Abstract
In children, sporadic nephrotic syndrome can be related to a genetic cause, but to what extent genetic alterations associate with resistance to immunosuppression is unknown. In this study, we designed a custom array for next-generation sequencing analysis of 19 target genes, reported as possible causes of nephrotic syndrome, in a cohort of 31 children affected by sporadic steroid-resistant nephrotic syndrome and 38 patients who exhibited a similar but steroid-sensitive clinical phenotype. Patients who exhibited extrarenal symptoms, had a familial history of the disease or consanguinity, or had a congenital onset were excluded. We identified a genetic cause in 32.3% of the children with steroid-resistant disease but zero of 38 children with steroid-sensitive disease. Genetic alterations also associated with lack of response to immunosuppressive agents in children with steroid-resistant disease (0% of patients with alterations versus 57.9% of patients without alterations responded to immunosuppressive agents), whereas clinical features, age at onset, and pathologic findings were similar in steroid‐resistant patients with and without alterations. These results suggest that heterogeneous genetic alterations in children with sporadic forms of nephrotic syndrome associate with resistance to steroids as well as immunosuppressive treatments. In these patients, a comprehensive screening using such an array may, thus, be useful for genetic counseling and may help clinical decision making in a fast and cost-efficient manner.
Keywords: genetic variant, proteinuria, high-throughput DNA sequencing, immunosuppression
Nephrotic syndrome represents one of the most common diagnoses in pediatric nephrology, with a prevalence of 16 per 100,000 children in Western countries.1–3 In most cases, the pathogenesis of nephrotic syndrome remains elusive, and the clinical phenotype of patients does not allow discrimination among different causes. Thus, children with nephrotic syndrome are usually treated with corticosteroids before any diagnostic procedure, and approximately 80% of them respond to such a treatment. According to this observation, pediatric nephrotic syndrome has been separated into two broad categories (i.e., steroid-sensitive nephrotic syndrome [SSNS] and steroid-resistant nephrotic syndrome [SRNS]). Although children affected by SSNS have good long-term prognosis, most patients with SRNS progress to ESRD within 2–10 years of diagnosis.1–3 Multiple putative causes of SRNS have been postulated, such as mutations in genes regulating the functional integrity of the podocyte as well as permeability factors or other environmental agents altering the podocyte membrane.4–6 Currently, at least 19 genes have been clearly identified as causes of SRNS,2,7–12 conferring a considerable genetic heterogeneity to the disorder. Extrapolations from selected child case series for single genes seemed to suggest that only a small percentage of total nephrotic syndrome cases results from defects of a single gene. Recent studies suggested that the frequency may be higher after several genes are analyzed at the same time,12 but they mostly included patients with a familial history, an early onset of the disease, or a syndromic picture, who are likely affected by genetic forms. Thus, in this study, we specifically analyzed the prevalence of genetic defects in children with sporadic forms of nephrotic syndrome (Supplemental Table 1), and we hypothesized that the presence of genetic alterations in podocyte genes may significantly associate with resistance to steroids as well as immunosuppressive treatments in these patients.
Results
To this aim, we analyzed 19 genes reported as possible causes of nephrotic syndrome (Supplemental Table 2A) using next-generation sequencing (NGS) technology in 31 children affected by sporadic SRNS as well as 38 additional patients who exhibited a similar clinical phenotype but were responsive to steroids (Supplemental Table 1). To focus the analysis on those patients who cannot be clinically distinguished except for their response to steroids or immunosuppressive treatments, patients who exhibited extrarenal symptoms, had a familial history, or had a congenital onset were excluded. To eventually identify novel genes associated with this disorder, 27 other candidate genes expressed in the glomerular filtration barrier were also included in this array (Supplemental Table 2B).
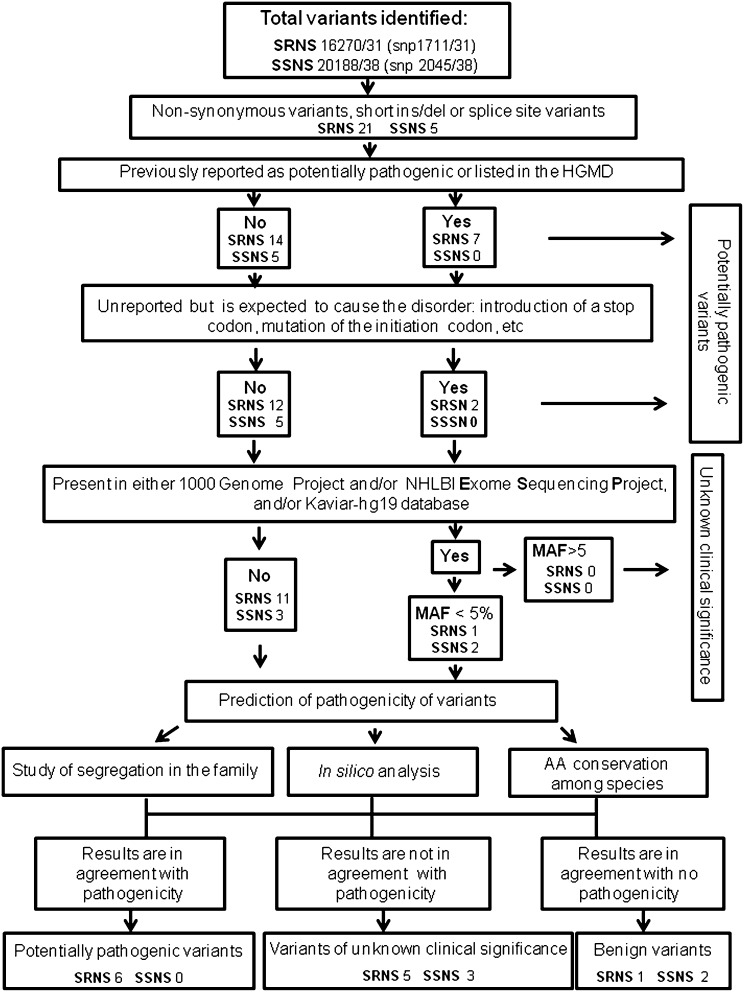
To prioritize the identified variants, we followed the algorithm that is shown in Figure 1 and detailed in the Supplemental Material. On the basis of the results, the variants were classified as potentially pathogenic variants, variants of unknown clinical significance, or benign variants in accordance with the American College of Medical Genetics and Genomics (ACMG) guidelines.13–15 Of 69 probands, 10 carried 17 mutated alleles that satisfied criteria for a molecular diagnosis (Table 1). More specifically, in five patients, we observed different variants in the NPHS2 gene (two homozygous and three compound heterozygous) that were already described as potentially pathogenic.16–20 In patient 1, we observed the non-neutral polymorphic variant –52 C>G, which was previously reported to reduce NPHS2 transcription by abolishing the binding of the regulatory factor upstream transcription factor 1 (USF1) to the NPHS2 promoter.16 This patient was homozygous, having inherited the two variants from each of two nonaffected parents, and displayed a significantly reduced podocin expression at the biopsy (Supplemental Figure 1), even if he was affected by a minimal change disease, where levels of podocin are usually normal.21 In patients 3–5, podocin was extremely reduced or virtually absent in the available bioptic specimens (Supplemental Figure 1). In two patients, we observed two unreported variants in the PLCE1 gene (both compound heterozygous). Because these variants resulted in the introduction of a stop codon and were consistent with the phenotype of the patients, they were recognized as potentially pathogenic in accordance with the interpretation of the ACMG guidelines. In all patients affected by NPHS2 or PLCE1 autosomal recessive disease, potentially pathogenic variants were inherited from each of two nonaffected parents. Finally, three other patients showed heterozygous variants in the dominantly transmitted ACTN4 and LMX1B genes. These variants were not previously reported, but they were localized in highly conserved domains of the protein (Supplemental Figure 2), predicted to be pathogenic by in silico analysis (Table 1), and consistent with the phenotype of the patients.13 More importantly, both the mutations in the ACTN4 gene had occurred de novo. By contrast, the variant in the LMX1B gene was inherited by the father, who did not have a clear history of renal disease and only showed microhematuria and mild proteinuria on medical check-up. However, after the LMX1B variant was identified, other possible clinical abnormalities of the nail-patella syndrome were carefully searched in the child. Although the nails and kneecaps (patellae) appeared normal and iliac horns were not found, on radiologic examination, the child revealed a bilateral lack of ossification of the proximal radial epiphysis (Supplemental Figure 3). This abnormal phenotype, even if asymptomatic, was previously reported as related to LMX1B mutations.22
Figure 1.
Flow diagram showing mechanisms for filtering variants to identify the variants of potential pathogenicity. AA, amino acid; HGMD, Human Gene Mutation Database; ins/del, insertion/deletion; MAF, minor allele frequency; NHLBI, National Heart, Lung, and Blood Institute; snp, single nucleotide polymorphism.
Table 1.
Potentially pathogenic variants identified in the patients included in the study
| Samples | Potentially Pathogenic Variants | State of Variants | Inheritance | Refs. | Prediction of Pathogenicity of Variants | ||
|---|---|---|---|---|---|---|---|
| In Silico Analysis | AA Conservation among Species | ||||||
| PolyPhen | PMut | ||||||
| Patient 1 | NPHS2 c.[-52C>G]+[-52C>G] | Homozygous | AR | Oleggini et al.16 | |||
| Patient 2 | NPHS2 c.[419delG]+[419delG]; p.[Gly140Aspfs*41]+[Gly140Aspfs*41] | Homozygous | AR | Boute et al.17 | |||
| Patient 3 | NPHS2 c.[413G>A]+[467_468insT]; p.[Arg138Gln]+[Leu156Phefs*11] | Compound heterozygous | AR | Boute et al.17; Caridi et al.18 | |||
| Patient 4 | NPHS2 c.[104insG]+[1143delC]; p.[Gly35Glyfs*35]+[Pro381Profs*5] | Compound heterozygous | AR | Boute et al.17; Berdeli et al.19 | |||
| Patient 5 | NPHS2 c. [771C>T]+[911C>T]; p.[Arg229Gln]+[Ser304Phe] | Compound heterozygous | AR | Tsukaguchi et al.20 | Pos dam | Path | ++a |
| Patient 6 | PLCE1 c.[4570_4571delAT]+[2277G>T]; p.[Met1524Alafs*5]+[Arg548Leu] | Compound heterozygous | AR | Prob dam | Path | +++b | |
| Patient 7 | PLCE1 c.[2038C>T]+[4327G>A]; p.[Gln680*]+[Gly1443Arg] | Compound heterozygous | AR | Prob dam | Path | +++b | |
| Patient 8 | ACTN4 c.[782C>A]+[=]; p.[Val261Glu]+[=] | Heterozygous | AD | Prob dam | Path | +++b | |
| Patient 9 | ACTN4 c.[464T>C]+[=]; p.[Ile155Thr]+[=] | Heterozygous | AD | Prob dam | Neu | +++b | |
| Patient 10 | LMX1B c.[833C>T]+[=]; p.[Ala278Val]+[=] | Heterozygous | AD | Prob dam | Path | +++b | |
AR, autosomal recessive; AD, autosomal dominant; Pos dam, possibly damaging; Path, pathologic; Prob dam, probably damaging; Neu, neutral; AA, amino acid.
Medium conservation.
High conservation.
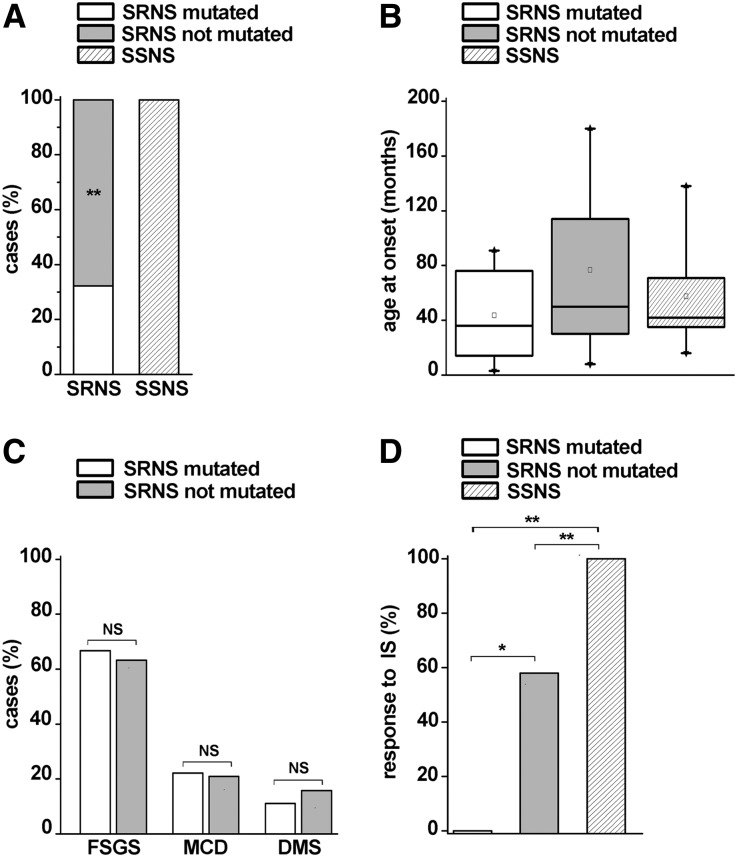
We then retrospectively collected clinicopathologic information about patients (Supplemental Tables 1 and 3), evaluating potential associations with the results of the genetic screening. As mentioned, we observed potentially pathogenic mutations in 10 of 69 patients: strikingly, all were steroid-resistant, underlying a prevalence of 32.3% of genetic forms among patients with sporadic SRNS (Figure 2A). By contrast, no genetic alterations were found in the group of patients with SSNS (Figure 2A) (chi-squared=11.85; P<0.001). The age at onset of nephrotic syndrome was comparable between the group of SRNS with potentially pathogenic mutations (median of 3 years and 6 months), the group of SRNS that was negative to the genetic screening (median of 4 years and 2 months), and the group of SSNS (median of 3 years and 9 months; Mann–Whitney U test: all not significant) (Figure 2B). Although differences in age distribution may become significant if a larger number of patients would be analyzed, they are extremely small, and age at onset largely overlaps in these three groups of patients, suggesting that it is difficult to distinguish sporadic patients carrying genetic mutations on the basis of this clinical criterion. In addition, pathologic diagnosis on renal biopsy was available in 28 patients with SRNS (Supplemental Table 3). No significant difference between patients with mutated and not mutated SRNS was found in the frequency of FSGS (66.7% versus 63.2%; NS), minimal change disease (22.1% versus 21%; NS), and diffuse mesangial sclerosis (11.1% versus 15.8%; NS) (Figure 2C), showing that neither the clinical features nor the pathologic phenotype can distinguish between these two groups. By contrast, response to immunosuppressive treatments was significantly different in mutated and not mutated SRNS patients. Indeed, six of 10 children with mutated SRNS were treated with immunosuppressive drugs, but none of them responded to the treatment (Supplemental Table 3). However, a response to immunosuppressors was observed in 57.9% of children affected by SRNS without mutations (chi-squared=4.08; P<0.05) (Figure 2D). As expected, all patients with SSNS responded to immunosuppressive drugs. Finally, 26 patients with SRNS were treated with renin-angiotensin system inhibitors (RAS-Is). Although too small to draw effective conclusions, the number of patients who responded to RAS-Is was not significantly different between patients with mutated and not mutated SRNS (33.3% versus 47.1%; NS). These results suggest that at least a subset of mutated children may benefit from treatment with RAS-Is.
Figure 2.
Analysis of patients’ genotypes in relation to age at onset of nephrotic syndrome and response to treatments. (A) Percentage of mutations in patients with SRNS or SSNS. Patients with mutated SRNS are represented in white, and patients with not mutated SRNS are represented in gray. Patients with SSNS are represented in the striped column. (B) Age at onset in patients affected by nephrotic syndrome. Patients with SRNS are classified as mutated (white box) or not mutated (gray box); Patients with SSNS are represented in the striped box. In each box, the bottom and top of the box are always the first and third quartiles, and the band inside the box is always the second quartile (the median). Squares inside each box represent the mean value for each group. The whiskers indicate variability outside the upper and lower quartiles, with the end of the whiskers representing fifth and 95th percentiles. Triangles represent first and 99th percentiles in each box. (C) Percentage of FSGS, minimal change disease (MCD), and diffuse mesangial sclerosis (DMS) pathologic findings in patients with mutated SRNS (white column) versus not mutated SRNS (gray column). (D) Percentage of responders to immunosuppressive treatment (IS) in patients with mutated SRNS (white column), not mutated SRNS (gray column), and SSNS (striped column). *P<0.05; **P<0.001.
Discussion
The advent of high-throughput DNA sequencing to find causal genes has lead us to better appreciate that many simple mendelian genetic diseases are anything but simple.23 This finding is particularly true for nephrotic syndrome, which is characterized by an extremely variable prognosis and response to treatment instead of a homogeneous clinical phenotype. However, the enormous potential of NGS technology represents “the reverse side of the medal,” leading to a sheer volume of new data that is overwhelming the capacity of institutions to manage it and limiting its use in clinical practice. Thus, in this study, we established an exhaustive strategy on the basis of screening through NGS of the largest number of genes involved in SRNS evaluated so far. This technique allowed us to analyze 46 genes in up to 12 patients in only 4 weeks with a cost of $830 per patient. In comparison, Sanger sequencing has a cost of $500 per gene, and therefore, a similar analysis would cost $10,000 per patient and require many months. Thus, this method has the advantage of a more feasible dataset for a bioinformatic analysis, and it is fast and cost-efficient, which makes it functionally interpretable and suitable for clinical practice.
This innovative strategy allowed us to establish the existence of potentially pathogenic variants in 32.3% of 31 patients with sporadic steroid-resistance, whereas using the same technical approach, zero of 38 additional patients who exhibited a similar clinical phenotype but were steroid-sensitive had genetic mutations in the analyzed genes. The identified genetic alterations included compound heterozygosity of recessive genes, which is known as a potential cause of FSGS,24 as well as de novo mutations or mutations in low-penetrance dominant genes. These types of mutations explain why mendelian disorders can be frequently observed in patients with sporadic SRNS, even in absence of consanguinity or familial history, and they suggest that, in a child who displays resistance to steroids, an extended genetic screening may help in providing appropriate genetic counseling to the family. More importantly, by specifically analyzing only sporadic cases of SRNS, this study shows that resistance to immunosuppressive treatments in children with nephrotic syndrome is frequently associated with heterogeneous genetic mutations in podocyte genes. Indeed, patients with sporadic SRNS carrying genetic mutations exhibited similar clinical features and pathologic findings compared with patients with not mutated SRNS but significantly differed for the rate of response to immunosuppressive drugs. Interestingly, although FSGS was the most frequent pathologic finding observed in patients with mutated SRNS, two thirds of cases of FSGS in patients with SRNS were observed in patients with not mutated SRNS. Considering that about 30% of FSGS in children is responsive to steroids,25,26 our results suggest that the predictive power of a bioptic finding of FSGS for a genetic cause is rather low. Consistently, a recent study that analyzed genetic mutations in children affected by sporadic FSGS without considering the response to treatment found a frequency of potentially pathogenic mutations of about 10%.27
Our study also has some limitations. Indeed, it is a retrospective study that analyzed all patients with sporadic nephrotic syndrome who were referred to our hospital and gave informed consent to the study. Although it reflects clinical practice, it may have generated bias among different groups that are not strictly matched. Thus, these conclusions will need to be confirmed through blinded, prospective studies. In addition, the number of patients studied is relatively small and cannot explain the reason for lack of response to immunosuppressive treatment in a relevant percentage of patients with not mutated SRNS. However, new causative genes for nephrotic syndrome are continuously discovered, and variants of unknown significance that could still act as disease modifiers were not considered in our analysis, suggesting that the role of genetic abnormalities in inducing resistance to treatments may still be underestimated. Interestingly, some reports showed that individuals with nephrotic syndrome caused by genetic mutations may occasionally be partially responsive to immunosuppressive agents.18,28–33 We suggest that, in a subset of patients carrying minor genetic weakness of the podocyte, the disorder may even sometimes recognize a combined etiopathogenesis, being initiated by environmental agents or permeability factors that can be occasionally modulated by immunosuppression.
Despite the limitations reported above, the results of this study suggest that this type of genetic analysis may improve the approach to children with sporadic nephrotic syndrome by promoting better genetic counseling and management of the treatment.
Concise Methods
In total, 69 patients affected by sporadic, nonsyndromic, and noncongenital nephrotic syndrome referred to the Meyer Children's Hospital of Florence, Italy from 2000 to 2013 and who provided informed consent were included in the study. Steroid-resistance was defined as failure to achieve remission after 8 weeks of prednisone (60 mg/m2 per day or 2 mg/kg per day for 4 weeks followed by 40 mg/m2 or 1.5 mg/kg on alternate days for 4 weeks) according to Kidney Disease Improving Global Outcomes (KDIGO) guidelines.34 None of these patients exhibited a phenotype of late responder to steroid treatment. Complete remission, partial remission, and absence of remission were defined according to KDIGO guidelines.34 On the basis of these criteria, we selected 31 patients with SRNS as well as 38 patients with the same clinical phenotype who were sensitive to steroid treatment (Supplemental Tables 1 and 3). All information about clinical features and response to treatment was collected retrospectively. DNA was extracted from peripheral blood using a QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), and the DNA libraries were constructed using Roche (Madison, WI) sample preparation protocol (Rapid Library Preparation Method Manual). A Roche NimbleGen sequence capture array in a solution-based method was used to capture all coding exons and flanking regions of 46 genes according to the manufacturer’s protocol (NimbleGen Arrays User’s Guide: 454 Optimized Sequence Capture). The array included 19 known genes responsible for nephrotic syndrome (Supplemental Table 2A); to potentially identify putative new genes, the other 27 candidate genes associated with proteinuria in animal models and expressed in the glomerular filtration barrier were included (Supplemental Table 2B). The enriched libraries were sequenced using the Roche 454 Sequencing FLX Platform according to the manufacturer’s protocols (emPCR Method Manual-Lib-L LV-Sequencing Method Manual). Reads were mapped to the reference human genome (UCSC build hg19) with the GS Mapper software, version 2.6 (Roche Diagnostic, Basel, Switzerland). Coverage statistics and mean depth were extracted from the mapping output files using custom scripts. To identify genetic variants with a possible pathogenic significance, the following criteria were applied. Variants were nonsynonymous, short insertion/deletion, or splice site variants (60-bp splice acceptor and 60-bp splice donor); variants were not included in dbSNP135 unless they were reported in the Human Gene Mutation Database (www.hgmd.cf.ac.uk), previously shown to alter the function of the mutated protein, or represented the second mutated allele in the presence of a surely pathogenic variant.35 Variants did not occur in healthy controls within the 1000 Genomes Project database (www.1000genomes.org) at a frequency >5%; variants did not occur within the Kaviar-hg19 database (http://db.systemsbiology.net/kaviar/) at a frequency >5%. Variants were not found in the Exome Variant Server of the National Heart, Lung, and Blood Institute Exome Sequencing Project (http://evs.gs.washington.edu/EVS/; release version: v.0.0.10) at a frequency >5%. Subsequently, a systematic classification scheme was applied using a combination of prediction programs. To distinguish potentially damaging variants from those variants predicted to have neutral effect, missense variants were analyzed with the dedicated softwares PolyPhen (http://genetics.bwh.harvard.edu/pph2/) and PMut (http://mmb.pcb.ub.es/PMut/PMut.jsp). Intronic variants localized ≤60 bp were analyzed with Netgene2 (http://genes.mit.edu/GENSCAN.html) and Berkeley Drosophila Genome Project splice site prediction (http://www.fruitfly.org/seq_tools/splice.html web server). The comparison of the amino acid sequence of all new identified variants with those of phylogenetically distant species was performed using CLUSTALW algorithm. On the basis of the results, variants were classified as potentially pathogenic variants, variants of unknown clinical significance, or benign variants in accordance with the interpretation guidelines of the ACMG.13–15 Identified variants were confirmed by Sanger sequencing (Supplemental Table 4). Parental samples, if available, were also analyzed to determine whether the mutated allele had been transmitted. Confocal microscopy was performed on 5-µm sections of renal biopsies from frozen tissues36 using a Leica TCS SP5-II laser confocal microscope (Leica, Milan, Italy) and the anti-podocin polyclonal antibody (Abcam, Inc., Cambridge, UK). Statistical analysis was performed using SPSS software (SPSS, Inc., Evanston, IL). Frequencies between groups were compared by chi-squared test, applying Yate correction when appropriate. Between-groups comparisons were made by means of t test for unpaired data or Mann–Whitney U test owing to normal or nonparametric distribution. A P value <0.05 was considered as statistically significant.
Disclosures
None.
Supplementary Material
Acknowledgments
We thank Dr. Michele Iacono (Application and Bioinformatics, Applied Science Roche Diagnostics SpA, Monza, Italy) for his valued contribution in the next-generation sequencing data analysis and Prof. Gian Marco Ghiggeri (Department of Nephrology, Gaslini Children Hospital, Genoa, Italy).
This work was supported by a contribution from the Tuscan Association for the Study of Childhood Renal Diseases (A.Ma.R.T.I.) and by the Meyer Children’s Hospital Foundation.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013111155/-/DCSupplemental.
References
- 1.Haraldsson B, Nyström J, Deen WM: Properties of the glomerular barrier and mechanisms of proteinuria. Physiol Rev 88: 451–487, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Hildebrandt F: Genetic kidney diseases. Lancet 375: 1287–1295, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benoit G, Machuca E, Antignac C: Hereditary nephrotic syndrome: A systematic approach for genetic testing and a review of associated podocyte gene mutations. Pediatr Nephrol 25: 1621–1632, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei C, Trachtman H, Li J, Dong C, Friedman AL, Gassman JJ, McMahan JL, Radeva M, Heil KM, Trautmann A, Anarat A, Emre S, Ghiggeri GM, Ozaltin F, Haffner D, Gipson DS, Kaskel F, Fischer DC, Schaefer F, Reiser J, PodoNet and FSGS CT Study Consortia : Circulating suPAR in two cohorts of primary FSGS. J Am Soc Nephrol 23: 2051–2059, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torban E, Bitzan M, Goodyer P: Recurrent focal segmental glomerulosclerosis: A discrete clinical entity. Int J Nephrol 2012: 246128, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cravedi P, Kopp JB, Remuzzi G: Recent progress in the pathophysiology and treatment of FSGS recurrence. Am J Transplant 13: 266–274, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tryggvason K, Patrakka J, Wartiovaara J: Hereditary proteinuria syndromes and mechanisms of proteinuria. N Engl J Med 354: 1387–1401, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Löwik MM, Groenen PJ, Levtchenko EN, Monnens LA, van den Heuvel LP: Molecular genetic analysis of podocyte genes in focal segmental glomerulosclerosis—a review. Eur J Pediatr 168: 1291–1304, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piscione TD, Licht C: Genetics of proteinuria: An overview of gene mutations associated with nonsyndromic proteinuric glomerulopathies. Adv Chronic Kidney Dis 18: 273–289, 2011 [DOI] [PubMed] [Google Scholar]
- 10.Machuca E, Benoit G, Antignac C: Genetics of nephrotic syndrome: Connecting molecular genetics to podocyte physiology. Hum Mol Genet 18[R2]: R185–R194, 2009 [DOI] [PubMed] [Google Scholar]
- 11.Saleem MA: New developments in steroid-resistant nephrotic syndrome. Pediatr Nephrol 28: 699–709, 2013 [DOI] [PubMed] [Google Scholar]
- 12.McCarthy HJ, Bierzynska A, Wherlock M, Ognjanovic M, Kerecuk L, Hegde S, Feather S, Gilbert RD, Krischock L, Jones C, Sinha MD, Webb NJ, Christian M, Williams MM, Marks S, Koziell A, Welsh GI, Saleem MA, RADAR the UK SRNS Study Group : Simultaneous sequencing of 24 genes associated with steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 8: 637–648, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE, Molecular Subcommittee of the ACMG Laboratory Quality Assurance Committee : ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet Med 10: 294–300, 2008 [DOI] [PubMed] [Google Scholar]
- 14.Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, Braxton A, Beuten J, Xia F, Niu Z, Hardison M, Person R, Bekheirnia MR, Leduc MS, Kirby A, Pham P, Scull J, Wang M, Ding Y, Plon SE, Lupski JR, Beaudet AL, Gibbs RA, Eng CM: Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med 369: 1502–1511, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacob HJ: Next-generation sequencing for clinical diagnostics. N Engl J Med 369: 1557–1558, 2013 [DOI] [PubMed] [Google Scholar]
- 16.Oleggini R, Bertelli R, Di Donato A, Di Duca M, Caridi G, Sanna-Cherchi S, Scolari F, Murer L, Allegri L, Coppo R, Emma F, Camussi G, Perfumo F, Ghiggeri GM: Rare functional variants of podocin (NPHS2) promoter in patients with nephrotic syndrome. Gene Expr 13: 59–66, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boute N, Gribouval O, Roselli S, Benessy F, Lee H, Fuchshuber A, Dahan K, Gubler MC, Niaudet P, Antignac C: NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet 24: 349–354, 2000 [DOI] [PubMed] [Google Scholar]
- 18.Caridi G, Bertelli R, Carrea A, Di Duca M, Catarsi P, Artero M, Carraro M, Zennaro C, Candiano G, Musante L, Seri M, Ginevri F, Perfumo F, Ghiggeri GM: Prevalence, genetics, and clinical features of patients carrying podocin mutations in steroid-resistant nonfamilial focal segmental glomerulosclerosis. J Am Soc Nephrol 12: 2742–2746, 2001 [DOI] [PubMed] [Google Scholar]
- 19.Berdeli A, Mir S, Yavascan O, Serdaroglu E, Bak M, Aksu N, Oner A, Anarat A, Donmez O, Yildiz N, Sever L, Tabel Y, Dusunsel R, Sonmez F, Cakar N: NPHS2 (podicin) mutations in Turkish children with idiopathic nephrotic syndrome. Pediatr Nephrol 22: 2031–2040, 2007 [DOI] [PubMed] [Google Scholar]
- 20.Tsukaguchi H, Sudhakar A, Le TC, Nguyen T, Yao J, Schwimmer JA, Schachter AD, Poch E, Abreu PF, Appel GB, Pereira AB, Kalluri R, Pollak MR: NPHS2 mutations in late-onset focal segmental glomerulosclerosis: R229Q is a common disease-associated allele. J Clin Invest 110: 1659–1666, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mao J, Zhang Y, Du L, Dai Y, Yang C, Liang L: Expression profile of nephrin, podocin, and CD2AP in Chinese children with MCNS and IgA nephropathy. Pediatr Nephrol 21: 1666–1675, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Sweeney E, Fryer A, Mountford R, Green A, McIntosh I: Nail patella syndrome: A review of the phenotype aided by developmental biology. J Med Genet 40: 153–162, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teer JK, Mullikin JC: Exome sequencing: The sweet spot before whole genomes. Hum Mol Genet 19[R2]: R145–R151, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huber TB, Kwoh C, Wu H, Asanuma K, Gödel M, Hartleben B, Blumer KJ, Miner JH, Mundel P, Shaw AS: Bigenic mouse models of focal segmental glomerulosclerosis involving pairwise interaction of CD2AP, Fyn, and synaptopodin. J Clin Invest 116: 1337–1345, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davin JC, Rutjes NW: Nephrotic syndrome in children: From bench to treatment. Int J Nephrol 2011: 372304, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fogo AB: The spectrum of FSGS: Does pathology matter? Nephrol Dial Transplant 25: 1034–1036, 2010 [DOI] [PubMed] [Google Scholar]
- 27.Laurin LP, Lu M, Mottl AK, Blyth ER, Poulton CJ, Weck KE: Podocyte-associated gene mutation screening in a heterogeneous cohort of patients with sporadic focal segmental glomerulosclerosis [published online ahead of print April 21, 2014]. Nephrol Dial Transplant [DOI] [PubMed] [Google Scholar]
- 28.Malina M, Cinek O, Janda J, Seeman T: Partial remission with cyclosporine A in a patient with nephrotic syndrome due to NPHS2 mutation. Pediatr Nephrol 24: 2051–2053, 2009 [DOI] [PubMed] [Google Scholar]
- 29.Hinkes B, Wiggins RC, Gbadegesin R, Vlangos CN, Seelow D, Nürnberg G, Garg P, Verma R, Chaib H, Hoskins BE, Ashraf S, Becker C, Hennies HC, Goyal M, Wharram BL, Schachter AD, Mudumana S, Drummond I, Kerjaschki D, Waldherr R, Dietrich A, Ozaltin F, Bakkaloglu A, Cleper R, Basel-Vanagaite L, Pohl M, Griebel M, Tsygin AN, Soylu A, Müller D, Sorli CS, Bunney TD, Katan M, Liu J, Attanasio M, O’toole JF, Hasselbacher K, Mucha B, Otto EA, Airik R, Kispert A, Kelley GG, Smrcka AV, Gudermann T, Holzman LB, Nürnberg P, Hildebrandt F: Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet 38: 1397–1405, 2006 [DOI] [PubMed] [Google Scholar]
- 30.Plank C, Kalb V, Hinkes B, Hildebrandt F, Gefeller O, Rascher W, Arbeitsgemeinschaft für Pädiatrische Nephrologie : Cyclosporin A is superior to cyclophosphamide in children with steroid-resistant nephrotic syndrome-a randomized controlled multicentre trial by the Arbeitsgemeinschaft für Pädiatrische Nephrologie. Pediatr Nephrol 23: 1483–1493, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ruf RG, Lichtenberger A, Karle SM, Haas JP, Anacleto FE, Schultheiss M, Zalewski I, Imm A, Ruf EM, Mucha B, Bagga A, Neuhaus T, Fuchshuber A, Bakkaloglu A, Hildebrandt F, Arbeitsgemeinschaft Für Pädiatrische Nephrologie Study Group : Patients with mutations in NPHS2 (podocin) do not respond to standard steroid treatment of nephrotic syndrome. J Am Soc Nephrol 15: 722–732, 2004 [DOI] [PubMed] [Google Scholar]
- 32.Gellermann J, Stefanidis CJ, Mitsioni A, Querfeld U: Successful treatment of steroid-resistant nephrotic syndrome associated with WT1 mutations. Pediatr Nephrol 25: 1285–1289, 2010 [DOI] [PubMed] [Google Scholar]
- 33.Büscher AK, Kranz B, Büscher R, Hildebrandt F, Dworniczak B, Pennekamp P, Kuwertz-Bröking E, Wingen AM, John U, Kemper M, Monnens L, Hoyer PF, Weber S, Konrad M: Immunosuppression and renal outcome in congenital and pediatric steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol 5: 2075–2084, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kidney Disease: Improving Global Outcomes (KDIGO) Glomerulonephritis Work Group. KDIGO Clinical Practice Guideline for Glomerulonephritis. Kidney Int Suppl 2: S139–S174, 2012 [Google Scholar]
- 35.Bell CJ, Dinwiddie DL, Miller NA, Hateley SL, Ganusova EE, Mudge J, Langley RJ, Zhang L, Lee CC, Schilkey FD, Sheth V, Woodward JE, Peckham HE, Schroth GP, Kim RW, Kingsmore SF: Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci Transl Med 3: ra4, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Angelotti ML, Ronconi E, Ballerini L, Peired A, Mazzinghi B, Sagrinati C, Parente E, Gacci M, Carini M, Rotondi M, Fogo AB, Lazzeri E, Lasagni L, Romagnani P: Characterization of renal progenitors committed toward tubular lineage and their regenerative potential in renal tubular injury. Stem Cells 30: 1714–1725, 2012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.