Abstract
Proteins that bind both DNA and RNA epitomize the ability to perform multiple functions by a single gene product. Such DNA- and RNA-binding proteins (DRBPs) regulate many cellular processes, including transcription, translation, gene silencing, microRNA biogenesis and telomere maintenance. Proteins that bind RNA were typically considered as functionally distinct from proteins that bind DNA and studied independently. This practice is becoming outdated, in part due to the discovery of long non-coding RNAs (lncRNAs) that target DNA-binding proteins. DRBPs have unique functional characteristics that stem from their specific structural attributes; these have evolved early in evolution and are widely conserved.
Proteins that bind DNA or RNA are often considered and studied independently. Transcription factors for example are usually modeled relatively simply: they bind to genomic promoters and control target gene expression by activating or repressing RNA polymerases. Following transcription, RNA binding proteins modulate protein expression by regulating the stability and translation of mRNAs. However, the consideration of DNA- and RNA-binding functions within proteins as separate entities is becoming outdated. The unappreciated, dual DNA- and RNA-binding capacity of a growing body of proteins plays a key role in modulating gene expression, cell survival and homeostasis. Recent studies have demonstrated that many transcription factors are capable of binding diverse types of RNA, which enables them to bind to the mRNA products of transcription to regulate their turnover and to integrate other signals, such as responses to stress1–7. Additionally, the prevalence and emerging functions of long non-coding RNAs (lncRNAs) have revealed that these RNAs target many types of proteins through direct interactions1,8–11.
In this analysis, we attempt to enumerate these DNA and RNA binding proteins (DRBPs) and describe their function, structure, and evolution. We will first broadly discuss the prevalence of DRBPs within the human genome. We highlight known functions of DRBPs with specific examples of how the simultaneous and serial RNA and DNA interaction allows for better gene targeting, finer control of gene expression, and integration of metabolic state or stress to modulate protein activity. We discuss the structural features of DRBPs that enable dual nucleic acid specificity, focusing on the limited number of solved structures that allow for direct comparison of a DRBP complexed to either DNA or RNA. Finally, we discuss the evolution of dual DNA and RNA binding domains within DRBPs, including both ancient domains, for which dual DNA and RNA binding conferred a selective advantage, and more modern domains which have recently been targeted by rapidly evolving lincRNAs.
Defining DNA and RNA binding proteins
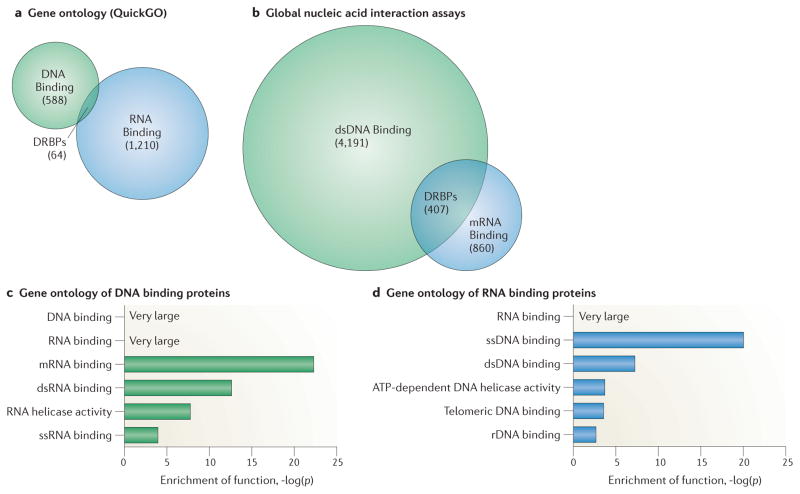
Defining the subset of human proteins that bind both DNA and RNA is a difficult task. Using gene ontology searches, only 64 human protein-coding genes in the QuickGO gene ontology database (European Bioinformatics Institute; ref. 12) are identified as having direct and specific experimental evidence for both RNA binding (GO:0003723) and DNA binding (GO:0003677) (Fig. 1a). The PROTEOME database (BioBase) returns 122 such proteins, although direct evidence is lacking for many of them.
Figure 1. Defining human DRBPs.
a. Venn diagram of DNA binding and of RNA binding proteins in the QuickGO database supported by low-throughput experimental evidence (as of July 2014)12. The overlap of these two sets represents human DNA and RNA interacting proteins (DRBPs), consisting of 64 proteins. b. Venn diagram of DNA binding and of RNA binding proteins identified in high-throughput studies defining the human mRNA2 and dsDNA3 interactomes13,14. There are 407 proteins found in both studies, indicating that they may bind both mRNA and dsDNA. c. Molecular function gene ontology analysis reveals that RNA binding is a potentially major function of the dsDNA binding proteins identified in REF 22. d. Gene ontology analysis reveals that DNA binding is potentially a major function of the mRNA binding proteins identified in REF 3. In parts a and b, circles are drawn to scale. In parts c and d, only selected molecular function attributes are shown for brevity. p-values in parts c and d indicate the probability that the over-representation of the stated ontology term in the selected 407 genes compared to all human genes is due to chance. These were calculated in the TRANSFAC + PROTEOME database (BIOBASE) using the hypergeometric distribution; “very large” indicates a p-value of less than 1 x 10−40 (−log(p) > 40).
An alternative approach involves combining evidence from studies that have separately attempted to catalog all human proteins that bind DNA or RNA (Fig. 1b). A study using protein microarrays and bioinformatics approaches identified over 4,000 human proteins that directly interact with double-stranded DNA (dsDNA) in vitro13. Gene ontology analysis of these proteins reveals that the term RNA binding is highly enriched for (p < 1x10−40), indicating that RNA binding may be a common feature of DNA binding proteins (Fig. 1c). Among these dsDNA-binding proteins, the ontology term ‘dsRNA binding’ is much more represented than ssRNA binding.
Another study used a crosslinking and mass spectrometry-based approach to identify 860 mRNA-binding proteins from HeLa cells termed the “mRNA interactome”14. Functional analysis of these proteins again indicates that dual nucleic acid binding is a widespread phenomenon (Fig. 1d), with both the terms ssDNA- and dsDNA-binding being significantly enriched for (p = 8.9 x 10−21 and 5.5 x 10−8, respectively). Notably, of the 860 proteins identified as mRNA binding, 407 (47.3%) were independently characterized as dsDNA binding in REF 2. Together, the two studies indicate that DRBPs are a widespread phenomenon, perhaps comprising 2% of the human proteome (407 proteins, Fig. 1b). This number would likely increase if proteins that are expressed in other cell types that require ligand binding-dependent signals for nucleic acid binding, or that bind other types of DNA or RNA, are included.
We note that many of the proteins identified in REFS 2 and 3 as DNA and/or RNA binding lack corroborating evidence from other studies, and thus these findings should be interpreted with caution. For example, many identified proteins such as polymerase subunits may bind nucleic acid-bound proteins without binding DNA and/or RNA directly. Additionally, many proteins that bind DNA or RNA in vitro may not bind them in vivo. However, we believe the two studies provide a reasonable estimate of potential human DRBPs due to their wide coverage of the human proteome, and we will discuss examples where the demonstration of protein-nucleic acid binding in vitro has preceded the discovery of such binding in vivo, sometimes by decades.
In Supplementary Table 1 we provide a detailed list of 149 human DRBPs, with comments on their nucleic acid binding properties, structure, and function. These proteins, drawn form the 271 proteins mentioned above, were selected based on experimental evidence demonstrating their ability to bind directly to both DNA and RNA, generally obtained from studies using more traditional experimental approaches than the high-throughput studies referenced above. While many of the proteins in Supplementary Table 1 have only been shown to bind DNA and/or RNA in vitro, the remainder of this review will focus on selected human DRBPs with known cellular roles.
Functions of DRBPs
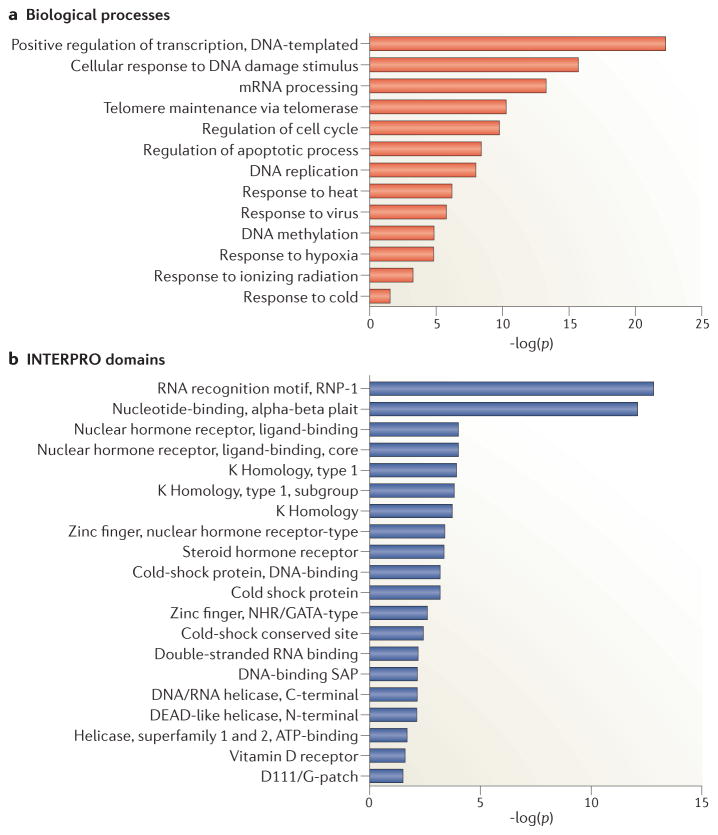
We performed gene ontology and domain enrichment analyses (Fig. 2) to illuminate the main biological functions of our list of human DRBPs (Supplementary Table 1). The gene ontology analysis revealed expected biological processes such as transcriptional regulation, mRNA processing, and DNA replication. However, several surprising functions are also implicated, including the DNA damage response, apoptosis, and responses to extreme temperatures (Fig. 2a).
Figure 2. Functional and structural properties of DRBPs.
The 149 DRBPs (Supplementary Table 1) were subjected to gene ontology enrichment of biological process (PROTEOME database, Biobase) and to INTERPRO domain enrichment (DAVID ontology77,78), to explore the biological functions of and protein domains commonly found in DRBPs. a. Gene ontology analysis. Biological processes such as transcriptional regulation and mRNA processing are expectedly prominent terms found enriched for DRBPs. However, unexpected functions are also enriched, including response to many cellular stresses (heat, viral, radiation, etc.). For brevity, only selected functions are shown. b. Domain enrichment analysis. All domains enriched in the set of 149 DRBPs that have p-values equal or smaller than p = 10−3 are shown. p-values in parts a and b indicate the probability that the over-representation of the stated term in the 149 DRBPs compared to all human genes is due to chance.
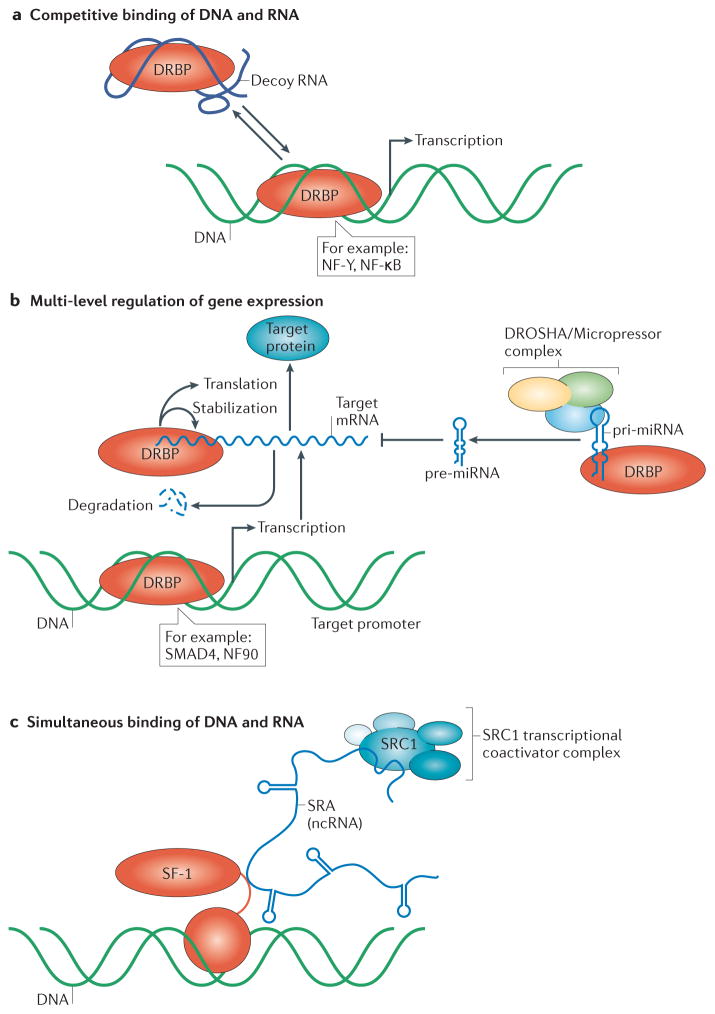
Ultimately, DRBP function is governed by their inherent structural and biochemical properties. One can envision DRBPs capitalizing on both RNA and DNA binding in a number of ways; for example, a transcription factor that binds DNA and RNA may interact orthogonally with RNAs that compete with DNA binding to repress transcription, or simultaneously with a promoter and an RNA coactivator to upregulate transcription. The following section will focus on DRBPs that bind DNA and RNA competitively (Fig. 3a).
Figure 3. Three archetypes of DRBP function.
a, RNA can compete with DNA for binding to DRBPs, typically at the same protein interface. In the case of transcription factors, this can reduce promoter occupancy and the transcription of target genes. b, DRBPs can regulate gene expression at multiple levels. In addition to binding to the promoters of genes to regulate their transcription, DRBPs can also affect miRNA processing and mRNA stability and translation. c, DRBPs, such as SF-1, can bind DNA and RNA simultaneously, whereby the RNA functions as scaffold to recruit other proteins to a specific DNA locus. Shown here is the DRBP, SF-1, binding to the lncRNA SRA to recruit the steroid receptor coactivator 1 (SRC-1) transcriptional complex in a ligand independent manner.
Binding DNA or decoy RNAs
The role of certain lncRNAs as decoys of genomic DNA is illustrated by the reduction in promoter occupancy by transcription factors, typically measured by chromatin immunoprecipitation (ChIP), in response to the overexpression of competing decoy RNAs. The glucocorticoid receptor (GCR), a steroid hormone receptor, is a classic example of a ligand-activated transcription factor (reviewed in 15). In its inactivated state, GCR is kept in the cytoplasm by chaperone proteins. Upon ligand binding, GCR translocates to the nucleus where it can bind to the promoters and regulate the transcription of hundreds of genes16. Given the anti-inflammatory role of the GCR, much effort has been put into developing modulators of GCR-driven transcription17. The lncRNA Growth Arrest Specific 5 (Gas5) was found to inhibit the transcriptional activity of GCR by competing directly with DNA for protein binding in vitro and in cells1; overexpression of Gas5 leads to a decrease in ChIP-detected GCR occupancy at its target promoters as well as decrease in the mRNA levels of glucocorticoid-activated genes1,18. Since cellular Gas5 levels are regulated by nonsense mediated decay18 in response to serum starvation and other stressors1, the transcriptional activity of the GCR is tuned by titrating the levels of Gas5 against the fixed number of genomic GCR binding sites in response to cellular stress. Three closely related steroid receptors that share the DNA specificity of the GCR, the androgen, progesterone, and mineralocorticoid receptors, are also susceptible to Gas5-mediated transcriptional repression1. Although steroid receptors have traditionally been thought of as DNA binding proteins, the affinity of the GCR for RNA and DNA is similar, as measured in vitro by GST pulldowns and fluorescence-based competition assays1. The most distantly related member of the steroid receptor family, the estrogen receptor, does not share the DNA specificity of the GCR and is not susceptible to Gas5-mediated transcriptional repression, indicating that the binding of steroid receptors to RNA is sequence specific1.
Additional examples of pairs of transcription factors and decoy RNAs are Nuclear Factor-Y, which binds also the lncRNA PANDA8, and NF-κB, which binds also a mouse pseudogene-derived RNA, Lethe9. The dual nucleic acid binding activity of NF-κB had been demonstrated in vitro many years prior to the discovery of an endogenous RNA target10, suggesting that transcription factors that are DRBPs such as AML1/RUNX1, whose RNA binding capacity has only been shown in vitro19, may have also endogenous RNA targets awaiting discovery. Although structural information on the interaction of human proteins with their RNA decoys is lacking, a recent study demonstrates an elaborate mechanism of an analogous bacterial system, the sequestration of RsmE by the ncRNA RsmZ11. Competitive DNA and RNA binding is not only a feature of transcription factors, but also of nucleic acid-modifying enzymes such as DNA methyltransferases. In humans, DNA methylation is initiated by DNMT3A and 3B; DNMT1 maintains this methylation by binding to hemimethylated DNA after replication (reviewed in 20). RNAs binding can inhibit the DNA binding and methylation activity of both DNMT3A21 and DNMT122. In vitro, DNMT1 binds RNA with a higher affinity than DNA as shown in electrophoretic mobility experiments21. In the cases of DNMT1 and likely DNMT3A, RNAs bind to the catalytic domain of the methyltransferase to inhibit DNA methylation21,22.
It is notable that several metabolic enzymes are DRBPs with competitive DNA and RNA binding capacity, such as the glycolytic enzymes lactate dehydrogenase (LDH)23–25, glyceraldehyde-3-phosphate dehydrogenase (GAPDH)26–28, and α-enolase (ENO1)14,29,30. In the case of GAPDH, both DNA and RNA compete for binding of the cofactor NAD+ to the enzyme26,28, suggesting that Rossmann fold-containing proteins such as GAPDH may be sensitive to cellular DNA and/or RNA levels. ENO1 binds RNA as a monomer30, which inhibits the formation of the catalytically active protein dimer31,32. NAD+ specific isocitrate dehydrogenase, which converts isocitrate to alpha-ketoglutarate, is allosterically inhibited by the 5′untranslated regions of yeast mitochondrial mRNA33. Binding of RNA and DNA to metabolic enzymes indicates that nucleic acids can modulate the function of proteins other than transcription factors to modulate cellular metabolism34.
DRBPs that regulate gene expression at multiple levels
Approximately half of the DRBPs we identified in our analysis are transcription factors. As discussed above, some such proteins have been shown to be the targets of RNA decoys. In contrast, several others bind both the DNA and the mRNA of their target genes (Fig. 3b). Regulating genes at both the DNA and RNA levels enables powerful, combinatorial control over protein expression, and may allow DRBPs to generate both immediate effects (through regulating RNA turnover) as well as long-lasting effects (through regulating transcription).
For the first example of such a DRBP, we return to GCR, which when activated can promote the transcription of anti-inflammatory genes35 as well as repress the transcription of pro-inflammatory genes36,37. Agonist-bound GCR destabilizes the mRNA of pro-inflammatory genes such as MCP-1 through direct RNA binding, perhaps by the recruitment of ribonucleases38. The identification, using RNA immunoprecipitation (RNA-IP), of a GCR binding motif in many immunogenic mRNAs suggests that GCR can accelerate the decay of a large number of mRNAs, broadening its role in the anti-inflammatory reponse2. Given that GCR also binds directly to pro-inflammatory transcription factors such as AP-1 and NF-κB39,40, it would appear that GCR takes advantage of its diverse DNA, RNA, and protein binding capacities to regulate inflammatory genes at the transcriptional and post-transcriptional level.
Transcription factors can also regulate gene expression post-transcriptionally through the regulation of microRNA (miRNA) biogenesis. miRNAs are small RNAs that facilitate gene silencing through sequence-specific pairing to and recruitment of the target mRNAs to the RNA induced silencing complex (RISC; reviewed in ref 41). Several transcription factors have been shown to regulate Drosha-mediated primary (pri-)miRNA processing, a key step in the biogenesis of functional miRNAs42. Smad proteins, the transducers of TGF-β signaling, activate transcription by forming a DNA-binding heterodimer (reviewed in 43). Smad proteins also increase the levels of several miRNAs, including miR-21 (ref. 44), which plays important roles and development and immunity45. Surprisingly, the increase in miR-21 levels is due not to increased transcription of pri-miR-21 transcripts, but due to increased Drosha-mediated processing of pri-miR-21 to precursor (pre-)miR-21 (ref. 44). Bioinformatics analysis identified a conserved RNA motif in TGF-β-regulated miRNAs that was shown by RNA-IP and electrophoretic mobility shift assays (EMSA) to bind directly to the Smad MH1 domain to mediate Drosha processing3. Interestingly, the RNA sequence motif that is bound by SMAD4 and mediates the regulation of miRNA expression post-transcriptionally, is identical to the DNA sequences that are bound by SMAD4 and mediate regulation of gene expression transcriptionally3.
NF90 (ILF3) is a particularly versatile DRBP that, along with its partner NF45, plays important roles in T cell activation46. Through the direct binding of DNA, mRNAs and miRNAs, NF90 controls transcription4,5, regulates mRNA turnover and translation47,48, and affects miRNA processing 49, respectively. All of these functions assist in its role in T cell activation: NF90 upregulates the mRNA levels of IL-2, a critical cytokine in T cell development50, by both binding its promoter and activating its transcription and by stabilizing the IL-2 mRNA through direct binding to its 3′ UTR, as was found by using EMSA and ribonucleoprotein immunoprecipitation analysis5,48. Additionally, using in vitro pri-miRNA processing assays and RNA-IP, NF90, when in complex with NF45, was shown to inhibit the processing of the pri-miRNA pri-let-7a binding it directly49. let-7a represses IL-6, a cytokine critical for T cell survival and proliferation51, which may link inflammation to cancer52, and let-7 downregulation following NF90 upregulation reduces survival in several cancer types53,54. In summary, these examples illustrate that DRBPs can utilize both transcriptional and post-transcriptional mechanisms to serve as potent controllers of gene expression.
Simultaneous binding of DNA and RNA
In contrast to DRBPs that target DNA or RNA serially to perform different or related functions, another class of DRBPs binds RNA and DNA simultaneously to perform a single function (Fig. 3c). Generally, transcription factors not only require DNA binding to target promoters but also bind to corepressors or coactivators to affect transcriptional regulation. There are several examples of RNA molecules acting as coactivators, by simultaneously binding DNA and various transcription factors. The lncRNA RMST (rhabdomyosarcoma 2-associated transcript), in particular, is required for binding of neurogenic gene promoters and subsequent upregulation by SOX255, a transcription factor with important roles in development, pluripotency, and cell fate56. RNA-IP and RNA pulldown experiments demonstrated that RMST interacts directly with SOX255,57, and DNA occupancy of SOX2 measured by ChIP-seq was reduced following RMST depletion55,57. The lncRNA Evf-2 serves as a transcriptional coactivator for Dlx-258 and recruits MECP2 to intergenic enhancers59. A direct interaction between Dlx-2 and Evf-2 has been demonstrated by the immunoprecipitation of Dlx-2 followed by RT-PCR of the Evf-2 lncRNA58, and MECP2 also has previously been shown to bind RNA60. It should be noted that RNA-mediated recruitment of a protein to a particular DNA locus might not require direct binding of both DNA and RNA by the protein, as lncRNAs could recruit transcription factors to a particular DNA locus to which the lncRNA is bound. Dual nucleic acid recognition also facilitates targeted gene repression, through RNA-guided DNA methylation. This phenomenon was first discovered in plants61, and some mammalian RNA guides of DNA methylation have since been found62,63, although their mechanisms of action are less clear. In mice, DNMT3A forms a complex with Tsix RNA to promote methylation of the Xist promoter64.
Several nuclear receptors, including SF-1, DAX-1, and the thyroid receptor α (TRα), bind directly to both gene promoters65,66 as well as to the RNA co-activator SRA in order to modulate the transcriptional activation6,7 (Fig. 3c). Using pulldown experiments, SF-1 and TRα have been shown to bind SRA through their hinges, which are flexible, disordered regions that connect their DNA and ligand binding domains6,67. Knockdown of SRA decreases the interaction of SF-1 with protein transcriptional activators as well as the transcription of SF-1-regulated genes6. Several other nuclear receptors associate with, but lack direct evidence for direct binding to, SRA, including the androgen, progesterone, and estrogen receptors as well as the retinoic acid receptor α (RARα), which may bind SRA and its target gene promoters simultaneously7,68–70. Crosslinking immunoprecipitation has demonstrated that RARα can bind to and regulate the translation of target mRNAs71 through a unique RNA binding motif at its C-terminus71.
Another example of simultaneous DNA and RNA binding that is required for DRBP function is the role of TRF2 (Telomeric Repeat binding Factor-2) at telomeres. Deletion of TRF2 leads to an arrest in cell division caused by the formation of chromosome end-fusions72. Crystal structures have revealed that TRF2 binds to telomeric DNA in a sequence-specific manner through a C-terminal DNA binding domain that resembles a homeodomain73. Part of the role of TRF2 at the telomere includes the recruitment, through its positively charged N-terminal GAR domain, of the origin recognition complex (ORC; a collection of proteins that serves as a scaffold for DNA replication factors, among other functions74), where it can assist in the maintenance of telomere structure75. Using biotinylated RNA pulldown experiments, RNA-IP, and EMSAs, the GAR domain responsible for ORC recruitment was later shown to bind telomere repeat-encoding RNA (TERRA)76. Depletion of TERRA hampers ORC recruitment to the telomere without affecting TRF2 binding to the telomere itself, suggesting a model in which TRF2 serves as a mediator between telomere DNA and TERRA, which in turn recruits factors required for telomere maintenance76.
Structural characteristics of DRBPs
For a protein such as TRF2 to coordinate telomeric DNA binding and recruit protein complexes by binding of RNA, it must have multiple nucleic acid binding motifs. Some DRBPs such as GCR and NF-κB have maintained domains capable of binding both DNA and RNA, allowing decoy RNAs to evolve and compete with DNA for protein binding. In this section, we will analyze the prevalence of structural domains in DRBPs and discuss examples of DRBPs that bind both single-stranded and double-stranded nucleic acids.
DRBP domains that enable DNA and RNA interactions
We performed INTERPRO domain enrichment analysis by DAVID77,78 on our 149 DRBPs in order to identify domains enriched in proteins that bind both DNA and RNA (Fig. 2b). The RNA recognition motif (RRM; also known as ribonucleoprotein domain {RNP} or RNA binding domain {RBD}) was the most highly enriched domain in DRBPs (p = 2 x 10−26). The RRM is an abundant, short (~100 amino acids-long) domain that generally recognizes single-stranded RNA (ssRNA) and is often present in proteins with other domains, such as zinc finger domains, WW domains, or additional RRMs79. Such multi-domain DRBPs may bind RNA and DNA simultaneously through separate domains, such as the two RRM-containing hnRNP A1 (ref. 80). Single RRM-containing proteins are also capable of binding both DNA and RNA, a function present for example in RBM3, TAF15, and TDP-43 (refs. 81–83) (Supplementary Table 1). Such bivalent domains may not have the same sequence specificity when binding DNA and RNA, highlighting the complexity of recognizing nucleotide bases in a sequence-dependent manner.
Nuclear receptor domains are also enriched in DRBPs: RARα binds mRNA through a unique C-terminal domain71, SF-1 binds the RNA coactivator SRA through its hinge and a unique Ftz-F1 domain, and TRα binds SRA through its hinge6,67 (Fig. 3c). The majority of nuclear receptors have two highly-conserved Cys4 zinc fingers through which they bind DNA, and some nuclear receptors, such as the GCR, can also bind RNA through these domains1. Other types of zinc fingers are also enriched in DRBPs, such as the RanBP2-type. Other notably enriched domains in DRBPs are the K homology (KH) domain, the double-stranded RNA binding domain (dsRBD), the cold shock domain (CSD), and various helicase domains. Each of these domains is capable of binding DNA and RNA, and here we will focus on the structural mechanisms underlying this dual specificity.
General properties of DRBPs
There are only two chemical differences between RNA and DNA. First, RNA, but not DNA, has a 2′OH group on the ribose sugar, which allows for an additional hydrogen bond and a greater diversity of secondary structure than is possible in DNA. Second, RNA contains uracil rather than thymine in DNA, which differs by the presence of a methyl group at the C5 position. A comparative analysis of known protein-nucleic acid structures revealed that the recognition of DNA occurs largely through electrostatics and direct base–protein interactions. RNA recognition by proteins, on the other hand, is dependent largely on shape complementarity and interaction with the 2′OH81. Given these general differences, one could expect that during evolution, highly selective protein interfaces would be generated that are optimized for either RNA or DNA, with minimal cross-binding. However, the most energetically favourable associations between proteins and nucleic acids relay on hydrophobic and charge–charge interactions. These interactions are less constrained than interactions with the sugar backbone or with the nucleotide base edge, which is capable of highly-specific Watson–Crick base pairing. Thus, DRBP domains that competitively bind DNA and RNA probably rely on the less specific hydrophobic and charge–charge interactions. For example, ssRNA-binding proteins are more likely to form hydrogen bonds with bases rather than with the phosphate–sugar backbone, compared to those that recognize folded RNA, such as ribosomal proteins and tRNA synthetases82. Because ssRNA-binding proteins do not rely heavily on sugar recognition, they are more likely to also bind DNA. This may explain why the RRM is the most enriched domain in our DRBPs (Fig. 2b)82.
The RNA recognition motif
The RRM is an extremely versatile domain that is capable of binding (mainly single-stranded) RNA and DNA, as well as proteins79. As stated above, RRMs preferentially interact with nucleic acid bases rather than with the phosphate-sugar backbone. The structural nature of ssRNA and ssDNA allows for much easier access to the exposed aromatic base faces, as opposed to hydrogen bonding to the base edges that occur frequently with double-stranded nucleic acid binding DRBPs. Additionally, stacking interactions with the faces of bases are more energetically favorable than recognition of the nucleotide edge. Therefore, stacking interactions between aromatic protein side chains and nucleic acid bases are often observed in single-stranded nucleic acid binding proteins.
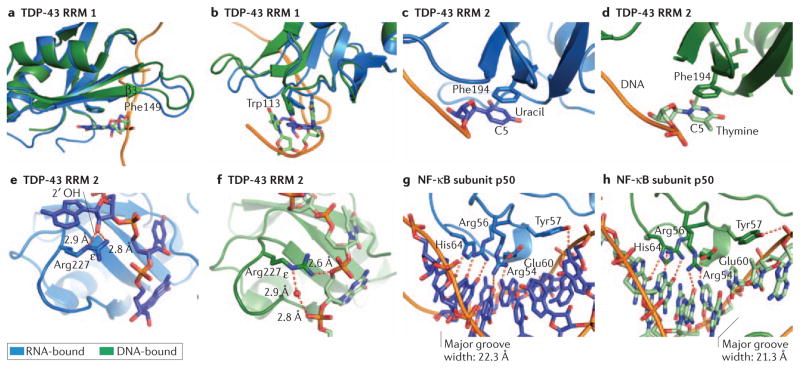
TDP-43 is a DRBP that plays important roles in mRNA splicing and miRNA biogenesis83,84. It contains two RRMs that are separated by a short loop, both capable of binding DNA and RNA. Crystal structures of both RRMs in complex with DNA and RNA have been reported85,86 (see also the currently unpublished structure of TDP43 RRMs: PDB4 4IUF), making TDP-43 an excellent case study for dual DNA and RNA recognition by RRMs. The DNA- and RNA-bound structures of TDP-43 reveal nearly identical modes of nucleic acid recognition. Aromatic side chains, such as Phe149 within the first RRM (RRM1), form stacking interactions with DNA or RNA bases (Fig 4a). Trp113, part of the more flexible loop 1, is able to shift conformations and base-stack slightly differently when bound to differing nucleic acid sequences (Fig. 4b), whereas Phe149, part of the rigid β3 sheet of the RRM1 fold, makes similar interactions with DNA and RNA (Fig. 4a). Relying on the more energetically favorable π-stacking interactions through the planar face of the DNA and RNA bases, results in less specificity than would be gained from hydrogen bonding with the base edge. Uracil and thymine interact with Phe194 of the second RRM in the RNA- and DNA-bound structures, respectively (Fig. 4c, d). Despite the additional methyl moiety at position C5 in the DNA, no TDP-43 residues recognize the edge of the nucleotide in order to interact or clash with the additional carbon (Fig. 4d). Thus, one of the chemical differences between DNA and RNA, the use of uracil in RNA, plays no role in nucleic acid discrimination in this example.
Figure 4. The structural basis for dual DNA and RNA recognition by TDP-43 and by the NF-κB subunit p50.
Protein-RNA structures are shown in blue and protein-DNA structures in green, with protein in the darker shade. π-stacking interactions play a prominent role in both the ssDNA and ssRNA binding activities of TDP-43: a–d. Phe149 (a) and Trp113 (b) within the first RRM of TDP43 stack with both RNA and DNA bases. c, d. In the second RRM of TDP-43, Phe194 is capable of recognizing both uracil in RNA and thymine in DNA; the additional methyl group at C5 in thymine does not contribute to nucleic acid specificity. e. When bound to RNA, both the terminal amine and ε nitrogen of Arg227 in the second RRM of TDP-43 contact a 2′OH on the RNA backbone. f. In contrast, these same groups can also make contacts with the DNA backbone, both directly and through water mediated hydrogen bonding. g, h. The p50 subunit of NF-κB makes strikingly similar base-specific contacts when bound to double-stranded DNA (g) or an RNA aptamer (h). This is due in large part to the similar secondary structure and chemical moieties presented by the RNA and DNA. Major groove width was calculated by 3DNA using phosphate-phosphate distances92.
In contrast, the second RRM of TDP-43 does make RNA-specific contacts with the 2′OH group. The majority of protein – 2′OH interactions are mediated through protein side chains81, and both the RRM2 Lys263 and Arg227 (Fig. 4e) side chains contact a 2′OH when bound to RNA. When it is bound to DNA, however, these same protein side chains contact the DNA backbone phosphates, demonstrating that amino acids are capable of reorienting to enable distinct types of interactions to support RNA and DNA binding (Fig. 4f). DNA binding is not a general property of all RRMs, however. For example, the RRM of PABP relies on a large number of RNA-specific 2′OH contacts for RNA interaction, and binding to DNA may be at low-affinity, if detectable at all87.
DRBPs that recognize double stranded nucleic acids
Crystal structures of protein – dsRNA complexes are less common than their single-stranded counterparts, but there are some examples that are instructive for dual nucleic acid recognition. NF-κB is a central transcription factor of the immune signaling, and is formed of homo- or heterodimers of Rel family proteins such as p50 (NFKB1) or p65 (RELA)88. High-affinity aptamers have been developed for both the p50 and p65 subunits of NF-κB, with affinity of RNA binding that approaches that of the transcription factor’s affinity for native DNA response elements10,89. DNA with an identical sequence to the p50-targeting aptamer will not bind p50 (ref. 10); p50 is therefore another DRBP that binds RNA and DNA is a sequence-specific manner, with different sequence specificities for each.
Crystal structures of p50 bound to DNA and to RNA have been solved, revealing that both bind at the same surface of the p50 Ig-like domain90,91. Although p50 binds to RNA as a monomer and to DNA as a dimer, similar networks of base-specific interactions occur in each structure between protein and nucleic acid (Fig. 4g, h). Not only do the DNA and RNA contacting residues of p50 maintain an equivalent position, but both DNA and RNA present similar interfaces for p50 recognition in charge distribution and in secondary structure (Fig. 4g, h)92. This is a seminal, structurally-confirmed example of “DNA mimicry” by RNA in order to bind to a transcription factor, and although the RNA in this case was artificial, DNA mimicry has been hypothesized to play a role in the endogenous regulation of several transcription factors1,93.
Structures have also been solved of the DRBP ADAR1, which binds both double stranded Z-DNA and double-stranded Z-RNA through its unique Zα domain94,95. The ability of the Zα domain to make sequence-independent interactions with the Z-form phosphate backbone of both DNA and RNA enables ADAR1 to sense nucleic acid secondary structure conformations. Thus, double-stranded nucleotide-binding DRBPs can recognize their DNA and RNA targets through sequence-specific interactions (in the case of NF-κB) or through non-specific interactions with the DNA and RNA backbone (in the case of ADAR1).
The evolution of DRBPs
The evolutionary forces driving the structure and function of DRBPs are complex, yet understanding them will help us identify new DRBPs and perhaps predict their susceptibility to interactions with lncRNAs. While the DRBPs identified in our analysis are members of many different structural classes, each with their own evolutionary history, we will focus on members of two very dissimilar DRBP families: cold-shock domain (CSD)-containing proteins and eukaryotic DNA methyltransferases. CSD-containing proteins, required to protect cells from low temperatures, are members of an ancient DRBP family that utilize weak selection criteria to interact with nucleic acids and therefore intrinsically bind to both DNA and RNA. Members of the eukaryotic DNA methyltransferase family represent DRBPs that have more recently evolved the ability to recognize both DNA and RNA: they prefer interactions with DNA, and only one family member (DNMT2) acquired the ability to bind and methylate tRNAs96. The discovery of a eukaryotic DNA methyltransferase with exquisite tRNA methylation activity and only modest DNA methylation activity showcases how evolution can rewire protein surfaces to create new functions of DRBPs.
The ancient cold shock domain DRBPs
The CSD is one of the most ancient nucleic acid binding domains, found in both prokaryotes and eukaryotes. All CSD-containing proteins bind DNA and RNA (Supplementary Table 1). In humans, several CSD-containing proteins exist, such as the three Y-box binding proteins, the lin-28 homologs LIN28A and LIN28B, and CSDE1 (also known as UNR). The Y-box protein 1 (YB-1) was originally named for its ability to bind and repress the “Y-box” of MHC Class II promoters97. YB-1 also binds RNA, with roles in alternative splicing98, translational control99, and RNA stabilization100. In addition, YB-1 binds to damaged DNA and is involved in the DNA damage response101–103 - it translocates to the nucleus following stresses such as UV radiation104,105.
In bacteria, CSDs exist in short proteins that contain one CSD with little flanking sequences. In Escherichia coli, there are nine such proteins (cspA-cspI), which are likely products of multiple gene duplication events106. Of these, cspA, cspB, cspG, and cspI are induced by cold stress, with cspA briefly comprising over 10% of all protein synthesized during cold shock107–110. Simultaneous deletion of the four genes results in lack of E. coli colony formation at 25°C or lower111. cspD is induced by nutrient stress112, but cspC and cspE are constitutively expressed at normal growth temperature113. Many (if not all) of the csp genes bind DNA and RNA114,115 and have similar roles as the human CSD-containing proteins: in maintaining RNA stability114, in translational regulation116, transcriptional control116,117, DNA replication and repair118,119, and chromosome folding120.
CSD-containing proteins are widespread in plants121, where they perform similar cellular functions. The first csp-like protein found in plants was the wheat WCSP1, which is upregulated specifically by cold stress and binds ssDNA, dsDNA, and RNA homopolymers122. WCSP1 was found to complement the cold-sensitive phenotype of the E. Coli cspA/B/G/I knockouts mentioned above123, exhibiting remarkable functional conservation. In addition, WCSP1 showed in E. Coli nucleic acid melting activity, critical to preventing inappropriate nucleic acid secondary structures that disrupt and terminate transcription. This activity is similar to the endogenous E. coli cspA, which also has transcription antitermination activity123. In Arabidopsis thaliana four CSD-containing proteins are found, AtCSP1-4, all of which can also complement the quadruple csp knockout in E. coli to varying degrees, suggesting that their DNA and RNA interactions are well conserved during evolution124–126.
Unlike their counterparts in bacteria, most plant and animal CSD-containing proteins have additional functional domains (including more CSDs), which expand their functions, protein-protein interactions, and/or nucleic acid binding specificities. For example, the human protein UNR has five CSDs, which serve to increase the protein’s affinity for target RNA sequences127. YB-1 has both an N- and a C-terminal domain flanking its CSD, which can support homomultimerization and interactions with many other protein partners (reviewed in 128). In addition to its CSD, WCSP1 has three CCHC zinc fingers through which most of its dsDNA binding is mediated122. Nevertheless, the exceptional sequence and functional conservation between eukaryotic CSD-containing proteins and bacterial csps proteins demonstrate a conserved, ancient role and origin of the domain. Most likely, a CSD fold capable of binding DNA and RNA was present in the last common ancestor of bacteria, archaea, and eukaryotes129.
The curious case of DNMT2
DNA methylation plays important roles in gene expression and in repressing transposable elements in eukaryotic cells. There are three eukaryotic proteins in the cytosine-C5 DNA methyltransferase family, DNMT1, DNMT2, and DNMT3. Whereas DNMT1 and DNMT3 play important roles in maintaining genome-wide methylation, DNMT2 has diminutive DNA methylation activity130 and instead is capable of methylating tRNAs96 (Fig. 5a). When this activity was discovered, it was speculated that the three eukaryotic DNMTs might have evolved from an RNA methyltransferase96. However, there is no evidence that DNMT2 is more closely related to the ancestral protein of the family members. In fact, the three eukaryotic DNMTs may not be monophyletic and may have evolved from separate prokaryotic DNA methylation restriction-modification enzymes131. Thus, it seems likely that DNMT2 shifted its nucleic acid specificity from DNA to RNA in the last common eukaryotic ancestor131 (Fig. 5b).
Figure 5. DNA methyltransferases target both DNA and RNA.
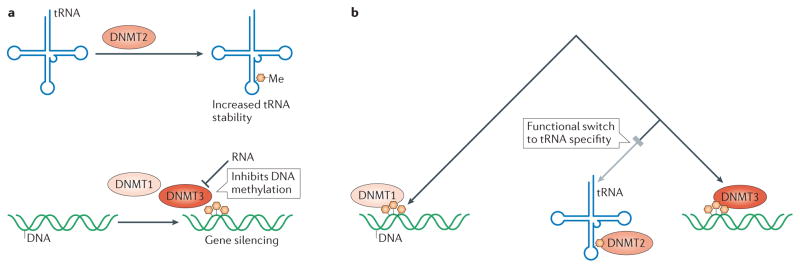
a, Best known for their role in gene silencing, all DNA methyltransferase family members are able to interact with both RNA and DNA21,22,96. DMNT1 and DMNT3 play a role in initiating and maintaining DNA methylation while DNMT2 methylates tRNAs. This tRNA modification is critical for maintaining tRNA stability and cell viability. b, Cladeogram depicting the evolution of the three major families of DNMTs131. DNMT2 likely diverged from its ancestral DNA methyltransferase activity to perform a critical role in methylating tRNAs, a function which it performs redundantly with NUSN2132. This radical change in DNMT’s substrate specificity highlights the ability of evolution to reshape a DNA-binding interface into one that preferentially recognizes RNA.
Despite the relatively narrow substrate specificity of DNMT2 compared to its family members, it is highly conserved and is the only extant DNMT in some species such as Saccharomyces pombe and Drosophila melanogaster131. This seems to indicate that DNMT2 has an important physiological role; however, DNMT2−/− mice are viable, fertile, and yield no obvious phenotype96. This apparent contradiction was resolved with the recent report that deleting DNMT2 in addition to the deletion of another tRNA methyltransferase, NSUN2, is lethal132. These mice exhibit defects in tRNA stability, protein synthesis, and differentiation132, implying that the DNA methylation activity of DNMT2 is dispensable whereas its tRNA methylation activity is not.
NSUN2 is a member of the NCL1 family of eukaryotic RNA cytosine-C5 methyltransferases, which are broadly distributed among eukaryotes133. Interestingly, NSUN2 itself is a DRBP, able to bind and methylate both tRNA and hemi-methylated DNA134. CLIP-based analyses showed that NSUN2 methylates also ncRNAs and mRNAs135. Given the distant evolutionary relationship between DNA and RNA cytosine-C5 methyltransferases131, NSUN2 and DNMT2 have most likely undergone convergent evolution from an RNA- and a DNA-binding protein family, respectively, to ensure proper tRNA modification. Furthermore, these evolutionary trajectories have bestowed on both proteins the ability (if residual) to bind and modify both DNA and RNA. Not only does this indicate that proteins with evolutionary conserved DNA-binding activities are capable of binding RNA (and vice-versa), but also that some nucleic acid substrates may be similar enough in sequence and structure to foment protein promiscuity. As mentioned above, this phenomenon is exploited by RNAs, both endogenous and artificial, that function as decoys in order to modulate DRBP function1,19,136.
Conclusion and perspectives
In this analysis, we have demonstrated that DRBPs comprise a significant fraction of cellular proteins – perhaps 2% of the human proteome – and play important cellular roles. Their functions include the control of transcription and translation, DNA repair, mediating responses to stress, splicing, apoptosis and more. These functions are intimately linked to their structure: orthogonal binding of DNA and RNA provides an opportunity for competitive regulation of transcription by decoy RNAs whereas simultaneous binding of DNA and RNA permits transcriptional activation by RNA coactivators or allows the recruitment of RNA-containing complexes to specific DNA loci. In turn, the structures underlying DRBP functions are linked to their evolution. Some DRBPs contain ancient domains that have long bound DNA or RNA; others contain multiple domains that separately confer DNA and RNA binding and mediate their functional roles.
The majority of RNA binding proteins have had remarkably similar motifs during evolution137, although individual members of protein families, such as the forkhead box transcription factors, can have diverse nucleic acid-sequence specificities arising from independent evolutionary events138. It is also worth noting that intrinsically disordered protein domains that do not fold into defined secondary structures may also play important roles in mediating nucleic acid binding14, as was found for RNA chaperones139. In addition to protein evolution, nucleic acid sequence evolution also plays important roles in the evolution of DRBP function. The emergence of lncRNAs has illuminated new cellular binding-targets for proteins previously thought of as DNA-specific binding proteins. Tens of thousands of human lncRNAs have been catalogued140, and it is likely that many of them have yet-undiscovered functions requiring binding to proteins that are currently considered as DNA-specific binding proteins or that have so far only been shown to bind RNA in vitro. For example, GCR and the estrogen receptor were shown to bind DNA and RNA competitively over 20 years before a physiological role for RNA-steroid receptor interactions was established141–143. Experimental selection techniques such as Systematic Evolution of Ligands by Exponential Enrichment (SELEX) have been used to develop inhibitory RNA aptamers for DNA binding proteins such as NF-κB. If such inhibitory RNA binding is functionally advantageous, the rapidly evolving sequences of lncRNAs144 could provide a platform for the evolution of an analogous, endogenous function, and many DRBPs may have species-specific RNA targets. For example, the RNA Lethe, which binds NF-κB, exists only in mice, and is not present even in the closely related rat genome145.
Proteins that bind both DNA and RNA could have several obvious functional advantages. By binding to both mRNAs and their encoding promoters, DRBPs can exert a powerful, amplified effect on gene expression. This also allows for greater flexibility in generating cellular responses, since these DRBPs could generate rapid effects on protein synthesis as well as impart long-acting changes on gene expression. At a cellular level, using one DRBP rather than two independent DNA- and RNA-binding proteins is more efficient, as it requires the transcription and translation of only one gene product. Finally, competitive RNA and DNA binding by some DRBPs allows for an additional level of transcription factor regulation through RNA decoys. These functional advantages, in addition to the rapid pace at which lncRNAs and their functions are being discovered, strongly indicate that more DRBPs and DRBP-mediated functions will be discovered in the coming years.
Supplementary Material
Acknowledgments
W.H.H. is supported by American Heart Association predoctoral fellowship 13PRE16920012 and was supported by a National Institutes of Health training grant to Emory University 5T32GM008602-14. Work in the lab of E.A.O. is supported by R01DK095750 from the National Institutes of Health (NIDDK) and 14GRNT20460124 from the American Heart Association.
Glossary
- Rossmann Fold
A common protein folding pattern that contains the topology of a β-α-β fold. It is found in many nucleotide binding proteins
- Drosha
Drosha is a nuclease of the RNaseIII family, which as part of a complex cleaves primary (pri-)miRNA transcripts into precursor (pre-)miRNAs in the nucleus
- Primary (pri-)miRNA
The primary transcripts of microRNA genes are stem-loop structures pr ocessed by the Drosha complex into precursor (pre-)miRNAs. Typical pri-miRNAs are hundreds of bp-long and can co ntain several pre-miRNAs
- Precursor (pre-)miRNA
The product of Drosha-mediated cleavage of primary microRNAs. The stem-loop structured pre-miRNA are exported to the cytoplasm and further processed by Dicer to fully mature miRNAs
- MAD Homology 1 (MH1) domain
The MH1 domain is evolutionarily conserved and found in SMAD proteins. It contains four alpha-helices, six short beta-strands, five loops, and recognizes specific DNA sequences
- Homeodomain fold
A protein structural domain that binds DNA and RNA and is found most commonly in transcription factors. It consists of a 60 amino acids-long helix-turn-helix structure
- GAR domain
A glycine- and arginine-rich motif that adopts a repeated beta-turn structure. Found most commonly in proteins that bind RNA
- Zinc Finger domains
A structural motif that is characterized by the coordination of one or more zinc ions. There are a number of different zinc finger motifs and each display different binding modes and structures
- WW domains
A small protein motif of 40 amino acids that mediates specific protein-protein interactions with short proline-rich or proline-containing motifs
- Ftz-F1 domain
A protein domain first identified in the Fushi tarazu factor 1 (FTZ-F1) nuclear receptor. The domain contains an evolutionarily conserved LXXLL motif that recognizes other LXXLL-related motifs
- K Homology domain
A conserved protein domain that interacts with both RNA and DNA through a binding cleft formed between two alpha helices, two beta sheets, and the GXXG loop
- Cold shock domain (CSD)
A small, ~70 kDa domain with high similarity to the RNP-1 RNA-binding motif. This domain is found in DNA and RNA binding proteins in prokaryotes and eukaryotes
- Interface
The solvent accessible portion of a protein that is capable of binding a ligand – including DNA and RNA – in a competitive or non-competitive manner
- π-stacking interactions
A non-covalent interaction between two aromatic molecules due to the attractive force originating from the opposing electrostatic potentials between two adjacent aromatic amino acid residues
- Aptamers
A single stranded DNA or RNA that selectively binds small molecules, proteins, and peptides with high affinity. Aptamers have dynamic tertiary structures, which contributes to their diversity of binding targets
- Z-DNA
DNA in the conformation of a left-handed double-helix
- Z-RNA
RNA in the conformation of a left-handed double helix
- Y-box
A DNA target sequence with the consensus CTGATTG, which is recognized and bound by certain proteins
- RNA homopolymers
A sequence of ribonucleotides consisting of a single base; for exampl.e, CCCCCCCC
- Systematic Evolution of Ligands by Exponential Enrichment (SELEX)
A technique to identify ligand-binding sequence specificity, based sequential rounds of binding of an oligonucleotides library to the ligand, followed by PCR amplification of the bound sequences
- Intrinsically disordered proteins
Proteins that do not have a well-ordered, three dimensional structure, such as proteins containing random coils or proteins containing multiple domains connected with flexible linkers
Biographies
William H. Hudson is a Ph. D. candidate in the Molecular Systems Pharmacology Program in the lab of Eric Ortlund at Emory University, Atlanta, GA. Will obtained his B.S. in Biomedical Engineering at the Georgia Institute of Technology. His thesis work aims to characterize how the glucocorticoid receptor, a central transcription factor controlling metabolism and inflammation, interacts with both newly discovered repressive DNA elements and with long intergenic noncoding RNAs,
Eric Ortlund received his Ph. D. from the University of South Carolina, USA, in 2002. Following a postdoctoral fellowship at the University of North Carolina in Chapel Hill, USA, he joined the faculty within the department of Biochemistry at Emory University as an Assistant Professor in 2008. Dr. Ortlund’s lab pursues structural, evolutionary, and biochemical studies of human nuclear receptors, which are ligand regulated transcription factors that play central roles in development, cancer, stress and metabolism. http://www.biochem.emory.edu/ortlund/
References
- 1.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. This study showed that a widely expressed lncRNA was able to accumulate during stress and act as an RNA decoy to prevent steroid receptors from binding to their target DNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ishmael FT, et al. The human glucocorticoid receptor as an RNA-binding protein: global analysis of glucocorticoid receptor-associated transcripts and identification of a target RNA motif. J Immunol. 2011;186:1189–1198. doi: 10.4049/jimmunol.1001794. Showed that the glucocorticoid receptor binds to mRNAs involved in inflamation to accelerate their degradation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Smad proteins bind a conserved RNA sequence to promote microRNA maturation by Drosha. Mol Cell. 2010;39:373–384. doi: 10.1016/j.molcel.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi L, et al. Dynamic binding of Ku80, Ku70 and NF90 to the IL-2 promoter in vivo in activated T-cells. Nucleic Acids Res. 2007;35:2302–10. doi: 10.1093/nar/gkm117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi L, Godfrey WR, Lin J, Zhao G, Kao PN. NF90 regulates inducible IL-2 gene expression in T cells. J Exp Med. 2007;204:971–7. doi: 10.1084/jem.20052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu B, et al. Dax-1 and steroid receptor RNA activator (SRA) function as transcriptional coactivators for steroidogenic factor 1 in steroidogenesis. Mol Cell Biol. 2009;29:1719–34. doi: 10.1128/MCB.01010-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lanz RB, et al. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. Discovered a long noncoding RNA invovled in coordinating steroid receptor-driven gene activation. [DOI] [PubMed] [Google Scholar]
- 8.Hung T, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. Describes a high-throughput approach to identify lncRNAs that interfere with transcripton factors in order to repress gene expersion, suggesting potentially widespread roles for promoter lncRNAs in cell-growth control. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rapicavoli NA, et al. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLife. 2013;2:e00762–e00762. doi: 10.7554/eLife.00762. Showed that expression of pseudogene lncRNAs is actively regulated and that these lncRNAs may interact directly with proteins to modulate cell signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lebruska LL, Maher LJ., 3rd Selection and characterization of an RNA decoy for transcription factor NF-κB. Biochemistry. 1999;38:3168–74. doi: 10.1021/bi982515x. [DOI] [PubMed] [Google Scholar]
- 11.Duss O, et al. Structural basis of the non-coding RNA RsmZ acting as a protein sponge. Nature. 2014;509:588–592. doi: 10.1038/nature13271. Shows how a bacterial lncRNA acts as a sponge to seqester and release proteins invlovled in translation. [DOI] [PubMed] [Google Scholar]
- 12.Binns D, et al. QuickGO: a web-based tool for gene ontology searching. Bioinformatics. 2009;25:3045–6. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu S, et al. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. Used a combined bioinformatics and protein microarray-based strategy to systematically characterize the human protein-DNA interactome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castello A, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell. 2012;149:1393–1406. doi: 10.1016/j.cell.2012.04.031. Identified the RNA interactome by using high-throughput covalent capture techniques coupled to proteomic-based detection strategies. [DOI] [PubMed] [Google Scholar]
- 15.Kadmiel M, Cidlowski JA. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol Sci. 2013;34:518–530. doi: 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy TE, et al. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark AR, Belvisi MG. Maps and legends: The quest for dissociated ligands of the glucocorticoid receptor. Pharmacol Ther. 2012;134:54–67. doi: 10.1016/j.pharmthera.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 18.Kanai A, Tani H, Torimura M, Akimitsu N. The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS ONE. 2013;8:e55684. doi: 10.1371/journal.pone.0055684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukunaga J, et al. The Runt domain of AML1 (RUNX1) binds a sequence-conserved RNA motif that mimics a DNA element. RNA. 2013;19:927–936. doi: 10.1261/rna.037879.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gros C, et al. DNA methylation inhibitors in cancer: Recent and future approaches. Biochimie. 2012;94:2280–2296. doi: 10.1016/j.biochi.2012.07.025. [DOI] [PubMed] [Google Scholar]
- 21.Holz-Schietinger C, Reich NO. RNA modulation of the human DNA methyltransferase 3A. Nucleic Acids Res. 2012;40:8550–8557. doi: 10.1093/nar/gks537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Ruscio A, et al. DNMT1-interacting RNAs block gene-specific DNA methylation. Nature. 2013;503:371–6. doi: 10.1038/nature12598. Described how RNA transcripts are able to regulate local DNA methylation by interacting with DNA methyltransferases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cattaneo A, Biocca S, Corvaja N, Calissano P. Nuclear localization of a lactic dehydrogenase with single-stranded DNA-binding properties. Exp Cell Res. 1985;161:130–140. doi: 10.1016/0014-4827(85)90497-5. [DOI] [PubMed] [Google Scholar]
- 24.Calissano P, Volontè C, Biocca S, Cattaneo A. Synthesis and content of a DNA-binding protein with lactic dehydrogenase activity are reduced by nerve growth factor in the neoplastic cell line PC12. Exp Cell Res. 1985;161:117–129. doi: 10.1016/0014-4827(85)90496-3. [DOI] [PubMed] [Google Scholar]
- 25.Pioli PA, Hamilton BJ, Connolly JE, Brewer G, Rigby WF. Lactate dehydrogenase is an AU-rich element-binding protein that directly interacts with AUF1. J Biol Chem. 2002;277:35738–35745. doi: 10.1074/jbc.M204002200. [DOI] [PubMed] [Google Scholar]
- 26.Demarse NA, et al. Direct binding of glyceraldehyde 3-phosphate dehydrogenase to telomeric DNA protects telomeres against chemotherapy-induced rapid degradation. J Mol Biol. 2009;394:789–803. doi: 10.1016/j.jmb.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh R, Green M. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 1993;259:365–368. doi: 10.1126/science.8420004. Identified glyceraldehyde-3-phosphate dehydrogenase as a tRNA binding protein, demonstrating in prinicple how tRNA levels may control metbolism and how metabolites may influcence protein synthesis. [DOI] [PubMed] [Google Scholar]
- 28.Nagy E, Rigby WFC. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold) J Biol Chem. 1995;270:2755–2763. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- 29.Ray R, Miller DM. Cloning and Characterization of a Human C-Myc Promoter-Binding Protein. Mol Cell Biol. 1991;11:2154–2161. doi: 10.1128/mcb.11.4.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hernandez-Perez L, et al. α-Enolase binds to RNA. Biochimie. 2011;93:1520–1528. doi: 10.1016/j.biochi.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Fletcher L, Rider CC, Taylor CB. Enolase isoenzymes. III. Chromatographic and immunological characteristics of rat brain enolase. Biochim Biophys Acta. 1976;452:245–52. doi: 10.1016/0005-2744(76)90077-2. [DOI] [PubMed] [Google Scholar]
- 32.Kato K, et al. Immunoassay of human muscle enolase subunit in serum: a novel marker antigen for muscle diseases. Clin Chim Acta. 1983;131:75–85. doi: 10.1016/0009-8981(83)90354-6. [DOI] [PubMed] [Google Scholar]
- 33.Anderson SL, Minard KI, McAlister-Henn L. Allosteric inhibition of NAD+-specific isocitrate dehydrogenase by a mitochondrial mRNA. Biochemistry. 2000;39:5623–5629. doi: 10.1021/bi000272e. [DOI] [PubMed] [Google Scholar]
- 34.Hentze MW, Preiss T. The REM phase of gene regulation. Trends Biochem Sci. 2010;35:423–426. doi: 10.1016/j.tibs.2010.05.009. [DOI] [PubMed] [Google Scholar]
- 35.Auphan N, DiDonato JA, Rosette C, Helmberg A, Karin M. Immunosuppression by glucocorticoids: inhibition of NF-κB activity through induction of IκB synthesis. Science. 1995;270:286–90. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- 36.Surjit M, et al. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–241. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 37.Jonat C, et al. Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell. 1990;62:1189–204. doi: 10.1016/0092-8674(90)90395-u. [DOI] [PubMed] [Google Scholar]
- 38.Dhawan L, Liu B, Blaxall BC, Taubman MB. A novel role for the glucocorticoid receptor in the regulation of monocyte chemoattractant protein-1 mRNA stability. J Biol Chem. 2007;282:10146–10152. doi: 10.1074/jbc.M605925200. [DOI] [PubMed] [Google Scholar]
- 39.Yang-Yen HF, et al. Transcriptional interference between c-Jun and the glucocorticoid receptor: Mutual inhibition of DNA binding due to direct protein-protein interaction. Cell. 1990;62:1205–1215. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]
- 40.Ray A, Prefontaine KE. Physical association and functional antagonism between the p65 subunit of transcription factor NF-κB and the glucocorticoid receptor. Proc Natl Acad Sci USA. 1994;91:752–756. doi: 10.1073/pnas.91.2.752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;2008:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 42.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 43.Massagué J, Wotton D. Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumarswamy R, Volkmann I, Thum T. Regulation and function of miRNA-21 in health and disease. RNA Biol. 2011;8:706–713. doi: 10.4161/rna.8.5.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kao PN, et al. Cloning and expression of cyclosporin A- and FK506-sensitive nuclear factor of activated T-cells: NF45 and NF90. J Biol Chem. 1994;269:20691–9. [PubMed] [Google Scholar]
- 47.Kuwano Y, et al. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 2009;38:225–238. doi: 10.1093/nar/gkp861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shim J, Lim H, Yates RJ, III, Karin M. Nuclear export of NF90 is required for interleukin-2 mRNA stabilization. Mol Cell. 2002;10:1331–1344. doi: 10.1016/s1097-2765(02)00730-x. [DOI] [PubMed] [Google Scholar]
- 49.Sakamoto S, et al. he NF90-NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol. 2009;29:3754–3769. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Malek TR, Castro I. Interleukin-2 Receptor Signaling: At the Interface between Tolerance and Immunity. Immunity. 2010;33:153–165. doi: 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dienz O, Rincon M. The effects of IL-6 on CD4 T cell responses. Clim Immunol. 2009;130:27–33. doi: 10.1016/j.clim.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takamizawa J, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 54.Guo NL, et al. Confirmation of gene expression-based prediction of survival in non-small cell lung cancer. Clin Cancer Res. 2008;14:8213–8220. doi: 10.1158/1078-0432.CCR-08-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ng SY, Bogu Gireesh K, Soh Boon S, Stanton Lawrence W. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell. 2013;51:349–359. doi: 10.1016/j.molcel.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 56.Sarkar A, Hochedlinger K. The Sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell. 2013;12:15–30. doi: 10.1016/j.stem.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ng S-Y, Johnson R, Stanton LW. Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J. 2012;31:522–533. doi: 10.1038/emboj.2011.459. Described how the lncRNA RMST physically interacts with the transcription factor SOX2 to co-regulate a large pool of downstream genes implicated in neurogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feng J, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bond AM, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeffery L, Nakielny S. Components of the DNA methylation system of chromatin control are RNA-binding proteins. J Biol Chem. 2004;279:49479–49487. doi: 10.1074/jbc.M409070200. [DOI] [PubMed] [Google Scholar]
- 61.Wassenegger M, Heimes S, Riedel L, Sänger HL. RNA-directed de novo methylation of genomic sequences in plants. Cell. 1994;76:567–576. doi: 10.1016/0092-8674(94)90119-8. [DOI] [PubMed] [Google Scholar]
- 62.Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 64.Sun BK, Deaton AM, Lee JT. A transient heterochromatic state in Xist preempts X inactivation choice without RNA stabilization. Mol Cell. 2006;21:617–628. doi: 10.1016/j.molcel.2006.01.028. [DOI] [PubMed] [Google Scholar]
- 65.Zazopoulos E, Lalli E, Stocco DM, Sassone-Corsi P. DNA binding and transcriptional repression by DAX-1 blocks steroidogenesis. Nature. 1997;390:311–5. doi: 10.1038/36899. [DOI] [PubMed] [Google Scholar]
- 66.Kabe Y, et al. The role of human MBF1 as a transcriptional coactivator. J Biol Chem. 1999;274:34196–34202. doi: 10.1074/jbc.274.48.34196. [DOI] [PubMed] [Google Scholar]
- 67.Xu B, Koenig RJ. An RNA-binding domain in the thyroid hormone receptor enhances transcriptional activation. J Biol Chem. 2004;279:33051–33056. doi: 10.1074/jbc.M404930200. [DOI] [PubMed] [Google Scholar]
- 68.Zhao X, et al. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol Cell. 2004;15:549–558. doi: 10.1016/j.molcel.2004.06.044. [DOI] [PubMed] [Google Scholar]
- 69.Watanabe M, et al. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor α coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J. 2001;20:1341–1352. doi: 10.1093/emboj/20.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 70.Deblois G, Giguere V. Ligand-independent coactivation of ERα AF-1 by steroid receptor RNA activator (SRA) via MAPK activation. J Steroid Biochem Mol Biol. 2003;85:123–31. doi: 10.1016/s0960-0760(03)00225-5. [DOI] [PubMed] [Google Scholar]
- 71.Poon MM, Chen L. Retinoic acid-gated sequence-specific translational control by RARα. Proc Natl Acad Sci USA. 2008;105:20303–20308. doi: 10.1073/pnas.0807740105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 73.Court R, Chapman L, Fairall L, Rhodes D. How the human telomeric proteins TRF1 and TRF2 recognize telomeric DNA: a view from high-resolution crystal structures. EMBO Rep. 2005;6:39–45. doi: 10.1038/sj.embor.7400314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chesnokov IN. Multiple functions of the origin recognition complex. In: Kwang WJ, editor. Int Rev Cytol. Vol. 256. Academic Press; 2007. pp. 69–109. [DOI] [PubMed] [Google Scholar]
- 75.Deng Z, Dheekollu J, Broccoli D, Dutta A, Lieberman PM. The origin recognition complex localizes to telomere repeats and prevents telomere-circle formation. Curr Biol. 2007;17:1989–1995. doi: 10.1016/j.cub.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 76.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35:403–413. doi: 10.1016/j.molcel.2009.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 78.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maris C, Dominguez C, Allain FHT. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 80.Fiset S, Chabot B. hnRNP A1 may interact simultaneously with telomeric DNA and the human telomerase RNA in vitro. Nucleic Acids Res. 2001;29:2268–75. doi: 10.1093/nar/29.11.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bahadur RP, Zacharias M, Janin J. Dissecting protein-RNA recognition sites. Nucleic Acids Res. 2008;36:2705–16. doi: 10.1093/nar/gkn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Allers J, Shamoo Y. Structure-based analysis of protein-RNA interactions using the program ENTANGLE. J Mol Biol. 2001;311:75–86. doi: 10.1006/jmbi.2001.4857. [DOI] [PubMed] [Google Scholar]
- 83.Buratti E, et al. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–84. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kawahara Y, Mieda-Sato A. TDP-43 promotes microRNA biogenesis as a component of the Drosha and Dicer complexes. Proc Natl Acad Sci USA. 2012;109:3347–52. doi: 10.1073/pnas.1112427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lukavsky PJ, et al. Molecular basis of UG-rich RNA recognition by the human splicing factor TDP-43. Nat Struct Mol Biol. 2013;20:1443–1449. doi: 10.1038/nsmb.2698. [DOI] [PubMed] [Google Scholar]
- 86.Kuo PH, Doudeva LG, Wang YT, Shen CKJ, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Deo RC, Bonanno JB, Sonenberg N, Burley SK. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 88.Hayden MS, Ghosh S. NF-κB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wurster SE, Maher LJ. Selection and characterization of anti-NF-κB p65 RNA aptamers. RNA. 2008;14:1037–1047. doi: 10.1261/rna.878908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang DB, et al. Crystal structure of NF-κB (p50)2 complexed to a high-affinity RNA aptamer. Proc Natl Acad Sci USA. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Müller CW, Rey FA, Sodeoka M, Verdine GL, Harrison SC. Structure of the NF-κB p50 homodimer bound to DNA. Nature. 1995;373:311–317. doi: 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- 92.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Barrick JE, Sudarsan N, Weinberg Z, Ruzzo WL, Breaker RR. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA. 2005;11:774–784. doi: 10.1261/rna.7286705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal Structure of the Zα Domain of the Human Editing Enzyme ADAR1 Bound to Left-Handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 95.Placido D, Brown BA, Lowenhaupt K, Rich A, Athanasiadis A. A left-handed RNA double helix bound by the Zα domain of the RNA-editing enzyme ADAR1. Structure. 2007;15:395–404. doi: 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goll MG, et al. Methylation of tRNAAsp by the DNA Methyltransferase Homolog Dnmt2. Science. 2006;311:395–398. doi: 10.1126/science.1120976. Showed that the DNA methylase Dnmt2 evovled the ability to methylate tRNA. [DOI] [PubMed] [Google Scholar]
- 97.Didier DK, Schiffenbauer J, Woulfe SL, Zacheis M, Schwartz BD. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci USA. 1988;85:7322–6. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stickeler E, et al. The RNA binding protein YB-1 binds A/C-rich exon enhancers and stimulates splicing of the CD44 alternative exon v4. EMBO J. 2001;20:3821–30. doi: 10.1093/emboj/20.14.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Skabkina OV, Lyabin DN, Skabkin MA, Ovchinnikov LP. YB-1 autoregulates translation of its own mRNA at or prior to the step of 40S ribosomal subunit joining. Mol Cell Biol. 2005;25:3317–23. doi: 10.1128/MCB.25.8.3317-3323.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Evdokimova V, et al. The major mRNA-associated protein YB-1 is a potent 5′ cap-dependent mRNA stabilizer. EMBO J. 2001;20:5491–5502. doi: 10.1093/emboj/20.19.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Marenstein DR, et al. Stimulation of human endonuclease III by Y box-binding protein 1 (DNA-binding protein B). Interaction between a base excision repair enzyme and a transcription factor. J Biol Chem. 2001;276:21242–9. doi: 10.1074/jbc.M101594200. [DOI] [PubMed] [Google Scholar]
- 102.de Souza-Pinto NC, et al. Novel DNA mismatch-repair activity involving YB-1 in human mitochondria. DNA Repair. 2009;8:704–719. doi: 10.1016/j.dnarep.2009.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ise T, et al. Transcription factor Y-box binding protein 1 binds preferentially to cisplatin-modified DNA and interacts with proliferating cell nuclear antigen. Cancer Res. 1999;59:342–6. [PubMed] [Google Scholar]
- 104.Stein U, et al. Hyperthermia-induced nuclear translocation of transcription factor YB-1 leads to enhanced expression of multidrug resistance-related ABC transporters. J Biol Chem. 2001;276:28562–9. doi: 10.1074/jbc.M100311200. [DOI] [PubMed] [Google Scholar]
- 105.Koike K, et al. Nuclear translocation of the Y-box binding protein by ultraviolet irradiation. FEBS Lett. 1997;417:390–4. doi: 10.1016/s0014-5793(97)01296-9. [DOI] [PubMed] [Google Scholar]
- 106.Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–55. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]
- 107.Etchegaray JP, Jones PG, Inouye M. Differential thermoregulation of two highly homologous cold-shock genes, cspA and cspB, of Escherichia coli. Genes Cells. 1996;1:171–8. doi: 10.1046/j.1365-2443.1996.d01-231.x. [DOI] [PubMed] [Google Scholar]
- 108.Wang N, Yamanaka K, Inouye M. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J Bacteriol. 1999;181:1603–9. doi: 10.1128/jb.181.5.1603-1609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Nakashima K, Kanamaru K, Mizuno T, Horikoshi K. A novel member of the cspA family of genes that is induced by cold shock in Escherichia coli. J Bacteriol. 1996;178:2994–7. doi: 10.1128/jb.178.10.2994-2997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Goldstein J, Pollitt NS, Inouye M. Major cold shock protein of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:283–7. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xia B, Ke H, Inouye M. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol Microbiol. 2001;40:179–188. doi: 10.1046/j.1365-2958.2001.02372.x. [DOI] [PubMed] [Google Scholar]
- 112.Yamanaka K, Inouye M. Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. J Bacteriol. 1997;179:5126–30. doi: 10.1128/jb.179.16.5126-5130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Phadtare S, Inouye M. Role of CspC and CspE in regulation of expression of RpoS and UspA, the stress response proteins in Escherichia coli. J Bacteriol. 2001;183:1205–1214. doi: 10.1128/JB.183.4.1205-1214.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 115.Phadtare S, Inouye M. Sequence-selective interactions with RNA by CspB, CspC and CspE, members of the CspA family of Escherichia coli. Mol Endocrinol. 1999;33:1004–1014. doi: 10.1046/j.1365-2958.1999.01541.x. [DOI] [PubMed] [Google Scholar]
- 116.Hanna MM, Liu K. Nascent RNA in transcription complexes interacts with CspE, a small protein in E. coli implicated in chromatin condensation. J Mol Biol. 1998;282:227–39. doi: 10.1006/jmbi.1998.2005. [DOI] [PubMed] [Google Scholar]
- 117.Bae W, Xia B, Inouye M, Severinov K. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc Natl Acad Sci USA. 2000;97:7784–9. doi: 10.1073/pnas.97.14.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Yamanaka K, Zheng W, Crooke E, Wang YH, Inouye M. CspD, a novel DNA replication inhibitor induced during the stationary phase in Escherichia coli. Mol Microbiol. 2001;39:1572–84. doi: 10.1046/j.1365-2958.2001.02345.x. [DOI] [PubMed] [Google Scholar]
- 119.Chen CS, et al. A proteome chip approach reveals new DNA damage recognition activities in Escherichia coli. Nat Methods. 2008;5:69–74. doi: 10.1038/NMETH1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hu KH, et al. Overproduction of three genes leads to camphor resistance and chromosome condensation in Escherichia coli. Genetics. 1996;143:1521–32. doi: 10.1093/genetics/143.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Karlson D, Imai R. Conservation of the cold shock domain protein family in plants. Plant Physiol. 2003;131:12–5. doi: 10.1104/pp.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Karlson D, Nakaminami K, Toyomasu T, Imai R. A cold-regulated nucleic acid-binding protein of winter wheat shares a domain with bacterial cold shock proteins. J Biol Chem. 2002;277:35248–35256. doi: 10.1074/jbc.M205774200. [DOI] [PubMed] [Google Scholar]
- 123.Nakaminami K, Karlson DT, Imai R. Functional conservation of cold shock domains in bacteria and higher plants. Proc Natl Acad Sci USA. 2006;103:10122–10127. doi: 10.1073/pnas.0603168103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Imai R, Kim MH, Sasaki K, Sato S, Sonoda Y. Plant and Microbe Adaptations to Cold in a Changing World. Springer; New York: 2013. pp. 131–142. [Google Scholar]
- 125.Juntawong P, Sorenson R, Bailey-Serres J. Cold shock protein 1 chaperones mRNAs during translation in Arabidopsis thaliana. Plant J. 2013;74:1016–1028. doi: 10.1111/tpj.12187. [DOI] [PubMed] [Google Scholar]
- 126.Kim JS, et al. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res. 2007;35:506–16. doi: 10.1093/nar/gkl1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Triqueneaux G, Velten M, Franzon P, Dautry F, Jacquemin-Sablon H. RNA binding specificity of Unr, a protein with five cold shock domains. Nucleic Acids Res. 1999;27:1926–34. doi: 10.1093/nar/27.8.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Mosc) 2012;76:1402–1433. doi: 10.1134/S0006297911130049. [DOI] [PubMed] [Google Scholar]
- 129.Mihailovich M, Militti C, Gabaldón T, Gebauer F. Eukaryotic cold shock domain proteins: highly versatile regulators of gene expression. BioEssays. 2010;32:109–118. doi: 10.1002/bies.200900122. [DOI] [PubMed] [Google Scholar]
- 130.Hermann A, Schmitt S, Jeltsch A. The human Dnmt2 has residual DNA-(cytosine-C5) methyltransferase activity. J Biol Chem. 2003;278:31717–31721. doi: 10.1074/jbc.M305448200. [DOI] [PubMed] [Google Scholar]
- 131.Lyko F, Jurkowski TP, Jeltsch A. On the Evolutionary Origin of Eukaryotic DNA Methyltransferases and Dnmt2. PLoS ONE. 2011;6:e28104. doi: 10.1371/journal.pone.0028104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Tuorto F, et al. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol. 2012;19:900–905. doi: 10.1038/nsmb.2357. [DOI] [PubMed] [Google Scholar]
- 133.Pavlopoulou A, Kossida S. Phylogenetic analysis of the eukaryotic RNA (cytosine-5)-methyltransferases. Genomics. 2009;93:350–357. doi: 10.1016/j.ygeno.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 134.Sakita-Suto S, et al. Aurora-B regulates RNA methyltransferase NSUN2. Mol Biol Cell. 2007;18:1107–1117. doi: 10.1091/mbc.E06-11-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hussain S, et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013;4:255–261. doi: 10.1016/j.celrep.2013.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Reiter NJ, Maher LJ, Butcher SE. DNA mimicry by a high-affinity anti-NF-κB RNA aptamer. Nucleic Acids Res. 2008;36:1227–1236. doi: 10.1093/nar/gkm1141. Showed how a rapidly evolved RNA aptamer targeted the DNA-binding interface of a transcrption factor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ray D, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Nakagawa S, Gisselbrecht SS, Rogers JM, Hartl DL, Bulyk ML. DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc Natl Acad Sci USA. 2013;110:12349–12354. doi: 10.1073/pnas.1310430110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 140.Volders PJ, et al. LNCipedia: a database for annotated human lncRNA transcript sequences and structures. Nucleic Acids Res. 2012;41:D246–D251. doi: 10.1093/nar/gks915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ali M, Vedeckis W. The glucocorticoid receptor protein binds to transfer RNA. Science. 1987;235:467–470. doi: 10.1126/science.3798121. [DOI] [PubMed] [Google Scholar]
- 142.Feldman M, Kallos J, Hollander VP. RNA inhibits estrogen receptor binding to DNA. J Biol Chem. 1981;256:1145–8. [PubMed] [Google Scholar]
- 143.Rossini GP. RNA-containing nuclear binding sites for glucocorticoid-receptor complexes. Biochem Biophys Res Commun. 1984;123:78–83. doi: 10.1016/0006-291x(84)90382-6. Together with Ref. 2, 143 and 144, revealed that RNA binding to transcrption factors may be a widespread phenomenon. [DOI] [PubMed] [Google Scholar]
- 144.Necsulea A, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–40. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 145.Flicek P, et al. Ensembl 2014. Nucleic Acids Res. 2013;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.