Abstract
Genomic aberrations affecting the genes encoding factor H (FH) and the five FH-related proteins (FHRs) have been described in patients with atypical hemolytic uremic syndrome (aHUS), a rare condition characterized by microangiopathic hemolytic anemia, thrombocytopenia, and ARF. These genomic rearrangements occur through nonallelic homologous recombinations caused by the presence of repeated homologous sequences in CFH and CFHR1-R5 genes. In this study, we found heterozygous genomic rearrangements among CFH and CFHR genes in 4.5% of patients with aHUS. CFH/CFHR rearrangements were associated with poor clinical prognosis and high risk of post-transplant recurrence. Five patients carried known CFH/CFHR1 genes, but we found a duplication leading to a novel CFHR1/CFH hybrid gene in a family with two affected subjects. The resulting fusion protein contains the first four short consensus repeats of FHR1 and the terminal short consensus repeat 20 of FH. In an FH-dependent hemolysis assay, we showed that the hybrid protein causes sheep erythrocyte lysis. Functional analysis of the FHR1 fraction purified from serum of heterozygous carriers of the CFHR1/CFH hybrid gene indicated that the FHR1/FH hybrid protein acts as a competitive antagonist of FH. Furthermore, sera from carriers of the hybrid CFHR1/CFH gene induced more C5b-9 deposition on endothelial cells than control serum. These results suggest that this novel genomic hybrid mediates disease pathogenesis through dysregulation of complement at the endothelial cell surface. We recommend that genetic screening of aHUS includes analysis of CFH and CFHR rearrangements, particularly before a kidney transplant.
Keywords: hemolytic uremic syndrome, genetic renal disease, kidney disease, complement, transplantation
Atypical hemolytic uremic syndrome (aHUS) is a rare condition characterized by microangiopathic hemolytic anemia, thrombocytopenia, and ARF.1,2 Genetic and acquired abnormalities causing dysregulation of the alternative pathway (AP) of complement have been found in 50%–60% of cases.1 About 20% of aHUS patients carry mutations in CFH encoding the major regulator of the AP: factor H (FH).3,4 Anti-FH autoantibodies have been reported in 10% of patients.5–7
The CFH gene and CFHR1-R5 encoding five FH-related (FHR) proteins are located in tandem in the regulators of complement activation cluster at chromosome 1q32.8 The high level of sequence identity among CFH and CFHRs suggests that this region is the result of genomic duplications that occurred during the evolution.9 The presence of repeated sequences favors genomic rearrangements through nonallelic homologous recombination (NAHR). The most frequently observed NAHR is the deletion of CFHR3-CFHR1 that is strongly associated with anti-FH autoantibodies and aHUS.7,10–12 Several hybrid genes deriving from NAHR in the CFH-CFHR region have been also identified in aHUS,13–15 including hybrid CFH/CFHR1 and CFH/CFHR3 genes. Recently, a hybrid CFHR1/CFH gene that encodes a fusion protein with the first three short consensus repeats (SCRs) of FHR1 and the last two SCRs of FH has been reported in a sporadic case of aHUS.16
Here, we describe a novel CFHR1/CFH hybrid gene in a familial form of aHUS. The resulting fusion protein contains the first four SCRs of FHR1 and the terminal SCR20 of FH. Functional studies revealed that the hybrid protein causes complement dysregulation at the cell surface by acting as a competitive antagonist of FH.
Results
Genomic CFH-CFHRs Rearrangements
Among 154 aHUS patients (67 patients with complement gene mutations) screened by multiplex ligation-dependent probe amplification (MLPA), we found genomic rearrangements involving CFH-CFHRs in seven patients (4.5%), of whom only one patient also had a heterozygous mutation (c.1429+1G>C in CFI). Two cousins with disease onset at the ages of 6 months and 20 years (the former also carried the CFI mutation)3 and two patients with sporadic aHUS (onset: 1 and 22 years) had a previously described heterozygous CFH/CFHR1 hybrid gene including exons 1–21 of CFH and exons 5 and 6 of CFHR1.13 In a sporadic patient (onset at 5 months), we found another previously reported hybrid CFH/CFHR114 including exons 1–22 of CFH and exon 6 of CFHR1. Four of five carriers of CFH/CFHR1 hybrid genes developed ESRD early after disease onset. Three of them manifested disease recurrence in the graft, whereas the fourth received post-transplant eculizumab prophylaxis and had good allograft function at 10 months follow-up.
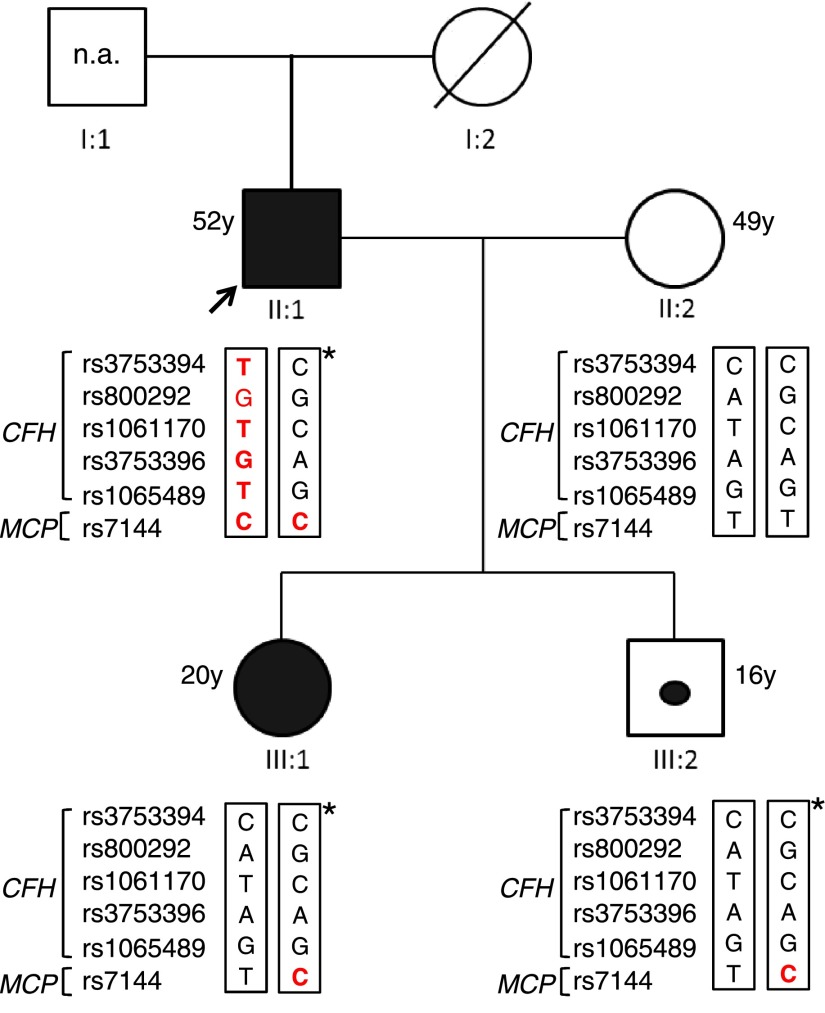
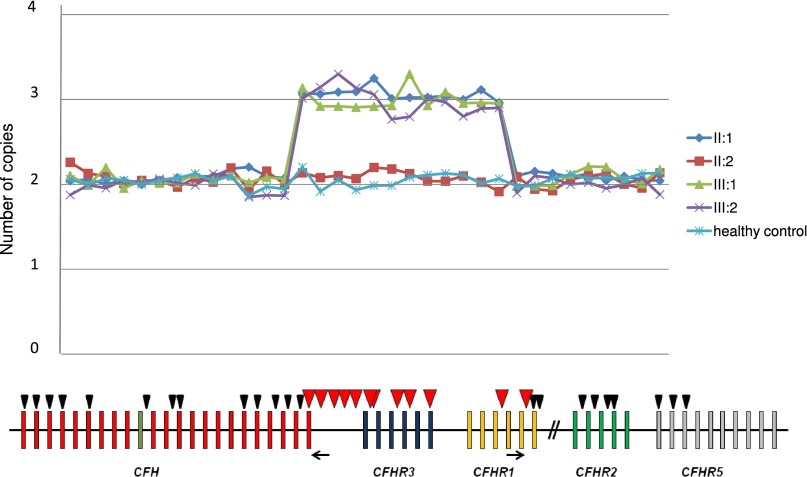
In addition, in two affected patients from a family (the proband and his daughter in Figure 1), MLPA revealed three copies from probes 14–25 (Table 1), suggesting a large heterozygous duplication starting from exon 23 of CFH, including CFHR3, and ending after exon 5 of CFHR1 (Figure 2). The duplication was confirmed by comparative genomic hybridization (CGH) array (Figure 3). The proband’s wife was normal, whereas the unaffected son carried the heterozygous duplication like his affected relatives (Figures 1 and 2).
Figure 1.
Italian family pedigree. The proband (black arrow) is patient II:1, his wife is II:2, his affected daughter is III:1, and the unaffected son is III:2. Patients II:1 and III:1 are represented as a black square and circle, respectively. The unaffected carrier III:2 is indicated by a black dot. Genotype of CFH single nucleotide polymorphisms targeting the CFH-H3 risk (TGTGT) haplotype (rs3753394 c.1–332 C>T, rs800292 c.184G>A p.V62I, rs1061170 c.1204T>C p.Y402H, rs3753396 c.2016A>G p.Q672Q, and rs1065489 c.2808 G>T p.E936D) and the MCP single nucleotide polymorphism (rs7144 c.*897 T>C) targeting the MCPggaac risk haplotype are reported in red. The age of all subjects is shown. n.a., samples not available. *Chromosome with the duplication.
Table 1.
MLPA probes used to determine CFH-CFHR copy number
| Probe No. | Probe Name | Hybridization Sequence (20 Nucleotides Adjacent to Ligation Site) |
|---|---|---|
| 1 | CFH exon 1 | TGCTACACAA-ATAGCCCATA |
| 2 | CFH exon 2 | GGTCTGACCA-AACATATCCA |
| 3 | CFH exon 3 | TCCTTTTGGT-ACTTTTACCC |
| 4 | CFH exon 4 | ATTACCGTGA-ATGTGACACA |
| 5 | CFH exon 6 | AAAGAGGAGA-TGCTGTATGC |
| 6 | CFH intron 10 | TAGGTAGTCA-TATTTGGAAC |
| 7 | CFH intron 12 | TGGACACATT-ATGATTGAGT |
| 8 | CFH exon 13 | AGTTGGACCT-AATTCCGTTC |
| 9 | CFH exon 18 | GGAACCATTA-ATTCATCCAG |
| 10 | CFH exon 19 | AGGATGTGTA-TAAGGCGGGG |
| 11 | CFH intron 20 | GAATTCTATT-TACACTTCCG |
| 12 | CFH intron 21 | TAATAGGGTA-TATTATTTTC |
| 13 | CFH intron 22 | GAAAAATCTC-TGTGATGAGT |
| 14a | CFH exon 23 | AGCTTTATTC-GAGAACAGGT |
| 15a | CFH intron 23 | TCAATACATA-AATGCACCAA |
| 16a | CFH intron 23 | CACTTATACA-TGCAATCCGT |
| 17a | CFH intron 23 | AGTCCGAGGT-AGAAAGGGAC |
| 18a | CFH intron 23 | GTGGTAATCT-TGGCTCTCAG |
| 19a | CFHR3 intron 1 | AGGTAAGTTA-AAAGAGATCT |
| 20a | CFHR3 intron 1 | CATTTTCTTG-TGGAATTACAGC |
| 21a | CFHR3 intron 3 | CGGACGACAG-TCTCAGACTT |
| 22a | CFHR3 intron 4 | GGGTTATATG-AATTCCTACA |
| 23a | CFHR3 exon 6 | TCCCTTCCCG-ACACACTGCTTG |
| 24a | CFHR1 intron 3 | AGAGTTTCAG-GTCCATGTGT |
| 25a | CFHR1 intron 5 | AATCTGTGAT-TATTTTGTTA |
| 26 | CFHR1 exon 6 | CCTGTTCTCA-AATAAAGCTTCT |
| 27 | CFHR1 exon 6 | TTTTCCAAGT-TTTAATATGG |
| 28 | CFHR2 intron 1 | TGTCTGTACT-TGGAGTTTCG |
| 29 | CFHR2 intron 2 | AGATCATAAA-CACTTGATAA |
| 30 | CFHR2 intron 3 | AATACCTGTG-TGTGGTTTATAG |
| 31 | CFHR2 exon 4 | ATATGCTCCAGG-TTCATCAGTT |
| 32 | CFHR5 exon 1 | TGGGTATCCA-CTGTTGGGGG |
| 33 | CFHR5 exon 2 | TGAAGAAGAT-TATAACCCTT |
| 34 | CFHR5 exon 3 | CTTCAGGACT-AATACATCTG |
Probes 10–14 were designed within our laboratory. The remaining probes are from the MRC Holland kit (SALSA MLPA P236-A3 ARMD Kit).
Probes spanning the duplication.
Figure 2.
Results of MLPA analysis showing a novel CFHR1/CFH hybrid gene. MLPA analysis over the CFH-CFHR region shows three copies in a large region beginning with CFH exon 23 and ending after CFHR1 exon 5 in patients II:1 (proband), III:1 (affected daughter), and III:2 (unaffected son) but not II:2 (unaffected wife). The analysis has been performed with the SALSA MLPA P236-A3 ARMD Kit (MCR Holland) implemented with homemade probes analyzed in a separate assay and covering the last exons and introns of the CFH gene (exon 19, intron 20, intron 21, intron 22, and exon 23). MLPA probes are represented by triangles. Red triangles indicate duplicated probes. The positions of primers for breakpoint mapping are shown by black arrows.
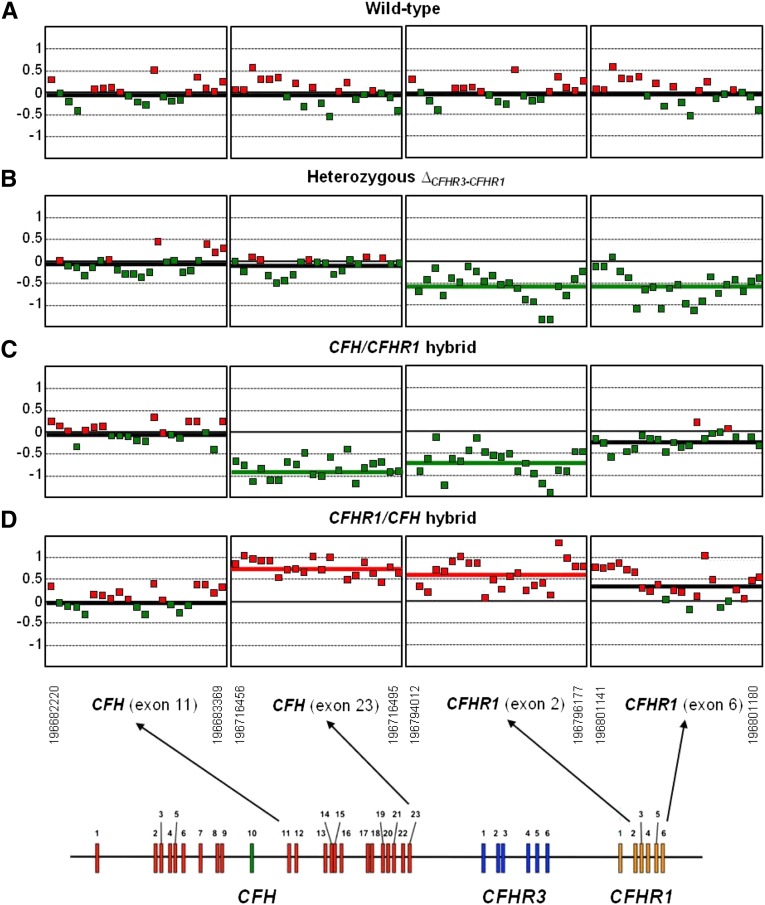
Figure 3.
CGH array analysis confirms the presence of the CFHR1/CFH hybrid gene. DNAs from four different individuals are compared: (A) a wild-type individual (carrying two copies of CFH, CFHR3, and CFHR1), (B) an individual heterozygous for the CFHR3-CFHR1 deletion, (C) a carrier in heterozygosis of the CFH/CFHR1 hybrid gene, and (D) the proband in this study carrying in heterozygosis the new CFHR1/CFH hybrid gene. As indicated in the diagram at the bottom, only the fragments of the array corresponding to CFH exon 11, CFH exon 23, CFHR1 exon 2, and CFHR1 exon 6 are shown. A thick colored line within each fragment indicates the average DNA gain (red) or loss (green). Notice that, in the carrier of the CFHR1/CFH hybrid gene, CFH exon 23 and CFHR1 exon 2 are overrepresented, confirming the heterozygous duplication in this region. For comparison, the reverse situation occurs in the carrier of the CFH/CFHR1 hybrid gene.
Breakpoint Mapping and Identification of a CFHR1/CFH Hybrid Gene
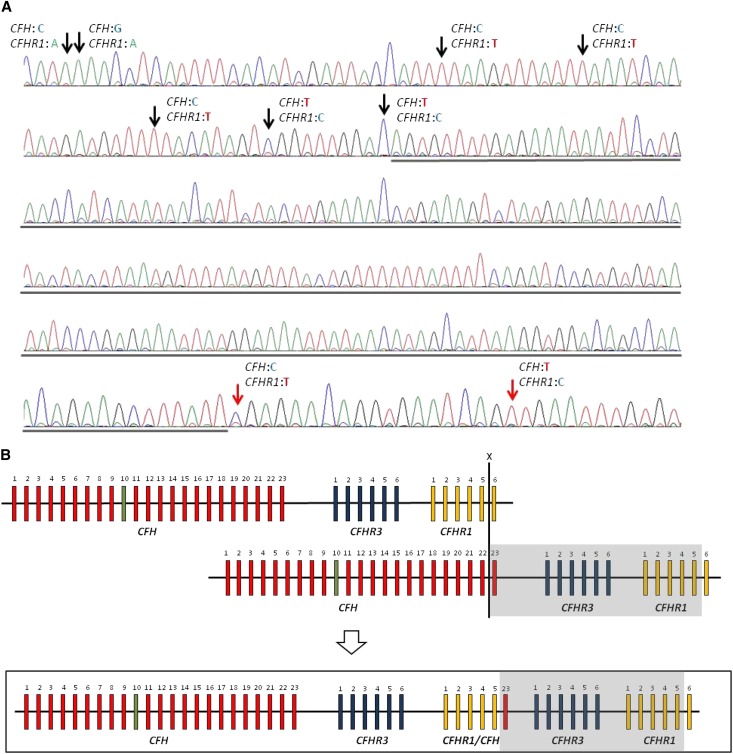
A 1955-bp PCR product was obtained from the DNA of the proband, his affected daughter, and his unaffected son using specific primers annealing CFHR1 intron 4 (forward) and CFH intron 23 (reverse) (Figure 2), whereas no PCR product was obtained with control DNA (Supplemental Figure 1A). Bidirectional sequencing of the long-range PCR product (primers are shown in Supplemental Table 1) revealed that the breakpoint is within a 193-bp region starting 999 bp after CFHR1 exon 5 (Figure 4). These results imply that an NAHR event occurred between the last exon of CFH and the last exon of CFHR1 (Figure 4B), resulting in a CFHR1/CFH hybrid and an extra copy each of CFHR3 and CFHR1 (Figure 4B). The CFHR1/CFH hybrid encodes for a protein with the first four SCRs of FHR1 and the last SCR20 of FH (Figure 5A). The rearrangement was confirmed by multiplex PCR-amplifying fragments with different sizes for the CFHR1/CFH hybrid and the wild-type CFHR1 in carriers of the duplication and only the wild-type CFHR1 fragment in noncarriers (Supplemental Figure 1B).
Figure 4.
Identification of the genomic breakpoint. (A) The breakpoint is located in a region of 193 bp (underlined) between intron 5 of CFHR1 and exon 23 of CFH. The limits of the region are identified by single nucleotide polymorphism differences between CFH and CFHR1 indicated by arrows. Black arrows indicate CFHR1-specific single nucleotide polymorphism variants, and red arrows indicate CFH-specific single nucleotide polymorphism variants. (B) NAHR occurring between CFH and CFHR1 at the positions indicated by the X results in the formation of the CFHR1/CFH hybrid gene consisting of the first five exons of CFHR1, exon 23 of CFH, and an extra copy each of CFHR3 and CFHR1 (boxed area). The duplicated region is highlighted in gray.
Figure 5.
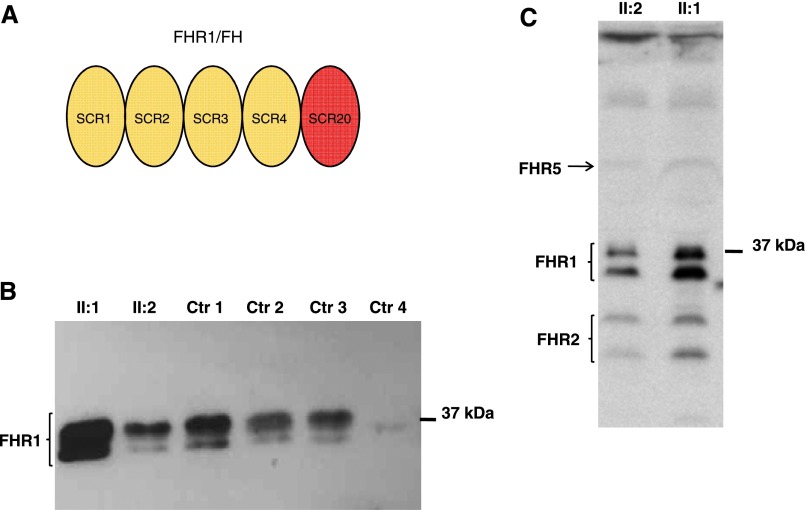
The FHR1/FH hybrid protein. (A) The protein product of the CFHR1/CFH hybrid gene is a five-SCR protein, where SCR1, -2, -3, and -4 are derived from FHR1 and SCR5 is from FH. (B) Western blot of the FHR1/FH hybrid protein was performed using a polyclonal goat anti-FH antibody against sera from the proband with the heterozygous duplication and the hybrid CFHR1/CFH gene (II:1), his wife (II:2) with two normal copies of CFHR1, a control with two copies of CFHR1 (Ctr 1), two controls with one copy of CFHR1 (Ctr 2 and Ctr 3), and a control with zero copies of CFHR1 (Ctr 4). The presence of the FHR1/FH hybrid protein accounts for the increased band density seen in the proband (II:1). (C) Western blot of the FHR1/FH hybrid protein was performed using a mouse anti–FHR1-FHR2-FHR5 mAb against sera from the proband (II:1) and his wife (II:2). The densities of the two bands of FHR1 isoforms in the proband serum are more pronounced than the densities in his wife’s serum, suggesting the presence of secreted FHR1/FH hybrid protein.
Clinical, Laboratory, and Genetic Data of the Family with the CFHR1/CFH Hybrid Gene
The proband (Figure 1) was hospitalized at the Molinette Hospital, Turin, Italy at the age of 49 years with malignant hypertension, severe ARF (serum creatinine=7.1 mg/dl), anemia (hemoglobin [Hb]=7.9 g/dl), and thrombocytopenia (platelets=109,000/μl) that manifested 1 week after an upper respiratory tract infection. His past medical history was uneventful. Laboratory examinations showed lactate dehydrogenase=2704 IU/L, serum haptoglobin<3 mg/dl, total bilirubin=2.4 mg/dl, and hematuria. Tests for autoantibodies (ANA, anti-DNA, ANCA, anticardiolipin, and lupus anticoagulant) were negative. The presence of Shiga toxin–producing Escherichia coli in stool culture was excluded, and ADAMTS13 activity was normal. A diagnosis of aHUS was made. Antihypertensive polytherapy with minoxidil, clonidine, doxazosin, and nifedipine was started. The patient was also treated with steroids, packed red blood cell transfusions, plasma infusions, and plasma exchanges. Despite treatment, the patient became anuric, and hemodialysis was started without recovering renal function. After 2 months, the patient was discharged under antihypertensive therapy in hematologic remission (Hb=11 g/dl, platelets=234,000/μl) but on chronic hemodialysis. At the last follow-up, 3 years after aHUS onset, the patient was in stable condition and waiting for renal transplantation. Serum C3, C4, and FH and plasma SC5b-9 levels were in the normal range; the test for anti-FH antibodies was negative (Table 2).
Table 2.
Complement assessment of all subjects of the family with hybrid gene
| Complement Parameters | II:1 (on Dialysis) | III:1 | II:2 | III:2 | |
|---|---|---|---|---|---|
| Before Eculizumab | After Eculizumab | ||||
| C3 (83–177 mg/dl) | 111 | 79 | 91 | n.a. | n.a. |
| C4 (15–45 mg/dl) | 26 | 19 | n.a. | n.a. | n.a. |
| CH50 (79–187 units Eq/ml) | n.a. | 226 | 5 | n.a. | n.a. |
| sC5b-9 (127–303 ng/ml) | 236 | 329 | 543 | 194 | 242 |
| FH (120–560 mg/L) | 164 | 242 | 156 | 136 | 133 |
| Ab anti-FH (AU/ml) | Absent | Absent | Absent | Absent | Absent |
The normal ranges were set as mean±2 SD of the values recorded in healthy subjects. The proband is subject II:1, his affected daughter is III:1, his wife is II:2, and the unaffected son is III:2. n.a., not available.
His daughter (Figure 1) was hospitalized at 20 years of age because of severe hemolytic anemia (Hb=5.1 g/dl, lactate dehydrogenase=2000 IU/L), thrombocytopenia (platelets=111,000/μl), schistocytes in the peripheral smear, hypertension, ARF (serum creatinine=6 mg/dl), microhematuria, and proteinuria (4.2 g/24 h). She was taking birth control pills. ADAMTS13 activity was normal, and Coombs test was negative. Shiga toxin–producing E. coli was not detected in stool culture. aHUS was suspected, and the patient was treated with daily plasma infusions, packed red blood cell transfusions, steroids, furosemide, and two Ig infusions. Hypertension was controlled by amlodipine and doxazosin. Because of persistence of hemolysis and severe renal dysfunction, seven daily plasma exchanges were performed without benefit. Serum C3 levels were slightly lower than normal, serum C4 and plasma SC5b-9 were normal, and anti-FH antibodies were absent (Table 2). On the thirteenth day of hospitalization, the patient started treatment with eculizumab (four 900-mg weekly doses and then 1200 mg every 2 weeks) after meningococcal vaccination and under prophylaxis with ciprofloxacin, obtaining hematologic remission and progressive renal function improvement. After 2 months, the patient was discharged, with serum creatinine=1.88 mg/dl, Hb=9 g/dl, platelets=270,000/μl, proteinuria=2.4 g/24 h, under 1200 mg eculizumab every 2 weeks, and antihypertensive therapy. At the last follow-up, 7 months after onset, the patient was in stable remission (serum creatinine=1.5 mg/dl, Hb=12.8 g/dl, platelets=294,000/μl, proteinuria=1.4 g/24 h) with eculizumab treatment every 2 weeks. Serum C3 was normal, whereas plasma SC5b-9 was slightly elevated (Table 2). Total complement CH50 activity was heavily depressed (Table 2) because of chronic eculizumab treatment. Family history did not disclose other relatives with aHUS.
Genotyping for the CFH single nucleotide polymorphisms rs3753394, rs800292, rs1061170, rs3753396, rs1065489, and rs7144 in the membrane cofactor protein (MCP) gene showed that the proband was heterozygous for the aHUS risk haplotype CFH-H3 (TGTGT) on the chromosome without the duplication and homozygous for the MCP allele c.*897 T>C (rs7144) that tags the MCPggaac risk haplotype. Neither the affected daughter nor the unaffected son, both carrying the duplication, showed the CFH-H3 risk haplotype, whereas both were heterozygous for the MCP c.*897 T>C risk allele (Figure 1).
FHR1 Western Blotting
Western blot analysis of the proband’s serum showed FHR1 bands with higher density than serum from his wife, consistent with the presence of an extra CFHR1 copy encoding the FHR1/FH fusion protein (Figure 5, B and C).
Hemolytic and Competition Assays
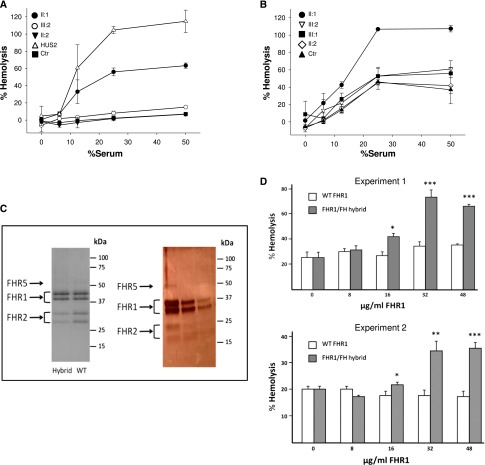
The FH C terminus is crucial for FH binding to cell surfaces. It is expected that, in the FHR1/FH hybrid protein reported here, the replacement of the FHR1 C-terminal region by the C terminus of FH will confer to the protein the capacity to compete with FH for binding to cell surfaces, thus antagonizing FH complement regulatory activity. To test this possibility, we evaluated whether the serum from heterozygous carriers of the CFHR1/CFH hybrid lyses sheep erythrocytes in an FH-dependent hemolysis assay. Serum from the proband lysed sheep cells in a dose-dependent manner, whereas normal serum and serum from the proband’s wife without the duplication did not (Figure 6A). A replica of this experiment, setting the lysis obtained with the proband serum as 100%, revealed a lower degree of lysis with serum from the affected daughter or the unaffected son, both carriers of the CFHR1/CFH hybrid (Figure 6B).
Figure 6.
Hemolytic and competition assays show that FHR1/FH antagonizes FH-dependent complement regulation on sheep erythrocytes. (A and B) Lysis of sheep erythrocytes by serum from an aHUS patient with the C-terminal mutation W1183L (HUS2) is compared with that obtained with sera from the proband (II:1; with the heterozygous duplication and the hybrid CFHR1/CFH gene), his affected daughter (III:1) and his unaffected son (III:2; both with the heterozygous duplication and the hybrid CFHR1/CFH gene), his wife (II:2; with two normal copies of CFHR1), and a healthy control (Ctr). Lysis is shown as a function of the volume of serum added and is represented as a percentage, setting to 100% either (A) the lysis obtained with 25% serum from the W1183L mutant patient or (B) the lysis obtained with 25% serum from the proband. Data are means±SDs of triplicates. (C, right panel) Coomassie-stained gel of the final purified FHR1 preparations from heterozygous carriers of the CFHR1/CFH hybrid gene (Hybrid) or normal wild-type controls (WT) used in the assays. (C, left panel) Silver-stained gel of the hybrid FHR1/FH-containing fractions eluted from the gel filtration column (containing FHR1, FHR2, and FHR5). In the Coomassie-stained gel, the purified FHR1 preparations from the carriers of the hybrid protein and healthy subjects were normalized to contain identical concentrations of FHR1 protein. Notice that the relative intensities of the FHR1, FHR2, and FHR5 proteins in this gel indicate that there is apparently a smaller amount of FHR2 and FHR5 compared with FHR1 in the carriers of the CFHR1/CFH hybrid gene. This result is consistent with these individuals carrying an extra copy of the CFHR1 gene. The silver-stained gel shows that the preparations are free from contaminants. (D) FHR1 fractions purified from normal controls (white) or heterozygous carriers of the CFHR1/CFH hybrid gene (gray) were added two different control sera (experiments 1 and 2), in which the native FHR1 protein was removed and functional FH was titrated to about 50% by using the anti–N-terminal FH antibody OX24 (6.5 μg/ml OX24 in 10% normal human serum). In both experiments, adding purified FHR1 from heterozygous carriers of the CFHR1/CFH hybrid gene resulted in dose-dependent sheep erythrocyte hemolysis, whereas purified FHR1 from normal serum had no effect. Data are means±SDs of triplicates. *P<0.05, **P<0.01, ***P<0.001 versus WT (purified FHR1 from normal serum).
To evaluate whether the lysis obtained with the proband’s serum was a consequence of the competition between FH and the FHR1/FH hybrid protein, we purified the FHR1 fraction from the serum of hybrid protein carriers and tested whether purified FHR1 added to normal human serum competed with FH and caused lysis in a modified FH-dependent sheep hemolysis assay. FHR1, FHR2, and FHR5 do not exist as individual molecular entities in human plasma. They are present as a complex mix of heterooligomeric proteins (dimers and tetramers), in which the oligomers containing FHR1 are mostly the major components.17 We, therefore, refer to the purified mix of FHR1 (including the hybrid FHR1/FH), FHR2, and FHR5 heterooligomers here as purified FHR1 (Figure 6C). Addition of purified FHR1 from heterozygous CFHR1/CFH hybrid carriers to two different control sera resulted in dose-dependent sheep erythrocyte hemolysis, whereas equal amounts of purified FHR1 from normal serum had no effect (Figure 6D).
C5b-9 Deposition on Endothelial Cells
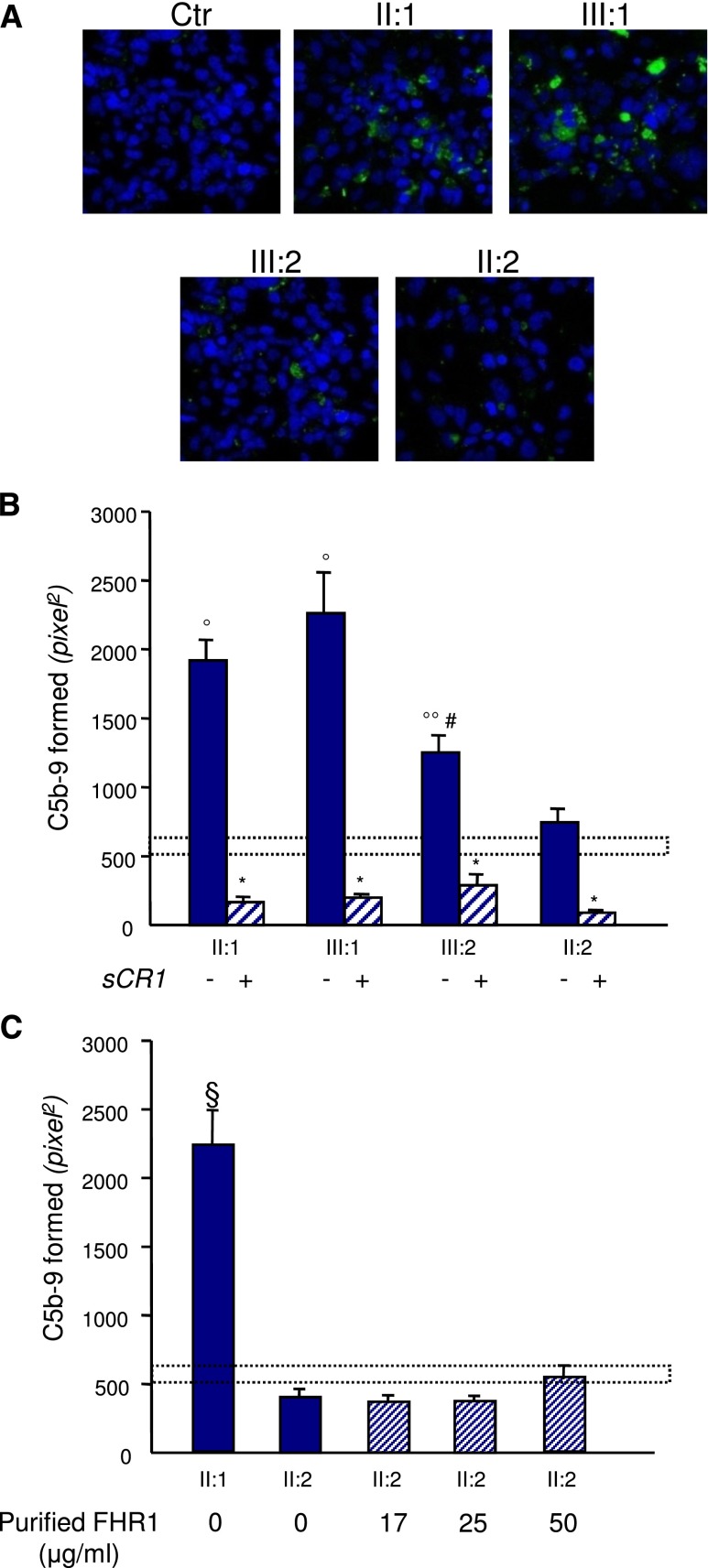
To determine whether the presence of the FHR1/FH hybrid protein in the circulation resulted in complement dysregulation at the endothelial cell level, the human microvascular endothelial cell line-1 (HMEC-1) preactivated with ADP was exposed to serum from the proband or his relatives, and the surface area covered by C5b-9 deposits was evaluated. We found more C5b-9 deposition on endothelial cells exposed to sera from the proband (in remission) and his affected daughter (with acute disease) versus cells incubated with control serum or serum from the proband’s wife (Figure 7, A and B). Interestingly, serum from the unaffected son who carries the FHR1/FH hybrid also induced higher than normal C5b-9 deposits on HMEC-1, although to a lesser extent than sera from his affected relatives (Figure 7, A and B). Addition of increasing amounts of purified FHR1 from normal serum to serum from the wild-type proband’s wife had no effect on C5b-9 deposits (Figure 7C).
Figure 7.
Sera from CFHR1/CFH carriers cause C5b-9 deposition on endothelial cells. Panel A shows representative confocal microscopy images of C5b-9 staining (green) of ADP-activated HMEC-1 exposed for 4 hours to serum (diluted 1:2 in test medium) from a healthy control (Ctr), the proband (II:1; with the heterozygous duplication and the hybrid CFHR1/CFH gene), his affected daughter (III:1) and his unaffected son (III:2; both with the heterozygous duplication and the hybrid CFHR1/CFH gene), and his wife (II:2; with two normal copies of CFHR1). Original magnification, ×400. Blue indicates the 4′,6-diamidino-2-phenylindole staining of cell nuclei. (B) The graph shows the quantification of HMEC-1 area covered by C5b-9 deposits after incubation with serum from a healthy control, the proband, and relatives of the proband in the presence or not of the complement inhibitor sCR1 (150 µg/ml). The dotted rectangle shows the range of C5b-9 deposits induced by a pool of control sera. Data are means±SEMs. *P<0.001 versus serum untreated; °P<0.001, °°P<0.05 versus control; #P<0.05 versus patients. (C) Quantification of HMEC-1 area covered by C5b-9 deposits after incubation with serum from the proband or the healthy wife of the proband with or without the addition of increasing amounts of purified FHR1 from normal serum to mimic the product of one (17 and 25 μg/ml) or two (50 μg/ml) extra CFHR1 gene copies (details are in Supplemental Material). FHR1 addition to serum from the proband’s wife had no effect on C5b-9 deposits. Data are means±SEMs. The dotted rectangle shows the range of C5b-9 deposits induced by a pool of control sera. §P<0.001 versus all the others and versus controls.
Discussion
In this study, we have found heterozygous genomic rearrangements among CFH and CFHR genes in 4.5% of patients with aHUS. CFH/CFHR rearrangements were associated with poor clinical prognosis and high risk of post-transplant recurrence.13,15,16
Of relevance, in a family where two individuals have been affected by aHUS, we found a novel duplication in the CFH-CFHR genomic area. The duplication results in an additional copy of CFHR3 and the formation of an extra hybrid copy of CFHR1 derived from the fusion of the first five exons of CFHR1 and the last exon 23 of CFH. The hybrid gene encodes an FHR1/FH protein, in which the last SCR5 has been replaced by SCR20 of FH. This rearrangement is similar but not identical to the rearrangement reported by Eyler et al.16 in a patient with sporadic aHUS in which the CFHR1/CFH hybrid included two exons of CFH.
The data reported here that sera from the patients with the genomic duplication and the hybrid CFHR1/CFH gene induced more sheep erythrocyte hemolysis and more C5b-9 deposition on endothelial cells ex vivo than control serum document that such rearrangement results in complement dysregulation at the cell surface.
FHR1 has a C-terminal SCR5 that is 97% identical to the C-terminal SCR20 of FH that contains the surface recognition domain, and it is capable of binding to C3b, C3d, and cell surfaces.18 Using a hemolysis assay with guinea pig erythrocytes as the complement-activating surface, two independent studies documented that FHR1, by forming homodimers or heterodimers with FHR2 or FHR5, acquires avidity for cell-bound complement fragments and competes with FH for ligand binding.17,19
The observation here that the addition of purified FHR1 from control subjects to human serum did not induce hemolysis of sheep erythrocytes and did not increase complement deposition on microvascular endothelial cells discloses that the capability of FHR1 to compete with FH-binding is restricted to only a subset of self-surfaces depending on the relative affinity of FHR1 and FH to their specific cell surface ligands. Published data that the Ser1191Leu/Val1197Ala FH mutant and the FH/FHR1 hybrid, in which the C terminus of FH is identical to the C terminus of FHR1, failed to bind and protect sheep erythrocytes from complement lysis and resulted in aHUS document that the two amino acidic differences between SCR5 (Leu290 and Ala296) of FHR1 and SCR20 (Ser1191 and Val1197) of FH are enough to dictate the affinity differences between the two proteins on aHUS-relevant surfaces. Thus, changing the C-terminal region of FH for that of FHR1 disables the ability of FH to bind and regulate complement properly on those surfaces. On the basis of these data, we made two hypotheses. (1) The reciprocal hybrid FHR1/FH protein in which the C-terminal region of FHR1 is replaced by the FH C terminus, in contrast to wild-type FHR1, should compete the binding of FH to aHUS-relevant surfaces. (2) Because FHR1 is devoid of the FH complement regulatory properties (i.e., cofactor and decay-accelerating activities),17,20,21 the hybrid FHR1/FH protein should antagonize the protection from complement damage conferred by FH. Results of competition experiments showing that purified FHR1 from heterozygous carriers of the FHR1/FH hybrid dose-dependently caused complement dysregulation on sheep erythrocytes when added to normal human serum show that the hypotheses are correct.
Altogether, the data presented here indicate that it is the mutant protein and not the excess of FHR1 that is pathogenetic and associates with aHUS.
The role of FHR3 in complement activation and FH function has not been investigated as thoroughly. FHR3 does not interact with FHR117; however, it can bind to surface-bound C3b.21 The functional consequences of the extra CFHR3 copy in carriers of the duplication and hybrid CFHR1/CFH gene remain to be established.
Of note in this family, we have identified one unaffected carrier. A combination of CFH and MCP polymorphic variants and environmental triggers has been shown to concur to aHUS penetrance in individuals with complement gene mutations.3,22–25 The unaffected carrier has the same genetic risk as his sister, because they share the MCPggaac risk haplotype on the chromosome with the duplication, but his sister, at the time of disease onset, was taking contraceptive pills, a known aHUS precipitant.1 The unaffected carrier is 16 years old, and he may still be at risk of developing the disease on exposure to environmental trigger(s) that will activate complement and/or the endothelium.3,26,27 This possibility is supported by finding that serum from this subject induced excessive C5b-9 deposition on activated endothelial cells compared with control serum.
The proband in this family developed ESRD and is waiting for a kidney transplant. However, because the FHR1/FH hybrid protein is secreted into the circulation, it will persist after transplantation, thus predisposing to developing recurrence in the graft.28 The fact that this predisposition may be the case is supported by a previously published report of a patient carrying a similar hybrid CFHR1/CFH gene who lost the first graft after a severe aHUS relapse.16 In the same patient, a subsequent kidney transplant performed under eculizumab prophylaxis was successful.16 This finding, together with the finding here that the affected proband’s daughter achieved full remission in the native kidneys on treatment with eculizumab, confirms the role of complement in the pathogenesis of aHUS in carriers of CFHR1/CFH hybrid genes.16
In summary, we describe a new CFHR1/CFH hybrid gene that adds to previously reported genomic rearrangements in the CFH-CFHR region.13–16 Such rearrangements are associated with post-transplant recurrence, which however, can be prevented by eculizumab prophylaxis.16,28 Because of the unaffordable economic burden of such an expensive drug, identification of patients who are the most likely to benefit from eculizumab is mandatory. Combined liver/kidney transplantation has been used in patients with CFH mutations29 and could be useful for patients with CFH/CFHR and CFHR1/CFH hybrid genes. This procedure provides surgical correction of the genetic abnormality29 and is less expensive than single kidney transplant with chronic eculizumab prophylaxis, but it is associated with higher short-term complications30 and requires highly experienced centers.
We recommend that genetic screening of aHUS includes analysis of CFH/CFHR rearrangements by specific tests, like MLPA or array CGH, particularly before programming a kidney transplant.
Concise Methods
Study Participants
aHUS was diagnosed in cases reported to have one or more episodes of nonimmune hemolytic anemia, thrombocytopenia, and renal impairment. One hundred fifty-four consecutive patients of the International Registry of Hemolytic-Uremic Syndrome/Thrombotic Thrombocytopenic Purpura analyzed for CFH, CFI, MCP, C3, CFB, and THBD genes were screened for genomic rearrangements affecting CFH, CFHR1, CFHR2, CFHR3, and CFHR5. The study was approved by the Ethics Committee of the Azienda Sanitaria Locale, Bergamo, Italy, and informed consent was obtained in accordance with the Declaration of Helsinki.
A detailed description of study participants, methods used for complement profile assessment, mutation screening and genotyping, MLPA, CGH arrays, breakpoint identification, and Western blot analysis of FHR1 is in Supplemental Material.
FH-Dependent Hemolytic Assay
Sheep erythrocytes (2% packed cell volume) in AP buffer (2.5 mM barbital, 1.5 mM sodium barbital, 144 mM NaCl, 7 mM MgCl2 and 10 mM EGTA, pH 7.4) were incubated with increasing amounts of serum. After 30 minutes at 37°C, the reaction was stopped by adding AP buffer with 20 mM EDTA. After centrifugation, supernatants were read at 414 nm. Erythrocytes diluted in distilled water were taken as 100% lysis, and erythrocytes diluted in AP buffer were used as blank for spontaneous lysis.
Purification of FHR1 and FHR1/FH Hybrid Protein and Competition Assays
FHR1 and FHR1/FH hybrid heterooligomers were purified from human plasma/serum by immunoaffinity chromatography. Briefly, filtered plasma/serum was loaded into an immunoaffinity column coupled to an in-house anti-human FHR1 mouse mAb (MBC125) (Supplemental Material). Bound protein was eluted at low pH, and fractions containing the protein were pooled and purified using a heparin column (GE Healthcare). Eluted proteins were further polished in a gel filtration column (GE Healthcare) equilibrated with 10 mM Hepes buffer (pH 7.0) and 150 mM NaCl. Before use, FHR1 and FHR1/FH concentrations were normalized to obtain preparations that contained identical concentrations of FHR1 protein. The capacity of the FHR1/FH hybrid protein to compete with the activity of FH was assessed in a modified sheep hemolytic assay. The human serum used in these assays was previously deprived from endogenous FHR1 using an immunoaffinity column with the anti-FHR1, FHR2, and FHR5 MBC125 mAb,17 and the FH activity decreased by approximately 50% (20% lysis in the sheep erythrocyte assay) by adding the anti–N-terminal FH antibody OX24 (Supplemental Figure 2). Sheep erythrocytes in AP buffer (2% packed cell volume) were incubated with 10% human serum in AP buffer containing increasing amounts of purified FHR1 from controls and carriers of the CFHR1/CFH hybrid for 1.5 hour at 37°C. The reaction was stopped by adding AP buffer with 20 mM EDTA; after centrifugation, supernatants were read at 414 nm, and lysis was plotted versus concentration of FHR1.
C5b-9 Deposition on Endothelial Cells
HMEC-1, activated with ADP, was incubated with serum from patients or controls. The staining on the endothelial cell surface with rabbit anti-human complement C5b-9 was evaluated by confocal inverted laser microscope. The area occupied by the fluorescent staining was evaluated and expressed as pixels2 per field analyzed. For each sample, the mean of 15 fields (excluding the lowest and the highest values) was calculated. Results are expressed as means±SEMs.
Data were analyzed by ANOVA. P values of <0.05 were considered to be statistically significant. Additional details are in Supplemental Material.
Disclosures
S.R.d.C. and M.N. have received honoraria from Alexion Pharmaceuticals for giving lectures and participating in advisory boards. None of these activities have had any influence on the results or interpretation in this article. Other authors declare no conflicts of interest.
Supplementary Material
Acknowledgments
The authors thank D. Serena Bettoni for SC5b-9 and CH50 evaluations and Drs. Caterina Mele, Paraskevas Iatropoulos, Rossella Piras, and Ramona Maranta for complement gene sequencing. We also thank Drs. Roberta Donadelli and Miriam Galbusera for helpful discussion of data and Drs. E. Beggiato, A. Borchiellini, L. Colla, and M. Burdese for clinical management of patients. Finally, we thank the patients and their relatives; without their contribution, this work could not have been done.
This work was supported by the Fondazione ART per la Ricerca sui Trapianti ART ONLUS (Milan, Italy), the Fondazione Aiuti per la Ricerca sulle Malattie Rare ARMR ONLUS (Bergamo, Italy), by grants from Fondazione Telethon (GGP09075) and European Union Seventh Framework Programme FP7-EURenOmics Project 305608. S.R.d.C. is supported by Spanish Ministerio de Economia y Competitividad Grant SAF2011-26583, Comunidad de Madrid Grant S2010/BMD-2316, the Fundación Renal Iñigo Alvarez de Toledo, and Seventh Framework Programme European Union Project EURenOmics Grant 305608.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013121339/-/DCSupplemental.
References
- 1.Noris M, Remuzzi G: Atypical hemolytic-uremic syndrome. N Engl J Med 361: 1676–1687, 2009 [DOI] [PubMed] [Google Scholar]
- 2.Kavanagh D, Goodship TH: Atypical hemolytic uremic syndrome. Curr Opin Hematol 17: 432–438, 2010 [DOI] [PubMed] [Google Scholar]
- 3.Bresin E, Rurali E, Caprioli J, Sanchez-Corral P, Fremeaux-Bacchi V, Rodriguez de Cordoba S, Pinto S, Goodship TH, Alberti M, Ribes D, Valoti E, Remuzzi G, Noris M, European Working Party on Complement Genetics in Renal Diseases : Combined complement gene mutations in atypical hemolytic uremic syndrome influence clinical phenotype. J Am Soc Nephrol 24: 475–486, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pérez-Caballero D, González-Rubio C, Gallardo ME, Vera M, López-Trascasa M, Rodríguez de Córdoba S, Sánchez-Corral P: Clustering of missense mutations in the C-terminal region of factor H in atypical hemolytic uremic syndrome. Am J Hum Genet 68: 478–484, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dragon-Durey MA, Loirat C, Cloarec S, Macher MA, Blouin J, Nivet H, Weiss L, Fridman WH, Frémeaux-Bacchi V: Anti-Factor H autoantibodies associated with atypical hemolytic uremic syndrome. J Am Soc Nephrol 16: 555–563, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Dragon-Durey MA, Sethi SK, Bagga A, Blanc C, Blouin J, Ranchin B, André JL, Takagi N, Cheong HI, Hari P, Le Quintrec M, Niaudet P, Loirat C, Fridman WH, Frémeaux-Bacchi V: Clinical features of anti-factor H autoantibody-associated hemolytic uremic syndrome. J Am Soc Nephrol 21: 2180–2187, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore I, Strain L, Pappworth I, Kavanagh D, Barlow PN, Herbert AP, Schmidt CQ, Staniforth SJ, Holmes LV, Ward R, Morgan L, Goodship TH, Marchbank KJ: Association of factor H autoantibodies with deletions of CFHR1, CFHR3, CFHR4, and with mutations in CFH, CFI, CD46, and C3 in patients with atypical hemolytic uremic syndrome. Blood 115: 379–387, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Díaz-Guillén MA, Rodríguez de Córdoba S, Heine-Suñer D: A radiation hybrid map of complement factor H and factor H-related genes. Immunogenetics 49: 549–552, 1999 [DOI] [PubMed] [Google Scholar]
- 9.Male DA, Ormsby RJ, Ranganathan S, Giannakis E, Gordon DL: Complement factor H: Sequence analysis of 221 kb of human genomic DNA containing the entire fH, fHR-1 and fHR-3 genes. Mol Immunol 37: 41–52, 2000 [DOI] [PubMed] [Google Scholar]
- 10.Józsi M, Licht C, Strobel S, Zipfel SL, Richter H, Heinen S, Zipfel PF, Skerka C: Factor H autoantibodies in atypical hemolytic uremic syndrome correlate with CFHR1/CFHR3 deficiency. Blood 111: 1512–1514, 2008 [DOI] [PubMed] [Google Scholar]
- 11.Zipfel PF, Edey M, Heinen S, Józsi M, Richter H, Misselwitz J, Hoppe B, Routledge D, Strain L, Hughes AE, Goodship JA, Licht C, Goodship TH, Skerka C: Deletion of complement factor H-related genes CFHR1 and CFHR3 is associated with atypical hemolytic uremic syndrome. PLoS Genet 3: e41, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abarrategui-Garrido C, Martínez-Barricarte R, López-Trascasa M, de Córdoba SR, Sánchez-Corral P: Characterization of complement factor H-related (CFHR) proteins in plasma reveals novel genetic variations of CFHR1 associated with atypical hemolytic uremic syndrome. Blood 114: 4261–4271, 2009 [DOI] [PubMed] [Google Scholar]
- 13.Venables JP, Strain L, Routledge D, Bourn D, Powell HM, Warwicker P, Diaz-Torres ML, Sampson A, Mead P, Webb M, Pirson Y, Jackson MS, Hughes A, Wood KM, Goodship JA, Goodship TH: Atypical haemolytic uraemic syndrome associated with a hybrid complement gene. PLoS Med 3: e431, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maga TK, Meyer NC, Belsha C, Nishimura CJ, Zhang Y, Smith RJ: A novel deletion in the RCA gene cluster causes atypical hemolytic uremic syndrome. Nephrol Dial Transplant 26: 739–741, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis NJ, McNicholas B, Awan A, Waldron M, Reddan D, Sadlier D, Kavanagh D, Strain L, Marchbank KJ, Harris CL, Goodship TH: A novel hybrid CFH/CFHR3 gene generated by a microhomology-mediated deletion in familial atypical hemolytic uremic syndrome. Blood 119: 591–601, 2012 [DOI] [PubMed] [Google Scholar]
- 16.Eyler SJ, Meyer NC, Zhang Y, Xiao X, Nester CM, Smith RJ: A novel hybrid CFHR1/CFH gene causes atypical hemolytic uremic syndrome. Pediatr Nephrol 28: 2221–2225, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tortajada A, Yébenes H, Abarrategui-Garrido C, Anter J, García-Fernández JM, Martínez-Barricarte R, Alba-Domínguez M, Malik TH, Bedoya R, Cabrera Pérez R, López Trascasa M, Pickering MC, Harris CL, Sánchez-Corral P, Llorca O, Rodríguez de Córdoba S: C3 glomerulopathy-associated CFHR1 mutation alters FHR oligomerization and complement regulation. J Clin Invest 123: 2434–2446, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Józsi M, Zipfel PF: Factor H family proteins and human diseases. Trends Immunol 29: 380–387, 2008 [DOI] [PubMed] [Google Scholar]
- 19.Goicoechea de Jorge E, Caesar JJ, Malik TH, Patel M, Colledge M, Johnson S, Hakobyan S, Morgan BP, Harris CL, Pickering MC, Lea SM: Dimerization of complement factor H-related proteins modulates complement activation in vivo. Proc Natl Acad Sci U S A 110: 4685–4690, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinen S, Hartmann A, Lauer N, Wiehl U, Dahse HM, Schirmer S, Gropp K, Enghardt T, Wallich R, Hälbich S, Mihlan M, Schlötzer-Schrehardt U, Zipfel PF, Skerka C: Factor H-related protein 1 (CFHR-1) inhibits complement C5 convertase activity and terminal complex formation. Blood 114: 2439–2447, 2009 [DOI] [PubMed] [Google Scholar]
- 21.Skerka C, Chen Q, Fremeaux-Bacchi V, Roumenina LT: Complement factor H related proteins (CFHRs). Mol Immunol 56: 170–180, 2013 [DOI] [PubMed] [Google Scholar]
- 22.Noris M, Caprioli J, Bresin E, Mossali C, Pianetti G, Gamba S, Daina E, Fenili C, Castelletti F, Sorosina A, Piras R, Donadelli R, Maranta R, van der Meer I, Conway EM, Zipfel PF, Goodship TH, Remuzzi G: Relative role of genetic complement abnormalities in sporadic and familial aHUS and their impact on clinical phenotype. Clin J Am Soc Nephrol 5: 1844–1859, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Córdoba SR, de Jorge EG: Translational mini-review series on complement factor H: Genetics and disease associations of human complement factor H. Clin Exp Immunol 151: 1–13, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caprioli J, Castelletti F, Bucchioni S, Bettinaglio P, Bresin E, Pianetti G, Gamba S, Brioschi S, Daina E, Remuzzi G, Noris M, International Registry of Recurrent and Familial HUS/TTP : Complement factor H mutations and gene polymorphisms in haemolytic uraemic syndrome: The C-257T, the A2089G and the G2881T polymorphisms are strongly associated with the disease. Hum Mol Genet 12: 3385–3395, 2003 [DOI] [PubMed] [Google Scholar]
- 25.Esparza-Gordillo J, Goicoechea de Jorge E, Buil A, Carreras Berges L, López-Trascasa M, Sánchez-Corral P, Rodríguez de Córdoba S: Predisposition to atypical hemolytic uremic syndrome involves the concurrence of different susceptibility alleles in the regulators of complement activation gene cluster in 1q32. Hum Mol Genet 14: 703–712, 2005 [DOI] [PubMed] [Google Scholar]
- 26.Kavanagh D, Goodship TH, Richards A: Atypical hemolytic uremic syndrome. Semin Nephrol 33: 508–530, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caprioli J, Noris M, Brioschi S, Pianetti G, Castelletti F, Bettinaglio P, Mele C, Bresin E, Cassis L, Gamba S, Porrati F, Bucchioni S, Monteferrante G, Fang CJ, Liszewski MK, Kavanagh D, Atkinson JP, Remuzzi G, International Registry of Recurrent and Familial HUS/TTP : Genetics of HUS: The impact of MCP, CFH, and IF mutations on clinical presentation, response to treatment, and outcome. Blood 108: 1267–1279, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valoti E, Alberti M, Noris M: Posttransplant recurrence of atypical hemolytic uremic syndrome. J Nephrol 25: 911–917, 2012 [DOI] [PubMed] [Google Scholar]
- 29.Saland JM, Ruggenenti P, Remuzzi G, Consensus Study Group : Liver-kidney transplantation to cure atypical hemolytic uremic syndrome. J Am Soc Nephrol 20: 940–949, 2009 [DOI] [PubMed] [Google Scholar]
- 30.Remuzzi G, Ruggenenti P, Colledan M, Gridelli B, Bertani A, Bettinaglio P, Bucchioni S, Sonzogni A, Bonanomi E, Sonzogni V, Platt JL, Perico N, Noris M: Hemolytic uremic syndrome: A fatal outcome after kidney and liver transplantation performed to correct factor H gene mutation. Am J Transplant 5: 1146–1150, 2005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.