Abstract
Embryonic stem cell maintenance, differentiation, and somatic cell reprogramming require the interplay of multiple pluripotency factors, epigenetic remodelers, and extracellular signaling pathways. RNA-binding proteins (RBPs) are involved in a wide range of regulatory pathways, from RNA metabolism to epigenetic modifications. In recent years we have witnessed more and more studies on the discovery of new RBPs and the assessment of their functions in a variety of biological systems, including stem cells. We review the current studies on RBPs and focus on those that have functional implications in pluripotency, differentiation, and/or reprogramming in both the human and mouse systems.
Keywords: RNA-binding protein, embryonic stem cell, pluripotency, differentiation, somatic cell reprogramming, lncRNA
Introduction
Embryonic stem cells (ESCs) can be derived from the inner cell mass of the preimplantation blastocyst and maintained in vitro under defined culture conditions without losing their pluripotency and self-renewal capacities (Evans and Kaufman, 1981; Martin, 1981; Bongso et al., 1994; Thomson et al., 1998). Pluripotency is defined as the potential to give rise to any cell type of the embryo proper, whereas self-renewal refers to the ability of a cell to propagate indefinitely without losing its pluripotent properties. Pluripotency maintenance as well as the exit and re-acquisition of pluripotency by differentiation and somatic cell reprogramming, respectively, require the interplay of transcriptional regulators, epigenetic modifiers, and extracellular signaling pathways (Boyer et al., 2006; Buganim et al., 2013). Tremendous efforts have been directed toward studying chromatin binding proteins such as DNA binding transcription factors and chromatin modifying proteins (Chambers and Tomlinson, 2009; Kashyap et al., 2009; Hanna et al., 2010; Apostolou and Hochedlinger, 2013; Saunders et al., 2013; Radzisheuskaya and Silva, 2014). However, much less is known about the roles of RNA-binding proteins (RBPs) in pluripotency, differentiation, and reprogramming. RBPs participate in every step of RNA biology, from transcription, splicing, and polyadenylation to RNA modification, transport, translation, and turnover. Moreover, RBPs can function as bridging factors between RNA molecules and protein complexes. The recent technical development for studying RBP properties and partners (Ule et al., 2005; Darnell, 2010; Li et al., 2014) has facilitated the discovery of new RBPs and has opened up new avenues for understanding their biological functions. This review focuses on RBPs that play roles in ESC maintenance, differentiation, and somatic cell reprogramming in the human and mouse settings.
Characterization of RNA-binding proteins
RNA-binding domains
Historically, RBPs were named as such because they possessed canonical RNA-binding domains for direct and specific interactions with their RNA targets. The specificity of these interactions can be sequence- and/or structure-mediated, giving rise to different modes of recognition. Post-translational modification of RBPs can also modify their RNA-binding affinity, function, and localization, generating additional layers of complexity (Reviewed in (Glisovic et al., 2008)). The main canonical RNA-binding domains are discussed below and are summarized in Table 1.
Table 1.
RNA binding domains and representative proteins with functions in development, differentiation, and reprogramming
| RNA binding domain | Example | Function | Reference |
|---|---|---|---|
| RRM | Rbfox2 | Regulation of alternative splicing of reprogramming and differentiation | Yeo et al., 2009; Venables et al., 2013b |
| SR | Son | Regulation of alternative splicing of pluripotency genes in hESC | Lu et al; 2013 |
| KH | hnRNP K and hnRNP E1/E2 | Translational control in erythroid differentiation | Ostareck et al., 1997 |
| RGG | hnRNPU | RNA/DNA binding, activation of OCT4 gene expression | Choi et al., 2011 |
| PAZ | Ago2 | Regulation of miRNA and siRNA pathways for proper ESC maintenance | Shekar et al., 2011 |
| dsRBD | Drosha | microRNA maturation | Kanellopoulou et al., 2005; Wang et al., 2007; Doyle et al., 2013 |
| DEAD box | DDX5 | Regulation of splicing and transcription in cell differentiation and proliferation | Caretti et al., 2006 |
| ZnF | Lin28 | Inhibition of pri-miR-let7 processing | Piskounova et al., 2011 |
| PuF | Pum1 | Transcriptional repression and activation by binding to 3′UTR of target mRNA | Galgano et al., 2008 |
| Non canonical | Ezh2 | Chromatin silencing through H3 K27 trimetylation | Zhao et al., 2010 |
RNA-recognition motif (RRM)
The RNA-recognition motif (RRM), also known as RNA-binding domain (RBD) or ribonucleoprotein (RNP) domain, is the most abundant (0.5%–1% of human genes) (Venter et al., 2001) and is by far the most extensively studied RNA-binding domain in higher vertebrates (Maris et al., 2005). This domain has been shown to interact not only with RNA, but also with DNA and protein partners. It is often found as multiple repeats within a single protein. A single RBD can bind 2–6 nucleotides, whereas multiple copies of the domain allow for the recognition of larger and more complex RNA targets, thus enhancing the affinity and specificity of RNA-binding (Maris et al., 2005).
Serine-arginine rich splicing factors (SR)
Serine-arginine (SR) rich splicing factors are a conserved family of RBPs essential for cell survival and exon-intron boundary recognition during spliceosome assembly (Manley and Tacke, 1996). SR proteins are not only involved in the regulation of constitutive and alternative splicing, but also in regulating a wider range of processes, from transcription to translation (Zhong et al., 2009). Examples of SR proteins are the large and poorly characterized Son and Srsf3, whose functions are discussed later in this review.
K-homology domain (KH)
The hnRNP K-homology (KH) domain is approximately 70 amino acids long and is found in proteins with different functions such as splicing, transcriptional regulation, and translational control (Valverde et al., 2008). The KH domain recognizes four nucleotides with rather weak affinity, but can act in synergy when present in multiple copies in RBPs.
RGG box
RGG domains, first discovered in some hnRNPs, consist of several Arg-Gly-Gly repeats (Kiledjian and Dreyfuss, 1992; Dreyfuss et al., 1993). RGG motifs can bind to their target RNAs directly or indirectly through other proteins (Godin and Varani, 2007). The arginine of the RGG is often dimethylated, which is an important modification in regulating the RNA-binding activity (Rajyaguru and Parker, 2012). The best known examples of RGG proteins are members of the hnRNP family of proteins such as hnRNPA1, hnRNPK, and hnRNPU.
Double-stranded RNA-binding domain (dsRBD)
Double-stranded RNA-binding domains (dsRBDs) were first described for their ability to bind RNA motifs by their 3D-shapes rather than by their sequences (Stefl et al., 2005). Typical dsRBDs are 70 amino acids long, have a conserved structure, and are involved in various processes such as RNA interference, localization, processing, translational control, and editing (Chang and Ramos, 2005). This module is often combined with auxiliary functional domains to allow for specialized functions. For example, Adar2 and PKR have similar dsRBDs, but their specificity is achieved by a deaminase domain and a kinase domain, respectively (Valente and Nishikura, 2005; García et al., 2007).
Piwi/Argonaute/Zwille (PAZ) domain
The PAZ domain is named after the proteins Piwi, Argonaute, and Zwille, which are involved in post-transcriptional gene silencing. The PAZ domain is composed of two subdomains, one of which is well characterized as a single-stranded nucleic acid binding domain (Yan et al., 2003). The post-transcriptional gene silencing mediated by PAZ-containing RBPs is achieved by binding of the PAZ domain to the two-base 3′ overhangs that characterize small interfering RNAs (siRNAs) (Borozdin et al., 2004; Simon et al., 2011; Tian et al., 2011).
RNA helicase DEAD-box
RNA helicases are abundant enzymes that utilize ATP to bind or remodel RNA and RNA-protein complexes (RNPs), and can be found in all organisms. The largest helicase family is comprised of the DEAD-box family of proteins, which are named due to their characteristic Asp-Glu-Ala-Asp motifs. DEAD-box proteins have essential roles in cellular RNA metabolism including transcription, pre-mRNA splicing, ribosome biogenesis, nucleo-cytoplasmatic transport, translation, and RNA decay (reviewed in (Cordin et al., 2006; Linder and Jankowsky, 2011)).
RNA-binding zinc finger (ZnF)
The zinc finger (ZnF) represents a classical dsDNA binding domain, but it can also interact with RNA (Pelham and Brown, 1980; Amarasinghe et al., 2000; Teplova and Patel, 2008). ZnFs can be found alone or in tandem in RBPs, and also in combination with other types of RNA-binding domains. The zinc finger-containing RBPs are classified according to the amino acids that interact with the Zn2+ ion, e.g. CCHH, CCCH, and CCCC, with the CCHH-type being the most frequent. Examples of ZnF proteins involved in RNA processes and that are reviewed here are Mbnl1 (Teplova and Patel, 2008) and Lin28 (Hagan et al., 2009).
PUF RNA-binding repeats
The PUF family of proteins (formed by Pumilio and FBF) are involved in the regulation of embryogenesis, development, and differentiation of most higher eukaryotes (Quenault et al., 2011). The PUF proteins contain a PUM-HD-type RNA-binding domain composed of eight repeats of around 36 amino acids, each one having the ability to bind a single nucleotide in the 3′ untranslated region (UTR) of a target mRNA (Edwards et al., 2001; Wang et al., 2001, 2002). The specificity and high affinity that characterize the RNA-binding PUF domain, together with its modular design, make it attractive for protein engineering. The specific binding of a protein to a given RNA of interest can be achieved by fusing PUF to any desired effector domain (Wang et al., 2002; Filipovska et al., 2011). Pum1 is one of the members of the PUF family of RBPs that was recently discovered as a pluripotency regulator, which will be discussed later in this review.
Non-canonical RNA-binding domains
Some RBPs, despite the fact of not possessing canonical RNA-binding domains, have been shown to interact and function with RNAs. The best studied example is the polycomb protein Ezh2. Ezh2 is one of the catalytic subunits of the Polycomb Repressive Complex 2 (PRC2) which is involved in epigenetic repression by chromatin compaction. Several studies have demonstrated how Ezh2 is able to interact with several RNA species that will affect its function, by either recruiting or impeding its access to its chromatin targets (Zhao et al., 2010; Davidovich et al., 2013). Other proteins containing non-canonical RNA-binding domains which function in pluripotency and/or differentiation are Lsd1, Suz12 and Wdr5 and will be discussed later in this review.
Several high-throughput analyses in ESCs (Baltz et al., 2012; Castello et al., 2012; Kwon et al., 2013) have identified RNA-binding candidates that do not contain any of the canonical and well-studied RNA-binding motifs described above. It remains to be determined if those proteins are actual RBPs or if their binding to RNA is mediated through intermediate proteins. Additional domain mapping experiments should be carried out to identify previously uncharacterized RNA-binding domains. That the increasing number of proteins shown to interact with RNA makes RNA-binding a more ubiquitous property than previously expected, and highlights not only the importance of RBPs but also of regulatory RNAs.
Approaches for studying RBPs
Understanding the physical association of RNA and protein in vivo requires the use of specific techniques that take into account the biochemistry of both molecules. Accordingly, a series of specific crosslinking procedures, followed by immunoprecipitation techniques have been developed. The first reported protocol was RNA immunoprecipitation (RIP) that allowed for the identification of RNA species associated to RNA-binding proteins (Niranjanakumari et al., 2002). By using formaldehyde as a reversible crosslinker agent combined with high-stringency IP conditions, specific RNAs associated with bait proteins could be determined. However, the sticky nature of RNA necessitated the use of many controls, and only a few targets identified via RIP could actually be validated. A breakthrough in mapping RNA–protein interactions in vivo arrived when CLIP (cross-linking and immunoprecipitation) strategies were developed (Ule et al., 2003; Ule et al., 2005; Jensen and Darnell, 2008). The main difference with RIP is that crosslinking of tissues or cultured cells is achieved by treatment with UV-B irradiation that specifically generates a covalent bond between RNA-RNA and RNA-protein complexes (Zwieb and Brimacombe, 1978; Brimacombe et al., 1988) without inducing protein–protein crosslinks. The formation of such bound complexes is restricted to short-distance interactions (on the order of Angstroms) and allows for the use of highly stringent washes, reducing the background and increasing the specificity of the co-immunoprecipitated RNA molecules. HITS-CLIP (also called CLIP-seq) or “high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation” includes RNA digestion (to reduce its size, typically to 20–100 nt) and ligation to RNA linkers for further reverse transcription and high throughput sequencing, in order to identify the exact binding sites of an RBP to its target RNAs (reviewed by Darnell in 2010). Some RBPs show a low efficiency UV crosslink, making it difficult to analyze by CLIP. In that case, an RNA-protein in tandem immunoprecipitation (RIPiT) protocol is recommended (Singh et al., 2014).
A novel technique was developed to overcome the low efficiency of crosslinks generated by 254 nm UV-B. PAR-CLIP or photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation relies on the incorporation of photoreactive ribonucleoside analogs (i.e., 4-thiouridine or 4-SU, and 6-thioguanosine or 6-SG) into nascent RNA transcripts by living cells (Hafner et al., 2010b). Crosslinking by irradiating cells with 365 nm UV light induces an efficient crosslink of photolabeled RNAs to their interacting RBPs. The precise position of the crosslinking can be determined by the appearance of mutations during sequencing of the co-immunoprecipitated (co-IPed) RNAs (4-SU results in cytidine conversions whereas 6-SG gives rise to adenosine mutations). Moreover, the presence of mutations is a hallmark for UV-crosslinked RNAs, which can be used to discriminate specifically co-IPed RNAs from background sequences belonging to abundant cellular RNAs.
A systematic analysis of preferential RNA sequences of RBPs can be achieved by RNAcompete experiments (Ray et al., 2009). This method involves an in vitro competitive binding reaction where an RBP is incubated with a high molar excess of a complex pool of RNAs, followed by affinity recovery of the protein and analysis of the co-IPed RNAs by microarray. Many human RBPs have been tested by RNAcompete, allowing for the identification of their consensus RNA-binding sites (Ray et al., 2013). This technique is very helpful for predicting the function of an RBP when combined with RNAseq experiments after knockdown of that specific RBP. For instance, an RBP whose consensus binding motif would map to the 3′ UTRs of RNAs, and whose depletion would lead to downregulation of its RNA targets, would be expected to be involved in RNA-binding and stabilization. On the other hand, a protein whose consensus binding motif would map to alternative exon boundaries of its target RNAs, and whose depletion results in differences in the inclusion/exclusion ratio of the alternative exon, would likely be considered an alternative splicing regulator.
A different approach can be taken to study RBPs if the main interest is to unveil the proteins bound to a given RNA species or RNA group (Castello et al., 2012). In that case the first step is the use of a tagged oligonucleotide as bait in order to capture all the RBPs covalently bound to it after UV crosslinking. The co-IPed proteins would then be identified by mass spectrometry analysis.
Technique developments in the last decade focusing on RBPs and their interacting RNAs have led to an exponential increase in the number of published studies on RBP function in various biological processes. Table 2 summarizes the published high-throughput studies of the target RNAs of selected RBPs reviewed in this paper.
Table 2.
High-throughput studies of RBPs with roles in ESC maintenance, differentiation, and/or reprogramming discussed in this review
| Protein | Cell line | Technique | Reference |
|---|---|---|---|
| Rbfox2 | hESCs | CLIP-seq | Yeo et al., 2009 |
| mESCs | CLIP | Jangi et al., 2014 | |
| Mouse brain | HITS-CLIP | Weyn-Vanhentenryck et al., 2014 | |
| Mbnl1/Rbfox2 | hESCs/human fibroblasts | High-throughput PCR for AS | Venables et al., 2013b |
| Son | hESCs | RIP-qPCR | Lu et al., 2013 |
| Srsf3 | P19 | CLIP-seq | Änkö et al., 2012 |
| Fip1 | mESCs | CLIP-seq | Lackford et al., 2014 |
| Unr | Drosophila | RIP-seq | Mihailovic et al., 2012 |
| Esrp1 | mESCs | RIP | Fagoonee et al., 2013 |
| Dazl | Mouse testis | Oligo dT captured | Tsui et al., 2000 |
| Mouse testis | UV-RIP | Reynolds et al., 2005 | |
| Pum1 | Drosophila | RIP-Chip | Gerber et al., 2006 |
| Tagged-AGO2/Tagged-PUM2 | HEK293 | PAR-CLIP | Hafner et al., 2010a |
| Ago2 | mESCs | RIP | Hanina et al., 2010 |
| mESCs | HITS-CLIP | Leung et al., 2011 | |
| Mouse brain/HeLa | HITS-CLIP | Chi et al., 2009 | |
| Dgcr8/Drosha | HEK293 | HITS-CLIP | Macias et al., 2012 |
| Lin28 | hESCs | CLIP-seq | Wilbert et al., 2012 |
| mESCs | CLIP-seq | Cho et al., 2012 | |
| Lsd1/Ezh2 | Foreskin fibroblasts (CRL-2091), lung fibroblasts (FL2), and HeLa (CCL-2) | RIP-qPCR | Tsai et al., 2010 |
| Ezh2 | mESCs | RIP-seq | Zhao et al., 2010 |
| mESCs | PAR-CLIP | Kaneko et al., 2013 | |
| Suz12 | CEM cells | RIP-qPCR | Kanhere et al., 2010 |
| Human foreskin and foot fibroblasts | RIP | Rinn et al., 2007 | |
| Ezh2/Suz12/CoREST | HeLa, human fetal lung fibroblast, and foreskin fibroblasts | RIP-Chip (native IP) | Khalil et al., 2009 |
| Jarid2 | mESCs | PAR-CLIP/CLIP-qPCR | Kaneko et al., 2014 |
| Wdr5 | mESCs | RIPiT | Yang et al., 2014 |
| Thoc2 | mESCs | RIP-seq | Wang et al., 2013 |
| Thoc5 | mESCs | RIP-qPCR | Wang et al., 2013 |
| Tagged-Rbm47 | mESCs | RIP-PCR | Yeganeh et al., 2013 |
Functions of RBPs in ESC maintenance, differentiation, and reprogramming
RBPs with important roles in ESC maintenance, differentiation, and somatic cell reprogramming can be divided into four categories according to their distinct functions. The largest group of known RBPs is implicated in the first and second categories, which correspond to co- and post-transcriptional regulation of mRNAs, respectively. The first group includes alternative splicing and alternative polyadenylation, whereas the second refers to microRNA (miR) recognition and pairing, RNA stability regulation, and RNA turnover. The third category, includes RBPs involved in the sequestration or transportation of RNAs in the nucleus or to the cytoplasm, respectively. Lastly, the fourth category, which mainly comprises long non-coding RNA (lncRNA)-interacting RBPs, contains epigenetic regulators with histone remodeling functions. All of these categories are discussed in detail below and summarized in Fig. 1 and Table 3.
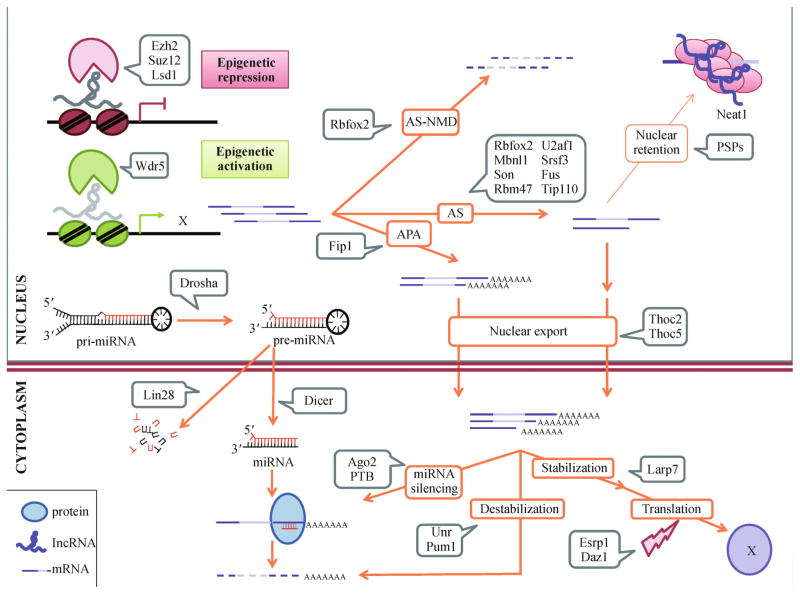
Figure 1.
RBPs with important roles in ESC maintenance and/or differentiation are depicted according to the RNA metabolism step in which they are implicated. Ezh2, Suz12 and Lsd1 repress transcription whereas Wdr5 activates it, binding to chromatin through lncRNAs. Rbfox2 is implicated both in AS and AS-NMD, whereas Mbnl1, Son, Rbm47, U2af1, Srsf3 and Fus have only been reported to regulate AS. Fip1 mediates APA. Once processed, transcripts can either be retained in paraspeckles by PSPs or exported to the cytoplasm. Thoc2 and Thoc5 regulate the transport of important pluripotency-related transcripts in ESC. In the cytoplasm, transcripts are stabilized and transcribed to proteins or targeted for destabilization and/or miRNA silencing by Unr and Pum1 or Ago2 and PTB, respectively. Translation efficiency can be also subjected to regulation, through proteins such as Larp7 (stabilizer) or Esrp1 and Dazl (reduce ribosomal loading). Processing of pri-miRNAs by Drosha and pre-miRNA by Dicer gives rise to mature miRNAs available in the cytoplasm for transcript silencing. Lin28 affects its targets through targeting pre-miRNAs for degradation, therefore reducing the steady-state levels of miRNAs. From top to bottom: AS-NMD: alternative splicing-coupled nonsense-mediated decay, AS: alternative splicing; APA: alternative polyadenylation; PSPs: paraspeckle proteins; pri-miRNA: primary microRNA transcript; pre-miRNA: precursor microRNA transcript; miRNA: microRNA.
Table 3.
Summary of the main RBPs with functions in ESC maintenance, differentiation or somatic cell reprogramming
| Protein | Functional process | Reference |
|---|---|---|
| Unr | Embryonic development | Boussadia et al., 1997 |
| ESC maintenance | Elatmani et al., 2011 | |
| Paraspeckle proteins | ESC differentiation | Prasanth et al., 2005; Chen and Carmichael, 2009 |
| Thoc2/Thoc5 | Embryonic development | Wang et al., 2006 |
| ESC maintenance and somatic cell reprogramming | Ivanova et al., 2006; Ding et al., 2009; Subramanian et al., 2009; Chia et al., 2010 | |
| Dicer | Embryonic development | Bernstein et al., 2003 |
| ESC proliferation and differentiation | Kanellopoulou et al., 2005; Murchison et al., 2005; Wang et al., 2007 | |
| Lin28 | Pluripotency maintenance | Schulman et al., 2005; Melton et al., 2010 |
| Somatic cell reprogramming | Yu et al., 2007 | |
| Ago2 | Post-implantation embryonic development | Morita et al., 2007 |
| ESC proliferation and differentiation | Shekar et al., 2011 | |
| Ezh2 | X-inactivation | Zhao et al., 2008;Kaneko et al., 2010; Kanhere et al., 2010 |
| ESC differentiation | Zhao et al., 2010 | |
| Suz12 | ESC differentiation | Pasini et al., 2007 |
| X-inactivation | Zhao et al., 2008;Kaneko et al., 2010; Kanhere et al., 2010 | |
| Jarid2 | ESC differentiation | Peng et al. 2009; Pasini et al. 2010 |
| X-inactivation | da Rocha et al., 2014; Kaneko et al. 2014 | |
| Rbfox2 | ESC viability | Yeo et al., 2009 |
| ESC differentiation | Venables et al., 2013b | |
| Lsd1 | ESC differentiation | Adamo et al., 2011; Whyte et al., 2012 |
| Wdr5 | ESC maintenance and iPSC generation | Ramakrishna et al., 2011; Li et al., 2012; Yang et al., 2014 |
| Fip1 | ESC self-renewal and differentiation | Ding et al., 2009; Hu et al., 2009; Lackford et al., 2014 |
| Son | ESC maintenance | Chia et al., 2010 |
| Somatic cell reprogramming | Lu et al., 2013 | |
| Tip110 | ESC maintenance | Liu et al., 2012; Liu et al., 2013 |
| PTB | Repression of a neural phenotype in non-neural cells | Yuanchao Xue et al. 2013 |
| U2af1 | Somatic cell reprogramming | Ohta et al., 2013b |
| Srsf3 | Somatic cell reprogramming | Ohta et al., 2013b |
| Mbnl1 | ESC differentiation | Venables et al., 2013b |
| Esrp1 | ESC maintenance – differentiation balance | Fagoonee et al., 2013 |
| Dazl | ESC maintenance – differentiation balance | Xu et al., 2013 |
| Larp7 | ESC maintenance | Dai et al., 2014 |
| Pum1 | ESC differentiation | Leeb et al., 2014 |
RBPs with co-transcriptional regulatory function
After their transcription, nascent pre-mRNAs mature into functional mRNAs. This maturation includes constitutive splicing and polyadenylation for intron removal and poly-A tail addition, respectively. In most cases, however, alternative splicing and/or alternative polyadenylation are required, impacting the final mRNA sequence, structure, and/or stability.
Alternative splicing
More than half of human and mouse genes express multiple mRNAs through alternative splicing (AS) (Lander et al., 2001; Modrek et al., 2001; Johnson et al., 2003). AS is the process by which a unique primary transcript gives rise to multiple mature mRNAs, most commonly by alternative inclusion or exclusion (“skipping”) of individual exons (Altschul et al., 1990; Florea et al., 1998; Hubbard et al., 2002). In the past few years, studies have been reported on the regulation of AS during differentiation and reprogramming of mouse and human ESCs (Pritsker et al., 2005; Sugnet et al., 2006; Yeo et al., 2007; Kunarso et al., 2008; Salomonis et al., 2009; Salomonis et al., 2010; Wu et al., 2010; Gabut et al., 2011; Ohta et al., 2013b). Pluripotency-associated transcripts such as Dnmt3b, Nanog, Sall4, and Oct4 have been shown to be themselves subjected to AS (Atlasi et al., 2008; Gopalakrishnan et al., 2009; Rao et al., 2010; Das et al., 2011; Tsai et al., 2014). The importance of the regulation of alternative splicing for the maintenance of the stem cell state and differentiation is exemplified by stage-specific splicing factors such as Rbfox2, Mbnl1, and Son in the pluripotency control process.
Rbfox2 (also known as Fox2 and Rbm9) was first described in mammals as a tissue-specific RBP of muscle and neuronal cells that is involved in splicing regulation (Underwood et al., 2005). More recently, it has been shown to be required for human ESC (hESC) viability, where its RNA targets have been determined by CLIP-seq (Yeo et al., 2009). Rbfox2 binds and regulates AS of transcripts coding for RBPs such as Lin28 and Rbfox2 itself, nuclear mRNA splicing factors, and serine/threonine kinases, indicative of a role for Rbfox2 in regulating RNA metabolism and signaling pathways in hESCs. Together with Mbnl1, Rbfox2 has also been implicated in regulating exon inclusion/exclusion during differentiation and reprogramming, influencing the structure of proteins involved in membrane dynamics, cell adhesion, migration, and polarity (Venables et al., 2013b). The post-transcriptional regulatory pathway NMD (nonsense-mediated decay) is often coupled to AS to prevent the production of truncated proteins from transcripts containing premature termination codons (Brogna and Wen, 2009). In mouse ESCs (mESCs), Rbfox2 has also been implicated in regulating RBP gene expression (including itself) through AS-NMD, resulting in the auto-regulation of splicing networks (Jangi et al., 2014). According to its role in regulating mesenchymal and epithelial-specific splicing events (Venables et al., 2013a; Braeutigam et al., 2014) and the requirement of MET during the early stage of reprogramming (Li et al., 2010; Samavarchi-Tehrani et al., 2010), a potential role for Rbfox2 in somatic cell reprogramming should be expected.
As stated above, Mbnl1 is another splicing factor that has been implicated in coordinating the inclusion and exclusion of differentiation-specific exons (Venables et al., 2013b). Mbnl1 represses ESC-specific alternative splicing events in differentiated cells, and its best-characterized target is the Foxp1 ES-specific exon (Foxp1-ES). The minimal expression of Mbnl1 in ESCs and its upregulation in somatic cells are compatible with specific roles of Foxp1-ES in ESC maintenance and somatic cell reprogramming (Gabut et al., 2011; Han et al., 2013).
Son was identified in an RNAi screen as an essential regulator of maintenance of hESC identity (Chia et al., 2010). Son is a DNA- and RNA-binding protein that has been previously implicated in maintaining genome stability and regulating RNA splicing for effective cell cycle progression in HeLa cells (Ahn et al., 2011). In hESCs, Son functions as a weak splicing regulator of many genes, among which are Oct4, Prdm14, Med24, and E4f1 pluripotency-related transcripts (Lu et al., 2013). Moreover, depletion of Son is detrimental for somatic cell reprogramming. Another Oct4 splicing regulator in hESCs is Tip110, which regulates the Oct4A splicing variant both in vitro and in vivo, and is necessary for hESC maintenance (Liu et al., 2012; Liu et al., 2013). Oct4A has been shown to be the Oct4 variant responsible for maintenance of pluripotency (Tsai et al., 2014). Son and Tip110 regulate AS of pluripotency transcripts in a gene specific manner. In line with that, mouse Nanog, but not Oct4 or Sox2, was recently found to be bound by another RBP, Rbm47, although the functional relevance of their interaction remains to be elucidated (Yeganeh et al., 2013).
Other splicing factors that have been implicated in regulating somatic cell reprogramming are the two RNA-binding proteins U2af1 and Srsf3 (Ohta et al., 2013b). U2af1 (also known as U2af35) is an auxiliary component of the U2 snRNP and participates in the exon definition step during the splicing process (Zhang et al., 1992). Its mutation has been implicated in splicing defects in different cancer cell types (Yoshida et al., 2011; Imielinski et al., 2012; Brooks et al., 2014). Srsf3, on the other hand, is not only a general and alternative splicing factor but has also been shown to be essential in RNA polyadenylation, RNA export, and protein translation (Zahler et al., 1992; Lou et al., 1998; Huang et al., 2003; Bedard et al., 2007). Knockdown of these RBPs reduces the efficiency of MEF reprogramming without affecting the phenotype, morphology, or proliferation of MEFs (Ohta et al., 2013a).
In mouse ESCs, the RBP Fus was identified as an Oct4 interactor by Lufkin and collaborators (Cheong et al., 2011). Fus is implicated in RNA metabolism, including splicing and miRNA processing (Ishigaki et al., 2012; Morlando et al., 2012). Whereas it has been shown to regulate gastrulation in frogs (Dichmann and Harland, 2012), Fus function in RNA regulation and the nature of its partnership with Oct4 in ESCs is currently unknown.
Additional RBPs have been shown to function in regulating AS in pluripotency and differentiation (Yeo et al., 2009; Han et al., 2013; Ohta et al., 2013b; Venables et al., 2013b). Nevertheless, their interplay with the pluripotency network remains elusive. Further study is needed to determine the interrelationship between the splicing factors and pluripotency factors and their functional contributions to ESC maintenance, somatic cell reprogramming, and differentiation of ESCs.
Alternative polyadenylation
mRNA alternative polyadenylation (APA) is very common (70% of mammalian genes produce APA mRNAs) and plays an important role in post-transcriptional gene regulation (Elkon et al., 2013; Tian and Manley, 2013). APA gives rise to different protein C-termini or 3′ UTRs that may impact the protein output of gene expression through affecting the stability, translation, and/or intracellular localization of mRNAs. APA is highly regulated in ESCs, during somatic cell reprogramming, and during development (Flavell et al., 2008; Ji et al., 2009; Ji and Tian, 2009; Shepard et al., 2011; Boutet et al., 2012). In contrast to more differentiated cells, ESCs favor proximal polyadenylation sites and produce shorter 3′-UTRs, therefore having an impact on microRNA or RBP regulatory pathways (Sandberg et al., 2008; Mueller et al., 2013). However, the mechanisms underlying these APA changes remain poorly understood.
A very recent report has identified the first factor involved in pluripotency-specific APA (Lackford et al., 2014). Fip1 is an essential mRNA 3′ processing factor that was identified in two genome-wide RNAi screens as a potential self-renewal factor in mouse ESCs (Ding et al., 2009; Hu et al., 2009). Lackford and colleagues showed that Fip1 depletion leads to partial differentiation of mESCs and inhibition of MEF reprogramming. Fip1-mediated maintenance of ESC-specific APA profiles is required to promote optimal expression of pluripotency and self-renewal factors, linking for the first time APA regulation to cell fate determination.
RBPs regulating post-transcriptional regulation
Gene expression in eukaryotic cells involves several steps of RNA processing that culminate with the translation of RNA into protein. Apart from the rates of transcription, pre-mRNA splicing, and polyadenylation, mRNAs can further be subjected to regulation by means of stabilization through interaction with RBPs or degradation via the NMD pathway or microRNA-interaction. All these steps, together with the regulation of mRNA export to the cytoplasm, will determine the steady-state amount of mRNAs, and as a result, the amount of protein that will be available to the cell.
RBPs regulating mRNA stability
In the mouse, the cytoplasmic protein Unr has been shown to play an essential role during development, as homozygous disruption of the Unr gene is embryonic lethal (Boussadia et al., 1997). Nevertheless, Unr−/− ESCs can be obtained, although with a spontaneous primitive endoderm differentiation propensity (Elatmani et al., 2011). Unr acts downstream of Nanog, contributing to ESC maintenance through the binding and destabilization of the Gata6 mRNA (Elatmani et al., 2011).
La is another RNA-binding protein shown to be important for preimplantation development and ESC derivation (Park et al., 2006). Although it has been implicated in many RNA-related pathways and has been well established as playing a role in protecting tRNAs and small RNAs from degradation (reviewed in (Wolin and Cedervall, 2002), its targets in ESCs have not been determined. By contrast, the La related protein Larp7 has been recently assigned a role in ESC maintenance (Dai et al., 2014). Larp7 binds and stabilizes Lin28 mRNA by recruiting the poly(A) polymerase Star-PAP, safeguarding ESCs from entering a primed-differentiation state.
From a genome-wide screen for genes that are down-regulated during differentiation of germline cell-derived pluripotent stem cells (GPSCs) (Fagoonee et al., 2010), three RBPs, including Esrp1, were identified. Esrp1, also known as Rbm35a, is a tumor suppressor for colorectal cancer that acts through binding to the 5′ UTRs of mRNAs and inhibiting their translation (Ivanov et al., 2007). More recently, its negative role in maintaining the balance of self-renewal and commitment to differentiation has been established (Fagoonee et al., 2013). Through its binding to Oct4 and Sox2 mRNAs, Esrp1 fine-tunes their polysomal loading, and therefore affects their translation efficiency, explaining why the knockdown of Esrp1 results in increased self-renewal and enhanced MEF reprogramming.
Dazl is an RBP involved in mRNA stability in mESCs (Xu et al., 2013). Dazl is expressed in mouse germ and embryonic stem cells, but has opposite functions in these two cell types. Whereas it functions as a translational stimulator in germ cells (Tsui et al., 2000), its role in ESCs is mainly repressive (Xu et al., 2013). Through binding to the 3′ UTR of Oct4, Sox2, and Mvh mRNAs, Dazl represses translation and decreases the steady-state levels of these pluripotency regulators in mESCs, balancing stem cell maintenance and differentiation.
Pum1 is another RBP that inhibits translation and promotes degradation of its target mRNAs through binding at a highly conserved eight-nucleotide motif located in their 3′ UTRs (Galgano et al., 2008). The naive pluripotency factor transcripts for Tfcp2l1, Sox2, Tbx3, and Esrrb are among the targets of Pum1 in mESCs (Leeb et al., 2014). Given their shared function in regulating translation of pluripotency-related transcripts such as Oct4 and Sox2, Dazl, Pum1, and Esrp1 are likely to be regulated by the same pathways in ESCs and/or during differentiation. Further studies are needed to understand how these post-transcriptional regulators are controlled by the pluripotency network.
RBPs involved in microRNA regulation
MicroRNAs (miRs) are small (20–25 nt) non-coding RNAs that promote specific destabilization and degradation of their target mRNAs by base-pairing to their 5′ or 3′ UTRs (Bartel, 2009; Lytle et al., 2007). Several miR families have been implicated in the control of ESC maintenance, differentiation, and somatic cell reprogramming (Marson et al., 2008; Sinkkonen et al., 2008; Tay et al., 2008; Judson et al., 2009; Melton et al., 2010; Tiscornia and Izpisúa Belmonte, 2010; Anokye-Danso et al., 2011). miR processing consists of successive maturation steps of the initially transcribed primary RNA (pri-miRNA), which can be subjected to regulation (Kim et al., 2008). First, pri-miRNAs are processed by the dsRNA-binding proteins Dgcr8 (Pasha) and RNase III type enzyme Drosha, giving rise to the pre-miRNAs. The pre-miRNAs will then be further cleaved by Dicer, another RNase III type enzyme, to finally give rise to the functional mature miRNAs (see Fig. 1) (Krol et al., 2010). Dicer-deficient mice die around day 7.5 of mouse embryonic development with serious morphological abnormalities (Bernstein et al., 2003). Nevertheless, Dicer and Dgcr8 null mESCs can be maintained, albeit with a defect in proliferation and differentiation (Kanellopoulou et al., 2005; Murchison et al., 2005; Wang et al., 2007). This phenotype can be explained by the incapacity for miRNA maturation due to the absence of either of these two RBPs and the subsequent improper downregulation and accumulation of transcripts involved in cell cycle control and pluripotency regulation. Both the defective proliferation and differentiation can be rescued by overexpression of the mature pluripotency-associated miR-290 and miR-302 and differentiation-promoting let-7 miRNA, respectively (Hutvágner et al., 2001; Wang et al., 2007; Kim and Choi, 2012).
Mature miRNAs and other silencing RNAs exert their function in the RNA-silencing pathway with the help of RBPs such as the Argonaute family of proteins (Hutvagner and Simard, 2008). The Argonaute family can be divided in two subfamilies: Piwi and Ago. In humans, the latter consists of Ago1, Ago2, Ago3, and Ago4, which are widely expressed and associate with miRNAs and siRNAs. Conversely, members of the Piwi subfamily are mainly restricted to the germline, where they associate with piwi-interacting RNAs (piRNAs). Ago2−/− null embryos arrest during development early after implantation (Morita et al., 2007) and Ago2−/− mESCs have defects in self-renewal and differentiation due to defective miRNA function (Shekar et al., 2011). Although it was previously thought that miRNAs and siRNAs were needed for guiding Ago proteins to their target mRNAs, Leung and colleagues showed by HITS-CLIP that in mESCs, Ago2 is also able to bind its targets in the absence of miRNAs (Leung et al., 2011).
Lin28 is an RBP that regulates the pri-miR let-7 family by promoting their degradation through uridylation (Heo et al., 2008; Heo et al., 2009; Hagan et al., 2009; Thornton et al., 2012). By reducing the mature miR let-7 levels, Lin28 contributes to maintenance of pluripotency and prevents differentiation of ESCs (Schulman et al., 2005; Melton et al., 2010). Lin28 has also been shown to promote reprogramming of human somatic cells (Yu et al., 2007). Besides the regulatory effect of Lin28 on let-7 biogenesis, Lin28 also has a let-7 independent function (Balzer et al., 2010). RIP and HITS-CLIP studies in human and mouse ESCs have shown that Lin28 is able to bind exons and 5′ and 3′ UTRs of mRNAs, and consequently regulate their expression. As a result, Lin28 not only controls pri-miRNA maturation, but also regulates many other genes encoding cell cycle regulators, secretory proteins, other RBP genes (including Lin28 itself), and even the pluripotency factor Oct4, by affecting their translational efficiency (Qiu et al., 2010; Peng et al., 2011; Cho et al., 2012; Wilbert et al., 2012; Hafner et al., 2013).
In the cytoplasm of hESCs, Lin28 interacts with L1td1 in P-bodies, a subcellular compartment where RNAs are directed either for translation or for silencing through miRNAs (Närvä et al., 2012). L1td1 (or Ecat11) is another RBP whose expression is rapidly downregulated upon differentiation (Wong et al., 2011). In contrast to mouse ESCs, L1td1 is required for maintenance of self-renewal in human ESCs (Iwabuchi et al., 2011; Närvä et al., 2012). Although its exact function alone, or together with Lin28 remains to be determined, L1td1’s association with proteins in P-bodies suggests a potential role in translational regulation.
The polypyrimidine-tract binding protein (PTB) is a known splicing regulator that has recently been shown to also regulate microRNA functions (Xue et al., 2013). PTB can both activate or repress gene expression by binding to the 3′ UTR of its targets, together or in proximity to Ago2. PTB influences the secondary structure of 3′ UTRs to enhance or inhibit miR recognition, and can also compete and influence Ago2 binding to their common targets. Similarly to PTB downregulation during neuronal differentiation, PTB knockdown is enough to cause transdifferentiation of different cell types to neuronal-like cells and functional neurons by affecting the stability of key neuronal genes and components of the REST complex (Xu et al., 2013). This is the first study to describe an RBP being able to cause transdifferentiation by its own, and highlights the importance of RBP regulation in cell-wide processes.
RBPs regulating nuclear retention and export of RNAs
Once RNAs are transcribed and processed, a large amount of them must be exported to the cytoplasm to exert their functions. Several specific processes such as nuclear retention or export are mediated and regulated by RBPs in ESCs and during differentiation.
Paraspeckle proteins (PSPs) are widely expressed RBPs that are located in subnuclear structures known as paraspeckles (Fox et al., 2002; Fox and Lamond, 2010). Paraspeckles are nuclear domains formed by RBPs organized around the scaffolding lncRNA Neat1 (Clemson et al., 2009; Souquere et al., 2010). Human ESCs do not possess paraspeckles. However, when they are induced to differentiate into trophectoderm (TE), increased Neat1 expression leads to the formation of paraspeckles, which bind and retain some transcripts with inverted repeats (IRAlus) in their UTRs (Prasanth et al., 2005; Chen and Carmichael, 2009). One such IRAlus-containing transcript is the pluripotency-associated Lin28, which is efficiently exported to the cytoplasm in hESCs, but becomes retained upon TE differentiation, resulting in reduced Lin28 protein levels. In mice, Neat1 is expressed in ESCs and is further upregulated during differentiation (Bond and Fox, 2009; Sunwoo et al., 2009). Although paraspeckles have been shown to not be essential for development (Nakagawa et al., 2011), some PSPs have been implicated in pluripotency and differentiation through transcriptional, co-transcriptional and export regulation (Hata et al., 2008; Park et al., 2013). The repertoire of PSP-interacting RNAs in ESCs remains to be determined in order to clarify the function of these proteins in pluripotency and differentiation processes.
The THO complex is a highly conserved protein complex that couples RNA splicing and export from the nucleus to the cytoplasm (Katahira, 2012). Thoc2 and 5 are RBPs within the THO complex that have been identified as potential self-renewal regulators in mESCs from genome-wide RNAi screens (Ivanova et al., 2006; Ding et al., 2009; Subramanian et al., 2009; Chia et al., 2010). THO knockout mice are embryonic lethal (Wang et al., 2006) and Thoc2 or Thoc5 knockdown in mESCs leads to loss of self-renewal (Wang et al., 2013). The importance of THO relies on its function in exporting pluripotency transcripts such as Nanog, Sox2, and Klf4 from the nucleus to the cytoplasm. In line with this finding, Thoc2 or Thoc5 knockdown abrogates somatic cell reprogramming and inner cell mass specification, highlighting the importance of RBP-controlled nuclear export regulation in pluripotency and reprogramming (Saunders and Wang, 2014).
RBPs with functions in epigenetic regulation
RBPs can potentially bind any RNA species. Whereas RBPs involved in co-transcriptional and post-transcriptional regulation mainly associate with mRNAs and siRNAs/miRNAs, those involved in epigenetic regulation preferentially interact with non-coding and long non-coding RNAs (ncRNAs/lncRNAs). LncRNAs are non-coding RNAs longer than 200 nucleotides with diverse functions (Wang and Chang, 2011). LncRNAs can function in cis (i.e. Kcnq1ot1) or in trans (i.e. HOTAIR) to the locus from which they are transcribed (Wang and Chang, 2011; Moran et al., 2012) as scaffolds for bringing together remodeling complexes that need to work in the same chromatin domain (Wang and Chang, 2011). Other ncRNAs work in cis as decoys to prevent epigenetic factors from binding to the loci from which they arise. Several lncRNAs are transcriptionally regulated in ESCs, and their functions in maintaining the pluripotent state have been reported (Dinger et al., 2008; Sheik Mohamed et al., 2010; Guttman et al., 2011). RBP interaction with lncRNAs can result in both chromatin activation and repression, the latter being the most extensively studied (Wang and Chang, 2011).
Epigenetic repression
The polycomb repressive complex 2 (PRC2) catalyzes the di-and trimethylation of H3K27 to mediate epigenetic silencing (Cao et al., 2002; Kuzmichev et al., 2002; Kirmizis et al., 2004). PRC2 is dispensable for maintenance of ESC pluripotency but is necessary for ESC differentiation and embryonic development (Pasini et al., 2007; Chamberlain et al., 2008). Polycomb components are chromatin remodelers that do not possess DNA-binding domains themselves; and therefore need intermediate proteins and ncRNAs to bind their targets (Margueron and Reinberg, 2011). Both catalytic subunits of PRC2, Ezh2 and Suz12, have been shown to interact with RNAs. The first study for determining PRC2-interacting RNAs by RIP-seq showed that between 10% and 25% of the ESC transcriptome was pulled-down by Ezh2 immunoprecipitation (Zhao et al., 2010). Although there was a concern regarding the specificity of the interactions due to the large number of transcripts pulled-down, recent studies support this finding and propose a new model. In this model, PRC2 would not only bind to lncRNAs to mediate silencing, but would also bind to actively transcribed loci where ncRNA species would serve as decoys for PRC2 binding, keeping these loci in a transcriptionally active state (Davidovich et al., 2013). A similar phenomenon has been described for the DNA methyltransferase Dnmt1, supporting the idea that an RNA-mediated decoy could serve as a more general co-transcriptional regulator than previously expected (Di Ruscio et al., 2013).
X chromosome inactivation (XCI) is a process that occurs in females during early development for gene dosage compensation, which can also be observed in vitro upon female ESC differentiation (Augui et al., 2011; Schulz and Heard, 2013). Moreover, it has recently been shown that maintenance of both X chromosomes active is a barrier for ESC differentiation (Schulz et al., 2014). XCI requires the recruitment of PRC2 to the chromosome that will be silenced by the lncRNA Xist (Zhao et al., 2008). Ezh2 and Suz12 are thought to be responsible for mediating Xist-PRC2 interaction, although they do not possess canonical RNA-binding domains (Zhao et al., 2008; Kaneko et al., 2010; Kanhere et al., 2010). Nevertheless, recently the direct interaction of PRC2 and Xist has been disputed by means of high super-resolution microscopy (Cerase et al., 2014). More experiments are needed to confirm either of the proposed models and clarify the seemingly contradictory results.
Jarid2 is a PRC2 cofactor that was shown to be critical for PRC2 binding to its target genes for regulation of ESC differentiation (Peng et al., 2009; Pasini et al., 2010; Shen et al., 2009). Two recent studies have identified Jarid2 as another Xist-interacting RBP (da Rocha et al., 2014; Kaneko et al., 2014), which may promote PRC2 recruitment for XCI.
Ezh2 and Suz12 also bind other lncRNAs with critical functions during embryonic development, such as HOTAIR and Kcnq1ot1 that mediate HOXD silencing and gene imprinting, respectively (Rinn et al., 2007; Kaneko et al., 2010; Tsai et al., 2010) (for a deeper review see (Brockdorff, 2013). In turn, HOTAIR lncRNA is also bound by the histone demethylase Lsd1. Lsd1 is another epigenetic modifier with important functions in ESC differentiation through its repressive H3 demethylase activity (Adamo et al., 2011; Whyte et al., 2012). HOTAIR may work in trans by bridging Lsd1 and PRC2 to allow their cooperativity in mediating gene repression (Tsai et al., 2010). HOTAIR expression is induced during differentiation and contributes to self-renewal of adult stem cells. It is also required for epithelial-to-mesenchymal (EMT) transition and metastasis in cancer cell lines (Gupta et al., 2010; Pádua Alves et al., 2013).
Epigenetic activation
Potential roles of lncRNA-binding RBPs in gene activation are poorly understood. Wdr5 is the first RBP that links lncRNAs to gene activation in ESCs. Wdr5 is a core subunit of the MLL1-4 and SET1A/1B complexes, which is involved in the deposition of histone 3 lysine 4 methylation, a mark of active gene expression. Binding of Wdr5 to lncRNAs recruits it to several loci to facilitate transcription (Wang et al., 2011; Gomez et al., 2013). In human foreskin fibroblasts, Wdr5 binding to HOTTIP lncRNA recruits the MLL complexes to the 5′ HOXA locus, generating a broad range of H3K4me3 and transcriptional activation (Wang et al., 2011). The mutual regulation of HOTTIP and Wdr5 generates a positive feedback loop that maintains the activity of the locus. Wdr5 contributes to self-renewal of ESCs through maintaining active chromatin and plays an important role in the generation of induced pluripotent stem cells from differentiated somatic cells (Ang et al., 2011). A recent high-throughput study has detected over a thousand Wdr5-bound RNA molecules in mESCs, including mRNAs, lncRNAs, pri-miRNAs, and small nucleolar RNAs (Yang et al., 2014). Some of the lncRNAs bound by Wdr5 were themselves known to be required for ESC pluripotency and differentiation (Guttman et al., 2011), partly explaining Wdr5’s function in ESC maintenance. Thus, Wdr5 connects lncRNAs with epigenetic remodelers to generate a transcriptional regulation output in ESCs.
Conclusions and perspectives
Even though RBPs have been extensively studied for decades, it is not until recently that they have been implicated in ESC maintenance, differentiation and somatic cell reprogramming. The development of specific techniques for the characterization of RNA-protein interactions has led to an increasing number of RBPs and their binding targets identified and characterized. In ESCs more than five hundred RBPs, almost half of them with previously uncharacterized functions, have been described (Kwon et al., 2013). The major challenge is to distinguish bona fide RBPs and target RNAs from potential false positives due to indirect protein associations and the “sticky” nature of RNAs. The use of UV-crosslinking and PAR-CLIP should be sufficient to overcome these problems, revealing only specific RBP targets. The presence of a canonical RBD also supports the potential RBP function of a particular protein, although it is not always required for RNA-protein interaction. Domain mapping and in vitro binding assays (such as RNA EMSA or REMSA) should help to clarify whether those non-canonical RBPs directly bind to or indirectly interact with RNA molecules.
After pre-mRNA transcription RBPs play important roles in all successive steps for RNA maturation, transport, and stabilization before its final translation into protein. It was historically believed that RBPs were playing merely housekeeping roles in RNA metabolism. Actually, most of the studies on RBPs were focused on their function in constitutive splicing or translational regulation. Nevertheless, an increasing number of studies are now demonstrating that RBPs not only play constitutive roles but are important for the maintenance of a specific cell-state or identity. A good example to illustrate this is the family of heterogeneous ribonucleoproteins (hnRNPs). hnRNPs constitute a diverse family of RBPs with well-known functions in preventing the folding of pre-mRNA into secondary structures, mRNA splicing and transporting to the cytoplasm (Dreyfuss et al., 1993). Nevertheless, some of the hnRNPs have also been recently shown to be essential for early mouse development and stem cell maintenance, such as hnRNPI/PTB discussed earlier in this review (Shibayama et al., 2009) or hnRNPA2 (Ji and Tulin, 2012) and hnRNPU/AUF1 (Choi et al., 2011). It is now evident that RBPs do not merely act as adaptors between RNAs and other proteins, but are implicated in the active maintenance of a defined cell state.
All the steps of RNA maturation can be subjected to RBP regulation as a fast way to respond to external signaling cues. In vivo, stem cells exist as a transient pluripotent population that arises from the totipotent morula, and that will give rise to every cell type of the embryo. In vitro, stem cells can be maintained indefinitely in culture under defined conditions although in a meta-stable transcriptional state that results in heterogeneous populations of cells coexisting in a dish (Chambers et al., 2007; Hayashi et al., 2008; Toyooka et al., 2008; Tanaka, 2009). Both in vivo and in vitro, the transition between pluripotent and primed states requires the action of fast-responding mechanisms. Likely, the RBP-mediated regulation of co-transcriptional regulation, nuclear export, mRNA stabilization or destabilization, and microRNA-mediated degradation is important to regulate the final steady-state levels of proteins needed for differentiation or reprogramming in those transitory states. As previously commented in this review, Oct4 levels, which need to be tightly regulated for ESC maintenance and commitment (Niwa et al., 2000; Karwacki-Neisius et al., 2013; Radzisheuskaya et al., 2013), are fine-tuned post-transcriptionally by many RBPs, which function to control its alternative splicing (Son, Fus, and Tip110) or translation efficiency (Esrp1, Dazl) for example. On the other hand, the regulation is transcript-specific, as other core pluripotency factors such as Esrrb, Sox2, and Nanog are regulated by different RBPs. It remains to be determined how these RBPs are incorporated into pluripotency networks and how those networks interact with the transcriptional regulatory machinery.
RBPs use RNAs as a platform for recruitment of protein complexes that will either modify the RNA itself or other nearby proteins, thereby serving as adaptors between the RNA and protein interfaces. Many RBPs have already been identified with potential functional roles in ESC maintenance, differentiation, and/or somatic cell reprogramming (summarized in Fig. 1 and Table 3). The high prevalence of RBPs in the ESC proteome (Kwon et al., 2013) suggests that many more are awaiting to be discovered. The recent technical improvement in RNA-immunoprecipitation protocols makes it feasible to determine new RBPs involved in ESC maintenance or differentiation in different ways. First, proteins bound to specific RNAs, such as mRNAs or ncRNAs with important roles in ESC maintenance or differentiation can be easily identified by RNA-IP coupled to mass spectrometry and further validated. Second, genome-wide assays of proteins with differential expression during reprogramming or differentiation are also valuable data sources for selecting and characterizing RBPs with potential important functions in stem cells. Furthermore, published interactomes of the core pluripotency protein network in ESCs include RBPs that could play an important role in coupling pluripotency maintenance and RNA metabolism. As a result, the integration of RBP function in the bigger picture of pluripotency and differentiation networks will lead to a much better understanding of molecular mechanisms underlying pluripotency and reprogramming, as well as directed differentiation of pluripotent cells.
Acknowledgments
We are grateful to Arven Saunders for critical reading and comments on the manuscript. The research in the Wang laboratory was funded by grants from the National Institutes of Health (NIH) and the Empire State Stem Cell Fund through New York State Department of Health (NYSTEM). J.W. is also a recipient of Irma T. Hirschl and Weill-Caulier Trusts Career Scientist Award.
Abbreviations
- ESC
Embryonic stem cell
- RBP
RNA-binding protein
- RBD
RNA-binding domain
- mRNA
mRNA
- PRC
Polycomb repressive complex
- XCI
X-chromosome inactivation
- siRNA
small interfering RNA
- RNP
ribonucleoprotein
- ds
double stranded
- UTR
untranslated region
- miR
micro RNA
- AS
alternative splicing
- NMD
nonsense-mediated decay
- lncRNA
long non-coding RNA
- ncRNA
non-coding RNA
- EMT
epitelial-to-mesenchymal
- PSP
paraspeckle proteins
- IRAlus
inverted repeat Alu elements
Footnotes
Compliance with ethics guidelines
Diana Guallar and Jianlong Wang declare that they have no conflict of interest. This manuscript is a review article and does not involve a research protocol requiring approval by the relevant institutional review board or ethics committee.
References
- Adamo A, Sesé B, Boue S, Castaño J, Paramonov I, Barrero MJ, Izpisua Belmonte JC. LSD1 regulates the balance between self-renewal and differentiation in human embryonic stem cells. Nat Cell Biol. 2011;13(6):652–659. doi: 10.1038/ncb2246. [DOI] [PubMed] [Google Scholar]
- Ahn EY, DeKelver RC, Lo MC, Nguyen TA, Matsuura S, Boyapati A, Pandit S, Fu XD, Zhang DE. SON controls cell-cycle progression by coordinated regulation of RNA splicing. Mol Cell. 2011;42 (2):185–198. doi: 10.1016/j.molcel.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amarasinghe GK, De Guzman RN, Turner RB, Chancellor KJ, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the psi-RNA packaging signal. Implications for genome recognition. J Mol Biol. 2000;301(2):491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- Ang YS, Tsai SY, Lee DF, Monk J, Su J, Ratnakumar K, Ding J, Ge Y, Darr H, Chang B, Wang J, Rendl M, Bernstein E, Schaniel C, Lemischka IR. Wdr5 mediates self-renewal and reprogramming via the embryonic stemcell core transcriptional network. Cell. 2011;145(2):183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Änkö ML, Müller-McNicoll M, Brandl H, Curk T, Gorup C, Henry I, Ule J, Neugebauer KM. The RNA-binding landscapes of two SR proteins reveal unique functions and binding to diverse RNA classes. Genome Biol. 2012;13(3):R17. doi: 10.1186/gb-2012-13-3-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8(4):376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature. 2013;502(7472):462–471. doi: 10.1038/nature12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atlasi Y, Mowla SJ, Ziaee SA, Gokhale PJ, Andrews PW. OCT4 spliced variants are differentially expressed in human pluripotent and nonpluripotent cells. Stem Cells. 2008;26(12):3068–3074. doi: 10.1634/stemcells.2008-0530. [DOI] [PubMed] [Google Scholar]
- Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011;12(6):429–442. doi: 10.1038/nrg2987. [DOI] [PubMed] [Google Scholar]
- Baltz AG, Munschauer M, Schwanhäusser B, Vasile A, Murakawa Y, Schueler M, Youngs N, Penfold-Brown D, Drew K, Milek M, Wyler E, Bonneau R, Selbach M, Dieterich C, Landthaler M. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol Cell. 2012;46(5):674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Balzer E, Heine C, Jiang Q, Lee VM, Moss EG. LIN28 alters cell fate succession and acts independently of the let-7 microRNA during neurogliogenesis in vitro. Development. 2010;137(6):891–900. doi: 10.1242/dev.042895. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard KM, Daijogo S, Semler BL. A nucleo-cytoplasmic SR protein functions in viral IRES-mediated translation initiation. EMBO J. 2007;26(2):459–467. doi: 10.1038/sj.emboj.7601494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35(3):215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186(5):637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongso A, Fong CY, Ng SC, Ratnam S. Isolation and culture of inner cell mass cells from human blastocysts. Hum Reprod. 1994;9(11):2110–2117. doi: 10.1093/oxfordjournals.humrep.a138401. [DOI] [PubMed] [Google Scholar]
- Borozdin W, Wright MJ, Hennekam RC, Hannibal MC, Crow YJ, Neumann TE, Kohlhase J. Novel mutations in the gene SALL4 provide further evidence for acro-renal-ocular and Okihiro syndromes being allelic entities, and extend the phenotypic spectrum. J Med Genet. 2004;41(8):e102. doi: 10.1136/jmg.2004.019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussadia O, Amiot F, Cases S, Triqueneaux G, Jacquemin-Sablon H, Dautry F. Transcription of unr (upstream of N-ras) down-modulates N-ras expression in vivo. FEBS Lett. 1997;420(1):20–24. doi: 10.1016/s0014-5793(97)01479-8. [DOI] [PubMed] [Google Scholar]
- Boutet SC, Cheung TH, Quach NL, Liu L, Prescott SL, Edalati A, Iori K, Rando TA. Alternative polyadenylation mediates microRNA regulation of muscle stem cell function. Cell Stem Cell. 2012;10(3):327–336. doi: 10.1016/j.stem.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Mathur D, Jaenisch R. Molecular control of pluripotency. Curr Opin Genet Dev. 2006;16(5):455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Braeutigam C, Rago L, Rolke A, Waldmeier L, Christofori G, Winter J. The RNA-binding protein Rbfox2: an essential regulator of EMT-driven alternative splicing and a mediator of cellular invasion. Oncogene. 2014;33(9):1082–1092. doi: 10.1038/onc.2013.50. [DOI] [PubMed] [Google Scholar]
- Brimacombe R, Stiege W, Kyriatsoulis A, Maly P. Intra-RNA and RNA-protein cross-linking techniques in Escherichia coli ribosomes. Methods Enzymol. 1988;164:287–309. doi: 10.1016/s0076-6879(88)64050-x. [DOI] [PubMed] [Google Scholar]
- Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19(4):429–442. doi: 10.1261/rna.037598.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogna S, Wen J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat Struct Mol Biol. 2009;16(2):107–113. doi: 10.1038/nsmb.1550. [DOI] [PubMed] [Google Scholar]
- Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, Pedamallu CS, Sivachenko A, Rosenberg M, Chmielecki J, Lawrence MS, DeLuca DS, Getz G, Meyerson M. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS ONE. 2014;9(1):e87361. doi: 10.1371/journal.pone.0087361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14(6):427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Caretti G, Schiltz RL, Dilworth FJ, Di Padova M, Zhao P, Ogryzko V, Fuller-Pace FV, Hoffman EP, Tapscott SJ, Sartorelli V. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev Cell. 2006;11:547–560. doi: 10.1016/j.devcel.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, Krijgsveld J, Hentze MW. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149(6):1393–1406. doi: 10.1016/j.cell.2012.04.031. [DOI] [PubMed] [Google Scholar]
- Cerase A, Smeets D, Tang YA, Gdula M, Kraus F, Spivakov M, Moindrot B, Leleu M, Tattermusch A, Demmerle J, Nesterova TB, Green C, Otte AP, Schermelleh L, Brockdorff N. Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proc Natl Acad Sci USA. 2014;111(6):2235–2240. doi: 10.1073/pnas.1312951111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26(6):1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450 (7173):1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chambers I, Tomlinson SR. The transcriptional foundation of pluripotency. Development. 2009;136(14):2311–2322. doi: 10.1242/dev.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KY, Ramos A. The double-stranded RNA-binding motif, a versatile macromolecular docking platform. FEBS J. 2005;272(9):2109–2117. doi: 10.1111/j.1742-4658.2005.04652.x. [DOI] [PubMed] [Google Scholar]
- Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35(4):467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong CY, Lon Ng PM, Ponnampalam R, Tsai HH, Bourque G, Lufkin T. In silico tandem affinity purification refines an Oct4 interaction list. Stem Cell Res Ther. 2011;2(3):26. doi: 10.1186/scrt67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460(7254):479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia NY, Chan YS, Feng B, Lu X, Orlov YL, Moreau D, Kumar P, Yang L, Jiang J, Lau MS, Huss M, Soh BS, Kraus P, Li P, Lufkin T, Lim B, Clarke ND, Bard F, Ng HH. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468(7321):316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- Cho J, Chang H, Kwon SC, Kim B, Kim Y, Choe J, Ha M, Kim YK, Kim VN. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151(4):765–777. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- Choi HS, Kim WT, Kim H, Kim JJ, Ko JY, Lee SW, Jang YJ, Kim SJ, Lee MJ, Jung HS, Kzhyshkowska J, Um SJ, Lee MY, Lee SH, Kim CH, Ryu CJ. Identification and characterization of adenovirus early region 1B-associated protein 5 as a surface marker on undifferentiated human embryonic stem cells. Stem Cells Dev. 2011;20 (4):609–620. doi: 10.1089/scd.2010.0265. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33(6):717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, Granier C, Matias NR, Sanulli S, Chow J, Schulz E, Picard C, Kaneko S, Helin K, Reinberg D, Stewart AF, Wutz A, Margueron R, Heard E. Jarid2 Is Implicated in the Initial Xist-Induced Targeting of PRC2 to the Inactive X Chromosome. Mol Cell. 2014;53(2):301–316. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- Dai Q, Luan G, Deng L, Lei T, Kang H, Song X, Zhang Y, Xiao ZX, Li Q. Primordial Dwarfism Gene Maintains Lin28 Expression to Safeguard Embryonic Stem Cells from Premature Differentiation. Cell Rep. 2014;7(3):735–746. doi: 10.1016/j.celrep.2014.03.053. [DOI] [PubMed] [Google Scholar]
- Darnell RB. HITS-CLIP: panoramic views of protein-RNA regulation in living cells. Wiley Interdiscip Rev RNA. 2010;1(2):266–286. doi: 10.1002/wrna.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Jena S, Levasseur DN. Alternative splicing produces Nanog protein variants with different capacities for self-renewal and pluripotency in embryonic stem cells. J Biol Chem. 2011;286(49):42690–42703. doi: 10.1074/jbc.M111.290189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidovich C, Zheng L, Goodrich KJ, Cech TR. Promiscuous RNA binding by Polycomb repressive complex 2. Nat Struct Mol Biol. 2013;20(11):1250–1257. doi: 10.1038/nsmb.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ruscio A, Ebralidze AK, Benoukraf T, Amabile G, Goff LA, Terragni J, Figueroa ME, De Figueiredo Pontes LL, Alberich-Jorda M, Zhang P, Wu M, D’Alò F, Melnick A, Leone G, Ebralidze KK, Pradhan S, Rinn JL, Tenen DG. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503(7476):371–376. doi: 10.1038/nature12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichmann DS, Harland RM. fus/TLS orchestrates splicing of developmental regulators during gastrulation. Genes Dev. 2012;26(12):1351–1363. doi: 10.1101/gad.187278.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Paszkowski-Rogacz M, Nitzsche A, Slabicki MM, Heninger AK, de Vries I, Kittler R, Junqueira M, Shevchenko A, Schulz H, Hubner N, Doss MX, Sachinidis A, Hescheler J, Iacone R, Anastassiadis K, Stewart AF, Pisabarro MT, Caldarelli A, Poser I, Theis M, Buchholz F. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4(5):403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Soldà G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18(9):1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle M, Badertscher L, Jaskiewicz L, Güttinger S, Jurado S, Hugenschmidt T, Kutay U, Filipowicz W. The double-stranded RNA binding domain of human Dicer functions as a nuclear localization signal. RNA. 2013;19(9):1238–1252. doi: 10.1261/rna.039255.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Matunis MJ, Piñol-Roma S, Burd CG. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62(1):289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105(2):281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- Elatmani H, Dormoy-Raclet V, Dubus P, Dautry F, Chazaud C, Jacquemin-Sablon H. The RNA-binding protein Unr prevents mouse embryonic stem cells differentiation toward the primitive endoderm lineage. Stem Cells. 2011;29(10):1504–1516. doi: 10.1002/stem.712. [DOI] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14 (7):496–506. doi: 10.1038/nrg3482. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Fagoonee S, Bearzi C, Di Cunto F, Clohessy JG, Rizzi R, Reschke M, Tolosano E, Provero P, Pandolfi PP, Silengo L, Altruda F. The RNA-binding protein ESRP1 fine-tunes the expression of pluripotency-related factors in mouse embryonic stem cells. PLoS ONE. 2013;8(8):e72300. doi: 10.1371/journal.pone.0072300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagoonee S, Hobbs RM, De Chiara L, Cantarella D, Piro RM, Tolosano E, Medico E, Provero P, Pandolfi PP, Silengo L, Altruda F. Generation of functional hepatocytes from mouse germ line cell-derived pluripotent stem cells in vitro. Stem Cells Dev. 2010;19(8):1183–1194. doi: 10.1089/scd.2009.0496. [DOI] [PubMed] [Google Scholar]
- Filipovska A, Razif MF, Nygård KK, Rackham O. A universal code for RNA recognition by PUF proteins. Nat Chem Biol. 2011;7(7):425–427. doi: 10.1038/nchembio.577. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60(6):1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 1998;8(9):967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: a novel nuclear domain. Curr Biol. 2002;12(1):13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- Fox AH, Lamond AI. Paraspeckles. Cold Spring Harb Perspect Biol. 2010;2(7):a000687. doi: 10.1101/cshperspect.a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O’Hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, Nedelec S, Wichterle H, Woltjen K, Hughes TR, Zandstra PW, Nagy A, Wrana JL, Blencowe BJ. An alternative splicing switch regulates embryonic stem cell pluripotency and reprogramming. Cell. 2011;147(1):132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Galgano A, Forrer M, Jaskiewicz L, Kanitz A, Zavolan M, Gerber AP. Comparative analysis of mRNA targets for human PUF-family proteins suggests extensive interaction with the miRNA regulatory system. PLoS ONE. 2008;3(9):e3164. doi: 10.1371/journal.pone.0003164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García MA, Meurs EF, Esteban M. The dsRNA protein kinase PKR: virus and cell control. Biochimie. 2007;89(6–7):799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Gerber AP, Luschnig S, Krasnow MA, Brown PO, Herschlag D. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc Natl Acad Sci USA. 2006;103(12):4487–4492. doi: 10.1073/pnas.0509260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582 (14):1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin KS, Varani G. How arginine-rich domains coordinate mRNA maturation events. RNA Biol. 2007;4(2):69–75. doi: 10.4161/rna.4.2.4869. [DOI] [PubMed] [Google Scholar]
- Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, Chang HY, Brahic M, Kirkegaard K. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell. 2013;152(4):743–754. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, Van Emburgh BO, Shan J, Su Z, Fields CR, Vieweg J, Hamazaki T, Schwartz PH, Terada N, Robertson KD. A novel DNMT3B splice variant expressed in tumor and pluripotent cells modulates genomic DNA methylation patterns and displays altered DNA binding. Mol Cancer Res. 2009;7(10):1622–1634. doi: 10.1158/1541-7786.MCR-09-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNAHOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477 (7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jr, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010a;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, Ulrich A, Wardle GS, Dewell S, Zavolan M, Tuschl T. PAR-CliP—a method to identify transcriptome-wide the binding sites of RNA binding proteins. J Vis Exp. 2010b;(41):2034. doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Max KE, Bandaru P, Morozov P, Gerstberger S, Brown M, Molina H, Tuschl T. Identification of mRNAs bound and regulated by human LIN28 proteins and molecular requirements for RNA recognition. RNA. 2013;19(5):613–626. doi: 10.1261/rna.036491.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16(10):1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Irimia M, Ross PJ, Sung HK, Alipanahi B, David L, Golipour A, Gabut M, Michael IP, Nachman EN, Wang E, Trcka D, Thompson T, O’Hanlon D, Slobodeniuc V, Barbosa-Morais NL, Burge CB, Moffat J, Frey BJ, Nagy A, Ellis J, Wrana JL, Blencowe BJ. MBNL proteins repress ES-cell-specific alternative splicing and reprogramming. Nature. 2013;498(7453):241–245. doi: 10.1038/nature12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanina SA, Mifsud W, Down TA, Hayashi K, O’Carroll D, Lao K, Miska EA, Surani MA. Genome-wide identification of targets and function of individual MicroRNAs in mouse embryonic stem cells. PLoS Genet. 2010;6(10):e1001163. doi: 10.1371/journal.pgen.1001163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna JH, Saha K, Jaenisch R. Pluripotency and cellular reprogramming: facts, hypotheses, unresolved issues. Cell. 2010;143(4):508–525. doi: 10.1016/j.cell.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Nishimura R, Muramatsu S, Matsuda A, Matsubara T, Amano K, Ikeda F, Harley VR, Yoneda T. Paraspeckle protein p54nrb links Sox9-mediated transcription with RNA processing during chondrogenesis in mice. J Clin Invest. 2008;118(9):3098–3108. doi: 10.1172/JCI31373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Lopes SM, Tang F, Surani MA. Dynamic equilibrium and heterogeneity of mouse pluripotent stem cells with distinct functional and epigenetic states. Cell Stem Cell. 2008;3(4):391–401. doi: 10.1016/j.stem.2008.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32(2):276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138(4):696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23(7):837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Gattoni R, Stévenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11(3):837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, Durbin R, Eyras E, Gilbert J, Hammond M, Huminiecki L, Kasprzyk A, Lehvaslaiho H, Lijnzaad P, Melsopp C, Mongin E, Pettett R, Pocock M, Potter S, Rust A, Schmidt E, Searle S, Slater G, Smith J, Spooner W, Stabenau A, Stalker J, Stupka E, Ureta-Vidal A, Vastrik I, Clamp M. The Ensembl genome database project. Nucleic Acids Res. 2002;30(1):38–41. doi: 10.1093/nar/30.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9(1):22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P, Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H, Banerji S, Zander T, Seidel D, Leenders F, Ansén S, Ludwig C, Engel-Riedel W, Stoelben E, Wolf J, Goparju C, Thompson K, Winckler W, Kwiatkowski D, Johnson BE, Jänne PA, Miller VA, Pao W, Travis WD, Pass HI, Gabriel SB, Lander ES, Thomas RK, Garraway LA, Getz G, Meyerson M. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki S, Masuda A, Fujioka Y, Iguchi Y, Katsuno M, Shibata A, Urano F, Sobue G, Ohno K. Position-dependent FUS-RNA interactions regulate alternative splicing events and transcriptions. Sci Rep. 2012;2:529. doi: 10.1038/srep00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I, Lo KC, Hawthorn L, Cowell JK, Ionov Y. Identifying candidate colon cancer tumor suppressor genes using inhibition of nonsense-mediated mRNA decay in colon cancer cells. Oncogene. 2007;26 (20):2873–2884. doi: 10.1038/sj.onc.1210098. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442(7102):533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Iwabuchi KA, Yamakawa T, Sato Y, Ichisaka T, Takahashi K, Okita K, Yamanaka S. ECAT11/L1td1 is enriched in ESCs and rapidly activated during iPSC generation, but it is dispensable for the maintenance and induction of pluripotency. PLoS ONE. 2011;6(5):e20461. doi: 10.1371/journal.pone.0020461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi M, Boutz PL, Paul P, Sharp PA. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes Dev. 2014;28(6):637–651. doi: 10.1101/gad.235770.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KB, Darnell RB. CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol. 2008;488:85–98. doi: 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Tulin AV. Poly(ADP-ribose) controls DE-cadherin-dependent stem cell maintenance and oocyte localization. Nat Commun. 2012;3:760. doi: 10.1038/ncomms1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Lee JY, Pan ZH, Jiang BJ, Tian B. Progressive lengthening of 3′ untranslated regions of mRNAs by alternative polyadenylation during mouse embryonic development. Proc Natl Acad Sci USA. 2009;106 (17):9535–9535. doi: 10.1073/pnas.0900028106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Tian B. Reprogramming of 3′ Untranslated Regions of mRNAs by Alternative Polyadenylation in Generation of Pluripotent Stem Cells from Different Cell Types. PLoS ONE. 2009;4(12):e8419. doi: 10.1371/journal.pone.0008419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302(5653):2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27(5):459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Bonasio R, Saldaña-Meyer R, Yoshida T, Son J, Nishino K, Umezawa A, Reinberg D. Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. 2014;53(2):290–300. doi: 10.1016/j.molcel.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Li G, Son J, Xu CF, Margueron R, Neubert TA, Reinberg D. Phosphorylation of the PRC2 component Ezh2 is cell cycle-regulated and up-regulates its binding to ncRNA. Genes Dev. 2010;24(23):2615–2620. doi: 10.1101/gad.1983810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko S, Son J, Shen SS, Reinberg D, Bonasio R. PRC2 binds active promoters and contacts nascent RNAs in embryonic stem cells. Nat Struct Mol Biol. 2013;20(11):1258–1264. doi: 10.1038/nsmb.2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araújo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, Pombo A, Fisher AG, Young RA, Jenner RG. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38(5):675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karwacki-Neisius V, Göke J, Osorno R, Halbritter F, Ng JH, Weiße AY, Wong FC, Gagliardi A, Mullin NP, Festuccia N, Colby D, Tomlinson SR, Ng HH, Chambers I. Reduced Oct4 expression directs a robust pluripotent state with distinct signaling activity and increased enhancer occupancy by Oct4 and Nanog. Cell Stem Cell. 2013;12(5):531–545. doi: 10.1016/j.stem.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap V, Rezende NC, Scotland KB, Shaffer SM, Persson JL, Gudas LJ, Mongan NP. Regulation of stem cell pluripotency and differentiation involves a mutual regulatory circuit of the NANOG, OCT4, and SOX2 pluripotency transcription factors with polycomb repressive complexes and stem cell microRNAs. Stem Cells Dev. 2009;18(7):1093–1108. doi: 10.1089/scd.2009.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katahira J. mRNA export and the TREX complex. Biochim Biophys Acta. 2012;1819(6):507–513. doi: 10.1016/j.bbagrm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106(28):11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11(7):2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BM, Choi MY. Non-canonical microRNAs miR-320 and miR-702 promote proliferation in Dgcr8-deficient embryonic stem cells. Biochem Biophys Res Commun. 2012;426(2):183–189. doi: 10.1016/j.bbrc.2012.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Saetrom P, Snøve O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105(42):16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18(13):1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- Kunarso G, Wong KY, Stanton LW, Lipovich L. Detailed characterization of the mouse embryonic stem cell transcriptome reveals novel genes and intergenic splicing associated with pluripotency. BMC Genomics. 2008;9(1):155. doi: 10.1186/1471-2164-9-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SC, Yi H, Eichelbaum K, Föhr S, Fischer B, You KT, Castello A, Krijgsveld J, Hentze MW, Kim VN. The RNA-binding protein repertoire of embryonic stem cells. Nat Struct Mol Biol. 2013;20 (9):1122–1130. doi: 10.1038/nsmb.2638. [DOI] [PubMed] [Google Scholar]
- Lackford B, Yao C, Charles GM, Weng L, Zheng X, Choi EA, Xie X, Wan J, Xing Y, Freudenberg JM, Yang P, Jothi R, Hu G, Shi Y. Fip1 regulates mRNA alternative polyadenylation to promote stem cell self-renewal. EMBO J. 2014;33(8):878–889. doi: 10.1002/embj.201386537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ the International Human Genome Sequencing Consortium . Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Leeb M, Dietmann S, Paramor M, Niwa H, Smith A. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell. 2014;14(3):385–393. doi: 10.1016/j.stem.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol. 2011;18(2):237–244. doi: 10.1038/nsmb.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA, Pei D. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7(1):51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z, Dou Y. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012;11(2):163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Song J, Yi C. Genome-wide mapping of cellular protein-RNA interactions enabled by chemical crosslinking. Genomics Proteomics Bioinformatics. 2014;12(2):72–78. doi: 10.1016/j.gpb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12(8):505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- Liu Y, Lee MR, Timani K, He JJ, Broxmeyer HE. Tip110 maintains expression of pluripotent factors in and pluripotency of human embryonic stem cells. Stem Cells Dev. 2012;21(6):829–833. doi: 10.1089/scd.2011.0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Timani K, Ou X, Broxmeyer HE, He JJ. C-MYC controlled TIP110 protein expression regulates OCT4 mRNA splicing in human embryonic stem cells. Stem Cells Dev. 2013;22(5):689–694. doi: 10.1089/scd.2012.0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou H, Neugebauer KM, Gagel RF, Berget SM. Regulation of alternative polyadenylation by U1 snRNPs and SRp20. Mol Cell Biol. 1998;18(9):4977–4985. doi: 10.1128/mcb.18.9.4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Göke J, Sachs F, Jacques PE, Liang H, Feng B, Bourque G, Bubulya PA, Ng HH. SON connects the splicing-regulatory network with pluripotency in human embryonic stem cells. Nat Cell Biol. 2013;15(10):1141–1152. doi: 10.1038/ncb2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci USA. 2007;104(23):9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias S, Plass M, Stajuda A, Michlewski G, Eyras E, Cáceres JF. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol. 2012;19(8):760–766. doi: 10.1038/nsmb.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes Dev. 1996;10(13):1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469(7330):343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272(9):2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134(3):521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78(12):7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463 (7281):621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihailovic M, Wurth L, Zambelli F, Abaza I, Militti C, Mancuso FM, Roma G, Pavesi G, Gebauer F. Widespread generation of alternative UTRs contributes to sex-specific RNA binding by UNR. RNA. 2012;18(1):53–64. doi: 10.1261/rna.029603.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29(13):2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran VA, Perera RJ, Khalil AM. Emerging functional and mechanistic paradigms of mammalian long non-coding RNAs. Nucleic Acids Res. 2012;40(14):6391–6400. doi: 10.1093/nar/gks296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita S, Horii T, Kimura M, Goto Y, Ochiya T, Hatada I. One Argonaute family member, Eif2c2 (Ago2), is essential for development and appears not to be involved in DNA methylation. Genomics. 2007;89(6):687–696. doi: 10.1016/j.ygeno.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Morlando M, Dini Modigliani S, Torrelli G, Rosa A, Di Carlo V, Caffarelli E, Bozzoni I. FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2012;31 (24):4502–4510. doi: 10.1038/emboj.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller AA, Cheung TH, Rando TA. All’s well that ends well: alternative polyadenylation and its implications for stem cell biology. Curr Opin Cell Biol. 2013;25(2):222–232. doi: 10.1016/j.ceb.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2005;102(34):12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Naganuma T, Shioi G, Hirose T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193(1):31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Närvä E, Rahkonen N, Emani MR, Lund R, Pursiheimo JP, Nästi J, Autio R, Rasool O, Denessiouk K, Lähdesmäki H, Rao A, Lahesmaa R. RNA-binding protein L1TD1 interacts with LIN28 via RNA and is required for human embryonic stem cell self-renewal and cancer cell proliferation. Stem Cells. 2012;30(3):452–460. doi: 10.1002/stem.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niranjanakumari S, Lasda E, Brazas R, Garcia-Blanco MA. Reversible cross-linking combined with immunoprecipitation to study RNA-protein interactions in vivo. Methods. 2002;26(2):182–190. doi: 10.1016/S1046-2023(02)00021-X. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24(4):372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Ohta S, Nishida E, Yamanaka S, Yamamoto T. Global splicing pattern reversion during somatic cell reprogramming. Cell Reports. 2013a;5 (2):357–366. doi: 10.1016/j.celrep.2013.09.016. [DOI] [PubMed] [Google Scholar]
- Ohta S, Nishida E, Yamanaka S, Yamamoto T. Global splicing pattern reversion during somatic cell reprogramming. Cell Reports. 2013b;5 (2):357–366. doi: 10.1016/j.celrep.2013.09.016. [DOI] [PubMed] [Google Scholar]
- Ostareck DH, Ostareck-Lederer A, Wilm M, Thiele BJ, Mann M, Hentze MW. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- Pádua Alves C, Fonseca AS, Muys BR, de Barros E, Lima Bueno R, Bürger MC, de Souza JE, Valente V, Zago MA, Silva WA., Jr Brief report: The lincRNA Hotair is required for epithelial-to-mesenchymal transition and stemness maintenance of cancer cell lines. Stem Cells. 2013;31(12):2827–2832. doi: 10.1002/stem.1547. [DOI] [PubMed] [Google Scholar]
- Park JM, Kohn MJ, Bruinsma MW, Vech C, Intine RV, Fuhrmann S, Grinberg A, Mukherjee I, Love PE, Ko MS, DePamphilis ML, Maraia RJ. The multifunctional RNA-binding protein La is required for mouse development and for the establishment of embryonic stem cells. Mol Cell Biol. 2006;26(4):1445–1451. doi: 10.1128/MCB.26.4.1445-1451.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Lee JM, Hwang MY, Son GH, Geum D. NonO binds to the CpG island of oct4 promoter and functions as a transcriptional activator of oct4 gene expression. Mol Cells. 2013;35(1):61–69. doi: 10.1007/s10059-013-2273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27(10):3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, Bak M, Tommerup N, Rappsilber J, Helin K. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(7286):306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- Pelham HR, Brown DD. A specific transcription factor that can bind either the 5S RNA gene or 5S RNA. Proc Natl Acad Sci USA. 1980;77(7):4170–4174. doi: 10.1073/pnas.77.7.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139 (7):1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Chen LL, Lei XX, Yang L, Lin H, Carmichael GG, Huang Y. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29(3):496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147 (5):1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123(2):249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Pritsker M, Doniger TT, Kramer LC, Westcot SE, Lemischka IR. Diversification of stem cell molecular repertoire by alternative splicing. Proc Natl Acad Sci USA. 2005;102(40):14290–14295. doi: 10.1073/pnas.0502132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C, Ma Y, Wang J, Peng S, Huang Y. Lin28-mediated post-transcriptional regulation of Oct4 expression in human embryonic stem cells. Nucleic Acids Res. 2010;38(4):1240–1248. doi: 10.1093/nar/gkp1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21(2):104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Radzisheuskaya A, Chia GB, dos Santos RL, Theunissen TW, Castro LF, Nichols J, Silva JC. A defined Oct4 level governs cell state transitions of pluripotency entry and differentiation into all embryonic lineages. Nat Cell Biol. 2013;15(6):579–590. doi: 10.1038/ncb2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzisheuskaya A, Silva JC. Do all roads lead to Oct4? the emerging concepts of induced pluripotency. Trends Cell Biol. 2014;24(5):275–284. doi: 10.1016/j.tcb.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajyaguru P, Parker R. RGG motif proteins: modulators of mRNA functional states. Cell Cycle. 2012;11(14):2594–2599. doi: 10.4161/cc.20716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishna S, Suresh B, Lim KH, Cha BH, Lee SH, Kim KS, Baek KH. PEST motif sequence regulating human NANOG for proteasomal degradation. Stem Cells Dev. 2011;20(9):1511–1519. doi: 10.1089/scd.2010.0410. [DOI] [PubMed] [Google Scholar]
- Rao S, Zhen S, Roumiantsev S, McDonald LT, Yuan GC, Orkin SH. Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol Cell Biol. 2010;30(22):5364–5380. doi: 10.1128/MCB.00419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Chan ET, Peña Castillo L, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, Hughes TR. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol. 2009;27(7):667–670. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LO, Lei EP, Fraser AG, Blencowe BJ, Morris QD, Hughes TR. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499(7457):172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds N, Collier B, Maratou K, Bingham V, Speed RM, Taggart M, Semple CA, Gray NK, Cooke HJ. Dazl binds in vivo to specific transcripts and can regulate the pre-meiotic translation of Mvh in germ cells. Hum Mol Genet. 2005;14(24):3899–3909. doi: 10.1093/hmg/ddi414. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N, Nelson B, Vranizan K, Pico AR, Hanspers K, Kuchinsky A, Ta L, Mercola M, Conklin BR. Alternative splicing in the differentiation of human embryonic stem cells into cardiac precursors. PLOS Comput Biol. 2009;5(11):e1000553. doi: 10.1371/journal.pcbi.1000553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS, Clark TA, Williams A, Blume JE, Samal E, Mercola M, Merrill BJ, Conklin BR. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci USA. 2010;107(23):10514–10519. doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7(1):64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–1647. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Faiola F, Wang J. Concise review: pursuing self-renewal and pluripotency with the stem cell factor Nanog. Stem Cells. 2013;31(7):1227–1236. doi: 10.1002/stem.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders A, Wang J. Export and expression: mRNAs deliver new messages for controlling pluripotency. Cell Stem Cell. 2014;14(5):549–550. doi: 10.1016/j.stem.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Schulman BR, Esquela-Kerscher A, Slack FJ. Reciprocal expression of lin-41 and the microRNAs let-7 and mir-125 during mouse embryogenesis. Dev Dyn. 2005;234(4):1046–1054. doi: 10.1002/dvdy.20599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz EG, Heard E. Role and control of X chromosome dosage in mammalian development. Curr Opin Genet Dev. 2013;23(2):109–115. doi: 10.1016/j.gde.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Schulz EG, Meisig J, Nakamura T, Okamoto I, Sieber A, Picard C, Borensztein M, Saitou M, Blüthgen N, Heard E. The two active X chromosomes in female ESCs block exit from the pluripotent state by modulating the ESC signaling network. Cell Stem Cell. 2014;14(2):203–216. doi: 10.1016/j.stem.2013.11.022. [DOI] [PubMed] [Google Scholar]
- Sheik Mohamed J, Gaughwin PM, Lim B, Robson P, Lipovich L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA (New York, NY. 2010;16:324–337. doi: 10.1261/rna.1441510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekar PC, Naim A, Sarathi DP, Kumar S. Argonaute-2-null embryonic stem cells are retarded in self-renewal and differentiation. J Biosci. 2011;36(4):649–657. doi: 10.1007/s12038-011-9094-1. [DOI] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17(4):761–772. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibayama M, Ohno S, Osaka T, Sakamoto R, Tokunaga A, Nakatake Y, Sato M, Yoshida N. Polypyrimidine tract-binding protein is essential for early mouse development and embryonic stem cell proliferation. FEBS J. 2009;276(22):6658–6668. doi: 10.1111/j.1742-4658.2009.07380.x. [DOI] [PubMed] [Google Scholar]
- Simon B, Kirkpatrick JP, Eckhardt S, Reuter M, Rocha EA, Andrade-Navarro MA, Sehr P, Pillai RS, Carlomagno T. Recognition of 2′-O-methylated 3′-end of piRNA by the PAZ domain of a Piwi protein. Structure. 2011;19(2):172–180. doi: 10.1016/j.str.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Singh G, Ricci EP, Moore MJ. RIPiT-Seq: a high-throughput approach for footprinting RNA:protein complexes. Methods. 2014;65(3):320–332. doi: 10.1016/j.ymeth.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkkonen L, Hugenschmidt T, Berninger P, Gaidatzis D, Mohn F, Artus-Revel CG, Zavolan M, Svoboda P, Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15(3):259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- Souquere S, Beauclair G, Harper F, Fox A, Pierron G. Highly ordered spatial organization of the structural long noncoding NEAT1 RNAs within paraspeckle nuclear bodies. Mol Biol Cell. 2010;21(22):4020–4027. doi: 10.1091/mbc.E10-08-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl R, Skrisovska L, Allain FH. RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle. EMBO Rep. 2005;6(1):33–38. doi: 10.1038/sj.embor.7400325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V, Klattenhoff CA, Boyer LA. Screening for novel regulators of embryonic stem cell identity. Cell Stem Cell. 2009;4(5):377–378. doi: 10.1016/j.stem.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Sugnet CW, Srinivasan K, Clark TA, O’Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, Ares M., Jr Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLOS Comput Biol. 2006;2(1):e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19(3):347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TS. Transcriptional heterogeneity in mouse embryonic stem cells. Reprod Fertil Dev. 2009;21(1):67–75. doi: 10.1071/rd08219. [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455(7216):1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol. 2008;15(12):1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Thornton JE, Chang HM, Piskounova E, Gregory RI. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7) RNA. 2012;18(10):1875–1885. doi: 10.1261/rna.034538.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem Sci. 2013;38(6):312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Simanshu DK, Ma JB, Patel DJ. Structural basis for piRNA 2′-O-methylated 3′-end recognition by Piwi PAZ (Piwi/Argonaute/Zwille) domains. Proc Natl Acad Sci USA. 2011;108(3):903–910. doi: 10.1073/pnas.1017762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiscornia G, Izpisúa Belmonte JC. MicroRNAs in embryonic stem cell function and fate. Genes Dev. 2010;24(24):2732–2741. doi: 10.1101/gad.1982910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka Y, Shimosato D, Murakami K, Takahashi K, Niwa H. Identification and characterization of subpopulations in undifferentiated ES cell culture. Development. 2008;135(5):909–918. doi: 10.1242/dev.017400. [DOI] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SC, Chang DF, Hong CM, Xia P, Senadheera D, Trump L, Mishra S, Lutzko C. Induced overexpression of OCT4A in human embryonic stem cells increases cloning efficiency. Am J Physiol Cell Physiol. 2014;306(12):C1108–C1118. doi: 10.1152/ajpcell.00205.2013. [DOI] [PubMed] [Google Scholar]
- Tsui S, Dai T, Warren ST, Salido EC, Yen PH. Association of the mouse infertility factor DAZL1 with actively translating polyribosomes. Biol Reprod. 2000;62(6):1655–1660. doi: 10.1095/biolreprod62.6.1655. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen K, Mele A, Darnell RB. CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods. 2005;37 (4):376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302 (5648):1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25(22):10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L, Nishikura K. ADAR gene family and A-to-I RNA editing: diverse roles in posttranscriptional gene regulation. Prog Nucleic Acid Res Mol Biol. 2005;79:299–338. doi: 10.1016/S0079-6603(04)79006-6. [DOI] [PubMed] [Google Scholar]
- Valverde R, Edwards L, Regan L. Structure and function of KH domains. FEBS J. 2008;275(11):2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- Venables JP, Brosseau JP, Gadea G, Klinck R, Prinos P, Beaulieu JF, Lapointe E, Durand M, Thibault P, Tremblay K, Rousset F, Tazi J, Abou Elela S, Chabot B. RBFOX2 is an important regulator of mesenchymal tissue-specific splicing in both normal and cancer tissues. Mol Cell Biol. 2013a;33(2):396–405. doi: 10.1128/MCB.01174-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Lapasset L, Gadea G, Fort P, Klinck R, Irimia M, Vignal E, Thibault P, Prinos P, Chabot B, Abou Elela S, Roux P, Lemaitre JM, Tazi J. MBNL1 and RBFOX2 cooperate to establish a splicing programme involved in pluripotent stem cell differentiation. Nat Commun. 2013b;4:2480. doi: 10.1038/ncomms3480. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigó R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291(5507):1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC, Yang YW, Liu B, Sanyal A, Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta RA, Wysocka J, Lei M, Dekker J, Helms JA, Chang HY. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Miao YL, Zheng X, Lackford B, Zhou B, Han L, Yao C, Ward JM, Burkholder A, Lipchina I, Fargo DC, Hochedlinger K, Shi Y, Williams CJ, Hu G. The THO complex regulates pluripotency gene mRNA export and controls embryonic stem cell self-renewal and somatic cell reprogramming. Cell Stem Cell. 2013;13(6):676–690. doi: 10.1016/j.stem.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chang Y, Li Y, Zhang X, Goodrich DW. Thoc1/Hpr1/p84 is essential for early embryonic development in the mouse. Mol Cell Biol. 2006;26(11):4362–4367. doi: 10.1128/MCB.02163-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McLachlan J, Zamore PD, Hall TM. Modular recognition of RNA by a human pumilio-homology domain. Cell. 2002;110(4):501–512. doi: 10.1016/s0092-8674(02)00873-5. [DOI] [PubMed] [Google Scholar]
- Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7(4):855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39(3):380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck SM, Mele A, Yan Q, Sun S, Farny N, Zhang Z, Xue C, Herre M, Silver PA, Zhang MQ, Krainer AR, Darnell RB, Zhang C. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Reports. 2014;6(6):1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Bilodeau S, Orlando DA, Hoke HA, Frampton GM, Foster CT, Cowley SM, Young RA. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature. 2012;482(7384):221–225. doi: 10.1038/nature10805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilbert ML, Huelga SC, Kapeli K, Stark TJ, Liang TY, Chen SX, Yan BY, Nathanson JL, Hutt KR, Lovci MT, Kazan H, Vu AQ, Massirer KB, Morris Q, Hoon S, Yeo GW. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48(2):195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Cedervall T. The La protein. Annu Rev Biochem. 2002;71 (1):375–403. doi: 10.1146/annurev.biochem.71.090501.150003. [DOI] [PubMed] [Google Scholar]
- Wong RC, Ibrahim A, Fong H, Thompson N, Lock LF, Donovan PJ. L1TD1 is a marker for undifferentiated human embryonic stem cells. PLoS ONE. 2011;6(4):e19355. doi: 10.1371/journal.pone.0019355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Habegger L, Noisa P, Szekely A, Qiu C, Hutchison S, Raha D, Egholm M, Lin H, Weissman S, Cui W, Gerstein M, Snyder M. Dynamic transcriptomes during neural differentiation of human embryonic stem cells revealed by short, long, and paired-end sequencing. Proc Natl Acad Sci USA. 2010;107(11):5254–5259. doi: 10.1073/pnas.0914114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Tan X, Lin Q, Schmidt B, Engel W, Pantakani DV. Mouse Dazl and its novel splice variant functions in translational repression of target mRNAs in embryonic stem cells. Biochim Biophys Acta. 2013;1829(5):425–435. doi: 10.1016/j.bbagrm.2012.12.010. [DOI] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, Jiang L, Cai Z, Sun H, Zhang K, Zhang Y, Chen J, Fu XD. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152(1–2):82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426(6965):468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- Yang YW, Flynn RA, Chen Y, Qu K, Wan B, Wang KC, Lei M, Chang HY. Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. eLife. 2014;3:e02046. doi: 10.7554/eLife.02046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeganeh M, Seyedjafari E, Kamrani FA, Ghaemi N. RNA-binding protein Rbm47 binds to Nanog in mouse embryonic stem cells. Mol Biol Rep. 2013;40(7):4391–4396. doi: 10.1007/s11033-013-2528-0. [DOI] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16(2):130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Xu X, Liang TY, Muotri AR, Carson CT, Coufal NG, Gage FH. Alternative splicing events identified in human embryonic stem cells and neural progenitors. PLOS Comput Biol. 2007;3(10):1951–1967. doi: 10.1371/journal.pcbi.0030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, Chalkidis G, Suzuki Y, Shiosaka M, Kawahata R, Yamaguchi T, Otsu M, Obara N, Sakata-Yanagimoto M, Ishiyama K, Mori H, Nolte F, Hofmann WK, Miyawaki S, Sugano S, Haferlach C, Koeffler HP, Shih LY, Haferlach T, Chiba S, Nakauchi H, Miyano S, Ogawa S. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478(7367):64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin II, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6(5):837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zamore PD, Carmo-Fonseca M, Lamond AI, Green MR. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci USA. 1992;89(18):8769–8773. doi: 10.1073/pnas.89.18.8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322(5902):750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35(1):1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C, Brimacombe R. RNA-protein cross-linking in Eschericia coli 30S ribosomal subunits: a method for the direct analysis of the RNA regions involved in the cross-links. Nucleic Acids Res. 1978;5(4):1189–1206. doi: 10.1093/nar/5.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]