Eco1p acetylates the cohesin subunit Smc3p to establish cohesion. It was believed that Smc3p-K113 acetylation promoted cohesion by antagonizing Wpl1p. We find that Eco1p acetylation promotes cohesion independently of antagonizing Wpl1p, likely by altering Smc3p head function. In addition, Eco1p targets other than Smc3-K113 help promote efficient establishment.
Abstract
Cohesin complex mediates cohesion between sister chromatids, which promotes high-fidelity chromosome segregation. Eco1p acetylates the cohesin subunit Smc3p during S phase to establish cohesion. The current model posits that this Eco1p-mediated acetylation promotes establishment by abrogating the ability of Wpl1p to destabilize cohesin binding to chromosomes. Here we present data from budding yeast that is incompatible with this Wpl1p-centric model. Two independent in vivo assays show that a wpl1∆ fails to suppress cohesion defects of eco1∆ cells. Moreover, a wpl1∆ also fails to suppress cohesion defects engendered by blocking just the essential Eco1p acetylation sites on Smc3p (K112, K113). Thus removing WPL1 inhibition is insufficient for generating cohesion without ECO1 activity. To elucidate how ECO1 promotes cohesion, we conducted a genetic screen and identified a cohesion activator mutation in the SMC3 head domain (D1189H). Smc3-D1189H partially restores cohesion in eco1∆ wpl1∆ or eco1 mutant cells but robustly restores cohesion in cells blocked for Smc3p K112 K113 acetylation. These data support two important conclusions. First, acetylation of the K112 K113 region by Eco1p promotes cohesion establishment by altering Smc3p head function independent of its ability to antagonize Wpl1p. Second, Eco1p targets other than Smc3p K112 K113 are necessary for efficient establishment.
INTRODUCTION
Cohesin is an essential, evolutionarily conserved four-subunit complex that tethers sister chromatids from their formation in S phase through metaphase (Guacci et al., 1997; Michaelis et al., 1997; Losada et al., 1998; Sumara et al., 2000; Tomonaga et al., 2000). This tethering (cohesion) enables each sister chromatid pair to achieve a bipolar attachment to the mitotic spindle and thereby promotes high-fidelity chromosome segregation at anaphase. In budding yeast, the cohesin subunits are named MCD1/SCC1, SMC1, SMC3, and SCC3/IRR1 (Figure 1A; Guacci et al., 1997; Michaelis et al., 1997). Cohesin also plays roles in chromosome condensation, transcriptional regulation, and DNA damage repair (Guacci et al., 1997; Rollins et al., 2004; Ström et al., 2004; Unal et al., 2004). These processes are temporally and spatially distinct. For example, cohesion is established during S phase, whereas condensation occurs during mitosis (Onn et al., 2008). Cohesin binds specific sites called cohesin-associated regions (CARs) for cohesion and condensation, whereas it binds any DNA adjacent to double-strand breaks to help facilitate damage repair (Blat and Kleckner, 1999; Megee et al., 1999; Laloraya et al., 2000; Kim et al., 2002; Ström et al., 2004; Unal et al., 2004). Elucidating the mechanisms underlying the temporal and spatial regulation of cohesin is crucial to understanding how cohesin performs its diverse functions and how it tethers chromatin.
FIGURE 1:
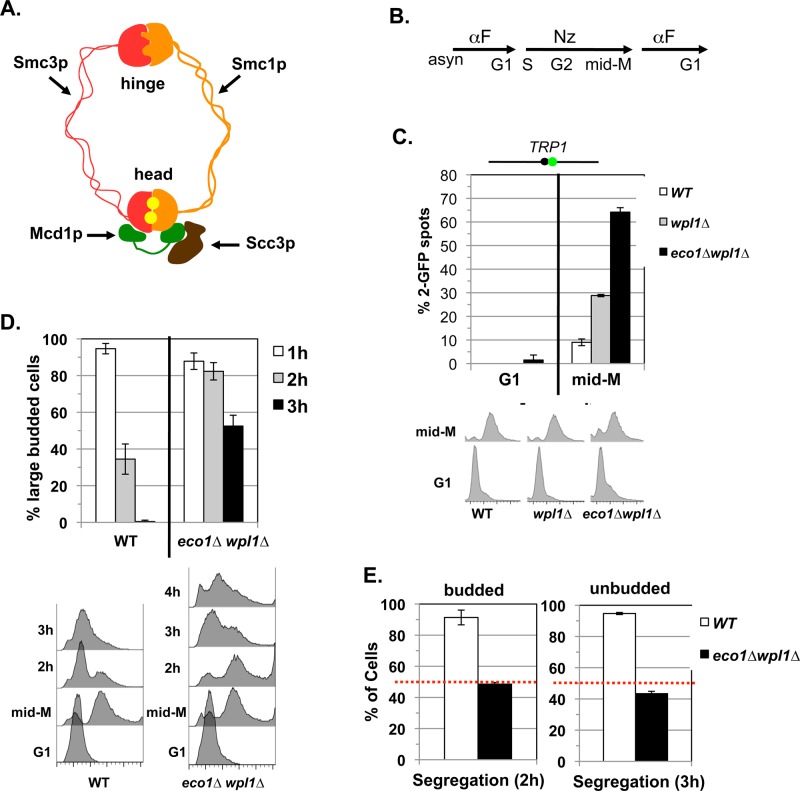
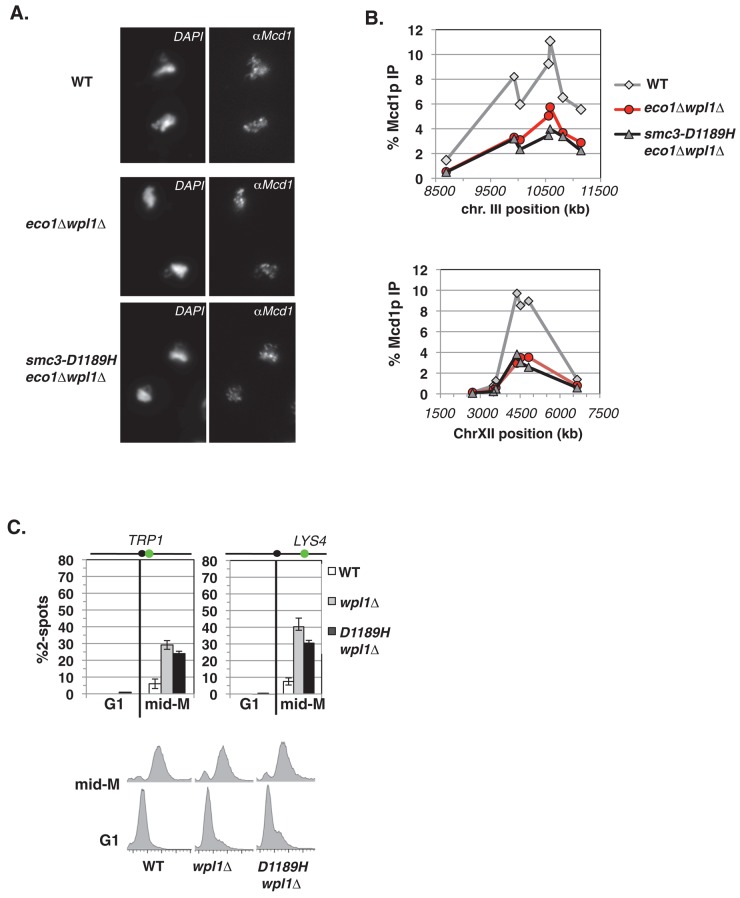
eco1∆ wpl1∆ cells lack segregation-competent cohesion. (A) Cartoon depicting the structure of the cohesin complex. Smc3p (red) interacts with Smc1p (orange) at the hinge and head domains. Mcd1p (green) binds to both the Smc1p and Smc3p heads. Scc3p (brown) binds to Mcd1p. Two Walker A/B ATPases (yellow balls) are in the heads. (B) Regimen used to assess segregation-competent segregation in cells. (C) Cohesion loss at CEN-proximal TRP1 locus at mid–M phase arrest. Haploid WT (VG3460-2A), eco1∆ wpl1∆ (VG3502-1C), and wpl1∆ (VG3513-1B) strains were arrested in G1 using αFactor at 23°C and then released and rearrested in mid–M phase at 23°C using nocodazole (Materials and Methods). The percentage of cells with two GFP spots (separated sister chromatids) is plotted (top) along with DNA content (bottom). The lack of G1 cells with two GFP spots demonstrates absence of preexisting aneuploidy. The green dot above the graph shows the position of LacO arrays relative to the centromere. (D, E) Assay for segregation-competent cohesion. Haploid WT (VG3460-2A) and eco1∆ wpl1∆ (VG3502-1C) cells were treated as in B. (D) Assessment of cell-cycle progression after release from mid–M (nocodazole) arrest. Percentage of large-budded cells remaining at 1, 2, and 3 h after release from mid–M phase arrest (top) and DNA content (bottom). (E) Segregation of chromosome IV sister chromatids after release from mid–M phase. Proper segregation in large-budded cells 2 h after release (left). Proper segregation in unbudded cells 3 h after release (right). Random segregation is 50% and is marked by a dotted red line. Data for C–E are from two independent experiments; 100–300 cells scored for each data point.
A simple model for the regulation of cohesion establishment was built around the cohesin inhibitor, termed WPL1/RAD61 in yeast (Rowland et al., 2009; Sutani et al., 2009; Chan et al., 2012). This WPL1-centric model emerged from three observations in budding yeast. First, the ECO1/CTF7 protein (Eco1p/Ctf7p) acetylates the Smc3p cohesin subunit at lysine residues 112 and 113 (K112, K113), and this K113 acetylation is required to allow chromatin-bound cohesin to establish cohesion (Skibbens et al., 1999; Tóth et al., 1999; Rolef Ben-Shahar et al., 2008; Unal et al., 2008; Zhang et al., 2008). Second, deleting WPL1 (wpl1∆) suppresses the inviability of cells deleted for ECO1 (eco1∆) or bearing the SMC3 acetylation-null smc3-K113R or smc3-K112R,K113R allele (lysine to arginine; Rolef Ben-Shahar et al., 2008; Rowland et al., 2009; Sutani et al., 2009). Third, the stability of cohesin binding to chromosomes is decreased in eco1 mutants relative to wild type but is more stable than wild type in either wpl1∆ or eco1∆ wpl1∆ cells (Chan et al., 2012). Based on these and other studies, it was proposed that a ring-like cohesin molecule topologically entraps both sister chromatids to generate cohesion (Onn et al., 2008; Chan et al., 2012). In this scenario, Wpl1p inhibits cohesion establishment by destabilizing cohesin binding (Supplemental Figure S1). Acetylation of Smc3p at K112, K113 promotes cohesion by antagonizing Wpl1p, thereby stabilizing cohesin binding/sister entrapment.
Recent data contradict the Wpl1p-centric view of cohesion regulation. Previously we and two other laboratories showed that although viable, eco1∆ wpl1∆ mutants have defects in cohesion as severe as that of eco1 mutants (Rowland et al., 2009; Sutani et al., 2009; Guacci and Koshland, 2012). We extended this result by showing that eco1∆ wpl1∆ cells are defective in the establishment of cohesion (Guacci and Koshland, 2012). These results led us to three conclusions (Guacci and Koshland, 2012). First, since budding yeast can be viable with very little, if any, sister chromatid cohesion, a second mechanism for bipolar attachment of sister chromatids to the spindle must exist. We provided evidence that this alternative mechanism in yeast results from the unusual precocious assembly of the spindle and attachment of microtubules to kinetochores on partially replicated chromosomes during S phase. Second, wpl1∆ must restore viability to eco1∆ cells by promoting an essential cohesin function distinct from sister chromatid cohesion. Several reports provide evidence that this function is condensation, as Wpl1p inhibits cohesin-dependent chromosome condensation in yeast (Guacci and Koshland, 2012; Lopez-Serra et al., 2013; Eng et al., 2014). Third, Eco1p must promote cohesion establishment independent of any need to antagonize Wpl1p inhibition. However, evidence showing that Eco1p mediates Wpl1p-independent generation of cohesion was lacking.
Our conclusions have been challenged by a disagreement in the field about the magnitude and the nature of the cohesion defect in the eco1∆ wpl1∆ cells. All groups used the gold standard in the field to assess cohesion—direct in vivo monitoring of discrete chromosome loci marked with green fluorescent protein (GFP). Three groups observed a 60–70% cohesion defect in eco1∆ wpl1∆ cells, which is as severe as seen in cohesin complex mutants (Rowland et al., 2009; Sutani et al., 2009; Guacci and Koshland, 2012). Two of these groups postulated that cohesion might still exist on chromosomes at some sites distinct from loci tagged by GFP (Rowland et al., 2009; Sutani et al., 2009). One group noted that a minor portion of minichromosomes isolated from eco1∆ wpl1∆ cells showed altered gel mobility compared with eco1 mutants, which they interpreted as evidence of cohesion (Rowland et al., 2009). If true, such residual cohesion could enable bipolar chromosome attachments and thereby promote segregation and viability. Although there is value in resolving this issue, it would not alter the fact that cohesion establishment is severely disrupted at multiple loci in eco1∆ wpl1∆ cells. Finally, one group observed only a 20% cohesion defect in eco1∆ wpl1∆ cells despite using the same genetic background as two groups observing the 60–70% defect (Rolef Ben-Shahar et al., 2008). This discrepancy might reflect secondary mutations arising during the outgrowth of eco1∆ wpl1∆ mutants. Any interpretation of cohesion in eco1∆ wpl1∆ cells is further complicated by the fact that Wpl1p plays a positive as well as an inhibitory role in cohesion. This positive role is revealed in budding yeast by a partial defect in cohesion establishment and reduced cohesin binding to chromosomes in wpl1∆ cells (Rowland et al., 2009; Sutani et al., 2009; Guacci and Koshland, 2012).
Here we provide in vivo evidence that eco1∆ wpl1∆ cells lack sufficient cohesion to promote chromosome segregation. We also conducted a genetic screen to identity mutations that restore cohesion in eco1∆ wpl1∆ cells. One such cohesion activator mutation in the Smc3p head domain changes the highly conserved aspartic acid residue 1189 to histidine (smc3-D1189H) and partially restores cohesion to eco1∆ wpl1∆ cells. The D1189H mutation restores nearly wild-type levels of cohesion to smc3-K112R,K113R cells when assayed in-cis (i.e., chimeric smc3-RR-D1189H allele). In contrast, a wpl1∆ has no effect on the major cohesion defect engendered by smc3-K112R,K113R. Our results provide strong evidence that Eco1p acetylation of Smc3p modulates its head domain to promote cohesion establishment. This modulation is independent of any antagonism of Wpl1p and occurs at a step distinct from cohesin binding to chromosomes. Our data also suggest that Eco1p has additional targets that promote efficient cohesion establishment other than the Smc3p K112, K113 residues that have been the prime focus of establishment studies.
RESULTS
eco1∆ wpl1∆ cells have little or no functional cohesion
Several groups, including ours, found major cohesion defects in eco1∆ wpl1∆ cells when directly monitoring cohesion at specific chromosomal loci, but one group reported significantly less of a defect (Rowland et al., 2009; Sutani et al., 2009; Guacci and Koshland, 2012; Rolef Ben-Shahar et al., 2008). To rule out the possibility that secondary mutations arose during outgrowth of our strains to generate the dramatic cohesion defect, we used a conditional eco1∆ wpl1∆ strain. For this purpose, we constructed a conditional ECO1 allele using the auxin-inducible degron system (AID), which entailed C-terminal addition of 3V5 and AID2, termed ECO1-AID (Materials and Methods; Eng et al., 2014). We then deleted WPL1 in the ECO1-AID background (ECO1-AID wpl1∆) and compared its phenotypes to wild-type, ECO1-AID alone, and eco1∆ wpl1∆ strains. Addition of auxin to media induces degradation of the essential Eco1-AIDp so the ECO1-AID strain becomes inviable (Supplemental Figure S2A). A wpl1∆ suppresses the auxin-dependent inviability of the ECO1-AID allele, recapitulating the ability of wpl1∆ to bypass an essential function of Eco1p (Supplemental Figure S2A). Moreover, the ECO1-AID wpl1∆ strain also exhibits an auxin-dependent benomyl sensitivity similar to that of eco1∆ wpl1∆ cells, consistent with a cohesion defect.
We next asked whether a severe cohesion defect is indeed generated in the first cell cycle after Eco1p depletion in a wpl1∆ background. After strains were arrested in G1, auxin was added to deplete Eco1-AIDp. Cells were released from G1 into media containing auxin and nocodazole to arrest them in mid–M phase under conditions in which Eco1-AIDp is depleted over this entire cell-cycle interval (Supplemental Figure S2B). Yeast cells are termed mid–M phase because sister chromatids have cohesion and are condensed (Guacci et al., 1994). Cohesion was scored at a CEN-distal (LYS4) locus. In this assay, a failure to establish or maintain cohesion leads to the presence of two GFP foci (spots) in cells. Wild-type cells have robust cohesion, as shown by few mid–M phase cells with two GFP spots, whereas ECO1-AID cells have a severe defect, as shown by the high percentage of cells with two GFP spots (Supplemental Figure S2C, left). Of importance, >70% of either ECO1-AID wpl1∆ or eco1∆ wpl1∆ cells have two GFP spots, indicating a cohesion defect as severe as seen in ECO1-AID cells (Supplemental Figure S2C, right). Few G1 cells from any strain had two GFP spots, demonstrating the absence of preexisting aneuploidy, which indicates that mid–M phase cells with two GFP spots arose due to defective cohesion. Thus the first cell-cycle depletion of Eco1p in the wpl1∆ background recapitulates studies in which eco1∆ wpl1∆ cells had both major cohesion defects and benomyl sensitivity (Sutani et al., 2009; Guacci and Koshland, 2012). This result makes it highly unlikely that secondary mutations are responsible for these eco1∆ wpl1∆ mutant phenotypes.
Observing major cohesion defects at specific loci in eco1∆ wpl1∆ cells does not rule out the possibility that cohesion exists at other chromosomal regions. If such residual cohesion exists, it should enable bipolar chromosome attachments and thereby promote sister segregation and viability. We assessed this possibility using a nocodazole-arrest release segregation assay (Figure 1B and Materials and Methods). Arrest in mid–M phase using nocodazole abrogates any early S-phase attachments that enable cohesion-independent segregation (Guacci and Koshland, 2012). Subsequent release from nocodazole arrest allows spindles to form, kinetochores to attach to microtubules, and cells to complete mitosis. We labeled the CEN4-proximal (TRP1) locus because chromosome IV is the second largest yeast chromosome. As such, it is an excellent substrate to assess whether cohesion exists at other loci along a chromosome and thereby enable bipolar attachment in mitosis to promote subsequent segregation (i.e., segregation-competent cohesion).
To assess segregation-competent cohesion, wild-type and eco1∆wpl1∆ strains were released from G1 into media containing nocodazole to induce arrest in mid–M phase as large-budded cells lacking spindles. Consistent with our previous analyses, cohesion was robust in wild-type cells and severely defective in eco1∆ wpl1∆ cells (Figure 1C). Mid–M phase–arrested cells were allowed to complete mitosis and arrest in G1 by removing nocodazole and adding α-factor (Figure 1B). We first monitored cell-cycle progression. By 2 h after release from nocodazole arrest, many wild-type cells completed anaphase and exited mitosis, as seen by a decrease in the percentage of large-budded cells and the appearance of cells with 1C DNA content (Figure 1D). In contrast, most eco1∆ wpl1∆ cells remained large budded and had 2C DNA content. By 3 h, virtually all wild-type cells had completed mitosis and arrested in G1, whereas only ∼50% of eco1∆ wpl1∆ cells had done so. This mitotic progression delay of eco1∆ wpl1∆ cells is characteristic of checkpoint-mediated delays after global cohesion abrogation via mutants in ECO1 or cohesin subunits (Skibbens et al., 1999; Stead et al., 2003; Noble et al., 2006).
We then scored chromosome IV segregation in large-budded cells 2 h after release from nocodazole arrest (Materials and Methods). More than 80% of large budded wild-type cells had one GFP spot in each daughter nucleus, indicating segregation of sister chromatids (Figure 1E, left). In contrast, only 50% of large-budded eco1∆ wpl1∆ cells showed segregation to the daughter nuclei. We also scored segregation in unbudded cells, which have completed mitosis and rearrested in G1 (Materials and Methods). Approximately 90% of unbudded wild-type cells exhibit proper chromosome IV segregation, whereas only ∼50% of unbudded eco1∆ wpl1∆ cells showed segregation (Figure 1E, right). Moreover, sub-1C and >1C peaks are observed in eco1∆ wpl1∆ cells at 4 h after release, consistent with a global defect in segregation (Figure 1D). Random segregation (50%) is expected if there is a complete lack of cohesion, leading us to conclude that eco1∆ wpl1∆ cells have little or no segregation-competent cohesion.
A genetic screen identified smc3-D1189H as a mutation that partially restores cohesion in eco1∆ wpl1∆ cells
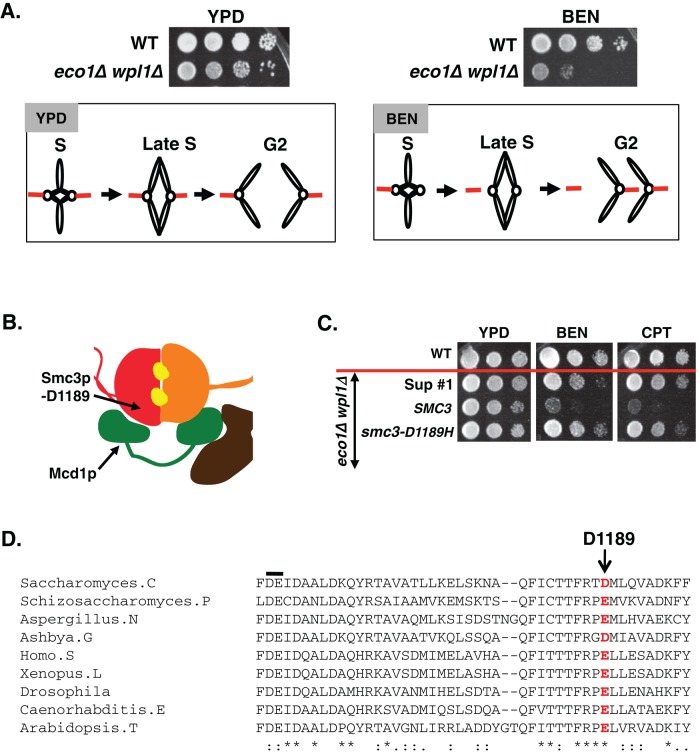
Our previous study indicated that cohesin mutants use an alternative mechanism to segregate their sister chromatids despite abrogation of cohesion (Guacci and Koshland, 2012). This cohesion-independent mechanism depends on the persistence of bipolar sister chromatid attachments to kinetochore microtubules generated in early S phase (Figure 2A). Our finding that eco1∆ wpl1∆ cells have little or no segregation-competent cohesion suggests that they also depend on this alternative pathway. This view is consistent with the observation that eco1∆ wpl1∆ cells are inviable when exposed to low levels of the microtubule-depolymerizing drug benomyl (Figure 2A; Sutani et al., 2009; Guacci and Koshland, 2012). These cells are also very sensitive to camptothecin, a topoisomerase I inhibitor that induces DNA damage, possibly because without cohesion, they are less competent to repair DNA damage (Guacci and Koshland, 2012). We reasoned that screening for eco1∆ wpl1∆ cells resistant to either drug should select for suppressor mutations that restore cohesion establishment in the absence of both Eco1p and Wpl1p (see Supplemental Figure S3 and Materials and Methods for screen details).
FIGURE 2:
Genetic screen identifies the smc3-D1189H mutation as a suppressor of eco1∆ wpl1∆ strain sensitivity to benomyl and camptothecin. (A) Rationale for screen. Haploid WT (VG3349-1B) and eco1∆ wpl1∆ (VG3503-4A) were grown to saturation at 23°C, plated at 10-fold serial dilution on YPD alone or containing 12.5 μg/ml BEN, and then incubated at 23°C for 3 d. Left, eco1∆ wpl1∆ cell viability on YPD and schematic showing sister segregation via early S-phase attachments despite failure to establish cohesion. Right, eco1∆ wpl1∆ cell inviability on BEN and schematic showing benomyl-induced loss of S-phase spindle attachments and consequent missegregation and inviability. (B) Cartoon showing localization of the D1189 residue in the SMC3 head domain. (C) Cross drug resistance of the smc3-D1189H suppressor. Haploid WT (3460-2A) and three eco1∆ wpl1∆ background strains, parent SMC3 (VG3502-1A), D1189H suppressor 1 (Sup #1), and rebuilt smc3-D1189H (VG3547-3B), were grown and plated as described in A onto YPD alone or containing 12.5 μg/ml BEN or 10 μg/ml CPT and incubated for 3 d at 23°C. Strains below the red line are eco1∆ wpl1∆ background. (D) Schematic showing evolutionary conservation of budding yeast smc3-D1189 residue (red letter). The black line above the sequence shows the conserved DE residues of the Smc3p Walker B box.
Only about 1 in 1 million eco1∆ wpl1∆ cells exhibited benomyl or camptothecin resistance. Suppressors were then tested for resistance to the other drug (i.e., benomyl-resistant clones were assayed for camptothecin resistance). Only a small number of clones exhibited dual drug resistance, suggesting that suppressing the cohesion defect of eco1∆ wpl1∆ cells required specific and rare changes in genes. All clones exhibiting dual drug resistance were subjected to whole-genome sequencing. The same amino acid change in Smc3p was found in four independent clones exhibiting dual drug resistance. This mutation is located in the Smc3p head domain and changed aspartic acid residue 1189 to histidine (D1189H; Figure 2B). To assess linkage between the smc3-D1189H mutation and the drug resistance, we reintroduced this allele in place of SMC3 in the parent eco1∆ wpl1∆ (Materials and Methods). We compared one of the screen-derived D1189H suppressors (sup #1) to the rebuilt smc3-D1189H eco1∆ wpl1∆ strain and found that they are phenotypically indistinguishable (Figure 2C). This result confirms that smc3-D1189H is responsible for the drug resistance. The D1189 residue is highly conserved evolutionarily, as the analogous position in all SMC3 family members is either an aspartic acid or a glutamic acid residue (Figure 2D).
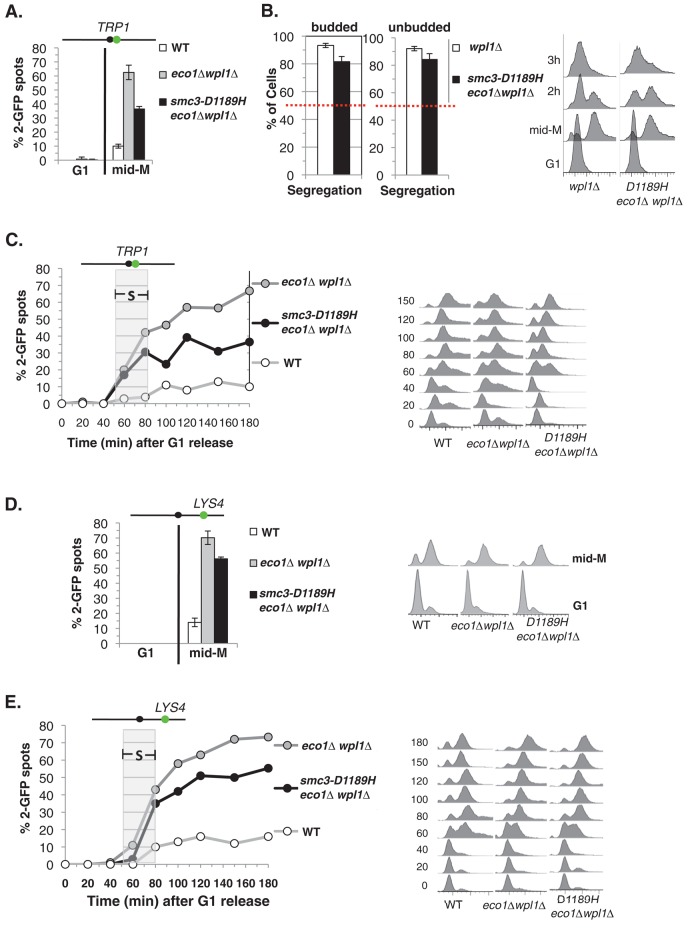
Cohesion around the centromere is essential to enable sister kinetochores to reform bipolar attachments after their transient detachment from spindle microtubules. Therefore we assumed that smc3-D1189H eco1∆ wpl1∆ cells acquired benomyl resistance because cohesion was restored at centromere-proximal regions. To test this idea, we began by comparing the level of cohesion at the centromere-linked TRP1 locus in wild-type, eco1∆ wpl1∆, and our reconstructed smc3-D1189H eco1∆ wpl1∆ strain. The smc3-D1189H eco1∆ wpl1∆ cells had restored cohesion relative to eco1∆ wpl1∆ but not fully restored to wild-type levels (Figure 3A). This level of cohesion is sufficient to explain the benomyl resistance, as it is similar to the cohesion level and benomyl resistance seen in a wpl1∆ strain (compare Figures 1C and 3A; Guacci and Koshland, 2012). We then tested whether the cohesion restored by smc3-D1189H as measured via the GFP-spot assay is functional, segregation-competent cohesion using our nocodazole-arrest release segregation assay (Figure 1B). Because smc3-D1189H eco1∆ wpl1∆ and wpl1∆ cells have similar levels of cohesion, we compared their ability to promote segregation. After release from nocodazole arrest, smc3-D1189H eco1∆ wpl1∆ segregated sister chromatids properly in 80% of cells, much better than the random 50% segregation observed in eco1∆ wpl1∆ cells and nearly as well as wpl1∆ or wild-type cells (Figure 3B). Therefore the cohesion restored by smc3-D1189H is segregation competent. Moreover, smc3-D1189H eliminates the sub-1C peak found in eco1∆ wpl1∆ cells, suggesting global restoration of cohesion (Figure 3B). Because smc3-D1189H restores cohesion to eco1∆ wpl1∆ cells, we term it a cohesion activator mutation.
FIGURE 3:
smc3-D1189H partially restores sister chromatid cohesion in eco1∆ wpl1∆ cells. (A) Cohesion loss at a CEN-proximal TRP1 locus. Haploid WT (VG3460-2A), eco1∆ wpl1∆ (VG3502-1C), and smc3-D1189H eco1∆ wpl1∆ (VG3547-3B) were arrested in mid–M phase as described in Figure 1C. The percentage of cells with two GFP signals (sister separation) is plotted. The lack of G1 cells with two GFP spots demonstrates absence of preexisting aneuploidy. Data are from four independent experiments; 100–300 cells were scored for each data point in each experiment. (B) Assay for segregation-competent cohesion. Haploid smc3-D1189H eco1∆ wpl1∆ (VG3549-7A) and wpl1∆ (VG3513-1B) cells were treated as depicted in Figure 1B and then assayed for chromosome segregation after release from mid–M arrest. Proper segregation of chromosome IV sister chromatids in large-budded cells 2 h after release (left) and in unbudded cells 3 h after release (middle) and DNA content (right). Random segregation will be 50% and is marked by a dotted red line. Data were generated simultaneously with strains in Figure 1, C–E, in two independent experiments in which 100–300 cells were scored for each data point. (C) Kinetics of cohesion loss at a CEN-proximal TRP1 locus. Strains in A released from G1 and arrested in mid–M phase as described in Figure 1C. The percentage of cells with two GFP spots is plotted (left) along with DNA content (right). Gray box shows S phase. Between 100 and 300 were cells scored for each data point. (D, E) Cohesion loss at a CEN-distal LYS4 locus. Haploid wild-type (VG3349-1B), eco1∆ wpl1∆ (VG3503-4A), and smc3-D1189H eco1∆ wpl1∆ (VG3549-7A) were arrested in mid–M phase arrest as described in Figure 1C. (D) Cohesion loss at mid–M phase arrest. The percentage of cells with two GFP spots is plotted (left) along with DNA content (right). Data were derived from two independent experiments, and 100–300 cells were scored for each data point. (E) Time course to assess kinetics of cohesion loss. The percentage of cells with two GFP spots is plotted (left) along with DNA content (right). Gray box shows S phase. Between 100 and 300 were cells scored for each data point. The green dot above the graph shows the position of LacO arrays relative to the centromere.
To further characterize the partial restoration of cohesion in smc3-D1189H eco1∆ wpl1∆ cells, we examined cohesion at TRP1 as cells progressed through the cell cycle as compared with wild-type and eco1∆ wpl1∆ cells. Cells were released from G1 into media containing nocodazole to induce mid–M phase arrest. Cell aliquots were fixed in G1 and at 20-min intervals after G1 release to assess cohesion and DNA content. As expected for wild-type cells, separated sister chromatids were rarely observed, indicative of robust cohesion (Figure 3C). The eco1∆ wpl1∆ cells exhibited separated sister chromatids (two GFP spots) beginning in S phase, consistent with a previously reported establishment defect (Guacci and Koshland, 2012). Fewer smc3-D1189H eco1∆ wp1∆ cells exhibited two GFP spots than eco1∆ wpl1∆ cells, but the kinetics of cohesion loss was similar, as it also begins during S phase. These results indicate that smc3-D1189H promotes cohesion establishment in the eco1∆ wp1∆ background, but the residual cohesion defect is due to incomplete restoration of establishment rather than a defect in cohesion maintenance. We next examined cohesion at the CEN-distal (LYS4) locus in these strain backgrounds. The smc3-D1189H allele also partially suppresses the cohesion establishment defect of eco1∆ wpl1∆ (Figure 3, D and E). Note that the suppression was less robust than at the CEN-distal locus compared with the CEN-proximal locus.
smc3-D1189H cohesin responds to regulators as well as wild-type cohesin
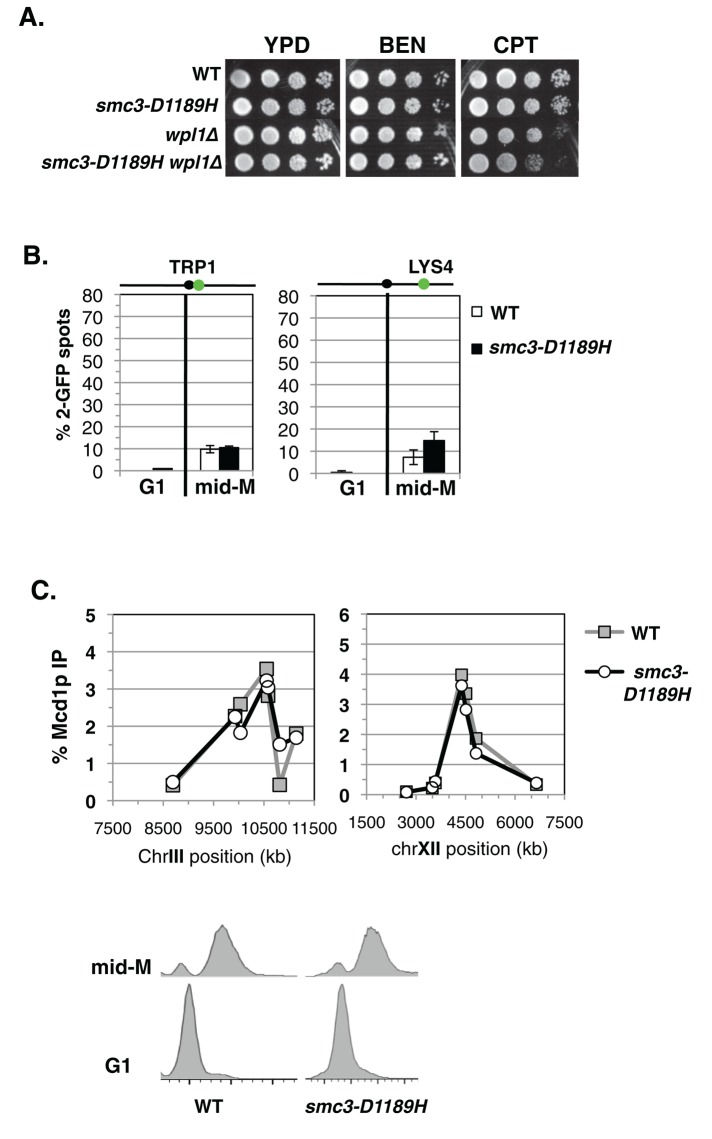
The partial restoration of cohesion by smc3-D1189H in the eco1∆ wpl1∆ background could reflect the inability of smc3-D1189H to fully compensate for cohesion defects caused by the absence of Eco1p and Wpl1p. Alternatively, it could be due to a cohesion defect caused by smc3-D1189H itself. To test whether the smc3-D1189H mutation has inherent defects in cohesion and cohesin function, we generated an otherwise wild-type yeast strain bearing smc3-D1189H as the sole SMC3 allele (Materials and Methods). Wild-type and smc3-D1189H cells were assayed for drug sensitivity and showed similar strong resistance to both benomyl and camptothecin (Figure 4A). We next assayed cohesion at CEN-proximal (TRP1) and CEN-distal (LYS4) loci, as well as cohesin binding to CARs via chromatin immunoprecipitation (ChIP). Wild-type and smc3-D1189H cells were released from G1 and arrested in mid–M phase (Materials and Methods). Few mid–M phase cells had two GFP spots in either wild-type or smc3-D1189H cells, indicating that both strains had robust cohesion (Figure 4B). Analysis of the chromosomal binding of cohesin subunits shows that they colocalize, making the analysis of any one a surrogate marker for cohesin binding (Glynn et al., 2004; Lengronne et al., 2004; Heidinger-Pauli et al., 2010). We used antibodies against Mcd1p (αMcd1p) as a marker to detect cohesin binding to chromosomes. ChIP showed no difference in Mcd1p binding at the CEN-proximal CARC1 or the CEN-distal CARL1 in these two strains (Figure 4C). Thus smc3-D1189H appears fully competent to promote cohesion and to respond normally to cohesion regulation by both Eco1p and Wpl1p. Finally, the Scc2p loader complex is required for cohesion and viability in wild-type cells (Ciosk et al., 2000). Smc3-D1189H cells also require Scc2p for both viability and cohesion (Supplemental Figure S4). These results show that smc3-D1189H cohesin functions as well as wild-type (WT) cohesin when normal cohesin regulation is present. Therefore the partial suppression of the cohesion defect of eco1∆ wpl1∆ cells by smc3-D1189H is not due to an inherent cohesion defect associated with smc3-D1189H. Instead, it indicates that smc3-D1189H restores only a subset of the Eco1p and/or Wpl1p activities.
FIGURE 4:
smc3-D1189H cohesin is fully functional in a WT background. (A) Assessing the drug sensitivity of smc3-D1189H cells. Haploid WT (VG3599-9C), smc3-D1189H (VG3600-13C), wpl1∆ (VG3604-4C), and smc3-D1189H wpl1∆ (VG3605-5D) were grown and plated onto YPD alone or containing BEN (12.5 μg/ml) or CPT (15 μg/ml) as described in Figure 2A and then incubated at 23°C for 3 d for YPD and CPT and 4 d for BEN. (B) Cohesion loss in mid–M phase cells. WT and smc3-D1189H cells were released from G1 and arrested in mid–M phase as described in Figure 1C. Left, cohesion loss at CEN-proximal TRP1 locus in haploid WT (VG3599-9C) and smc3-D1189H (VG3600-13C) cells. Right, cohesion loss at CEN-distal LYS4 locus in haploid WT (VG3557-2A) and smc3-D1189H (VG3558-2D) strains. Cohesion loss is the percentage of cells with two GFP spots. Data were derived from two independent experiments. Between 100 and 300 cells were scored for each data point in each experiment. (C) ChIP of Mcd1p in mid–M phase–arrested cells. Haploid WT (VG3599-9C) and smc3-D1189H (VG3600-13C) mid–M phase–arrested cells from B were subjected to ChIP using αMcd1p antibodies (top) and DNA content determined (bottom). Mcd1p binding was assessed by quantitative PCR. Data are presented as percentage of total DNA assayed using the same primer pairs at each site. Left, chromosome III pericentric CARC1. Seven primer pairs used to assay Mcd1p binding at loci spanning an ∼2.6-kb region including CARC1 of chromosome III. Right, chromosome XII CEN-distal CARL1. Seven primer pairs used to assay Mcd1p binding at loci spanning an ∼4.5-kb region including CARL1 of chromosome XII. WT (gray line, gray squares) and smc3-D1189H (black line, open circles).
smc3-D1189H does not suppress the cohesion defect of a wpl1∆
The residual cohesion defect in smc3-D1189H eco1∆ wpl1∆ (35% sister chromatid separation at TRP1) is very similar to that of a wpl1∆ mutant alone (compare Figures 1C and 3A; Guacci and Koshland, 2012). This similarity could reflect that smc3-D1189H robustly suppresses the eco1∆ without affecting the cohesion defect caused by a wpl1∆. Alternatively, it could reflect complete suppression of wpl1∆, but only partial suppression of the eco1∆. We first addressed whether smc3-D1189H suppresses a wpl1∆. One signature of a wpl1∆ is a reduction in cohesin bound at CARs as compared with WT cells (Rowland et al., 2009; Sutani et al., 2009). In contrast, an eco1 mutant does not reduce cohesin binding at CARs (Noble et al., 2006). Therefore, if smc3-D1189H in eco1∆ wpl1∆ suppressed the loss of Wpl1p, we would expect it to increase cohesin binding to WT levels. We examined whether smc3-D1189H affects cohesin binding to chromosomes in the eco1∆ wpl1∆ background. Cells were released from G1 and then rearrested in mid–M phase (Materials and Methods). Mid–M phase cells were processed to assess cohesin binding to chromosomes using both chromosome spreads and ChIP.
We first used chromosome spreads to qualitatively assess cohesin binding to chromosomes. We detected robust Mcd1p staining on chromosomal DNA in wild-type, eco1∆ wpl1∆, and smc3-D1189H eco1∆ wpl1∆ cells, indicative of broad cohesin binding (Figure 5A). We then used ChIP to perform a quantitative analysis of cohesin binding at two CARs—one in the pericentric region of chromosome III (CARC1) and one at a CEN-distal locus (CARL1). Mcd1p binding at either CAR site was reduced by twofold to threefold in the eco1∆ wpl1∆ cells compared with wild type (Figure 5B). This degree of reduction was previously reported for wpl1∆ and eco1∆ wpl1∆ cells (Sutani et al., 2009). The smc3-D1189H eco1∆ wpl1∆ cells exhibit the same reduced cohesin binding as the parent eco1∆ wpl1∆ cells (Figure 5B). Thus smc3-D1189H did not suppress the cohesin-loading defect characteristic of a wpl1∆. Moreover, this result indicates that the improved sister chromatid cohesion engendered by smc3-D1189H in eco1∆ wpl1∆ cells occurs via a mechanism distinct from increasing cohesin localization to CARs.
FIGURE 5:
Smc3-D1189H fails to suppress the characteristic wpl1∆ defects of reduced cohesin binding and partial cohesion loss. (A–C) Haploid wild-type (WT; VG3349-1B), eco1∆ wpl1∆ (VG3503-4A), and smc3-D1189H eco1∆ wpl1∆ (VG3549-7A) cells were arrested in mid–M phase as described in Figure 1C. (A) Chromosome spreads of mid–M phase cells. WT (top), eco1∆ wpl1∆ (middle), and smc3-D1189H eco1∆ wpl1∆ (bottom). Cells were processed to detect chromosomal DNA (4′,6-diamidino-2-phenylindole) and cohesin using αMcd1p antibodies. (B) ChIP of Mcd1p in mid–M phase cells. Cells were fixed and processed for ChIP using αMcd1p antibodies. WT (gray line, gray diamonds), eco1∆ wpl1∆ (red line, red circles), and smc3-D1189H eco1∆ wpl1∆ (black line, gray triangles). Mcd1p binding was assessed as described in Figure 4C. Top, chromosome III pericentric CARC1. Bottom, chromosome XII CEN-distal CARL. (C) Effect of smc3-D1189H on cohesion loss in a wpl1∆ background. Haploid WT, wpl1∆, and smc3-D1189H wpl1∆ cells were arrested in mid–M phase as described in Figure 1C. Left, cohesion loss at CEN-proximal TRP1 locus assayed in haploid WT (VG3460-2A), wpl1∆ (VG3604-4C), and smc3-D1189H wpl1∆ (VG3605-5D) strains. Right, cohesion loss at CEN-distal LYS4 locus assessed in haploid WT (VG3349-1B), wpl1∆ (VG3626-2E), and smc3-D1189H wpl1∆ (VG3627-3C) strains. Bottom, DNA content. The percentage of cells with two GFP signals (sister separation) is plotted. Data were derived from two independent experiments; 100–300 cells were scored for each data point in each experiment.
To assess specifically whether smc3-D1189H affects the wpl1∆ cohesion defect, we analyzed its effect when Eco1p is present. For this purpose, we constructed an smc3-D1189H wpl1∆ strain and compared its phenotype to that of a wpl1∆ strain. The smc3-D1189H is unable to suppress the slight camptothecin sensitivity of a wpl1∆ (Figure 4A). In addition, the smc3-D1189H wpl1∆ strain exhibited a very similar cohesion defect as the wpl1∆ strain (Figure 5C). These results suggest that smc3-D1189H partially restores cohesion in eco1∆ wpl1∆ cells by recapitulating Eco1p activity rather than compensating for the loss of Wpl1p.
smc3-D1189H suppresses the requirement for Eco1p-mediated Smc3p K112 K113 acetylation in cohesion generation
Eco1p acetylates sites on cohesin subunits and cohesin regulators (Ivanov et al., 2002; Rolef Ben-Shahar et al., 2008; Unal et al., 2008; Zhang et al., 2008). Smc3p is acetylated at both K112 and K113, but K113 had been shown to be critical for cohesion (Unal et al., 2008; Zhang et al., 2008). The function and identity of other sites remain largely untested. Therefore it was possible that smc3-D1189H suppressed the eco1∆ wpl1∆ cohesion defect by providing the function of all the Eco1p substrates or of only a subset of them.
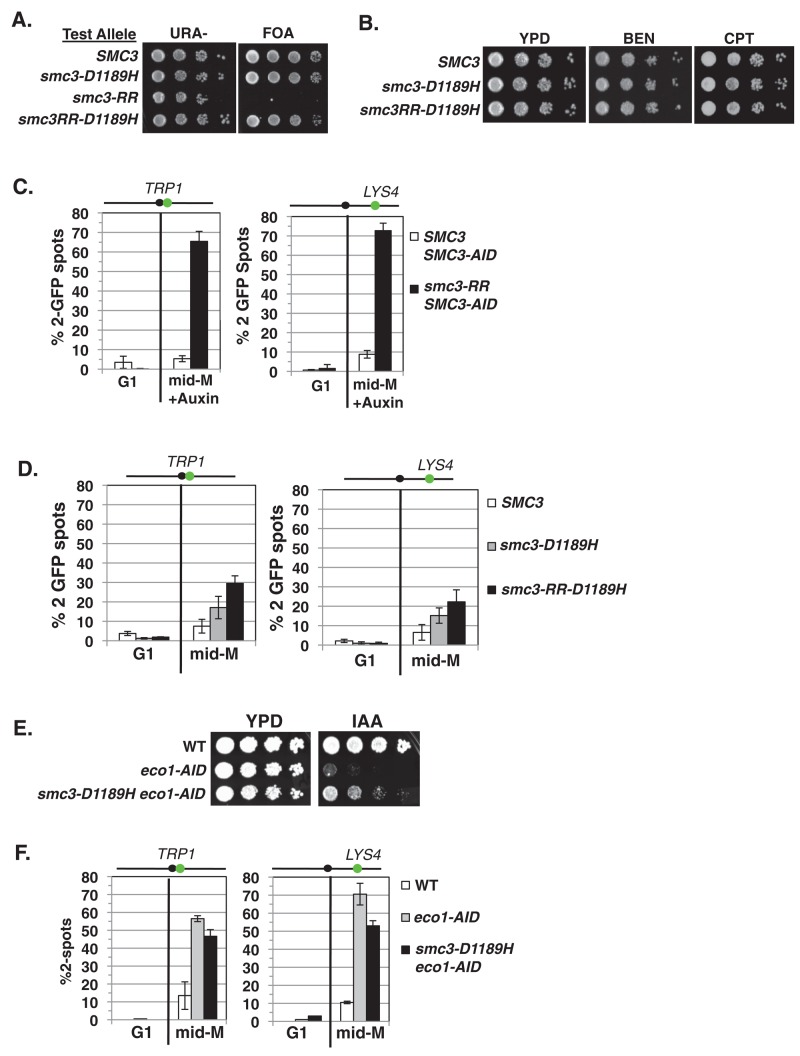
To distinguish between these possibilities, we first tested whether smc3-D1189H suppressed inviability and major cohesion defects associated with the failure to acetylate Smc3p K112 and K113 (Unal et al., 2008; Zhang et al., 2008). To assess bypass of the smc3-K112R, K113R acetyl-null (smc3-RR) inviability, we used an SMC3 shuffle strain in which the endogenous SMC3 is deleted but cells are kept viable by the presence of plasmid pEU42 (SMC3 URA3 CEN). We integrated a second “test SMC3 allele” —wild-type SMC3, smc3-D1189H, the K112R, K113R acetyl-null (smc3-RR), or a chimeric smc3-RR-D1189H allele—into the shuffle strain at the LEU2 locus. These test alleles were assayed for ability to support viability as the sole SMC3 source by growth on media containing 5-fluoroorotic acid (FOA; Materials and Methods). FOA selectively kills URA3 cells, thereby selecting for cells that had lost plasmid pEU42. As expected, all strains grew well on URA– medium because of the presence of WT SMC3 on pEU42 (Figure 6A). On FOA medium, cells with WT SMC3 and smc3-D1189H test alleles grew well, whereas the smc3-RR allele could not support viability. Cells bearing the chimeric smc3-RR-D1189H allele grew as well as on FOA medium as WT and smc3-D1189H cells, demonstrating that the smc3-D1189H mutation suppresses the inviability of the RR mutation (Figure 6A). We then compared the drug sensitivity of cells bearing only the WT SMC3, smc3-D1189H, or chimeric smc3-RR-D1189H test alleles as the sole SMC3. All strains showed equal resistance to both benomyl and camptothecin, suggesting that chimeric allele efficiently generated cohesion (Figure 6B).
FIGURE 6:
smc3-D1189H robustly suppresses the cohesion defect of the smc3-K112R, K113R (RR) mutation. (A) Plasmid shuffle to assess viability of the chimeric smc3-RR-D1189H allele. Haploid shuffle strain VG3464-16C bearing plasmid pEU42 (SMC3 CEN URA3) and a second SMC3 “test allele,” SMC3, smc3-D1189H, smc3-RR, or chimeric smc3RR-D1189H, was grown and plated as described in Figure 2A onto URA–dropout or FOA-containing media. Plates were incubated 3 d at 23°C. (B) Assessment of drug sensitivity. Haploid SMC3 (MB45-1A), smc3-D1189H (MB46-1A), or chimeric smc3-RR-D1189H (MB47-1A) strains grown and plated as described in Figure 2A onto YPD, BEN, and CPT and incubated at 30°C for 2 d. (C). Cohesion loss of in smc3-RR cells. Haploids bearing SMC3-AID and a second SMC3 allele, either WT or smc3-RR, were depleted for SMC3-AID from G1 through mid–M phase arrest. Left, cohesion loss at CEN-proximal TRP1 locus assessed in haploid SMC3 SMC3-AID (MB84-1A) and smc3-RR SMC3-AID (MB83-1A) strains. Right, cohesion loss at CEN-distal LYS4 locus assessed in haploid SMC3 SMC3-AID (MB81-1A) and smc3-RR SMC3-AID (MB79-1A) strains. The percentage of cells with two GFP spots (sister separation) is plotted. (D) Cohesion loss of smc3-RR-D1189H in mid–M phase–arrested cells. Haploid SMC3, smc3-D1189H and chimeric smc3-RR-D1189H were arrested in mid–M phase as described in Figure 1B. Left, cohesion loss at CEN-proximal TRP1 locus assessed in haploid wild-type (SMC3; MB65-1A), smc3-D1189H (MB66-1A), or chimeric smc3-RR-D1189H (MB67-1A) strain. Right, cohesion loss at CEN-distal LYS4 locus assessed in haploid strains from B. The percentage of cells with two GFP spots (sister separation) is plotted. (E, F) Assessing smc3-D1189H ability to suppress Eco1p depletion. (E) Viability after Eco1p depletion. Haploid WT (VG3620-4C), ECO1-AID (VG3662-1D), and smc3-D1189H ECO1-AID (VG3663-2E) were grown and plated as described in Figure 2A onto YPD alone or containing auxin (750 μM) and then incubated at 23°C for 2 d. (F) Cohesion loss after auxin-mediated Eco1p depletion from G1 cells through mid–M phase arrest. Left, cohesion loss at CEN-proximal TRP1 locus assayed in haploid strains WT (VG3460-2A), ECO1-AID2 (VG3659-1A) and smc3-D1189H ECO1-AID2 (VG3663-2E) strains. Right, cohesion loss at CEN-distal LYS4 locus assessed in haploid WT (VG3620-4C), ECO1-AID2 (VG3646-1A) and smc3-D1189H ECO1-AID2 (VG3650-1E) strains. The percentage of cells with two GFP signals (sister separation) is plotted. (C, D, F) Data were derived from two independent experiments; 100–300 cells were scored for each data point in each experiment.
To test the suppression of the smc3-RR cohesion defect by smc3-D1189H, we compared cohesion at CEN-proximal and CEN-distal loci in cells expressing only chimeric smc3-RR-D1189H allele with cells expressing only the smc3-RR allele. Making this comparison was complicated by the fact that the smc3-RR allele cannot support viability (Unal et al., 2008; Zhang et al., 2008). Consequently, the smc3-RR allele was assayed in a strain bearing an smc3 temperature-sensitive allele at nonpermissive temperature and shown to have a major cohesion defect (Unal et al., 2008; Zhang et al., 2008). However, we wanted to assess the smc3-RR allele at the low temperature (23°C) used for our previous assays. Therefore we constructed a parent strain bearing a conditional SMC3-AID allele, which enables the SMC3-AIDp to be rapidly degraded at 23°C upon auxin addition, rendering cells inviable and severely defective for cohesion (Supplemental Figure S5, A–C). We integrated a second SMC3 allele, either wild-type SMC3 or smc3-RR, into the SMC3-AID–bearing strain. As expected, the SMC3 allele suppressed the SMC3-AID inviability on auxin, whereas the smc3-RR failed to do so (Supplemental Figure S5C). We next assessed cohesion at CEN-proximal and CEN-distal loci in cells released from G1 and arrested in mid–M phase under conditions in which SMC3-AIDp was depleted over this entire cell-cycle window (Supplemental Figures S2B and S5D). The SMC3 allele enabled robust cohesion, whereas the smc3-RR cells were severely compromised for cohesion, as ∼70% of cells had two GFP spots (Figure 6C), a value similar to that for strains bearing only SMC3-AID as the sole SMC3 (Supplemental Figure S5B). We then examined cohesion at mid–M phase in strains in which the sole SMC3 allele is wild-type SMC3, smc3-D1189H, or the chimeric smc3-RR-D1189H allele (Supplemental Figure S5E). Only 20–30% of cells in the chimeric smc3-RR-D1189H strain exhibited two GFP spots, a dramatic improvement over the cohesion defect of the smc3-RR strain and similar to that seen in either WT SMC3 or smc3-D1189H cells (Figure 6D). These results indicate that the smc3-D1189H mutation strongly suppresses the cohesion defect generated by blocking K112, K113 acetylation.
Evidence that Eco1-dependent acetylation of sites other than Smc3-K112, K113 are required for efficient cohesion establishment
We next addressed whether smc3-D1189H could suppress loss of all Eco1p acetylation targets in cells as well as it suppressed the smc3-K112, K113 acetyl null. Because eco1∆ cells are not viable, we used strains bearing a conditional ECO1-AID allele and either wild-type SMC3 or smc3-D1189H as the sole SMC3 alleles in cells. As expected, wild-type haploids grew on yeast extract/peptone/dextrose (YPD) medium either alone or containing auxin (Figure 6E). Auxin-containing medium prevented growth of ECO1-AID cells, but a low-level of viability was detected when the smc3-D1189H allele was present, indicating at best a weak suppression of the essential ECO1 function (Figure 6E).
We then examined cohesion at CEN-proximal and CEN-distal loci in these three strains after auxin-mediated ECO1-AIDp depletion from G1 through arrest in mid–M phase (Supplemental Figures S2B and S5F). As expected, the wild-type cells had robust cohesion (10% two GFP spots), whereas the ECO1-AID strain had a severe cohesion defect (60–70% two GFP spots; Figure 6F). The smc3-D1189H ECO1-AID strain also showed a strong cohesion defect, with only modest improvement in cohesion at CEN-proximal and CEN-distal loci, respectively. Thus the chimeric smc3-RR-D1189H allele in a WT background generated cohesion more efficiently than the smc3-D1189H allele did in an ECO1-AID background: 15 and 30% better at TRP1 and at LYS4, respectively (compare Figure 6, D and F). Therefore, whereas the smc3-RR and ECO1-AID alone have a similar ∼70% cohesion defect, smc3-D1189H suppresses the smc3-RR cohesion defect much better than that of an Eco1-AIDp depletion. These results suggest that the D1189H mutation effectively provides the K112, K113 acetylation function but that other Eco1p acetylation targets are necessary for efficient cohesion generation.
smc3-D1189H activates cohesion by a Wpl1p-independent mechanism
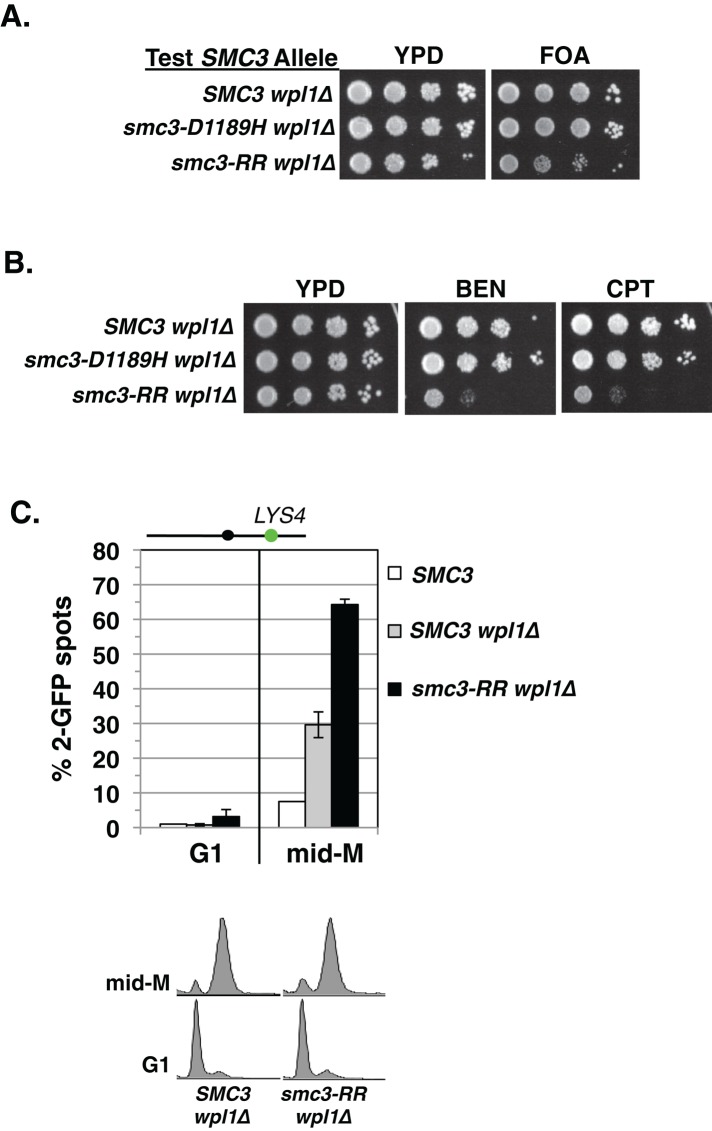
Previous work showed that a wpl1∆ suppresses the smc3-RR allele lethality (Rolef Ben-Shahar et al., 2008). This result raised the prospect that in the chimeric smc3-RR-D1189H allele, the D1189H mutation merely blocked the anticohesion function of Wpl1p. If true, then a wpl1∆ smc3-RR strain should be phenotypically identical to the chimeric smc3-RR-D1189H strain. Therefore we compared wild-type and smc3-RR wpl1∆ strains for viability, drug sensitivity, and cohesion generation. We first used a wpl1∆ SMC3 shuffle strain to assess smc3-RR wpl1∆ viability and drug sensitivity compared with SMC3 wpl1∆. As previously reported, the wpl1∆ suppresses smc3-RR allele inviability (Figure 7A). However, unlike the chimeric smc3-RR-D1189H strain, the smc3-RR wpl1∆ strain is highly sensitive to both benomyl and camptothecin, suggestive of a major cohesion defect (Figure 7B). Indeed, when we examined cohesion in mid–M phase–arrested cells, smc3-RR wpl1∆ cells had as dramatic a defect (∼70% two GFP spots) as the smc3-RR cells (Figure 7C). Thus wpl1∆ failed to suppress the cohesion defect engendered by smc3-RR, whereas smc3-D1189H suppressed it very well. These results demonstrate that smc3-D1189H bypasses the smc3-RR defect in cohesion by a mechanism distinct from antagonizing Wpl1p. Moreover, it demonstrates that a Wpl1p-independent step is required for sister chromatid cohesion.
FIGURE 7:
A wpl1∆ suppresses the inviability of smc3-RR–bearing cells but not the cohesion defect. (A) Plasmid shuffle to assess viability of smc3-RR wpl1∆ cells. Haploid wpl1∆ SMC3 shuffle strain (VG3578-1A) bearing pEU42 (SMC3 CEN URA3) and a second integrated test SMC3 allele—WT (SMC3 wpl1∆), smc3-D1189H (smc3-D1189H wpl1∆), or smc3-RR (smc3-RR wpl1∆)—were grown and plated as described in Figure 2A onto URA–dropout or FOA-containing media. Plates were incubated 2 d at 30°C. (B) Assessment of smc3-RR wpl1∆ strain drug sensitivity. Haploid SMC3 wpl1∆ (MB48-1A), smc3-D1189H wpl1∆ (VG3627-3C), and smc3-RR wpl1∆ (MB50-1A) strains were grown and plated as described in Figure 2A onto YPD alone or containing BEN (10 μg/ml) or CPT (10 μg/ml) and incubated at 23°C for 3 d for YPD and 4 d for BEN and CPT plates. (C) Cohesion loss at CEN-proximal locus LYS4. Haploid wild-type (WT; VG3627-3C), wpl1∆ (SMC3 wpl1∆; MB48-1A), and smc3-RR wpl1∆ (MB50-1A) strains arrested in mid–M phase as described in Figure 1B. Cells were scored for cohesion loss (top) and DNA content (bottom). Data are derived from two independent experiments; 100–300 cells were scored for each data point in each experiment.
DISCUSSION
The prevailing Wpl1p-centric model posits that the Eco1p acetyltransferase promotes cohesion establishment simply by antagonizing Wpl1p-mediated inhibition (Rowland et al., 2009; Sutani et al., 2009; Chan et al., 2012). Contrary to this model, we previously showed that eco1∆ wpl1∆ cells have little or no cohesion, based on in vivo monitoring of specific chromosomal GFP-tagged loci (Guacci and Koshland, 2012). We suggested that Eco1p promotes cohesion establishment by a Wpl1p-independent mechanism. Here we provide additional in vivo metrics to support the absence of cohesion in eco1∆ wpl∆ cells. First, we used a nocodazole arrest–release assay, which uses proper chromosome segregation as a sensitive readout for the presence of residual cohesion on a chromosome. We find that eco1∆ wpl1∆ cells, like cohesin subunit mutants, lack segregation competent cohesion (this study; Guacci and Koshland, 2012).
The lack of cohesion in eco1∆ wpl1∆ is also supported by published cytological observations from other laboratories. Before anaphase, yeast cells have a short mitotic spindle overlaid by a short bilobed barrel of centromeres and a single DNA mass (Pearson et al., 2004; Yeh et al., 2008). However, cohesion-defective yeast precociously elongate their spindle to generate two widely spaced centromere clusters and two separated DNA masses (Yeh et al., 2008). Both an eco1 mutant and an eco1∆ wpl1∆ double mutant exhibit the same phenotype of widely spaced centromere clusters and two separated DNA masses (Skibbens et al., 1999; Chan et al., 2012), indicative of an equivalent global failure in spindle-restraining cohesion.
We also identified smc3-D1189H as a mutation that significantly restores cohesion in eco1∆ wpl1∆ cells as measured by both the GFP-spot assay and presence of segregation-competent cohesion. Taken together, these independent in vivo measures of cohesion strongly corroborate our previous conclusion that wpl1∆ poorly, if at all, suppresses the cohesion defect of eco1∆. Moreover, because both eco1∆ wpl1∆ and wpl1∆ smc3-RR cells exhibit major defects in cohesion and are very sensitive to both benomyl and camptothecin, we propose that these dual drug sensitivities represent signatures of cohesion-defective cells. Thus Eco1p promotes cohesion establishment by a Wpl1p- independent mechanism that was missed by previous studies.
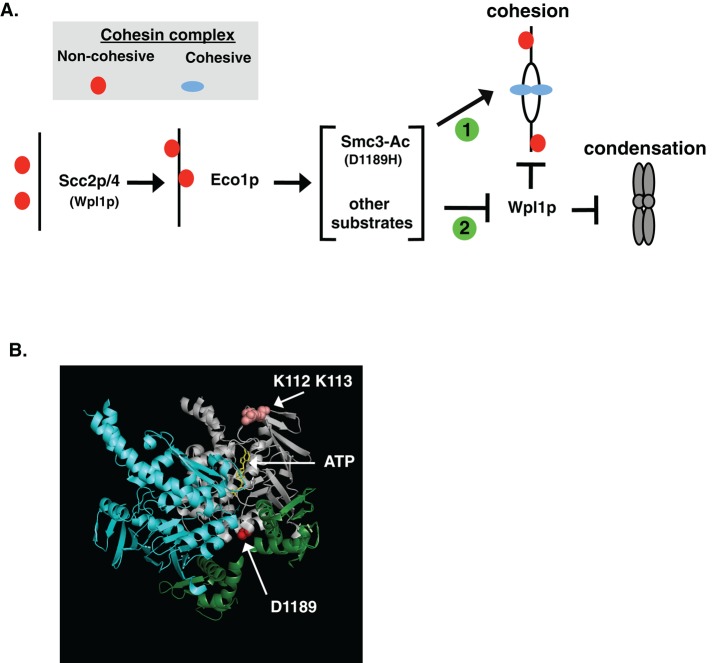
The characterization of the smc3-D1189H cohesion activator mutation provides important clues about this unrecognized Wpl1p-independent mechanism for cohesion generation. Smc3-D1189H robustly suppresses the cohesion defect of an acetylation-null smc3-K112R, K113R, whereas a wpl1∆ fails to suppress the K112R, K113R cohesion defect. This stark difference between smc3-D1189H and wpl1∆ further validates that Eco1p acetylation of Smc3p K112, K113 promotes cohesion by Wpl1p-independent mechanism (Figure 8A, arrow 1). The K112, K113, and D1189 and residues are in the Smc3p head domain, suggesting a common mechanism for modulating cohesin. No crystal structure of the Smc3p head is available, and so we used the structure of the related Smc1p head bound to the Mcd1p C-terminus as a surrogate for Smc3p head binding to the Mcd1p N-terminus (Haering et al., 2004). Although D1189 does not appear to be in close proximity to K112, K113 residues, it is in a region that could alter interactions with Mcd1p and or affect Smc3p ATPase function (Figure 8B). The latter is intriguing, as we previously proposed a connection between K112, K113 acetylation and Smc3 ATPase function based on genetic interactions between mutations in K112, K113 and mutations of Walker B residues required for hydrolysis residues (Heidinger-Pauli et al., 2010).
FIGURE 8:
Regulatory and structural model for Eco1p-mediated regulation of cohesin. (A) Model of cohesin regulation. Eco1p-mediated acetylation of Smc3p and other targets play a Wpl1p-independent role to promote cohesion establishment (arrow 1) and overcome Wpl1p inhibition of condensation and cohesion (T-bar 2). Cohesin in the noncohesive form (red circles) and a cohesive form (blue ovals) are shown. (B) smc3-D1189 modeled on Smc1p crystal structure. Shown are the head domains of Smc3p (gray) and Smc1p (blue) bound as a dimer to Mcd1p N-terminus and C-terminus (green). Indicated by arrows are the D1189 residue (red) at the bottom of Smc3p, the ATP molecule in the Smc3p Walker A/B box (yellow), and the Smc3-K112, K113 residues (salmon colored).
The smc3-D1189H allele suppresses the cohesion defect of smc3-K112R, K113R much better than the defect when ECO1 function is lost. This difference suggests that Eco1p acetylates other targets besides Smc3p K113 to promote cohesion (Figure 8A, arrow 1). The view is supported by the observations that Smc3 K113 and K112, K113 acetyl mimics (glutamine or asparagine residues) only partially suppress the cohesion defect of reduced Eco1p activity (Unal et al., 2008). Eco1p acetylates itself, Mcd1p (Scc1p), Scc3p, and the cohesin regulator, Pds5p (Ivanov et al., 2002). Of these, only the Mcd1p substrate has been examined, and its acetylation has no role during S-phase cohesion but instead promotes DNA damage repair cohesion (Heidinger-Pauli et al., 2009). Further studies of other Eco1p targets are needed to elucidate how cohesion is regulated.
We find that although a wpl1∆ does restore viability to smc3-RR or eco1∆ cells, it fails to restore cohesion. Similarly, the smc3-K112R, K113Q acetyl-mimic allele restores viability without restoring cohesion (Guacci and Koshland, 2012). We previously showed that the smc3-K112R, K113Q allele, like a wpl1∆ in eco1∆ background, restores condensation (Guacci and Koshland, 2012). This led to the idea that Smc3p acetylation overcomes Wpl1p inhibition of an essential cohesin function, likely condensation (Figure 8A, T-bar 2; Guacci and Koshland, 2012). smc3-D1189H restores viability to smc3-RR cells without restoring cohesion, which fits with the idea that D1189H mimics acetylation to promote condensation. The limited ability of smc3-D1189H to suppress the inviability of ECO1-AID depletion suggests that other Eco1p targets besides Smc3-K112, K113, possibly Wpl1p itself, also contribute to condensation (Figure 8A, T-bar 2).
The fact that cohesion is defective in both eco1∆ wpl1∆ and smc3-K112R,K113R cells provides insights into the mechanism of cohesion (this study; Guacci and Koshland, 2012). Cohesin binding to chromosomes is as stable in smc3-K112R, K113R cells as in wild-type cells but is more stable in eco1∆ wpl1∆ cells (Rowland et al., 2009; Chan et al., 2012). Therefore the failure to acetylate Smc3p does not compromise cohesin binding to chromatin but likely mediates a subsequent step necessary for cohesion. Consistent with this conclusion, Smc3-D1189H does not alter cohesin binding levels on chromosomes in eco1∆ wpl1∆ cells despite partially restoring cohesion. We suggest that Smc3p acetylation/smc3-D1189Hp promote cohesion establishment by converting the stably bound cohesin to a tethering form (Figure 8A, arrow 1). Recent studies support the idea that a second step distinct from stable cohesin binding to DNA is required for cohesion maintenance (Eng et al., 2014; Tong and Skibbens, 2014).
The nature of this second step remains to be determined. A new study on the related Smc condensin complex provided evidence for a two-step mode of DNA binding (Piazza et al., 2014). One can envision cohesin having two sites of different DNA binding that are differentially regulated—one used for cohesin binding and the second for sister tethering. Alternatively, models from studies of the Smc-like Rad50 or bacterial Smc complexes suggest that Smc complexes oligomerize, potentially by changes in the coiled-coil domain conformation (Hopfner et al., 2002; Woo et al., 2009; Bürmann et al., 2013). Finally, a recent study of bacterial Smc complexes revealed that its kleisin (Mcd1p) subunit binds the coiled coils as well as the Smc heads (Bürmann et al., 2013). This structure provides a potential mechanism by which Smc3p acetylation and/or ATPase could alter kleisin (Mcd1p) binding to the coiled-coil domain. Whether this alters Mcd1p-Smc head binding, Smc3p head conformation, or longer-range cohesin structure remains an open question. Testing these ideas will require in vitro biochemical assays for which the D1189H mutation provides a powerful new reagent.
Do other organisms require additional Eco1p acetylation sites and the Wpl1p-independent function of Eco1p for cohesion establishment? One functional study in Schizosaccharomyces pombe suggested that additional sites besides the Smc3p K112, K113 equivalent lysines (Psm3-K105R, K106R) are needed for centromere-proximal cohesion (Feytout et al., 2011). In contrast, the existence of a Wpl1p-independent function for Eco1p might appear less likely, as the cohesion defect associated with reduced Eco1p activity in S. pombe and vertebrate cells appears to be significantly suppressed by reduction in WPL1 activity (Gandhi et al., 2006; Feytout et al., 2011; Vaur et al., 2012). However, closer scrutiny suggests that this conclusion may be premature. In vertebrates, WAPL (Wpl1p orthologue) removes ∼95% of chromosomally bound cohesin from prophase to metaphase (Losada et al., 1998; Sumara et al., 2000; Gandhi et al., 2006; Kueng et al., 2006). The remaining 5% of cohesin is sufficient to tether sisters, although cohesion is less robust and sisters become more separated and resolved. Small interfering RNA (siRNA) depletion of the ECO1 orthologue ESCO2 generates metaphase chromosomes with significantly defective cohesion, whereas codepletion of WAPL and ESCO2 was reported to reduce this cohesion defect (Gandhi et al., 2006). However, these experiments compared distinct chromosomal states—prophase (siRNA both WAPL and ESCO2) versus metaphase (siRNA of just ESCO2)—that have greatly different levels of cohesin bound. The extra bound cohesin on the prophase-like chromosomes could obscure a significant cohesion defect. Therefore the existence of a WAPL-independent function of Eco1p remains an open question in vertebrates.
In S. pombe, unlike budding yeast and vertebrates, acetylation of Smc3p at K112, K113 equivalents (Psm3-K105R,K106R) is not essential (Feytout et al., 2011). Similarly, the cohesin regulator Pds5p is not essential in S. pombe but is essential in budding yeast and metazoans (Hartman et al., 2000; Tanaka et al., 2001; Dorsett, 2005; Vaur et al., 2012). This difference remains despite the fact that physical interactions between Pds5p and Eco1p have been demonstrated in both yeasts (Tanaka et al., 2001; Noble et al., 2006). Perhaps cohesin regulation in S. pombe has been simplified during evolution so as to contain natural variants that function analogous to budding yeast smc3-D1189H, bypassing the need for Eco1p-mediated activation of cohesion establishment. Indeed, our initial characterization of other cohesion activators in eco1∆ wpl1∆ cells suggests multiple targets for potential natural variants.
Although additional studies will be necessary to tease out these potential differences of cohesin regulation between species, the underlying mechanism of its cohesion activity is undoubtedly conserved. Further studies of the D1189H mutation and other eco1∆ wpl1∆ cohesion activators will provide powerful new reagents to elucidate cohesin regulation and function.
MATERIALS AND METHODS
Yeast strains and media
Yeast strains used in this study are A364A background, and their genotypes are listed in Supplemental Table S1. SC minimal and YPD media were prepared as described (Guacci et al., 1997). Benomyl (a gift from Dupont, Wilmington, DE) and camptothecin (Sigma-Aldrich, St. Louis, MO) plates used to assess drug sensitivity were prepared as previously described (Guacci and Koshland, 2012). Preparation of media containing auxin (Sigma-Aldrich) for depletion of AID tagged proteins was as previously described (Eng et al., 2014).
Dilution plating assays
Cells were grown to saturation in YPD medium at 23°C (or 30°C when listed) and then plated in 10-fold serial dilutions. Cells were incubated on plates at relevant temperatures or containing drugs as described. For plasmid shuffle assays, cells were grown to saturation in YPD medium to allow loss of plasmid pEU42 (SMC3 CEN URA3) and then plated in 10-fold serial dilutions.
G1 arrest and release into mid–M phase arrest
G1 arrest.
Asynchronous cultures of cells were grown to mid log phase at 23°C in YPD medium, and then αFactor (Sigma-Aldrich) was added to 10−8 M. Cells were incubated for 3 h to induce arrest in G1 phase. This incubation time was increased to 3.5 h for all strains in any experiment in which an eco1∆ wpl1∆ background strain was used. For depletion of AID tagged proteins, auxin was added (500 μM final) and cells incubated an additional 1 h in αFactor-containing medium.
Release from G1 into mid–M phase arrest.
G1 arrested cells were washed three times in YPD containing 0.1 mg/ml Pronase E (Sigma-Aldrich) and once in YPD and then resuspended in YPD containing nocodazole (Sigma-Aldrich) at 15 μg/ml final. Cells were incubated at 23°C for 3 h to arrest in mid–M phase. For depletion of AID-tagged proteins, auxin was added (500 μM final) in all wash media and in resuspension media containing nocodazole to ensure depletion at all times.
Nocodazole arrest–release assay for chromosome IV segregation
Cells were arrested in G1 phase and then released and rearrested in mid–M phase using nocodazole as described. Cells were washed three times with YPD and then resuspended in YPD containing αFactor (10−8 M) and incubated 4 h at 23°C
Scoring segregation in large-budded (telophase) cells.
Large-budded cells after nocodazole arrest–release were scored for proper chromosome IV sister segregation. Only cells with two GFP signals were scored. Cells were scored as exhibiting segregation when there was one GFP signal in each daughter bud. The percentage of chromosome IV sister chromatids scored as having segregated was calculated as (large-budded cells with segregated sisters/total number of large-budded cells) × 100.
Scoring segregation in unbudded (G1 phase) cells.
After cell division, segregation generates two cells with one GFP spot each, whereas missegregation generates one cell with two GFP spots and one cell with no GFP spots. Therefore the percentage of unbudded cells showing segregation was calculated as (unbudded cells with one GFP signal/total number of unbudded cells) × 100.
Genetic screen of eco1∆ wpl1∆ cells for cohesion activator suppressors
Our results indicate that eco1∆ wpl1∆ cells have little or no cohesion and are sensitive to benomyl (BEN) and camptothecin (CPT; Guacci and Koshland, 2012). BEN destabilizes microtubules and so induces detachment of early S-phase kinetochore–microtubule attachments essential for cohesin-independent segregation (Figure 2A). CPT inhibits topoisomerase I, which induces single-strand nicks that can become double-strand DNA breaks (DSBs) during replication in S phase. Because cohesin/cohesion play a role in DSB repair (Ström et al., 2004; Unal et al., 2004), CPT lethality is likely due to a combination of reduced repair of DSBs and DNA-damage checkpoint–induced cell-cycle delays, increasing the likelihood of loss kinetochore–microtubule attachments formed in S phase.
We wanted to identify mutations that restore cohesion to eco1∆ wpl1∆ cells. We reasoned that suppressors that restore cohesion in eco1∆ wpl1∆ cells will be resistant to both drugs, and so we conducted a genetic screen on this basis. To set the parameter for the screen, we assayed haploid eco1∆ wpl1∆ cells for the BEN and CPT concentrations that induce complete lethality when 106 cells are plated (Supplemental Figure S3A). For the screen, haploid eco1∆ wpl1∆ cells were dilution streaked on YPD plates and grown at 23°C to enable formation of colonies from single cells (Supplemental Figure S3B). A small amount of one single colony was inoculated into YPD liquid medium and grown to saturation at 23°C. Aliquots containing ∼2 × 107 cells were plated onto YPD containing either BEN or CPT at 12.5–15 μg/ml and then incubated 4–5 d at 23°C. Twenty-four different single colonies were subjected to this regimen to generate a pool of suppressors with independent origins. A few drug-resistant colonies (suppressors) arose on each plate. Suppressor clones were retested for resistance to the same drug and for cross-resistance—that is, BEN-resistant clones tested for CPT resistance and vice versa. Suppressors that exhibited cross drug-resistance were subjected to whole-genome sequencing using TruSeq DNA Sample v2 Kit (Illumina, San Diego, CA).
Monitoring cohesion using LacO-GFP assay
Cohesion was monitored using the LacO-LacI system, in which cells contained a GFP-LacI fusion and tandem LacO repeats integrated at one chromosomal locus, which recruits the GFP-LacI (Straight et al., 1996). CEN-distal cohesion was monitored by integrating LacO repeats at LYS4, located 470 kb from CEN4. CEN-proximal cohesion is monitored by integrating LacO at TRP1, located 10 kb from CEN4. Cells were fixed and processed to allow the number of GFP signals in each cell to be scored and the percentage of cells with two GFP spots determined as previously described (Guacci and Koshland, 2012). Bulk chromosomal DNA for imaging was visualized as previously described (Guacci and Koshland, 2012).
Plasmid constructs
Site-directed mutagenesis using the QuikChange Site Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) was used to generate the smc3-D1189H allele on URA3 or LEU2 integration plasmid pVG441 D1189H (smc3-D1189H URA3) or pVG419 D1189H (smc3-D1189H LEU2), respectively. The smc3-D1189H mutation was confirmed by sequencing the entire open reading frame (ORF), as well as the promoter region, to ensure that it was the only change.
Strain construction
SMC3 shuffle strain construction.
Haploids containing plasmid pEU42 (SMC3 URA3 CEN) had their endogenous SMC3 gene deleted and replaced by the HPH cassette (encodes resistance to hygromycin B [Roche Biologicals, Indianapolis, IN]) using standard PCR-mediated, homology-based recombination.
Assessment of SMC3 test alleles by integration at the LEU2 locus.
A second SMC3 “test allele” was cloned onto an integrating vector (pVG419; SMC3 LEU2) and linearized within LEU2 by BstEII digestion. Linearized plasmid pVG419 bearing WT or smc3 mutant alleles was transformed into shuffle strains to integrate them at the LEU2 locus, and LEU+ transformants were selected. These “test alleles” were assayed for their ability to support viability as the sole SMC3 source as follows. LEU+ clones were grown to saturation in YPD medium at 23°C to allow loss of plasmid pEU42 and then plated in 10-fold serial dilutions on medium containing FOA (US Biologicals, Salem, MA). FOA selectively kills URA3 cells, thereby selecting for loss of pEU42, which allows assessment of test allele ability to support viability as the sole SMC3 in cells. As a control, cells were also plated on either YPD or URA–medium. This shuffle strategy was used to create WT, wpl1∆, and eco1∆ wpl1∆ cells with smc3-D1189H alleles integrated at the LEU2 locus as the sole SMC3.
Insertion of SMC3 alleles at the endogenous locus.
Two different strategies were used. One used SMC3 shuffle strains described earlier for one step-gene replacement. A linear DNA fragment containing the desired SMC3 ORF allele, SMC3 promoter, and 3′ untranslated region were transformed into shuffle strains, plated on YPD, and grown overnight. Plates were replica plated to FOA, and FOA-resistant clones were selected and tested for sensitivity to hygromycin B, which occurs when smc3∆::HPH is replaced by the transformed linear SMC3 allele. Transplacement alleles were confirmed by PCR screening and PCR sequencing.
The second strategy to replace the SMC3 allele with smc3-D1189H allele in haploid eco1∆ wpl1∆ cells or to replace SMC3 with SMC3-AID in WT cells bearing TIR1 was as follows. Plasmid pVG441 D1189H (smc3-D1189H URA3) or pVG465 (SMC3-AID URA3) was linearized within the SMC3 ORF by PshAI digestion. Linearized pVG441 or pVG465 was transformed into haploid eco1∆ wpl1∆ strains (VG3502-1A and VG3503-4C) or a wild-type TIR1 strain (VG3620-4C), respectively. URA+ colonies contain the SMC3 URA3 plasmid integrated at the SMC3 locus to create tandem SMC3 genes. URA+ transformants were replica plated onto YPD and then dilution streaked on FOA to excise the URA3 marker and thereby select for loss of one SMC3 allele. PCR-mediated sequencing was used to identify clones containing only the smc3-D1189H or SMC3-AID allele.
Strains containing AID-tagged proteins
Details of the auxin-mediated destruction of AID-tagged proteins in yeast was previously described (Eng et al., 2014). Briefly, the TIR1 E3-ubiquiting ligase placed under control of the GPD promoter and marked by Candida albicans TRP1 replaced the TRP1 gene on chromosome IV. ECO1 and SCC2 were C-terminally tagged with 3V5-AID2 sequences by standard PCR techniques and transformed into yeast strains bearing TIR1 to generate ECO1-AID or SCC2-AID allele, respectively. For SMC3-AID, a BglII site was inserted after Smc3p amino acid residue N607 and then the 3V5-AID1 cassette inserted on a BamHI/BglII fragment via standard cloning techniques. We replaced SMC3 with SMC3-AID at the endogenous locus as described. PCR screening and auxin-mediated sensitivity were used to identify clones containing AID-tagged genes.
Chromosome spreads
Chromosome spreads were performed as previously described, except primary that antibody was diluted in 5% bovine serum albumin, 0.2% milk, 1× phosphate-buffered saline, and 0.2% Triton X-100 (Hartman et al., 2000). Mcd1p was detected using rabbit anti-Mcd1p antibodies (Rb#555; αMcd1p) at a 1:10,000 dilution.
Chromatin immunoprecipitation
ChIP was performed as previously described (Wahba et al., 2013; Eng et al., 2014).
Microscopy
Images were acquired with a Zeiss Axioplan2 microscope (100× objective, numerical aperture, 1.40) equipped with a Quantix charge-coupled device camera (Photometrics).
Flow cytometry analysis
Flow cytometry analysis was performed as previously described (Eng et al., 2014).
Supplementary Material
Acknowledgments
We thank Anjali Zimmer, Gamze Çamdere, Thomas Eng, Lorenzo Constantino, Liam Holt, Brett Robison, Rebecca Lamothe, and Elçin Ünal for critical reading of the manuscript. We thank Thomas Eng, Gamze Çamdere, and Jessica Poon for technical support and all other members of the Koshland lab for support and helpful comments. This work was funded by National Institutes of Health Grant R01GM092813.
Abbreviations used:
- BEN
benomyl
- CPT
camptothecin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-08-1268) on November 5, 2014.
REFERENCES
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. doi: 10.1016/s0092-8674(00)81019-3. [DOI] [PubMed] [Google Scholar]
- Bürmann F, Shin H-C, Basquin J, Soh Y-M, Giménez-Oya V, Kim Y-G, Oh B-H, Gruber S. An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat Struct Mol Biol. 2013;20:371–379. doi: 10.1038/nsmb.2488. [DOI] [PubMed] [Google Scholar]
- Chan K-L, Roig MB, Hu B, Beckouet F, Metson J, Nasmyth K. Cohesin's DNA exit gate is distinct from its entrance gate and is regulated by acetylation. Cell. 2012;150:961–974. doi: 10.1016/j.cell.2012.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Nasmyth K. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- Dorsett D. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development. 2005;132:4743–4753. doi: 10.1242/dev.02064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng T, Guacci V, Koshland D. ROCC, a conserved region in cohesin's Mcd1 subunit, is essential for the proper regulation of the maintenance of cohesion and establishment of condensation. Mol Biol Cell. 2014;25:2351–2364. doi: 10.1091/mbc.E14-04-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feytout A, Vaur S, Genier S, Vazquez S, Javerzat J-P. Psm3 acetylation on conserved lysine residues is dispensable for viability in fission yeast but contributes to Eso1-mediated sister chromatid cohesion by antagonizing Wpl1. Mol Cell Biol. 2011;31:1771–1786. doi: 10.1128/MCB.01284-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16:2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glynn EF, Megee PC, Yu H-G, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2004;2:E259. doi: 10.1371/journal.pbio.0020259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125:517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D. Cohesin-independent segregation of sister chromatids in budding yeast. Mol Biol Cell. 2012;23:729–739. doi: 10.1091/mbc.E11-08-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haering CH, Schoffnegger D, Nishino T, Helmhart W, Nasmyth K, Löwe J. Structure and stability of cohesin's Smc1-kleisin interaction. Mol Cell. 2004;15:951–964. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Hartman T, Stead K, Koshland D, Guacci V. Pds5p is an essential chromosomal protein required for both sister chromatid cohesion and condensation in Saccharomyces cerevisiae. J Cell Biol. 2000;151:613–626. doi: 10.1083/jcb.151.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Onn I, Koshland D. Genetic evidence that the acetylation of the Smc3p subunit of cohesin modulates its ATP-bound state to promote cohesion establishment in Saccharomyces cerevisiae. Genetics. 2010;185:1249–1256. doi: 10.1534/genetics.110.116871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidinger-Pauli JM, Unal E, Koshland D. Distinct targets of the Eco1 acetyltransferase modulate cohesion in S phase and in response to DNA damage. Mol Cell. 2009;34:311–321. doi: 10.1016/j.molcel.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner K-P, Craig L, Moncalian G, Zinkel RA, Usui T, Owen BAL, Karcher A, Henderson B, Bodmer JL, McMurray CT, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- Ivanov D, Schleiffer A, Eisenhaber F, Mechtler K, Haering CH, Nasmyth K. Eco1 is a novel acetyltransferase that can acetylate proteins involved in cohesion. Curr Biol. 2002;12:323–328. doi: 10.1016/s0960-9822(02)00681-4. [DOI] [PubMed] [Google Scholar]
- Kim J-S, Krasieva TB, LaMorte V, Taylor AMR, Yokomori K. Specific recruitment of human cohesin to laser-induced DNA damage. J Biol Chem. 2002;277:45149–45153. doi: 10.1074/jbc.M209123200. [DOI] [PubMed] [Google Scholar]
- Kueng S, Hegemann B, Peters BH, Lipp JJ, Schleiffer A, Mechtler K, Peters J-M. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127:955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- Laloraya S, Guacci V, Koshland D. Chromosomal addresses of the cohesin component Mcd1p. J Cell Biol. 2000;151:1047–1056. doi: 10.1083/jcb.151.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengronne A, Katou Y, Mori S, Yokobayashi S, Kelly GP, Itoh T, Watanabe Y, Shirahige K, Uhlmann F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–578. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Serra L, Lengronne A, Borges V, Kelly G. Budding yeast Wapl controls sister chromatid cohesion maintenance and chromosome condensation. Curr Biol 23, 64–69. 2013 doi: 10.1016/j.cub.2012.11.030. [DOI] [PubMed] [Google Scholar]
- Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev. 1998;12:1986–1997. doi: 10.1101/gad.12.13.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee PC, Mistrot C, Guacci V, Koshland D. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol Cell. 1999;4:445–450. doi: 10.1016/s1097-2765(00)80347-0. [DOI] [PubMed] [Google Scholar]
- Michaelis C, Ciosk R, Nasmyth K. Cohesins: chromosomal proteins that prevent premature separation of sister chromatids. Cell. 1997;91:35–45. doi: 10.1016/s0092-8674(01)80007-6. [DOI] [PubMed] [Google Scholar]
- Noble D, Kenna MA, Dix M, Skibbens RV, Unal E, Guacci V. Intersection between the regulators of sister chromatid cohesion establishment and maintenance in budding yeast indicates a multi-step mechanism. Cell Cycle. 2006;5:2528–2536. doi: 10.4161/cc.5.21.3405. [DOI] [PubMed] [Google Scholar]
- Onn I, Heidinger-Pauli JM, Guacci V, Unal E, Koshland DE. Sister chromatid cohesion: a simple concept with a complex reality. Annu Rev Cell Dev Biol. 2008;24:105–129. doi: 10.1146/annurev.cellbio.24.110707.175350. [DOI] [PubMed] [Google Scholar]
- Pearson CG, Yeh E, Gardner M, Odde D, Salmon ED, Bloom K. Stable kinetochore-microtubule attachment constrains centromere positioning in metaphase. Curr Biol. 2004;14:1962–1967. doi: 10.1016/j.cub.2004.09.086. [DOI] [PubMed] [Google Scholar]
- Piazza I, Rutkowska A, Ori A, Walczak M, Metz J, Pelechano V, Beck M, Haering CH. Association of condensin with chromosomes depends on DNA binding by its HEAT-repeat subunits. Nat Struct Mol Biol. 2014;21:560–568. doi: 10.1038/nsmb.2831. [DOI] [PubMed] [Google Scholar]
- Rolef Ben-Shahar T, Heeger S, Lehane C, East P, Flynn H, Skehel M, Uhlmann F. Eco1-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science. 2008;321:563–566. doi: 10.1126/science.1157774. [DOI] [PubMed] [Google Scholar]
- Rollins RA, Korom M, Aulner N, Martens A, Dorsett D. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol. 2004;24:3100–3111. doi: 10.1128/MCB.24.8.3100-3111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland BD, Roig MB, Nishino T, Kurze A, Uluocak P, Mishra A, Beckouet F, Underwood P, Metson J, Imre R, et al. Building sister chromatid cohesion: smc3 acetylation counteracts an antiestablishment activity. Mol Cell. 2009;33:763–774. doi: 10.1016/j.molcel.2009.02.028. [DOI] [PubMed] [Google Scholar]
- Skibbens RV, Corson LB, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stead K, Aguilar C, Hartman T, Drexel M, Meluh P, Guacci V. Pds5p regulates the maintenance of sister chromatid cohesion and is sumoylated to promote the dissolution of cohesion. J Cell Biol. 2003;163:729–741. doi: 10.1083/jcb.200305080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Ström L, Lindroos HB, Shirahige K, Sjögren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16:1003–1015. doi: 10.1016/j.molcel.2004.11.026. [DOI] [PubMed] [Google Scholar]
- Sumara I, Vorlaufer E, Gieffers C, Peters BH, Peters JM. Characterization of vertebrate cohesin complexes and their regulation in prophase. J Cell Biol. 2000;151:749–762. doi: 10.1083/jcb.151.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T, Kawaguchi T, Kanno R, Itoh T, Shirahige K. Budding yeast Wpl1(Rad61)-Pds5 complex counteracts sister chromatid cohesion-establishing reaction. Curr Biol. 2009;19:492–497. doi: 10.1016/j.cub.2009.01.062. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Hao Z, Kai M, Okayama H. Establishment and maintenance of sister chromatid cohesion in fission yeast by a unique mechanism. EMBO J. 2001;20:5779–5790. doi: 10.1093/emboj/20.20.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomonaga T, Nagao K, Kawasaki Y, Furuya K, Murakami A, Morishita J, Yuasa T, Sutani T, Kearsey SE, Uhlmann F, et al. Characterization of fission yeast cohesin: essential anaphase proteolysis of Rad21 phosphorylated in the S phase. Genes Dev. 2000;14:2757–2770. doi: 10.1101/gad.832000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong K, Skibbens RV. Cohesin without cohesion: a novel role for Pds5 in Saccharomyces cerevisiae. PLoS One. 2014;9:e100470. doi: 10.1371/journal.pone.0100470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tóth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13:320–333. doi: 10.1101/gad.13.3.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, Koshland D. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16:991–1002. doi: 10.1016/j.molcel.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, Koshland DE. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321:566–569. doi: 10.1126/science.1157880. [DOI] [PubMed] [Google Scholar]
- Vaur S, Feytout A, Vazquez S, Javerzat J-P. Pds5 promotes cohesin acetylation and stable cohesin-chromosome interaction. EMBO Rep. 2012;13:645–652. doi: 10.1038/embor.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahba L, Gore SK, Koshland D. The homologous recombination machinery modulates the formation of RNA-DNA hybrids and associated chromosome instability. Elife. 2013;2:e00505. doi: 10.7554/eLife.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J-S, Lim JH, Shin HC, Suh MK, Ku B, Lee KH, Joo K, Robinson H, Lee J, Park SY, et al. Structural studies of a bacterial condensin complex reveal ATP-dependent disruption of intersubunit interactions. Cell. 2009;136:85–96. doi: 10.1016/j.cell.2008.10.050. [DOI] [PubMed] [Google Scholar]
- Yeh E, Haase J, Paliulis LV, Joglekar A, Bond L, Bouck D, Salmon ED, Bloom KS. Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr Biol. 2008;18:81–90. doi: 10.1016/j.cub.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Shi X, Li Y, Kim BJ, Jia J, Huang Z, Yang T, Fu X, Jung SY, Wang Y, et al. Acetylation of Smc3 by Eco1 is required for S phase sister chromatid cohesion in both human and yeast. Mol Cell. 2008;31:143–151. doi: 10.1016/j.molcel.2008.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.