Abstract
The guinea pig (Cavia porcellus) provides a useful animal model for studying the pathogenesis of many infectious diseases, and for preclinical evaluation of vaccines. However, guinea pig models are limited by the lack of immunological reagents required for characterization and quantification of antigen-specific T cell responses. To address this deficiency, an enzyme-linked immunospot (ELISPOT) assay for guinea pig interferon (IFN)-γ was developed to measure antigen/epitope-specific T cell responses to guinea pig cytomegalovirus (GPCMV) vaccines. Using splenocytes harvested from animals vaccinated with a modified vaccinia virus Ankara (MVA) vector encoding the GPCMV GP83 (homolog of human CMV pp65 [gpUL83]) protein, we were able to enumerate and map antigen-specific responses, both in vaccinated as well as GPCMV-infected animals, using a panel of GP83-specific peptides. Several potential immunodominant GP83-specific peptides were identified, including one epitope, LGIVHFFDN, that was noted in all guinea pigs that had a detectable CD8+ response to GP83. Development of a guinea pig IFN-γ ELISPOT should be useful in characterization of additional T cell-specific responses to GPCMV, as well as other pathogens. This information in turn can help focus future experimental evaluation of immunization strategies, both for GPCMV as well as for other vaccine-preventable illnesses studied in the guinea pig model.
Keywords: ELISPOT, Interferon gamma, IFN-γ, Guinea pig, Cytomegalovirus vaccine, pp65, Glycoprotein B, Congenital CMV infection, CMV pp65, Guinea pig immunology, Guinea pig cytokine, Placenta, TORCH infection
1. Introduction
Guinea pigs have provided a useful model for vaccines against many pathogens [1,2], including tuberculosis [3,4], influenza [5], and viral hemorrhagic fevers [6,7]. Since human cytomegalovirus (HCMV) will not complete its infectious cycle in non-human cells, animal models of CMV must be employed. In particular, the guinea pig cytomegalovirus (GPCMV) model is uniquely useful for studying CMV vaccine strategies [8–12]. Vaccines based on the recombinant GPCMV glycoprotein B (gB) homolog, a target of neutralizing antibody produced following HCMV infection [13], have shown varying degrees of effectiveness in preventing maternal and placental infection, and in limiting congenital viral transmission [8,14]. The GPCMV homolog of HCMV ppUL83 (pp65) [15], GP83, elicits both antibody and specific CD4+ and CD8+ cellular responses following vectored immunization with a recombinant alphavirus replicon [16]. Unfortunately, no studies have examined effector function or cytokine profile of T-cells following GP83 vaccination. A possible effector of HCMV vaccine-mediated protection is the interferon gamma (IFN-γ) response [17]. Expression of IFN-γ by T cells correlates with protection against CMV disease following transplantation [18,19] and in AIDS patients [20]. IFN-γ expression by T cells is also critical in vaccine-induced protection for other herpes viruses [21,22]. For murine CMV (MCMV) and rhesus CMV (RhCMV), DNA vaccines elicit robust expression of IFN-γ by T cells, and these responses correlate with protection upon subsequent viral challenge [23,24]. IFN-γ ELISPOT assays have also been valuable in identifying the viral gene products that serve to elicit T cell responses in the context of HCMV infection [25–27]. To better identify potential correlates of protective immunity following vaccination in the GPCMV model, the present study sought to develop an IFN-γ ELISPOT assay to quantify peptide-specific stimulation of GP83-specific splenocytes, following vaccination of guinea pigs with a modified vaccinia virus Ankara (MVA)-vectored GP83 vaccine.
2. Materials and methods
2.1. Guinea pigs
Twenty-two, 200 g Hartley guinea pigs (Elm Hill Laboratories, Chelmsford, MA) were housed under conditions approved by the University of Minnesota Institutional Animal Use Committee. Guinea pigs were all documented to be GPCMV-seronegative prior to study [14].
2. Antibodies
Mouse monoclonal anti-guinea pig IFN-γ antibodies (N-G3 and V-E4) [28,29] were a generous gift from Hubert Schäfer. This antibody combination was optimal for IFN-γ sandwich ELISA when V-E4 was used as the capture antibody and N-G3 was used as the detecting antibody [28]. These antibodies recognize conformation-specific epitopes in the N-terminal 132 amino acids (aa) of the guinea pig (Cavia porcellus) IFN-γ open reading frame (ORF) [30]. N-G3 was biotinylated using a biotin (type 1) conjugation kit (LNK041B, AbD serotech, Raleigh, NC) and verified by a Biotin Quantitation Kit (28005, Thermo Fisher Scientific, Rockford, IL).
2.3. Peptide pools
Peptides (Sigma–Aldrich, St. Louis, MO) were designed to span GP83 in 9 aa long fragments with 5 aa overlaps (140 total peptides; Supplemental Table 1). Peptides in DMSO (10 mg/ml) were allocated into pools containing 11 or 12 peptides. Every peptide was in two different pools, allowing identification of individual immunoreactive peptides (Supplemental Table 2).
2.4. Recombinant MVA
Recombinant MVAs were generated using MVA transfer vector ZWIIA as previously described [31]. Plasmids pKTS 404 and pKTS 437, expressing a truncated, secreted form of GPCMV gB and a full-length form of GP83, respectively, were used as templates for PCR-mediated insertion of each ORF into ZWIIA. Recombinant MVAs were generated on chicken embryo fibroblasts via homologous recombination by transfection/infection method as described previously [32,33]. Viruses were subjected to plaque purification by limiting dilution. PCR and DNA sequence analyses of PCR-generated fragments confirmed the predicted insertion and orientation of the gB and GP83 ORFs into the recombinant MVA genome.
2.5. Vaccines and vaccination
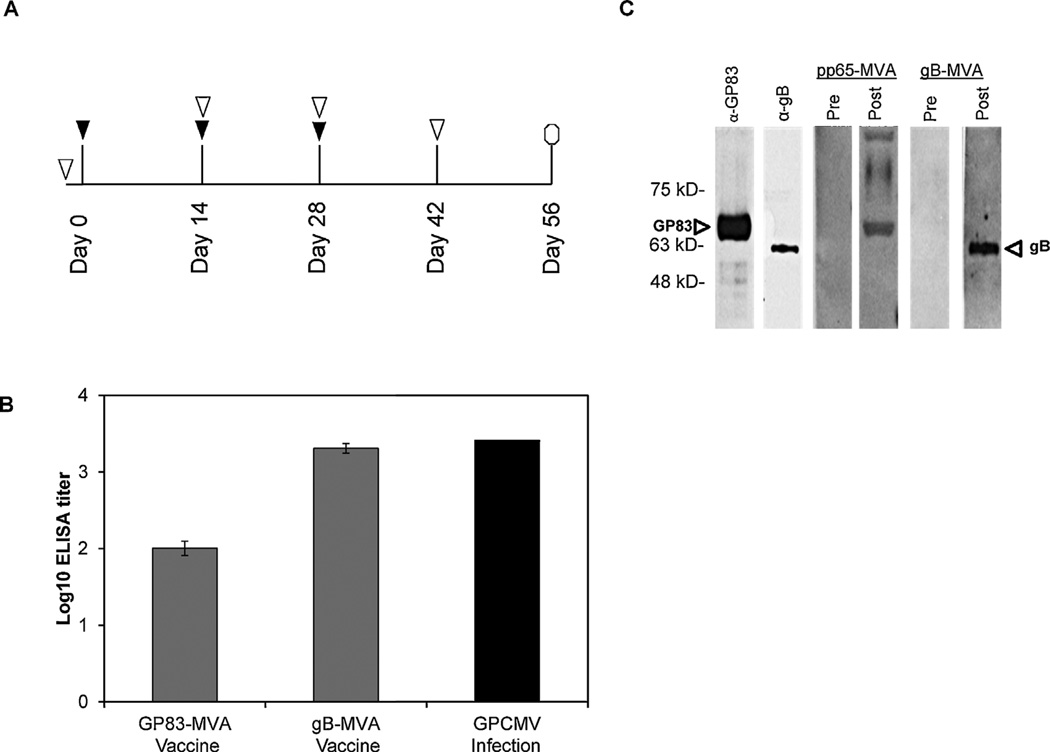
Animals were initially injected with 1 × 108 plaque forming units of either MVA-gB (4 animals) or MVA-GP83 (12 animals). Two additional doses were administered at two-week intervals (Fig. 1A ▼). Blood was collected prior to the first vaccination, at the time of subsequent boosts, and at two weeks post-final vaccination (Fig. 1A ▽).
Fig. 1.
Antibody response to MVA vaccination. (A) Timeline of vaccinations and blood draws. Animals were initially injected with 1 × 108 pfu of either MVA-gB (4 animals) or MVA-GP83 (12 animals), and two additional boosts of the same dose were given at two-week intervals (▼). Blood was collected prior to the first vaccination, at the time of each subsequent boost, and at two weeks post-final vaccination (▽). Antibody titers expressed as log transformed data from end-point dilution ELISA assay as described in Section 2.7. (B) Mean ELISA titers of MVA-GP83 (n = 12) and MVA-gB (n = 4) groups are expressed as log-transformed data from the 3rd bleed post-vaccination (14 days following the third dose). Data depicted are results of end-point dilution ELISA assay as described in Section 2.7. Animals below the cut-off of the ELISA assay (1:80) were assigned a titer of 1:40 for statistical analyses. No detectable response was observed in pre-immune sera (data not shown). ELISA titers were significantly higher following vaccination with MVA-gB construct (p < 0.0005). ELISA responses from 4 GPCMV naïve animals challenged with GPCMV (1 × 105 pfu inoculated sc) were also measured at day 28 (black bar). (C) Specificity of the immune response was determined by Western blot analysis. Antibody responses to GP83 and gB were measured using purified virions followed by immune blotting with rabbit polyclonal anti-GP83, mouse monoclonal anti-gB, pre-immune sera, and sera from the fourth bleed (representative animals shown for MVA-gB and MVA-GP83 vaccinated animals).
2.6. GPCMV infection
To measure T cell responses in naturally infected animals, four guinea pigs were infected by subcutaneous (sc) injection with 1 × 105 plaque forming units (pfu) of salivary gland passaged GPCMV. Two additional uninfected animals served as negative controls. Serum was collected prior to infection and at 21 days post-infection (dpi) to document seroconversion. Animals were sacrificed and spleens harvested at 28 dpi for splenocyte isolation and ELISPOT analyses.
2.7. ELISA
Humoral responses were monitored by ELISA as previously described [14]. Virus specific antibodies bound to viral proteins in the wells were detected using 100 µl of rabbit anti-guinea pig HRP IgG (A55455-1 ml, Sigma) diluted 1:1000 in PBS. Secondary antibody binding was detected by incubating 100 µl of the HRP sub-strate, chromogen tetramethyl benzidine (Life Technologies, Grand Island, NY), measured using a SpectraMax M2 Spectrophotometer (Molecular Devices, Sunnyvale, CA) and the ScanMax Pro program. Titers <80 were assigned a value of 40 for statistical comparisons.
2.8. Western blot analysis
For immune blotting, proteins were detected with polyclonal antibodies to GP83 and gB generated by conjugating peptides (CGRRTGNADRHRRDRDGGDDDDDE and GQLGEDNEILLGTHRMET, respectively) to keyhole limpet hemocyanin, followed by injection with incomplete Freund’s adjuvant into rabbits [34]. Antibody binding was detected using goat anti-rabbit antibodies conjugated to horseradish peroxidase (Cell Signaling, Boston, MA) followed by enhanced chemiluminescence detection (GE Biosciences, Pitts-burgh, PA).
2.9. Splenocyte isolation
Spleens were harvested from guinea pigs at 28–32 days following the third vaccination. Spleens were placed in 10 ml of cold PBS with 10% fetal bovine serum (FBS) and 2× antibiotic–antimycotic (Life Technologies, Carlsbad, CA). Spleens were minced finely and sequentially passed through a 100 µm screen and a 70 µm screen (BD Biosciences, San Jose, CA). Cells were then pelleted via centrifugation at 400 × g for 10 min at 4°C using an Eppendorf 5810R 15-amp version centrifuge (Hamburg, Germany). Splenocytes were isolated over Ficoll gradients by centrifugation at 800 × g for 15 min at 20°C. Isolated splenocytes were washed twice and passed through another 70 µm screen and diluted to 1 × 106 viable cells per ml in RPMI media with 10% (v/v) FBS and 2 × antibiotic–antimycotic. Viability was determined by trypan blue staining.
2.10. ELISPOT assay for IFN-γ
All reagents used were filtered through a 0.22 µm filter. Wells of 96-well Multiscreen HTS Plates (Millipore, Billerica, MA) were coated with 100 µl primary anti-IFN-γ antibody solution (5 µg/ml in PBS, pH 7.4, V-E4) for 24 h at 4°C. Nonspecific binding was blocked with 200 µl of RPMI media with 10% (v/v) FBS for 2 h at room temperature. After blocking and washing, 1 × 105 splenocytes in 100 µl of RPMI were mixed with 50 µl stimulant (no stimulation DMSO control, positive control concanavalin A at 20 µg/ml, or peptide pools at 20 µg/ml) in triplicate. After incubation in humidified 5% CO2at 37°C for 18 h, cells were removed by washing and 100 µl of biotinylated secondary anti-IFN-γ antibody (2 µg/ml, N-G3) in blocking buffer was added to each well. Following a 2 hr incubation and washing, alkaline phosphatase-conjugated streptavidin (SEL002, R&D Systems Inc., Minneapolis, MN) was diluted 1:100 and wells were incubated with 100 µl for 1 h at room temperature. Following washes, wells were incubated for 1 h at room temperature with 100 µl of BCIP/NBT detection reagent (SEL002, R & D Systems) and spots counted with an AID Elispot Reader System using Elispot 6.0-iSpot (Autoimmune Diagnostika GmbH, Straßberg, Germany).
2.11. Statistical analyses
For all data, triplicate samples were analyzed for mean and standard error of the mean (SEM). To control for background in ELISPOT studies, the mean number of spots obtained in the presence of medium alone (no cells) was subtracted from the mean number of spots counted in each of the control or experimental conditions. To assess if a response was significantly different compared to a negative control (DMSO), Dunnett’s multiple comparison test was applied using Prism 6 software (GraphPad Software Inc., La Jolla, CA). A response to peptide pools was considered to be significantly higher than negative control when p ≤ 0.05. Group comparisons were performed using t-tests.
3. Results
3.1. Generation of recombinant MVA expressing gB and GP83
Recombinant MVA was generated using the vector pZWIIA as previously described [31] in the MVA deletion 2 site. Correct, directional insertion of each ORF was confirmed by PCR and sequencing. For GP83, the full-length ORF was expressed; for gB [which served as a negative control for the GP83 ELISPOT], a truncated version encoding a 687 aa protein (I687) was expressed. A silent aa mutation was noted by sequencing at position 451 of the gB ORF. Expression of gB by Western blot identified a variety of isoforms migrating at 100–130 kDa (data not shown). GP83 expression was also detected in cells infected with the MVA-GP83 construct (data not shown).
3.2. In vivo immunogenicity comparison of GPCMV MVA-gB and MVA-GP83 vaccines
Twenty-two guinea pigs (~200 g) were separated into four groups. These groups consisted of animals immunized with 1 × 108 pfu of MVA-gB vaccine (4 animals); MVA-GP83 vaccine (12 animals); animals inoculated with 1 × 105 of GPCMV (4 animals); and 2 uninoculated negative controls. For the animals immunized with MVA-GP83, 6 animals were immunized by sc route, while 6 animals were immunized by intramuscular route; however, no differences were observed in immune response when these two routes were compared (data not shown). All of the MVA-gB animals were immunized by sc route. For all MVA vaccine groups, two additional boosts were administered at two-week intervals (Fig. 1A ▼). For vaccinated animals, blood was collected prior to the first vaccination, at each subsequent boost, and at two weeks post-final vaccination (Fig. 1A ▽). The ELISA titers of the two groups of animals vaccinated with MVA-GP83 (n = 12) or MVA-gB (n = 4) were compared. For most animals that seroconverted to MVA-GP83 immunization, an ELISA response was identified after the third dose. Animals below the cut-off of the ELISA assay (1:80) were assigned a titer of 1:40 for statistical analyses, including determination of the group mean ELISA response. The group mean antibody titer was 2.0 log10 ± 0.08 (Fig. 1B). An anti-GPCMV antibody response was detectable following the first MVA-gB vaccination (data not shown). Following the third vaccination, MVA-gB vaccinated animals had approximately twenty-fold higher ELISA titers when compared to the animals vaccinated with MVA-GP83 (3.3 log10 ± 0.1; p < 0.0005, t-test, Fig. 1B). The ELISA titers observed following MVA-gB vaccination were comparable to those demonstrated following experimental infection with GPCMV-22122 (3.4 log10; Fig. 1B). To further characterize antibody responses, immunoblots were performed. These results demonstrated both gB and GP83-specific bands (Fig. 1C). In total, we observed that 10/12 of MVA-GP83 immunized animals responded to vaccination, either with a humoral response (1/12), a cellular response (2/12), or both (7/10; described below).
3.3. IFN-γ ELISPOT response to MVA vaccination
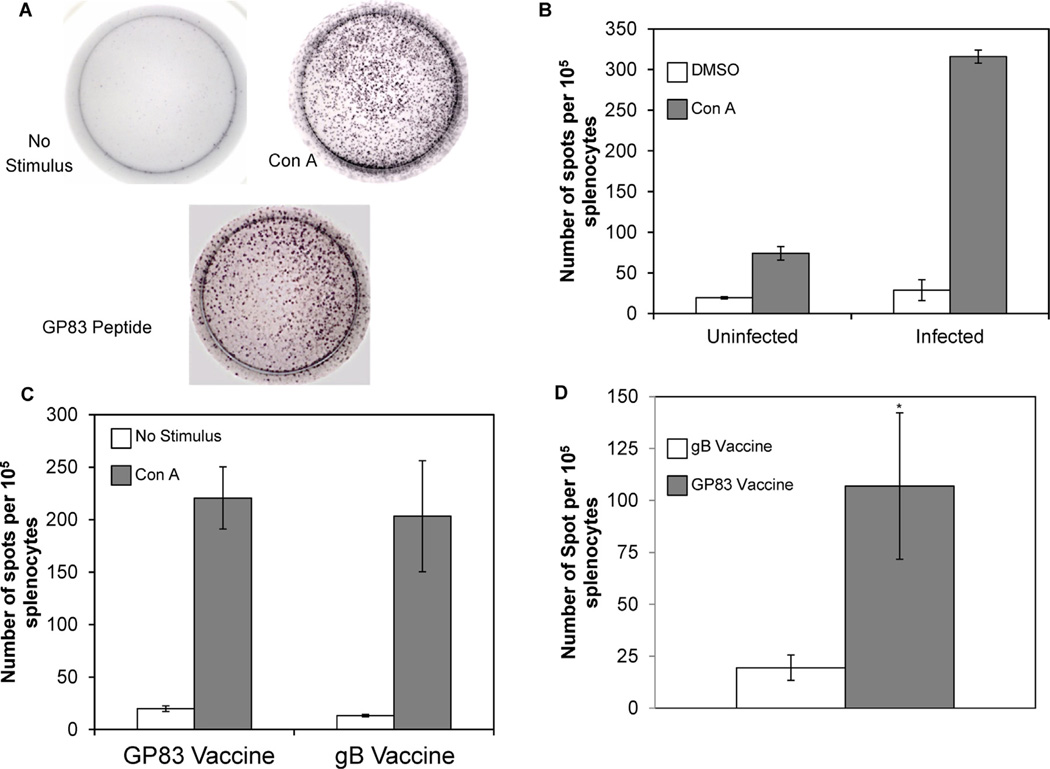
As a test of the ELISPOT assay’s ability to measure IFN-γ responses to infection, spleens were collected from guinea pigs following inoculation with salivary gland-passaged GPCMV at 28 dpi (n = 4), or from uninfected animals (n = 2). Splenocytes (105 cells per well) were stimulated with the mitogen ConA, 20 µg/ml, or with the DMSO control (Fig. 2A). There were a small number of background spots present in DMSO control-treated splenocytes in both uninfected and infected animals (19.5/105 splenocytes ± 1.5 SEM and 28.8/105 splenocytes ± 12.3 SEM respectively; Fig. 2B white bars). Infection led to an expansion of cells capable of secreting IFN-γ, since uninfected animals had a smaller pool (74 cells/105 splenocytes ± 8 SEM) of cells responding to ConA treatment compared to infected animals (315/105 splenocytes ± 8 SEM; Fig. 2B, gray bars).
Fig. 2.
Enumeration of IFN-γ excreting splenocytes in response to mitogen and peptide stimulation. Splenocytes were isolated from uninfected or infected animals at 28 dpi using a Ficoll gradient (A) Splenocytes were either treated with DMSO (no stimulus control), the mitogen ConA, or GP83 peptides and enumerated using ELISPOT assays specific for IFN-γ. Representative wells are shown. (B) Triplicate wells for each sample were counted and averaged for each animal. Results are shown in mean number of IFN-γ positive cells per 100,000 splenocytes for group (DMSO treated cells are shown in white and ConA treated cells are shown in gray; error bars = SEM). Splenocytes were isolated from animals at 28–32 days after the third vaccination using a Ficoll gradient. Results were quantitated as above for each vaccinated group. (C) DMSO treated cells are shown in white and ConA treated cells are shown in gray (error bars = SEM). (D) Harvested splenocytes were stimulated with pooled peptides that span the entire GP83 protein, as described in Section 2.10. Triplicate wells for each sample were counted and the mean for all animals (including non-responders) in both vaccine groups determined. Mean number of IFN-γ positive cells per 100,000 splenocytes for each vaccinated group is shown. Animals vaccinated with MVA-gB are shown in white and MVA-GP83 vaccinated animals are shown in gray (error bars = SEM; *p < 0.05).
IFN-γ response was next measured in MVA-gB and MVA-GP83 vaccinated animals. Animals were sacrificed approximately 30 days following the third vaccination and splenocytes isolated. A small number of background IFN-γ producing cells were observed in both gB and GP83 groups following DMSO treatment (13.2 ± 1.1 SEM and 19.8 ± 2.7 SEM respectively; Fig. 2C white bars). Similar to GPCM Vinfection, large numbers of IFN-γ producing cells (220 ± 29 SEM and 203 ± 53 SEM) were found in splenocytes stimulated with ConA from animals vaccinated with gB or GP83, respectively (Fig. 2C gray bars).
To detect antigen specific responses, splenocytes from uninfected and infected animals as well as gB and GP83 vaccinated animals were stimulated with overlapping 9 aa peptides that spanned GP83. Animals from the gB vaccinated group showed no significant response to the GP83 peptides compared to DMSO controls (19 ± 6 SEM spots; Fig. 2D white bar). The GP83 vaccinated group, however, showed a significant response to GP83 peptide stimulation compared to DMSO controls (107 ± 35 SEM spots; p < 0.05; Fig. 2D gray bar). This result was comparable to the response exhibited by animals experimentally infected by sc inoculation with GPCMV (169.5 ± 71 SEM spots). In total, 75% of GP83 vaccinated animals had a significant response to peptide stimulation (compared to no stimulus controls, p < 0.05, t-test; Table 1).
Table 1.
Summary of humoral and cell mediated immune responses to MVA-gB and MVA-GP83 vaccination.
| Vaccine group | # of ELISA-positive guinea pigs |
# of WB-positive guinea pigs |
# of ELISPOT- positive guinea pigs |
|---|---|---|---|
| gB | 4/4 | 4/4 | 0/4 |
| GP83 | 8/12 | 2/12 | 9/12 |
3.4. Mapping of GP83 epitopes
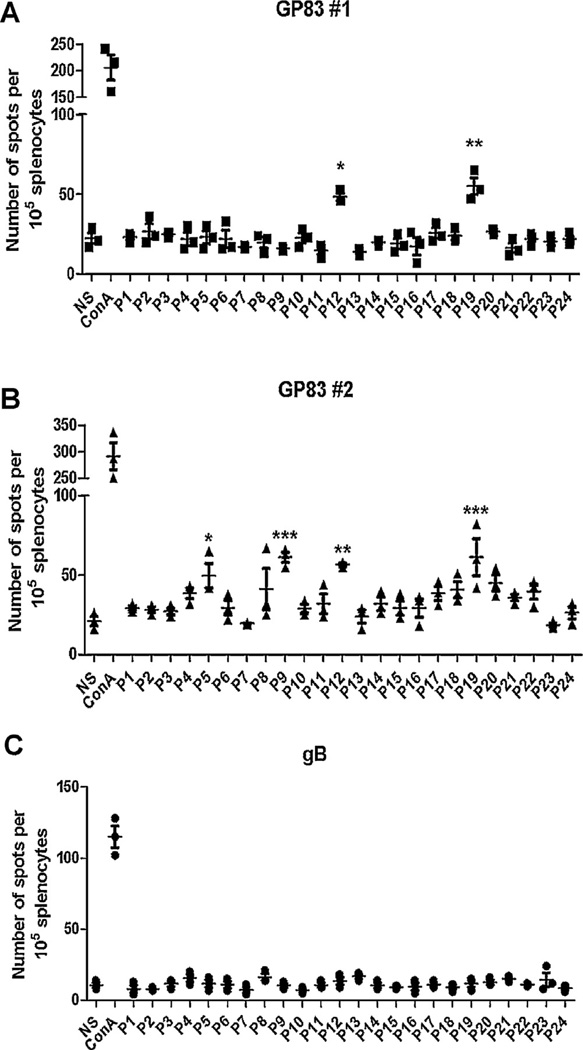
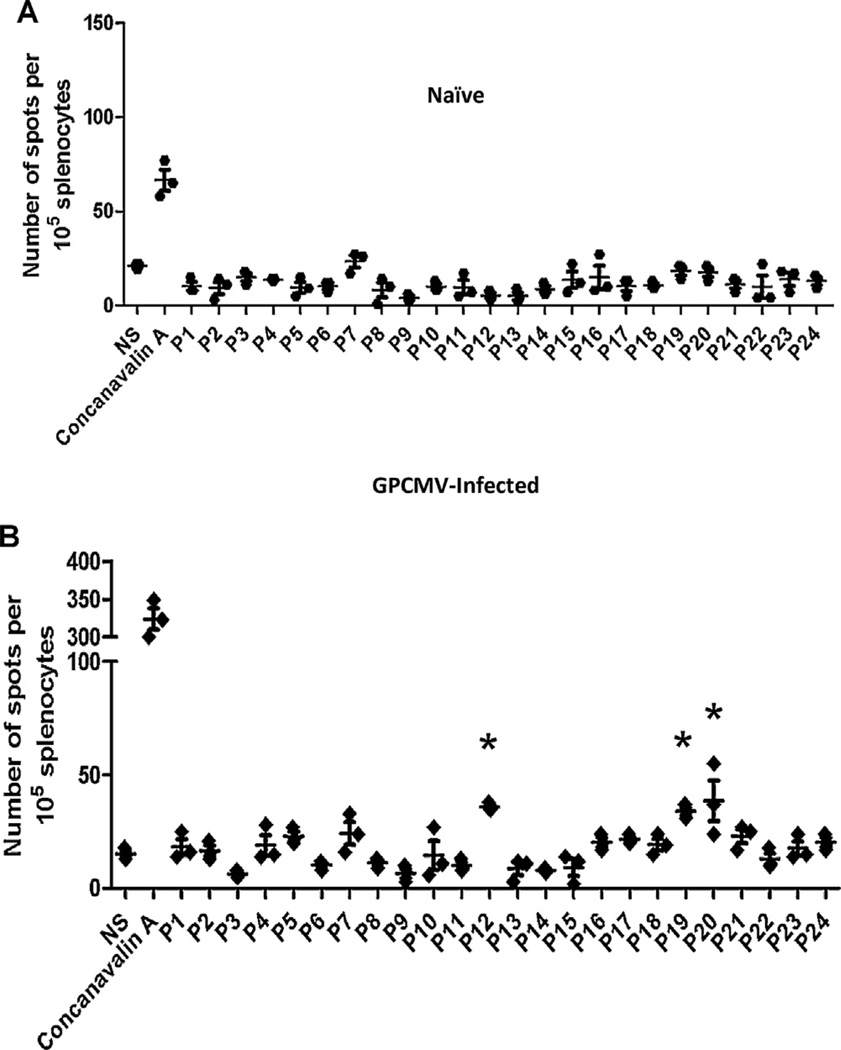
GP83 epitopes recognized by vaccinated guinea pigs were mapped using peptide pools. As described above, the library of 140 total GP83 peptides were combined into 24 pools of 11–12 with each peptide being in two unique pools (Supplemental Table 2). Immunoreactive peptide pools were classified as any pool that displayed a significant response (p < 0.05) compared to unstimulated DMSO controls. Individual peptides were identified as the shared peptide between two pools with similar response. In 9 out of the 12 MVA-GP83 vaccinated animals, T cell responses to one or more peptides were detected (Fig. 3A and B). There was no detectable IFN-γ response in 3 out of 12 animals vaccinated with MVA-GP83 (data not shown) or, as expected, in any animals vaccinated with MVA-gB (Fig. 3C). Peptide pools corresponding to peptides 46, 60, and 106 (Supplemental Table 1) had significant responses compared to DMSO controls (p < 0.05) in two guinea pigs each. Interestingly, all animals that had a detectable response responded to peptide 84 (LGIVHFFDN; Table 2). Splenocytes from all infected animals had a significant response to at least one of the pools of peptides (Fig. 4B, representative animal shown). Peptide 84 elicited responses in 3 of 4 naturally infected animals (Table 2), suggesting this might be an immunodominant peptide following GP83 vaccination as well as natural infection.
Fig. 3.
Mapping of T cell epitopes of GP83 using peptide pools. Splenocytes were stimulated with one of 24 different pools of peptides that contained 11–12 9-mers that span the GP83 protein. IFN-γ producing cells were counted in triplicate for each animal for each pool using ELISPOT. Peptides were considered to stimulate T cells if the response differed significantly from unstimulated cells (DMSO control). (A) Six animals vaccinated with MVA-GP83 responded to a single peptide contained in both pool 12 and pool 19. Representative data from one animal is shown. (B) The remaining 3 animals vaccinated with the MVA-GP83 construct displayed stimulation of T cells by multiple peptides (representative from one animal data shown). (C) No response was observed to any pool in any animals vaccinated with MVA-gB vaccine. Among the MVA-GP83 vaccinated animals, 3/12 were non-responders by this assay. A representative example from one animal is shown. *p < 0.05, **p < 0.005, ***p < 0.001.
Table 2.
Summary of immunoreactive GP83 peptides identified as epitopes using IFN-γ ELISPOT.
| Peptide # | Amino acid sequence | Location | GP83 | WT |
|---|---|---|---|---|
| 46 | ACMTHVDSL | 181–189 | 2/12 | 0/4 |
| 60 | LASHAQVVM | 237–245 | 2/12 | 1/4 |
| 84 | LGIVHFFDN | 333–341 | 9/12 | 3/4 |
| 106 | GDAKDDGSE | 421–429 | 2/12 | 2/4 |
Fig. 4.
Characterization of the anti-GP83 T cell response generated by GPCMV infection. Splenocytes from uninfected and GPCMV infected animals were stimulated with one of 24 different pools of peptides that contained 11–12 9-mers that spanned the GP83 protein. IFN-γ producing cells were counted in triplicate for each animal for each pool using ELISPOT. Peptides were considered to stimulate T cells if the response differed significantly from unstimulated cells (DMSO control). (A) No response was observed to any pool in uninfected animals (representative data from one animal is shown). (B) Significant responses compared to unstimulated cells were engendered by multiple peptide pools followings stimulation of splenocytes isolated from GPCMV infected animals (representative data from one animal is shown; *p < 0.05).
4. Discussion
The aim of this study was to develop an ELISPOT assay to facilitate GPCMV vaccine studies, using murine monoclonal antibodies specific for guinea pig IFN-γ [28]. Using these same antibodies, one previous report measured systemic expression of IFN-γ by ELISA in a guinea pig model of Mycobacterium tuberculosis infection [35]. Another study demonstrated differential expression of IFN-γ mRNA induced by attenuated and virulent M. tuberculosis in guinea pig cells in a BCG vaccination model [30]. This is to our knowledge the first IFN-γ assay measuring functional responses of individual cells in a guinea pig model. Since guinea pigs serve as a model for multiple clinically relevant diseases, such as M. tuberculosis [3,4], foot and mouth disease [36,37], Ebola, and hemorrhagic arenaviruses [7,38], the IFN-γ ELISPOT assay described in our study could be useful in the experimental characterization of the cellular immune response in these settings.
Both MVA-vectored vaccines (gB and GP83) elicited an immune response in these studies. Since the chief protective effect of gB vaccine is believed to be related to the antibody (and not the CD8+) response to gB (reviewed in [39]), IFN-γ ELISPOTS with gB-specific peptides were not performed in this study. However, since gB has been shown to elicit cytotoxic T cell responses in HCMV [40,41], additional work to elucidate the gB peptide response in guinea pigs is warranted. These studies focused on GP83, the GPCMV homolog of the immunodominant HCMV pp65 protein [15,42]. The majority (75%) of MVA-GP83 immunized guinea pigs demonstrated ELISPOT responses to various combinations of GP83 peptides. Moreover, the stimulation of T cells by MVA-GP83 appeared to be similar in strength and diversity to the anti-GP83 T cell response generated by GPCMV infection. While previous research has shown that GP83 vaccines can elicit a cell-mediated immune response [16], this study is the first to quantitate antigen specific effector T cell function on a per cell basis. The induction of guinea pig IFN-γ likely has intrinsic anti-GPCMV properties. The antiviral activity of recombinant guinea pig IFN-γ (expressed in Escherichia coli) was demonstrated with a guinea pig fibroblast cell line (104C1) infected with encephalomyocarditis virus [43]. Guinea pig IFN-γ was also shown to induce upregulation of MHC class I expression in guinea pig fibroblasts, compatible with this cytokine mediating a protective role against infectious agents [29]. IFN-γ demonstrates substantial antiviral activity in a number of CMV systems [44,45]. The number of IFN-γ secreting T cells is a positive predictor of HCMV disease control following solid tissue and hematopoietic transplantation [18,19,46,47] and in AIDS patients [20]. Moreover, IFN-γ has been shown to be a predictor of immunoprotection following vaccination in MCMV and RhCMV models [23,24]. Thus, the combined results of this study and the protective effects of IFN-γ listed above support the hypothesis that the T cell response elicited by MVA-GP83 vaccination may be protective against congenital CMV infection. Studies are underway to determine if MVA-GP83 vaccine is protective, with or without concomitant MVA-gB vaccine, in a congenital GPCMV challenge model, and to determine the magnitude of the IFN-γ response associated with improved pup outcomes in the setting of maternal viral challenge during pregnancy.
Wide variation in response to individual GP83 peptides was observed in this study, with 75% of animals responding to at least one GP83 peptide and 25% of the animals responding to multiple peptides. Similar variability in the T cell repertoire to individual epitopes has been observed in both humans and rhesus macaques naturally infected with CMV [48–50]. The diversity of guinea pig responses observed is likely due to genetic variability in the animals, since the Hartley strain animals used in this study are outbred animals. One epitope, LGIVHFFDN was recognized by all guinea pigs (9/12) that had a detectable GP83 ELISPOT response. Additionally, three other epitopes of GP83 (ACMTHVDSL, aa 181–189; LASHAQVVM, aa 237–245; and GDAKDDGSE, aa 422–430) were recognized by some animals immunized with the MVA-GP83 construct. Not surprisingly, these epitopes were not conserved with pp65 peptides known to elicit CD8+ responses in the context of HCMV infection [27,49,51–55]. Our analysis is limited by the relatively small number of outbred animals studied in this report. A larger study would be necessary to identify if these results apply more broadly and to determine if other GP83 epitopes are recognized. We chose in this study to use 9-mer peptides, since our major interest was in identifying CD8+ T cell IFγ responses engendered through class I presentation. 9-mer peptides are known to be optimal for this application [56]. Longer peptides may have allowed class II presentation and the induction of CD4+ T cell responses, and this will be evaluated in future studies. Efforts are also underway to identify other GPCMV T cell antigens/epitopes that could be exploited for vaccines against congenital infection. These studies in turn can help inform and direct future vaccine trials of subunit vaccines capable of eliciting responses to T-cell targets against HCMV, in particular those strategies that may be useful in protecting the fetus against congenital viral transmission.
Supplementary Material
Acknowledgements
This work was supported by National Institute of Health Grants AI-063356 and AI-103960 (D.J.D.), HD-044864 and HD-038416 (M.R.S.), and a City of Hope cancer center core services award (P30CA033572). Assistance from K. Yeon Choi (Texas A&M Health Sciences Center, College Station) in protocol development is acknowledged. We thank Jason Zabeli for technical assistance. We are grateful to Hubert Schäfer (Robert Koch-Institute, Berlin, Germany) for his generous gift of guinea pig IFN-γ antibodies, N-G3 and V-E4.
Abbreviations
- HCMV
human cytomegalovirus
- GPCMV
guinea pig cytomegalovirus
- gB
glycoprotein B antigen
- pp65
phosphoprotein 65
- GP83
guinea pig CMV UL83 homolog
- ELISPOT
enzyme-linked immunosorbent spotassay
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.vaccine.2014.05.011.
Footnotes
Contributors: Mark R. Schleiss and Don J. Diamond developed and designed the study concept. Mark R. Schleiss generated recombinant GPCMV plasmids and Felix Wussow generated the MVA vaccines, under the guidance of Donald J. Diamond. Peter Gillis and Nelmary Hernandez-Alvarado performed immunizations and carried out the ELISPOT assays with guidance from Mark R. Schleiss. Peter Gillis, Nelmary Hernandez-Alvarado, Josephine S. Gnanandarajaha, and Felix Wussow acquired the data. All authors were involved in the interpretation of the data. Peter Gillis wrote the first draft of the manuscript and all authors were involved in the further drafting of the manuscript or revising it critically for important intellectual content. All authors approved the manuscript, before it was submitted by the corresponding author. All authors had full access to the data and had final responsibility to submit for publication.
Conflicts of interest: The authors report no conflicts of interest with respect to the work described in this manuscript.
Contributor Information
Peter A. Gillis, Email: gill0221@umn.edu.
Nelmary Hernandez-Alvarado, Email: hernande@umn.edu.
Josephine S. Gnanandarajah, Email: gnan0007@gmail.com.
Felix Wussow, Email: fwussow@coh.org.
Don J. Diamond, Email: DDiamond@coh.org.
Mark R. Schleiss, Email: schleiss@umn.edu.
References
- 1.Hickey AJ. Guinea pig model of infectious disease - viral infections. Curr Drug Targets. 2011;12:1018–1023. doi: 10.2174/138945011795677827. [DOI] [PubMed] [Google Scholar]
- 2.Padilla-Carlin DJ, McMurray DN, Hickey AJ. The guinea pig as a model of infectious diseases. Comp Med. 2008;58:324–340. [PMC free article] [PubMed] [Google Scholar]
- 3.Kaufmann SHE, Gengenbacher M. Recombinant live vaccine candidates against tuberculosis. Curr Opin Biotechnol. 2012;23:900–907. doi: 10.1016/j.copbio.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Skerry C, Pokkali S, Pinn M, Be NA, Harper J, Karakousis PC, et al. Vaccination with recombinant mycobacterium tuberculosis PknD attenuates bacterial dissemination to the brain in guinea pigs. PLOS ONE. 2013:8. doi: 10.1371/journal.pone.0066310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin F, Shen XF, McCoy JR, Mendoza JM, Yan J, Kemmerrer SV, et al. A novel prototype device for electroporation-enhanced DNA vaccine delivery simultaneously to both skin and muscle. Vaccine. 2011;29:6771–6780. doi: 10.1016/j.vaccine.2010.12.057. [DOI] [PubMed] [Google Scholar]
- 6.Lan SY, Schelde LM, Wang JL, Kumar N, Ly H, Liang YY. Development of infectious clones for virulent and avirulent pichinde viruses: a model virus to study arenavirus-induced hemorrhagic fevers. J Virol. 2009;83:6357–6362. doi: 10.1128/JVI.00019-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLay L, Lan S, Ansari A, Liang Y, Ly H. Identification of virulence determinants within the L genomic segment of the pichinde arenavirus. J Virol. 2013;87:6635–6643. doi: 10.1128/JVI.00044-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashimoto K, Yamada S, Katano H, Fukuchi S, Sato Y, Kato M, et al. Effects of immunization of pregnant guinea pigs with guinea pig cytomegalovirus glycoprotein B on viral spread in the placenta. Vaccine. 2013;31:3199–3205. doi: 10.1016/j.vaccine.2013.04.078. [DOI] [PubMed] [Google Scholar]
- 9.Crumpler MM, Choi KY, McVoy MA, Schleiss MR. A live guinea pig cytomegalovirus vaccine deleted of three putative immune evasion genes is highly attenuated but remains immunogenic in a vaccine/challenge model of congenital cytomegalovirus infection. Vaccine. 2009;27:4209–4218. doi: 10.1016/j.vaccine.2009.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi YC, Hsiung GD. Cytomegalovirus-infection in guinea-pigs. 2. Transplacental and horizontal transmission. J Infect Dis. 1978;138:197–202. doi: 10.1093/infdis/138.2.197. [DOI] [PubMed] [Google Scholar]
- 11.Schleiss MR, Bourne N, Jensen NJ, Bravo F, Bernstein DI. Immunogenicity evaluation of DNA vaccines that target guinea pig cytomegalovirus proteins glycoprotein B and UL83. Viral Immunol. 2000;13:155–167. doi: 10.1089/vim.2000.13.155. [DOI] [PubMed] [Google Scholar]
- 12.Olejniczak MJ, Choi KY, McVoy MA, Cui X, Schleiss MR. Intravaginal cytomegalovirus (CMV) challenge elicits maternal viremia results in congenital transmission in a guinea pig model. Virol J. 2011:8. doi: 10.1186/1743-422X-8-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cranage MP, Kouzarides T, Bankier AT, Satchwell S, Weston K, Tomlinson P, et al. Identification of the human cytomegalovirus glycoprotein-B gene and induction of neutralizing antibodies via its expression in recombinant vaccinia virus. EMBO J. 1986;5:3057–3063. doi: 10.1002/j.1460-2075.1986.tb04606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schleiss MR, Bourne N, Stroup G, Bravo FJ, Jensen NJ, Bernstein DI. Protection against congenital cytomegalovirus infection and disease in guinea pigs, conferred by a purified recombinant glycoprotein B vaccine. J Infect Dis. 2004;189:1374–1381. doi: 10.1086/382751. [DOI] [PubMed] [Google Scholar]
- 15.Wills MR, Carmichael AJ, Mynard K, Jin X, Weekes MP, Plachter B, et al. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol. 1996;70:7569–7579. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schleiss MR, Lacayo JC, Belkaid Y, McGregor A, Stroup G, Rayner J, et al. Preconceptual administration of an alphavirus replicon UL83 (pp65 homolog) vaccine induces humoral and cellular immunity and improves pregnancy outcome in the guinea pig model of congenital cytomegalovirus infection. J Infect Dis. 2007;195:789–798. doi: 10.1086/511982. [DOI] [PubMed] [Google Scholar]
- 17.Kallas EG, Reynolds K, Andrews J, Fitzgerald T, Kasper M, Menegus M, et al. Cytomegalovirus-specific IFN gamma and IL-4 are produced by antigen expanded human blood lymphocytes from seropositive volunteers. Immunol Lett. 1998;64:63–69. doi: 10.1016/s0165-2478(98)00080-7. [DOI] [PubMed] [Google Scholar]
- 18.Eid AJ, Brown RA, Hogan WJ, Lahr BD, Eckel-Passow JE, Litzow MR, et al. Kinetics of interferon-gamma producing cytomegalovirus (CMV)-specific CD4+ and CD8+T lymphocytes and the risk of subsequent CMV viremia after allogeneic hematopoietic stem cell transplantation. Transpl Infect Dis. 2009;11:519–528. doi: 10.1111/j.1399-3062.2009.00446.x. [DOI] [PubMed] [Google Scholar]
- 19.Hodson EM, Jones CA, Strippoli GFM, Webster AC, Craig JC. Immunoglobulins, vaccines or interferon for preventing cytomegalovirus disease in solid organ transplant recipients. Cochrane Database Syst Rev. 2007;(2):CD005129. doi: 10.1002/14651858.CD005129.pub2. [DOI] [PubMed] [Google Scholar]
- 20.Sinclair E, Tan QX, Sharp M, Girling V, Poon C, Van Natta M, et al. Protective immunity to cytomegalovirus (CMV) retinitis in AIDS is associated with CMV-specific T cells that express interferon-G and interleukin-2 and have a CD8(+) cell early maturational phenotype. J Infect Dis. 2006;194:1537–1546. doi: 10.1086/508997. [DOI] [PubMed] [Google Scholar]
- 21.Bettahi I, Zhang X, Afifi RE, BenMohamed L. Protective immunity to genital herpes simplex virus type 1 and type 2 provided by self-adjuvanting lipopeptides that drive dendritic cell maturation and elicit a polarized Th1 immune response. Viral Immunol. 2006;19:220–236. doi: 10.1089/vim.2006.19.220. [DOI] [PubMed] [Google Scholar]
- 22.van Rooij EMA, de Bruin MGM, de Visser YE, Middel WGJ, Boersma WJA, Bianchi ATJ. Vaccine-induced T cell-mediated immunity plays a critical role in early protection against pseudorabies virus (suid herpes virus type 1) infection in pigs. Vet Immunol Immunopathol. 2004;99:113–125. doi: 10.1016/j.vetimm.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Yue Y, Kaur A, Eberhardt MK, Kassis N, Zhou SS, Tarantal AF, et al. Immunogenicity and protective efficacy of DNA vaccines expressing rhesus cytomegalovirus glycoprotein B, phosphoprotein 65-2, and viral interleukin-10 in rhesus macaques. J Virol. 2007;81:1095–1109. doi: 10.1128/JVI.01708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morello CS, Kelley LA, Munks MW, Hill AB, Spector DH. DNA immunization using highly conserved murine cytomegalovirus genes encoding homologs of human cytomegalovirus UL54 (DNA polymerase) and UL105 (helicase) elicits strong CD8 T-cell responses and is protective against systemic challenge. J Virol. 2007;81:7766–7775. doi: 10.1128/JVI.00633-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson L, Piccinini G, Lilleri D, Revello MG, Wang ZD, Markel S, et al. Human cytomegalovirus proteins pp65 and immediate early protein 1 are common targets for CD8(+) T cell responses in children with congenital or postnatal human cytomegalovirus infection. J Immunol. 2004;172:2256–2264. doi: 10.4049/jimmunol.172.4.2256. [DOI] [PubMed] [Google Scholar]
- 26.Khan N, Cobbold M, Keenan R, Moss PAH. Comparative analysis of CD8(+) T cell responses against human cytomegalovirus proteins pp65 and immediate early 1 shows similarities in precursor frequency, oligoclonality, and phenotype. J Infect Dis. 2002;185:1025–1034. doi: 10.1086/339963. [DOI] [PubMed] [Google Scholar]
- 27.Schalich J, Vytvytska O, Zauner W, Fischer MB, Buschle M, Aichinger G, et al. Analysis of the human cytomegalovirus pp65-directed T-cell response in healthy HLA-A2-positive individuals. Biol Chem. 2008;389:551–559. doi: 10.1515/bc.2008.065. [DOI] [PubMed] [Google Scholar]
- 28.Schäfer H, Kilem G, Kropp B, Burger R. Monoclonal antibodies to guinea pig interferon-gamma: tools for cytokine detection and neutralization. J Immunol Methods. 2007;328:106–117. doi: 10.1016/j.jim.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 29.Schäfer H, Burger R. Tools for cellular immunology and vaccine research the in the guinea pig: monoclonal antibodies to cell surface antigens and cell lines. Vaccine. 2012;30:5804–5811. doi: 10.1016/j.vaccine.2012.07.012. [DOI] [PubMed] [Google Scholar]
- 30.Jeevan A, Yoshimura T, Lee KE, McMurray DN. Differential expression of gamma interferon mRNA induced by attenuated and virulent Mycobacterium tuberculosis in guinea pig cells after Mycobacterium bovis BCG vaccination. Infect Immun. 2003;71:354–364. doi: 10.1128/IAI.71.1.354-364.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, La Rosa C, Li Z, Ly H, Krishnan A, Martinez J, et al. Vaccine properties of a novel marker gene-free recombinant modified vaccinia Ankara expressing immunodominant CMV antigens pp65 and IE1. Vaccine. 2007;25:1132–1141. doi: 10.1016/j.vaccine.2006.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang ZD, La Rosa C, Mekhoubad S, Lacey SF, Villacres MC, Markel S, et al. Attenuated poxviruses generate clinically relevant frequencies of CMV-specific T cells. Blood. 2004;104:847–856. doi: 10.1182/blood-2003-10-3469. [DOI] [PubMed] [Google Scholar]
- 33.Wang ZD, La Rosa C, Maas R, Ly H, Brewer J, Mekhoubad S, et al. Recombinant modified vaccinia virus Ankara expressing a soluble form of glycoprotein B causes durable immunity and neutralizing antibodies against multiple strains of human cytomegalovirus. J Virol. 2004;78:3965–3976. doi: 10.1128/JVI.78.8.3965-3976.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schleiss MR. Cloning and characterization of the guinea-pig cytomegalovirus glycoprotein-B gene. Virology. 1994;202:173–185. doi: 10.1006/viro.1994.1333. [DOI] [PubMed] [Google Scholar]
- 35.Tree JA, Patel J, Thom RE, Elmore MJ, Schaefer H, Williams A, et al. Temporal changes in the gene signatures of BCG-vaccinated guinea pigs in response to different mycobacterial antigens. Vaccine. 2010;28:7979–7986. doi: 10.1016/j.vaccine.2010.09.061. [DOI] [PubMed] [Google Scholar]
- 36.Chen HT, Liu YS. Immunity of foot-and-mouth disease serotype asia 1 by sub-lingual vaccination. PLOS ONE. 2013:8. doi: 10.1371/journal.pone.0063839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohana Subramanian B, Madhanmohan M, Sriraman R, Chandrasekhar Reddy RV, Yuvaraj S, Manikumar K, et al. Development of foot-and-mouth disease virus (FMDV) serotype O virus-like-particles (VLPs) vaccine and evaluation of its potency. Antiviral Res. 2012;96:288–295. doi: 10.1016/j.antiviral.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 38.Paessler S, Walker DH. Pathogenesis of the viral hemorrhagic fevers. Annu Rev Pathol. 2013;8:411–440. doi: 10.1146/annurev-pathol-020712-164041. [DOI] [PubMed] [Google Scholar]
- 39.Schleiss MR. Comparison of vaccine strategies against congenital CMV infection in the guinea pig model. J Clin Virol. 2008;41:224–230. doi: 10.1016/j.jcv.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 40.Hopkins JI, Fiander AN, Evans AS, Delchambre M, Gheysen D, Borysiewicz LK, Cytotoxic T. cell immunity to human cytomegalovirus glycoprotein B. J Med Virol. 1996;49:124–131. doi: 10.1002/(SICI)1096-9071(199606)49:2<124::AID-JMV9>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 41.Borysiewicz LK, Hickling JK, Graham S, Sinclair J, Cranage MP, Smith GL, et al. Human cytomegalovirus-specific cytotoxic T cells relative frequency of stage-specific CTL recognizing the 72-kD immediate early protein and glycoprotein B expressed by recombinant vaccinia viruses. J Exp Med. 1988;168:919–931. doi: 10.1084/jem.168.3.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walter EA, Greenberg PD, Gilbert MJ, Finch RJ, Watanabe KS, Thomas ED, et al. Reconstitution of cellular immunity against cytomegalovirus in recipients of allogeneic bone marrow by transfer of T-cell clones from the donor. N Engl J Med. 1995;333:1038–1044. doi: 10.1056/NEJM199510193331603. [DOI] [PubMed] [Google Scholar]
- 43.Jeevan A, McFarland CT, Yoshimura T, Skwor T, Cho H, Lasco T, et al. Production and characterization of guinea pig recombinant gamma interferon and its effect on macrophage activation. Infect Immun. 2006;74:213–224. doi: 10.1128/IAI.74.1.213-224.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davignon JL, Castanie P, Yorke JA, Gautier N, Clement D, Davrinche C. Anti-human cytomegalovirus activity of cytokines produced by CD4+ T-cell clones specifically activated by IE1 peptides in vitro. J Virol. 1996;70:2162–2169. doi: 10.1128/jvi.70.4.2162-2169.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vescovini R, Biasini C, Fagnoni FF, Telera AR, Zanlari L, Pedrazzoni M, et al. Massive load of functional effector CD4+ and CD8+ T cells against cytomegalovirus in very old subjects. J Immunol. 2007;179:4283–4291. doi: 10.4049/jimmunol.179.6.4283. [DOI] [PubMed] [Google Scholar]
- 46.Zhou W, Longmate J, Lacey SF, Palmer JM, Gallez-Hawkins G, Thao L, et al. Impact of donor CMV status on viral infection and reconstitution of multifunction CMV-specific T cells in CMV-positive transplant recipients. Blood. 2009;113:6465–6476. doi: 10.1182/blood-2009-02-203307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cantisan S, Lara R, Montejo M, Redel J, Rodriguez-Benot A, Gutierrez-Aroca J, et al. Pretransplant interferon- secretion by CMV-specific CD8+T cells informs the risk of CMV replication after transplantation. Am J Transplant. 2013;13:738–745. doi: 10.1111/ajt.12049. [DOI] [PubMed] [Google Scholar]
- 48.Yue Y, Kaur A, Zhou SS, Barry PA. Characterization and immunological analysis of the rhesus cytomegalovirus homologue (Rh112) of the human cytomegalovirus UL83 lower matrix phosphoprotein (pp65) J Gen Virol. 2006;87:777–787. doi: 10.1099/vir.0.81516-0. [DOI] [PubMed] [Google Scholar]
- 49.Elkington R, Walker S, Crough T, Menzies M, Tellam J, Bharadwaj M, et al. Ex vivo profiling of CD8(+)-T-cell responses to human cytomegalovirus reveals broad and multispecific reactivities in healthy virus carriers. J Virol. 2003;77:5226–5240. doi: 10.1128/JVI.77.9.5226-5240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, et al. Broadly targeted human cytomegalovirus-specific CD4(+) and CD8(+) T cells dominate the memory compartments of exposed subjects. J Exp Med. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akiyama Y, Maruyama K, Mochizuki T, Sasaki K, Takaue Y, Yamaguchi K. Identification of HLA-A24-restricted CTL epitope encoded by the matrix protein pp65 of human cytomegalovirus. Immunol Lett. 2002;83:21–30. doi: 10.1016/s0165-2478(02)00073-1. [DOI] [PubMed] [Google Scholar]
- 52.Ohnishi M, Sakurai T, Heike Y, Yamazaki R, Kanda Y, Takaue Y, et al. Evaluation of cytomegalovirus-specific T-cell reconstitution in patients after various allogeneic haematopoietic stem cell transplantation using interferon-gamma-enzyme-linked immunospot and human leucocyte antigen tetramer assays with an immunodominant T-cell epitope. Br J Haematol. 2005;131:472–479. doi: 10.1111/j.1365-2141.2005.05800.x. [DOI] [PubMed] [Google Scholar]
- 53.Zaia JA, Gallez-Hawkins G, Li XL, Yao ZQ, Lomeli N, Molinder K, et al. Infrequent occurrence of natural mutations in the pp65(495–503) epitope sequence presented by the HLA A*0201 allele among human cytomegalovirus isolates. J Virol. 2001;75:2472–2474. doi: 10.1128/JVI.75.5.2472-2474.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Provenzano M, Mocellin S, Bettinotti M, Preuss J, Monsurro V, Marincola FM, et al. Identification of immune dominant cytomegalovirus epitopes using quantitative real-time polymerase chain reactions to measure interferon-gamma production by peptide-stimulated peripheral blood mononuclear cells. J Immunother. 2002;25:342–351. doi: 10.1097/00002371-200207000-00006. [DOI] [PubMed] [Google Scholar]
- 55.Britten CM, Meyer RG, Graf C, Huber C, Wolfel T. Identification of T cell epi-topes by the use of rapidly generated mRNA fragments. J Immunol Methods. 2005;299:165–175. doi: 10.1016/j.jim.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 56.Schumacher TN, De Bruijn ML, Vernie LN, Kast WM, Melief CJ, Neefjes JJ, et al. Peptide selection by MHC class I molecules. Nature. 1991;350:703–706. doi: 10.1038/350703a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.