Abstract
Cysteine is a uniquely reactive amino acid, capable of undergoing both nucleophlilic and oxidative post-translational modifications. One such oxidation reaction involves the covalent modification of cysteine via the gaseous second messenger nitric oxide (NO), termed S-nitrosylation (SNO). This dynamic post-translational modification is involved in the redox regulation of proteins across all phylogenic kingdoms. In mammals, calcium-dependent activation of nitric oxide synthase triggers the local release of nitric oxide, which activates nearby guanylyl cyclases and cGMP-dependent pathways. In parallel, diffusible nitric oxide can locally modify redox active cellular thiols, functionally modulating many redox sensitive enzymes. Aberrant S-nitrosylation is implicated in the pathology of many diseases, including neurodegeneration, inflammation, and stroke. In this review, we discuss current methods to label sites of S-nitrosylation for biochemical analysis. The most popular method involves a series of biochemical steps to mask free thiols followed by selective nitrosothiol reduction and capture. Other emerging methods include mechanism-based phosphine probes and mercury enrichment chemistry. By bridging new enrichment approaches with high-resolution mass spectrometry, large-scale analysis of protein nitrosylation has highlighted new pathways of oxidative regulation.
Introduction
Sulfur is the lightest element that can produce stable exceptions to the octet rule due to the presence of “d” orbitals. Typical cysteine residues in proteins have a side chain pKa values of 8.01, and thus ~10% of cysteine thiols are in their reactive thiolate form at physiological pH. However, many redox active or catalytic cysteine residues have dramatically reduced pKa values. Such thiolates have evolved to promote catalysis or redox regulation. For example, the pKa of the catalytic cysteine in methionine sulfoxide reductase is reduced to 5.7 upon substrate binding2,3. Similarly the active site thiol of glutaredoxin has a low pKa near 3.54,5. Such altered acid-dissociation constants enhance thiol reactivity, which in turn promote reactions with electrophilic oxidants to produce distinct post-translational modifications.
In this review, we focus on the chemistry and dynamics of protein S-nitrosylation. This unique oxidative modification directly modulates the localization and activity of cellular proteins involved in cellular growth and regulation6,7. In neurons, stimulus-dependent depolarization leads to calcium influx, calmodulin activation, and stimulation of nitric oxide (NO) synthases8–10. Local nitric oxide release induces spatially restricted S-nitrosylation of channels, phosphatases, and other redox active thiols6,11,12. Emerging proteomics studies implicate hundreds of endogenous sites of nitrosylation13–16, although the stoichiometry and functional consequences of these post-translational remains an active research area.
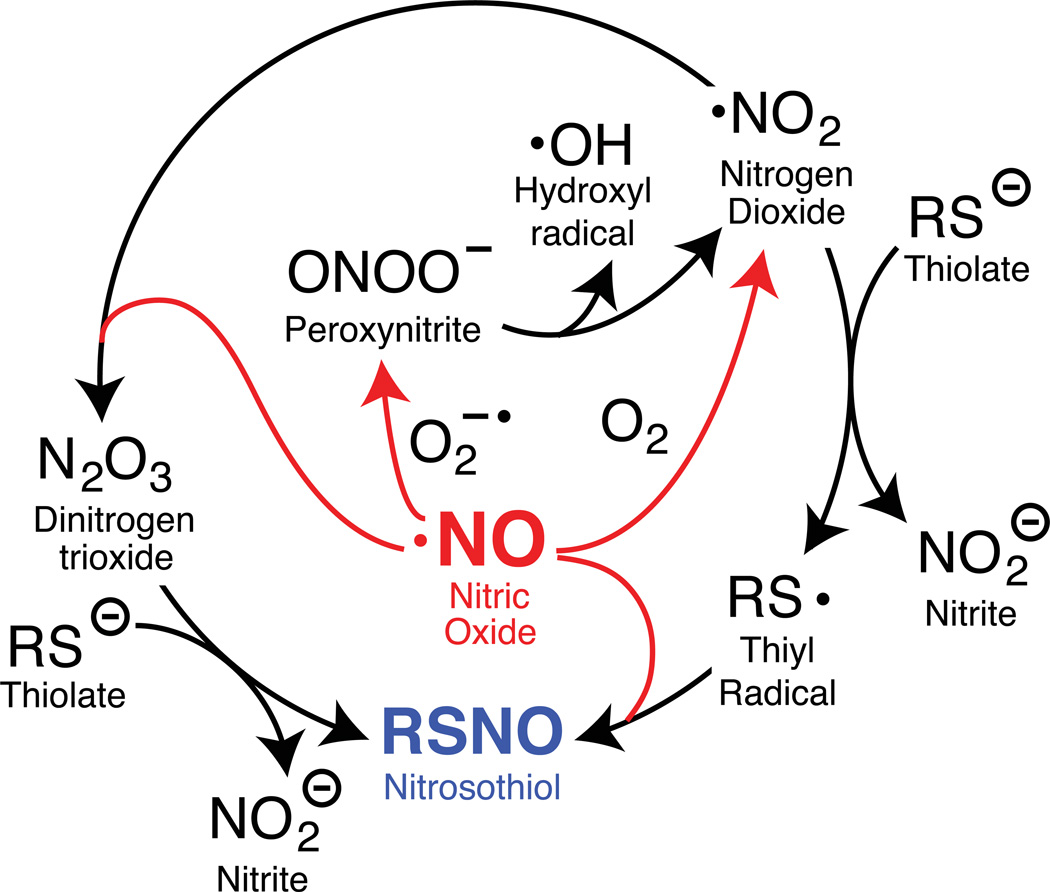
There are several mechanistic routes leading to protein S-nitrosylation (Figure 1). Heme-dependent nitric oxide synthases generate nitric oxide via a two-step, five-electron oxidation of L-arginine17–20. This reaction uses two moles of molecular oxygen and 3/2 moles of NADPH per mole of nitric oxide formed21. Nitric oxide itself is not especially reactive towards protonated cellular thiols, particularly under aerobic conditions22–24. In order to generate nitrosothiols, nitric oxide must first undergo secondary oxidation to nitrogen dioxide, which occurs via at least two distinct pathways25. In the first pathway, nitric oxide reacts with a superoxide radical to make peroxynitrite. Peroxynitrite (pKa of 6.526) converts to peroxynitrous acid at physiological pH, which undergoes hemolytic cleavage to form the hydroxyl and the nitrogen dioxide radicals. The nitrogen dioxide radical can in turn react with nitric oxide to form dinitrogen trioxide, which reacts with thiolates to form nitrosothiols24. In a second pathway, nitrogen dioxide reacts with a thiolate to generate nitrite and a thiyl radical27. The resulting thiyl radical is the only species able to react directly with nitric oxide to generate nitrosothiols. Additionally, metal-dependent formation of thiyl radicals28 promotes S-nitrosylation by one-electron oxidation of thiols to thiyl radicals, or through metal-nitrosyl complex intermediates29,30. Furthermore, iron and nitric oxide spontaneously react to form dinitrosyliron complexes (DNIC), which can be intermediates in nitrosothiol formation31–34. All of these routes generate diffusible reactive radicals with enhanced reactivity towards thiols with reduced pKa values, such as catalytic or redox-active thiols.
Figure 1.
Formation of nitrosothiols from nitric oxide (NO) occurs through distinct oxidative pathways27,82–85, each involving two molecules of NO for each nitrosothiol formed. Superoxide radical = O2−•. Oxygen = O2.
S-nitrosylation is reversible, either by nitric oxide release or by direct transfer to other cellular thiols35,36. Such trans-nitrosylation reactions mobilize the exchange of nitric oxide from one protein to another, relaying nitroso-oxidation through multiple carriers37–40. Trans-nitrosylation provides another route for the dynamic exchange of nitrosothiols. Millimolar glutathione levels maintain an intracellular reducing environment that protects proteins from oxidative modifications41. Abundant glutathione scavenges nitrosothiols by trans-nitrosylation, yielding a reduced protein thiol and nitrosoglutathione, which is reduced either by S-nitrosoglutathione reductase (GSNOR) or thioredoxin cascades. Such thiol exchange reactions are prevalent at physiological pH and predominate in comparison to hydrolysis reactions42, and are driven by the levels of reduced thiols in a given environment43. Several studies have demonstrated trans-nitrosylation cascades relay the nitroso adduct from one protein thiol to another, eventually nitrosylating and inactivating select enzymes, for example nuclear chromatin-modifying enzymes44. This model suggests stable S-nitrosylation sites are protected from the cellular environment, masked inside proteins or membranes. These studies demonstrate the surprising resilience of certain nitrosylated proteins in face of millimolar glutathione, and hints at orchestrated pathways of nitrosothiol transfer in cellular regulation. Indeed, hydrophobicity does enhance the rate of reaction between NO and oxygen by several fold45, suggesting that thiols in hydrophobic environments, either in membranes or in hydrophobic protein domains, may be more prone to stable S-nitrosylation6. Overall, protein S-nitrosylation is modulated by thiol pKa, vicinal hydrophobicity6, proximity to NOS enzymes and by activities of redox enzymes such as thioredoxin, GSNOR, and accessibility to reduced glutathione.
Protein nitrosylation functionally regulates protein activities by transiently occupying thiol residues. Functional cysteines often reside in the active sites of enzymes, such as phosphatases, proteases, acyl-transferases, and ubiquitin ligases46. These thiols reside in environments that promote thiolate formation by reducing the side-chain pKa, leading to a more redox-active cysteine. Importantly, the active site of any enzyme is more likely to be protected from the environment, which likely prevents exchange with bulkier thiols. Accordingly, more stable S-nitrosylation is inversely correlated with thiolate exposure, and stabilized in protected environments. Indeed, nitrosylation of cellular phosphatases potentiates kinase cascades to promote cell growth, or inactivates lipid phosphatases during ischemic stroke47–49. Due to the selective targeting of functional cysteines, nitrosylation may serve as a general redox switch important for the reversible inactivation of functional cysteine residues. For example, nitrosylation of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) recruits the ubiquitin ligase Siah1, and the GAPDH-Siah1 complex translocates to the nucleus where Siah1 targets nuclear proteins for degradation50–52. Similarly, argininosuccinate synthase, the enzyme that generates arginine from citrulline53, is inhibited by S-nitrosylation at Cys13254. Elevated nitric oxide levels induce argininosuccinate synthase nitrosylation and inactivation, providing an autoregulatory loop that prevents excess oxidant production.
S-nitrosylation can also directly compete other reversible cysteine post-translational modifications, such as protein palmitoylation. Upon synaptic stimulation, protein palmitoyl thioesterases remove the membrane anchoring lipids from the neuronal scaffolding protein PSD-95, promoting egress from the post-synaptic density55. Simultaneous activity-dependent calcium influx activates neuronal nitric oxide synthase (nNOS), which is stably associated with PSD-95 via PDZ-dependent interactions. This locally generated flux of NO directly nitrosylates the newly de-acylated thiols, thus blocking further palmitoylation and membrane association. The mutually competitive modification of PSD-95 cysteines orchestrates synaptic release through an exchange of specific cysteine post-translational modifications55. Furthermore, S-nitrosylation of the NMDA receptor leads to channel desensitization and channel closing, preventing excitotoxicity48. This cascade of post-translational events ensures proper membrane release of PSD-95, and ensures proper channel desensitization. This example raises the question if such mutually competitive modifications are unique for PSD-95, or if many proteins undergo dynamic exchange of cysteine PTMs. Furthermore, PSD-95 nitrosylation is channeled by protein-protein interactions, which provides selectivity for the deacylated thiols. The breadth of cysteine modifications competition is unknown. Interestingly, several palmitoylated proteins, including Ras and G-proteins, are rapidly de-palmitoylated after receptor stimulation56–58, which coincides activation of NADPH oxidases and NO synthases.
Ascorbate-dependent enrichment strategies
Sensitive and selective labeling tools are critical for the precise detection and annotation of S-nitrosylation. Despite the acceptance of S-nitrosylation as an important protein regulatory modification, there remains a lack of direct methods to study this reversible redox modification. Early approaches used chemiluminescent, colorimetric or electrochemical methods to detect total nitric oxide liberated from nitrosylated thiols in a sample59. These methods measure bulk release, and do not distinguish between heme, metabolite, or protein sources. Furthermore, these approaches eliminate any information about the sites and dynamics of nitrosylation on select proteins.
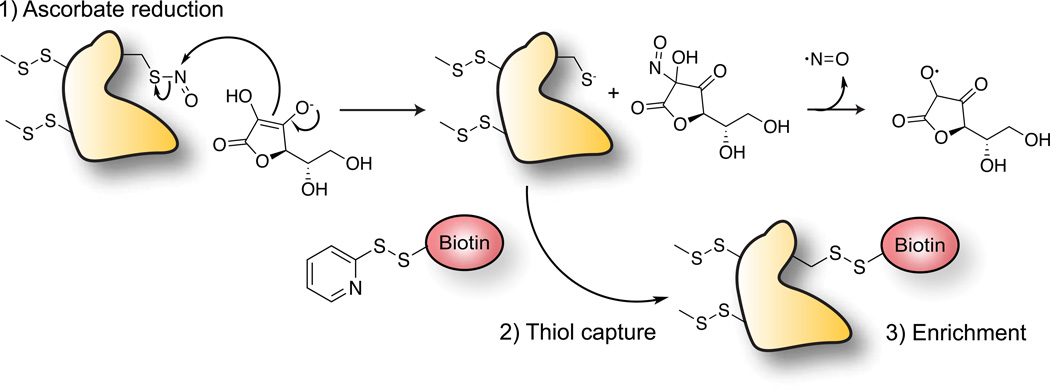
The “biotin-switch” technique was a major advance in the study and annotation of S-nitrosylated proteins60. This widely adopted method involves a series of biochemical steps, beginning with addition of the alkylating agent 2-iodacetamide, or by methyl methanethiosulfonate (MMTS) to block all free thiols. After removing the thiol capture reagents, the sample is treated with ascorbate, which reduces nitrosothiols to generate free sulfhydryl groups. This approach is selective for S-nitrosylation over other oxidative modifications, largely due to the unique mechanism of indirect reduction (Figure 2)61,62. In the presence of nitrosothiols, ascorbate undergoes a trans-nitrosation reaction involving nitrosonium (NO+) transfer to generate nitrosoascorbate, which decomposes to nitric oxide and the semi-dehydroascorbate radical. Therefore, ascorbate does not directly donate an electron for nitrosothiol reduction. This distinct mechanism is thought to provide chemical orthogonality to other oxidative modifications, making ascorbate an ideal nitrosothiol-selective reducing agent62. Following ascorbate-mediated reduction, the newly unmasked thiols groups are then captured using a pyridyldithiol-activated, sulfhydryl-reactive biotin-linked probes for affinity enrichment60. While these reagents are commonly available, several commercial kits are available that include ascorbate, metal-chelating buffers, MMTS, and iodoacetamide detection reagents. Recent adaptations for quantitative proteomics labeling strategies are also commercially available, and have been used to quantify individual sites of S-nitrosylation63. Thiol resin assisted capture (RAC) replaces biotin and streptavidin purification with direct disulfide capture to activated thiol resin immediately after ascorbate treatment64,65. This approach simplifies the procedure, and eliminates non-specific enrichment of endogenous biotinylated carboxylases.
Figure 2.
Biotin-switch technique for ascorbate-dependent reduction of nitrosothiols. Enriched proteins are either analyzed by SDS-PAGE or annotated by mass spectrometry.
Ultimately, the biotin-switch purification method is indirect, and highly dependent on the complete alkylation of all free thiols. Protein nitrosylation is a low abundance modification, so even low levels of uncapped thiols can lead to a high false positive rate. Interestingly, a heating step is generally included during thiol capping, often to 50° C. S-nitrosothiols are known to undergo thermal decomposition via homolytic cleavage of the S-N bond to yield the corresponding disulfides and nitric oxide, the latter is then oxidized to nitrogen dioxide66,67. This step may introduce later complications, as the thermal stability of distinct nitrosothiols has not been thoroughly evaluated. Clearly, prolonged heating will promote nitric oxide release, and potentially suppress detection of labile nitrosothiols. Furthermore, after ascorbate reduction, newly free thiols are able to exchange with existing disulfide bonds, scrambling native sites of nitrosylation. Resin assisted capture is likely to reduce the extent of scrambling by providing high excess of activated disulfides for immediate capture65. Another source of false-positives is sunlight driven disulfide reduction, which can be eliminated by performing all procedures in complete darkness68. This unfortunate restraint makes sample preparation more tedious, but is essential to eliminate non-specific disulfide reduction. Overall, biotin-switch technique is the current standard for nitrosothiol labeling, enrichment, and analysis.
Caveats of nitric oxide donors
Thousands of S-nitrosylated proteins have been reported from ascorbate-dependent enrichment using the biotin-switch method and mass spectrometry13. Unfortunately, nearly all reported proteomics data was collected from biological samples after the addition of exogenous or physiological nitrosylating agents. Upon donor release, gaseous NO is oxidized by molecular oxygen to form a peroxynitrite radical, which then reacts with a second NO molecule to generate two molecules of nitrogen dioxide. This reaction is limited by an apparent third-order rate law (k = 2.5×106 M−2s−1)69–71, which means the reaction rate depends on the product of square of the nitric oxide concentration and molecular oxygen69. Therefore, at very low nitric oxide concentrations the reaction is very slow, but at high nitric oxide concentrations the reaction is extremely rapid. Thus, depending on the concentration of nitric oxide released, the half-life of the reaction can range from 0.5 seconds to 50 hours69. When donors release nitric oxide at higher than physiological concentrations, it is likely that non-physiological nitrosothiols are formed at less activated thiols. Therefore, it is difficult to interpret the results of proteomics experiments performed on donor treated samples72.
In a recent study, two nitric oxide donors (spermine NONOate and CysNO) were compared to understand their ability to form nitrosothiols73. Surprisingly, spermine NONOate (t1/2 = 39 min and 230 min) produces a high amount of dinitrosyliron complexes (DNIC) and very low amount of nitrosothiols. Conversely, Cys-NO (t1/2 ≤ 2min)73,74 efficiently produced nitrosothiols. Since the majority of nitrosylation proteomics experiments rely on nitric oxide donor-treated samples, much of the current literature should be carefully interpreted. While nitric oxide donors are important tools for many experiments, it is important in the future to focus on detecting endogenous nitrosothiols.
Emerging nitrosothiol enrichment strategies
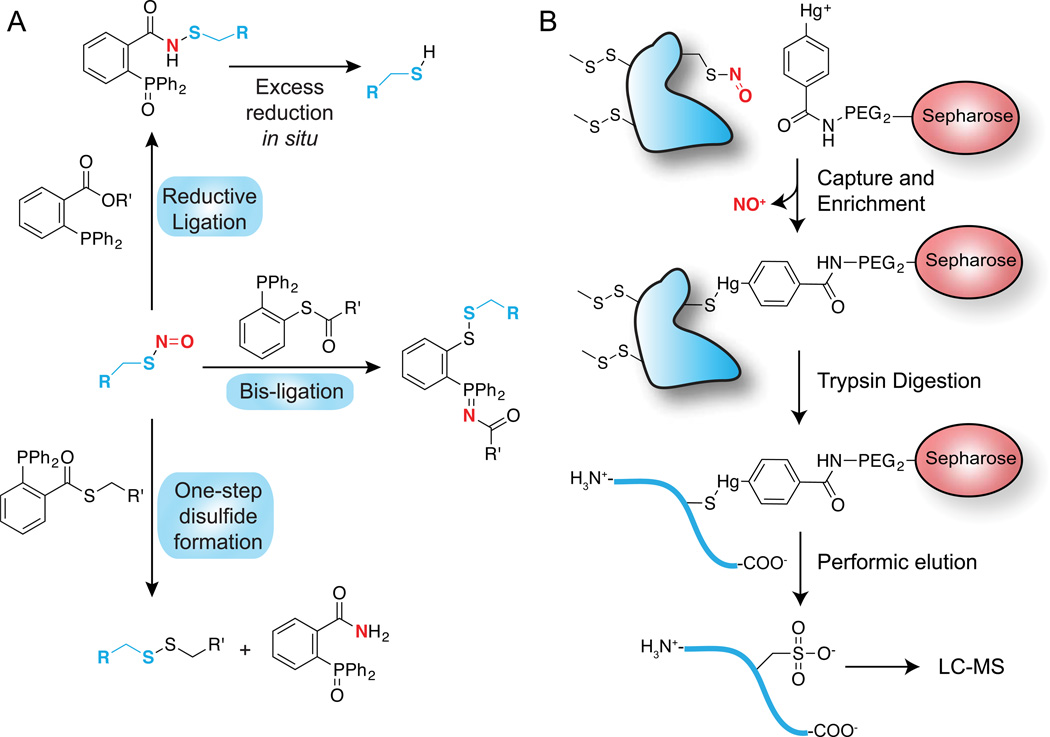
Given the limitations of the biotin-switch method, several alternative approaches have recently been reported (Figure 3A). In a series of innovative reports, Xian presents triaryl-substituted phosphines as a novel chemoselective reaction for conversion of nitrosothiols to a stable substituted thiobenzamide75,76. This reductive ligation reaction mechanism is similar to the Staudinger ligation, and is initiated by nitrosothiol reaction with the phosphine to form an azaylide, which then undergoes an intramolecular reaction and hydrolysis to yield the substituted thiobenzamide adduct. Additional variants of this reaction proceed by a similar azaylide intermediate, but undergo distinct rearrangements to yield varying products. The bis-ligation reaction uses phosphine-thioester probes to form disulfide-iminophosphorane products77. Importantly, the nitrogen originating from nitric oxide transforms to the iminophosphorane, providing an analytical linkage to both the originating nitric oxide and thiol. This methodology has been used to quantify the formation of S-nitrosoglutathione in activated macrophages by mass spectrometry, and allowed sensitive profiling of other nitrosylated metabolites in cell lysates78. The one-step disulfide method similarly uses a phosphine-thioester to first form the thiobenzamide adduct and thiolate, followed by intermolecular thioester exchange with the released thiolates to generate a disulfide linkage77. The reaction results in disulfide formation and elimination of the phosphine oxide. Finally, alkyl-aryl phosphines react with SNO generation of the azaylide, followed by reductive elimination to generate dehydroalanine79. Dehydrolalanine is an electrophilic Michael acceptor, and can be linked to a variety of tagged nucleophiles for straightforward chemoselective enrichment.
Figure 3.
Chemoselective nitrosothiol labeling methods. In both approaches, free thiols are first blocked by addition of MMTS. (A) Triaryl-phosphine ligation methods. Three reactions are shown that describe recent reports of nitrosothiol-selective phosphine reactions. The reductive ligation approach was demonstrated on fixed cells, but led to over-reduction to the free thiol. The one-step disulfide formation reaction was demonstrated on cell lysates after nitric oxide donor treatment. (B) Phenyl-mercury enrichment of nitrosothiols for proteomic annotation. Sepharose beads or biotin are linked to phenyl-mercury for nitrosothiol enrichment, followed by trypsin digestion. Nitrosylated peptides are released from the resin by perfomic acid oxidation to the sulfonic acid for mass spectrometry annotation.
In cell lysates, excess phosphine reagent led to complete reduction of nitrosothiols to free thiols75, suggesting these methods may require careful optimization. To avoid this pitfall, free thiols were first alkylated with NEM, followed by selective nitrosothiol reduction with triaryl-phosphines. The resulting free thiols were labeled using biotin or fluorophore-linked maleimide reagents for nitrosothiol detection. In gel-based experiments, phosphine reduction demonstrated superior selectivity to dithiothreitol, which non-selectively reduced disulfides and sulfenylated thiols75. This methodology was used in fixed cells to detect nitrosothiols after lipopolysaccharide stimulation in macrophages. Overall, these mechanistic phosphine probes show early promise as an alternative to ascorbate-dependent enrichment and proteomic analysis.
Organo-mercury enrichment methods have emerged as an alternative approach for direct nitrosothiol labeling and enrichment80,81 (Figure 3B). Nitroso-cysteines react directly with phenylmercury to yield a stable thiol-mercury bond. This reaction is direct, selective, and highly efficient. By coupling the phenylmercury to biotin or agarose beads, nitrosothiols can be directly labeled and enriched from tissue lysates. Bound peptides are then released from the resin by mild performic acid treatment, which oxidizes the thiol to the sulfonic acid for selective detection by mass spectrometry81. The fate of MMTS-capped thiols is not reported, although they are presumably simultaneously oxidized. This suggests MMTS should be replaced with iodoacetamide reagent to block free thiols before analysis. Furthermore, methionine is similarly oxidized by perfomic acid to the sulfone, adding additional complexity to the proteomic analysis. Nonetheless, this method lead to the identification of nearly a thousand nitrosylation sites summed across mouse brain, heart, liver, kidney, lung, and thymus tissues. Furthermore, about half or more of these sites were absent in eNOS knockout mice, suggesting the majority of nitrosylation originates from nitric oxide synthases80. Interestingly, more than 70% of the nitrosylation sites in heart tissue were found on mitochondrial proteins80. Further analysis suggests widespread regulation of metabolic enzymes in involved in glycolysis, gluconeogenesis, pyruvate metabolism, the Kreb’s cycle, oxidative phosphorylation, amino acid metabolism, ketone body formation, and fatty acid pathways. The simplicity of this method offers several advantages over ascorbate-dependent capture methods, particularly since it is a direct enrichment followed by a unique oxidation to sulfonate for direct proteomic annotation. Furthermore, this method is the first to report robust differences by comparative proteomic analysis of eNOS knockout mice. This data provides key evidence that nitric oxide synthase activation contributes to nitrosylation in vivo, and suggests supplementation with nitric oxide donors could have broader implications on cellular metabolism.
Conclusions
Protein S-nitrosylation has emerged as an important oxidative post-translational modification. Several labeling methods have emerged that take advantage of the unique reactivity, either by reduction and capture, or by direct chemical labeling. Importantly, such methodologies have enabled proteomic analysis of S-nitrosylation in cells and tissues. In conjunction with isotopic labeling methods, quantitative proteomic profiling of S-nitrosylation will enable in-depth global profiling, independent of nitric oxide donors. With the development of selective methods for each distinct cysteine post-translational modification, multiplexed analysis has the potential to discover new cellular pathways orchestrated post-translational exchanges. Such analysis will integrate palmitoylation dynamics, and explore the mutual competition at distinct cysteine residues between nitric oxide and hydrogen peroxide. Importantly, each of these modifications are labile, and rapidly hydrolyzed in the presence of free thiols, such as glutathione. Therefore, we anticipate such tools will enable a greater understanding of the role of compartmentalization in thiol modification stability and function.
Acknowledgements
We would like to thank Mi Hee Lim (U. Michigan) and members of the Martin lab for helpful discussions. Funding is provided by the National Institutes of Health (R00CA151460) and the University of Michigan.
References
- 1.Cannan RK, Knight B. Biochemical Journal. 1927;21:1384–1390. doi: 10.1042/bj0211384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lim JC, Gruschus JM, Kim G, Berlett BS, Tjandra N, Levine RL. Journal of Biological Chemistry. 2012;287:25596–25601. doi: 10.1074/jbc.M112.369116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoine M, Gand A, Boschi-Muller S, Branlant G. Journal of Biological Chemistry. 2006;281:39062–39070. doi: 10.1074/jbc.M608844200. [DOI] [PubMed] [Google Scholar]
- 4.Jao S-C, Ospina English SM, Berdis AJ, Starke DW, Post CB, Mieyal J. J Biochemistry. 2006;45:4785–4796. doi: 10.1021/bi0516327. [DOI] [PubMed] [Google Scholar]
- 5.Gan ZR, Polokoff MA, Jacobs JW, Sardana MK. Biochemical and Biophysical Research Communications. 1990;168:944–951. doi: 10.1016/0006-291x(90)91120-h. [DOI] [PubMed] [Google Scholar]
- 6.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Nature Reviews Molecular Cell Biology. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 7.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Nature Cell Biology. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 8.Bredt DS, Snyder SH. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:682–685. doi: 10.1073/pnas.87.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busse R, Mulsch A. Febs Letters. 1990;265:133–136. doi: 10.1016/0014-5793(90)80902-u. [DOI] [PubMed] [Google Scholar]
- 10.Hope BT, Michael GJ, Knigge KM, Vincent SR. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:2811–2814. doi: 10.1073/pnas.88.7.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lima B, Forrester MT, Hess DT, Stamler JS. Circulation Research. 2010;106:633–646. doi: 10.1161/CIRCRESAHA.109.207381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seth D, Stamler JS. Current Opinion in Chemical Biology. 2011;15:129–136. doi: 10.1016/j.cbpa.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee T-Y, Chen Y-J, Lu C-T, Ching W-C, Teng Y-C, Huang H-D, Chen Y-J. Bioinformatics. 2012;28:2293–2295. doi: 10.1093/bioinformatics/bts436. [DOI] [PubMed] [Google Scholar]
- 14.Maron BA, Tang SS, Loscalzo J. Antioxidants & Redox Signaling. 2013;18:270–287. doi: 10.1089/ars.2012.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murray CI, Uhrigshardt H, O'Meally RN, Cole RN, Van Eyk JE. Molecular & Cellular Proteomics. 2012;11 doi: 10.1074/mcp.M111.013441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiktorowicz JE, Stafford S, Rea H, Urvil P, Soman K, Kurosky A, Perez-Polo JR, Savidge TC. Biochemistry. 2011;50:5601–5614. doi: 10.1021/bi200008b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakuma I, Stuehr DJ, Gross SS, Nathan C, Levi R. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:8664–8667. doi: 10.1073/pnas.85.22.8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palmer RMJ, Ashton DS, Moncada S. Nature. 1988;333:664–666. doi: 10.1038/333664a0. [DOI] [PubMed] [Google Scholar]
- 19.Marletta MA, Yoon PS, Iyengar R, Leaf CD, Wishnok JS. Biochemistry. 1988;27:8706–8711. doi: 10.1021/bi00424a003. [DOI] [PubMed] [Google Scholar]
- 20.Palmer RMJ, Moncada S. Biochemical and Biophysical Research Communications. 1989;158:348–352. doi: 10.1016/s0006-291x(89)80219-0. [DOI] [PubMed] [Google Scholar]
- 21.Leone AM, Palmer RMJ, Knowles RG, Francis PL, Ashton DS, Moncada S. Journal of Biological Chemistry. 1991;266:23790–23795. [PubMed] [Google Scholar]
- 22.Padgett CM, Whorton AR. Cell Biochemistry and Biophysics. 1995;27:157–177. doi: 10.1007/BF02738108. [DOI] [PubMed] [Google Scholar]
- 23.Pryor WA, Church DF, Govindan CK, Crank G. Journal of Organic Chemistry. 1982;47:156–159. [Google Scholar]
- 24.Kharitonov VG, Sundquist AR, Sharma VS. Journal of Biological Chemistry. 1995;270:28158–28164. doi: 10.1074/jbc.270.47.28158. [DOI] [PubMed] [Google Scholar]
- 25.Stamler JS, Singel DJ, Loscalzo J. Science. 1992;258:1898–1902. doi: 10.1126/science.1281928. [DOI] [PubMed] [Google Scholar]
- 26.Logager T, Sehested K. Journal of Physical Chemistry. 1993;97:6664–6669. [Google Scholar]
- 27.Jourd'heuil D, Jourd'heuil FL, Feelisch M. Journal of Biological Chemistry. 2003;278:15720–15726. doi: 10.1074/jbc.M300203200. [DOI] [PubMed] [Google Scholar]
- 28.Saez G, Thornalley PJ, Hill HAO, Hems R, Bannister JV. Biochimica Et Biophysica Acta. 1982;719:24–31. doi: 10.1016/0304-4165(82)90302-6. [DOI] [PubMed] [Google Scholar]
- 29.Stubauer G, Giuffre A, Sarti P. Journal of Biological Chemistry. 1999;274:28128–28133. doi: 10.1074/jbc.274.40.28128. [DOI] [PubMed] [Google Scholar]
- 30.Vanin AF, Malenkova IV, Serezhenkov VA. Nitric Oxide-Biology and Chemistry. 1997;1:191–203. doi: 10.1006/niox.1997.0122. [DOI] [PubMed] [Google Scholar]
- 31.Boese M, Mordvintcev PI, Vanin AF, Busse R, Mulsch A. Journal of Biological Chemistry. 1995;270:29244–29249. doi: 10.1074/jbc.270.49.29244. [DOI] [PubMed] [Google Scholar]
- 32.Scott MJ, Holm RH. Angewandte Chemie-International Edition in English. 1993;32:564–566. [Google Scholar]
- 33.Harrop TC, Song DT, Lippard SJ. Journal of the American Chemical Society. 2006;128:3528–3529. doi: 10.1021/ja060186n. [DOI] [PubMed] [Google Scholar]
- 34.Tinberg CE, Tonzetich ZJ, Wang H, Do LH, Yoda Y, Cramer SP, Lippard SJ. Journal of the American Chemical Society. 2010;132:18168–18176. doi: 10.1021/ja106290p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gundimeda U, Chen ZH, Gopalakrishna R. Faseb Journal. 1993;7:A1120–A1120. [Google Scholar]
- 36.Arstall MA, Bailey C, Gross WL, Balligand JL, Smith TW, Kelly RA. Circulation. 1996;94:3041–3041. [Google Scholar]
- 37.Whiteside LM, Young PR. Biophysical Journal. 2001;80:280A–280A. [Google Scholar]
- 38.Tsikas D, Sandmann J, Luessen P, Savva A, Rossa S, Stichtenoth DO, Frolich JC. Biochimica Et Biophysica Acta-Protein Structure and Molecular Enzymology. 2001;1546:422–434. doi: 10.1016/s0167-4838(01)00166-2. [DOI] [PubMed] [Google Scholar]
- 39.Wu CG, Parrott AM, Liu T, Jain MR, Yang YF, Sadoshima J, Li H. Journal of Proteomics. 2011;74:2498–2509. doi: 10.1016/j.jprot.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakamura T, Lipton SA. Antioxidants & Redox Signaling. 2013;18:239–249. doi: 10.1089/ars.2012.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hwang C, Sinskey AJ, Lodish HF. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 42.Bracher PJ, Snyder PW, Bohall BR, Whitesides GM. Origins of Life and Evolution of Biospheres. 2011;41:399–412. doi: 10.1007/s11084-011-9243-4. [DOI] [PubMed] [Google Scholar]
- 43.Tsikas D, Sandmann J, Rossa S, Gutzki FM, Frolich JC. Analytical Biochemistry. 1999;270:231–241. doi: 10.1006/abio.1999.4084. [DOI] [PubMed] [Google Scholar]
- 44.Nakamura T, Wang L, Wong CCL, Scott FL, Eckelman BP, Han X, Tzitzilonis C, Meng F, Gu Z, Holland EA, Clemente AT, Okamoto S-i, Salvesen GS, Riek R, Yates JR, III, Lipton SA. Molecular Cell. 2010;39:184–195. doi: 10.1016/j.molcel.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moller MN, Li Q, Vittur DA, Robinson JM, Lancaster JR, Jr, Denicola A. Chemical Research in Toxicology. 2007;20:709–714. doi: 10.1021/tx700010h. [DOI] [PubMed] [Google Scholar]
- 46.Brandes N, Schmitt S, Jakob U. Antioxidants & Redox Signaling. 2009;11:997–1014. doi: 10.1089/ars.2008.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pei D-S, Sun Y-F, Song Y-J. Neurochemical Research. 2009;34:1507–1512. doi: 10.1007/s11064-009-9938-3. [DOI] [PubMed] [Google Scholar]
- 48.Shi Z-Q, Sunico CR, McKercher SR, Cui J, Feng G-S, Nakamura T, Lipton SA. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3137–3142. doi: 10.1073/pnas.1215501110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwak Y-D, Ma T, Diao S, Zhang X, Chen Y, Hsu J, Lipton SA, Masliah E, Xu H, Liao F-F. Molecular Neurodegeneration. 2010;5 doi: 10.1186/1750-1326-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Padgett CM, Whorton AR. American Journal of Physiology-Cell Physiology. 1995;269:C739–C749. doi: 10.1152/ajpcell.1995.269.3.C739. [DOI] [PubMed] [Google Scholar]
- 51.Kornberg MD, Sen N, Hara MR, Juluri KR, Nguyen JVK, Snowman AM, Law L, Hester LD, Snyder SH. Nature Cell Biology. 2010;12:U1094–U1089. doi: 10.1038/ncb2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hara MR, Agrawal N, Kim SF, Cascio MB, Fujimuro M, Ozeki Y, Takahashi M, Cheah JH, Tankou SK, Hester LD, Ferris CD, Hayward SD, Snyder SH, Sawa A. Nature Cell Biology. 2005;7:U665–U640. doi: 10.1038/ncb1268. [DOI] [PubMed] [Google Scholar]
- 53.Hattori Y, Campbell EB, Gross SS. Journal of Biological Chemistry. 1994;269:9405–9408. [PubMed] [Google Scholar]
- 54.Hao G, Xie LJ, Gross SS. Journal of Biological Chemistry. 2004;279:36192–36200. doi: 10.1074/jbc.M404866200. [DOI] [PubMed] [Google Scholar]
- 55.Ho GPH, Selvakumar B, Mukai J, Hester LD, Wang YX, Gogos JA, Snyder SH. Neuron. 2011;71:131–141. doi: 10.1016/j.neuron.2011.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Nat Methods. 2012;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tom CT, Martin BR. ACS Chem Biol. 2013;8:46–57. doi: 10.1021/cb300607e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wedegaertner PB, Bourne HR. Cell. 1994;77:1063–1070. doi: 10.1016/0092-8674(94)90445-6. [DOI] [PubMed] [Google Scholar]
- 59.Wink DA, Kim S, Coffin D, Cook JC, Vodovotz Y, Chistodoulou D, Jourd'heuil D, Grisham MB. Nitric Oxide, Pt C. 1999;301:201–211. doi: 10.1016/s0076-6879(99)01083-6. [DOI] [PubMed] [Google Scholar]
- 60.SR J, SH S. Science Signaling. 2001;2001:1. [Google Scholar]
- 61.Kirsch M, Buescher A-M, Aker S, Schulz R, de Groot H. Organic & Biomolecular Chemistry. 2009;7:1954–1962. doi: 10.1039/b901046g. [DOI] [PubMed] [Google Scholar]
- 62.Holmes AJ, Williams DLH. Journal of the Chemical Society-Perkin Transactions 2. 2000:1639–1644. [Google Scholar]
- 63.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kohr MJ, Sun JH, Aponte A, Wang GH, Gucek M, Murphy E, Steenbergen C. Circulation Research. 2011;108:U418–U450. doi: 10.1161/CIRCRESAHA.110.232173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Nature Biotechnology. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams DLH. Accounts of Chemical Research. 1999;32:869–876. [Google Scholar]
- 67.Grossi L, Montevecchi PC. Chemistry-a European Journal. 2002;8:380–387. doi: 10.1002/1521-3765(20020118)8:2<380::AID-CHEM380>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 68.Forrester MT, Foster MW, Stamler JS. Journal of Biological Chemistry. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- 69.Goldstein S, Czapski G. Journal of the American Chemical Society. 1996;118:3419–3425. doi: 10.1021/ja00269a020. [DOI] [PubMed] [Google Scholar]
- 70.Goldstein S, Czapski G. Journal of the American Chemical Society. 1995;117:12078–12084. doi: 10.1021/ja00269a020. [DOI] [PubMed] [Google Scholar]
- 71.Czapski G, Goldstein S. Free Radical Biology and Medicine. 1995;19:785–794. doi: 10.1016/0891-5849(95)00081-8. [DOI] [PubMed] [Google Scholar]
- 72.Zhang YH, Hogg N. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2004;287:L467–L474. doi: 10.1152/ajplung.00350.2003. [DOI] [PubMed] [Google Scholar]
- 73.Hickok JR, Vasudevan D, Thatcher GRJ, Thomas DD. Antioxidants & Redox Signaling. 2012;17:962–968. doi: 10.1089/ars.2012.4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Singh SP, Wishnok JS, Keshive M, Deen WM, Tannenbaum SR. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:14428–14433. doi: 10.1073/pnas.93.25.14428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li S, Wang H, Xian M, Whorton AR. Nitric Oxide. 2012;26:20–26. doi: 10.1016/j.niox.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang JM, Wang H, Xian M. Organic Letters. 2009;11:477–480. doi: 10.1021/ol802663q. [DOI] [PubMed] [Google Scholar]
- 77.Zhang JM, Wang H, Xian M. Journal of the American Chemical Society. 2009;131 doi: 10.1021/ja905558w. 3854-+. [DOI] [PubMed] [Google Scholar]
- 78.Seneviratne UI, Godoy LC, Wishnok JS, Wogan GN, Tannenbaum SR. J Am Chem Soc. 2013 doi: 10.1021/ja401565w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang H, Zhang JM, Xian M. Journal of the American Chemical Society. 2009;131 doi: 10.1021/ja905558w. 13238-+. [DOI] [PubMed] [Google Scholar]
- 80.Doulias P-T, Tenopoulou M, Greene JL, Raju K, Ischiropoulos H. Science Signaling. 2013;6 doi: 10.1126/scisignal.2003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Doulias P-T, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beckman JS, Koppenol WH. American Journal of Physiology-Cell Physiology. 1996;271:C1424–C1437. doi: 10.1152/ajpcell.1996.271.5.C1424. [DOI] [PubMed] [Google Scholar]
- 83.Fukuto JM, Switzer CH, Miranda KM, Wink DA. Annual Review of Pharmacology and Toxicology. 2005;45:335–355. doi: 10.1146/annurev.pharmtox.45.120403.095959. [DOI] [PubMed] [Google Scholar]
- 84.Radi R, Beckman JS, Bush KM, Freeman BA. Journal of Biological Chemistry. 1991;266:4244–4250. [PubMed] [Google Scholar]
- 85.Halliwell B, Gutteridge JM. Free Radicals in Biology and Medicine. Oxford University Press; 2007. [Google Scholar]