Abstract
Heterotrophic nitrifiers synthesize nitrogenous gasses when nitrifying ammonium ion. A Cupriavidus pauculus, previously thought an Alcaligenes sp. and noted as an active heterotrophic nitrifier-denitrifier, was examined for its ability to produce nitrogen gas (N2) and nitrous oxide (N2O) while heterotrophically nitrifying the organic substrate pyruvic oxime [CH3–C(NOH)–COOH]. Neither N2 nor N2O were produced. Nucleotide and phylogenetic analyses indicated that the organism is a member of a genus (Cupriavidus) known for its resistance to metals and its metabolism of xenobiotics. The microbe (a Cupriavidus pauculus designated as C. pauculus strain UM1) was examined for its ability to perform heterotrophic nitrification in the presence of Cu2+ and Ni2+ and to metabolize the xenobiotic phenol. The bacterium heterotrophically nitrified well when either 1 mM Cu2+ or 0.5 mM Ni2+ was present in either enriched or minimal medium. The organism also used phenol as a sole carbon source in either the presence or absence of 1 mM Cu2+ or 0.5 mM Ni2+. The ability of this isolate to perform a number of different metabolisms, its noteworthy resistance to copper and nickel, and its potential use as a bioremediation agent are discussed.
1. Introduction
Heterotrophic nitrification is the oxidation of nitrogenous compounds, both organic and inorganic, to more oxidized products. Organic nitrogenous compounds, for example, may be oxidized to organic nitrocontaining compounds or can be converted to NO2 − or NO3 − [1, 2]. Denitrification, the reduction of the nitrogen oxides NO3 − and NO2 − to N2O or N2 [3], is performed by a diverse group of prokaryotes, including members of the Archaea and the Bacteria [4, 5]. Castignetti and Hollocher [6] described a bacterium, provisionally identified as an Alcaligenes sp., which was capable of the two processes, that is, both vigorous heterotrophic nitrification and denitrification. They also noted that a number of different denitrifiers, representing some of the more commonly encountered soil denitrifying microbes [4, 7], heterotrophically nitrify [1].
Heterotrophic nitrification is performed by chemoheterotrophs [2, 7]. The process has the potential to result in nitrogen oxidation [2, 8] and to effect bioremediation, especially with respect to the removal of excess nitrogen [2, 9]. Jetten et al. [2] noted that Thiosphera pantotropha (now Paracoccus pantotrophus) and other heterotrophic nitrifiers could simultaneously oxidize and reduce N compounds with gaseous N products being formed. Similarly, heterotrophic nitrifiers have been noted to aerobically denitrify [10]. The simultaneous processes of heterotrophic nitrification and denitrification were postulated to have resulted in the underestimation of the significance of heterotrophic nitrification [2, 11]. Rather than inorganic N sources, organic N sources were the substrates for heterotrophic nitrification, which, in some environments, was more significant than autotrophic nitrification [11, 12].
The role of heterotrophic nitrifiers is the subject of current research [13–17] as such microbes may be of consequence to both natural (pastures and grasslands) [8, 9, 11, 12] and artificial (wastewater treatment) [2, 10, 13, 18] environments. Some of these organisms have significant capacities to both heterotrophically nitrify and denitrify [2, 10, 19] and some synthesize N2O or N2 during the course of heterotrophic nitrification [17, 20] or heterotrophic nitrification-aerobic denitrification metabolism [10, 14–16, 18, 21]. To date, studies have focused on inorganic N compounds as the sources of nitrogenous gas production, with either NH4 +, NH2OH, or NO2 − serving as the substrate for either N2 or N2O production [10, 14, 16–19]. Whether organic-N substrates, such as pyruvic oxime (PO; H3C–C(NOH)–COOH), serve as sources of N2 or N2O synthesis during heterotrophic nitrification or heterotrophic nitrification coupled to aerobic denitrification has received little, if any, attention.
The current study was undertaken to examine whether the heterotrophic nitrification of PO by the organism previously known as an Alcaligenes sp. (1) resulted in the gaseous nitrogen products N2 or N2O, (2) supported heterotrophic nitrification-aerobic denitrification such that either N2 or N2O was produced, and (3) occurred when the organism was cultured in the presence of the heavy metals Cu and Ni. In addition, (1) whether a representative xenobiotic, phenol, was used as a carbon source, (2) an analysis of the bacterium's phylogenetic identity, and (3) the identity of the crucial gene necessary for denitrification (nitrite reductase) were conducted. At the time of its original characterization, the Alcaligenes sp.'s taxonomic classification was based on phenotypic traits and its G+C content [66.1 mol%-D. Castignetti, D. (1980) Characterization of a soil heterotrophic nitrifier and its synergistic interactions with Nitrobacter agilis. Dissertation, University of Massachusetts, Amherst, MA]. In the current study, the bacterium was characterized with respect to its 16S rRNA gene sequence and nitrite reductase (nir) gene via cloning and nucleotide sequencing. Identification of the bacterium as a Cupriavidus pauculus spurred an examination of its Cu2+ and Ni2+ resistance capacities, its ability to heterotrophically nitrify PO in the presence of these metals, and its use of the xenobiotic phenol in the absence or presence of Cu2+ and Ni2+ as this genus is known for its tolerance of metals and use of xenobiotics. (To avoid confusion, the provisionally identified Alcaligenes sp. will be referred to as C. pauculus UM1 in the remainder of this report.)
2. Materials and Methods
2.1. Resting Cell Experiments
To examine the ability of C. pauculus UM1 to produce nitrogenous gasses while heterotrophically nitrifying PO, the bacterium was grown to mid-to-late logarithmic phase in the PO-mineral salts medium described below, harvested by centrifugation (10,000 ×g for 10 min) and washed twice in 50 mM pH 7.0 phosphate buffer to make resting cells. Cells were divided into control (boiled for 5 minutes) and experimental groups and 5 mL was placed into a 33 mL vial along with a stirring bar. Vials were then sealed with a rubber septum and flushed, for 5 min, with the appropriate gas (see below) to remove the air. At t = 0, 1 mL of 60 mM PO in the phosphate buffer was added via syringe and needle to yield a final concentration of 10 mM PO. Gasses used to flush the vials were either 100% helium (He) or 100% O2. Three sets of conditions were examined: no O2 (anaerobic), low O2 (3.6% O2), and 100% O2. Helium was used to flush the vials either when an anaerobic environment was to be tested or when 1 mL of pure O2 was added, via syringe and needle, to yield an approximately 3.6% O2 atmosphere in the vials. During PO metabolism by the organism, the oxidation of PO-nitrogen results in its conversion to NO2 − [22]. NO2 − synthesis from PO was therefore used to monitor PO use throughout these experiments. At t = 0, approximately 3 and 22 hours, 0.11 mL of cells was removed from each vial (via a syringe and needle) and used to measure the consequent production of NO2 − from PO. 40 μL of gaseous headspace was simultaneously removed to measure the synthesis of N2, NO, N2O, and CO2 by the cells. As C. pauculus UM1 is a respiratory microbe, it was not expected that cells without O2 (He only) would nitrify. Hence, to vials that received only He, 10 mM NaNO2 was added to ensure the presence of an oxidant. This condition allowed for assessment of heterotrophic nitrification of the PO at the expense of NO2 − reduction, in which case the NO2 − would be the oxidant and substitute for O2 gas.
2.2. Growth and Metal Tolerance
To determine the bacterium's tolerance of either Cu2+ or Ni2+, growth was performed using mid-to-late log cultures as the inoculum (0.5% (v/v)) of the bacterium grown in nutrient broth (NB-Sigma Chemical Co., St. Louis, MO) without any metal added. To perform a growth curve with Cu and Ni present, the medium used was NB to which the metals were added from 2 M stocks of either CuCl2–2H2O or NiCl2–6H2O to result in the desired final concentrations [23]. Controls consisted of NB devoid of metal additions. Minimum inhibitory concentrations (MIC) experiments were conducted as described by Chess [24] using NB supplemented with Cu2+ or Ni2+ rather than antibiotics. For both growth and MIC experiments, samples were run in triplicate and each experiment was performed at least twice. For all growth experiments, cultures were incubated at 28–30°C and shaken at 300 r.p.m.
To determine C. pauculus UM1's ability to heterotrophically nitrify PO when either Cu2+ or Ni2+ was present, three different types of growth experiments were conducted. For the first, the bacterium was cultured in NB with 10–12 mM PO with either 1 mM Cu2+ or 0.5 mM Ni2+ and PO use was monitored as described above (via conversion to NO2 −). Controls consisted of identical cultures but without PO. To determine if the organism could heterotrophically nitrify PO without supplemental carbon nutrients (i.e., NB), a second experiment type was used where the C. pauculus UM1 was grown in a mineral salts medium, previously described [25], containing either 1 mM Cu2+ or 0.5 mM Ni2+, in which 10–12 mM PO served as the sole C source and where the uninoculated medium was the control. The minerals salts medium was made and autoclaved without 0.5 g L−1 MgSO4-7H2O, 0.5 g L−1 NH4Cl, and the 10–12 mM PO, which were filter-sterilized and added to the cooled, sterile medium. To determine the effect of Cu2+ and Ni2+ on the relative rate of C. pauculus UM1's ability to grow and nitrify PO, the third experiment type was conducted in which C. pauculus UM1 was grown in the above mineral salts medium without metals (control) or with either 1 mM Cu2+ or 0.5 mM Ni2+. To determine if C. pauculus UM1 could use phenol as a sole source of carbon, the organism was grown to midlog in NB and used at a 1 : 100 dilution to inoculate the above mineral salts medium containing filter-sterilized 0.5 g L−1 MgSO4-7H2O, 0.5 g L−1 NH4Cl, and phenol (2 mM) as the sole C source. Culturing of C. pauculus UM1 into this medium lacking phenol showed that “carryover” growth of the bacterium due to residual NB did not occur as growth measurements noted a decline of culture turbidity with time as opposed to an increase if phenol was present.
To determine if C. pauculus UM1 could use phenol as a sole C source when cultured in the presence of either 1 mM Cu2+ or 0.5 mM Ni2+, the bacterium was grown in ~2 mM phenol-mineral salts medium and used at a 1 : 100 dilution to inoculate the same medium containing either no metals, 1 mM Cu2+, or 0.5 mM Ni2+. As the medium with 1 mM Cu2+ had an initial absorbance above 0 and growth with phenol was fairly sparse, growth was spurred in the cultures by adding an aliquot of filter-sterilized 200 mM phenol solution after 7 days, thereby reestablishing the cultures to a phenol concentration of ~2 mM.
2.3. Nucleotide Cloning, Sequencing, and Gene Identifications
The subject of investigation is a bacterium originally isolated from an agricultural soil [22, 25]. The 16S rRNA gene sequence of the bacterium was analyzed by conducting PCR using the Escherichia coli equivalents from base pair 8 to base pair 926 and then from base pair 906 to 1512. A total of 1501 nucleotides of the bacterium's 16S rRNA gene were cloned and sequenced.
Genomic DNA from the bacterium, for both 16S rRNA gene and nirS and nirK gene analyses (see below), was isolated using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI) and used as template DNA. The first primer pair employed was 8F and 926R, the 8F primer sequence being 5′ AGAGTTTGATCCTGGCTCAG 3′ [26]. The complement to the 8F primer is the 926R primer which has the sequence of 5′ CCGTCAATTCCTTTRAGTTT 3′ [27]. The second primer pair consists of 906F and 1512R. The 1512R sequence was 5′ ACGGYTACCTTGTTACGACTT 3′ [28] while the 906F sequence is the 5′ to 3′ reverse complement of the 926R primer (16S rDNA). PCR reactions were performed using either a Hybaid PCR Sprint Thermocycler (Cole-Parmer, Vernon Hills, IL) or a Techne-412 (Scie-Plas Ltd., Cambridge, UK) thermocycler. PCR Master Mix (Promega, Madison, WI) was used, as described by the manufacturer to make 50 μL samples. PCR conditions were one cycle of 94°C for 4 min followed by 33 cycles of 94°C for 0.75 min, 56.4°C for 1 min, and 72°C for 1.5 min followed by one cycle of 72°C for 7 min. Samples were then kept at either 4 or 10°C until removed for analysis. Gel analysis was routinely performed using a sodium borate system [29]. Negative controls, with sterile water substituted for template DNA (water controls), were performed to ensure that the DNA copied was from the bacterium being analyzed. Genomic DNA of stock bacteria (e.g., Pseudomonas spp., E. coli) was used as a positive control to insure that amplicons of the proper size were made.
Amplicons of interest were cloned as described in the instructions for the TOPO TA Cloning Kit for Sequencing system (Invitrogen, Carlsbad, CA). The resulting pCR4 plasmids, containing the amplicons of interest, were isolated using the Wizard Plus SV Minipreps DNA Purification System (Promega, Madison, WI), verified that they contained the clone of interest by using the plasmid to produce the appropriate PCR product, and then sequenced by the University of Chicago Cancer Research Center DNA Sequencing Facility (Chicago, IL) using either an Applied Biosystems 3730XL 96-capillary or a 3130 16-capillary automated DNA sequencer. Sequences were analyzed using Chromas Lite (http://www.technelysium.com.au/chromas_lite.html Technelysium Pty, Ltd.), to make an initial decision as to the quality and acceptability of the sequence, followed by a more thorough analysis to align and append sequences using Lasergene (DNASTAR, Inc. Madison, WI) software. Analysis of the 16S rRNA gene nucleotide sequence allowed for the construction of appended nucleotide sequences of the E. coli equivalent of base pairs 8-1512. Sequences were aligned, appended, and analyzed via BLAST-N (GenBank-National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/) to search for matches and to identify the closest sequenced relatives. The data generating the best matches were used to characterize the bacterium. In addition, the validity and the quality of the GenBank 16S rRNA gene sequences were investigated via submitting the sequences to Greengenes [30]. Sequence alignments and phylogenetic analysis of the bacterium were performed via the use of the SeaView (http://pbil.univ-lyon1.fr/software/seaview.html) software [31]. Phylogenetic tree construction was done via the SeaView program using the parameters of 100 bootstrap replicates and the NJ algorithm. The dissimilarity bar and accession numbers of the sequences used for comparison are included in the figure.
Denitrifying bacteria contain the crucial key enzyme (nitrite reductase), which is necessary to reduce nitrite to nitric oxide [3, 32, 33]. A denitrifying bacterium contains nitrite reductase as either the cytochrome cd1 (NirS) or the copper-containing (NirK) enzyme and an individual organism contains a single type of the nitrite reductase gene, either nirS or nirK [3, 32, 33]. As C. pauculus UM1 had previously been identified as an active denitrifier [6], we examined it for the presence of either the nirS or nirK gene by using the primer pairs and amplification conditions described by either Braker et al. [32] or Throbäck et al. [33]. The only modification was that the annealing temperature of the Throbäck et al. [33] procedure was adjusted to 57°C. For the examination of the presence of the nirS gene, the primer sets were nirS1F and nirS6R [32] or cd3aF and Rc3d [33]. The former yields a theoretical 873 bp amplified product while the latter yields a theoretical amplified product of 425 bp. Primer sets for the examination of the presence of the nirK gene were nirK1F and nirK5R [32] or FlaCu and R3Cu [33]. The former yields a theoretical 514 bp amplified product while the latter yields a theoretical amplified product of 475 bp. DNA from Pseudomonas stutzeri (ATCC 11607) served as a positive control for the amplification of the nirS amplicon while DNA from Achromobacter xylosoxidans (ATCC 15173) served as a positive control for the amplification of the nirK amplicon. Once the amplicon of the correct size for nirS was identified (the bacterium contains only the nirS gene—see below), it was cloned using the TOPO TA Cloning Kit for Sequencing system noted above. The resultant plasmid was isolated, verified via PCR as containing a nirS amplicon, and sequenced. Aligned and appended sequences were analyzed by BLAST-N to identify the closest matches.
2.4. Nucleotide Sequences
The partial 16S rRNA sequence of the bacterium and the nirS gene sequences have been deposited in GenBank and are listed below.
2.5. Chemicals and Analytic Methods
All chemicals used to make the above media were of reagent grade or better. The sodium salt of PO was synthesized via the method of Quastel et al. [34]. NO2 − synthesis was measured to monitor PO usage and was determined by the sulfanilamide-N-(1-naphthyl)ethylenediamine method [35]. Molecular sieve (Linde-Union Carbide, South Plainfield, NJ) and Poropak Q (Waters, Milford, MA) columns, in a Shimadzu GC-8A (Kyoto, Japan) gas chromatograph, were used to determine CO2, N2, NO, and N2O as described [36]. Growth was measured via absorbance at 600 nm and phenol was quantified via the method of Slinkard and Singleton [37], adjusted to accommodate 1 mL samples.
2.6. Statistical Analyses
Comparison of mean values (Tables 1 and 2 and Figure 3) was performed via t-testing using SYSTAT 13 or Excel. Comparison of the curves and of the individual points (means of three replicates) for a given time in Figures 2 and 3 was done using the ANOVA and Tukey's honestly significantly different tests in SYSTAT 13.
Table 1.
Growth and nitrification of PO by C. pauculus UM1 in the presence of either 1 mM Cu2+ or 0.5 mM Ni2+ in either NB or PO-mineral salts mediuma,b.
| Growth condition | Time (d) | |||
|---|---|---|---|---|
| 0 | 7 | 0 | 7 | |
| Growth (Abs600) | Growth (Abs600) | NO2 − (mM) | NO2 − (mM) | |
| NB-1 mM Cu2+ | 0.01 (0.00) | 2.04 (0.09) | 0.00 (0.00) | 0.01 (0.02) |
| NB-1 mM Cu2+-PO | 0.07 (0.00) | 2.50 (0.26) | 0.00 (0.00) | 13.62d (0.48) |
|
| ||||
| NB-0.5 mM Ni2+ | 0.00 (0.00) | 1.77 (0.04) | 0.00 (0.00) | 0.03 (0.01) |
| NB-0.5 mM Ni2+-PO | 0.01 (0.00) | 2.07d (0.06) | 0.00 (0.00) | 6.61d (1.29) |
|
| ||||
| PO-1 mM Cu2+ (uninoculated) | 0.01 (0.01) | 0.00 (0.00) | 0.00 (0.00) | −0.04 (0.00) |
| PO-1 mM Cu2+ | 0.00 (0.00) | 0.14d (0.01) | 0.03d (0.01) | 10.39d (0.32) |
|
| ||||
| PO-0.5 mM Ni2+ (uninoculated) | 0.00 (0.00) | 0.00 (0.00) | −0.01 (0.01) | −0.01 (0.00) |
| PO-0.5 mM Ni2+ | 0.00 (0.00) | 0.23c (0.06) | 0.12d (0.01) | 12.36d (0.98) |
aInocula for the NB-1 mM Cu2+ and the NB-0.5 mM Ni2+ experiments were grown in the NB-1 mM Cu2+ and NB-0.5 mM Ni2+ medium, respectively, until late logarithmic growth and used at 0.5% (v/v). Inocula for the PO-1.0 mM Cu2+ medium and for the PO-0.5 mM Ni2+ medium were grown in 0.2% PO medium until late logarithmic growth and used at 0.5% (v/v).
bStandard deviation values are in parentheses.
cStatistically different from the corresponding control at P≤ 0.05.
dStatistically different from the corresponding control at P≤ 0.01.
Table 2.
Growth of C. pauculus UM1 when in the presence of either no metals, 1 mM Cu2+, or 0.5 mM Ni2+ in the phenol-mineral salts mediuma,b.
| Growth condition | Time (d) | |||||
|---|---|---|---|---|---|---|
| 0 | 3 | 6 | 0 | 3 | 6 | |
| Growth (Abs600) | Growth (Abs600) | Growth (Abs600) | Phenol (mM) | Phenol (mM) | Phenol (mM) | |
| Phenol | 0.0018 (0.0005) | 0.0800d (0.0028) | 0.0715d,e (0.0038) | 1.89 (0.12) | 0.27d (0.01) | 0.20d,e (0.01) |
|
| ||||||
| Phenol with 1 mM Cu2+ | 0.0920 (0.0094) | 0.1417d (0.0118) | 0.1385 (0.0324) | 1.89 (0.12) | 0.27d (0.02) | 0.22d,e (0.00) |
|
| ||||||
| Phenol with 0.5 mM Ni2+ | 0.0025 (0.0005) | 0.0769c (0.0252) | 0.1793c (0.0616) | 1.93 (0.18) | 1.30 (0.57) | 0.20d (0.00) |
aInocula were a 1 : 100 dilution of C. pauculus UM1 grown to midlogarithmic phase in 2 mM phenol-mineral salts medium. The relatively large t = 0 Abs600 value of the phenol with 1 mM Cu2+ cultures was due to the blue color imparted to the medium by Cu2+.
bStandard deviation values are in parentheses.
cStatistically different from the initial (time 0) measurement at P≤ 0.05.
dStatistically different from the initial (time 0) measurement at P≤ 0.01.
eStatistically different from the day 3 measurement at P≤ 0.05.
Figure 3.
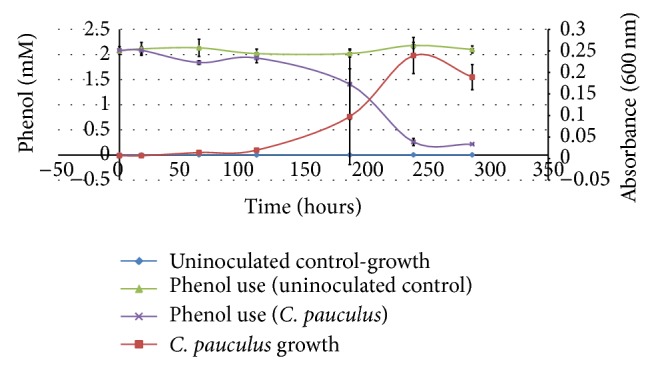
Growth of C. pauculus UM1 with phenol as the sole carbon source. (For both the growth of C. pauculus UM1 and the amount of phenol remaining in the media, significant differences (P ≤ 0.01) were noted for 244 and 288 hours.) Error bars represent the standard deviation of a point.
Figure 2.
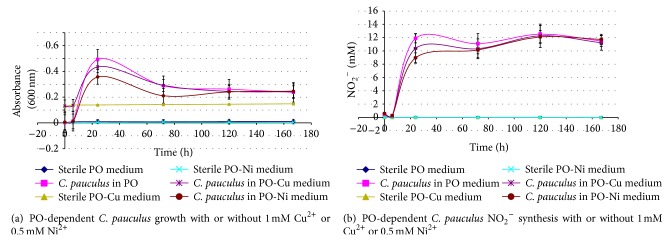
Growth (a) and nitrification (b) of C. pauculus UM1 in PO-mineral salts medium with or without either 1.0 mM Cu2+ or 0.5 mM Ni2+. (The inoculum for the cultures was grown in 0.2% PO-mineral salts medium until mid-to-late logarithmic growth and used at 0.5% (v/v). The growth curves for C. pauculus UM1 either with 1.0 mM Cu2+ (P ≤ 0.05) or 0.5 mM Ni2+ (P ≤ 0.01) or without Cu2+ or Ni2+ (P ≤ 0.01) were significantly different from their corresponding uninoculated controls. Similarly, the NO2 − synthesis curves either with 1.0 mM Cu2+, 0.5 mM Ni2+ or without either Cu2+ or Ni2+ were significantly different (P ≤ 0.01) from their corresponding uninoculated controls. At the T = 24 and 72 hours, growth of all inoculated C. pauculus UM1 cultures was significantly different (P ≤ 0.01) than the corresponding uninoculated controls. In addition, at T = 120 and 167 hours, C. pauculus UM1 cultures grown either without Cu2+ or Ni2+ or with 0.5 mM Ni2+ were significantly different (P ≤ 0.01) than their corresponding uninoculated controls. At T = 24 hours, the growth of C. pauculus UM1 with 0.5 mM Ni2+ was significantly less (P ≤ 0.01) than C. pauculus UM1 grown without Cu2+ or Ni2+. At T = 120 and 167 hours, the growth of all C. pauculus UM1 cultures was not significantly different than the uninoculated 1 mM Cu2+ control. At all measurements, NO2 − concentrations were significantly different (P ≤ 0.01) between the inoculated cultures and their corresponding uninoculated controls. It is presumed that, at the early time points (T = 0 and perhaps 6 hours), this was due to NO2 − carryover with the inocula. At T = 24 hours, NO2 − concentrations were significantly different (P ≤ 0.05) between the inoculated PO-0.5 mM Ni+ cultures and inoculated PO cultures without either Cu2+ or Ni2+.) Error bars represent the standard deviation of a point.
3. Results and Discussion
To determine if the bacterium produces either N2 or N2O while heterotrophically nitrifying, we attempted to grow the bacterium in sealed test tubes from which gasses could be sampled. Due to insufficient growth of the bacterium, these experiments were discontinued. C. pauculus UM1 was thus grown in the mineral salts-PO medium aerobically at 30°C, harvested at mid-to- late logarithmic growth and resting cells were prepared. Under either 0% (He), 3.6%, or 100% O2 atmospheres, resting cells of the bacterium produced neither N2, NO nor N2O. Although control (boiled) vials failed to produce either NO2 − or CO2, experimental vials clearly produced NO2 − by the 3 hours and CO2 was noted by the 22-hour time point.
A number of organisms produce either N2 or N2O while heterotrophically nitrifying or during heterotrophic nitrification-aerobic denitrification [2, 14, 16, 17, 20]. When PO was present under anaerobic conditions, with NO2 − present to serve as an oxidant, neither N2 nor N2O was synthesized. With PO as a nitrogen source and under full or limited O2 conditions, C. pauculus UM1 produced neither N2 nor N2O but did make NO2 −, indicating that while heterotrophic nitrification had occurred, aerobic, or anaerobic, denitrification had not. While previously shown to be clearly capable of heterotrophic nitrification ([22, 25] and this study) and denitrification [6], the current experiments indicate that C. pauculus UM1 produced neither N2 nor N2O via heterotrophic nitrification or via heterotrophic nitrification-aerobic denitrification.
NH3 or NH4 + are the likely sources of N2 and N2O produced via heterotrophic nitrification [2, 16, 17, 20]. The failure of C. pauculus UM1 to make either of these gasses likely reflects that PO oxidation to NO2 − probably occurs via an organic route, that is, via a carbon intermediate and not via NH2OH [34, 38, 39]. While C. pauculus UM1 vigorously denitrifies [6] and avidly oxidizes both the organic-N source PO and the inorganic N-source hydroxylamine [22, 25], the inability of C. pauculus UM1 to synthesize either N2 or N2O during the oxidation of PO limits its ability to remove N as a gas from aerobic environments.
As the bacterium was originally described prior to today's nucleotide sequencing techniques, the phylogenetic identities of the bacterium and the nitrite reductase gene of the organism were of interest. PCR primers [40] specifically designed to amplify 16S rRNA genes from Ralstonia (i.e., Ralstonia and Cupriavidus, see below) resulted in an amplicon of the proper size from genomic DNA of the bacterium but not from genomic DNA from a non-Cupriavidus (Pseudomonas) bacterium. The cloning, sequencing, and BLAST-N analysis of 1501 nucleotides of C. pauculus UMI's 16S rRNA gene resulted in the greatest similarity (98.0% identity) match to Cupriavidus pauculus strain KPS201 and Ralstonia sp. HB1 and HB2. A similar match (97.9%) to C. pauculus strain KPS201 was also obtained from Greengenes. A multiple alignment of similar 16S ribosomal DNA sequences indicates that the bacterium is most closely related to C. pauculus KPS201 and other Cupriavidus species (Figure 1). The nucleotide sequence of the 1501 nucleotides of C. pauculus UM1's 16S rRNA gene has been deposited in GenBank under the accession number GQ504718. Of the first 24 GenBank matches, including 20 of which were noted as being members of the genera Cupriavidus and Ralstonia or the family Burkholderiaceae (of which Cupriavidus (Ralstonia) are included genera—the other matches were to unidentified isolates), the sequence is a perfect match between nucleotides 906–926, strongly indicating that the primers used for this region did not result in errant sequence data.
Figure 1.
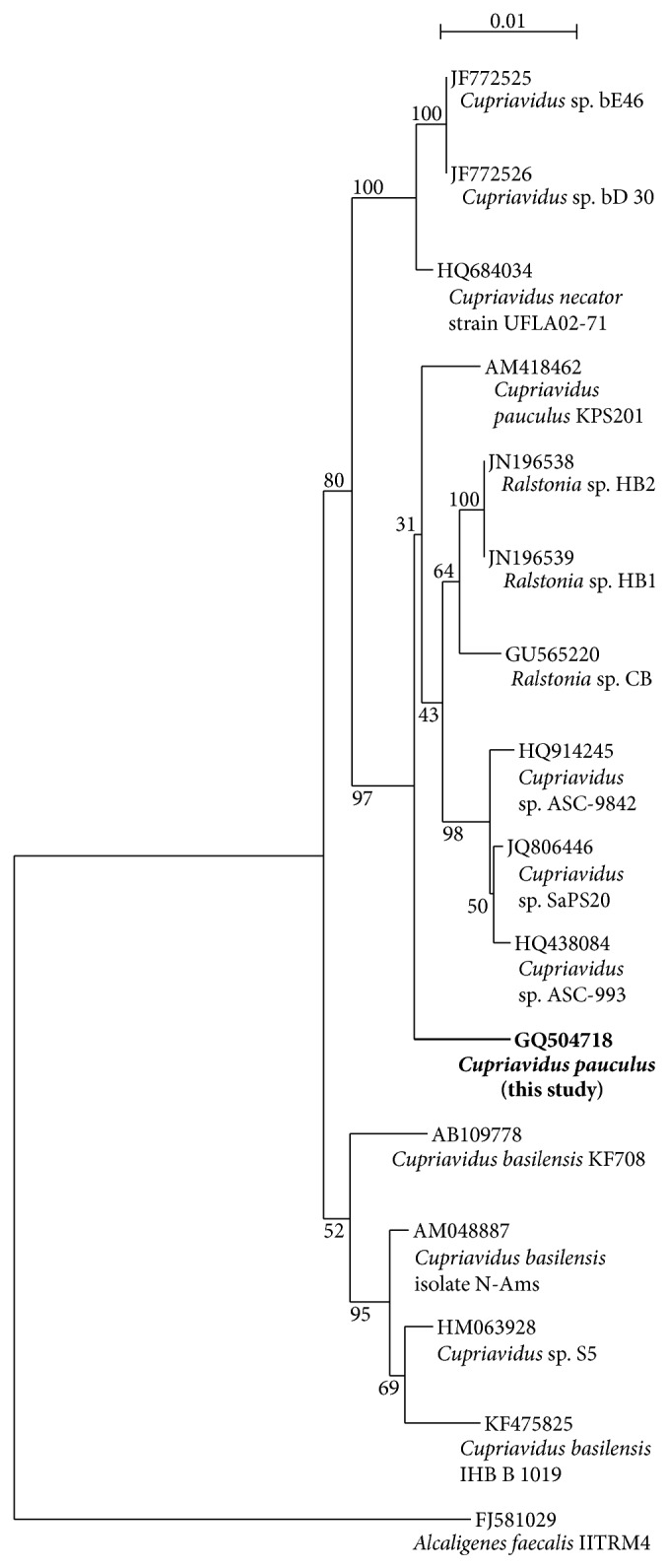
Phylogenetic analysis of the bacterium of this study based on 16S rRNA gene homologies. (The species/strain designations used for comparison are included in the figure. Alcaligenes faecalis IITRM4 served as the outgroup.)
A 98% identity match to a species' 16S rRNA nucleotide sequence is considered species identification [41, 42]. Analyses of the 16S rRNA gene sequence of C. pauculus UM1 revealed that its greatest homology was to the microbes C. pauculus strain KPS201 and Ralstonia sp. HB1 or HB2, all of which had a GenBank 98.0% similarity match and E values of 0.0. Nucleotide analysis by Greengenes (the more reliable database as it screens for chimeras and performs standard alignments and taxonomic classifications based on multiple published taxonomies [30]) indicated a 97.9% match to Cupriavidus pauculus strain KPS201, with the second closest match being to Ralstonia eutropha strain H16 (97.4%).
We note that Alcaligenes eutrophus has been reclassified as Ralstonia eutrophus, then as Wautersia eutropha, and finally as Cupriavidus necator, the type species [43]. Cupriavidus pauculus is now the accepted nomenclature for the microbes previously known as Ralstonia paucula and Wautersia paucula [43]. As the Ralstonia spp. bear no species identification and are thus of less consequence than the bacterium (C. pauculus KPS201) identified as being the closest match in Greengenes, we propose that the identity of our isolate is a C. pauculus strain (C. pauculus UM1). Phylogenetic analysis (Figure 1) suggests a close association with C. pauculus KPS201 and with organisms originally classified as either Ralstonia or Cupriavidus. The claim is supported by the bacterium's phenotypic traits [25] and its G+C content (66.1 mol%) as the genus has a G+C content ranging from 63 to 69 mol% [43].
Both of the primer sets used to generate nitrite reductase amplicons yielded highly similar matches to sequences of the nirS gene, that is, the cytochrome cd1 (nirS) nitrite reductase gene (data not shown). nirS gene sequences GQ504717, generated from the Braker et al. [32] amplicon, and GQ504716, from the Throbäck et al. [33] amplicon, have been deposited in GenBank.
The ability to denitrify is widespread and precludes identifying denitrifiers by using 16S rRNA gene analyses [3, 44, 45]. That the genus Cupriavidus is capable of denitrification is supported by work [46] that noted that members of the genus were among the most frequently isolated denitrifiers obtained from a luvisol, that the type species of the genus (C. necator) was specifically identified as a denitrifier, and that a number of previously recognized genera capable of denitrification are now part of the Cupriavidus genus. GenBank lists 20 Cupriavidus nirS entries, 9 norB entries, and 6 nosZ entries and Schwartz et al. [47] noted that Ralstonia eutropha (C. necator) H16 contained the megaplasmid pHG1, which had a gene cluster for denitrification. The cloned sequences of C. pauculus UM1's nitrite reductase gene were good matches (87–95%) to known nirS nucleotide sequences (87% Ralstonia eutropha JMP134 chromosome 2, complete sequence, 95% Pseudomonas chloritidismutans partial nirS gene for nitrite reductase, isolate AW-1). As denitrifiers contain a single type of nitrite reductase [3, 32, 33] and since our isolate contained a nirS-like gene, we conclude that C. pauculus UM1 denitrifies via its cytochrome cd1 nitrite reductase. While an earlier study [48] reported that R. paucula (now C. pauculus) was unable to denitrify, considering our isolate's resistance to Cu and Ni (see below), its sharing of a denitrification capacity with other members of the genus [46, 47, 49], its 16S rRNA gene homology to C. pauculus KPS201, and other Cupriavidus spp. and the above G+C content and phenotypic similarities, the data support that C. pauculus UM1 denitrifies via the use of a nirS-like gene. Of particular interest, and revealed in this study, is that even though C. pauculus UM1 sp. contains a nirS-like gene which it uses to denitrify [6], the organism did not invoke its use when heterotrophically nitrifying as evidenced by the lack of either N2O or N2 synthesis under the heterotrophic nitrification or heterotrophic nitrification-aerobic denitrification conditions of the current study.
Makkar and Casida [50] were the first to characterize the genus Cupriavidus. They noted that the type strain (C. necator) grew more quickly in media containing 0.4, 0.6, or 0.8 mM Cu2+ than with no copper. The genus is noted for its metal resistance [43] and members have been studied for their ability to tolerate metals [51, 52]. Like C. necator, C. pauculus UM1 tolerated copper (and nickel) well, demonstrating robust growth in media containing 1 mM Cu2+ and 0.5 mM Ni2+ (Tables 1 and 2 and Figure 2). MIC values were determined as 3 mM Cu2+ and 1 mM Ni2+ (data not shown). C. pauculus UM1 also grew readily and heterotrophically nitrified well, when in NB-PO with either 1 mM Cu2+ or 0.5 mM Ni2+ (Table 1). Confirmation of C. pauculus UM1's ability to withstand Cu2+ was noted as it grew readily in the 1 and 2 mM Cu2+ synthetic medium used to study the related bacterium Cupriavidus metallidurans CH34 [51].
The ability of C. pauculus UM1 to heterotrophically nitrify PO in the absence of other carbon sources (no NB present) when either Cu2+ or Ni2+ was present was investigated. When in the presence of either 1 mM Cu2+ or 0.5 mM Ni2+, the bacterium readily used PO as its sole C source, nitrifying it vigorously (Table 1). The relative ability of the bacterium to use PO and to nitrify it with and without the metals was also examined (Figure 2). C. pauculus UM1 grew well and nitrified readily under these conditions, demonstrating growth and nitrification when in the presence of either 1 mM Cu2+ or 0.5 mM Ni2+ that was commensurate with the metals being absent. The higher Abs600 of either the control or experimental samples containing Cu2+ was due to the blue color imparted to the clear medium by the 1 mM Cu2+.
C. pauculus KPS 201 [23] was able to withstand notably high concentrations of nickel (MIC of 28.9 mM). More common metal resistance values of approximately 0.5–12 mM Cu [53–55] and 2–25 mM Ni [53, 54, 56, 57] have been noted for a number of metal-resistant microbes, with Ni-sensitivity cited as tolerating no more than 0.1 mM Ni [57]. C. pauculus UM1 readily grew in media containing either 1 mM Cu2+ or 0.5 mM Ni2+ (Tables 1 and 2 and Figure 2) and tolerated higher copper than nickel concentrations (see above). C. pauculus UM1 is thus Cu and Ni tolerant when compared to other microbes [53–56]. Of interest is the observation that C. pauculus UM1's ability to use PO, as both a carbon and heterotrophic nitrification source, was not impeded by the metals (Table 1 and Figure 2). When an inoculum was grown in a nutrient-rich medium (NB) containing either 1 mM Cu2+ or 0.5 mM Ni2+ and was used to inoculate media containing either NB-PO, NB-PO-1 mM Cu2+, or NB-PO-0.5 mM Ni2+, C. pauculus UM1 grew as fast in the presence of either 1 mM Cu2+ or 0.5 mM Ni2+ as without the metals being present (data not shown) and yielded higher amounts of cells (Table 1). When C. pauculus UM1 was grown in a PO-mineral salts medium and used to inoculate either PO, PO-1 mM Cu2+, or PO-0.5 mM Ni2+ mineral salts media, the organism grew as readily in the metal-containing media and nitrified to the same extent, producing equal amounts of NO2 − (Figure 2).
Xenobiotics metabolized and degraded by C. pauculus include benzene, toluene [58], 3-nitrophenol [59], the herbicide 2,4-dichlorophenoxyacetic acid [60, 61], and the pesticides chlorpyrifos (O,O-diethyl O-(3,5,6-trichloro-2-pyridyl)phosphorothioate) and TCP (3,5,6-trichloro-2-pyridinol) [62, 63]. As a representative xenobiotic, phenol (2 mM) supported the growth of C. pauculus UM1 (Figure 3), using virtually all of the phenol by the end of the experiment. A shorter induction (about 2-3 days) was observed if the concentration of phenol was reduced to 1 mM (data not shown). C. pauculus UM1 also demonstrated the ability to use phenol in the presence of either Cu2+ or Ni2+ (Table 2) which suggests that it may be an effective bioremediation agent in environments where these metals are present. Preliminary experiments show that C. pauculus UM1 grows in media with catechol as the sole C source (data not shown). That C. pauculus UM1 uses phenol as a sole carbon source (Table 2 and Figure 3) suggests that catechol is an intermediate as phenol is known to be metabolized via one of two catechol-dependent pathways [64, 65].
This report is the first, to our knowledge, to describe a member of the genus Cupriavidus as being a heterotrophic nitrifier. C. pauculus UM1 is a potent heterotrophic nitrifier and denitrifier and is able to resist the toxic effects of copper and nickel while heterotrophically nitrifying (Table 1, Figure 2 and MIC values of 3 mM Cu2+ and 1.0 mM Ni2+). Its use of phenol as a carbon source further expands the remarkable metabolic abilities of the genus and the species. Given C. pauculus UM1's metabolic capacities, it will be of interest to determine whether the microbe contains the metal resistance genes noted in other Cupriavidus spp. as well as the genes necessary to metabolize xenobiotics such as those noted above. We are investigating the use of such xenobiotics by C. pauculus UM1. If C. pauculus UM1 does contain such genes, it may be employed as a bioremediation agent. Given the bacterium's resistance to copper and nickel, its metabolism of such xenobiotics may be beneficial in those environments where metal tolerance is either an advantage or a necessity.
4. Conclusions
A soil microbe was examined for its ability to produce nitrogenous gasses while heterotrophically nitrifying. The organism nitrified the organic substrate pyruvic oxime to nitrite well but produced neither N2 nor N2O. Nucleotide and phylogenetic analyses resulted in classifying the bacterium as a member of the genus Cupriavidus, thus establishing that this genus can heterotrophically nitrify. The microbe, C. pauculus UM1, performed heterotrophic nitrification in the presence of Cu2+ and Ni2+, nitrifying well when either 1 mM Cu2+ or 0.5 mM Ni2+ was present. C. pauculus UM1 also metabolized the xenobiotic phenol, using it as substrate for growth in either the absence or presence of 1 mM Cu2+ or 0.5 mM Ni2+.
Subsequent to the submission of this paper, 16S rRNA gene cloning of C. pauculus UM1 was performed using the primer pair of Yu et al. (Yu, C, Hongwei, G, Yanming, Z, Mingjun, D, Zhenxying, W, Laihua, Z, Qing, D, Biao, X, Chengzhu, L, Zhiquin, Y, Xizhi X (2012), analysis of the bacterial diversity existing on animal hide and wool: development of a preliminary pcr-restriction fragment length polymorphism fingerprint database for identifying isolates, Journal of AOAC International, 95(6):1750–1754). These primers are designed to clone from base pair 27 to base pair 1492 of the E. coli 16S rRNA gene. With respect to base pairs 906–926, the sequence obtained was a perfect match to that previously obtained for C. pauculus UM1's 16S rRNA gene, further supporting the conclusion that the match between nucleotides 906 and 926 was not the result of primer sequence resulting in errant sequence data.
Acknowledgments
The authors are indebted to Martin Berg, John Kelly, Catherine Putonti, and Samantha Lindemann for their helpful discussions and assistance concerning this work. The support of a Loyola University Chicago Provost Award and of Loyola University Chicago Mulcahy Scholarships is acknowledged as funding part of this work.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Castignetti D., Hollocher T. C. Heterotrophic nitrification among denitrifiers. Applied and Environmental Microbiology. 1984;47(4):620–623. doi: 10.1128/aem.47.4.620-623.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jetten M. S. M., Logemann S., Muyzer G., Robertson L. A., De Vries S., Van Loosdrecht M. C. M., Kuenen J. G. Novel principles in the microbial conversion of nitrogen compounds. Antonie van Leeuwenhoek. 1997;71(1-2):75–93. doi: 10.1023/A:1000150219937. [DOI] [PubMed] [Google Scholar]
- 3.Zumft W. G. Cell biology and molecular basis of denitrification? Microbiology and Molecular Biology Reviews. 1997;61(4):533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gamble T. N., Betlach M. R., Tiedje J. M. Numerically dominant denitrifying bacteria from world soils. Applied and Environmental Microbiology. 1977;33(4):926–939. doi: 10.1128/aem.33.4.926-939.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Philippot L. Denitrifying genes in bacterial and Archaeal genomes. Biochimica et Biophysica Acta: Gene Structure and Expression. 2002;1577(3):355–376. doi: 10.1016/S0167-4781(02)00420-7. [DOI] [PubMed] [Google Scholar]
- 6.Castignetti D., Hollocher T. C. Vigorous denitrification by a heterotrophic nitrifier of the genus Alcaligenes. Current Microbiology. 1981;6(4):229–231. doi: 10.1007/BF01566978. [DOI] [Google Scholar]
- 7.Verstraete W. Heterotrophic nitrification in soils and aqueous media. Izvestiya Akademii Nauk SSSR, Seriya Biologicheskaya. 1975;4:541–558. (Rus). [Google Scholar]
- 8.Cookson W. R., Müller C., O'Brien P. A., Murphy D. V., Grierson P. F. Nitrogen dynamics in an Australian semiarid grassland soil. Ecology. 2006;87(8):2047–2057. doi: 10.1890/0012-9658(2006)87[2047:NDIAAS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 9.Du R., Lu D., Wang G. Diurnal, seasonal, and inter-annual variations of N2O fluxes from native semi-arid grassland soils of inner Mongolia. Soil Biology & Biochemistry. 2006;38(12):3474–3482. doi: 10.1016/j.soilbio.2006.06.012. [DOI] [Google Scholar]
- 10.Joo H.-S., Hirai M., Shoda M. Piggery wastewater treatment using Alcaligenes faecalis strain No. 4 with heterotrophic nitrification and aerobic denitrification. Water Research. 2006;40(16):3029–3036. doi: 10.1016/j.watres.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 11.Islam A., Chen D., White R. E. Developing a technique to quantify heterotrophic and autotrophic nitrification in acidic pasture soils. Communications in Soil Science and Plant Analysis. 2007;38(17-18):2309–2321. doi: 10.1080/00103620701588437. [DOI] [Google Scholar]
- 12.Islam A., Chen D., White R. E. Heterotrophic and autotrophic nitrification in two acid pasture soils. Soil Biology and Biochemistry. 2007;39(4):972–975. doi: 10.1016/j.soilbio.2006.11.003. [DOI] [Google Scholar]
- 13.Bai Y., Sun Q., Zhao C., Wen D., Tang X. Microbial degradation and metabolic pathway of pyridine by a Paracoccus sp. strain BW001. Biodegradation. 2008;19(6):915–926. doi: 10.1007/s10532-008-9193-3. [DOI] [PubMed] [Google Scholar]
- 14.Chen Q., Ni J. Ammonium removal by Agrobacterium sp. LAD9 capable of heterotrophic nitrification-aerobic denitrification. Journal of Bioscience and Bioengineering. 2012;113(5):619–623. doi: 10.1016/j.jbiosc.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 15.Yao S., Ni J., Chen Q., Borthwick A. G. L. Enrichment and characterization of a bacteria consortium capable of heterotrophic nitrification and aerobic denitrification at low temperature. Bioresource Technology. 2013;127:151–157. doi: 10.1016/j.biortech.2012.09.098. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Q.-L., Liu Y., Ai G.-M., Miao L.-L., Zheng H.-Y., Liu Z.-P. The characteristics of a novel heterotrophic nitrification-aerobic denitrification bacterium, Bacillus methylotrophicus strain L7. Bioresource Technology. 2012;108:35–44. doi: 10.1016/j.biortech.2011.12.139. [DOI] [PubMed] [Google Scholar]
- 17.Zhao B., An Q., He Y. L., Guo J. S. N2O and N2 production during heterotrophic nitrification by Alcaligenes faecalis strain NR. Bioresource Technology. 2012;116:379–385. doi: 10.1016/j.biortech.2012.03.113. [DOI] [PubMed] [Google Scholar]
- 18.Chen P., Li J., Li Q. X., Wang Y., Li S., Ren T., Wang L. Simultaneous heterotrophic nitrification and aerobic denitrification by bacterium Rhodococcus sp. CPZ24. Bioresource Technology. 2012;116:266–270. doi: 10.1016/j.biortech.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 19.Su J.-J., Yeh K.-S., Tseng P.-W. A strain of Pseudomonas sp. isolated from piggery wastewater treatment systems with heterotrophic nitrification capability in Taiwan. Current Microbiology. 2006;53(1):77–81. doi: 10.1007/s00284-006-0021-x. [DOI] [PubMed] [Google Scholar]
- 20.Papen H., von Berg R., Hinkel I., Thoene B., Rennenberg H. Heterotrophic nitrification by Alcaligenes faecalis: NO2 −, NO3 −, N2O, and NO production in exponentially growing cultures. Applied and Environmental Microbiology. 1989;55(8):2068–2072. doi: 10.1128/aem.55.8.2068-2072.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Q., Ni J. Heterotrophic nitrification-aerobic denitrification by novel isolated bacteria. Journal of Industrial Microbiology and Biotechnology. 2011;38(9):1305–1310. doi: 10.1007/s10295-010-0911-6. [DOI] [PubMed] [Google Scholar]
- 22.Castignetti D., Gunner H. B. Nitrite and nitrate synthesis from pyruvic-oxime by an Alcaligenes sp. Current Microbiology. 1981;5(6):379–384. doi: 10.1007/BF01566754. [DOI] [Google Scholar]
- 23.Pal A., Wauters G., Paul A. K. Nickel tolerance and accumulation by bacteria from rhizosphere of nickel hyperaccumulators in serpentine soil ecosystem of Andaman, India. Plant and Soil. 2007;293(1-2):37–48. doi: 10.1007/s11104-007-9195-7. [DOI] [Google Scholar]
- 24.Chess B. Laboratory Applications in Microbiology, A Case Study Approach. Boston, Mass, USA: McGraw Hill; 2009. Chapter 20: antimicrobial sensitivity testing: Kirby-Bauer, tube dilution, and E-test methods; pp. 163–176. [Google Scholar]
- 25.Castignetti D., Gunner H. B. Sequential nitrification by an Alcaligenes sp. and Nitrobacter agilis . Canadian Journal of Microbiology. 1980;26(9):1114–1119. doi: 10.1139/m80-184. [DOI] [PubMed] [Google Scholar]
- 26.Dicksved J., Flöistrup H., Bergström A., Rosenquist M., Pershagen G., Scheynius A., Roos S., Alm J. S., Engstrand L., Braun-Fahrländer C., Von Mutius E., Jansson J. K. Molecular fingerprinting of the fecal microbiota of children raised according to different lifestyles. Applied and Environmental Microbiology. 2007;73(7):2284–2289. doi: 10.1128/AEM.02223-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu W.-T., Marsh T. L., Cheng H., Forney L. J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Applied and Environmental Microbiology. 1997;63(11):4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fry N. K., Fredrickson J. K., Fishbain S., Wagner M., Stahl D. A. Population structure of microbial communities associated with two deep, anaerobic, alkaline aquifers. Applied and Environmental Microbiology. 1997;63(4):1498–1504. doi: 10.1128/aem.63.4.1498-1504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brody J. R., Kern S. E. Sodium boric acid: a Tris-free, cooler conductive medium for DNA electrophoresis. BioTechniques. 2004;36(2):214–216. doi: 10.2144/04362BM02. [DOI] [PubMed] [Google Scholar]
- 30.DeSantis T. Z., Hugenholtz P., Larsen N., et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Applied and Environmental Microbiology. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gouy M., Guindon S., Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Molecular Biology and Evolution. 2010;27(2):221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 32.Braker G., Fesefeldt A., Witzel K.-P. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Applied and Environmental Microbiology. 1998;64(10):3769–3775. doi: 10.1128/aem.64.10.3769-3775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Throbäck I. N., Enwall K., Jarvis Å., Hallin S. Reassessing PCR primers targeting nirS, nirK and nosZ genes for community surveys of denitrifying bacteria with DGGE. FEMS Microbiology Ecology. 2004;49(3):401–417. doi: 10.1016/j.femsec.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 34.Quastel J. H., Scholefield P. G., Stevenson J. W. Oxidation of pyruvic acid oxime by soil organisms. The Biochemical journal. 1952;51(2):278–286. doi: 10.1042/bj0510278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van'T Riet J., Stouthamer A. H., Planta R. J. Regulation of nitrate assimilation and nitrate respiration in Aerobacter aerogenes . Journal of Bacteriology. 1968;96(5):1455–1464. doi: 10.1128/jb.96.5.1455-1464.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.St John R. T., Hollocher T. C. Nitrogen 15 tracer studies on the pathway of denitrification in Pseudomonas aeruginosa . The Journal of Biological Chemistry. 1977;252(1):212–218. [PubMed] [Google Scholar]
- 37.Slinkard K., Singleton V. Total phenol analysis: automation and comparison with manual methods. The American Journal of Enology and Viticulture. 1977;28:49–55. [Google Scholar]
- 38.Castignetti D., Petithory J. R., Hollocher T. C. Pathway of oxidation of pyruvic oxime by a heterotrophic nitrifier of the genus Alcaligenes: evidence against hydrolysis to pyruvate and hydroxylamine. Archives of Biochemistry and Biophysics. 1983;224(2):587–593. doi: 10.1016/0003-9861(83)90246-1. [DOI] [PubMed] [Google Scholar]
- 39.Ono Y., Enokiya A., Masuko D., Shoji K., Yamanaka T. Pyruvic oxime dioxygenase from the heterotrophic nitrifier Alcaligenes faecalis: purification, and molecular and enzymatic properties. Plant and Cell Physiology. 1999;40(1):47–52. doi: 10.1093/oxfordjournals.pcp.a029473. [DOI] [Google Scholar]
- 40.Coenye T., Spilker T., Reik R., Vandamme P., LiPuma J. J. Use of PCR analyses to define the distribution of Ralstonia species recovered from patients with cystic fibrosis. Journal of Clinical Microbiology. 2005;43(7):3463–3466. doi: 10.1128/JCM.43.7.3463-3466.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Madigan M., Martinko J., Dunlap P., Clark D. Biology of Microorganisms. 12th. chapter 14. San Francisco, Calif, USA: Pearson-Benjamin Cummings; 2009. Microbial evolution and systematics; pp. 387–389. [Google Scholar]
- 42.Rosselló-Mora R., Amann R. The species concept for prokaryotes. FEMS Microbiology Reviews. 2001;25(1):39–67. doi: 10.1016/S0168-6445(00)00040-1. [DOI] [PubMed] [Google Scholar]
- 43.Vandamme P., Coenye T. Taxonomy of the genus Cupriavidus: a tale of lost and found. International Journal of Systematic and Evolutionary Microbiology. 2004;54(6):2285–2289. doi: 10.1099/ijs.0.63247-0. [DOI] [PubMed] [Google Scholar]
- 44.Braker G., Ayala-Del-Río H. L., Devol A. H., Fesefeldt A., Tiedje J. M. Community structure of denitrifiers , bacteria, and archaea along redox gradients in pacific northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Applied and Environmental Microbiology. 2001;67(4):1893–1901. doi: 10.1128/AEM.67.4.1893-1901.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mosier A. C., Francis C. A. Denitrier abundance and activity across the San Francisco Bay estuary. Environmental Microbiology Reports. 2010;2(5):667–676. doi: 10.1111/j.1758-2229.2010.00156.x. [DOI] [PubMed] [Google Scholar]
- 46.Verbaendert I., Boon N., de Vos P., Heylen K. Denitrification is a common feature among members of the genus Bacillus . Systematic and Applied Microbiology. 2011;34(5):385–391. doi: 10.1016/j.syapm.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 47.Schwartz E., Henne A., Cramm R., Eitinger T., Friedrich B., Gottschalk G. Complete nucleotide sequence of pHG1: a Ralstonia eutropha H16 megaplasmid encoding key enzymes of H2-based lithoautotrophy and anaerobiosis. Journal of Molecular Biology. 2003;332(2):369–383. doi: 10.1016/S0022-2836(03)00894-5. [DOI] [PubMed] [Google Scholar]
- 48.Vandamme P., Goris J., Coenye T., Hoste B., Janssens D., Kersters K., De Vos P., Falsen E. Assignment of centers for disease control group IVc-2 to the genus Ralstonia as Ralstonia paucula sp. nov. International Journal of Systematic Bacteriology. 1999;49(2):663–669. doi: 10.1099/00207713-49-2-663. [DOI] [PubMed] [Google Scholar]
- 49.Cramm R. Genomic view of energy metabolism in Ralstonia eutropha H16. Journal of Molecular Microbiology and Biotechnology. 2009;16(1-2):38–52. doi: 10.1159/000142893. [DOI] [PubMed] [Google Scholar]
- 50.Makkar N. S., Casida L. E. Cupriavidus necator gen. nov., sp. nov.: a nonobligate bacterial predator of bacteria in soil. International Journal of Systematic Bacteriology. 1987;37:323–326. [Google Scholar]
- 51.Monchy S., Benotmane M. A., Janssen P., Vallaeys T., Taghavi S., Van Der Lelie D., Mergeay M. Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals. Journal of Bacteriology. 2007;189(20):7417–7425. doi: 10.1128/JB.00375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryan M. P., Adley C. C. Specific PCR to identify the heavy-metal-resistant bacterium Cupriavidus metallidurans . Journal of Industrial Microbiology and Biotechnology. 2011;38(9):1613–1615. doi: 10.1007/s10295-011-1011-y. [DOI] [PubMed] [Google Scholar]
- 53.Ryan R. P., Ryan D. J., Dowling D. N. Multiple metal resistant transferable phenotypes in bacteria as indicators of soil contamination with heavy metals. Journal of Soils and Sediments. 2005;5(2):95–100. doi: 10.1065/jss2004.10.120. [DOI] [Google Scholar]
- 54.Rajkumar M., Ma Y., Freitas H. Characterization of metal-resistant plant-growth promoting Bacillus weihenstephanensis isolated from serpentine soil in Portugal. Journal of Basic Microbiology. 2008;48(6):500–508. doi: 10.1002/jobm.200800073. [DOI] [PubMed] [Google Scholar]
- 55.Vanneste J. L., Cornish D. A., Yu J., Boyd R. J., Morris C. E. Isolation of copper and streptomycin resistant phytopathogenic Pseudomonas syringae from lakes and rivers in the central north island of New Zealand. New Zealand Plant Protection. 2008;61:80–85. [Google Scholar]
- 56.Patel J. S., Patel P. C., Kalia K. Isolation and characterization of nickel uptake by nickel resistant bacterial isolate (NiRBI) Biomedical and Environmental Sciences. 2006;19(4):297–301. [PubMed] [Google Scholar]
- 57.Tripathi V. N., Srivastava S. Extracytoplasmic storage as the nickel resistance mechanism in a natural isolate of Pseudomonas putida S4. Canadian Journal of Microbiology. 2006;52(4):287–292. doi: 10.1139/W05-133. [DOI] [PubMed] [Google Scholar]
- 58.Lee E.-H., Park H., Cho K.-S. Biodegradation of methane, benzene, and toluene by a consortium MBT14 enriched from a landfill cover soil. Journal of Environmental Science and Health Part A: Toxic/Hazardous Substances and Environmental Engineering. 2013;48(3):273–278. doi: 10.1080/10934529.2013.726812. [DOI] [PubMed] [Google Scholar]
- 59.Chi X.-Q., Zhang J.-J., Zhao S., Zhou N.-Y. Bioaugmentation with a consortium of bacterial nitrophenol-degraders for remediation of soil contaminated with three nitrophenol isomers. Environmental Pollution. 2013;172:33–41. doi: 10.1016/j.envpol.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 60.Kraiser T., Stuardo M., Manzano M., Ledger T., González B. Simultaneous assessment of the effects of an herbicide on the triad: rhizobacterial community, an herbicide degrading soil bacterium and their plant host. Plant and Soil. 2013;366(1-2):377–388. doi: 10.1007/s11104-012-1444-8. [DOI] [Google Scholar]
- 61.Sakai Y., Ogawa N., Fujii T., Sugahara K., Miyashita K., Hasabe A. 2,4-dichlorophenoxyacetic acid-degrading genes from bacteria isolated from soil in Japan: spread of Burkholderia cepacia RASC-type degrading genes harbored on large plasmids. Microbes and Environments. 2007;22:145–156. [Google Scholar]
- 62.Cao L., Liu H., Zhang H., Huang K., Gu T., Ni H., Hong Q., Li S. Characterization of a newly isolated highly effective 3,5,6-trichloro-2-pyridinol degrading strain Cupriavidus pauculus P2. Current Microbiology. 2012;65(3):231–236. doi: 10.1007/s00284-012-0150-3. [DOI] [PubMed] [Google Scholar]
- 63.Lu P., Li Q., Liu H., Feng Z., Yan X., Hong Q., Li S. Biodegradation of chlorpyrifos and 3,5,6-trichloro-2-pyridinol by Cupriavidus sp. DT-1. Bioresource Technology. 2013;127:337–342. doi: 10.1016/j.biortech.2012.09.116. [DOI] [PubMed] [Google Scholar]
- 64.Pérez-Pantoja D., de la Iglesia R., Pieper D. H., González B. Metabolic reconstruction of aromatic compounds degradation from the genome of the amazing pollutant-degrading bacterium Cupriavidus necator JMP134. FEMS Microbiology Reviews. 2008;32(5):736–794. doi: 10.1111/j.1574-6976.2008.00122.x. [DOI] [PubMed] [Google Scholar]
- 65.Powlowski J., Shingler V. Genetics and biochemistry of phenol degradation by Pseudomonas sp. CF600. Biodegradation. 1994;5(3-4):219–236. doi: 10.1007/BF00696461. [DOI] [PubMed] [Google Scholar]


