Abstract
Targeted, capture-based DNA sequencing is a cost-effective method to focus sequencing on a coding region or other customized region of the genome. There are multiple targeted sequencing methods available, but none has been systematically investigated and compared. We evaluated four commercially available custom-targeted DNA technologies for next-generation sequencing with respect to on-target sequencing, uniformity, and ability to detect single-nucleotide variations (SNVs) and copy number variations. The technologies that used sonication for DNA fragmentation displayed impressive uniformity of capture, whereas the others had shorter preparation times, but sacrificed uniformity. One of those technologies, which uses transposase for DNA fragmentation, has a drawback requiring sample pooling, and the last one, which uses restriction enzymes, has a limitation depending on restriction enzyme digest sites. Although all technologies displayed some level of concordance for calling SNVs, the technologies that require restriction enzymes or transposase missed several SNVs largely because of the lack of coverage. All technologies performed well for copy number variation calling when compared to single-nucleotide polymorphism arrays. These results enable laboratories to compare these methods to make informed decisions for their intended applications.
Next-generation sequencing technologies have enabled cancer genomics discovery by providing a high-throughput and cost-effective strategy to sequence thousands of cancer genomes.1 Next-generation sequencing has helped identify novel genomic alterations in cancer, including, but not limited to, single-nucleotide variations (SNVs), copy number variations (CNVs), and gene fusions that may serve as predictive biomarkers for matching patients to targeted therapies in trials.2 Although whole genome and exome sequencing enables discovery of novel genomic alterations, clinical-grade applications must consider constraints, including cost per patient, time to results, and depth of coverage. Consequently, clinical implementation of sequencing has focused on customized sequencing of actionable genes, exons, or regions.3,4 A targeted-capture next-generation sequencing strategy is clinically pragmatic, because it is scalable, is economical, and allows for deeper sequencing coverage compared to whole genome or whole exome approaches. Thus, many laboratories are using or considering custom capture gene panels for diverse applications, including discovery, validation testing, or clinical-grade assay development.3,5–8 To this end, we compared the proficiency of four methods to capture and sequence a custom gene panel.
Practical considerations for targeted gene sequencing include cost of sequencing and wanted depth of coverage. Thus, ideal methods provide high on-target and uniform sequencing. Amplicon-based assays for cancer gene hotspots offer high on-target specificity, but can only cover a limited scope of regions.2 Beyond such assays in cancer,5,6,8 hybridization-based capture strategies can focus on larger regions of interest and additional types of alteration, such as CNVs.3,7,9
Although earlier studies compared the strengths and weaknesses of three methods for whole exome capture using in-solution hybridization,10–14 several new clinically oriented methods have been developed that offer rapid, simplified sample preparation and lower requirements for DNA input. Herein, we describe a comprehensive comparison of four DNA capture technologies for a panel of 257 cancer-related genes. We evaluated the ability of four methods for each to capture targeted regions in 16 samples, including six pairs of tumor and normal tissue and four cell lines, in terms of alignment rate, sequencing uniformity, GC content bias, SNV concordance, and CNV-calling ability.
Materials and Methods
Cell Lines and Tissue DNA Samples
BT-20 and MCF-7 breast cancer cell lines (with PIK3CA hotspot mutations), HCC-2218 (with ERBB2 gene amplification), and HCC-2218's matching lymphoblastoid cell line, HCC-2218BL, were obtained from ATCC (Manassas, VA). De-identified paired tumor and matched-normal DNA samples of breast, melanoma, lung, and colon were obtained from Origene Technologies Inc. (Rockville, MD) (Supplemental Table S1). DNA was extracted from log phase growing cell lines using Blood and Cell Culture DNA Mini-kit (Qiagen, Valencia, CA). The quantity and quality of DNA were measured with NanoDrop 2000c (Thermo Scientific, Waltham, MA) (optical density ratio: 260:280 = 1.8 to 2.0; 260:230 = 2.0 to 2.2) and TapeStation 2200 (Genomic DNA Screen Tape; Agilent Technologies, Santa Clara, CA). The double-stranded DNA was further quantified using Quant-iT dsDNA BR Assay kit by Qubit 2.0 fluorometer (A260/A280 is 1.8 to 2.0) (Invitrogen, Carlsbad, CA). All 16 sample DNAs were captured and sequenced with SureSelect (Agilent Technologies), HaloPlex (Agilent Technologies), Nextera (Illumina, San Diego, CA), and SeqCap (Roche Nimblegen, Madison, WI), methods, except for tissue samples 3, 4, 5, and 6, for which there was enough DNA input for HaloPlex and Nextera only.
SureSelect Custom Target Enrichment Library Preparation
The genomic DNA (gDNA; 2 μg) was diluted with 1× low Tris-EDTA buffer and sheared using Covaris S2 sonicator to achieve target peak of 150 to 200 bp (SonoLab 7 settings: Duty Factor, 10%; Peak Incident Power, 175; cycles per burst, 200; DNA treatment time, 360 seconds; water bath temperature, 4°C) (Covaris, Woburn, MA). Agilent's SureSelectXT Target Enrichment protocol version 1.5 was followed for library preparation without modification. Fourteen cycles of PCR were performed for amplification of the post-captured library, and the quality of the final DNA library was assessed using the High Sensitivity D1K ScreenTape and TapeStation 2200 (Agilent Technologies, Santa Clara, CA). Per manufacturer's protocol, library peak size was in the range of 300 to 400 nucleotides.
HaloPlex Custom Exome Library Preparation
gDNA (200 ng; split among eight different restriction reactions) plus 25 ng of excess gDNA was used for a total of 225 ng of gDNA, as described in the protocol. Agilent's HaloPlex Target Enrichment Protocol version D.3 was followed for library preparation without modification. Eighteen cycles of PCR were completed for amplification of the captured library. The quality of the final DNA library was assessed with the High Sensitivity D1K ScreenTape (TapeStation 2200). Per manufacturer's protocol, library peak size was in the range of 225 to 525 nucleotides.
Nextera Custom Enrichment Library Preparation
gDNA (50 ng) was used, and Nextera Custom Enrichment sample was followed for the preparation protocol without modification (Illumina). Ten cycles of PCR were completed for amplification of the final library, as recommended. The enriched DNA library was quantitated using real-time quantitative PCR, as described in the Illumina Sequencing Library real-time quantitative PCR quantification guide (Illumina).
SeqCap EZ Choice Library Preparation
Standard genomic libraries were initially prepared using 1 μg of gDNA Illumina TruSeq DNA (Illumina). Next, the library was amplified by ligation-mediated PCR (eight cycles) and hybridized to custom probes, and final amplification was completed of the post-captured library by ligation-mediated PCR (14 cycles), according to the manufacturer's protocol following the SeqCap EZ library preparation guide (NimbleGen, Madison, WI). Roche Nimblegen's policy prohibited release of SeqCap's probe coordinates, but the design is available on request.
Sequencing of Libraries
Index-tagged libraries were quantified using HS Qubit dsDNA assay (Invitrogen). SureSelect (Agilent Technologies), Haloplex (Agilent Technologies), and SeqCap (Roche Diagnostics, Basel, Switzerland) library samples were pooled and diluted to 2 nmol/L stocks for multiplexed (four-plex) sequencing on Illumina's MiSeq (2 × 100 bp). For Nextera samples, the library pooling guideline was followed to pool 16 samples at once for sequencing on a MiSeq (2 × 100 bp). Although we used 100-bp reads for detecting SNVs and CNVs, users may select an alternate read length, depending on wanted downstream goal of identifying SNVs, CNVs, or structural variants (http://www.ncbi.nlm.nih.gov/gap; Accession number phs000811.v1.p1).
SNP Array
Affymetrix Genome-Wide Human SNP Array 6.0 was performed using DNA isolated from breast cancer cell lines HCC-2218 and HCC-2218BL through Case Western Reserve University's (Cleveland, OH) Genomic Sequencing Core to determine CNV (Affymetrix, Santa Clara, CA). The Core followed the Affymetrix Genome-Wide Human SNP Array 6.0 method for sample preparation. Briefly, 500 ng gDNA was digested with NspI and StyI restriction enzymes and then ligated to adaptors. The adaptor-ligated DNA fragments were amplified, fragmented, labeled, and hybridized to SNP Array 6.0.
Mycoplasma Testing and Authentication
The cell lines BT-20, MCF-7, HCC-2218BL, and HCC-2218 were tested negative for Mycoplasma and authenticated (DNA Diagnostics Center, Cincinnati, OH).
Alignment
Adapters were removed from sequencing data using Illumina's MiSeq Reporter software version 2.2.29 on SureSelect, Nextera, and SeqCap libraries and Agilent's SureCall software version 1.1.0.15 on HaloPlex libraries. Raw paired-end sequencing FASTQ files were aligned to the human genome (hg19) using the Burrows-Wheeler Aligner (BWA-0.6.2)15 under the default parameters. The two resulting suffix array index files were merged and converted to Sequence Alignment/Map (SAM); the resulting SAM file was converted to Binary Alignment/Map (BAM) using the SAMtools16 view command, and the results were sorted by chromosome and position using SAMtools-0.1.1816 sort command. After alignment, we removed duplicate reads from SureSelect, Nextera, and SeqCap data using the Picard-1.84 MarkDuplicates command with default parameters (Broad Institute, http://broadinstitute.github.io/picard, last accessed September 22, 2014). Duplicates were not removed from HaloPlex data, per manufacturer's instructions. Afterward, the reads in all samples were realigned around known indels in dbSNP file hg19 snp13717 using Genome Analysis Toolkit-2.4.7 (GATK)18 using the RealignerTargetCreator and IndelRealigner, followed by the Picard FixMateInformation command. Finally, a quality score recalibration was performed for all samples using the GATK BaseRecalibrator and PrintPreads commands under the default parameters. Last, we used SAMtools sort to sort the final BAM files by name to generate a name-sorted BAM file.
We generated alignment statistics and percentage of targeted bases covered at various depths. We used Browser Extensible Data (BED)Tools19 to calculate three different alignment percentages: percentage of raw sequenced reads that aligned to the human genome, percentage of raw sequenced reads that aligned to targeted regions for the respective technology, and percentage of aligned reads that aligned to targeted regions for the respective technology. These three different alignment statistics were calculated for the final BAM file (ie, after removing duplicates, realigning around indels, and recalibrating quality score). The BEDTools bamtobed command was used to convert the name-sorted BAM files to bedpe format. By using the previously mentioned bedpe file, we calculated percentage of raw FASTQ reads to align to hg19. To calculate alignment to targeted regions for each technology, we used the BEDTools pairtobed commands using the previously mentioned bedpe file and each technology's targeted regions. A read was considered on target if at least one base from a paired-end read intersected a target region. To calculate percentage of targeted bases covered at various depths, we used mpileup files generated by SAMtools on the final BAM files and a custom Python script to calculate depth in target bases (Supplemental Code S1). We also used Picard's CollectInsertSizeMetrics under the default parameters, except for assuming the validation stringency to be lenient.
SNV Calling
By using the final BAM files after raw data post-processing, we called variants. Tumor-normal analyses followed a different pipeline from calling single-sample SNVs relative to the reference genome. For tumor-normal analyses, SAMtools was used to convert final BAM files into dual mpileup files. Then, the somatic function in VarScan2 version 2.3.320 performed SNV calls, assuming the tumor sample was pure and using default parameters. Taking only the somatic SNVs (ie, where the tumor and normal genotypes differed and the normal genotype matched the reference genome at the position in question) and germ-line SNVs (ie, where the tumor and normal genotyped matched, but differed from the reference genome at the position in question), we estimated tumor purity using PurityEst and custom scripts (for HCC-2218, purity was set to 1).21 With this calculated tumor purity, we recalculated SNVs using VarScan2. We also called tumor-normal variants using MuTect-1.1.422 under the default parameters (MuTect does not use tumor purity estimates), while inputting each technology's respective BED file. Furthermore, we used Strelka-1.0.1123 to call tumor-normal variants under the default parameters, except we set isSkipDepthFilters to 1 and maxInputDepth to ≤0; we quantified read count for each SNV using tier2 reads (Strelka does not use tumor purity). Regardless of SNV caller, somatic SNVs were annotated using ANNOVAR24 to remove intronic and synonymous SNVs and to associate exonic SNVs with known genes and their associated amino acid change, or stopgain, or stoploss.
In addition to the tumor-normal analysis, we also performed single-sample SNV calls relative to the reference genome for all samples (including cell lines, tumor samples, and normal samples). BAM files were used to generate mpileup files using SAMtools' mpileup function under the default parameters. SNVs were called using VarScan2′s mpileup2snp command under the default parameters. We also used GATK-2.4.7's18 HaplotypeCaller under the default parameters and Mpileup (SAMtools-0.1.18)16 (http://samtools.sourceforge.net/mpileup.shtml, last accessed August 8, 2014) under the default parameters to make single-sample SNV calls, except no maximum depth to call an SNV. GATK required each technology's respective BED file as input. Although the tumor purity cannot be calculated for single-sample analyses, downstream processing of single-sample SNVs was the same as for tumor-normal SNVs. We also used Agilent's SureCall software version 1.1.0.15, a platform-specific tool, for the analysis of HaloPlex data sets under the default parameters, according to the manufacturer's instructions (Agilent Technologies).
We compared SNV-calling concordance between technologies using the following equation:
| (1) |
Venn diagrams were generated using Venny (http://bioinfogp.cnb.csic.es/tools/venny, last accessed August 8, 2014).
CNV Calling
VarScan2's copynumber function was used to call CNVs for tumor-normal pairs using the dual mpileup file generated by SAMtools. This command included a data ratio to correct for uneven sequencing between a tumor and a normal pair; our data ratio was the number of reads in the normal mpileup file/the number of reads in the tumor mpileup file. The CNV output included chromosomal segments and their associated tumor-normal log2 ratio.
The results from sequencing-derived CNV calls were compared for the cell line HCC-2218 (using HCC-2218BL as a reference) against CNV calls produced by SNP6.0 array using Affymetrix's Genotyping Console software version 4.1.4.840 according to the manufacturer's instructions (Affymetrix). CNV log2 ratios were generated at individual base positions in our panel for HCC-2218 and HCC-2218BL. To directly compare CNV data from the SNP array and VarScan2-produced calls, for all base positions of both cell lines in the SNP array data, 2 to the respective log2 CNV ratio was raised to get an absolute CNV. Next, for each base position in the SNP array data, the absolute CNV for HCC-2218 was divided by that of the HCC-2218BL and took log2 of the quotient.
Log2 ratio CNV calls were correlated from each technology against the log2 CNV calls from the SNP array for the HCC-2218 cell line. For each technology, by using raw CNV output from VarScan2, for each CNV segment, all bases were assumed to have the log2 CNV ratio of the whole segment. Then, for all bases in the VarScan2 output, CNV calls were paired from VarScan2 and the SNP array (for all bases in which a call from the respective technology and SNP array were available). Before calculating correlation of CNV calls, any bases that had CNV log2 ratio between −0.5 and 0.5 were first removed from either VarScan2 or the SNP array to exclude low-amplitude copy alterations from our analysis. Then, the correlation of CNV calls between VarScan2 and the SNP array was calculated for each technology.
Statistical Tests
For data on which we performed significance tests, the two-sided U-test was performed, except when sequencing-estimated tumor purity was compared, for which a two-sided sign test was used. P values for all U-tests are listed in Supplemental Table S2.
Collaboration with GenomOncology
To ensure the reproducibility of some of our results in an independent analysis, GenomOncology (Westlake, OH) was contracted to perform analyses using its own computational pipeline. Paired-end sequencing was aligned to hg19 using bwa-0.7.5a. By using the SAMtools 0.1.18 view command, resulting SAM files were converted to BAM. BAM files were sorted using Picard 1.97 SortSam. Next, read groups were added using Picard AddOrReplaceReadGroups. Duplicates were removed from SureSelect, Nextera, and SeqCap using Picard MarkDuplicates. Then, a base quality recalibration was performed using GATK 2.2, followed by realignment around indels from dbSNP file hg19 snp135 using GATK. All commands were run using default parameters.
After alignment and post-processing, raw sequencing statistics were calculated and sequencing depth in mutually targeted regions was compared. Insert size was calculated using Picard CollectInsertSizeMetrics. By using Picard CollectAlignmentSummaryMetrics, alignment statistics and strandedness were calculated. Complexity was calculated, using Picard CollectAlignmentSummary, by counting the number of read pairs in the final BAM without duplicates (including HaloPlex) and dividing by the number of read pairs in the initially aligned BAM file. Raw sequencing depth was calculated using GATK DepthOfCoverage in mutually targeted regions. Normalized coverage and GC content at a base position were calculated as described earlier herein.
Results
Library Construction and Probe Design Vary between Targeted DNA Capture Methods
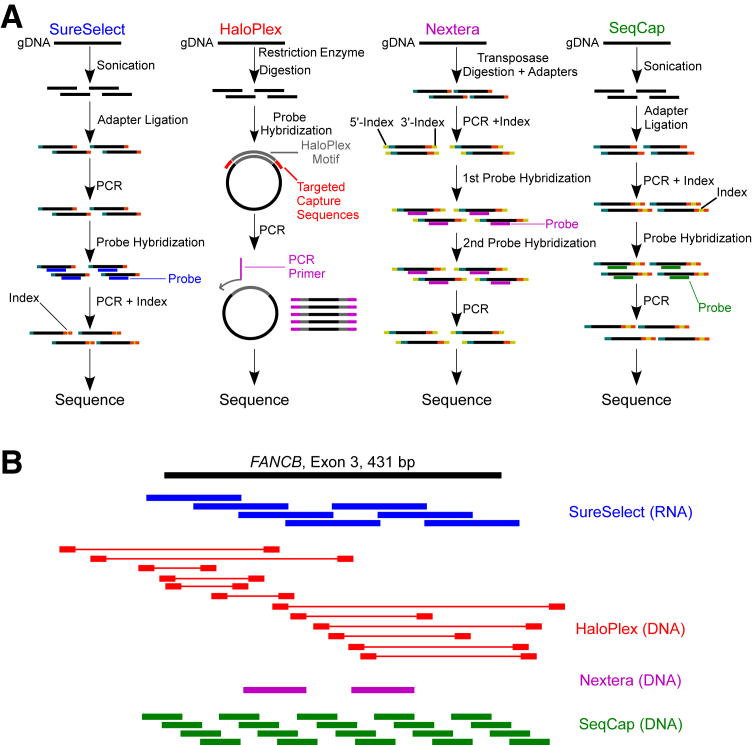
We assessed four commercial methods, Agilent's SureSelect and HaloPlex, Illumina's Nextera Custom Enrichment, and Roche Diagnostics/NimbleGen's SeqCap EZ Choice (Figure 1, A and B, and Table 1) for customized DNA capture and sequencing for a panel of 257 cancer-related genes (Supplemental Tables S3–S11) (note that Roche Diagnostics/Nimblegen's policy prohibits release of probe coordinates). Procedurally, these technologies contrast in several aspects that may be expected to affect results from targeted sequencing (Figure 1A and Table 1). First, SureSelect and SeqCap use sonication, whereas HaloPlex relies on restriction enzymes and Nextera uses transposase to generate DNA fragments. These differences can potentially affect the diversity of inserts generated for library construction. Second, HaloPlex uses a unique probe design in which the probes are not directly complementary to targeted regions, unlike the three other technologies. Instead, HaloPlex probe ends are complimentary only to the 5′- and 3′- ends of target regions, whereas the middle of the probe is a HaloPlex-specific motif that leads to the formation of a circular molecule during hybridization to DNA (Figure 1A). Third, library preparation for HaloPlex and Nextera requires only 48 hours, SureSelect needs 72 hours, and SeqCap needs 72 to 96 hours. Shortened sample preparation time is accomplished in HaloPlex through combining steps for adapter ligation with probe hybridization and in Nextera through combining adapter ligation with transposase-mediated fragmentation.
Figure 1.
Methods summary and capture methods for targeted DNA sequencing. A: Graphical depiction of four experimental protocols for library construction and capture between the four technologies. SureSelect and SeqCap have similar strategies for fragmentation and hybridization, whereas HaloPlex and Nextera use restriction enzyme and transposase to fragment DNA, respectively. B: Distribution of probes relative to an example target exon (FANCB, chrX: 14,862,977 to 14,863,408). SeqCap is an approximation (design was not released), whereas the remaining probes are from the actual design provided. HaloPlex's probes are only complimentary to sequences near the end of the probe, whereas the middle of the probe is a HaloPlex-specific motif (denoted by thick and thin red lines, respectively). SureSelect (blue) and SeqCap (green) use overlapping probes, whereas Nextera (magenta) uses gapped probes.
Table 1.
General Features of Targeted Exome Capture Strategies
| Approach | SureSelect | HaloPlex | Nextera | SeqCap |
|---|---|---|---|---|
| Fragmentation method | Sonication | Restriction enzymes | Transposase enzymes | Sonication |
| Probe type | RNA | DNA (molecular inversion probe) | DNA | DNA |
| Length of probes (bp) | 120 | 25–30 | 70 | 50–105 |
| Probe strategy | Tiled | Multiple amplicons | Gapped | Tiled, dense |
| No. of probes (in this study) | 14,368 | 44,860 | 7096 | 2.1 × 10−6 |
| Range of capture recommended | 1 kb to 24 Mb | Up to 25 Mb | 500 kb to 25 Mb | 1 kb to 50 Mb |
| Cost per library (USD) | 568 | 607 | 452 | 759 |
Manufacturers’ locations are given in Materials and Methods.
USD, United States dollars.
In addition to experimental differences, probe content and strategy vary widely between each capture method. Each technology uses distinctive probe lengths, genetic material, density, and layouts around target regions (Table 1). An example of probe design for each technology using an exon of the FANCB gene is shown in Figure 1B. Although SureSelect uses RNA probes, the other methods use DNA probes. Both SureSelect and SeqCap use a tiled probe design to ensure overlapping capture, but they differ in that SeqCap uses more shorter probes, whereas SureSelect uses fewer longer probes. Meanwhile, Nextera uses evenly spaced, gapped probes and relies on paired-end sequencing to fill the resultant gaps between probes (Figure 1B).12 Furthermore, the Nextera strategy uses two sequential probe-hybridization steps and has a requirement for sample pooling. In contrast, HaloPlex's probes are complimentary to the restriction enzyme digestion sites flanking the target region. The restriction enzyme digest allows simultaneous fragmentation of multiple samples compared to sonication that must occur serially.
Comparison of Libraries and On-Target Sequencing Metrics
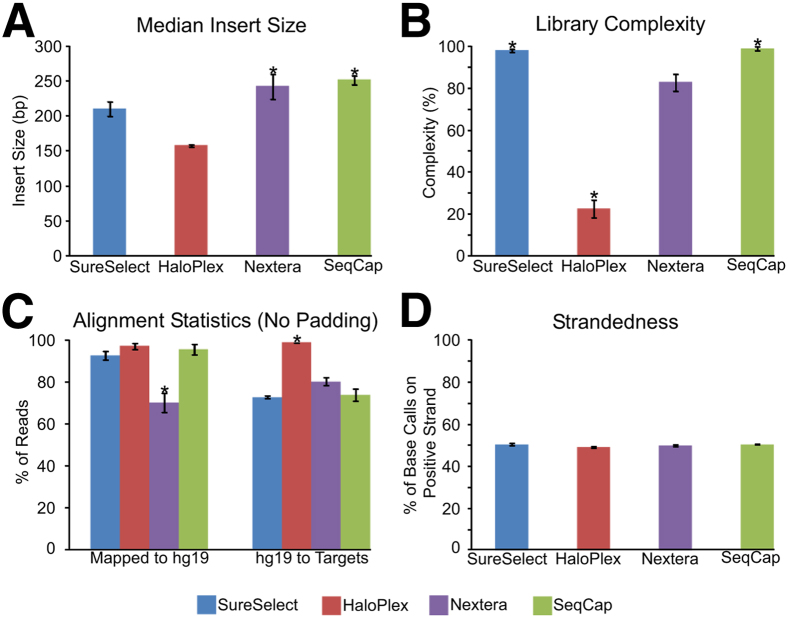
We used four well-characterized cancer cell lines and six matched tumor-normal tissue pairs (Supplemental Table S1) to appraise each technology's ability to capture and sequence target regions. Nextera and SeqCap libraries had the largest median insert sizes (P = 3.82 × 10−4) compared to other technologies (Figure 2A and Supplemental Figure S1A). Methods that used sonication for DNA fragmentation, SureSelect and SeqCap, displayed the highest library complexity (ie, percentage of unique reads) (P = 9.24 × 10−6 compared to other technologies), whereas HaloPlex displayed the lowest complexity (P = 9.24 × 10−6 compared to other technologies; GenomOncology's calculation of complexity is shown) (Figure 2B and Supplemental Figure S1B).
Figure 2.
Comparison of various library metrics. A: Library insert size (median ± median absolute deviation throughout figure). B: Library complexity, as measured by percentage of unique reads (of the total reads) in each library. C: Alignment metrics for each technology for reads (ie, after removing duplicates for SureSelect, Nextera, and SeqCap) mapping to hg19 and to each technology's targeted regions. D: Strandedness for each technology, as measured by the number of base calls on the positive strand. ∗P = 3.82 × 10−4 (U-test), Nextera and SeqCap have a significantly higher insert size than SureSelect and HaloPlex (A); ∗P < 9.24 × 10−6, SureSelect and SeqCap have significantly greater complexity than HaloPlex and Nextera, whereas HaloPlex has the lowest complexity (B); ∗P < 10−5, Nextera has the lowest alignment rate to the genome and HaloPlex has the highest alignment rate to target regions (C).
Next, we assessed each technology's on-target alignment rate, including percentage of sequenced reads that aligned to the reference genome (hg19) and aligned reads that mapped to targeted regions (Figure 2C, Supplemental Tables S12 and S13, and Supplemental Figure S1C). SureSelect, HaloPlex, and SeqCap showed >90% alignment from raw sequencing files to the human genome, whereas Nextera showed (median ± median absolute deviation, herein) 70.28% ± 4.67% alignment to the genome (P = 9.28 × 10−6 when compared to each technology; alignment is 97.16% ± 0.64%, when Nextera duplicates were included). HaloPlex displayed the highest on-target specificity by aligning 99.06% ± 0.19% of its mapped reads to targeted regions, significantly greater than 72.98% ± 0.74% for SureSelect, 80.36% ± 1.89% for Nextera, and 74.10% ± 2.75% for SeqCap (P = 9.24 × 10−6 when compared to each technology; HaloPlex showed 98.70% ± 0.53% duplicates). All trends for alignment rates were similar when we introduced 100- and 500-bp padding around the target regions (Supplemental Table S12 and Supplemental Figure S2). In addition to alignment rate, we also calculated strand specificity and found a relatively even distribution of alignments between both DNA strands for all methods (Figure 2D) (the base calls in mpileup files were used to calculate strandedness) (Supplemental Figure S1D) (for GenomOncology's calculation of strandedness).
Depth and Uniformity of On-Target Sequencing
Given the preceding alignment rates, we next considered the normalized sequencing depth (read count per million sequenced reads) in bases that were targeted by all methods (Figure 3A and Supplemental Figure S3A) (for GenomOncology's calculation of sequencing depth). HaloPlex had the greatest average normalized coverage (defined as means ± SD herein) (111.76 ± 102.43 reads per million sequenced reads; P < 10−323 when compared to each technology), whereas Nextera and SeqCap had the lowest average normalized coverage (62.94 ± 36.07 and 63.22 ± 16.67 reads per million sequenced reads, respectively; P < 10−323 when compared to each technology). However, HaloPlex also displayed the greatest average global SD of normalized coverage (42.97 ± 42.26 reads per million sequenced reads; P < 10−323 when compared to each technology), whereas SeqCap showed the smallest average global SD of normalized coverage (14.80 ± 16.17 reads per million sequenced reads; P < 10−323 when compared to each technology), suggesting that SeqCap might be the most uniform of the tested capture methods. Although each of the method’s designs differed somewhat in its target regions, we observed a similar trend for uniformity when evaluating on either commonly or respective target regions (Supplemental Figure S4A).
Figure 3.
Comparison of sequencing depth and variation in mutually targeted regions. A: Normalized coverage was calculated (reads per million sequenced reads) (y axis) for each mutually targeted base and plotted average (blue) and SD (red) versus genomic positions (x axis). Read count was obtained from Sequence Alignment/MAP (SAMtools) mpileup files. B: Plot of percentage of mutually targeted bases that were covered at average minimum normalized coverages (solid lines). An ideal curve is included for each technology (dotted lines), where the average normalized sequencing is uniformly distributed for each technology. Different technologies have different average normalized coverages, so ideal curves will be different between technologies. Solid lines should be compared to dotted lines for the respective technology. C: Average normalized coverage is plotted against the percent GC content in 100-bp windows. Darker colors indicate higher density of points, whereas lighter colors indicate lower density of points.
Although average depth of coverage is an important metric, ideal on-target sequencing should be equally distributed across all regions of interest. To assess uniformity of on-target sequencing, we plotted the percentage of commonly targeted bases versus minimum average normalized coverage (Figure 3B and Supplemental Figure S3B). Although none of the technologies is expected to provide completely uniform capture and sequencing, we sought to define an ideal plot if one assumed that capture and normalized sequencing was perfect. In scenarios leading to the ideal plots, all bases have a normalized coverage equal to the average normalized coverage for the technology (Figure 3B). Given the computed ideal curves, we compared the deviation of each technology from true uniformity. We found that HaloPlex had the greatest average distance to its corresponding ideal curve, whereas SeqCap had the lowest (P < 10−323, for both when compared to each technology). We observed the same uniformity calculations using each technology's respective targeted regions (Supplemental Figure S4B).
Impact of GC Content on Target Capture
Regions with low or high GC content can adversely affect targeted DNA sequencing through affecting probe hybridization and PCR amplification steps.10,12 Therefore, we investigated how target base composition affected the performance of each technology for commonly targeted bases (Figure 3C and Supplemental Figures S5 and S6). HaloPlex and Nextera exhibited a significant peak in normalized coverage near 60% GC, whereas coverage decreased sharply before and after 60% GC content. SureSelect and SeqCap, on the other hand, performed consistently with respect to GC content and lacked a clear peak at any GC percentage.
Having examined uniformity with respect to overall GC content, we also compared coverage of each technology for extremes of high-GC (≥80%) and low-GC (≤25%) regions in commonly targeted bases (Supplemental Table S14). In high-GC areas, SureSelect showed the highest median coverage (P = 1.10 × 10−47). In low-GC areas, SureSelect and SeqCap performed the best (P < 10−323). A similar analysis of GC content in regions targeted specifically by each technology is shown (Supplemental Table S14).
Performance of SNV Calling Across Technologies
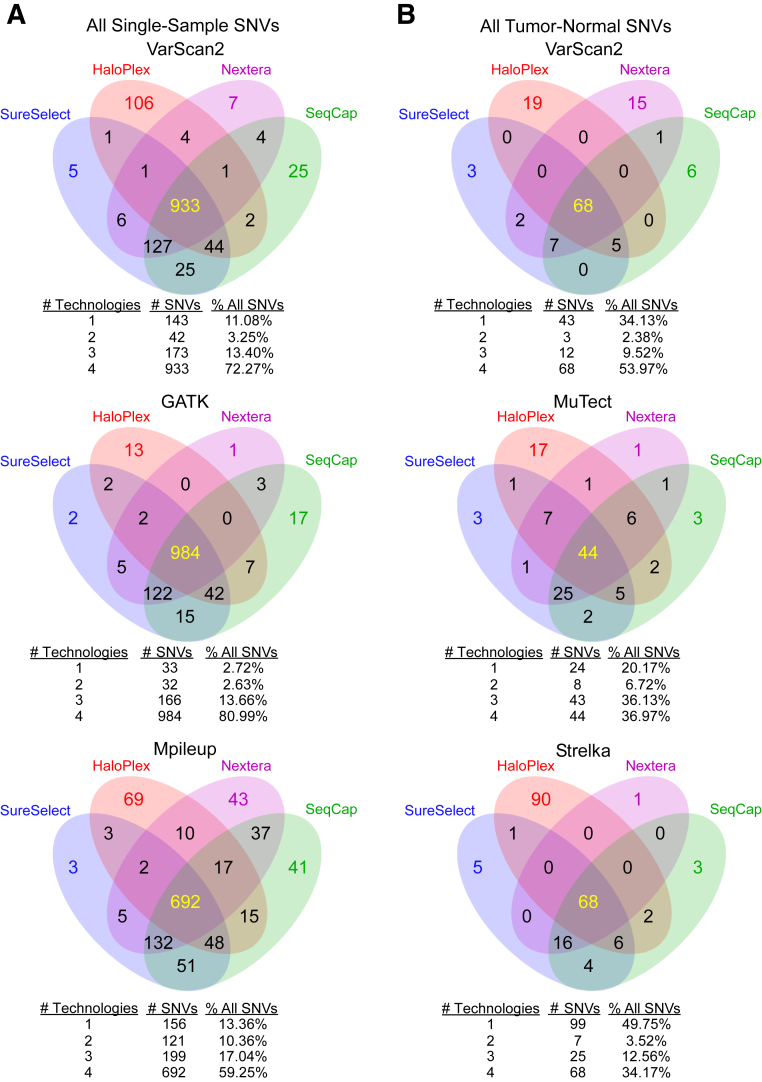
Identification of SNVs is a major goal of targeted DNA sequencing in cancer. For each cell line, tumor sample, and matched normal sample (limited to 12 samples because there was insufficient DNA for samples 3 to 6 on SureSelect and SeqCap), we generated single-sample SNV calls relative to the human genome (hg19) across the four technologies using VarScan2,20 GATK,18 and Mpileup.16 For concordance analysis, we limited our variant calls only to targeted regions common to all methods so we could directly compare the ability of each technology to call SNVs at identical genomic positions (Supplemental Tables S15–S17). Figure 4A shows the single-sample SNV concordance of the technologies using the previously mentioned variant callers (for concordance calculations among technologies) (Supplemental Table S18). We also called variants on HaloPlex data using Agilent's SureCall software version 1.1.0.15 and found a similar level of concordance with the other three technologies and three separate variant callers (Supplemental Figure S7 and Supplemental Tables S19 and S20) (two-tailed U-tests for numbers were P = 0.86, P = 0.96, and P = 0.96 for VarScan2, GATK, and Mpileup, respectively) (Figure 4A and Supplemental Figure S7).
Figure 4.
Summary of single-nucleotide variation (SNV) concordances among technologies. We determined SNVs using three different variant callers through single-sample (A) and paired tumor-normal (B) analyses. To get a summary of concordance among technologies, we pooled all SNVs from all samples together. Tables below each Venn diagram indicate the total and percentage of SNVs called by only one technology, only two technologies, only three technologies, and all four technologies. Samples 3 to 6 were sequenced using only HaloPlex and Nextera and excluded from the four-way comparison.
In addition, we assessed the four technologies' abilities to support SNV calling in cancer samples relative to a matched normal. Therefore, for each normal-matched cancer sample (Supplemental Table S1), we used VarScan2, MuTect,22 and Strelka23 to call tumor-normal SNVs in regions common to all technologies (Figure 4B and Supplemental Tables S21–S24). The concordance among technologies was generally lower in calling tumor-normal SNVs than in calling single-sample SNVs. The percentages of tumor-normal SNVs nominated by at least three technologies varied between 46.73% and 73.10%, depending on variant caller (Figure 4B). On the other hand, the equivalent for single-sample SNVs ranged between 76.29% and 94.65% for SNVs nominated by at least three technologies (Figure 4B).
Although there was a reasonable amount of agreement with respect to the percentage of SNVs called by all technologies independent of the SNV caller (between 34.17% and 80.99%), we observed some discordance between technologies (ie, an SNV missed by at least one technology). Thus, we investigated and found several possible causes for SNV discordance by inspecting aligned reads across all four methods using the Integrative Genomics Viewer.25 We focused on single-sample SNVs called in the four cell lines, because we had abundant DNA available for validation (Supplemental Figures S8 and S9 and Supplemental Table S25). Of 131 discordant SNVs, 72 (56.49%) that were not called in one or more technologies could, in most cases, be explained by a lack of coverage (Supplemental Figure S10) or the SNV caller failed to detect the SNV, because of an insufficient number of quality reads (Supplemental Figure S11). (Various studies have found 30% to 60% concordance between SNV callers applied to the same data.26,27) Furthermore, we observed that HaloPlex consistently produced unique outlier SNVs compared to other technologies, which Agilent's proprietary SureCall software version 1.1.0.15 failed to resolve (Supplemental Figures S8A and S9). Almost all SNVs called solely by HaloPlex data were caused by certain motifs known to cause Illumina sequencing errors [24 (18.32%) such SNVs of 131 discordant SNVs; we found that GATK correctly excludes SNVs in such error-prone reads]28,29 (Supplemental Figure S12 and Supplemental Table S26). Furthermore, several SNVs were missed by HaloPlex libraries, possibly because of their location in the vicinity of a HaloPlex restriction-enzyme digestion site, causing a bias toward wild-type base calls (Supplemental Figure S13).
For the remaining five discordant SNVs, we used Sanger sequencing to determine whether the SNV was actually present in the cell line or whether it was being called in error by one or more of the technologies (Supplemental Figure S14). Sanger sequencing confirmed three SNVs called by all technologies, except HaloPlex, and one SNV called by all technologies, except Nextera. Sanger sequencing refuted one SNV called only by HaloPlex.
Using SNV Variant Fractions to Calculate Tumor Purity
Considering that tumor samples generally contain admixtures of cancer and normal cells, variant fractions can be used to estimate or correlate with expected tumor purity.21 We used an ad hoc–rendered version of PurityEst21 to calculate approximate tumor purity for all tumor samples. Our computational estimates of tumor purity on all four technologies for the four tumor samples were concordant with available histology-estimated tumor purities (Supplemental Table S1). Sign tests between sequencing- and histology-estimated tumor purities showed no significant difference for each technology (P = 0.13, P = 0.47, P = 0.31, and P = 0.25 for SureSelect, HaloPlex, Nextera, and SeqCap, respectively).
Comparison of CNV Calling with SNP Array
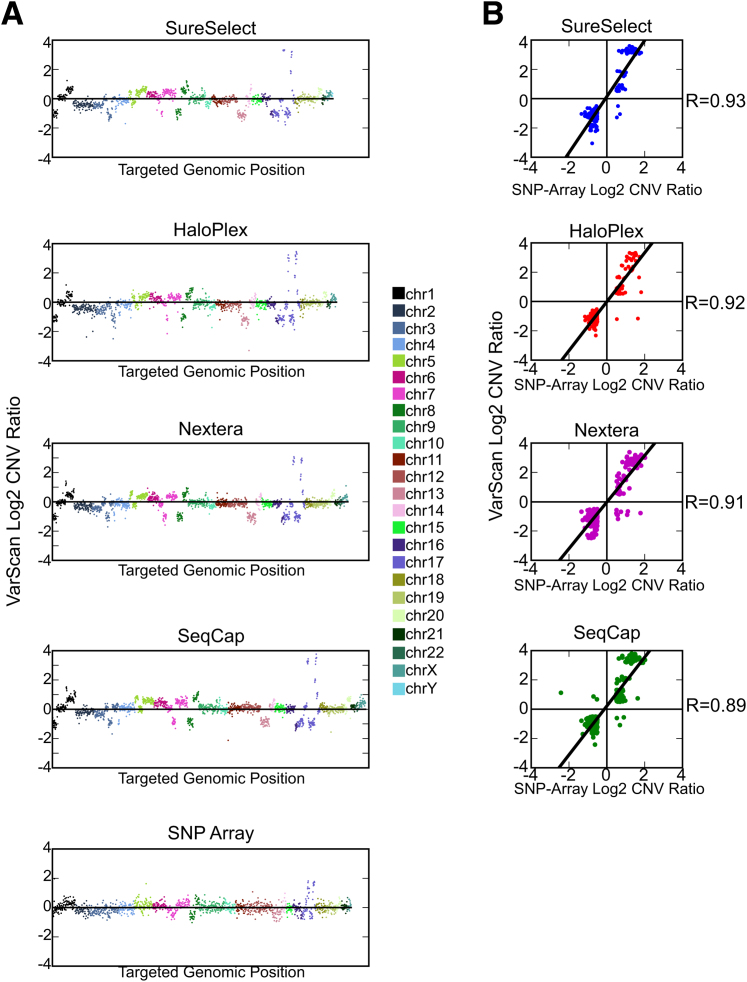
In addition to SNV detection, another application for targeted sequencing is the determination of CNVs on the basis of read depth.20 We calculated CNVs in the HCC-2218 cancer cell line relative to the matched normal HCC-2218BL for each capture method using VarScan2. Independently, we determined CNVs using a SNP array as a gold standard comparison30 on the same samples (Figure 5A and Supplemental Table S27). For each capture platform, we compared the log2 CNV ratios to the SNP array CNV ratios for the bases that were common to the respective technology and the SNP array (Figure 5B). We focused on high-amplitude and clinically significant CNV gains or losses.3,7 When we excluded positions whose sequencing or SNP array log2 CNV ratio fell between −0.5 and 0.5 (such positions we deemed copy neutral), correlation coefficients were 0.93, 0.92, 0.91, and 0.89 for SureSelect, HaloPlex, Nextera, and SeqCap (P < 0.001 for all technologies), respectively.
Figure 5.
Comparison of copy number variation (CNV) detection. A: Copy number ratio (log2 scale) plotted against genomic position for technology-specific targeted bases demonstrating overall comparable copy number calling between four technologies. A similar graph for single-nucleotide polymorphism (SNP) array is included for bases in any technology's target region. B: Correlation between CNV detection for each technology and SNP array was determined for high absolute threshold copy number gains and losses. Any base whose absolute value of log2 CNV ratio for either sequencing or SNP array was <0.5 (ie, between −0.5 and 0.5) was excluded from these graphs, because they were deemed copy neutral. Chr, chromosome.
Discussion
We examined the ability of four targeted DNA capture technologies to enrich and identify SNVs and CNVs in selected genes that are known to play important roles in cancer. Newer methods, such as HaloPlex and Nextera, require less experimental time and lower DNA input than older methods, such as SureSelect and SeqCap (Table 2). We sought to assess the advantages and disadvantages that may be inherent to differences in experimental procedures and probe design.
Table 2.
Comparison of Four Methods for Targeted DNA Sequencing
| Variable | SureSelect | HaloPlex | Nextera | SeqCap |
|---|---|---|---|---|
| Time (hours) | 72 | 48 | 48 | 72–96 |
| Recommended input DNA (μg) | 2 | 0.2 | 0.2 | 2 |
| Pooling | Optional | Optional | Required | Optional |
| Alignment (%) | ||||
| Manufacturer specified | 30–70 | 30–70 | >65 | 70–80 |
| This study | 73.25 | 98.98 | 80.18 | 73.73 |
| Library complexity (%) | 97.45 | 21.82 | 83.33 | 98.34 |
| Base calls on positive strand (%) | 50.89 | 49.26 | 49.88 | 50.46 |
| Uniformity | High | Low | Low | High |
| SNV | Yes | Yes | Yes | Yes |
| CNV | Yes | Yes | Yes | Yes |
Alignment herein means percentage of reads that aligned to the genome that aligned or should align to target regions.
CNV, copy number variation; SNV, single-nucleotide variation.
We expected some degree of variation in library complexity, on-target enrichment, and capture uniformity between protocols. Methods involving sonication for DNA fragmentation displayed the highest complexity in libraries compared to restriction or transposase enzymes (Figure 2). Although HaloPlex had the highest alignment percentage to the targeted region, most aligned data were, in fact, duplicates involving highly oversequenced regions and thereby contributing to lower uniformity (Figure 3B). HaloPlex and Nextera had the highest SD of normalized coverage (Figure 2B) and divergence from their respective ideal curves compared to SureSelect and SeqCap. In addition, HaloPlex and Nextera were adversely affected by variations in GC content, with coverage peaking at approximately 60% GC, whereas SureSelect and SeqCap displayed more evenly distributed performance.
In addition to library complexity, on-target alignment, uniformity, and variant calling, we noted several method-specific nuances. An experimental limitation of Nextera is its requirement for sample pooling during hybridization, unlike other methods, in which individual samples are hybridized to probes separately. Although this can save on costs of probe manufacturing, it can make ensuring equal or wanted sequencing depth difficult to achieve (Supplemental Table S13). The relative costs for each method are similar (Table 1). Because the HaloPlex design is dependent on restriction enzyme digest sites for capture, it functions like a PCR amplicon, leading to its high duplicate rate. Thus, SNVs occurring in or near digest sites may physically limit targeted capture (Supplemental Figure S13). More important, SureSelect is the only method that uses RNA probes for hybridization capture.31 For clinical laboratories developing custom gene panels, long-term storage of RNA probes is an important limitation, because these laboratories must consider quality control and there is an advantage to lot-tested reagents that can withstand long-term storage.
In summary, we have compared four custom-targeted DNA capture methods with respect to uniform sequencing and variant calling. This study demonstrates the potential biases of capture strategies due to differences in experimental procedures and probe design that may affect performance, including alignment and uniformity. Given the results herein, our laboratory prefers SeqCap or SureSelect approaches, mainly due to capture uniformity. This report may be used by other laboratories to select their preferred approach on the basis of their objectives.
Acknowledgments
We thank GenomOncology, the Ohio Supercomputer Center, and the Comprehensive Cancer Center at The Ohio State University Wexner Medical Center for supporting this study; Brianna Hunstad for administrative support; and Agilent Technologies and Alex Siebold for helping implement SureCall and reviewing our schematic of the HaloPlex protocol.
E.S., J.D., and S.R. conceived the research study; J.D, B.M.J., and J.M.L. prepared samples and sequenced libraries; E.S., R.H., and J.M. developed the computational pipeline for analysis of the sequencing data; E.S., J.M., and M.R.W. analyzed the sequencing data; J.M. and M.R.W. analyzed CNV data; J.M. and M.R.W evaluated alignment data for single-nucleotide variations; M.R.W. and J.D. performed Sanger sequencing and analysis; C.D.T. assisted with SureCall analyses; E.S. and J.M. generated figures and tables; E.S., J.D., B.M.J., M.R.W., J.M., S.D., J.W.R., and C.D.T. reviewed the data used to generate figures and tables; E.S., J.D., and S.R. wrote the manuscript; S.D. and J.R. edited the manuscript; and D.B. and S.R. supervised the research.
Footnotes
Supported by American Cancer Society grant MRSG-12-194-01-TBG (S.R.), the Prostate Cancer Foundation Young Investigator Award (S.R.), National Human Genome Research Institute grant UM1HG006508-01A1 (S.R.), Fore Cancer Research (S.R.), Pelotonia (S.R.), and a Pelotonia Postdoctoral fellowship (E.S.).
E.S. and J.D. contributed equally to this work.
Disclosures: None declared.
Supplemental Data
GenomOncology's analysis of exome data: raw statistics. By using their own pipeline, GenomOncology analyzed our exome sequencing data to verify some of our results. They aligned paired-end sequencing data to hg19 using bwa-0.7.5a. By using the Sequence Alignment/MAP (SAMtools) 0.1.18 view command, resulting SAM files were converted to Binary Alignment/Map (BAM). BAM files were sorted using Picard 1.97 SortSam. Then, read groups were added using Picard AddOrReplaceReadGroups. Then, duplicates were removed from SureSelect, Nextera, and SeqCap libraries using Picard MarkDuplicates. A base quality recalibration was then performed using Genome Analysis Toolkit (GATK) 2.2. Reads were realigned around indels from dbSNP file hg19 snp135 using GATK. All commands were run using default parameters. A: Library insert size (median ± median absolute deviation) throughout figure. B: Library complexity as measured by percentage of unique reads (of the total reads) in each library. C: Alignment metrics for each technology for reads after removing duplicates for SureSelect, Nextera, and SeqCap mapping to hg19 and mapped reads mapping to each technology's targeted regions. D: Strandedness for each technology as measured by the number of base calls on the positive (Pos.) strand. Pct., percentage.
Alignment rates of each technology with padding. Shown are the two different alignment rates for each technology (median ± median absolute deviation). Mapped to hg19 denotes the percentage of raw sequencing reads that mapped to the hg19 genome. hg19 to Targets signifies the percentage of mapped reads that aligned to each technology's respective target regions, extending all target regions 100 bp (A) or 500 bp (B) upstream of each 5′- end and downstream of each 3′- end. This extra 100 or 500 bp is denoted as padding.
GenomOncology's analysis: comparison of sequencing depth in mutually targeted regions. A: Normalized coverage was calculated (reads per million sequenced reads) (y axis) for each mutually targeted base and plotted average (blue) and SD (red) versus genomic positions (x axis). Read count was obtained by using the Genome Analysis Toolkit (GATK) DepthOfCoverage command. B: Plot of percentage of mutually targeted bases that were covered at certain average minimum normalized coverages.
Comparison of sequencing depth in regions targeted by each technology. A: We used all the genomic bases that each technology, specifically, targeted (x axis). For each targeted base, for each sample in each technology, we calculated the normalized coverage (reads per million sequenced reads) (y axis). We have plotted, for each base and method, the average (blue) and standard deviation (St.Dev., red) of normalized coverage. HaloPlex showed the greatest average normalized coverage over its target area (109.55 reads per million sequenced reads; P < 10−320 when compared to each technology). However, HaloPlex evidenced the greatest average global SD (43.00 reads per million sequenced reads; P < 10−320 when compared to each technology), whereas SeqCap showed the smallest global SD (14.92 reads per million sequenced reads; P < 10−320 when compared to each technology). This analysis confirms the uniformity analysis in commonly targeted regions. B: For all technologies, in each one's specific target region, we plotted the percentage of bases that were covered at various average minimum normalized coverages (solid lines). Furthermore, for each technology, we calculated the global average normalized coverage in technology-targeted regions. With these global averages, we plotted an ideal curve for each technology (dotted lines), in which the normalized coverage of each base is the average normalized coverage for each individual technology. We found that HaloPlex had the highest (P < 10−320, when compared to each technology) and SeqCap had the lowest (P < 10−320, when compared to each technology) average distance to its corresponding ideal curve.
Effects of GC content in regions targeted by each technology. For every technology, in each one's specific target region, we plotted average normalized coverage against the GC content. Darker colors indicate higher density of points, whereas lighter colors indicate lower density of points. GC content was calculated on the basis of percentage of Gs or Cs in a 100-bp window.
GenomOncology's analysis: how GC affects depth in mutually targeted regions. For every technology, in mutually targeted bases, GenomOncology plotted average normalized coverage against the GC content. Darker colors indicate higher density of points, whereas lighter colors indicate lower density of points. GC content was calculated on the basis of percentage of Gs or Cs in a 100-bp window.
Single-nucleotide variation (SNV) concordances among technologies using SureCall on HaloPlex. For single-sample analyses, SNVs were called from all libraries using the indicated SNV caller, except HaloPlex, for which we used SureCall. The latter is designed to perform single-sample analyses, but not tumor-normal analyses.
Single-nucleotide variation (SNV) concordances among technologies in cell lines. We determined SNVs using three different variant callers for all cell lines through single-sample analyses (A) and paired tumor-normal analyses (in this case, HCC-2218 versus HCC-2218BL) (B). To get a summary of concordance among technologies, we pooled all SNVs from all cell lines together and gave each SNV a unique identification on the basis of the sample, chromosome, reference allele, and alternate allele.
Single-nucleotide variation (SNV) concordances among technologies in cell lines using SureCall on HaloPlex. For single-sample analyses, SNVs were called from all libraries using the indicated SNV caller, except HaloPlex, for which we used SureCall. The latter is designed to perform single-sample analyses, but not tumor-normal analyses.
Discordant single-nucleotide variations (SNVs) due to lack of coverage. Shown are views from Integrative Genomics Viewer (IGV) of read alignment at an SNV in the BT-20 cell line, chr13:48,947,576 T→G in SureSelect (A), HaloPlex (B), Nextera (C), and SeqCap (D). All technologies, except HaloPlex, called this SNV. HaloPlex missed it, because of lack of coverage.
Discordant single-nucleotide variations (SNVs) due to insufficient quality reads. Shown are views in Integrative Genomics Viewer (IGV) of chr2:212,248,542 T→C in the MCF-7 cell line in SureSelect (A), HaloPlex (B), Nextera (C), and SeqCap (D). VarScan2 called this mutation in all technologies, except HaloPlex. VarScan2 failed to report this SNV in HaloPlex, because, after removal of low-quality base calls, variant frequency decreased to 18.94%, lower than VarScan2's minimum cutoff of 20%. Genome Analysis Toolkit (GATK) called this mutation in all four technologies.
Discordant single-nucleotide variations (SNVs) due to Illumina sequencing errors. The presence of some GG motifs causes sequencing errors, resulting in many mismatches in a short read span.28,29 Shown are read alignment views in Integrative Genomics Viewer (IGV) around chr2:29,448,410 T→G in BT-20 in SureSelect (A), HaloPlex (B), Nextera (C), and SeqCap (D). VarScan2 called this SNV in the HaloPlex library, but no other library.
Discordant single-nucleotide variation (SNV) due to a HaloPlex restriction site. Shown are views from Integrative Genomics Viewer (IGV) of read alignment of an SNV in the BT-20 cell line, chr13:48,955,428 C→T in SureSelect (A), HaloPlex (B), Nextera (C), and SeqCap (D). VarScan2 and Genome Analysis Toolkit (GATK) called this SNV in all technologies, except HaloPlex. This appears to be due to the SNV's presence within a HaloPlex restriction site, which biases this technology toward the wild-type allele. VarScan2, after removing low-quality base calls, finds variant frequency equals 6.08%, lower than VarScan2's minimum of 20% to report. Similarly, GATK, after poor base-quality filtration, also finds variant frequency equals 6.08%.
Sanger sequencing analysis of select single-nucleotide variations (SNVs). Occasionally, observing read alignments of an SNV from various technologies produced conflicting results on the existence of the SNV. In such cases, we used Sanger sequencing to verify or refute the existence of five chosen SNVs: MCF-7 cell line, chr10:62,547,870 T→G on the positive (pos) strand (A), MCF-7 cell line, chr6:94,066,507 G→T on the negative (neg) strand (B), MCF-7 cell line, chr3:178,936,091 G→A on the positive strand (note that Sanger sequencing called G, but there was a signal for A too; C), BT-20 cell line, chr12:18,649,057 C→T on the positive strand (D), and BT-20 cell line, chr13:48,955,428 C→T on the positive strand (E).
References
- 1.Koboldt D.C., Steinberg K.M., Larson D.E., Wilson R.K., Mardis E.R. The next-generation sequencing revolution and its impact on genomics. Cell. 2013;155:27–38. doi: 10.1016/j.cell.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyerson M., Gabriel S., Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 3.Frampton G.M., Fichtenholtz A., Otto G.A., Wang K., Downing S.R., He J. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wagle N., Berger M.F., Davis M.J., Blumenstiel B., Defelice M., Pochanard P., Ducar M., Van Hummelen P., Macconaill L.E., Hahn W.C., Meyerson M., Gabriel S.B., Garraway L.A. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beadling C., Neff T.L., Heinrich M.C., Rhodes K., Thornton M., Leamon J., Andersen M., Corless C.L. Combining highly multiplexed PCR with semiconductor-based sequencing for rapid cancer genotyping. J Mol Diagn. 2013;15:171–176. doi: 10.1016/j.jmoldx.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 6.Luthra R., Patel K.P., Reddy N.G., Haghshenas V., Routbort M.J., Harmon M.A., Barkoh B.A., Kanagal-Shamanna R., Ravandi F., Cortes J.E., Kantarjian H.M., Medeiros L.J., Singh R.R. Next generation sequencing based multi-gene mutational screen for acute myeloid leukemia using miseq: applicability for diagnostics and disease monitoring. Haematologica. 2014;99:465–473. doi: 10.3324/haematol.2013.093765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pritchard C.C., Salipante S.J., Koehler K., Smith C., Scroggins S., Wood B., Wu D., Lee M.K., Dintzis S., Adey A., Liu Y., Eaton K.D., Martins R., Stricker K., Margolin K.A., Hoffman N., Churpek J.E., Tait J.F., King M.C., Walsh T. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn. 2014;16:56–67. doi: 10.1016/j.jmoldx.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singh R.R., Patel K.P., Routbort M.J., Reddy N.G., Barkoh B.A., Handal B., Kanagal-Shamanna R., Greaves W.O., Medeiros L.J., Aldape K.D., Luthra R. Clinical validation of a next-generation sequencing screen for mutational hotspots in 46 cancer-related genes. J Mol Diagn. 2013;15:607–622. doi: 10.1016/j.jmoldx.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Wagle N., Van Allen E.M., Treacy D.J., Frederick D.T., Cooper Z.A., Taylor-Weiner A., Rosenberg M., Goetz E.M., Sullivan R.J., Farlow D.N., Friedrich D.C., Anderka K., Perrin D., Johannessen C.M., McKenna A., Cibulskis K., Kryukov G., Hodis E., Lawrence D.P., Fisher S., Getz G., Gabriel S.B., Carter S.L., Flaherty K.T., Wargo J.A., Garraway L.A. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61–68. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asan, Xu Y., Jiang H., Tyler-Smith C., Xue Y., Jiang T., Wang J., Wu M., Liu X., Tian G., Yang H., Zhang X. Comprehensive comparison of three commercial human whole-exome capture platforms. Genome Biol. 2011;12:R95. doi: 10.1186/gb-2011-12-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bainbridge M.N., Wang M., Burgess D.L., Kovar C., Rodesch M.J., D'Ascenzo M., Kitzman J., Wu Y.Q., Newsham I., Richmond T.A., Jeddeloh J.A., Muzny D., Albert T.J., Gibbs R.A. Whole exome capture in solution with 3 Gbp of data. Genome Biol. 2010;11:R62. doi: 10.1186/gb-2010-11-6-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark M.J., Chen R., Lam H.Y., Karczewski K.J., Euskirchen G., Butte A.J., Snyder M. Performance comparison of exome DNA sequencing technologies. Nat Biotechnol. 2011;29:908–914. doi: 10.1038/nbt.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parla J.S., Iossifov I., Grabill I., Spector M.S., Kramer M., McCombie W.R. A comparative analysis of exome capture. Genome Biol. 2011;12:R97. doi: 10.1186/gb-2011-12-9-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sulonen A.M., Ellonen P., Almusa H., Lepisto M., Eldfors S., Hannula S., Miettinen T., Tyynismaa H., Salo P., Heckman C., Joensuu H., Raivio T., Suomalainen A., Saarela J. Comparison of solution-based exome capture methods for next generation sequencing. Genome Biol. 2011;12:R94. doi: 10.1186/gb-2011-12-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su X., Zhang L., Zhang J., Meric-Bernstam F., Weinstein J.N. PurityEst: estimating purity of human tumor samples using next-generation sequencing data. Bioinformatics. 2012;28:2265–2266. doi: 10.1093/bioinformatics/bts365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saunders C.T., Wong W.S., Swamy S., Becq J., Murray L.J., Cheetham R.K. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 24.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S.Y., Speed T.P. Comparing somatic mutation-callers: beyond Venn diagrams. BMC Bioinformatics. 2013;14:189. doi: 10.1186/1471-2105-14-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O'Rawe J., Jiang T., Sun G., Wu Y., Wang W., Hu J., Bodily P., Tian L., Hakonarson H., Johnson W.E., Wei Z., Wang K., Lyon G.J. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med. 2013;5:28. doi: 10.1186/gm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meacham F., Boffelli D., Dhahbi J., Martin D.I., Singer M., Pachter L. Identification and correction of systematic error in high-throughput sequence data. BMC Bioinformatics. 2011;12:451. doi: 10.1186/1471-2105-12-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakamura K., Oshima T., Morimoto T., Ikeda S., Yoshikawa H., Shiwa Y., Ishikawa S., Linak M.C., Hirai A., Takahashi H., Altaf-Ul-Amin M., Ogasawara N., Kanaya S. Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res. 2011;39:e90. doi: 10.1093/nar/gkr344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gnirke A., Melnikov A., Maguire J., Rogov P., LeProust E.M., Brockman W., Fennell T., Giannoukos G., Fisher S., Russ C., Gabriel S., Jaffe D.B., Lander E.S., Nusbaum C. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GenomOncology's analysis of exome data: raw statistics. By using their own pipeline, GenomOncology analyzed our exome sequencing data to verify some of our results. They aligned paired-end sequencing data to hg19 using bwa-0.7.5a. By using the Sequence Alignment/MAP (SAMtools) 0.1.18 view command, resulting SAM files were converted to Binary Alignment/Map (BAM). BAM files were sorted using Picard 1.97 SortSam. Then, read groups were added using Picard AddOrReplaceReadGroups. Then, duplicates were removed from SureSelect, Nextera, and SeqCap libraries using Picard MarkDuplicates. A base quality recalibration was then performed using Genome Analysis Toolkit (GATK) 2.2. Reads were realigned around indels from dbSNP file hg19 snp135 using GATK. All commands were run using default parameters. A: Library insert size (median ± median absolute deviation) throughout figure. B: Library complexity as measured by percentage of unique reads (of the total reads) in each library. C: Alignment metrics for each technology for reads after removing duplicates for SureSelect, Nextera, and SeqCap mapping to hg19 and mapped reads mapping to each technology's targeted regions. D: Strandedness for each technology as measured by the number of base calls on the positive (Pos.) strand. Pct., percentage.
Alignment rates of each technology with padding. Shown are the two different alignment rates for each technology (median ± median absolute deviation). Mapped to hg19 denotes the percentage of raw sequencing reads that mapped to the hg19 genome. hg19 to Targets signifies the percentage of mapped reads that aligned to each technology's respective target regions, extending all target regions 100 bp (A) or 500 bp (B) upstream of each 5′- end and downstream of each 3′- end. This extra 100 or 500 bp is denoted as padding.
GenomOncology's analysis: comparison of sequencing depth in mutually targeted regions. A: Normalized coverage was calculated (reads per million sequenced reads) (y axis) for each mutually targeted base and plotted average (blue) and SD (red) versus genomic positions (x axis). Read count was obtained by using the Genome Analysis Toolkit (GATK) DepthOfCoverage command. B: Plot of percentage of mutually targeted bases that were covered at certain average minimum normalized coverages.
Comparison of sequencing depth in regions targeted by each technology. A: We used all the genomic bases that each technology, specifically, targeted (x axis). For each targeted base, for each sample in each technology, we calculated the normalized coverage (reads per million sequenced reads) (y axis). We have plotted, for each base and method, the average (blue) and standard deviation (St.Dev., red) of normalized coverage. HaloPlex showed the greatest average normalized coverage over its target area (109.55 reads per million sequenced reads; P < 10−320 when compared to each technology). However, HaloPlex evidenced the greatest average global SD (43.00 reads per million sequenced reads; P < 10−320 when compared to each technology), whereas SeqCap showed the smallest global SD (14.92 reads per million sequenced reads; P < 10−320 when compared to each technology). This analysis confirms the uniformity analysis in commonly targeted regions. B: For all technologies, in each one's specific target region, we plotted the percentage of bases that were covered at various average minimum normalized coverages (solid lines). Furthermore, for each technology, we calculated the global average normalized coverage in technology-targeted regions. With these global averages, we plotted an ideal curve for each technology (dotted lines), in which the normalized coverage of each base is the average normalized coverage for each individual technology. We found that HaloPlex had the highest (P < 10−320, when compared to each technology) and SeqCap had the lowest (P < 10−320, when compared to each technology) average distance to its corresponding ideal curve.
Effects of GC content in regions targeted by each technology. For every technology, in each one's specific target region, we plotted average normalized coverage against the GC content. Darker colors indicate higher density of points, whereas lighter colors indicate lower density of points. GC content was calculated on the basis of percentage of Gs or Cs in a 100-bp window.
GenomOncology's analysis: how GC affects depth in mutually targeted regions. For every technology, in mutually targeted bases, GenomOncology plotted average normalized coverage against the GC content. Darker colors indicate higher density of points, whereas lighter colors indicate lower density of points. GC content was calculated on the basis of percentage of Gs or Cs in a 100-bp window.
Single-nucleotide variation (SNV) concordances among technologies using SureCall on HaloPlex. For single-sample analyses, SNVs were called from all libraries using the indicated SNV caller, except HaloPlex, for which we used SureCall. The latter is designed to perform single-sample analyses, but not tumor-normal analyses.
Single-nucleotide variation (SNV) concordances among technologies in cell lines. We determined SNVs using three different variant callers for all cell lines through single-sample analyses (A) and paired tumor-normal analyses (in this case, HCC-2218 versus HCC-2218BL) (B). To get a summary of concordance among technologies, we pooled all SNVs from all cell lines together and gave each SNV a unique identification on the basis of the sample, chromosome, reference allele, and alternate allele.
Single-nucleotide variation (SNV) concordances among technologies in cell lines using SureCall on HaloPlex. For single-sample analyses, SNVs were called from all libraries using the indicated SNV caller, except HaloPlex, for which we used SureCall. The latter is designed to perform single-sample analyses, but not tumor-normal analyses.
Discordant single-nucleotide variations (SNVs) due to lack of coverage. Shown are views from Integrative Genomics Viewer (IGV) of read alignment at an SNV in the BT-20 cell line, chr13:48,947,576 T→G in SureSelect (A), HaloPlex (B), Nextera (C), and SeqCap (D). All technologies, except HaloPlex, called this SNV. HaloPlex missed it, because of lack of coverage.
Discordant single-nucleotide variations (SNVs) due to insufficient quality reads. Shown are views in Integrative Genomics Viewer (IGV) of chr2:212,248,542 T→C in the MCF-7 cell line in SureSelect (A), HaloPlex (B), Nextera (C), and SeqCap (D). VarScan2 called this mutation in all technologies, except HaloPlex. VarScan2 failed to report this SNV in HaloPlex, because, after removal of low-quality base calls, variant frequency decreased to 18.94%, lower than VarScan2's minimum cutoff of 20%. Genome Analysis Toolkit (GATK) called this mutation in all four technologies.
Discordant single-nucleotide variations (SNVs) due to Illumina sequencing errors. The presence of some GG motifs causes sequencing errors, resulting in many mismatches in a short read span.28,29 Shown are read alignment views in Integrative Genomics Viewer (IGV) around chr2:29,448,410 T→G in BT-20 in SureSelect (A), HaloPlex (B), Nextera (C), and SeqCap (D). VarScan2 called this SNV in the HaloPlex library, but no other library.
Discordant single-nucleotide variation (SNV) due to a HaloPlex restriction site. Shown are views from Integrative Genomics Viewer (IGV) of read alignment of an SNV in the BT-20 cell line, chr13:48,955,428 C→T in SureSelect (A), HaloPlex (B), Nextera (C), and SeqCap (D). VarScan2 and Genome Analysis Toolkit (GATK) called this SNV in all technologies, except HaloPlex. This appears to be due to the SNV's presence within a HaloPlex restriction site, which biases this technology toward the wild-type allele. VarScan2, after removing low-quality base calls, finds variant frequency equals 6.08%, lower than VarScan2's minimum of 20% to report. Similarly, GATK, after poor base-quality filtration, also finds variant frequency equals 6.08%.
Sanger sequencing analysis of select single-nucleotide variations (SNVs). Occasionally, observing read alignments of an SNV from various technologies produced conflicting results on the existence of the SNV. In such cases, we used Sanger sequencing to verify or refute the existence of five chosen SNVs: MCF-7 cell line, chr10:62,547,870 T→G on the positive (pos) strand (A), MCF-7 cell line, chr6:94,066,507 G→T on the negative (neg) strand (B), MCF-7 cell line, chr3:178,936,091 G→A on the positive strand (note that Sanger sequencing called G, but there was a signal for A too; C), BT-20 cell line, chr12:18,649,057 C→T on the positive strand (D), and BT-20 cell line, chr13:48,955,428 C→T on the positive strand (E).