region is highly conserved across several mammalian species, suggesting an important role for this genomic region in the Nrf2-dependent induction ABCC3. Furthermore, functional characterization of these novel elements revealed that one of these AREs is functionally important for the Nrf2-dependent induction of the ABCC3 gene. These findings are the first to our knowledge to identify an Nrf2-regulatory element within an intronic region of a gene. This novel finding may promote the future identification of AREs within nonflanking regions of genes and further elucidate nontraditional mechanisms of gene regulation.
Supplementary Material
Abbreviations
- ABC
ATP-binding cassette family
- ARE
antioxidant response element
- ChIP
chromatin immunoprecipitation
- GST
glutathione S-transferase
- Keap1
Kelch-like ECH-associated protein
- NASH
nonalcoholic steatohepatitis
- PCR
polymerase chain reaction
- RIPA
radioimmunoprecipitation assay buffer
- TSS
transcription start site
- wt
wild-type
Authorship Contributions
Participated in research design: Canet, Merrell, Cherrington.
Conducted experiments: Canet, Merrell, Harder, Maher, Jackson, Lickteig, Wu.
Contributed new reagents or analytic tools: Yamamoto, Zhang.
Performed data analysis: Canet, Merrell, Cherrington.
Wrote or contributed to the writing of the manuscript: Canet, Merrell, Cherrington.
Footnotes
This work was supported by the National Institutes of Health [Grants AI083927, ES006694, and HD062489], National Institute of Environmental Health Science Toxicology Training [Grant ES007091], and the National Science Foundation of Arizona.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Adachi T, Nakagawa H, Chung I, Hagiya Y, Hoshijima K, Noguchi N, Kuo MT, Ishikawa T. (2007) Nrf2-dependent and -independent induction of ABC transporters ABCC1, ABCC2, and ABCG2 in HepG2 cells under oxidative stress. J Exp Ther Oncol 6:335–348. [PubMed] [Google Scholar]
- Aleksunes LM, Klaassen CD. (2012) Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARα-, and Nrf2-null mice. Drug Metab Dispos 40:1366–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksunes LM, Slitt AL, Maher JM, Augustine LM, Goedken MJ, Chan JY, Cherrington NJ, Klaassen CD, Manautou JE. (2008) Induction of Mrp3 and Mrp4 transporters during acetaminophen hepatotoxicity is dependent on Nrf2. Toxicol Appl Pharmacol 226:74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthanarayanan M, Li Y, Surapureddi S, Balasubramaniyan N, Ahn J, Goldstein JA, Suchy FJ. (2011) Histone H3K4 trimethylation by MLL3 as part of ASCOM complex is critical for NR activation of bile acid transporter genes and is downregulated in cholestasis. Am J Physiol Gastrointest Liver Physiol 300:G771–G781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P, de Wolf C, van de Wetering K. (2007) Multidrug resistance-associated proteins 3, 4, and 5. Pflugers Arch 453:661–673. [DOI] [PubMed] [Google Scholar]
- Hardwick RN, Fisher CD, Canet MJ, Scheffer GL, Cherrington NJ. (2011) Variations in ATP-binding cassette transporter regulation during the progression of human nonalcoholic fatty liver disease. Drug Metab Dispos 39:2395–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo H, Magro PG, Pietsch EC, Patel BB, Scotto KW. (2010) Histone methyltransferase MLL1 regulates MDR1 transcription and chemoresistance. Cancer Res 70:8726–8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H, Serria MS, Kakizaki I, Hatayama I, Satoh K, Tsuchida S, Muramatsu M, Nishi S, Sakai M. (2002) Activation of mouse Pi-class glutathione S-transferase gene by Nrf2(NF-E2-related factor 2) and androgen. Biochem J 364:563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal AK. (2004) Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic Biol Med 36:1199–1207. [DOI] [PubMed] [Google Scholar]
- Jigorel E, Le Vee M, Boursier-Neyret C, Parmentier Y, Fardel O. (2006) Differential regulation of sinusoidal and canalicular hepatic drug transporter expression by xenobiotics activating drug-sensing receptors in primary human hepatocytes. Drug Metab Dispos 34:1756–1763. [DOI] [PubMed] [Google Scholar]
- Klaassen CD, Aleksunes LM. (2010) Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev 62:1–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy N, Tatomer D, Herber CB, Zhao X, Tang H, Sargeant T, Ball LJ, Summers J, Speed TP, Leitman DC. (2008) Differential regulation of native estrogen receptor-regulatory elements by estradiol, tamoxifen, and raloxifene. Mol Endocrinol 22:287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffey CM, Mahaffey NC, Holland W, Zhang H, Gandara DR, Mack PC, Forman HJ. (2012) Aberrant regulation of the MRP3 gene in non-small cell lung carcinoma. J Thorac Oncol 7:34–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffey CM, Zhang H, Rinna A, Holland W, Mack PC, Forman HJ. (2009) Multidrug-resistant protein-3 gene regulation by the transcription factor Nrf2 in human bronchial epithelial and non-small-cell lung carcinoma. Free Radic Biol Med 46:1650–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher JM, Aleksunes LM, Dieter MZ, Tanaka Y, Peters JM, Manautou JE, Klaassen CD. (2008) Nrf2- and PPAR alpha-mediated regulation of hepatic Mrp transporters after exposure to perfluorooctanoic acid and perfluorodecanoic acid. Toxicol Sci 106:319–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher JM, Cheng X, Slitt AL, Dieter MZ, Klaassen CD. (2005) Induction of the multidrug resistance-associated protein family of transporters by chemical activators of receptor-mediated pathways in mouse liver. Drug Metab Dispos 33:956–962. [DOI] [PubMed] [Google Scholar]
- Maher JM, Dieter MZ, Aleksunes LM, Slitt AL, Guo G, Tanaka Y, Scheffer GL, Chan JY, Manautou JE, Chen Y, et al. (2007) Oxidative and electrophilic stress induces multidrug resistance-associated protein transporters via the nuclear factor-E2-related factor-2 transcriptional pathway. Hepatology 46:1597–1610. [DOI] [PubMed] [Google Scholar]
- Malhotra D, Portales-Casamar E, Singh A, Srivastava S, Arenillas D, Happel C, Shyr C, Wakabayashi N, Kensler TW, Wasserman WW, et al. (2010) Global mapping of binding sites for Nrf2 identifies novel targets in cell survival response through ChIP-Seq profiling and network analysis. Nucleic Acids Res 38:5718–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrell MD, Augustine LM, Slitt AL, Cherrington NJ. (2008) Induction of drug metabolism enzymes and transporters by oltipraz in rats. J Biochem Mol Toxicol 22:128–135. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Rushmore TH, Pickett CB. (1994) Transcriptional regulation of a rat liver glutathione S-transferase Ya subunit gene. Analysis of the antioxidant response element and its activation by the phorbol ester 12-O-tetradecanoylphorbol-13-acetate. J Biol Chem 269:13656–13662. [PubMed] [Google Scholar]
- Nies AT, König J, Pfannschmidt M, Klar E, Hofmann WJ, Keppler D. (2001) Expression of the multidrug resistance proteins MRP2 and MRP3 in human hepatocellular carcinoma. Int J Cancer 94:492–499. [DOI] [PubMed] [Google Scholar]
- Okada K, Shoda J, Taguchi K, Maher JM, Ishizaki K, Inoue Y, Ohtsuki M, Goto N, Takeda K, Utsunomiya H, et al. (2008) Ursodeoxycholic acid stimulates Nrf2-mediated hepatocellular transport, detoxification, and antioxidative stress systems in mice. Am J Physiol Gastrointest Liver Physiol 295:G735–G747. [DOI] [PubMed] [Google Scholar]
- Ros JE, Libbrecht L, Geuken M, Jansen PL, Roskams TA. (2003) High expression of MDR1, MRP1, and MRP3 in the hepatic progenitor cell compartment and hepatocytes in severe human liver disease. J Pathol 200:553–560. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Shitara M, Yokota K, Hikosaka Y, Moriyama S, Yano M, Fujii Y. (2012) MRP3 gene expression correlates with NRF2 mutations in lung squamous cell carcinomas. Mol Med Rep 6:705–708. [DOI] [PubMed] [Google Scholar]
- Shen G, Kong AN. (2009) Nrf2 plays an important role in coordinated regulation of phase II drug metabolism enzymes and Phase III drug transporters. Biopharm Drug Dispos 30:345–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A. (2008) Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol 20:341–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slitt AL, Cherrington NJ, Maher JM, Klaassen CD. (2003) Induction of multidrug resistance protein 3 in rat liver is associated with altered vectorial excretion of acetaminophen metabolites. Drug Metab Dispos 31:1176–1186. [DOI] [PubMed] [Google Scholar]
- Sun Y, Ng L, Lam W, Lo CK, Chan PT, Yuen YL, Wong PF, Tsang DS, Cheung WT, Lee SS. (2008) Identification and characterization of a novel mouse peroxisome proliferator-activated receptor alpha-regulated and starvation-induced gene, Ppsig. Int J Biochem Cell Biol 40:1775–1791. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Aleksunes LM, Cui YJ, Klaassen CD. (2009) ANIT-induced intrahepatic cholestasis alters hepatobiliary transporter expression via Nrf2-dependent and independent signaling. Toxicol Sci 108:247–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang M, Zhang L, Cai H, Zhou S, Zhang J, Wang Y. (2010) Correlation of Nrf2, HO-1, and MRP3 in gallbladder cancer and their relationships to clinicopathologic features and survival. J Surg Res 164:e99–e105. [DOI] [PubMed] [Google Scholar]
- Young LC, Campling BG, Cole SP, Deeley RG, Gerlach JH. (2001) Multidrug resistance proteins MRP3, MRP1, and MRP2 in lung cancer: correlation of protein levels with drug response and messenger RNA levels. Clin Cancer Res 7:1798–1804. [PubMed] [Google Scholar]
- Zella LA, Kim S, Shevde NK, Pike JW. (2006) Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. Mol Endocrinol 20:1231–1247. [DOI] [PubMed] [Google Scholar]
- Zhang DD. (2006) Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev 38:769–789. [DOI] [PubMed] [Google Scholar]
- Zhang DD, Hannink M. (2003) Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol 23:8137–8151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, Shen R, Goodman OB, Jr, Nanus DM. (2006) Multiple androgen response elements cooperate in androgen regulated activity of the type 1 neutral endopeptidase promoter. Mol Cell Endocrinol 259:10–21. [DOI] [PubMed] [Google Scholar]
- Zollner G, Fickert P, Silbert D, Fuchsbichler A, Marschall HU, Zatloukal K, Denk H, Trauner M. (2003) Adaptive changes in hepatobiliary transporter expression in primary biliary cirrhosis. J Hepatol 38:717–727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.