Abstract
Burkitt lymphoma (BL) and diffuse large B-cell lymphoma (DLBCL) are aggressive tumors of mature B cells that are distinguished by a combination of histomorphological, phenotypic, and genetic features. A subset of B-cell lymphomas, however, has one or more characteristics that overlap BL and DLBCL, and are categorized as B-cell lymphoma unclassifiable, with features intermediate between BL and DLBCL (BCL-U). Molecular analyses support the concept that there is a biological continuum between BL and DLBCL that includes variable activity of MYC, an oncoprotein once thought to be only associated with BL, but now recognized as a major predictor of survival among patients with DLBCL treated with R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone). We tested whether a targeted expression profiling panel could be used to categorize tumors as BL and DLBCL, resolve the molecular heterogeneity of BCL-U, and capture MYC activity using RNA from formalin-fixed, paraffin-embedded biopsy specimens. A diagnostic molecular classifier accurately predicted pathological diagnoses of BL and DLBCL, and provided more objective subclassification for a subset of BCL-U and genetic double-hit lymphomas as molecular BL or DLBCL. A molecular classifier of MYC activity correlated with MYC IHC and stratified patients with primary DLBCL treated with R-CHOP into high- and low-risk groups. These results establish a framework for classifying and stratifying MYC-driven, aggressive, B-cell lymphomas on the basis of quantitative molecular profiling that is applicable to fixed biopsy specimens.
CME Accreditation Statement: This activity (“JMD 2015 CME Program in Molecular Diagnostics”) has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint sponsorship of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“JMD 2015 CME Program in Molecular Diagnostics”) for a maximum of 36 AMA PRA Category 1 Credit(s)™. Physicians should only claim credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
The World Health Organization classification of tumors defines neoplastic diseases according to unique clinical and biological characteristics.1 Burkitt lymphoma (BL) and diffuse large B-cell lymphoma (DLBCL) are aggressive tumors of mature B cells categorized as individual tumor types. The reliable differentiation of BL from DLBCL is important, because these tumors are treated with distinct chemotherapeutic regimens.2,3
BL is a neoplasm composed of monomorphic, intermediate-sized lymphocytes that are positive for markers of mature, germinal-center B cells and negative for the anti-apoptotic protein BCL2. Most cells (>95%) are positive for the proliferation marker, Ki-67/MIB1. The genetic hallmark of BL is a balanced translocation involving the MYC oncogene and, most commonly, the immunoglobulin heavy chain locus (IGH).1,4 Mutations in TCF3 and ID3 are also common.5,6 In contrast, DLBCL is composed of pleomorphic, large lymphoid cells and, in general, less apoptosis and a lower proliferation index than BL. DLBCLs express markers of mature B cells, with or without evidence of germinal center cell derivation, and often express BCL2. Genetically, only a small subset of DLBCLs have a MYC translocation, and mutations in TCF3 or ID3 are rare. However, mutations in genes encoding the components of the NF-κB and B-cell receptor signaling pathways are common.1,7–11
Most cases of BL and DLBCL are diagnosed with high confidence using traditional histopathological, immunophenotypic, and targeted genetic analyses. However, it is not uncommon to encounter tumors with one or more features overlapping BL and DLBCL. The 2008 World Health Organization Classification of Lymphoid Tumors recognized these cases with the novel diagnostic category, B-cell lymphoma unclassifiable, with features intermediate between DLBCL and BL (BCL-U).1 BCL-U is, by definition, a heterogeneous group, and its diagnosis requires that pathologists make subtle distinctions in histomorphological features, immunophenotype, and genetics that may not be highly reproducible.
Molecular classification of aggressive B-cell lymphomas using comprehensive gene-expression profiles (GEPs) of RNA isolated from frozen tumor samples accurately differentiates BL from DLBCL and confirms that a subset of cases has transcriptional signatures intermediate between BL and DLBCL.12,13 However, the pathological diagnoses corresponding to these biologically intermediate tumors have been inconsistent.13
Complicating the evaluation of aggressive lymphomas is the recognition that high MYC expression and biological activity, once thought to be only associated with BL, are major, independent predictors of poor clinical outcome among patients with primary DLBCL treated with R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone).14–18 In some series, the prognostic value of MYC is enhanced among tumors that co-express BCL2.14,19–21 Indeed, recent evidence suggests that high co-expression of MYC and BCL2 in tumor cells provides a biological basis for the inferior outcome among patients with the activated B-cell (ABC) type DLBCL when treated with standard chemotherapy.21
DLBCL with high MYC activity cannot be identified with certainty by morphological or genetic studies alone.15 The detection of MYC in fixed tumor biopsy specimens by immunohistochemistry (IHC) has the potential to identify DLBCLs with high MYC protein that corresponds to high MYC biological activity.15 However, IHC methods are difficult to standardize between institutions, and the interpretation of IHC staining is subjective.22
These data highlight a need for quantitative methods that capture the phenotypic, genetic, and molecular heterogeneity of aggressive B-cell lymphomas in clinical practice. Molecular classification on the basis of the unique GEPs of BL, DLBCL, and MYC-driven B-cell lymphomas has the potential to satisfy this need, but, until recently, GEP has not been amenable to formalin-fixed, paraffin-embedded (FFPE) tissues.23–26 Herein, we report a method of targeted expression profiling, followed by a two-stage molecular classifier of aggressive mature B-cell lymphomas that is applicable to FFPE biopsy specimens.
Materials and Methods
Tumor and Patient Cohorts
This study was performed with approval from the Institutional Review Boards (IRBs) of Brigham and Women's Hospital (Boston, MA; IRB number 2010P002736) and Massachusetts General Hospital (Boston, MA; IRB number 2007P001458). For each case, one or both of the corresponding pathologists of this study (S.J.R. and A.R.S.) reviewed H&E-stained slides and the original diagnostic reports to ensure that the final diagnosis fulfilled 2008 World Health Organization diagnostic criteria.
The training set (n = 41) comprises 12 BLs and 29 DLBCLs (one additional DLBCL later failed analytical quality control). The BLs were selected on the basis of the quality of available tissue and include all BL subtypes, as well as pediatric and adult patients (median age of diagnosis, 30.5 years; range, 3 to 62 years) (Supplemental Table S1). The DLBCLs were selected from a previously published larger series of adult patients15 who had all been diagnosed as having DLBCL not otherwise specified. Previously, MYC IHC-High was defined as >50% expression in tumor cells, and MYC IHC-Low was defined as ≤50% expression in tumor cells.15 For training, cases were deliberately selected to represent the extremes of MYC IHC-High (median, 70%; n = 13) and MYC IHC-Low (median, 20% to 30%; n = 16) to assist development of the MYC activity classifier. DLBCLs were not selected with regard to cell of origin (COO27) subtype, but previously classified using GEP as 10 ABC-type (34.5%), 13 germinal center B-cell type (GBC; 44.8%), 5 type 3 (17.2%), and 1 unclassified (3.4%) (Supplemental Table S2).
The test set (n = 55) comprised 9 BLs (all adult patients, 8 sporadic and 1 immunodeficiency associated), 41 DLBCLs, and 5 BCL-Us (Supplemental Tables S1, S2, and S3). Four additional cases failed analytical quality control. Eight of these tumors were genetic double-hit lymphomas (DHLs), for the purposes of this study defined as the combination of a MYC rearrangement and either a BCL2 or BCL6 rearrangement, and these were divided into three tumors with a pathological diagnosis of DLBCL and five tumors with a pathological diagnosis of BCL-U (Supplemental Table S3). The DLBCLs included these three DHLs, which were characterized by a combination of MYC and BCL2 rearrangements, as well as one single-hit lymphoma (SHL), characterized by a MYC rearrangement in isolation. The DLBCLs were chosen on the basis of the quality of available biopsy material and to represent a full range of MYC IHC expression. DLBCLs for the test set were not selected on the basis of COO subtype.27 COO classification data, using GEP (if available) and/or Han's IHC criteria,28,29 showed a distribution of 16 ABC/non-GCB types (39%), 17 GCB-types (41.5%), 3 type 3 (7.3%), and 5 unclassified (12.2%) (Supplemental Table S2). The five BCL-Us were selected on the basis of available cases and were all DHLs. Four of the five BCL-Us were characterized by a combination of MYC and BCL2 rearrangements and the remaining case had concurrent MYC and BCL6 rearrangements.
Patients included in an outcome cohort (outcome series; n = 40, 22 patients from the training set and 18 from the test set) were derived from a single institution (Brigham and Women's Hospital). All had confirmed primary DLBCLs and received standard immunochemotherapy (R-CHOP), as previously reported.15 All clinical data were collected before, and independent of, the reference and index tests reported in this study.
IHC and Cytogenetic Analysis
MYC IHC was performed on 96 tumors using a rabbit monoclonal antibody (clone Y69, catalog number ab32072; Epitomics/Abcam, Burlingame, CA), as described.15 The status of the MYC locus was determined by fluorescence in situ hybridization analysis for 96 tumors using a Vysis LSI MYC break-apart probe set (catalog number 05-J91-001), as described.15 Fluorescence in situ hybridization analyses were performed on indicated cases using the BCL2-IgH dual-fusion (catalog number 05-J71-001) and BCL6-IgH break-apart (catalog number 01N23-020; Abbott Laboratories, Abbott Park, IL) probe sets, respectively, following manufacturer's recommendations. For a few cases, a karyotype was obtained as part of the original diagnostic evaluation.15
RNA Extraction and Profiling
FFPE tissue blocks were divided into sections immediately before the RNA extraction. For each block, the initial section (10 μm thick) was discarded and three subsequent sections (10 μm thick) were taken for analysis. If the estimated surface area of lesional tissue was <5 mm2, an extra section (10 μm thick) was taken. Total RNA was isolated using the Qiagen RNeasy kit (catalog number 73504; Qiagen, Hilden, Germany) and quantified using a Nanodrop spectrophotometer (NanoDrop Products, Thermo Scientific, Wilmington, DE). RNA was diluted to 150 to 200 ng/5 μL, aliquoted, and stored at −80°C until use.
For the multiplexed, digital gene expression analysis, 150 to 200 ng of RNA for each sample was hybridized with 20 μL of reporter probes/reaction buffer and 5 μL of capture probes at 65°C for 20 hours. The hybridized samples were then processed on the NanoString nCounter preparation station for 2.5 hours and expression data were subsequently generated on the NanoString nCounter digital analyzer (NanoString Technologies, Seattle, WA) using the 600 fields of view setting over 4 hours.30 In total, tumors from 96 patients were profiled, with a further 5 tumors (5%) failing analytical quality control.
Target Selection for the Initial and Final Profiling Panels
Candidate gene targets were initially selected from published GEPs of BL and DLBCL,12,13 with preference given to genes within the transcription factor 3 (TCF3)/inhibitor of DNA binding 3 (ID3) signaling pathway,5 published MYC targets,31–37 and GEPs of frozen tissue corresponding to DLBCL samples in the training set.38 These were supplemented by additional targets of interest, including housekeeping (HK) genes (Supplemental Figure S1).
The initial panel of 200 probes included 37 unique transcripts distilled from a previously published TCF3 signature.5 These were subsequently validated by in silico differential analysis (DA) as best distinguishing BL from DLBCL in two independent series of B-cell non-Hodgkin lymphomas12,13 (Supplemental Figure S1). The panel also included transcripts from seven published data sets of MYC targets (101 targets selected)31–37 that were validated [false-discovery rate (FDR), <0.25; fold change, >1.3] by DA against Affymetrix U133 microarray GEPs of frozen DLBCLs. There were corresponding MYC IHC scores from matched FFPE tissue in the training cohort15,38 and differentially expressed genes, suggested by DA, of the GEPs of frozen DLBCLs, with corresponding MYC IHC scores (FDR, <0.25; fold change, >2.0). Finally, they were supplemented with BCL2 and related family members (5 targets), housekeeping control transcripts (15 targets), and select markers of specific cell lineages (CD3e, CD68, CD19, CD79a, and CD20) (Supplemental Table S4).
The final profiling panel, targeting 80 transcripts, was derived by analyzing data from the training set, both ranking the importance of each included gene and estimating how to exclude many without compromising the predicted accuracy (Supplemental Table S5). The predicted accuracy of each classifier was assessed on the training set using leave-one-out cross validation (LOO-CV), as well as on an independent test data set.
HK Gene Transcripts
Six HK genes were selected on the basis of the following criteria: i) low variation across samples; ii) even coverage along the expression range; iii) exclusion of the most highly expressed HK genes, because at high levels, the variation level of the HK genes is comparable to the variation of the other genes; and iv) exclusion of genes within regions of known recurrent copy number alteration in lymphoma.38 On the basis of these criteria, we selected the following six gene targets: AAMP, HMBS, KARS, PSMB3, TUBB, and H3F3A.
Data Normalization
Data from the preliminary targeted profiling panel (200 genes) and the final profiling panel (80 genes) were cross normalized using expression data from six cases tested with both panels. Normalization of the NanoString data was performed using the R package NanoStringNorm (http://cran.rproject.org/web/packages/NanoStringNorm; last accessed August 21, 2014). We used the sum of the expression values to estimate the technical assay variation, the mean to estimate background count levels, and the sum of the six HK genes to normalize for the RNA sample content. In addition, the data were log2 transformed.
Unsupervised Clustering of the Normalized Training Data Set
Unsupervised clustering of the data derived from the training set was performed using Gene-e (Broad Institute, http://www.broadinstitute.org/cancer/software/GENE-E, last accessed August 21, 2014) (Supplemental Table S6). These data were normalized using Nanostring's nSolver software package (version 1.1; NanoString) and then transformed to Log2 using Gene-e before one minus Pearson hierarchical clustering.
Classification Models
Classification models were selected on the basis of the training cohort using a bootstrapping scheme, in which 75% of the samples were drawn to train a classification model, which was then tested on the remaining 25% of the samples, with the train/test split repeated 100 times. Elastic nets,39 linear and polynomial support vector machines, shrunken centroids,40 and a random forest algorithm41 were evaluated as candidate prediction models. An elastic net39 prediction model was selected for both classifiers, on the basis of a bootstrapping evaluation scheme on the training set. For the development of the diagnostic classifier, cases with a pathological diagnosis of BL and DLBCL were used. For the MYC activity classifier, only DLBCLs were used, excluding BLs. DLBCLs with MYC IHC >50% and ≤50% were classified as MYC IHC-High and IHC-Low, respectively, and these labels were used in the training of the MYC activity classifier.15
Features were selected on the basis of differential expression, and their number was determined on the basis of LOO-CV performed on the training cohort; on the basis of this procedure, 21 genes were used for the diagnostic classifier and 61 for the MYC classifier (Supplemental Table S7). On the basis of their performance on the training data set, we selected the elastic net with an α parameter of 0.1 and a λ parameter of 0.1 as the classifier of choice for both stages. Classification accuracy of the final elastic net models was assessed on the training cohort using LOO-CV and comparing the predictions with the outcome of the IHC staining. Unbiased validation was then performed by training elastic net models on the entire training data set and applying them to the classification of cases in the test cohort.
Diagnostic and MYC Activity Scores
Elastic net models output class probabilities between 0 and 1 for each class (probability of class BL in the diagnostic classifier, and of class MYC IHC-High in the MYC transcriptional activity classifier), reflecting the confidence of a sample prediction. Before analysis, and to reflect the concept of a biological intermediate between BL and DLBCL, we defined diagnostic scores of >0.75 as representing molecular BL (mBL), <0.25 as representing molecular DLBCL (mDLBCL), and 0.25 to 0.75 as representing molecularly intermediate. MYC activity scores of 1 and 0 corresponded to tumors with high MYC and low MYC (as modeled on IHC expression15) with greatest probability, respectively. During development of the MYC activity classifier, 0.5 was optimized as the cutoff with the highest estimated accuracy to classify tumors with high and low MYC activity. Therefore, 0.5 is used for statistics regarding the efficacy of the classifier and for correlation to clinical outcome.
Reproducibility of the Assay
The test set and outcome series were profiled using two builds (independently constructed probe sets) of the 80-gene profiling panel. The binding efficiency of probes varies between builds and, therefore, the final data set was compiled by normalizing to both housekeepers and then between builds, using the expression profiles of tumor RNA that were profiled on both. RNA from a subset of cases was profiled multiple times over the course of the study to determine the reproducibility of the assay (Supplemental Figure S2).
Results
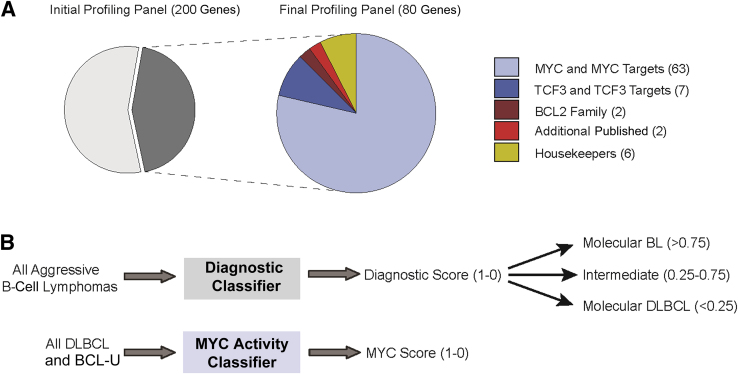
RNA was isolated from FFPE tissue corresponding to 41 aggressive B-cell lymphomas (training set) and was profiled using an initial panel of probes targeting 200 unique transcripts (Figure 1 and Supplemental Table S4). The resulting data were used to derive a pair of molecular classifiers, first to distinguish BL from DLBCL and second to distinguish high and low MYC activity in DLBCL using a parsimonious, 80-gene signature (Figure 1 and Supplemental Tables S5 and S7).
Figure 1.
Target gene selection and the generation of molecular classifiers. A: Schematic showing the distribution of gene transcripts that were assayed in the initial and final profiling panels. B: Schematic outlining the protocols for the molecular classification of all aggressive B-cell lymphomas and cases with the pathological diagnosis of diffuse large B-cell lymphoma (DLBCL) and B-cell lymphoma unclassifiable (BCL-U). Twenty-one genes were used for the Diagnostic classifier, and 61 genes were used for the MYC activity classifier. In addition, eight genes were common to both classifiers, and there were six housekeeping genes (HK).
Unsupervised Clustering of Targeted Expression Profiles of Select Lymphomas
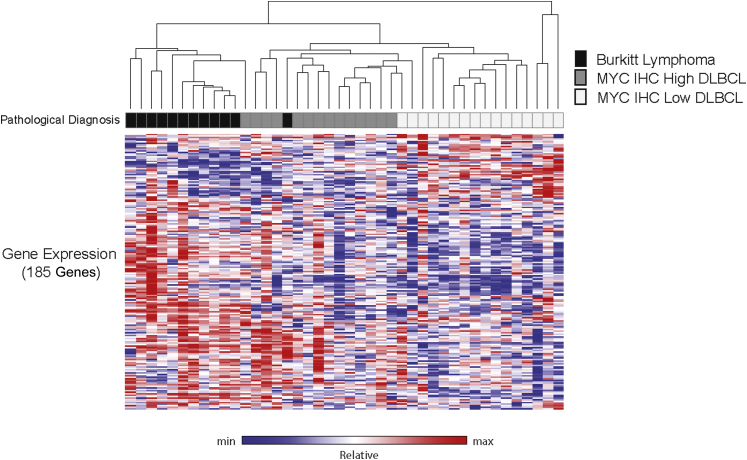
Unsupervised clustering of the normalized expression data from the 200-gene signature segregated the training set tumors into distinct groups that showed a close correlation with the original pathological diagnoses of BL, DLBCL MYC IHC-High, and DLBCL MYC IHC-Low (Figure 2). One DLBCL later failed classification quality control and was not used in subsequent analysis. One case, diagnosed as BL, clustered with DLBCL MYC IHC-High cases. Central review of this case confirmed that the tumor was originally diagnosed correctly. These data support the in silico methods used to develop the initial profiling panel and demonstrate the technical feasibility of the approach to broadly group aggressive lymphomas into clinically relevant categories.
Figure 2.
Unsupervised clustering of the normalized transcript values from 42 tumors comprising the training cohort and including all target probes in the initial profiling panel (one case later failed quality control during classification). The original pathological diagnosis and relative gene expression for the 185 genes comprising the initial profiling panel (heatmap) are shown (15 housekeeping genes were excluded). Max, maximum; min, minimum.
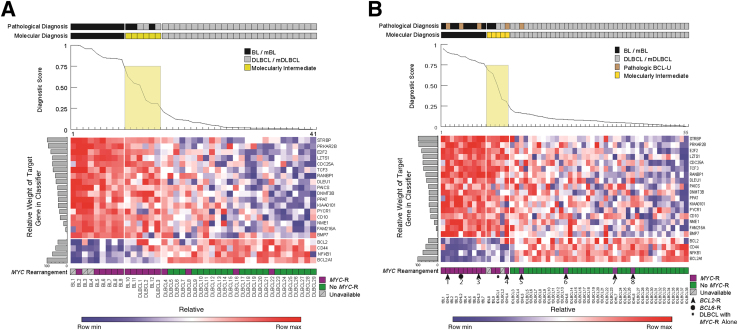
Performance of the Diagnostic Molecular Classifier on the Training and Test Sets
We tested the diagnostic molecular classifier against data derived from the training set in a LOO-CV (Figure 3A). When ranked by the diagnostic classifier scores, these data largely recapitulated the results obtained from the original unsupervised clustering analysis using the 200-gene panel. Of 41 cases, 35 (85%) were classified as mBL or mDLBCL with high confidence and correctly matched the pathological diagnoses of BL or DLBCL, respectively (Figure 3A and Table 1). Six cases had diagnostic scores of >0.25 and <0.75 and, thus, were not assigned to the categories of mBL or mDLBCL, respectively. Nevertheless, three of the molecularly intermediate cases had a pathological diagnosis of BL, and two of these had a diagnostic score >0.5; three molecularly intermediate cases had a pathological diagnosis of DLBCL, and two had a diagnostic score <0.5. We conclude that, in our training cohort, a 21-gene classifier can be used to distinguish most (85%) of pathological BL from DLBCL.
Figure 3.
A: Leave-one-out cross-validation of the final profiling panel and diagnostic classifier for the training cohort: Burkitt lymphoma (BL) and diffuse large B-cell lymphoma (DLBCL). B: Results of the diagnostic classifier for the test cohort: BL, B-cell lymphoma unclassifiable (BCL-U), and DLBCL cases categorized according to the original pathological diagnosis, the assigned molecular diagnosis (diagnostic scores of 0.25 to 0.75 categorized as molecularly intermediate), diagnostic score (line graph, intermediate values shaded), the relative expression of the indicated transcripts (heat map), including the relative contribution of each to the classifier (horizontal shaded bar graphs), and MYC-rearrangement status. The cases of genetic double hit lymphomas are numbered, and additional gene rearrangements are indicated by arrowheads (BCL2-) or a circle (BCL6-). The single-hit DLBCL, with MYC-rearrangement only, is indicated by an asterisk. Max, maximum; min, minimum.
Table 1.
Performance Statistics of Molecular Classifiers
| Variable | Diagnostic classifier∗ |
MYC activity classifier† |
|||||
|---|---|---|---|---|---|---|---|
| Training | Test | Training, all | Training, non-BL | Test, all | Test, non-BL | Outcome series | |
| Cases classified (%) | 85 | 92 | 100 | 100 | 100 | 100 | 100 |
| Accuracy | 1 | 1 | 0.93 | 0.90 | 0.80 | 0.80 | 0.87 |
| Sensitivity (95% CI) | 1 (0.66–1.0) | 1 (0.59–1.0) | 0.92 (0.73–0.99) | 0.85 (0.55–0.98) | 0.77 (0.55–0.92) | 0.69 (0.41–0.89) | 0.75 (0.35–0.96) |
| Specificity (95% CI) | 1 (0.87–1.0) | 1 (0.91–1.0) | 0.94 (0.70–0.99) | 0.94 (0.70–0.99) | 0.83 (0.64–0.94) | 0.86 (0.67–0.96) | 0.90 (0.73–0.98) |
| PPV | 1 | 1 | 0.96 | 0.92 | 0.77 | 0.73 | 0.67 |
| NPV | 1 | 1 | 0.88 | 0.88 | 0.83 | 0.83 | 0.93 |
NPV, negative predictive value; PPV, positive predictive value.
Only cases classified with high confidence (as mBL or mDLBCL) were included. The sensitivity refers to the ability of the test to identify pathological BL as mBL.
Only cases with matched MYC IHC and MYC activity scores were included. The sensitivity refers to the ability of the test to identify tumors with high MYC IHC expression (>50%) as having MYC activity score >0.5.
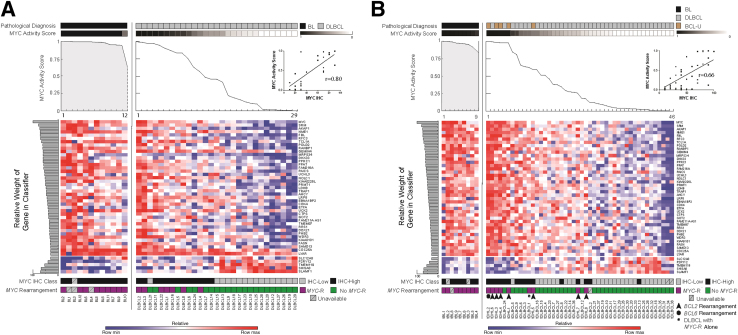
We next profiled and classified a test set of 55 cases that included 9 BLs, 41 DLBCLs, and 5 with the pathological diagnosis of BCL-U (Figure 3B). Among the non-BLs were one genetic single-hit lymphoma [genetic SHL, with isolated MYC translocation (tDLBCL1)] and eight genetic DHLs, all with MYC translocations. Seven DHLs had coexistent BCL2 translocations, and one DHL had a coexistent BCL6 translocation, tDHL1-8 (Figure 3B).
The diagnostic classifier successfully segregated all pathological BLs from all DLBCLs (Figure 3B and Table 1). Of 50, 46 (92%) of BLs and DLBCLs were classified with high confidence. Two BLs and two DLBCLs had intermediate diagnostic scores, but among these, the diagnostic scores for the BL were >0.5 and for the DLBCL, ≤0.5. The DLBCL with the highest diagnostic score (case tDLBCL1, score = 0.5) was the genetic SHL. The diagnostic classifier demonstrated a sensitivity of 1.0 (95% CI, 0.59–1.0) and a specificity of 1.0 (95% CI, 0.91–1.0) in the test set, for all tumors classified as mBL or mDLBCL (Table 1). We conclude that in our test cohort, a 21-gene classifier can be used to distinguish most (92%) of pathological BLs from DLBCLs.
Molecular classification segregated subsets of non-BLs with the pathological diagnosis of BCL-U and/or genetic evidence for MYC rearrangements into all three diagnostic categories (Figure 3B). Three BCL-U/DHLs (tDHL1, tDHL2, and tDHL3) had high diagnostic scores (0.90, 0.85, and 0.77, respectively) and were classified as mBL. One DLBCL/SHL (tDLBCL1) and one BCL-U/DHL (tDHL4) had lower diagnostic scores (0.50 and 0.31, respectively) and were classified as molecularly intermediate. Finally, one BCL-U/DHL (tDHL5) and three DLBCL/DHLs (tDHL6, tDHL7, and tDHL8) had low diagnostic scores (0.12, 0.05, 0.02, and 0.015, respectively) and were classified as mDLBCL. We conclude that the diagnostic molecular classifier reveals molecular heterogeneity among BCL-Us and DLBCLs with MYC translocations.
Molecular and Histopathological Features of BCL-U/DHL
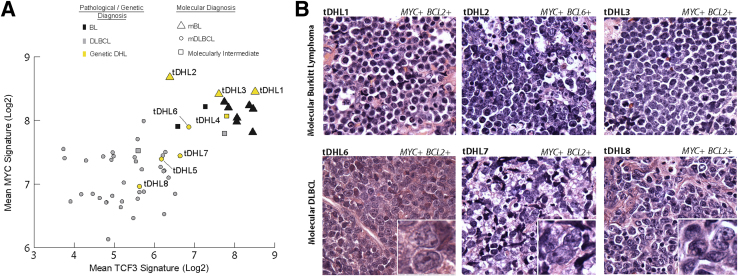
We next examined the molecular signatures and histopathological features of the BCL-Us and DLBCLs with MYC translocations in more detail (Figure 4). BCL-U/DHLs classified as mBL expressed both TCF3-associated transcripts and MYC-associated transcripts at levels that were comparable to BL (Figure 4A). DLBCL/DHLs classified as mDLBCL expressed TCF3-associated transcripts at low levels and MYC-associated transcripts at intermediate levels, which were comparable to many DLBCLs lacking a MYC translocation (Figure 4A). Additional transcripts (BCL2, CD44, NFKB1, and BCL2A1), differentially expressed between BL and DLBCL, also showed differential expression among the DHLs, and with the TCF3 and MYC signatures, resulted in the final classification indicated in Figure 3B.
Figure 4.
A: Scatterplot shows the mean TCF3 signature (seven genes, x axis) and mean MYC signature (10 genes, y axis) for each tumor from the test cohort. The mean values for each signature are derived from transcript counts from these genes, as originally used in the diagnostic classifier. Colors indicate the pathological/genetic diagnoses [black for Burkitt lymphoma (BL), gray for diffuse large B-cell lymphoma (DLBCL), and yellow for genetic double hit lymphomas (DHLs)]. Shapes indicate the molecular classification assigned by the diagnostic classifier [triangle for molecular (m)BL, circle for molecular DLBCL (mDLBCL), and square for molecularly intermediate]. B: Histomorphological features of lymphomas with a MYC rearrangement and either a BCL2- or BCL6- rearrangement (genetic DHL). H&E-stained sections of DHL classified as mBL and mDLBCL. Unique identifiers and details of relevant translocations are shown. The tumors classified as mDLBCL have insets highlighting nuclear morphological characteristics. Original magnification, ×1000 (main images and insets).
Further review of the histopathological features of the DHLs revealed distinct features between those that classify with high confidence as mBL and those that classify with high confidence as mDLBCL (Figure 4B). Cases classified as mBL were composed of sheets of tightly packed, intermediate- to large-sized cells with homogeneous, round nuclei and scant cytoplasm, which resembles the morphological features of classic BL. In contrast, cases classified as mDLBCL were composed of large-sized lymphoid cells with marked pleomorphism and nuclear irregularity typical of DLBCL. We conclude that the final molecular classifications of DHLs are supported by multiple molecular signatures, and correlate with distinct histopathological characteristics.
Performance of the MYC Activity Classifier on the Training and Test Sets
The MYC activity classifier was tested in the training cohort by LOO-CV. BLs were not used to build the classifier, but as expected, had high MYC activity scores (Figure 5A). In addition, all non-BLs with MYC translocation had MYC activity scores >0.5. The sensitivity and specificity of the molecular classifier for identifying MYC IHC-High among all cases in the training set were 0.92 (95% CI, 0.73–0.997) and 0.94 (95% CI, 0.70–0.99), respectively (Table 1). Overall, the correlation between the optimized, molecular MYC activity score and MYC IHC score among non-BLs in the training set was high (Spearman r = 0.80, P < 0.0001, 95% CI, 0.6–0.9) (Figure 5A).
Figure 5.
A: Leave-one-out cross-validation of the final profiling panel and MYC activity classifier for the training cohort. Burkitt lymphoma (BL) and diffuse large B-cell lymphoma (DLBCL) are segregated by pathological diagnosis, MYC activity score (line graph), the relative expression of the indicated transcripts (heat map), including the relative contribution of each to the classifier (horizontal, shaded bar graphs), MYC IHC class (MYC IHC-Low ≤50%, IHC-High >50), and MYC rearrangement status. Inset: The correlation between MYC IHC and MYC activity score for DLBCL only (Spearman r = 0.80; 95% CI, 0.6–0.9). B: Results of the final profiling panel and MYC activity classifier for the test cohort, BL, DLBCL, and B-cell lymphoma unclassifiable (BCL-U), are segregated by pathological diagnosis, MYC activity score (line graph), the relative expression of the indicated transcripts (heat map), including the relative contribution of each to the classifier (horizontal, shaded bar graphs), MYC IHC class (MYC IHC-Low ≤50%, IHC-High >50%), and MYC rearrangement status. Genetic double hit lymphomas are numbered and additional gene rearrangements are indicated by arrowheads (BCL2-) or a circle (BCL6-). The single-hit DLBCL, with MYC-rearrangement only, is indicated by an asterisk. Inset: The correlation between MYC IHC and MYC activity score for non-BL only (Spearman r = 0.66; 95% CI, 0.44–0.8). Max, maximum; min, minimum.
We next applied the MYC activity classifier to expression data from the test set. Again, BL cases showed high MYC activity scores (Figure 5B). The sensitivity and specificity of the molecular classifier identifying MYC IHC-High among all cases were 0.77 (95% CI, 0.55–0.92) and 0.83 (95% CI, 0.64–0.94), respectively (Table 1). The correlation between the molecular MYC score and the MYC IHC score for the test set (non-BLs) was lower than for the LOO-CV of the training set, but with overlapping CIs, thus preventing definitive comparison (Spearman r = 0.66, P < 0.0001, 95% CI, 0.44–0.8).
Non-BLs with a MYC translocation were expected to have up-regulated MYC activity, and for five of nine cases, tDHL1-4 and tDHL6, the MYC activity scores were high and comparable to those seen for BL (range, 0.98 to 1.00). There was a range of values among the remaining cases. For tDLBCL1 (genetic SHL) and tDHL5, the MYC activity scores were 0.63 and 0.60; and for tDHL7 and tDHL8, the scores were lower, at 0.26 and 0.18, respectively. Non-BLs with MYC translocations and high MYC activity scores had a pathological diagnosis of BCL-U, whereas those with other MYC activity scores had a pathological diagnosis of DLBCL. We conclude that the MYC activity classifier captures a spectrum of MYC biological activity in BCL-U and DLBCL that shows good correlation with MYC IHC and reveals heterogeneity in MYC biological activity among non-BL with MYC translocations.
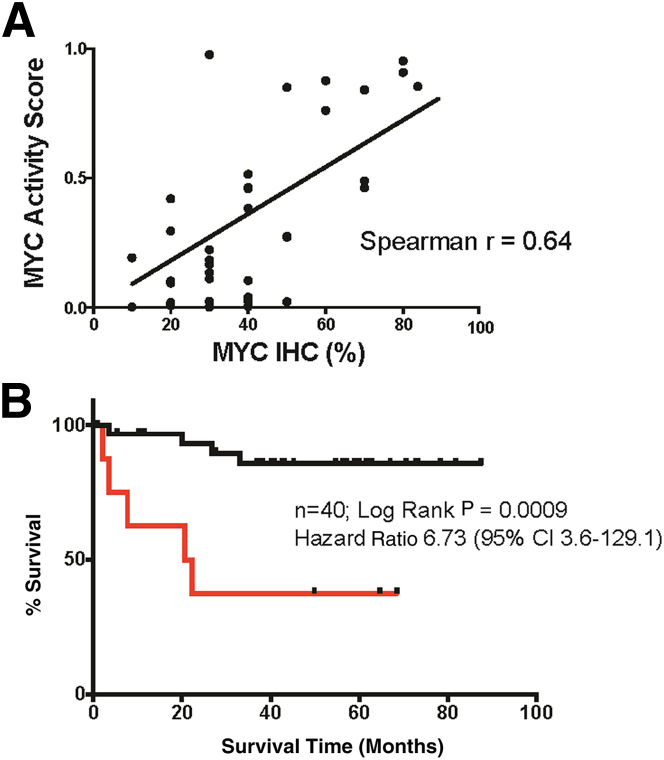
Clinical Significance of the MYC Activity Score in DLBCL
The MYC activity classifier was constructed to categorize aggressive B-cell lymphomas, according to MYC biological activity, rather than to predict clinical outcome. The MYC activity scores showed good, but not perfect, correlation with MYC IHC scores in the training and test sets. Therefore, we wanted to determine whether the results of the MYC classifier were sufficient to predict clinical outcome in a series for which MYC IHC has prognostic value.15 DLBCLs with MYC activity scores in excess of the optimized classifier cut point of 0.5 identified a patient population with inferior overall survival that was highly significant (nominal P = 0.0009, log-rank test; hazard ratio = 6.73) (Figure 6B). The correlation between MYC activity and MYC IHC scores was similar to the training and test sets, as expected (r = 0.64, P < 0.0001, 95% CI, 0.4–0.8) (Figure 6A). We conclude that the MYC activity classifier, built on MYC IHC data (Supplemental Figure S3), is capable of dividing patients into high- and low-risk categories.
Figure 6.
Results of the MYC classifier and overall survival among patients with primary diffuse large B-cell lymphoma treated with R-CHOP (rituximab, cyclophosphamide, doxorubicin, vincristine, prednisone)–based chemotherapy. A: The correlation between MYC score and MYC IHC for the outcome series (Spearman r = 0.64; 95% CI, 0.4–0.8). B: Kaplan-Meier curve shows OS for the outcome series with a MYC score >0.5 (red line) and a MYC score <0.5 (black line).
Discussion
The World Health Organization currently considers histomorphological, immunophenotypic, and genomic data to categorize aggressive B-cell lymphomas.1 However, the interpretation of histomorphological and IHC data remains subjective and requires expert review. Molecular profiling has the potential to aid diagnostic categorization by providing objective data from normalized gene expression signatures, but until recently, the degradation of RNA due to formalin compromised the ability to use fixed biopsy specimens.23–26
We have described a framework for the molecular classification of MYC-driven B-cell lymphomas using targeted expression profiling of RNA isolated from FFPE tissue. The approach described has several features that make it appealing. First, the assay requires only small amounts of FFPE tissue. We and others find that RNA isolated from the equivalent of 2× to 6× FFPE tissue sections (5 μm thick) is sufficient for analysis.24 Second, the assay is robust. We successfully profiled 96 FFPE tumor biopsy samples ranging from 0.5 to 13 years old, with only an additional 5 (5%) failing analytical quality control, and repeat testing of the same samples yielding nearly identical results. Third, the step-wise application of the diagnostic and MYC activity classifiers mimics the diagnostic approach used to evaluate aggressive B-cell lymphomas in clinical practice. Fourth, the molecular scores provide quantitative outputs that can be interpreted objectively. Thus, the assessment of defined molecular signatures from FFPE tissue, by using the methods described herein, has the potential to provide important additional biological information alongside traditional diagnostic techniques, to facilitate lymphoma classification.
We framed our definition of BL in terms of high MYC and TCF3 transcriptional activity, because these are known major determinants of tumor behavior.4–6 DLBCL was defined by variable MYC activity, low TCF3 activity, and high BCL2 and targets of NF-κB.12 This limited signature was sufficient to categorize >90% of BL and DLBCL in the test set with high confidence and with perfect accuracy (Table 1). The results are comparable to those reported in a prior exploratory study comparing categorization of BL and non-BL using targeted GEP against a gold standard global GEP,25 and validate a molecular diagnostic classification for cases of well-defined BL and DLBCL.
BCL-Us are intermediate tumors that share features with BL and DLBCL, according to traditional diagnostic evaluation, but intermediate tumors are also identified by molecular analyses.1,12,13 Histomorphologically intermediate and molecularly intermediate are nonsynonymous terms and will categorize mature, aggressive B-cell lymphomas in different ways.42 For example, in our test cohort, three BCL-Us were classified as mBL. This must be considered inaccurate in the context of World Health Organization classification but is consistent with prior molecular characterization of B-cell lymphomas in which most atypical BLs and a proportion of unclassifiable aggressive B-cell lymphomas were classified as mBL.13 Similarly, few BLs, BCL-Us, and DLBCLs in our series had diagnostic molecular scores intermediate between mBL and mDLBCL. This result is also consistent with prior analyses in which subsets of atypical BL, unclassifiable aggressive B-cell lymphoma, and DLBCL were classified as molecularly intermediate.13 These results support the concept that BCL-U is not a discrete diagnostic category, but includes tumors with molecular profiles of mBL, mDLBCL, and intermediate between mBL and mDLBCL.
Non-BL with MYC rearrangement is also a heterogeneous group that includes tumors with the pathological diagnoses of BCL-U and DLBCL by World Health Organization criteria.1,42–46 We found DHLs that classified as mBL, molecularly intermediate, and mDLBCL. This result also has precedence. A comprehensive GEP analysis of aggressive B-cell lymphomas highlighted groups of DHLs that classified as mBL and MYC-rearranged DLBCLs that classified as mDLBCL.12
Our results were further supported by the examination of the molecular subsignatures and the histomorphological features of the DHLs. We found that a subset of DHLs have a TCF3 signature that is comparable to or exceeds that of BL. This result was surprising, given recent reports that the TCF3 signature is specific for BL,5,6 although a recent study found that ID3 mutations can occur in DHL.45 It will be of interest to correlate TCF3/ID3 mutation status with molecular diagnosis in future studies.
DHLs that classified as mBL were histomorphologically typical of BL and cases that classified as mDLBCL were histomorphologically typical of DLBCL. Morphological heterogeneity among DHLs is recognized and may have clinical significance.47 It will be important to determine, by using larger cohorts, whether the molecular classifier reliably identifies subsets of DHL with distinct histomorphological characteristics, and to relate these data to clinical outcomes.
The prognostic role of MYC in DLBCL is well established, especially in the context of BCL2 expression, and an assessment of MYC activity has been proposed to be an important part of the diagnostic workup.14–16,19–21,48 MYC IHC is a single biomarker that serves as a surrogate for MYC activity. The threshold for MYC IHC that separates low- from high-risk disease varies between studies, from 10% to 50%, with most suggesting 40%.14–16,19–21,49 However, IHC is difficult to standardize between centers, even if an automated platform is used.22 Therefore, we anticipated that the MYC activity scores would show good, but not perfect, correlation with MYC IHC scores, which we observed. There are several pre-analytical and analytical variables that we must consider when reviewing MYC IHC data, such as time to tissue fixation and intraobserver and interobserver variability in assessment. A potential advantage of expression profiling is that the analysis of many gene transcripts provides redundancy to the assay and captures a transcriptional signature of MYC activity that IHC for MYC alone cannot offer. However, comparing between a single data point (MYC protein expression by IHC) and the combination of a broad set of data (MYC activity score) is also likely to contribute to the observed imperfect correlation between the two methods of assessment. It is also possible that additional MYC targets, not included in our final profiling panel, would improve the validity of the MYC activity score.
The MYC activity classifier was trained using the GEPs of DLBCLs alone, excluding BLs. Its subsequent application to BLs in the training and test sets revealed high MYC activity scores for all cases, which supports the validity of the classifier. Moreover, five of six non-BLs with the highest MYC activity scores in the test set had MYC translocations. Yet, we also observed tumors with MYC translocations and intermediate/low scores, indicating variable MYC activity among SHLs and DHLs.13,44
To evaluate the clinical relevance of these data, we correlated the MYC activity scores to clinical outcome in a small series of R-CHOP–treated patients with primary, de novo DLBCL. Segregating tumors into those with high (>0.5) and low (<0.5) MYC activity scores identified patient populations that differed significantly with respect to overall survival (nominal P = 0.0009). The results provide evidence that the MYC activity score, while showing imperfect correlation with IHC and genetics, captures a biological signature of clinical significance. The limited number of primary DLBCLs with documented treatment and outcome required that we include cases from the training and test sets; therefore, a more formal validation of the MYC classifier using an independent case series is needed. Ideally, such a study would compare the interinstitutional reproducibility and the prognostic value of the MYC classifier with MYC IHC in a large, multi-institutional cohort.
In summary, we have developed a quantitative method for classifying and stratifying aggressive B-cell lymphomas that is applicable to FFPE tissue samples. The molecular classifiers are robust, but likely to improve with further testing and with the inclusion of additional, select gene signatures.24 In addition to distinguishing BL from DLBCL, the diagnostic classifier provides unique data regarding the further classification of BCL-Us and DHLs that inform the standard diagnostic methods and warrant further investigation. This platform will allow for the standardized analysis of an expanded cohort of BCL-Us and DHLs, from which correlations between GEP and traditional pathological, genetics, and somatic mutational analysis can be further examined. The MYC activity classifier captures a key biological and prognostic hallmark of DLBCL and also has the potential to standardize assessment across institutions. The ability of this classifier to predict outcome requires further validation, initially in a large independent cohort in which MYC IHC expression is known to be predictive of outcome, and then in the context of a clinical trial.
Acknowledgments
C.D.C., D.G., and S.M. designed and performed experiments, reviewed and analyzed data, and cowrote the manuscript. A.R.S. and M.J.K. designed experiments and reviewed and analyzed data. A.E.K., C.B., R.E.C., and D.N. reviewed and analyzed data. B.C. and M.A.S. reviewed and analyzed data and co-edited the manuscript. H.H.S., A.L., P.D.C., and L.P.L. performed experiments. S.J.R. designed experiments, reviewed and analyzed data, and cowrote the manuscript.
Footnotes
Supported, in part, by the Leukemia and Lymphoma Society Translational Research Award, NIH grant P01CA092625 (M.A.S.), Leukemia and Lymphoma Society grant 6446-13 (S.M.), a Bright Red grant (C.D.C.), and a Clinical Research Training Fellowship (reference number 13015) funded by Leukaemia and Lymphoma Research (UK) (C.D.C.).
Disclosures: None declared.
Supplemental Data
Schematic depicting target gene selection. TCF3 genes were derived from Schmitz et al5 and then validated in silico by differential analysis against gene-expression profiles (GEPs) of Burkitt lymphoma (BL) and diffuse large B-cell lymphoma (DLBCL) from prior publications.12,13 Transcripts with false discovery rates (FDR) <0.25 were ranked by signal/noise ratio, and the top 37 within the union of the two analyses were selected for the initial profiling panel. Analysis of expression data from the training cohort and the construction of molecular classifiers resulted in the inclusion of seven gene targets in the final probe set that was tested in the test cohort. MYC biological activity signature was derived from the DA of 457 published MYC targets31–37 against the global GEP of frozen DLBCLs with corresponding MYC IHC class (MYC IHC-High versus MYC IHC-Low15) from the training cohort. DA of the entire 18,400 transcripts within the GEP of the frozen tissue with respect to the corresponding MYC IHC class was also performed.
Unsupervised hierarchical clustering of data for three tumors (DLBCL20, DLBCL3, and DLBCL10) tested on more than one occasion during the test study. Heat map data are normalized to the six housekeeping (HK) genes but are not normalized between profiling panel builds. The profiling panel build and experiment number, the relative expression of the transcripts used in the final profiling panel, are shown (heat map and HK gene data not shown). The asterisk indicates mean MYC activity score. Bar charts of respective MYC activity scores by build and experiment number use the final classifier output, after normalization of data between profiling panel builds.
Kaplan-Meier curves showing overall survival (OS) for the outcome series where matched MYC IHC and MYC activity score data were available. Two cases lacked MYC IHC score; therefore, n = 38, rather than n = 40, as in Figure 6. A: Segregated by MYC IHC: MYC IHC-High >50% (red line) and MYC IHC-Low ≤50% (black line). B: Segregated by MYC activity score: MYC activity high (>0.5, red line) and MYC activity low (<0.5, black line).
References
- 1.Swerdlow SH, World Health Organization. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Geneva, Switzerland, WHO Press, 2008
- 2.Magrath I., Adde M., Shad A., Venzon D., Seibel N., Gootenberg J., Neely J., Arndt C., Nieder M., Jaffe E., Wittes R.A., Horak I.D. Adults and children with small non-cleaved-cell lymphoma have a similar excellent outcome when treated with the same chemotherapy regimen. J Clin Oncol. 1996;14:925–934. doi: 10.1200/JCO.1996.14.3.925. [DOI] [PubMed] [Google Scholar]
- 3.Habermann T.M. Rituximab-CHOP versus CHOP alone or with maintenance rituximab in older patients with diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:3121–3127. doi: 10.1200/JCO.2005.05.1003. [DOI] [PubMed] [Google Scholar]
- 4.Hecht J.L., Aster J.C. Molecular biology of Burkitt's lymphoma. J Clin Oncol. 2000;18:3707–3721. doi: 10.1200/JCO.2000.18.21.3707. [DOI] [PubMed] [Google Scholar]
- 5.Schmitz R., Young R.M., Ceribelli M., Jhavar S., Xiao W., Zhang M. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490:116–120. doi: 10.1038/nature11378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Love C., Sun Z., Jima D., Li G., Zhang J., Miles R., Richards K.L., Dunphy C.H., Choi W.W.L., Srivastava G., Lugar P.L., Rizzieri D.A., Lagoo A.S., Bernal-Mizrachi L., Mann K.P., Flowers C.R., Naresh K.N., Evens A.M., Chadburn A., Gordon L.I., Czader M.B., Gill J.I., Hsi E.D., Greenough A., Moffitt A.B., McKinney M., Banerjee A., Grubor V., Levy S., Dunson D.B., Dave S.S. The genetic landscape of mutations in Burkitt lymphoma. Nat Genet. 2012;44:1321–1325. doi: 10.1038/ng.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang J., Grubor V., Love C.L., Banerjee A., Richards K.L., Mieczkowski P.A. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morin R.D., Mungall K., Pleasance E., Mungall A.J., Goya R., Huff R., Scott D.W., Ding J., Roth A., Chiu R., Corbett R.D., Chan F.C., Mendez-Lago M., Trinh D.L., Bolger-Munro M., Taylor G., Hadj Khodabakhshi A., Ben-Neriah S., Pon J., Meissner B., Woolcock B., Farnoud N., Rogic S., Lim E., Johnson N.A., Shah S., Jones S., Steidl C., Holt R., Birol I., Moore R., Connors J.M., Gascoyne R.D., Marra M.A. Mutational and structural analysis of diffuse large B-cell lymphoma using whole genome sequencing. Blood. 2013;122:1256–1265. doi: 10.1182/blood-2013-02-483727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lohr J.G., Stojanov P., Lawrence M.S., Auclair D., Chapuy B., Sougnez C., Cruz-Gordillo P., Knoechel B., Asmann Y.W., Slager S.L. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Savage K.J., Johnson N.A., Ben-Neriah S., Connors J.M., Sehn L.H., Farinha P., Horsman D.E., Gascoyne R.D. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009;114:3533–3537. doi: 10.1182/blood-2009-05-220095. [DOI] [PubMed] [Google Scholar]
- 11.Barrans S., Crouch S., Smith A., Turner K., Owen R., Patmore R., Roman E., Jack A. Rearrangement of MYC is associated with poor prognosis in patients with diffuse large B-cell lymphoma treated in the era of rituximab. J Clin Oncol. 2010;28:3360–3365. doi: 10.1200/JCO.2009.26.3947. [DOI] [PubMed] [Google Scholar]
- 12.Dave S.S., Fu K., Wright G.W., Lam L.T., Kluin P., Boerma E.-J., Greiner T.C., Weisenburger D.D., Rosenwald A., Ott G. Molecular diagnosis of Burkitt's lymphoma. N Engl J Med. 2006;354:2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 13.Hummel M., Bentink S., Berger H., Klapper W., Wessendorf S., Barth T.F., Bernd H.-W., Cogliatti S.B., Dierlamm J., Feller A.C. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 14.Johnson N.A., Slack G.W., Savage K.J., Connors J.M., Ben-Neriah S., Rogic S., Scott D.W., Tan K.L., Steidl C., Sehn L.H., Chan W.C., Iqbal J., Meyer P.N., Lenz G., Wright G., Rimsza L.M., Valentino C., Brunhoeber P., Grogan T.M., Braziel R.M., Cook J.R., Tubbs R.R., Weisenburger D.D., Campo E., Rosenwald A., Ott G., Delabie J., Holcroft C., Jaffe E.S., Staudt L.M., Gascoyne R.D. Concurrent expression of MYC and BCL2 in diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30:3452–3459. doi: 10.1200/JCO.2011.41.0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kluk M.J., Chapuy B., Sinha P., Roy A., Cin P.D., Neuberg D.S., Monti S., Pinkus G.S., Shipp M.A., Rodig S.J. Immunohistochemical detection of MYC-driven diffuse large B-cell lymphomas. PLoS One. 2012;7:e33813. doi: 10.1371/journal.pone.0033813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou K., Xu D., Cao Y., Wang J., Yang Y., Huang M. C-MYC aberrations as prognostic factors in diffuse large B-cell lymphoma: a meta-analysis of epidemiological studies. PLoS One. 2014;9:e95020. doi: 10.1371/journal.pone.0095020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cook J.R., Goldman B., Tubbs R.R., Rimsza L., Leblanc M., Stiff P., Fisher R. Clinical significance of MYC expression and/or “high-grade” morphology in non-Burkitt, diffuse aggressive B-cell lymphomas: a SWOG S9704 correlative study. Am J Surg Pathol. 2014;38:494–501. doi: 10.1097/PAS.0000000000000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perry A.M., Alvarado-Bernal Y., Laurini J.A., Smith L.M., Slack G.W., Tan K.L., Sehn L.H., Fu K., Aoun P., Greiner T.C., Chan W.C., Bierman P.J., Bociek R.G., Armitage J.O., Vose J.M., Gascoyne R.D., Weisenburger D.D. MYC and BCL2 protein expression predicts survival in patients with diffuse large B-cell lymphoma treated with rituximab. Br J Haematol. 2014;165:382–391. doi: 10.1111/bjh.12763. [DOI] [PubMed] [Google Scholar]
- 19.Green T.M., Young K.H., Visco C., Xu-Monette Z.Y., Orazi A., Go R.S., Nielsen O., Gadeberg O.V., Mourits-Andersen T., Frederiksen M., Pedersen L.M., Moller M.B. Immunohistochemical double-hit score is a strong predictor of outcome in patients with diffuse large B-cell lymphoma treated with rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone. J Clin Oncol. 2012;30:3460–3467. doi: 10.1200/JCO.2011.41.4342. [DOI] [PubMed] [Google Scholar]
- 20.Horn H., Ziepert M., Becher C., Barth T.F.E., Bernd H.W., Feller A.C., Klapper W., Hummel M., Stein H., Hansmann M.L., Schmelter C., Moller P., Cogliatti S., Pfreundschuh M., Schmitz N., Trumper L., Siebert R., Loeffler M., Rosenwald A., Ott G., German High-Grade Non-Hodgkin Lymphoma Study Group MYC status in concert with BCL2 and BCL6 expression predicts outcome in diffuse large B-cell lymphoma. Blood. 2013;121:2253–2263. doi: 10.1182/blood-2012-06-435842. [DOI] [PubMed] [Google Scholar]
- 21.Hu S., Xu-Monette Z.Y., Tzankov A., Green T., Wu L., Balasubramanyam A. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121:4021–4031. doi: 10.1182/blood-2012-10-460063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jong D., Rosenwald A., Chhanabhai M., Gaulard P., Klapper W., Lee A., Sander B., Thorns C., Campo E., Molina T., Norton A., Hagenbeek A., Horning S., Lister A., Raemaekers J., Gascoyne R.D., Salles G., Weller E. Immunohistochemical prognostic markers in diffuse large B-cell lymphoma: validation of tissue microarray as a prerequisite for broad clinical applications–a study from the Lunenburg Lymphoma Biomarker Consortium. J Clin Oncol. 2007;25:805–812. doi: 10.1200/JCO.2006.09.4490. [DOI] [PubMed] [Google Scholar]
- 23.Rimsza L.M., Wright G., Schwartz M., Chan W.C., Jaffe E.S., Gascoyne R.D., Campo E., Rosenwald A., Ott G., Cook J.R., Tubbs R.R., Braziel R.M., Delabie J., Miller T.P., Staudt L.M. Accurate classification of diffuse large B-cell lymphoma into germinal center and activated B-cell subtypes using a nuclease protection assay on formalin-fixed, paraffin-embedded tissues. Clin Cancer Res. 2011;17:3727–3732. doi: 10.1158/1078-0432.CCR-10-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott D.W., Wright G.W., Williams P.M., Lih C.-J., Walsh W., Jaffe E.S., Rosenwald A., Campo E., Chan W.C., Connors J.M., Smeland E.B., Mottok A., Braziel R.M., Ott G., Delabie J., Tubbs R.R., Cook J.R., Weisenburger D.D., Greiner T.C., Glinsmann-Gibson B.J., Fu K., Staudt L.M., Gascoyne R.D., Rimsza L.M. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin embedded tissue. Blood. 2014;123:1214–1217. doi: 10.1182/blood-2013-11-536433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Masqué-Soler N., Szczepanowski M., Kohler C.W., Spang R., Klapper W. Molecular classification of mature aggressive B-cell lymphoma using digital multiplexed gene expression on formalin-fixed paraffin-embedded biopsy specimens. Blood. 2013;122:1985–1986. doi: 10.1182/blood-2013-06-508937. [DOI] [PubMed] [Google Scholar]
- 26.Linton K., Howarth C., Wappett M., Newton G., Lachel C., Iqbal J., Pepper S., Byers R., Chan W.J., Radford J. Microarray gene expression analysis of fixed archival tissue permits molecular classification and identification of potential therapeutic targets in diffuse large B-cell lymphoma. J Mol Diagn. 2012;14:223–232. doi: 10.1016/j.jmoldx.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 27.Alizadeh A.A., Eisen M.B., Davis R.E., Ma C., Lossos I.S., Rosenwald A., Boldrick J.C., Sabet H., Tran T., Yu X., Powell J.I., Yang L., Marti G.E., Moore T., Hudson J., Lu L., Lewis D.B., Tibshirani R., Sherlock G., Chan W.C., Greiner T.C., Weisenburger D.D., Armitage J.O., Warnke R., Levy R., Wilson W., Grever M.R., Byrd J.C., Botstein D., Brown P.O., Staudt L.M. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 28.Hans C.P. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 29.Bogusz A.M., Baxter R.H.G., Currie T., Sinha P., Sohani A.R., Kutok J.L., Rodig S.J. Quantitative immunofluorescence reveals the signature of active B-cell receptor signaling in diffuse large B-cell lymphoma. Clin Cancer Res. 2012;18:6122–6135. doi: 10.1158/1078-0432.CCR-12-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geiss G.K., Bumgarner R.E., Birditt B., Dahl T., Dowidar N., Dunaway D.L., Fell H.P., Ferree S., George R.D., Grogan T., James J.J., Maysuria M., Mitton J.D., Oliveri P., Osborn J.L., Peng T., Ratcliffe A.L., Webster P.J., Davidson E.H., Hood L. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 31.Zeller K.I., Jegga A.G., Aronow B.J., O'Donnell K.A., Dang C.V. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mori S., Rempel R.E., Chang J.T., Yao G., Lagoo A.S., Potti A., Bild A., Nevins J.R. Utilization of pathway signatures to reveal distinct types of B lymphoma in the E-myc model and human diffuse large B-cell lymphoma. Cancer Res. 2008;68:8525–8534. doi: 10.1158/0008-5472.CAN-08-1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schuhmacher M., Kohlhuber F., Hölzel M., Kaiser C., Burtscher H., Jarsch M., Bornkamm G.W., Laux G., Polack A., Weidle U.H., Eick D. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y.H., Girard L., Giacomini C.P., Wang P., Hernandez-Boussard T., Tibshirani R., Minna J.D., Pollack J.R. Combined microarray analysis of small cell lung cancer reveals altered apoptotic balance and distinct expression signatures of MYC family gene amplification. Oncogene. 2005;25:130–138. doi: 10.1038/sj.onc.1208997. [DOI] [PubMed] [Google Scholar]
- 35.Chapuy B., McKeown M.R., Lin C.Y., Monti S. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–790. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlosser I., Hölzel M., Hoffmann R., Burtscher H., Kohlhuber F., Schuhmacher M., Chapman R., Weidle U.H., Eick D. Dissection of transcriptional programmes in response to serum and c-Myc in a human B-cell line. Oncogene. 2004;24:520–524. doi: 10.1038/sj.onc.1208198. [DOI] [PubMed] [Google Scholar]
- 37.Yu D., Cozma D., Park A., Thomas-Tikhonenko A. Functional validation of genes implicated in lymphomagenesis: an in vivo selection assay using a Myc-induced B-cell tumor. Ann N Y Acad Sci. 2005;1059:145–159. doi: 10.1196/annals.1339.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Monti S., Chapuy B., Takeyama K., Rodig S.J., Hao Y., Yeda K.T., Inguilizian H., Mermel C., Currie T., Dogan A., Kutok J.L., Beroukhim R., Neuberg D., Habermann T.M., Getz G., Kung A.L., Golub T.R., Shipp M.A. Integrative analysis reveals an outcome-associated and targetable pattern of p53 and cell cycle deregulation in diffuse large B cell lymphoma. Cancer Cell. 2012;22:359–372. doi: 10.1016/j.ccr.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou H., Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc B. 2005;67:301–320. [Google Scholar]
- 40.Tibshirani R., Hastie T., Narasimhan B., Chu G. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci U S A. 2002;99:6567–6572. doi: 10.1073/pnas.082099299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breiman L. Random Forests. Machine Learning. 2001;45:5–32. [Google Scholar]
- 42.Salaverria I., Siebert R. The gray zone between Burkitt's lymphoma and diffuse large B-cell lymphoma from a genetics perspective. J Clin Oncol. 2011;29:1835–1843. doi: 10.1200/JCO.2010.32.8385. [DOI] [PubMed] [Google Scholar]
- 43.Snuderl M., Kolman O.K., Chen Y.-B., Hsu J.J., Ackerman A.M., Cin P.D., Ferry J.A., Harris N.L., Hasserjian R.P., Zukerberg L.R., Abramson J.S., Hochberg E.P., Lee H., Lee A.I., Toomey C.E., Sohani A.R. B-cell lymphomas with concurrent IGH-BCL2 and MYC rearrangements are aggressive neoplasms with clinical and pathologic features distinct from Burkitt lymphoma and diffuse large B-cell lymphoma. Am J Surg Pathol. 2010;34:327–340. doi: 10.1097/PAS.0b013e3181cd3aeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aukema S.M., Kreuz M., Kohler C.W., Rosolowski M., Hasenclever D., Hummel M., Küppers R., Lenze D., Ott G., Pott C., Richter J., Rosenwald A., Szczepanowski M., Schwaenen C., Stein H., Trautmann H., Wessendorf S., Trümper L., Loeffler M., Spang R., Kluin P.M., Klapper W., Siebert R. Biologic characterization of adult MYC-translocation positive mature B-cell lymphomas other than molecular Burkitt lymphoma. Haematologica. 2013;99:726–735. doi: 10.3324/haematol.2013.091827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gebauer N., Bernard V., Feller A.C., Merz H. ID3 mutations are recurrent events in double-hit B-cell lymphomas. Anticancer Res. 2013;33:4771–4778. [PubMed] [Google Scholar]
- 46.Gebauer N., Bernard V., Gebauer W., Thorns C., Feller A.C., Merz H. TP53 mutations are frequent events in double-hit B-cell lymphomas with MYC and BCL2 but not MYC and BCL6 translocations. Leuk Lymphoma. 2014 doi: 10.3109/10428194.2014.907896. http://dx.doi.org/10.3109/10428194.2014.907896 [Epub ahead of print May 7, 2014] [DOI] [PubMed] [Google Scholar]
- 47.Johnson N.A., Savage K.J., Ludkovski O., Ben-Neriah S., Woods R., Steidl C., Dyer M.J.S., Siebert R., Kuruvilla J., Klasa R., Connors J.M., Gascoyne R.D., Horsman D.E. Lymphomas with concurrent BCL2 and MYC translocations: the critical factors associated with survival. Blood. 2009;114:2273–2279. doi: 10.1182/blood-2009-03-212191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedberg J.W. Double-hit diffuse large B-cell lymphoma. J Clin Oncol. 2012;30:3439–3443. doi: 10.1200/JCO.2012.43.5800. [DOI] [PubMed] [Google Scholar]
- 49.Valera A., Lopez-Guillermo A., Cardesa-Salzmann T., Climent F., Gonzalez-Barca E., Mercadal S., Espinosa I., Novelli S., Briones J., Mate J.L., Salamero O., Sancho J.M., Arenillas L., Serrano S., Erill N., Martinez D., Castillo P., Rovira J., Martinez A., Campo E., Colomo L. MYC protein expression and genetic alterations have prognostic impact in diffuse large B-cell lymphoma treated with immunochemotherapy. Haematologica. 2013;98:1554–1562. doi: 10.3324/haematol.2013.086173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic depicting target gene selection. TCF3 genes were derived from Schmitz et al5 and then validated in silico by differential analysis against gene-expression profiles (GEPs) of Burkitt lymphoma (BL) and diffuse large B-cell lymphoma (DLBCL) from prior publications.12,13 Transcripts with false discovery rates (FDR) <0.25 were ranked by signal/noise ratio, and the top 37 within the union of the two analyses were selected for the initial profiling panel. Analysis of expression data from the training cohort and the construction of molecular classifiers resulted in the inclusion of seven gene targets in the final probe set that was tested in the test cohort. MYC biological activity signature was derived from the DA of 457 published MYC targets31–37 against the global GEP of frozen DLBCLs with corresponding MYC IHC class (MYC IHC-High versus MYC IHC-Low15) from the training cohort. DA of the entire 18,400 transcripts within the GEP of the frozen tissue with respect to the corresponding MYC IHC class was also performed.
Unsupervised hierarchical clustering of data for three tumors (DLBCL20, DLBCL3, and DLBCL10) tested on more than one occasion during the test study. Heat map data are normalized to the six housekeeping (HK) genes but are not normalized between profiling panel builds. The profiling panel build and experiment number, the relative expression of the transcripts used in the final profiling panel, are shown (heat map and HK gene data not shown). The asterisk indicates mean MYC activity score. Bar charts of respective MYC activity scores by build and experiment number use the final classifier output, after normalization of data between profiling panel builds.
Kaplan-Meier curves showing overall survival (OS) for the outcome series where matched MYC IHC and MYC activity score data were available. Two cases lacked MYC IHC score; therefore, n = 38, rather than n = 40, as in Figure 6. A: Segregated by MYC IHC: MYC IHC-High >50% (red line) and MYC IHC-Low ≤50% (black line). B: Segregated by MYC activity score: MYC activity high (>0.5, red line) and MYC activity low (<0.5, black line).