Abstract
The immune system plays key roles in determining the fate of developing cancers by not only functioning as a tumour promoter facilitating cellular transformation, promoting tumour growth and sculpting tumour cell immunogenicity1–6, but also as an extrinsic tumour suppressor that either destroys developing tumours or restrains their expansion1,2,7. Yet clinically apparent cancers still arise in immunocompetent individuals in part as a consequence of cancer induced immunosuppression. In many individuals, immunosuppression is mediated by Cytotoxic T-Lymphocyte Associated Antigen-4 (CTLA-4) and Programmed Death-1 (PD-1), two immunomodulatory receptors expressed on T cells8,9. Monoclonal antibody (mAb) based therapies targeting CTLA-4 and/or PD-1 (checkpoint blockade) have yielded significant clinical benefits—including durable responses—to patients with different malignancies10–13. However, little is known about the identity of the tumour antigens that function as the targets of T cells activated by checkpoint blockade immunotherapy and whether these antigens can be used to generate vaccines that are highly tumour-specific. Herein, we use genomics and bioinformatics approaches to identify tumour-specific mutant proteins as a major class of T cell rejection antigens following αPD-1 and/or αCTLA-4 therapy of mice bearing progressively growing sarcomas and show that therapeutic synthetic long peptide (SLP) vaccines incorporating these mutant epitopes induce tumour rejection comparably to checkpoint blockade immunotherapy. Whereas, mutant tumour antigen-specific T cells are present in progressively growing tumours, they are reactivated following treatment with αPD-1- and/or αCTLA-4 and display some overlapping but mostly treatment-specific transcriptional profiles rendering them capable of mediating tumour rejection. These results reveal that tumour-specific mutant antigens (TSMA) are not only important targets of checkpoint blockade therapy but also can be used to develop personalized cancer-specific vaccines and to probe the mechanistic underpinnings of different checkpoint blockade treatments.
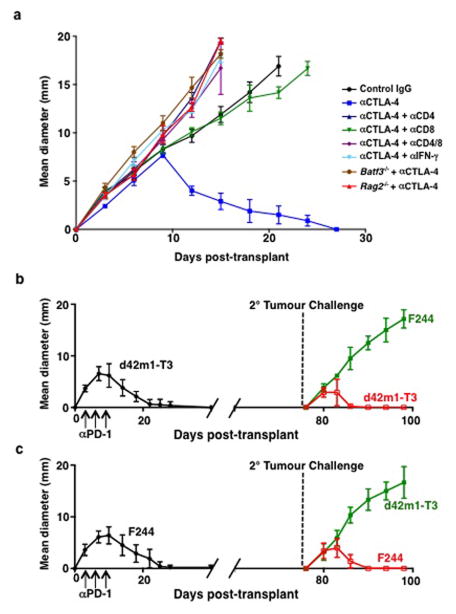
In this study, we used two distinct progressor MCA sarcoma cell lines (d42m1-T3 and F244) and asked whether they expressed sufficient immunogenicity to be controlled by checkpoint blockade immunotherapy. Both sarcoma lines were rejected in wild type (WT) mice treated therapeutically with αPD-1- and/or αCTLA-4 (Fig. 1a). Rejection was immunologic since it (a) was ablated by administration of mAbs that either deplete CD4+ or CD8+ cells or neutralize IFN-γ; (b) did not occur in Rag2−/− mice lacking T, B, and NKT cells or Batf3−/− mice lacking CD8α+/CD103+ dendritic cells required for tumour antigen cross-presentation to CD8+ T cells (Extended Data Fig. 1a); and (c) induced a memory response that protected mice against rechallenge with the same tumour cells that had been injected into naïve mice (Extended Data Fig. 1b,c).
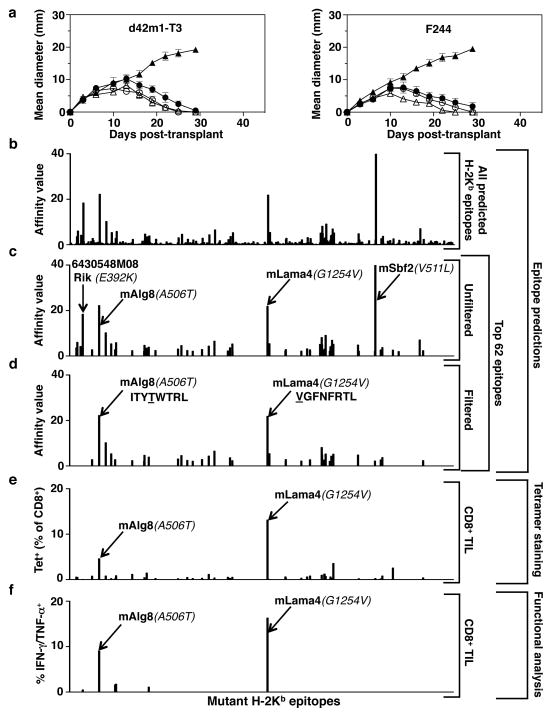
Figure 1. Mutations in Lama4 and Alg8 form top predicted d42m1-T3 epitopes.
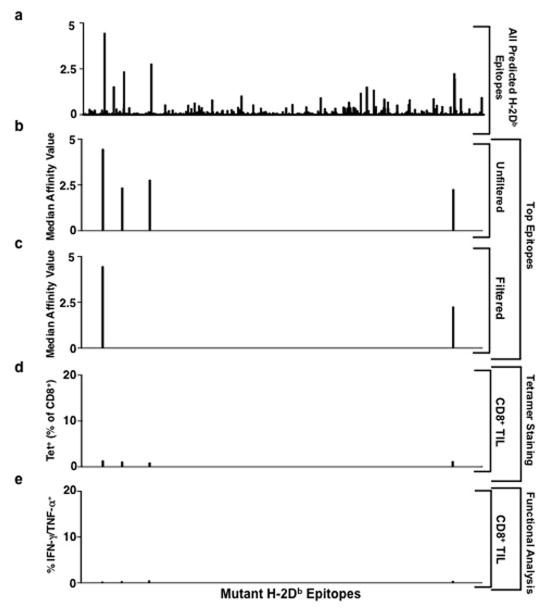
a, Growth of d42m1-T3 or F244 tumours in 5-mouse cohorts treated with αPD-1 (closed circles), αCTLA-4 (open circles), αPD-1+αCTLA-4 (open triangle) or control mAb (closed triangle). b, Potential H-2Kb binding epitopes predicted by in silico analysis of all missense mutations in d42m1-T3. c, Median affinity values for the top 62 predicted H-2Kb epitopes. d, Median affinity values of H-2Kb epitopes after filtering. e, Screening for specificities of CD8+ TILs from αPD-1 treated, d42m1-T3 tumour bearing mice using H-2Kb tetramers loaded with top 62 H-2Kb epitopes. f, IFN-γ and TNF-α induction in CD8+ TILs from αPD-1 treated, d42m1-T3 tumour bearing mice following culture with irradiated splenocytes pulsed with the top 62 H-2Kb peptides. Data are presented as per cent CD8+ TILs expressing IFN-γ, TNF-α or for both. Data are representative of two independent experiments.
Based on our previous success using genomics approaches to identify TSMA responsible for the spontaneous rejection of highly immunogenic, unedited MCA sarcomas14, we asked whether a similar approach could identify antigens responsible for αPD-1-mediated rejection of d42m1-T3 progressor tumours. To increase the robustness and accuracy of our epitope predictions, we modified our method as follows: (1) mutation calls from cDNA Capture Sequencing14 were translated to corresponding protein sequences, pipelined through three MHC class I epitope-binding algorithms and a median binding affinity calculated for each predicted epitope; (2) epitopes were prioritized based on predicted median binding affinities; and (3) filters were applied to the prioritized epitope list to (a) eliminate those predicted to be poorly processed by the immunoproteasome and (b) deprioritize those from hypothetical proteins or those that displayed lower binding affinity to class I than their corresponding WT sequences. Using this approach, many epitopes were predicted for H-2Db (49,677 9- and 10-mer epitopes) (Extended Data Fig. 2a) and H-2Kb (44,215 8- and 9-mer epitopes) (Fig. 1b) based on the 2,796 non-synonymous mutations expressed in d42m1-T314. Focussing on epitopes with the highest predicted binding affinity to H-2Db or H-2Kb, we narrowed the list down to four H-2Db-binding epitopes (Extended Data Fig. 2b) and 62 H-2Kb-binding epitopes (Fig. 1c). Applying the aforementioned filters eliminated two predicted strong-binding H-2Db epitopes (Extended Data Fig. 2c) and 20 predicted strong-binding H-2Kb epitopes (Fig. 1d) (epitope binding affinity distributions to different class I alleles are distict15). Based on the resulting in silico generated epitope landscape, two predominant H-2Kb restricted mutant epitopes were identified by their predicted binding affinities: an A506T mutation (ITYAWTRL→ITYTWTRL) in Asparagine-linked glycosylation 8 (alpha-1,3-glucosyltransferase) (Alg8) and a G1254V mutation (GGFNFRTL→VGFNFRTL) in Laminin alpha subunit 4 (Lama4). Based on H-2Kb consensus binding, the mutations that produce these epitopes occur at positions p4 (Alg8) and p1 (Lama4). Neither functions as an anchor residue for H-2Kb (these occur at p5 and p8). The mAlg8 and mLama4 epitopes are predicted to bind 1.2- and 12.8-fold stronger to H-2Kb respectively compared to WT sequences.
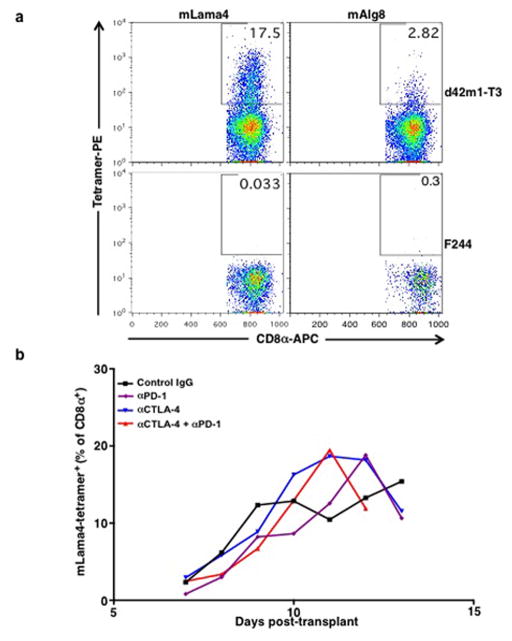
To identify which of the predicted d42m1-T3 neoepitopes functioned as targets for CD8+ T cells in αPD-1 treated, tumour bearing mice, freshly explanted CD8+ TILs were isolated just prior to tumour rejection (d11) and stained with fluorescently labelled H-2Kb or H-2Db tetramers loaded with their corresponding strong-binding 66 predicted mutant epitopes. The only tetramer-positive T cells consistently identified in the CD8+ TIL population were those reacting with mLama4-H-2Kb tetramers (13.1% of CD8+ TILs in experiment shown; 15.6 ± 2.7% as the mean of 6 experiments) or mAlg8-H-2Kb tetramers (4.2% of CD8+ TILs in experiment shown; 2.8 ± 1.1% as the mean of 6 experiments) (Fig. 1e and Extended Data Fig. 2d). Similar results were obtained when freshly explanted CD8+ TILs from the same mice were co-cultured with naïve irradiated splenocytes pulsed with each of the 66 predicted H-2Kb and H-2Db epitopes. The mLama4 and mAlg8 epitopes were, again, the only significant hits, inducing IFN-γ and TNF-α production (Fig. 1f and Extended Data Fig. 2e). These results demonstrate that d42m1-T3 expresses two dominant TSMA epitopes for CD8+ T cells following αPD-1 immunotherapy.
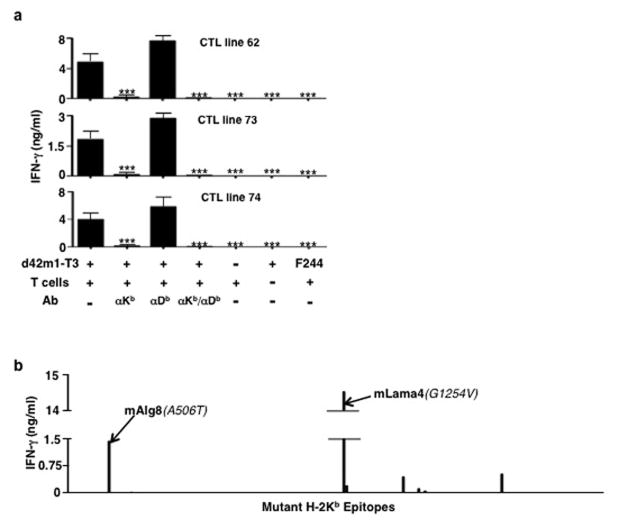
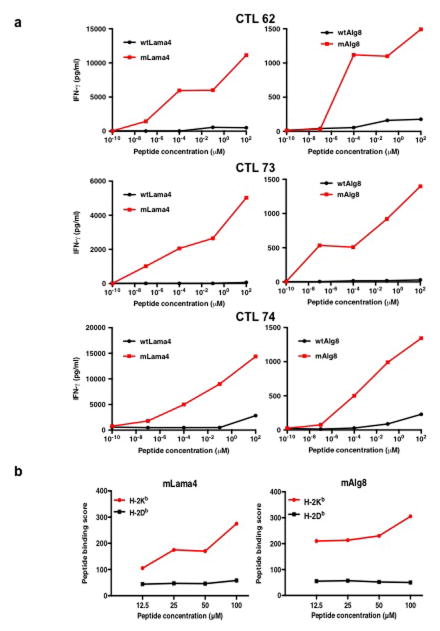
To independently validate these observations, we established CD8+ T cell lines from spleens of mice that had rejected d42m1-T3 tumours after αPD-1 treatment. These T cells produced IFN-γ when co-cultured with d42m1-T3 but not F244 or other independent sarcoma lines. Stimulation was restricted by H-2Kb but not H-2Db (Extended Data Fig. 3a) and the only predicted epitopes that stimulated these T cell lines were mLama4 and mAlg8 (Extended Data Fig. 3b) and not their WT forms (Extended Data Fig. 4a).
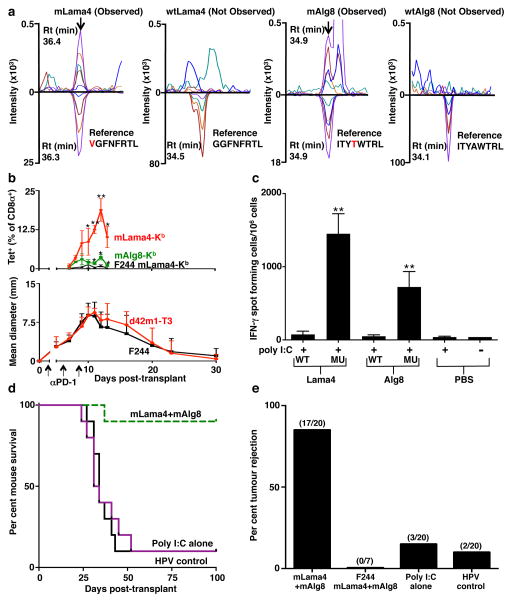
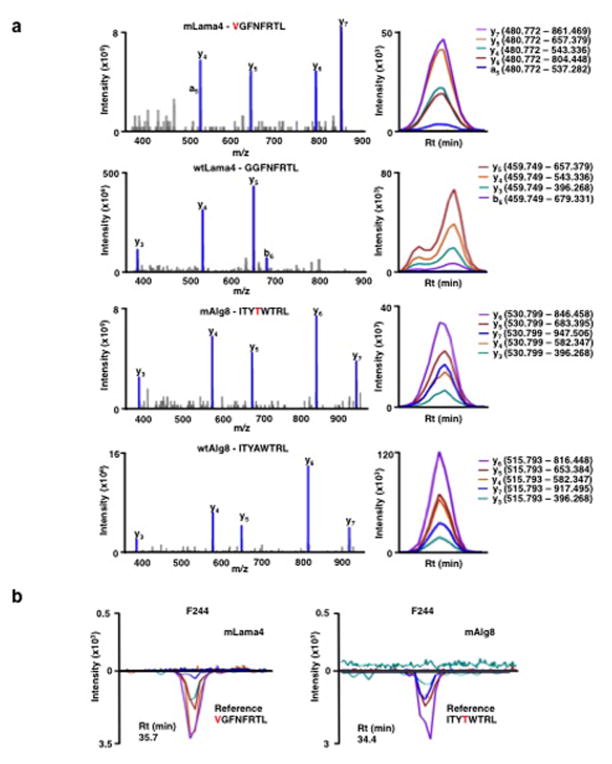
Four subsequent findings supported the conclusion that mLama4 and mAlg8 were the relevant antigens responsible for αPD-1-induced rejection of d42m1-T3. First, mLama4 or mAlg8 epitopes stabilized H-2Kb expression on RMA-S cells, which lack a functional antigen transporter and thus fail to stably express MHC class I proteins on the cell surface (Extended Data Fig. 4b). Second, both epitopes were detected by mass spectrometry (MS) in eluates of affinity purified H-2Kb isolated from d42m1-T3 tumours. Using a discovery MS approach, we identified mLama4 in the H-2Kb eluate (Extended Data Fig. 5a) and verified its identity using an isotope-labelled synthetic mLama4 peptide (Extended Data Fig. 5b). We also found more than 200 WT peptides associated with H-2Kb (Supplementary Table 1), but we have no evidence that any of these function as d42m1-T3 antigens. Mutant Alg8, wtLama4 and wtAlg8 peptides were not detected (Supplementary Table 1). In contrast, using the more sensitive targeted selected reaction monitoring (SRM) MS method, both mLama4 and mAlg8 peptides were identified in the H-2Kb eluate (Fig. 2a, Extended Data Fig. 6a and Supplementary Data 1). Notably, mLama4 and mAlg8 were the only predicted strong-binding mutant epitopes found. Peptides from wtLama4 or wtAlg8 were not detected. Neither mLama4 nor mAlg8 were detected in H-2Kb eluates from F244 cells (Extended Data Fig. 6b). Third, as detected by staining with H-2Kb -mLama4 or -mAlg8 tetramers, CD8+ T cells specific for either antigen accumulated temporally in d42m1-T3 tumours of αPD-1 treated mice, reaching maximal values just prior to tumour rejection (Fig. 2b and Extended Data Fig. 7a). No mLama4- or mAlg8-tetramer positive TILs were observed in F244 sarcomas from αPD-1 treated mice. Fourth, the two H-2Kb-restricted epitopes induced antigen-specific CD8+ T cell responses in naïve mice when injected together with poly I:C (pIC) as assessed by ELISPOT (Fig. 2c).
Figure 2. Mutant Lama4 and mAlg8 are therapeutically relevant d42m1-T3 TSMA.
a, Detection of mLama4 and mAlg8 bound to cellular H-2Kb by mass spectrometry. b, Time dependent tumour infiltration of mLama4- and mAlg8-specific CD8+ T cells (n=5), (top). Data represent means ± s.e.m of 5 independent experiments. Growth kinetics of d42m1-T3 and F244 during αPD-1 immunotherapy (n=5), (bottom). Data represent average tumour diameter ± s.e.m. and are representative of at least three independent experiments. c, IFN-γ ELISPOT analysis of peptide stimulated splenocytes from mice immunized with mLama4 or mAlg8 SLP plus polyI:C (n=3 mice per group). Data are means ± s.e.m. Representative of two independent experiments. Samples were compared using unpaired, two-tailed Student’s t test (*p<0.05, **p<0.01). d, Kaplan-Meier survival curves of d42m1-T3 tumour bearing mice (10 mice per group) therapeutically vaccinated with SLP vaccines plus poly I:C. mLama4 plus mAlg8 compared to HPV control: p=0.0002 [log-rank (Mantel-Cox) test]. Representative of two independent experiments. e, Cumulative data from two independent SLP therapeutic vaccine experiments using mice (7–10 per group) with d42m1-T3 or F244 tumours.
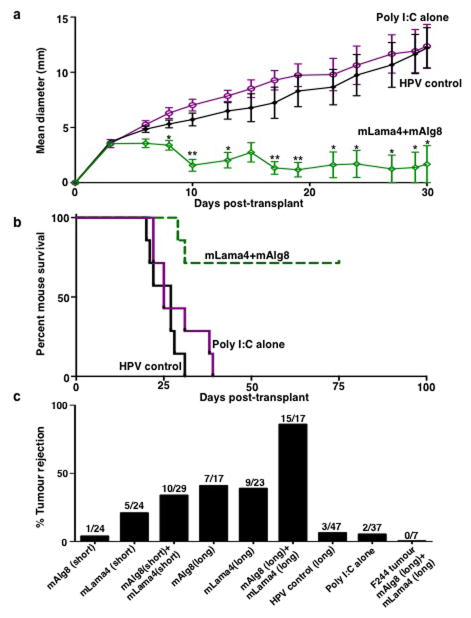
Since mLama4- and mAlg8-specific T cells were linked to αPD-1-induced d42m1-T3 rejection we asked whether a therapeutic vaccine comprised of these antigens could protect against tumour outgrowth. When 10-member groups of mice bearing established d42m1-T3 tumours were vaccinated with the combination of mLama4 (28-mer) and mAlg8 (21-mer) SLPs with pIC, 9 rejected their tumours compared to control mice vaccinated with irrelevant HPV (30-mer) SLP plus pIC (1/10 mice survived) or pIC alone (1/10 mice survived) (Fig. 2d and Extended Data Fig. 8a). In multiple experiments, mice vaccinated with mLama4 + mAlg8 SLP + pIC displayed an 85% survival (17/20) while those treated with HPV SLP + pIC or pIC alone showed 10% (2/20) and 15% (3/20) survival, respectively (Fig. 2e). Prophylactic administration of the combined mLama4 and mAlg8 SLP vaccine induced 88% survival (15/17) (Extended Data Fig. 8b,c). The combined mLama4 and mAlg8 prophylactic SLP vaccine induced superior protection compared to either SLP alone (Extended Data Fig. 8c) or compared to vaccines comprised of the minimal 8 amino acid epitopes (Extended Data Fig. 8c). The d42m1-T3 specific vaccines did not prevent outgrowth of unrelated F244 sarcomas (Fig. 2e and Extended Data Fig. 8c). These results not only demonstrate that mLama4 and mAlg8 are major antigenic targets that mediate checkpoint blockade-induced rejection of d42m1-T3 tumours but also show that αPD-1 or therapeutic SLP vaccines consisting of the TSMA targeted by αPD-1 are similarly efficacious.
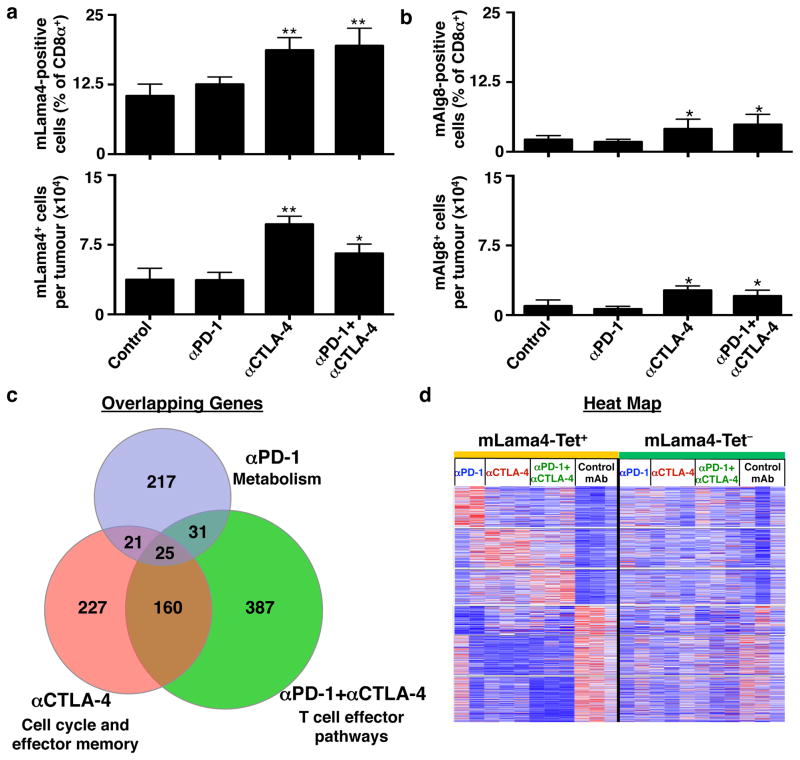
Since the aforementioned analyses were conducted using αPD-1 treated mice bearing d42m1-T3 tumours, we asked whether the presence of mLama4- and mAlg8-specific T cells in the TIL population was dependent on checkpoint blockade therapy. T cells specific for mLama4 or mAlg8 were detected in mice treated with control mAb or αPD-1 ± αCTLA-4 (Fig. 3a,b and Extended Data Fig. 7b). The percentage and total number of mLama4- or mAlg8-specific CD8+ TILs were similar in tumours from control mAb and αPD-1 treated mice but were elevated in mice treated with either αCTLA-4 or αCTLA-4 + αPD-1.
Figure 3. Differential effects of checkpoint blockade therapy on tumour antigen-specific CD8+ T cells.
a, b, Per cent of CD8+ TILs specific for mLama4 (a) or mAlg8 (b) following checkpoint blockade therapy (top). Mean number of mLama4- (a) or mAlg8- (b) specific CD8+ TILs per tumour following checkpoint blockade therapy (bottom). N=5 mice per group pooled. Data are means ± s.e.m. of at least three independent experiments. Samples were compared to control mAb treatment using unpaired, two-tailed Student’s t test (*p<0.05, **p<0.01). c, Venn diagram revealing relationships between differentially expressed genes (p<0.05) in mLama4-specific CD8+ TILs from mice treated with checkpoint blocking mAbs versus control mAb. d, Heatmap showing differentially expressed genes (p<0.05) in mLama4-specific CD8+ TILs from mice treated with checkpoint blocking versus control mAbs. Colour pattern is relative with respect to the row, with red indicating gene upregulation and blue indicating gene downregulation.
The observation that mLama4- and mAlg8-specific T cells were found in tumours from mice treated with either control mAb or checkpoint blockade mAbs prompted us to assess the resultant changes in the TIL population following αPD-1 and/or αCTLA-4 treatment. We used RNA-Sequencing (RNA-Seq) to assess gene expression in freshly isolated, mLama4-H-2Kb-tetramer+ TILs from groups of tumour bearing mice treated with control mAb, αPD-1, αCTLA-4, or αPD-1 + αCTLA-4. Since mLama4-specific T cells were 7-times more abundant in d42m1-T3 tumours than mAlg8-specific T cells in this series of experiments, we restricted our analysis to the former. Only a subset of 25 genes was commonly regulated (either up or down) by treatment with αPD-1 and/or αCTLA-4 (Fig. 3c and Extended Data Table 1a). This group included a subset of genes whose enhanced expression is similar to that observed in CD8+ T cells from mice during acute secondary viral infection and depressed in a manner similar to that of exhausted CD8+ T cells in chronic viral infection16 (Fig. 3c, Extended Data Table 1a, and Supplementary Table 2). In contrast, antigen-specific CD8+ TILs isolated from αPD-1 and/or αCTLA-4 treated mice displayed mostly treatment-specific alterations of non-overlapping sets of genes involved in CD8+ T cell effector functions (Supplementary Table 2). The effects of checkpoint blockade on gene expression were predominately observed on TSMA-specific T-cells and not in other CD8+ TILs (Fig. 3d).
To determine which pathways were regulated by the different checkpoint blockade therapies, we performed Gene Set Enrichment Analysis (GSEA) using canonical pathway- and immunological signature-databases. When compared to mLama4-specific TILs from control mAb-treated mice, tumour antigen-specific TILs from mice treated with αPD-1 and/or αCTLA-4 displayed a common set of alterations involving effector function, MAPK-, chemokine-, and cytokine receptor-signalling (Extended Data Table 1b and Supplementary Table 3). In contrast, mLama4-specific TILs from mice treated with αPD-1, αCTLA-4 or both mAbs displayed profound treatment-specific pathway alterations (Extended Data Table 1b and Supplementary Table 3). Treatment with αPD-1 produced metabolic changes including those involving oxidative phosphorylation, glycolysis, respiratory electron transport, TCA cycle and pentose phosphate pathways, as well as in pathways involved in IL-2 signalling. These cells also displayed a profile consistent with response to type I IFN. Treatment with αCTLA-4 increased NFAT and JAK-STAT signalling pathway activity, cellular proliferation/cell cycle, and activation of effector T cells. Treatment with both αCTLA-4 and αPD-1 induced a synergistic pattern of metabolic and effector T cell-specific functions including those involving T cell mediated anti-tumour activity. This was reflected in the most significant enhancement of effector molecules such as IFN-γ, Granzyme A, Granzyme B, and Fas ligand (Supplementary Table 2). Thus, whereas blockade of different inhibitory costimulators leads to a common biological outcome—tumour eradication—the precise mechanisms differ by which this outcome is achieved.
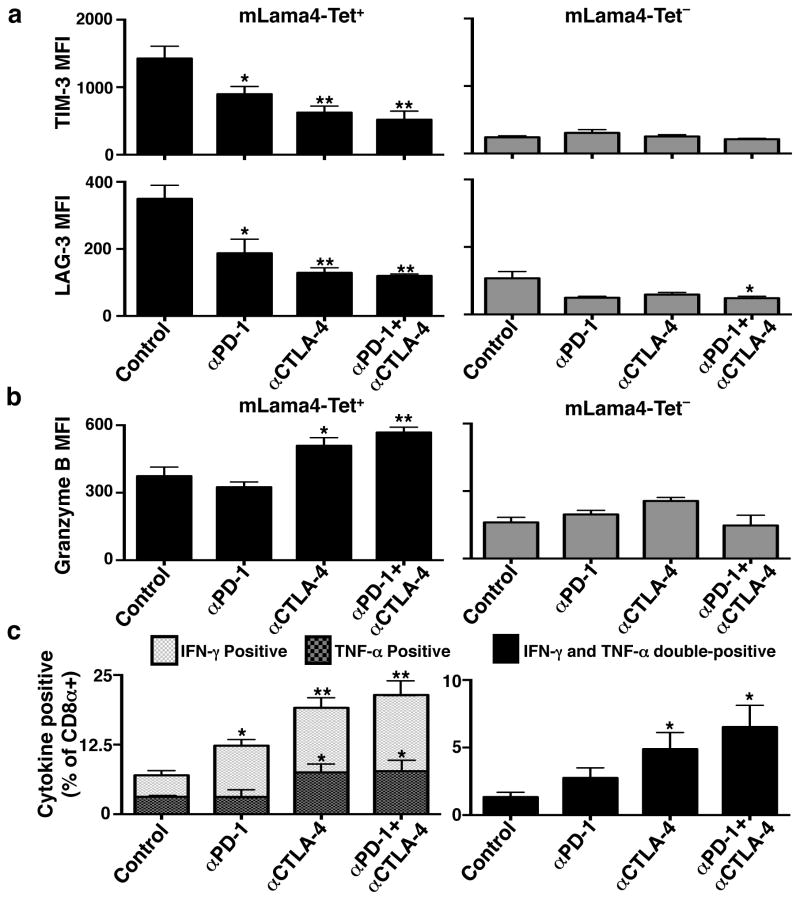
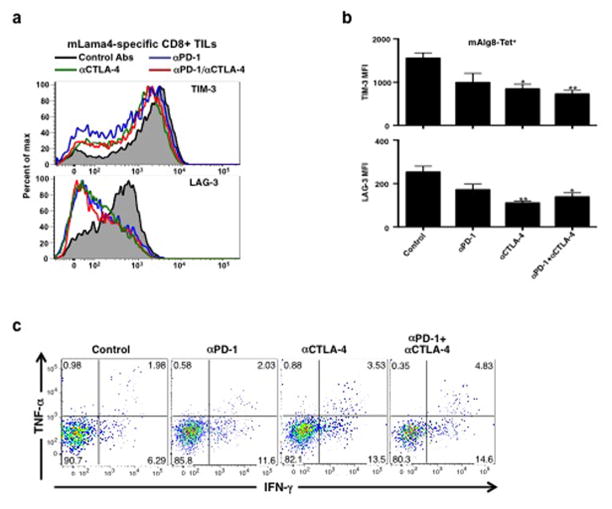
We also assessed changes in expression of functionally relevant proteins on/in CD8+ TILs in mice undergoing treatment with different checkpoint blocking mAbs. TILs specific for mLama4 or mAlg8 from mice treated with αPD-1 and/or αCTLA-4 displayed lower cell surface expression of Lymphocyte-Activation Gene 3 (LAG-3) and T Cell Immunoglobulin and Mucin Protein 3 (TIM-3) than those in progressively growing tumours in control mAb treated mice (Fig. 4a and Extended Data Fig. 9a,b). Elevated LAG-3 and TIM-3 expression is known to mark antigen-experienced, dysfunctional (i.e., exhausted) CD8+ T cells16,17 in chronic viral infection. Conversely, TILs specific for mLama4 or mAlg8 from αCTLA-4 or αCTLA-4 + αPD-1 treated mice displayed significantly higher levels of Granzyme B than antigen-specific TILs from mice treated with either αPD-1 alone or control mAb (Fig. 4b). Consistent with the RNA-Seq analysis, these changes were observed predominantly in antigen-specific TILs. In addition, whereas a low percentage of CD8+ TILs from mice treated with control mAb produced IFN-γ and TNF-α (Fig. 4c and Extended Data Fig. 9c), the percentage of IFN-γ producing TILs increased following treatment of the mice with αPD-1 and particularly with αCTLA-4 or the combination of αCTLA-4 + αPD-1. TILs expressing both cytokines, which likely represent the most potent anti-tumour effectors, were most highly represented following treatment of tumour bearing mice with the combination of αCTLA-4 + αPD-1.
Figure 4. Checkpoint blockade therapy alters the functional phenotypes of tumour antigen-specific CD8+ T cells.
a, TIM-3 or LAG-3 expression on CD8+ TILs following checkpoint blockade therapy. b, Granzyme B expression in CD8+ TILs following checkpoint blockade therapy. c, Per cent of CD8+ TILs positive for IFN-γ and/or TNF-α following checkpoint blockade therapy. N=5 mice per group pooled. Data are means ± s.e.m. of at least three independent experiments. Samples were compared to control mAb treated mice using an unpaired, two-tailed Student’s t test (*p<0.05, **p<0.01).
This report documents that TSMA are targets of checkpoint blockade immunotherapy and can be used in vaccines that therapeutically induce tumour rejection as effectively as checkpoint blockade therapy. The ability to rapidly and accurately identify TSMA using genomics and bioinformatics approaches18–20 and use them to generate MHC tetramers to identify tumour-specific T cells provides a significant advantage to the fields of tumour immunology and cancer immunotherapy. This approach has not only facilitated the identification of the antigenic targets of T cells affected by checkpoint blockade therapy21 but also has provided insights into the molecular changes that occur within the tumour antigen-specific T cell population that give rise to the anti-tumour effects of αPD-1 and/or αCTLA-4. Our findings provide some of the first experimental support for the clinical observations that (a) checkpoint blockade therapy amplifies, in some cases, pre-existing anti-tumour T cell responses8,9,21,22; (b) whereas αCTLA-4 treatment eliminates Tregs, promotes T cell priming, and renders the host more susceptible to autoimmunity13,22,23; αPD-1 promotes T cell activation acting as a rheostat of immune effector function9,22,24,25; and (c) dual blockade of CTLA-4 and PD-1 is particularly effective in promoting enhanced anti-tumour effector functions10,22,26.
The mutational loads of the MCA sarcomas used in this study are high and similar to those of UV- and carcinogen-induced human cancers. For these types of tumours, it is likely that TSMA vaccines targeting multiple antigens will be possible thereby providing better coverage of the tumour cell population in part due to dealing more effectively with tumour heterogeneity27,28. Additionally, the combination of a TSMA vaccine and checkpoint blockade may facilitate the immune system’s ability to recognize less immunogenic TSMA as well as shared, tumour-associated antigens (TAA) via mechanisms that mimic epitope-spreading29. However, recent studies have shown that human tumours containing far fewer mutations (e.g., 26 mutations) can be sensitive to TSMA based immunotherapy even for tumor specific antigens that are targets for class II restricted CD4+ T cells30. Our study thus provides a strong argument to actively pursue the use of TSMA as targets for cancer immunotherapy; as a means to identify patients who would best benefit from such therapy; and as components of MHC tetramers that can be used to identify tumour-specific T cells as biomarkers of successful anti-tumour responses.
METHODS
Mice
Wild type and Rag2−/− mice were purchased from Taconic Farms. Batf3−/− mice31 on a 129S6 background were provided by K.M. Murphy and were bred in our specific pathogen-free animal facility. All in vivo experiments used 8–12 week old male, 129S6 background mice (to match the sex and strain of the tumours) housed in our specific pathogen-free animal facility. All studies were performed in accordance with procedures approved by the AAALAC accredited Animal Studies Committee of Washington University in St. Louis.
Tumour transplantation
MCA-induced sarcomas used in this study were generated in male 129S6 strain wild type or Rag2−/− mice and were banked as low-passage tumour cells as previously described1. All cell lines used in this study were tested for mycoplasma. Tumour cells derived from frozen stocks and propagated in vitro in RPMI media (Hyclone) supplemented with 10% FCS (Hyclone) were washed extensively, resuspended at a density of 6.67 x 106 cells ml−1 in endotoxin-free PBS and then 150 μl injected subcutaneously into the flanks of recipient mice. Tumour cells were >90% viable at the time of injection as assessed by trypan blue exclusion. Tumour growth was quantified by caliper measurements and expressed as the average of two perpendicular diameters. Tumour growth measurements were performed blinded. Sample size was chosen based on extensive previous work with this animal model. No randomization was used. For antibody depletion studies, 250 μg of control mAb (PIP), anti-CD4 (GK1.5) or anti CD8α (YTS169.4) was injected intraperitoneally into mice at day –1 and every 7 days thereafter.
MHC class I epitope prediction
All missense mutations for d42m1-T3 were analysed for the potential to form MHC class I epitopes that bind to either H-2Db or H-2Kb molecules. The Stabilized Matrix Method (SMM) algorithm32, the Artificial Neural Network (ANN) algorithm33, and the NetMHCpan algorithm34 provided by the Immune Epitope Database and Analysis Resource (http://www.immuneepitope.org) was used to predict epitope processing and binding affinities and the results were ultimately expressed as affinity values (1/IC50 × 100). The median affinity value was calculated by taking the median of the predicted affinity values from SMM, ANN, and NetMHCpan algorithms. For the prediction of epitope processing, the NetChop algorithm35 available from http://www.immuneepitope.org was used and potential epitopes were filtered with those with a NetChop score of 0.6 or higher being prioritized. Filters were also applied to eliminate mutations occurring in hypothetical Riken proteins and to de-prioritize mutations resulting in affinity values that were less than that predicted for the wild type sequence.
Antibodies
Anti-H-2Kb (B8-24-3) and anti-H-2Db (B22/249) antibodies were provided by T. H. Hansen (Washington University School of Medicine). Anti-PD-1 (rat chimeric murine IgG1 clone 4H2) and anti-CTLA-4 (murine IgG2b clone 9D9) antibodies were provided by A.J. Korman (Bristol-Myers Squibb). Isotype control antibodies (mouse IgG2a OKT3 and mouse IgG1 GIR-208) were purchased from Bio X Cell and Leinco Technologies, respectively. Anti-CD4 (GK1.5), anti-CD8α (YTS169.4), anti-IFN-γ (H22) monoclonal antibodies and control immunoglobulin (PIP, a monoclonal antibody specific for bacterial glutathione S-transferase) were produced from hybridoma supernatants and purified in endotoxin-free form by Protein G affinity chromatography (Leinco Technologies). Fluorescently conjugated antibodies to CD3ε, CD4, CD8, CD45, LAG-3, TIM-3, PD-1, IFN-γ, and TNF-α were purchased from BioLegend. For tetramer staining, an antibody to CD8 was purchased from Accurate Chemical (clone CT-CD8α) as recommended for use with tetramer staining by the NIH Tetramer Core Facility at Emory. Fc block (anti-CD16/32) was purchased from BD Bioscience.
Peptides
All 8-, 9-, and 10-mer peptides were purchased from Peptide 2.0. All peptides were HPLC purified to > 95% purity. Peptides for vaccine experiments were synthesized at ISA Therapeutics B.V. The peptides were synthesized using solid phase Fmoc/tBu chemistry on a PTI Prelude peptide synthesizer and purified on a Gilson preparative HPLC system to > 95% purity. The identity and purity of the peptides for vaccine experiments were confirmed with UPLC-MS on a Waters Acquity UPLC/TQD system.
Checkpoint Blockade Immunotherapy
Tumour cells were subcutaneously transplanted into mice at 1 x 106 cells in 150 μl into the flank. Mice were treated intraperitoneally with 200 μg anti-PD-1 or anti-CTLA-4, used alone or in combination on days 3, 6, and 9 post-tumour transplant. For controls, mice were injected with 200 μg each of IgG2a and IgG1 isotype control antibodies.
Generation of CTL lines
To generate the d42m1-T3-specific CTL lines, wild-type mice were injected with 1×106 d42m1-T3 tumour cells and treated with αPD-1 that induced tumour rejection. Alternatively, wild-type mice were injected with 1×106 d42m1-T3 tumour cells and were not treated with αPD-1 leading to formation of progressively growing tumours that were then surgically resected. Fifty days later, these mice were subsequently rechallenged with 1×106 d42m1-T3 tumour cells, which then rejected. In the case of either protocol, spleens from independent mice that rejected the tumour were harvested two weeks after rejection and CTL lines were established by stimulating 40×106 splenocytes with 2×106 d42m1-T3 tumour cells pre-treated for 48 h with 100 U ml−1 of recombinant murine IFN-γ and then irradiated (100 Gy). Cultures were stimulated two more times with irradiated, IFN-γ stimulated tumour cells plus irradiated splenocytes and then CD8+ T cells were purified using magnetic beads (Miltenyi Biotec).
Peptide binding assay
The peptide-MHC class I binding assay with RMA-S cells36 was performed by incubating RMA-S cells with serial dilutions of peptides for 24 h. Cells were stained with mAbs to H-2Kb and H-2Db followed by secondary PE-conjugated goat anti-mouse Ig (BD Biosciences) and analysed by flow cytometry.
Tetramers
For determining kinetics of mLama4- or mAlg8-specific T cell infiltration into tumours, H-2Kb tetramers conjugated to PE were prepared with mutant Lama4 or Alg8 peptides and produced by the NIH Tetramer Core Facility (Emory University). For screening H-2Kb or H-2Db predicted epitopes, we generated peptide-MHC Class I complexes in house. The peptide-MHC class I complexes refolded with a UV-cleavable conditional ligand were prepared as described with modifications37. Briefly, recombinant H-2Kb and H-2Db heavy chains and human β2 microglobulin light chain were produced in Escherichia coli, isolated as inclusion bodies, and dissolved in 4M urea, 20 mM Tris pH 8.0. MHC Class I refolding reactions were performed by dialyzing a molar ratio of heavy chain: light chain: peptide of 1:1:8 against 10 mM potassium phosphate, pH 7.4 for 48 h. The UV-cleavable peptide SIINFEJL used to refold H-2Kb was purchased from Peptide 2.0. Refolded peptide-MHC class I complexes were captured by ion exchange (HiTrap Q HP, GE), biotinylated, and purified by gel filtration FPLC. UV-induced ligand exchange and combinatorial encoding of MHC Class I multimers was performed as described38, except that the peptide-MHC multimers used for flow cytometry staining were prepared by the addition of titrated amounts of streptavidin-fluorochrome in a 10 μL format39.
Measurement of IFN-γ production
To generate target cells, tumour cells were treated with 100 U ml−1 IFN-γ for 48 h and irradiated with 100 Gy before use. CTL cells (20,000 cells) were co-cultured with tumour target cells (20,000 cells) in 96-well round-bottomed plates for 48 h in a total volume of 200 μl. For peptide stimulation of CTL lines, 100,000 naïve irradiated (30 Gy) splenocytes were pulsed with various 8- or 9-mer peptides at 37° C for 1 h. Splenocytes were washed and 20,000 CTLs were added and the culture was incubated for 48 h. IFN-γ in supernatants was quantified using an IFN-γ ELISA kit (eBioscience). For blocking assays, 25 μg ml−1 of anti-H-2Kb (B8-24-3) or anti-H-2Db (B22/249) were added to the target cells (tumours) for 1 h prior to addition of CTL cells. For IFN-γ ELISPOT, pre-coated 96-well PVDF filtration plates (Millipore) were washed with PBS and conditioned with medium containing 10% FCS. Erythrocyte-free single-cell suspensions from the spleen were added in triplicate and incubated for 20 h with 1 μM mutant or wild type peptide. After extensive washes, 1 μg ml−1 biotinylated detection antibody was added. Streptavidin-alkaline phosphatase and 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium (BCIP/NBT) (Moss Substrates) were subsequently used for colour development. Plates were scanned and analysed on an ImmunoSpot reader (C.T.L.). Reagents for ELISPOT were purchased from Mabtech.
Tumour and spleen harvest
Established tumours were excised from mice, minced and treated with 1 mg ml−1 type IA collagenase (Sigma) in HBSS (Hyclone) for 1 h at 37 °C. Spleens were also harvested, crushed and vigorously resuspended to make single-cell suspensions. To remove aggregates and clumps, cells were filtered through a 40-micron strainer.
CD8+ TIL peptide restimulation
Cells from tumours were enriched for CD8+ cells using Miltenyi mouse CD8+ enrichment kit by following manufacturer’s protocol. Splenocytes harvested from naïve mice were labelled with CFSE and irradiated at 30 Gy. 100,000 labelled irradiated splenocytes were then pulsed with 1 μM of peptide and 100,000 CD8+ TILs were subsequently added and incubated at 37 °C. After 1 h, BD GolgiPlug (BD Bioscience) was added and cells were incubated for an additional 4 h. Surface staining was performed and cells were then fixed and permeabilized with BD Cytofix/Cytoperm kit. Cells were then stained for IFN-γ and TNF-α.
Isolation of H-2Kb presented peptides
For H-2Kb isolation, sarcoma cell lines d42m1-T3 and F244 were each expanded to 5 x 108 cells. Prior to harvesting, cells were stimulated with 300 U ml−1 IFN-γ for 48 hours to increase MHC expression. Detachment of cells was facilitated by incubation with 100 U ml−1 Collagenase IV (Gibco) in PBS for 10 min at 37° C in a 5% CO2 incubator. Cells were washed twice with PBS and cell pellets were subsequently snap frozen. MHC class I molecules were isolated as previously described40. In brief, cell pellets were taken up in 1 ml of lysis buffer (1.2% CHAPS (Applichem, Darmstadt, Germany), 1x Protease Inhibitor Cocktail (Roche) in PBS) and homogenized by sonication. Lysates were cleared from remaining cell debris by centrifugation (2,500g, 30 min) and passing through a 0.2 μm filter (Sartorius, Goettingen, Germany). MHC class I molecules were isolated by immunoaffinity purification using H-2Kb-specific antibody Y3 covalently coupled to cyanogen bromide-activated sepharose 4B (GE Healthcare). MHC molecules were eluted with 0.2% trifluoroacetic acid (TFA) and released peptides were further isolated by ultrafiltration through centricons with a 10 kDa cut-off membrane (Millipore, Schwalbach, Germany). Prior to LC-MS analysis peptides were desalted using C18 Zip Tips (Millipore) according to manufacturer’s instructions and volumes were adjusted by vacuum centrifugation.
Identification of H-2Kb peptides by discovery MS
For the discovery mass spectrometry41, mass spectrometry analysis was performed on an LTQ OrbitrapXL hybrid mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) equipped with a nanoelectron spray ion source coupled to a reverse-phase liquid chromatography UHPLC system (Ultimate 3000 RSLCnano, Dionex, Sunnyvale, CA, USA). Samples were injected onto a 75 μm x 2 cm trapping column (Acclaim PapMap RSLC, Dionex) at a flow rate of 4 μl min−1 for 5.75 min. Peptides were then separated on a 50 μm x 50 cm separation column (Acclaim PapMap RSLC, Dionex) at 50°C in a column oven with a flow rate of 175 nl min−1 and a gradient ranging from 2.5 to 32% Acetonitrile over 140 min. Data dependent acquisition was enabled using a top five method (the five most abundant ions with charges of 2+ or 3+ were selected for fragmentation during each scan cycle). Survey scans were performed in the orbitrap at a resolution of 60,000 and a mass range of 350–600 m z−1. Peptides were fragmented and analysed in the ion trap using collision-induced dissociation (CID, normalized collision energy 35%, activation time 30 ms, isolation width, 2 m z−1). Doubly charged masses for mLama4 and mAlg8 were prioritized for fragmentation using an inclusion list.
Data processing was performed with Proteome Discoverer 1.3 (Thermo Fisher) and Mascot search engine (Mascot 2.2.04, Matrix Science). Peak lists were searched against swissprot mus musculus database (exported September 10th 2013, 16,633 reviewed protein sequences) extended with the 62 mutated H-2Kb candidate sequences. Search restrictions were 5 ppm precursor mass tolerance and 0.5 Da fragment mass tolerance with no enzymatic cleavage specificity selected. Percolator algorithm was used to evaluate false discovery rate with a target value of 0.05 (5% FDR). Additional post processing filters were an ion score ≥ 20, search engine rank 1 and peptide length between 8 and 12 amino acids.
Identification of H-2Kb peptides by targeted SRM
To generate the SRM42 assay library, the synthetic peptides were first analysed on an AB Sciex QTRAP 5500 mass spectrometer in SRM-triggered MS2 mode. Peptides (PEPotec SRM Grade 2) were synthesized by Thermo Fischer Scientific GmbH (Ulm, Germany). 51 of the 62 strong binding H-2Kb peptides were selected for analysis because they had physiochemical properties that would allow their detection by MS if present. Chromatographic separations of peptides were performed by a nanoLC ultra 2Dplus system (Eksigent) coupled to a 15 cm fused silica emitter, 75 μm inner diameter, packed with a Magic C18 AQ 5 μm resin (Michrom BioResources). Peptides were loaded on the column from a cooled (4°C) nanoLC-AS2 autosampler (Eksigent) and separated in 60 minutes by a linear gradient of acetonitrile (5 – 35%) and water, containing 0.1% formic acid at a flow rate of 300 nl min−1. The mass spectrometer was operated in SRM mode, triggering acquisition of a full fragment ion spectrum upon detection of an SRM trace (threshold 1000 ion counts). SRM acquisition was performed with Q1 and Q3 operated at unit resolution (0.7 m z−1 half maximum peak width) with a dwell time of 20 ms for each transition. For each peptide the first fragment ion of the y-series with an m z−1 above the m z−1 precursor + 20 Thomson (Th), for the doubly charged peptide precursors, were used as triggering transitions. MS/MS spectra were acquired in enhanced product ion mode, using quadrupole (q2) fragmentation, low Q1 resolution, scan speed 10000 Da s−1, and an m z−1 range of 250–1000. Collision energies (CEs) were calculated according to the formulas: CE = 0.044 x m z−1 precursor + 5.5 for doubly charged precursor ions. Raw data files (wiff) were converted into mzXML format using msConvert from ProteoWizard (version 1.6.1455). Qtrap MS2 spectra were assigned to peptide sequences using the SEQUEST algorithm and PeptideProphet. The software Skyline43 was used to generate a spectral library from the Qtrap mzXML files. Optimal SRM transition conditions (collision energy and fragment ion selection) were determined for individual peptides as previously described by using synthetic peptides44. Retention times were extracted from the spectra and converted into a system-independent retention time (iRT) using spiked-in calibration peptides (Biognosys)45. For each target mutant peptides, a heavy isotope-labelled reference peptides was spiked in the H-2Kb peptide mixture for identification. For each peptide, three to seven transitions were monitored for the heavy and light version and the resulting SRM data were visualized and analysed manually by using the software Skyline. The unequivocal detection of mutant MHC Class I H-2Kb-associated peptides was achieved by comparing three properties between the reference heavy isotope-labelled peptide and its endogenous version: (a) their retention time must be consistent, (b) they must trigger all SRM transitions concurrently and (c) SRM transitions must be in the correct abundance hierarchy.
Fluorescence-activated cell sorting analysis
For flow cytometry, cells were stained for 5 min at room temperature with 500 ng of Fc block (anti-CD16/32) and then stained with 200 ng of antibodies to CD45, CD4 or CD8α in 100 μl of staining buffer (PBS with 2% FCS and 0.05% NaN3 (Sigma)) for 20 min at 4 °C. Propidium iodide (PI) (Sigma) was added at 1 μg ml−1 immediately before FACS analysis. For tetramer staining, cells were stained for 5 min at room temperature with 500 ng of Fc block (anti-CD16/32). Cells were then stained with anti-CD8 antibody (clone CT-CD8α from Accurate Chemical) for 20 min at 4 °C at a 1:100 dilution. Antibodies to CD45 and CD3ε along with H-2Kb tetramers conjugated to PE were then added at a concentration of 1:100 for 30 min at 4 °C. For intracellular cytokine staining of tumour infiltrating lymphocytes, cells were isolated incubated for 5 hours with GolgiPlug (BD bioscience). Cells were then harvested, washed, stained with anti-CD3ε and CD8α for 15 min, and then fixed and permeabilized with BD fixation and permeabilization kit (BD Biosciences). Cells with then stained with anti-IFN-γ or anti-TNF-α. For sorting of mLama4-specific cells, tumour-infiltrating cells were enriched for CD45+ cells using CD45 cell purification magnetic beads (Miltenyi Biotec). CD45 enriched cells were then sorted gating for PI− CD3ε+ CD8α+ mLama4-tetramer-PE+ or PI− CD3ε+ CD8α+ mLama4-tetramer-PE− cells. Sorting was performed on a BD FACSAria II (BD Biosciences). Sorted cells were pelleted and processed for RNA analysis. The purity of the sorted cells for input RNA was greater than 90% as assessed during post-sort cellular analysis. All flow cytometry was performed on the FACSCalibur (BD Biosciences) or LSR Fortessa (BD Biosciences) and analysed using FlowJo software (TreeStar).
Vaccination
Mice were vaccinated s.c. with 50 μg peptides corresponding to mLama4 or mAlg8 in combination with 100 μg poly I:C (Sigma-Aldrich). For SLP, the mLama4 peptide sequence used was QKISFFDGFEVGFNFRTLQPNGLLFYYT (epitope underlined) and the mAlg8 peptide sequence used was AVGITYTWTRLYASVLTGSLV (epitope underlined). Two weeks later spleens were harvested and splenocytes were restimulated with wild type or mutant Lama4 or Alg8 8-mer peptides. Cells were then collected for ELISPOT analysis. For prophylactic vaccination, mice were subcutaneously injected with 50 μg of peptides corresponding to mLama4 or mAlg8, used alone or in combination, along with 100 μg of poly I:C in a total volume of 200 μl diluted in HBSS on day -10, -3, and +4 post-tumour transplant. d42m1-T3 tumour cells were transplanted at 1x106 tumour cells on day 0. As controls, mice were immunized with either HBSS alone, poly I:C alone or a long peptide vaccine corresponding to human papilloma virus (HPV) with poly I:C. Therapeutic vaccination experiments were performed in identical manner except vaccines were administered on day +3, +9, and +15 post-tumour transplant. Lack of survival was defined as mouse death or tumour size of 20 mm.
RNA sequencing
The purity of the sorted cell preparations were greater than 90% as assessed by post-sort cellular analysis. mRNA was extracted from cell lysates using oligo-dT beads (Invitrogen). For cDNA synthesis, we used custom oligo-dT primers with a barcode and adapter-linker sequence (CCTACACGACGCTCTTCCGATCT—XXXXXXXX-T15). After first strand synthesis, samples were pooled together based on Actb qPCR values and RNA-DNA hybrids were degraded using consecutive acid-alkali treatment. Then, a second sequencing linker (AGATCGGAAGAGCACACGTCTG) was ligated using T4 ligase (NEB) followed by SPRI clean-up. The mixture then was enriched by 16 cycles of PCR and SPRI purified to yield final strand specific RNA-seq libraries. The libraries were sequenced on a HiSeq 2500 instrument using 50bpX25bp pair-end sequencing. Second read-mate was used for sample demultiplexing, at which point individual single-end fastqs files were aligned to mm9 genome using STAR aligner with the following options --runThreadN 8 --outFilterMultimapNmax 15 --outFilterMismatchNmax 6 --outReadsUnmapped Fastx --outSAMstrandField intronMotif --outSAMtype BAM SortedByCoordinate. Gene expression was quantitated using ht-seq and differentially expressed genes were defined using DESeq2 R package at p value <0.01. Gene Set Enrichment Analysis (GSEA) using canonical pathway- and immunological signature-databases was performed as previously described46. RNA-sequencing data is available at Gene Expression Omnibus (GEO) repository at http://www.ncbi.nlm.nih.gov/geo/.
Statistical analysis
Samples were compared using an unpaired, two-tailed Student’s t-test, unless specified.
Extended Data
Extended Data Figure 1. Innate and adaptive immune components are required for rejection of d42m1-T3 after checkpoint blockade therapy.
a, Cohorts of Rag2−/−, Batf3−/−, or wild type mice were treated with control mAb, αCD4, αCD8α, or αIFN-γ mAbs and then were injected with 1 x 106 d42m1-T3 tumour cells s.c. and subsequently treated with αCTLA-4 on days 3, 6, and 9 post-transplant. b, c, d42m1-T3 (b) or F244 (c) tumour cells were injected s.c. into WT mice (n=5) that were subsequently treated with αPD-1 on days 3, 6, and 9. Fifty days after tumours were rejected, mice were rechallenged with d42m1-T3 or F244 tumour cells. Data are presented as average tumour diameter ± s.e.m. of 5 mice per group and are representative of at least two independent experiments.
Extended Data Figure 2. H-2Db mutant epitopes of d42m1-T3 tumours.
a, Missense mutations in d42m1-T3 were subjected to in silico analysis for the potential to form H-2Db binding epitopes using three epitope prediction algorithms. The median predicted epitope binding affinity for each peptide was calculated and expressed as “median affinity value” where affinity value = 1/IC50 x 100. Predicted epitopes are arrayed along the X-axis in alphabetical order based on their protein of origin b, Unfiltered median affinity values for the 4 predicted H-2Db epitopes. c, Median affinity values of remaining 2 H-2Db epitopes after filtering. d, Tetramer staining of CD8+ TILs from tumour bearing mice treated with αPD-1 using H-2Db tetramers loaded with top 4 H-2Db synthetic peptides. e, IFN-γ and TNF-α intracellular cytokine staining of CD8+ TILs from tumour bearing mice treated with αPD-1 immunotherapy following co-culture with naïve irradiated splenocytes pulsed with the top 4 H-2Db synthetic peptides added at 1 μM final concentration. Data are presented as per cent of CD8+ TILs positive for IFN-γ, TNF-α, or both cytokines. Data are representative of two independent experiments.
Extended Data Figure 3. mLama4 and mAlg8 stimulate CD8+ T cell lines generated against d42m1-T3 following αPD-1.
a, CD8+ T cell lines generated from splenocytes of individual d42m1-T3 tumour bearing mice that rejected their tumours after αPD-1 therapy were incubated with irradiated d42m1-T3 tumour cells (or F244 tumour cells) treated with blocking mAb specific for H-2Kb, and/or H-2Db and IFN-γ production quantitated. Data are presented as means ± s.e.m and are representative of two independent experiments. Samples were compared using an unpaired, two-tailed Student’s t test (***p<0.001). b, IFN-γ release by the CTL 74 T cell line following co-culture with naïve irradiated splenocytes pulsed with the top 62 H-2Kb synthetic peptides added at 1 μM final concentration.
Extended Data Figure 4. mLama4 and mAlg8 bind H-2Kb and stimulate CD8+ T cell lines generated against d42m1-T3 following αPD-1 immunotherapy.
a, IFN-γ release by CTL 62, CTL 73, or CTL 74 T cell lines following stimulation with naïve irradiated splenocytes pulsed with WT or mutant forms of Lama4 or Alg8 peptides. b, RMA-S cells were incubated with 8 amino acid peptides of mLama4 or mAlg8 and surface expression of H-2Kb or H-2Db was assessed by flow cytometry. Mean fluorescent intensity of H-2Kb and H-2Db was expressed as peptide binding score. Data presented are representative of at least two independent experiments.
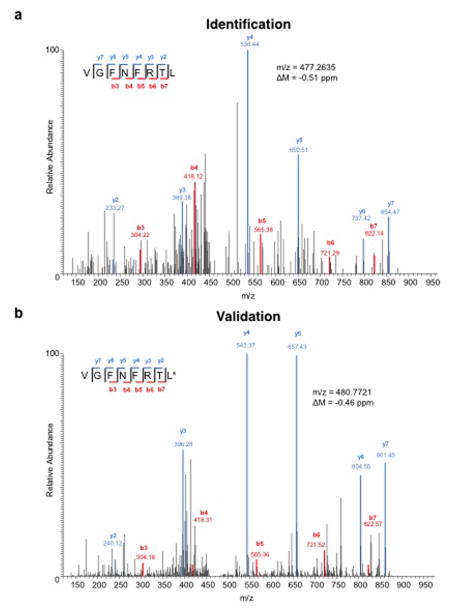
Extended Data Figure 5. Identification of a peptide bound to H-2Kb on d42m1-T3 tumour cells corresponding to mutant Lama4.
a, Identification of the peptide, VGFNFRTL, corresponding to mLama4 by discovery MS. b, Validation of the mLama4 peptide using an isotope-labelled synthetic peptide (VGFNFRTL (13C6, 15N1)).
Extended Data Figure 6. Generation of SRM assay library for the detection of mutant H-2Kb peptides on d42m1-T3.
a, SRM transitions were optimized for 51 of the 62 top predicted H-2Kb peptides. The 51 peptides chosen were selected based on having physiochemical properties that would allow their detection by MS if present. Only Lama4 and Alg8 are shown here for simplicity. The 51 peptides were synthesized and LC-MS/MS acquisition was performed on each peptide to determine the best collision energy and to obtain the full fragment ion spectrum (left panel); three to seven of the highest intensity peaks were selected to be built into SRM transitions. Optimal SRM transitions displayed as extracted ion chromatograms are shown (right panel). Q1 – Q3 transitions are indicated in parenthesis. The mutated amino acid in the peptide sequence is marked in red. b, F244 tumour cells, which lack the mLama4 and mAlg8 d42m1-T3 epitope, lack detectable mLama4 or mAlg8 in complex with H-2Kb as assessed by SRM.
Extended Data Figure 7. CD8+ T cells specific for mutant forms of Lama4 and Alg8 infiltrate d42m1-T3, but not F244, tumours.
a, Detection of tumour infiltrating mLama4- or mAlg8-specific T cells infiltrating d42m1-T3 or F244 tumours of mice treated with αPD-1. Tumours were harvested on day 12 post-transplant. Cells were gated on live CD45+ and CD8α+ tumour infiltrating lymphocytes. Detection of mLama4- or mAlg8-specific T cells was achieved by staining with peptide-MHC H-2Kb PE-labelled tetramers. Data are representative of at least five independent experiments. b, Detection of mLama4-specific tumour infiltrating T cells from tumour-bearing mice treated with αPD-1, αCTLA-4, both αPD-1 plus αCTLA-4 or control mAb. Detection of mLama4-specific T cells was achieved by staining with mLama4-MHC H-2Kb PE-labelled tetramers. Data presented are plotted as the mean mLama4 tetramer-positive as a percent of CD8α+ tumour infiltrating cells and are representative of at least three independent experiments.
Extended Data Figure 8. mAlg8 and mLama4 SLP vaccine control d42m1-T3 tumour outgrowth when administered therapeutically or prophylactically.
a, Tumour growth of d42m1-T3 tumours from mice therapeutically vaccinated with mLama4 and mAlg8 SLP plus Poly I:C, HPV control SLP plus Poly I:C or Poly I:C alone. Data shown are mean ± s.e.m. Mutant Lama4 and mAlg8 SLP vaccine group was compared to HPV control SLP vaccine group using an unpaired, two-tailed Student’s t test (*p<0.05 and **p<0.01) b, Kaplan-Meier survival curves of d42m1-T3 tumour bearing mice (7 per group) prophylactically vaccinated with SLP vaccines plus poly I:C. mLama4 plus mAlg8 compared to HPV control: p=0.0003 [log-rank (Mantel-Cox) test]. Representative of two independent experiments. c, Cumulative number of mice (7–10 per group) from at least two independent experiments rejecting d42m1-T3 or F244 tumours as a consequence of SLP or minimal epitope peptide prophylactic vaccination.
Extended Data Figure 9. Detection of TIM-3, LAG-3, IFN-γ and TNF-α expression in tumour infiltrating CD8+ T cells.
a, Representative histogram of TIM-3 or LAG-3 expression on mLama4-specific CD8+ tumour infiltrating T cells from tumour bearing mice treated with αPD-1, αCTLA-4, both αPD-1 and αCTLA-4 or control mAbs. b, TIM-3 and LAG-3 are reduced in mAlg8-specific CD8+ TILs from tumour-bearing mice treated with αPD-1, αCTLA-4, or both αPD-1 and αCTLA-4 compared to mice treated with control mAb. N=5 mice per group pooled. Data are presented as mean ± s.e.m. of at least three independent experiments. Samples were compared using an unpaired, two-tailed Student’s t test (*p<0.05, **p<0.01). c, Representative dot plots of IFN-γ and TNF-α stained CD8+ tumour-infiltrating T cells from tumour-bearing mice following treatment with αPD-1, αCTLA-4, both αPD-1 and αCTLA-4 or control mAbs. Data presented are representative of at least three independent experiments.
Extended Data Table 1. Commonly differentially regulated genes and GSEA pathways in mLama4+ TILs during checkpoint blockade therapy.
a, 25 differentially expressed genes (p<0.05) that are commonly up-regulated (red) or down-regulated (blue) in mLama4-specific CD8+ TILs from mice treated with all checkpoint blockade mAbs versus TILs from control mAb treated mice. Genes underlined and in bold are those involved in CD8+ T effector cells in acute verses chronic infection. b, GSEA pathway analysis showing selected pathways (and their p values) altered in mLama4-specific CD8+ TILs during different checkpoint blockade treatments.
| a | ||||||
|---|---|---|---|---|---|---|
| Commonly Altered Genes in Checkpoint Blockade Treated Groups | ||||||
| Gene | anti-PD-1 | anti-CTLA-4 | anti-PD-1 + anti-CTLA-4 | |||
| Fold Change | p value | Fold Change | p value | Fold Change | p value | |
| Abcf1 | 1.8498 | 0.0009 | 1.6563 | 0.0010 | 1.6133 | 0.0091 |
| Arhgap11a | 2.2469 | 0.0085 | 2.5201 | 0.0003 | 2.1815 | 0.0004 |
| Brca1 | 3.0675 | 0.0050 | 4.1248 | 0.0004 | 3.6843 | 0.0003 |
| Cep128 | 2.2903 | 0.0020 | 2.6134 | <10−4 | 2.1577 | 0.0079 |
| Clspn | 2.6332 | 0.0003 | 2.0996 | 0.0002 | 2.5427 | <10−4 |
| Golga4 | 2.1608 | 0.0046 | 1.6866 | 0.0089 | 2.1701 | 0.0003 |
| Kif20b | 2.6334 | 0.0003 | 2.6786 | <10−4 | 2.2927 | 0.0001 |
| Lef1 | 2.0932 | 0.0033 | 2.0713 | 0.0019 | 2.1006 | 0.0012 |
| Lmnb1 | 2.6944 | <10−4 | 2.6335 | 0.0002 | 2.1587 | 0.0002 |
| Nusap1 | 2.3610 | 0.0011 | 2.2774 | <10−4 | 1.8717 | 0.0036 |
| Pbdc1 | 2.2750 | 0.0002 | 1.8901 | 0.0038 | 1.7994 | 0.0086 |
| Ripk1 | 2.7563 | 0.0003 | 2.3822 | 0.0022 | 2.9873 | 0.0002 |
| Sclt1 | 2.8084 | 0.0013 | 2.4172 | 0.0035 | 2.4687 | 0.0044 |
| Smc1a | 1.6027 | 0.0070 | 1.5141 | 0.0043 | 1.5802 | 0.0033 |
| Smc2 | 2.4076 | 0.0005 | 2.3176 | <10−4 | 1.7604 | 0.0013 |
| Smc3 | 2.1347 | 0.0008 | 1.7558 | 0.0001 | 1.7829 | 0.0030 |
| 5330439K02Rik | 7.0531 | 0.0040 | 7.7751 | 0.0060 | 8.5834 | 0.0079 |
| Atp2b2 | 5.4570 | 0.0021 | 7.6665 | 0.0008 | 37.4543 | <10−4 |
| Dnajb1 | 1.9973 | 0.0061 | 2.0345 | 0.0007 | 3.1415 | <10−4 |
| Ephx1 | 2.8845 | 0.0059 | 3.3558 | 0.0006 | 6.8748 | 0.0001 |
| Errfi1 | 1.8630 | 0.0054 | 2.0856 | 0.0018 | 2.7131 | 0.0006 |
| Ifitm1 | 2.8256 | 0.0002 | 2.1070 | 0.0002 | 2.4488 | 0.0001 |
| Klhdc8a | 6.9301 | 0.0044 | 8.7394 | 0.0036 | 9.7055 | 0.0048 |
| Tctn2 | 8.3409 | 0.0012 | 13.5967 | 0.0004 | 14.7772 | 0.0007 |
| Xcl1 | 2.8113 | 0.0001 | 2.9306 | <10−4 | 2.2410 | <10−4 |
| b | |||
|---|---|---|---|
| p value | |||
| GSEA Pathway | anti-PD-1 | anti-CTLA-4 | anti-PD-1 + anti-CTLA-4 |
| REACTOME_IL_2_SIGNALING | 0.0353 | 0.2631 | 0.0562 |
| REACTOME_GLYCOLYSIS | 0.0487 | 0.8937 | 0.0586 |
| BIOCARTA_TNFR1_PATHWAY | 0.0395 | 0.3855 | 0.0564 |
| REACTOME_INTERFERON_SIGNALING | 0.0350 | 0.5374 | 0.0311 |
| ST_TYPE_I_INTERFERON_PATHWAY | 0.0173 | 0.0519 | 0.0063 |
| KEGG_OXIDATIVE_PHOSPHORYLATION | <10−4 | 0.3731 | <10−4 |
| REACTOME_TCA_CYCLE_AND_RESPIRATORY_ELECTRON_TRANSPORT | <10−4 | 0.9532 | <10−4 |
| REACTOME_GLUCOSE_METABOLISM | 0.0115 | 0.9055 | 0.0114 |
| KEGG_PENTOSE_PHOSPHATE_PATHWAY | 0.0167 | 0.6182 | 0.0278 |
| REACTOME_CELL_CYCLE_CHECKPOINTS | 0.0534 | <10−4 | 0.0730 |
| SA_G2_AND_M_PHASES | 0.1285 | 0.0023 | 0.1081 |
| KEGG_JAK_STAT_SIGNALING_PATHWAY | 0.1608 | <10−4 | 0.1568 |
| REACTOME_SYNTHESIS_OF_DNA | 0.0619 | <10−4 | 0.0563 |
| PID_E2F_PATHWAY | 0.0813 | <10−4 | 0.0670 |
| KEGG_CELL_CYCLE | 0.0530 | <10−4 | 0.0454 |
| REACTOME_INTEGRIN_CELL_SURFACE_INTERACTIONS | 0.0619 | 0.0066 | 0.0461 |
| PID_NFAT_TFPATHWAY | 0.0693 | 0.0054 | 0.0486 |
| PID_GMCSF_PATHWAY | 0.0875 | 0.0678 | 0.0467 |
| BIOCARTA_CXCR4_PATHWAY | 0.0525 | 0.9422 | 0.0462 |
| GOLDRATH_NAIVE_VS_EFF_CD8_TCELL_UP | <10−4 | <10−4 | 0.0052 |
| GSE10239_KLRG1INT_VS_KLRG1HIGH_EFF_CD8_TCELL_DN | <10−4 | <10−4 | <10−4 |
| KEGG_MAPK_SIGNALING_PATHWAY | <10−4 | <10−4 | <10−4 |
| KEGG_CHEMOKINE_SIGNALING_PATHWAY | <10−4 | <10−4 | <10−4 |
| KEGG_CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION | 0.0054 | <10−4 | <10−4 |
Supplementary Material
Acknowledgments
We are grateful to, Kenneth Murphy (Washington University) for the Batf3−/− mice, Ted Hansen (Washington University) for providing MHC class I antibodies and the H-2Kb construct, Daved Fremont (Washington University) for the human β2m construct, and the National Institutes of Health (NIH) Tetramer Core Facility for producing MHC class I tetramers. We thank Ariel Bensimon, Olga Schubert and Petri Kouvonen (ETH Zurich, Switzerland) for instrument maintenance and for technical support with the MS measurements and Rangan Vanganipuram, Mark Selby, and Jose Valle (Bristol-Meyers Squibb) for generating and supplying anti-PD-1 and anti-CTLA-4 in endotoxin free sterile form. We also thank Kathleen Sheehan (Washington University), Paul Allen (Washington University), Gavin Dunn (Washington University), and Ruby Chan (Washington University) for constructive criticisms and comments, all members of the Schreiber laboratory for discussions, and the many members of The Genome Institute at Washington University School of Medicine. We would also like to thank Wilbur Song (Washington University) for his assistance with the bioinformatics approaches, Pai Kvistborg (The Netherlands Cancer Institute) for assistance with tetramer combinatorial coding, and Christopher Nelson (Washington University) for advice with peptide-MHC monomer purification. This work was supported by grants to R.D.S. from the National Cancer Institute (RO1 CA043059, U01 CA141541), the Cancer Research Institute, and the WWWW Foundation; to R.D.S. and W.E.G. from The Siteman Cancer Center/Barnes-Jewish Hospital (Cancer Frontier Fund); to W.E.G. from Susan G. Komen for the Cure (Promise grant); to E.R.M. from the National Human Genome Research Institute; to G.J.F. from the National Institute of Health (P50 CA101942, P01 AI054456, P50 CA101942); to A.H.S from the National Institute of Health (P50 CA101942); and to T.N.S. from the Dutch Cancer Society (Queen Wilhelmina Research Award). E.C. is supported by a Marie Curie Intra-European Fellowship within the Seventh Framework Programme of the European Community for Research. M.M.G. was supported by a post-doctoral training grant (T32 CA00954729) from the National Cancer Institute and is currently supported by a post-doctoral training grant (Irvington Postdoctoral Fellowship) from the Cancer Research Institute. Aspects of studies at Washington University were performed with assistance by the Immunomonitoring Laboratory of the Center for Human Immunology and Immunotherapy Programs and the Siteman Comprehensive Cancer Center.
Footnotes
AUTHOR CONTRIBUTIONS:
M.M.G. and R.D.S. were involved in all aspects of this study including planning and performing experiments, analysing and interpreting data, and writing the manuscript. X.Z. performed peptide binding experiments, helped design and perform the vaccine experiments. H.S., E.C., R.A. and H-G.R planned and performed the mass spectrometry analyses, interpreted the data and were involved in writing the manuscript. T.N., J.P.W., C.D.A., M.D.V., S.K.L., and E.L.P, participated in assessing the phenotypes of the tumour-specific T cell lines, interpreting the data and in writing the manuscript. M.T. helped generate MHC class I multimers. A.J.K., J.P.A., G.J.F. and A.H.S. provided mAbs, helped plan the checkpoint blockade therapy experiments, and contributed to writing the manuscript. T.N.S helped generate MHC class I multimers, analysed data and was involved in writing the manuscript. WJ.K., G.E.M. and C.J.M.M. produced and purified the synthetic long peptides, participated in the planning of the vaccine experiments, analysed data and were involved in writing the manuscript. J.H. and E.R.M. were responsible for genomic analyses and epitope prediction and participated in writing the manuscript. W.E.G. contributed to the design and analysis of peptide binding and vaccine experiments and in writing the manuscript. Y.I. and M.N.A were responsible for optimizing the epitope prediction method, performing the RNA-sequencing analyses, analysing data and writing the manuscript. R.D.S. oversaw all the work performed.
LITERATURE CITED
- 1.Shankaran V, et al. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 2.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nature immunology. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 3.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 4.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trinchieri G. Cancer and inflammation: an old intuition with rapidly evolving new concepts. Annual review of immunology. 2012;30:677–706. doi: 10.1146/annurev-immunol-020711-075008. [DOI] [PubMed] [Google Scholar]
- 6.Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339:286–291. doi: 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koebel CM, et al. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 8.Quezada SA, Peggs KS, Simpson TR, Allison JP. Shifting the equilibrium in cancer immunoediting: from tumor tolerance to eradication. Immunological reviews. 2011;241:104–118. doi: 10.1111/j.1600-065X.2011.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolchok JD, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamid O, et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N Engl J Med. 2013;369:134–144. doi: 10.1056/NEJMoa1305133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. The New England journal of medicine. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hodi FS, et al. Improved survival with ipilimumab in patients with metastatic melanoma. The New England journal of medicine. 363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsushita H, et al. Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature. 482:400–404. doi: 10.1038/nature10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paul S, et al. HLA class I alleles are associated with peptide-binding repertoires of different size, affinity, and immunogenicity. Journal of immunology. 2013;191:5831–5839. doi: 10.4049/jimmunol.1302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.West EE, et al. Tight regulation of memory CD8(+) T cells limits their effectiveness during sustained high viral load. Immunity. 2011;35:285–298. doi: 10.1016/j.immuni.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 18.Castle JC, et al. Exploiting the mutanome for tumor vaccination. Cancer research. 72:1081–1091. doi: 10.1158/0008-5472.CAN-11-3722. [DOI] [PubMed] [Google Scholar]
- 19.Robbins PF, et al. Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nature medicine. 2013;19:747–752. doi: 10.1038/nm.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fritsch EF, et al. HLA-binding properties of tumor neoepitopes in humans. Cancer immunology research. 2014;2:522–529. doi: 10.1158/2326-6066.CIR-13-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Rooij N, et al. Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2013;31:e439–442. doi: 10.1200/JCO.2012.47.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curran MA, Montalvo W, Yagita H, Allison JP. PD-1 and CTLA-4 combination blockade expands infiltrating T cells and reduces regulatory T and myeloid cells within B16 melanoma tumors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:4275–4280. doi: 10.1073/pnas.0915174107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brunner MC, et al. CTLA-4-Mediated inhibition of early events of T cell proliferation. Journal of immunology. 1999;162:5813–5820. [PubMed] [Google Scholar]
- 24.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annual review of immunology. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okazaki T, Chikuma S, Iwai Y, Fagarasan S, Honjo T. A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nature immunology. 14:1212–1218. doi: 10.1038/ni.2762. [DOI] [PubMed] [Google Scholar]
- 26.Duraiswamy J, Kaluza KM, Freeman GJ, Coukos G. Dual blockade of PD-1 and CTLA-4 combined with tumor vaccine effectively restores T-cell rejection function in tumors. Cancer research. 2013;73:3591–3603. doi: 10.1158/0008-5472.CAN-12-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiotto MT, Rowley DA, Schreiber H. Bystander elimination of antigen loss variants in established tumors. Nature medicine. 2004;10:294–298. doi: 10.1038/nm999. [DOI] [PubMed] [Google Scholar]
- 28.Wolkers MC, Brouwenstijn N, Bakker AH, Toebes M, Schumacher TN. Antigen bias in T cell cross-priming. Science. 2004;304:1314–1317. doi: 10.1126/science.1096268. [DOI] [PubMed] [Google Scholar]
- 29.Corbiere V, et al. Antigen spreading contributes to MAGE vaccination-induced regression of melanoma metastases. Cancer research. 2011;71:1253–1262. doi: 10.1158/0008-5472.CAN-10-2693. [DOI] [PubMed] [Google Scholar]
- 30.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hildner K, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters B, Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics. 2005;6:132. doi: 10.1186/1471-2105-6-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lundegaard C, et al. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 2008;36:W509–512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoof I, et al. NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics. 2009;61:1–13. doi: 10.1007/s00251-008-0341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nielsen M, Lundegaard C, Lund O, Kesmir C. The role of the proteasome in generating cytotoxic T-cell epitopes: insights obtained from improved predictions of proteasomal cleavage. Immunogenetics. 2005;57:33–41. doi: 10.1007/s00251-005-0781-7. [DOI] [PubMed] [Google Scholar]
- 36.Esquivel F, Yewdell J, Bennink J. RMA/S cells present endogenously synthesized cytosolic proteins to class I-restricted cytotoxic T lymphocytes. The Journal of experimental medicine. 1992;175:163–168. doi: 10.1084/jem.175.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toebes M, et al. Design and use of conditional MHC class I ligands. Nature medicine. 2006;12:246–251. doi: 10.1038/nm1360. [DOI] [PubMed] [Google Scholar]
- 38.Andersen RS, et al. Parallel detection of antigen-specific T cell responses by combinatorial encoding of MHC multimers. Nature protocols. 2012;7:891–902. doi: 10.1038/nprot.2012.037. [DOI] [PubMed] [Google Scholar]
- 39.Kvistborg P, et al. TIL therapy broadens the tumor-reactive CD8(+) T cell compartment in melanoma patients. Oncoimmunology. 2012;1:409–418. doi: 10.4161/onci.18851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kowalewski DJ, Stevanovic S. Biochemical large-scale identification of MHC class I ligands. Methods in molecular biology. 2013;960:145–157. doi: 10.1007/978-1-62703-218-6_12. [DOI] [PubMed] [Google Scholar]
- 41.Thommen DS, et al. Two preferentially expressed proteins protect vascular endothelial cells from an attack by peptide-specific CTL. Journal of immunology. 2012;188:5283–5292. doi: 10.4049/jimmunol.1101506. [DOI] [PubMed] [Google Scholar]
- 42.Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nature biotechnology. 2010;28:710–721. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- 43.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lange V, Picotti P, Domon B, Aebersold R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Escher C, et al. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012;12:1111–1121. doi: 10.1002/pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.