Abstract
Protein palmitoylation describes the hydrophobic post-translational modification of cysteine residues in certain proteins, and is required for the spatial organization and composition of cellular membrane environments. Certain palmitoylated proteins are processed by acyl protein thioesterase (APT) enzymes, which catalyze thioester hydrolysis of palmitoylated cysteine residues. Inhibiting APT enzymes disrupts Ras trafficking and attenuates oncogenic growth signaling, highlighting these enzymes as potential therapeutic targets. As members of the serine hydrolase enzyme family, APT enzymes can be assayed by fluorophosphonate activity-based protein profiling (ABPP) methods, allowing rapid profiling of inhibitor selectivity and potency. In this review, we discuss recent progress in the development of potent and selective inhibitors to APT enzymes, including both competitive and non-competitive chemotypes. These examples highlight how ABPP methods integrate with medicinal chemistry for the discovery and optimization of inhibitors in complex proteomes.
Introduction
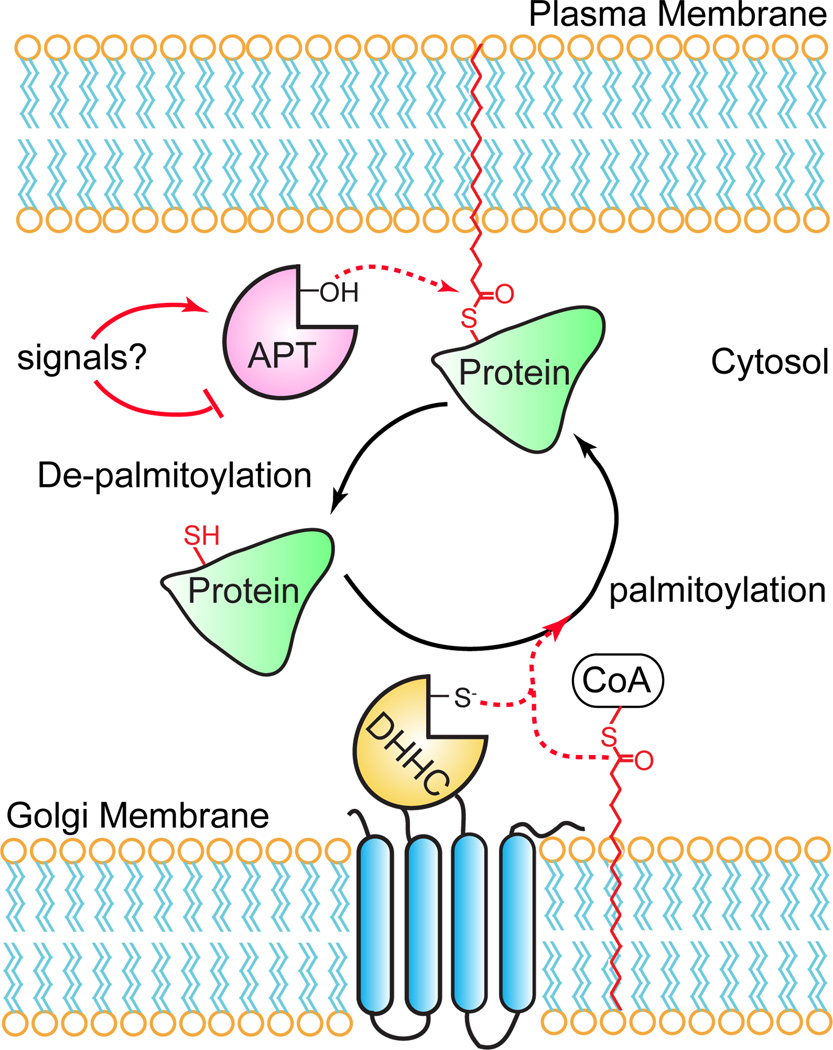
Protein palmitoylation describes the post-translational attachment of a long-chain fatty acid to a cysteine residue1. This hydrophobic modification is critical for the function, trafficking, and localization of diverse proteins with central roles in adhesion, signaling, and organelle structure. Recent proteomics studies have annotated hundreds of palmitoylated proteins2–8, uncovering a widespread role for such a hydrophobic modification. Due to the unstable thioester linkage, protein palmitoylation undergoes steady hydrolysis in cells, generally with a half-life of less than a few hours. Unlike static modifications like protein prenylation or myristoylation, protein palmitoylation provides a unique mechanism to reversibly regulate the membrane association of modified proteins. Indeed, dynamic palmitoylation of oncogenic HRas is required to mediate cell transformation9. Furthermore, activation of Ras or the G-protein Gαs stimulates palmitoylation turnover10, 11, suggesting palmitoylation dynamics may be an essential trigger in the activation or deactivation of distinct signaling pathways. In this model, protein palmitoyl thioesterases catalyze the de-palmitoylation and release of certain proteins from the plasma membrane, potentially attenuating membrane-coupled signaling pathways (Figure 1). The released proteins can then undergo re-palmitoylation by endomembrane-bound DHHC palmitoyl-transferases, restoring palmitoylation and resetting the cycle.
Figure 1. A dynamic S-palmitoylation cycle regulates the membrane association and activity of certain palmitoylated proteins.
Certain stimuli cause an acyl protein thioesterase to catalyze thioester hydrolysis of palmitoylated cysteine residues. De-palmitoylation liberates the protein from the plasma membrane, allowing it to segregate to the cytosol disrupting interactions with membrane-associated signaling complexes. The protein can then undergo re-palmitoylation by Golgi-resident DHHC palmitoyl transferases, transport back to the plasma membrane, and re-associate with signaling partners.
Given the evidence that HRas participates in an acylation/deacylation cycle12, a putative HRas thioesterase termed acyl protein thioesterase (APT1) was identified from soluble rat liver fractions13. This enzyme was previously characterized as lysophospholipase I (LYPLA1)14, but has several hundred-fold higher activity as a protein thioesterase13. APT1 removes palmitate from diverse palmitoyl protein substrates in vitro13, 15. Genetic deletion of the yeast homologue of APT1 shows a minor decrease in the turnover of palmitate on Gαs, yet has no defects in growth, mating, or deacylation of other palmitoylated proteins16. Despite these results, lysates from APT1 mutant yeast lack in vitro Gαs palmitoyl protein thioesterase activity16. Importantly, deletion of APT1 has no effect on lysophosphatidylcholine (LPC) hydrolysis activity, suggesting it is not a functional lysophospholipase16.
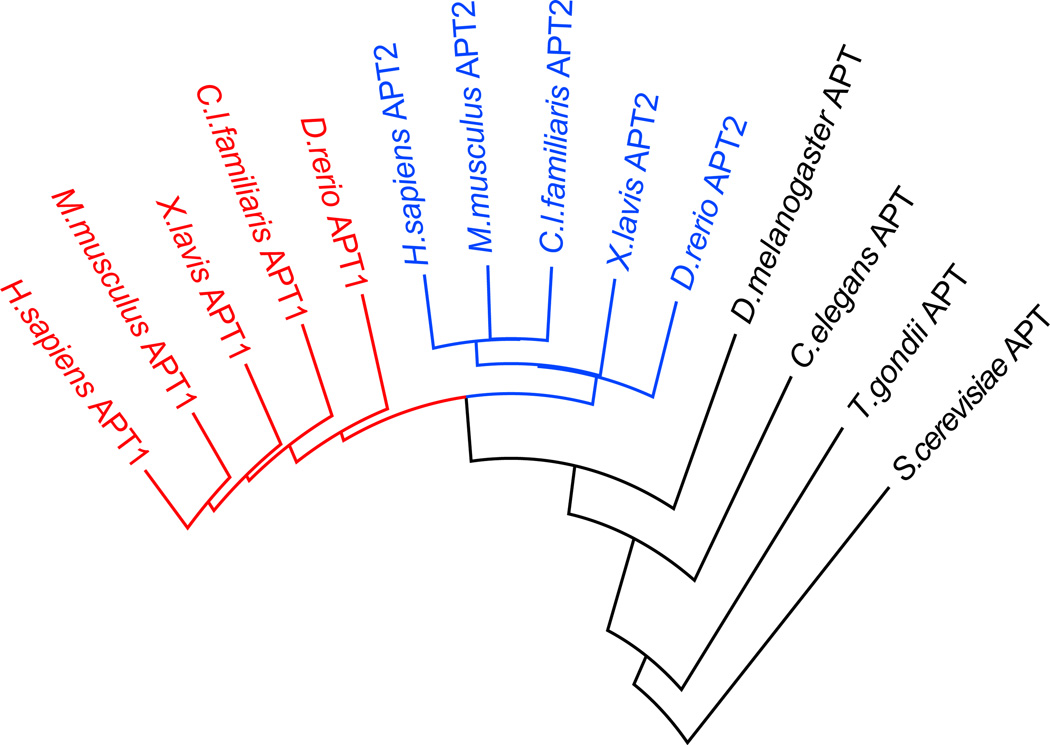
A second lysophospholipase II (APT2), is 68% homologous to APT117, but surprisingly is slightly more efficient than APT1 at de-palmitoylating semi-synthetic NRas18. The axonal protein GAP-43 or HRas are more rapidly de-palmitoylated after APT2 over-expression19. Phylogenic analysis shows that invertebrates evolved only one APT enzyme, while vertebrates evolved two (APT1 and APT2). (Figure 2). Both APT1 and APT2 have themselves been identified as palmitoylated proteins7, 20, although the predicted palmitoylation site is not conserved in canine APT1 or invertebrate APTs. In mammalian neurons, APT1-mRNA is translationally repressed by a synaptic microRNA complex21, 22. Upon glutamate stimulation, the repressive complex is degraded to allow local translation of APT1 in synaptic spines22. APT1 expression induces a reduction in spine volume potentially by de-palmitoylating Gα13 and reducing synaptic RhoA activation21. In addition, a recent in vivo RNAi screen identified APT2 as a candidate regulator of oncogenic HRas-dependent malignancy23. A third more distant homologue lysophospholipase-like 1 (LYPLAL1) contains a narrow substrate-binding pocket unable to accommodate long-chain fatty acids24, and genetic studies support a role in obesity25. Overall, APT enzymes represent candidate protein palmitoyl thioesterases, but little is known about their substrates or dependence on upstream signals.
Figure 2. Shared homology between vertebrate and invertebrate acyl protein thioesterases.
Vertebrates evolved two distinct APT enzymes, whereas invertebrates evolved a single APT enzyme. Phylogeny alignments were generated using DIALIGN, anchored at the catalytic serine residue.
To address the proteome-wide dynamics and the contribution of enzymatic de-palmitoylation, we recently applied non-radioactive bioorthogonal labeling approaches to selectively enrich palmitoylated proteins with enhanced turnover dynamics. This approach uses the commercially available fatty acid analogue 17-octadecynoic acid, which upon exogenous addition, is efficiently incorporated into endogenous sites of protein palmitoylation2, 26. After sufficient labeling, harvested cell lysates are conjugated to biotin-azide by copper-catalyzed click chemistry for subsequent streptavidin enrichment and mass spectrometry annotation. Using “pulse-chase” labeling methods, we profiled de-palmitoylation dynamics in mouse T-cells. Cells were first labeled with 17-ODYA, then after some time “chased” with excess palmitic acid with or without the non-selective lipid-like serine hydrolase inhibitor hexadecylfluorophosphonate (HDFP)5. This hydrophobic mechanism-based covalent inhibitor targets approximately 20 serine hydrolases characterized by their common preference for lipid-like substrates, including APT1 and APT2, but not LYPLAL1. We hypothesized that any candidate thioesterase is likely a serine hydrolase inhibited by HDFP. Quantitative proteomic analysis identified a subpopulation (<10%) of palmitoylated proteins that undergo rapid cycling mediated by one or more serine hydrolases targeted by HDFP5. This subset includes Ras family GTPases, G-proteins, and scaffolding proteins implicated in malignancy. This data confirms that enzymes functionally contribute to palmitoylation hydrolysis in cells, and HDFP inhibition is sufficient to block any enzyme-mediated palmitoylation turnover. Furthermore, given the relatively small subset of oncogenic signaling and scaffolding proteins, inhibiting de-palmitoylation may perturb key signaling pathways involved in malignancy.
APT enzymes are members of the α/β-hydrolase family of serine hydrolases27. The catalytic serine nucleophile forms a covalent acyl-intermediate with substrates, providing the opportunity to develop covalent mechanism-based inhibitors. Over the last decade several small molecule inhibitors of APTs have been described1. These include both competitive and non-competitive inhibitors, which have been useful tools in several studies supporting a functional role for APTs in enzymatic depalmitoylation. APT inhibition is reported to suppress MAPK signaling and attenuate growth of certain Ras-transduced hematopoietic cells28, 29, as well as modulating the membrane association of the estrogen receptor in hippocampal slices30. APT inhibition also enhances invasion of the parasitic protozoan T.gondii into human cells31, 32, suggesting unique regulation across evolution. In this review, we describe each reported APT inhibitor chemotype, including details regarding potency, selectivity, and biological evaluation. This includes a discussion on the application of competitive activity-based protein profiling (ABPP) methods to profile inhibitor selectivity and target occupancy in biological systems33.
Benzodiazepinediones
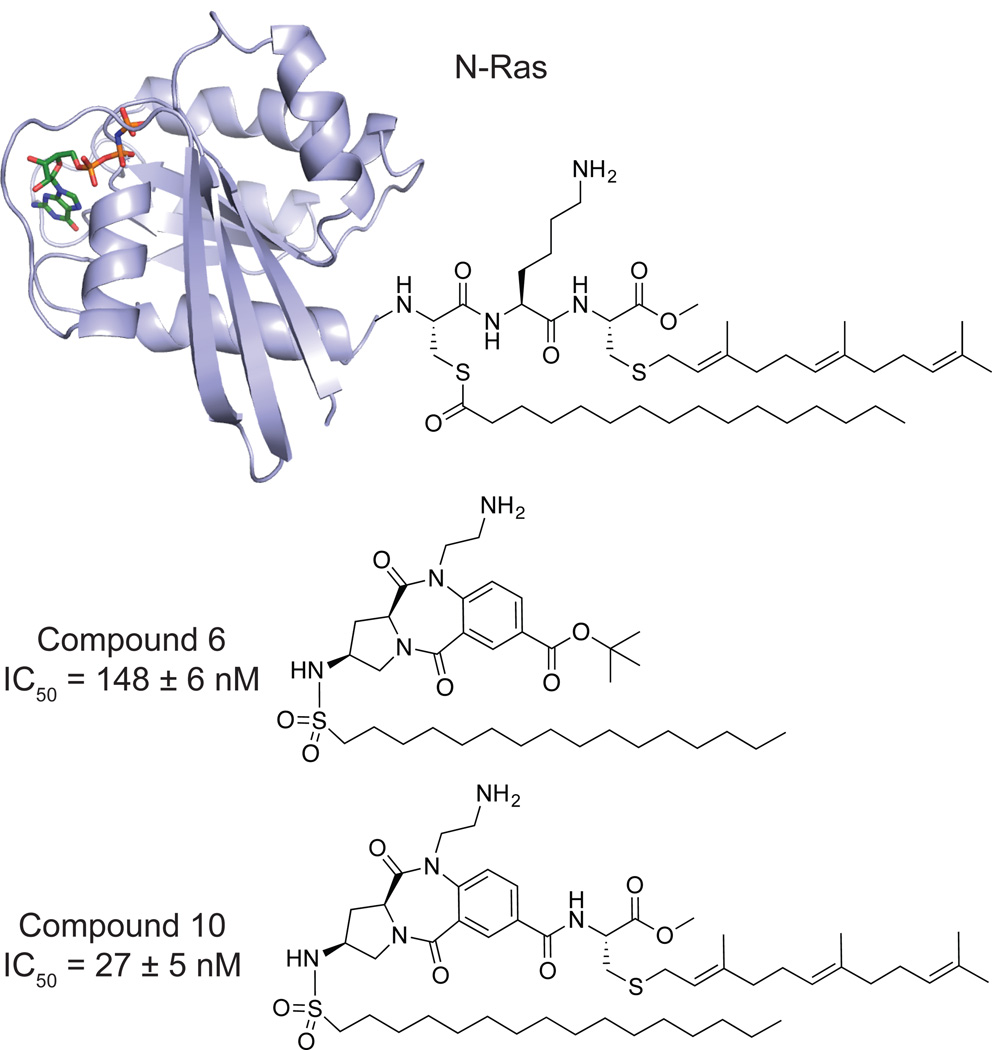
Early efforts towards the development of APT1 inhibitors were rationally designed based on substrate mimicry of a dually palmitoylated and farnesylated peptide (Figure 3). A peptide-imitating benzodiazepinedione core was fused to S-farnesyl cysteine methyl ester and an acyl-sulfonamide to mimic the serine hydrolase tetrahedral intermediate34, yielding a low nanomolar APT1 inhibitor (Compound 10, IC50 = 27 nM). Intermediates lacking the farnesyl moiety were 5-times less potent (Compound 6, IC50 = 148 nM), but presumably more soluble, and were tested in cells. Cells microinjected with the inhibitor showed complete mis-localization of NRas away from the plasma membrane, disrupting NRas-dependent PC12 cell neuronal differentiation. Adding 30 µM inhibitor to the cell growth media caused a similar block of neurite outgrowth, suggesting efficient cellular uptake. These impressive initial compounds confirmed the importance of APT enzymes in modulating Ras dynamics in cells, and inspired further efforts to develop more drug-like APT inhibitors.
Figure 3. Lipidated N-Ras and peptido-mimetic benzodiazepinediones.
Lipopeptide mimics for targeting APT1 are based on the C-terminal portion of lipidated N-Ras (Top, PDB 5P21). A peptide-resembling benzodiazepinedione core serves as a central scaffold equipped with both a primary amine to mirror the lysine side-chain and an essential hydrolysis-resistant hexadecylsulfonamide moiety imitating a palmitoyl thioester tetrahedral intermediate. (Middle, 6). 6 was further functionalized with a farnesylated cysteine methyl ester to yield a mimic of the C terminus of a fully processed N-Ras (Bottom, 10) which exhibited potent inhibitory activity.
β-lactones
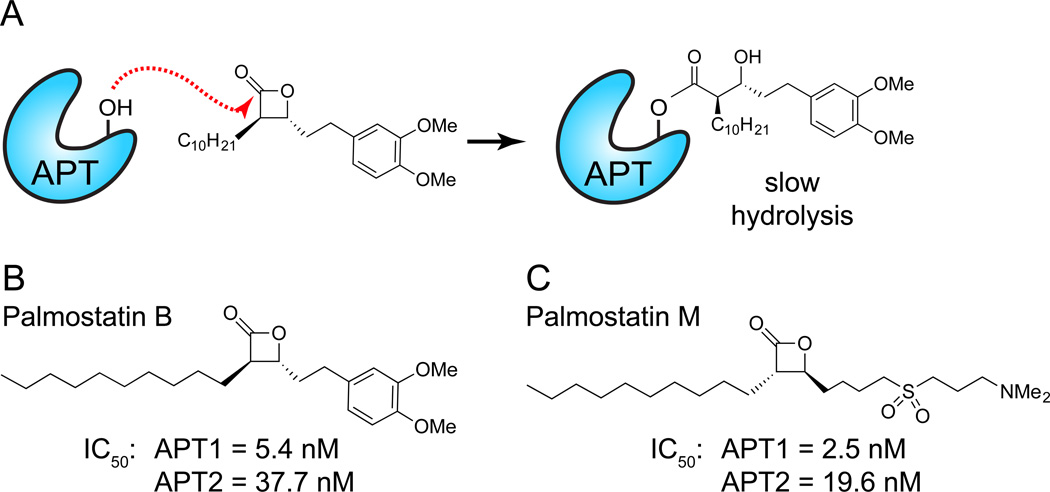
β-lactone serine hydrolase inhibitors are best characterized by the over-the-counter weight loss drug tetrahydrolipstatin (THL or Orlistat)35. Originally identified as a natural product from Streptomyces toxytricini36, THL is representative of diverse bacterial β-lactones with functional roles as bacterial effectors. Upon oral delivery, THL is a mechanism-based inhibitor of several digestive lipases that acts by preventing fatty acid intestinal absorption. After nucleophilic attack of the lactone ester and ring opening, the subsequent acyl-enzyme hydrolysis is slow, rendering the enzyme predominantly in an inactive state37 (Figure 4A). In order to discover novel APT1 inhibitors, a series of β-lactone derivatives were synthesized and profiled against APT1, leading to the discovery of the chiral acyl-β-lactone Palmostatin B (IC50 = 5.4 nM)28, 38 (Figure 4B). Palmostatin B is readily membrane-permeable, and directly reacts with APT1 and APT2 as both a substrate and an inhibitor in live cells28.
Figure 4. Mechanism-based β-lactones inhibit APT enzymes.
(A) APT competitive enzyme inhibition by Palmostatin B. Attack of the electrophilic lactone by the catalytic serine hydroxyl leads to a transient acyl-enzyme adduct, which slowly hydrolyzes to regenerate the active enzyme. (B) Structure of Palmostatin B. This inhibitor was discovered through the synthesis and screening of a small library of compounds on the basis of active-site similarity between APT1 and gastric lipase, and the fact that β-lactones are known lipase acylating reagents. (C) Structure of Palmostatin M. This inhibitor was developed based on the structural similarity of lysophosphatidyl choline. A common recognition motif was identified, consisting of an electropositive dimethylamino tail imitating lysine and choline, an electronegative sulfoxide to mirror the phosphate, a central electrophilic trans-β-lactone and a lipophilic tail resembling the palmitate moiety to create affinity to the lipid-binding pocket of the enzyme.
To study the effects of APT inhibition, Palmostatin B treated cells were imaged after microinjection of fluorescently labeled NRas, or following transfection of YFP-NRas in MDCK cells28. Palmostatin B addition led to YFP-NRas mis-localization and decreased compartmentalized NRas activation. These experiments likely required high nanomolar concentrations of labeled NRas for fluorescence imaging, which may encroach on the APT1 Km for palmitoylated NRas (~1 µM)18 and potentially enhance any observed phenotypes. Further experiments showed that Palmostatin B (50 µM) altered the morphology of oncogenic HRas transformed cells, restoring them to a less spindly appearance with increased E-Cadherin membrane localization. Palmostatin B displays poor stability in cell culture, and was added to cells at effectively 104-times the IC50, which may diminish selectivity. Interestingly, siRNA knockdown of APT1 did not statistically alter the ratio of golgi to plasma membrane NRas. This suggests either insufficient APT1 knockdown, or that Palmostatin B inhibits other protein thioesterases, such as APT2 (Ki = 34 nM)18 that may contribute to NRas processing. Further experiments showed that addition of 30 - 100 µM of Palmostatin B dose-dependently impedes the growth of HRas or NRas transduced hematopoietic cells, but does not affect cells transduced with a non-palmitoylated isoform of KRas29. Such high inhibitor concentrations may inhibit other intracellular hydrolases, but clearly define a role for de-palmitoylation in Ras regulation. Overall, Palmostatin B has provided important pharmacological evidence of protein palmitoylation dynamics in live cells.
In efforts to selectively target APT isoforms, a polar sulfone moiety was added the β-lactone scaffold to mimic the polarity of lysophosphatidylcholine (Figure 4C), yielding the improved analogue Palmostatin M (APT1 IC50 = 2.5 nM)38. Despite synthesis of a diverse library of greater than 50 compounds, no APT1 or APT2 isoform selective inhibitors were identified, which may signal a structural limitation of the β-lactone scaffold. Palmostatin M similarly induces NRas mis-localization from the plasma membrane and partially restores E-Cadherin localization at cell-cell junctions in HRas-transformed MDCK cells38. Because β-lactones form a semi-stable covalent complex with their targets, an alkynyl Palmostatin M analogue was synthesized to annotate potential cellular targets18. Lysates were labeled with the alkynyl probes, conjugated to a tri-functional rhodamine/biotin-azide by copper-catalyzed click chemistry, enriched with streptavidin, and analyzed by in-gel fluorescence or mass spectrometry to identify candidate target proteins. These experiments confirmed APT1 and APT2 as major Palmostatin M targets, but also identified other serine hydrolases including the lysosomal thioesterase PPT1 and retinoid-inducible serine carboxypeptidase (RISC). While these compounds are exceptionally potent, non-specific inhibition and poor drug-like properties limit the application of these probes for in vivo studies.
Boronic Acids
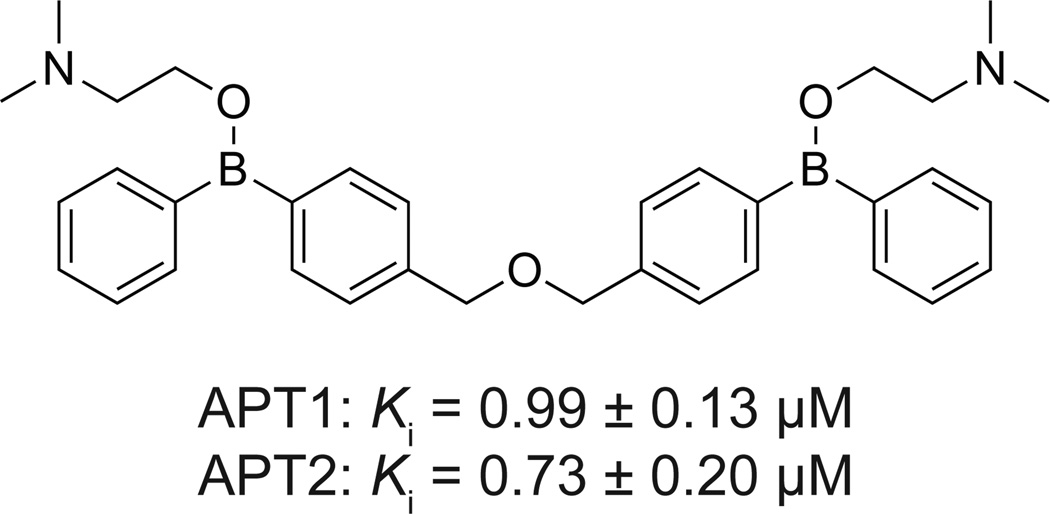
In order to discover new chemotypes that inhibit APT enzymes, an immobilized library of <15,000 natural products and inhibitors was assayed for APT binding39. In this approach, inhibitors were covalently attached to glass using a trifluoromethylaryldiazirine photo-crosslinking system, and then incubated with recombinant GST-fusions of APT1 or APT2. Binding was detected after incubation with anti-GST antibodies visualization of fluorescent secondary antibodies. Four similar bis-borinic acid and bisborinate ester structures were identified (Figure 5), which were further validated by surface plasmon resonance. Boron-based inhibitors of serine hydrolases act by boronate coordination of the side chain hydroxyl nucleophile of the active serine residue, mimicking the tetrahedral intermediate40. Several boron-containing drugs are in clinical use, including the proteasome inhibitor Bortezomib (Velcade), the DPP4 inhibitor Januvia, β-lactamase inhibitors, and many more under development40. Kinetic analysis of the diphenylboronic acids showed potent inhibition of APT1 (Ki = 0.99 ± µM) and APT2 (Ki = 0.73 ± 0.20 µM)39. Other analogues were slightly less potent, but showed a slight preference towards APT2. Similar to Palmostatin B, the diphenylboronate ester inhibitors demonstrated a dose-dependent inhibition of Erk1/2 phosphorylation and induced a less spindly phenotype in HRas transduced MDCK cells. While this study does not provide any data on the selectivity of these analogues, it does validate boron-containing inhibitors as a new APT inhibitor chemotype, and provides further phenotypic support that APT enzymes functionally regulate Ras-dependent signaling pathways.
Figure 5. Bis-boronic acid inhibitor of APT enzymes.
Representative bis-boronic acid-based inhibitor (Compound 4) and reported inhibitory values for both APT1 and APT2.
Triazole Ureas
Significant progress has been made in the development of selective serine hydrolase inhibitors through optimization of carbamates, leading to analogues with exquisite selectivity and potency as covalent probes41. Despite considerable effort, carbamates fail to reach a broad subset of serine hydrolases, including APT enzymes42. During efforts to explore novel covalent inhibitor scaffold of serine hydrolases, N-heterocyclic ureas were identified as a new chemotype for serine hydrolase inhibitor development43. Both carbamates and N-heterocyclic ureas undergo nucleophilic attack by the active-site nucleophilic serine, leading to formation of a non-hydrolysable carbamylated acyl-intermediate. These inhibitors are bioavailable, selective within the serine hydrolase enzyme family, and maintain activity both in vitro and in vivo43, 44. Furthermore, a facile, convergent synthesis of triazole ureas accelerated the development of selective sub-nanomolar, in vivo active, selective inhibitors for acyl peptide hydrolase (APEH), as well as low nanomolar inhibitors of PAFAH2 and ABHD1143 with limited medicinal chemistry efforts.
The development of triazole urea inhibitors is reinforced by guided medicinal chemistry using competitive fluorosphosphonate (FP) ABPP probes33, 45. In this assay, a complex proteome is mixed with a triazole urea analogue to achieve enzyme inhibition. Next, a reporter-tagged FP-probe is added that irreversibly reacts with all active serine hydrolases. Accordingly, any reduction in probe labeling corresponds to inhibition. Typically, the FP-tetramethylrhodamine (FP-TAMRA) is used for rapid gel-based analysis of both the potency and selectivity of a candidate inhibitor across all active serine hydrolases in a specific cell line or tissue isolate. For less abundant enzymes, quantitative analysis is performed using FP-biotin, followed by streptavidin enrichment and mass spectrometry annotation43. Rather than optimize for potency towards a specific target, this approach provides a simple assay to guide medicinal chemistry efforts primarily on selectivity.
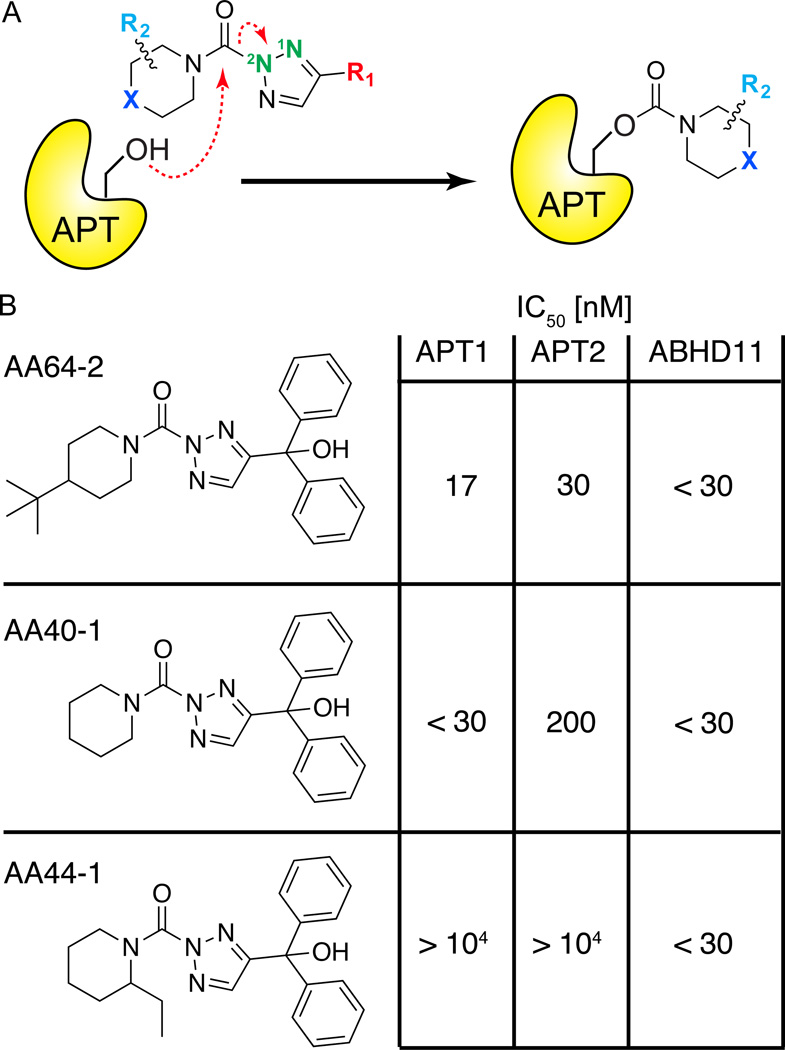
Triazole urea libraries were synthesized by click chemistry of alkynyl variants with azide, followed by conjugation to diverse carbamoyl chlorides43. This two-step approach leads to regioisomers at the N2 (major) and N1 (minor) positions, which may potentially have differential inhibitory properties44. After identification of a candidate triazole urea lead for APT1, an additional 19 analogs with variable structures at three positions: the substituent at 4-position of the triazole (R1), the substituent of the piperidine ring (R2), and the identity of the atom at the 4-position of the piperidine ring (X)46 (Figure 6A). Each round of medicinal chemistry was guided by gel-based competitive ABPP profiling. Optimization led to the identification of a highly potent tertbutylpiperidine (R2, X), and improved selectivity by addition of a bulky diphenyl methanol (R1)46. These inhibitors retain potency and selectivity in live cells with no observed toxicity. Despite significant effort, little selectivity was achieved against the unannotated hydrolase ABHD11. This is less of a concern, since highly selective analogues targeting ABHD11 can be used as control probes43.
Figure 6. Triazole urea covalent APT inhibitors.
(A) Mechanism of triazole urea inactivation of APT enzymes. The APT nucleophilic serine hydroxyl attacks the urea, releasing the triazole and leaving a stable carbamate adduct, inactivating the enzyme. (B) Selectivity and potency of optimized triazole urea inhibitors determined by gel-based competitive ABPP. AA64-2 is highly potent and selective for APT enzymes, with the exception of the uncharacterized hydrolase ABHD11. AA44-1 is highly selective for ABHD11, and can be used as a control probe.
The parasitic protozoan T.gondii is a close relative to the malaria parasite P.falciparum, where proteomics studies have annotated hundreds of palmitoylated proteins including many required for infectivity47. In order to explore the potential of APT inhibition in the parasitic life cycle, Palmostatin M was compared side by side with a piperidine (X), diphenyl methanol (R1), triazole urea (AA401)31. This analogue inhibits human APT1 (IC50 < 30 nM) and APT2 (IC50 = 200 nM), as well as ABHD11, Esterase D, and APEH46. These compounds were assayed for inhibition of T.gondii invasion, egress, and intracellular growth. By competitive ABPP methods, the T.gondii APT homologue TgASH1 was completely inhibited by 10 µM Palmostatin M or as little as 100 nM of AA401. The parasite’s lytic cycle is severely inhibited by incubation with 10 µM of either Palmostatin M or AA401, but this result is eliminated when the inhibitor concentration is lowered to 1 µM. Additional analysis showed neither of the compounds had any effect on parasitic egress. Interestingly, Palmostatin M reduces parasitic invasion, while AA401 inhibition may slightly enhance invasion. Furthermore, deletion of TgASH1 had no quantifiable effect on the membrane localization of known T.gondii palmitoylated proteins by immunofluorescence and no effect on the growth or life cycle. Since P.falciparum does not encode a TgASH1 homologue, it is unclear if TgASH1 accounts for all protein thioesterase activity in apicomplexans31. Overall, the potent triazole urea inhibitor AA401 appears to have increased selectivity and less off-target effects, providing a promising alterative to Palmostatin B for in vivo studies.
Chloroisocoumarins
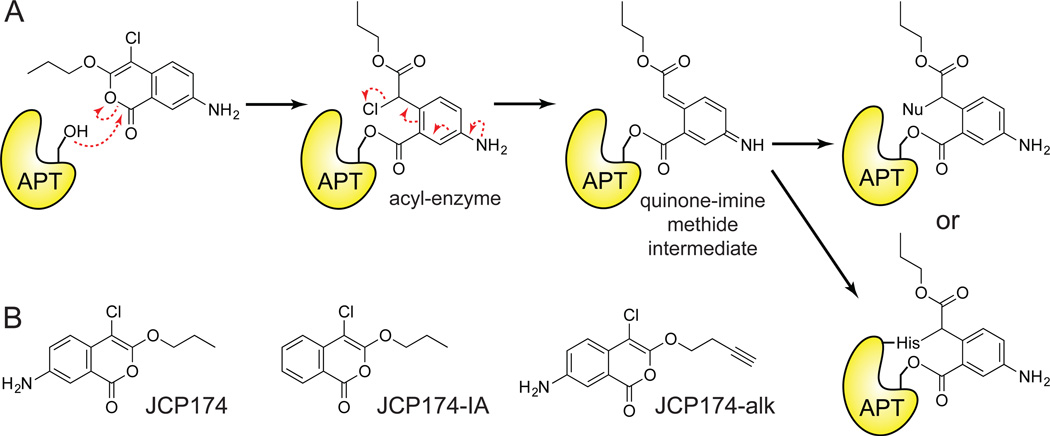
In a parallel T.gondii study, a forward chemical genetic screen was assayed to discover modulators of parasitic invasion. This study led to the discovery of compounds that attenuate invasion, including compounds targeting the redox chaperone TgDJ-148. In addition, the chloroisocoumarin JCP174 and similar derivatives were identified as enhancers of parasitic invasion32, and removing the aromatic amine in the 7-position eliminated activity, providing an inactive analogue (Figure 7B). Enhanced invasion was dose-dependent, and correlates with enhanced gliding motility, an established indicator of enhanced invasion. Invasion was enhanced within 5 minutes of inhibitor treatment with no observable effects on viability or intracellular replication.
Figure 7. Mechanism-based chloroisocoumarin JCP174 inhibits T.gondii APT.
(A) Putative mechanism of APT irreversible enzyme inactivation by JCP174. Attack of the electrophilic carbonyl by the catalytic serine hydroxyl leads to the formation of an acyl-enzyme derivative, unmasking a reactive α-chloro carbonyl group. Elimination of chlorine yields a quinone-imine-methide intermediate that is susceptible to nucleophilc attack by either the solvent or the enzyme's catalytic histidine side-chain producing a labile acyl-enzyme derivative or a stable alkylated acyl-enzyme adduct, respectively. (B) Structures of the active chloroisocoumarin inhibitor JCP174 (Left), the inactive control lacking 7-amino substitution JCP174-IA (Middle) and the click-compatible analog derivatized with an alkyne functional group JCP174-alk (Right).
Chloroisocoumarins were first identified as mechanism-based covalent inhibitors of serine proteases49. Chloroisocoumarins have limited stability and are susceptible to both serine and cysteine nucleophiles, leading to enhanced reactivity and significant potential off-targets. Indeed, JCP174 originated as a human elastase inhibitor (Kobs/[I] (M−1 s−1) = 54,000)50, but may also be reactive towards other proteases and hydrolases. This class of double-hit irreversible inhibitors first react with the enzyme’s serine nucleophile, and followed by elimination the chlorine on the acyl-enzyme intermediate to produce a electrophilic quinone-imine methide, which is susceptible to attack by accessible nucleophiles including the activated histidine of the catalytic triad (Figure 7A). While this mechanism was not explicitly demonstrated for TgASH1, the same chlorine elimination is predicted to produce a reactive electrophile.
In order to determine the functional target(s) of JCP174, parasite proteomes were treated with FP activity-based probes to profile potential serine hydrolase targets32. In-gel competitive ABPP analysis reported a single serine hydrolase target of JCP174. Additionally, an alkynyl analogue, JCP174-alk, was used to directly enrich targets after copper-catalyzed click chemistry to a biotin-azide and streptavidin enrichment. Mass spectrometry proteomics identified several proteins where JCP174-alk enrichment was reduced by competition with JCP174, including the APT homologue TgASH1 (IC50 = 1.32 µM). This finding corroborates the enhanced invasion trend reported after treatment with AA40131, although JCP174 induces a stronger and more significant enhancer phenotype. Furthermore, incomplete conditional knockdown of TgASH1 enhanced invasion, while a genetic knockout showed no effect and was unresponsive to JCP174 treatment. Recombinant TgASH1 was able to catalyze the in vitro depalmitoylation of established palmitoylated proteins with roles in invasion, and JCP174 treatment similarly enhanced their co-fractionation with membranes, suggestive of altered palmitoylation dynamics. In summary, chloroisocoumarins represent a new chemotype targeting APT enzymes, and highlight how forward chemoprotomics screens can identify new regulatory pathways in parasitic invasion.
Piperazine amides
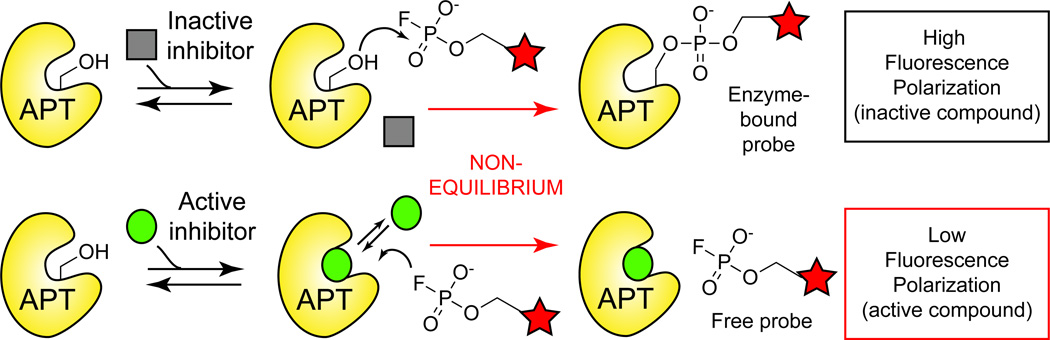
While several covalent chemotypes have emerged for APT enzymes, none are selective for either isoform. In the absence of an APT2 crystal structure, rational design of isoform selective inhibitors has been challenging. In collaboration with the NIH Molecular Libraries Production Center Network (MLPCN), both human APT1 and APT2 were assayed against a library of 315,004 compounds51 to identify new inhibitor chemotypes and explore the potential to identify isoform-selective leads. This high-throughput screening (HTS) effort used a fluorescence polarization-based competitive ABPP assay, termed FluoPol-ABPP52 (Figure 8). In this approach, recombinant APT enzyme is incubated with 10 µM of a candidate inhibitor, followed by a short, time-dependent addition of fluorophosphonate-rhodamine (FP-TAMRA). Upon covalent alkylation of the nucleophilic serine, the fluorescence polarization increases relative to enhanced anisotropy of the enzyme-fluorophore complex. Based on these parameters, any loss of polarization identifies potent inhibitors able to effectively compete with the irreversible covalent reaction. This assay routinely yields Z-factors >0.8, providing a robust approach to identify novel isoform selective APT inhibitors. Importantly, the FluoPol-ABPP assay occurs under time-dependent non-equilibrium conditions, providing a primary selection for inhibitors with potent Koff constants, which correlate with enhanced residency time and improved in vivo pharmacokinetics53.
Figure 8. FluoPol-ABPP high-throughput competitive assay to discover new APT inhibitor chemotypes.
Inhibitor discovery by active site occupancy profiling with the activity-based probe FP-TAMRA. APT enzyme is pre-incubated with inhibitor, and then under non-equilibrium conditions, treated with FP-TAMRA, which irreversibly competes with candidate inhibitors. Activity is reported by a change in fluorescence polarization induced by the increased anisotropy of the enzyme-probe complex.
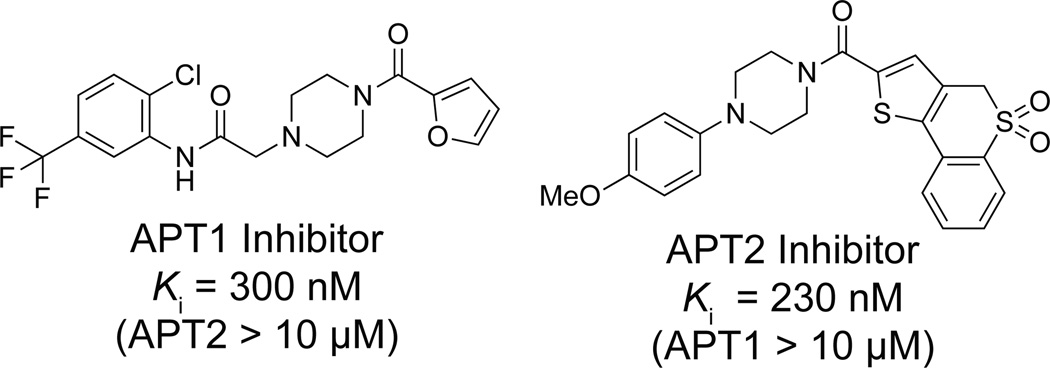
Separate HTS screens were performed for APT1 and APT2, yielding hit rates of 0.156% and 0.380%, respectively46, 51. Each hit compound was re-tested in a confirmatory screen, which validated 69% APT1 inhibitors, and 66% APT2 inhibitors. This list was then manually filtered to exclude non-selective compounds identified in similar HTS assays for the serine hydrolases RBBP9 and PME1, and the cysteine-dependent enzyme GSTO54. In total, 91 APT1, 61 APT2, and 95 dual APT1/APT2 inhibitors were selected for in-gel ABPP confirmatory analysis. This filtered group of inhibitors included one triazole urea, but largely converged to a common (>1/3) piperazine amide chemotype (Figure 9). This motif bears striking similarity to triazole ureas, since each contain a five-membered ring (furan, thiophene, or triazole) linked across an amide/urea with a N-heterocycle. Furthermore, due to the large diversity of the library, a structure-activity relationship emerged, identifying APT1 inhibitors with extensions from the piperazine arm, and APT2 inhibitors with hydrophobic extensions off the thiophene arm. Surprisingly, many of the inhibitors were selective for a specific APT isoform, an elusive property not found with other chemotypes. Overall, the most potent inhibitors were characterized in a fluorogenic substrate assay, providing Ki values in the 200 – 300 nM range, a surprising success for leads taken directly from the screening library. Interestingly, gel-based competitive ABPP shows stark differences in potency between the APT1 and APT2 compounds, yet steady-state kinetic analyses yielded effectively similar Ki values. These findings suggest APT inhibitor’s potency is derived primarily from its small Koff value, and highlight how such non-equilibrium assays can improve selection for compounds with improved residency times.
Figure 9. Lead isoform-selective inhibitors for APT1 and APT2 identified by FluoPol-ABPP.
Competitive assays with aliphatic FP-TAMRA proceeded to completion quickly under standard conditions, making it difficult to capture the optimal kinetic window for reversible inhibition competition51. To circumvent this problem, competitive ABPP assays were performed using a less reactive hydrophilic probe, FP-PEG-TAMRA. This probe is a less optimal substrate for APT enzymes, slowing the reaction kinetics enough to assay active site competition. Such findings have significant implications for competitive ABPP selectivity assays with reversible inhibitors, since different enzymes may react with ABPP probe variants with distinct kinetic windows.
For both APT1 and APT2, a small library of <10 analogues were assayed to establish a preliminary structure-activity relationship51. For the APT2 lead inhibitor, the para-(methoxyphenyl)piperazine arm was intolerant to substitutions in the ortho and meta positions. Further modifications on the thiophene arm were more tolerant, suggesting opportunities for further optimization. Importantly, the (o-methoxyphenyl)piperazine analogue completely lost activity, providing an non-reactive control analogue. For the APT1 inhibitor, a similar negative control compound was identified with an α-methyl substitution at the amide linkage. Inhibitor selectivity was confirmed by gel-based competitive ABPP, but also by SILAC-ABPP, a quantitative mass spectrometry method to quantify inhibitor occupancy with increased sensitivity43. This approach confirmed isoform selective inhibition across the serine hydrolase enzyme family for each lead inhibitor. These experiments demonstrate how competitive ABPP methods can be used to profile reversible inhibitors in complex proteomes, although the relative reactivity of each enzyme towards the probe may vary. Therefore, when using competitive ABPP methods to assay reversible inhibitors, it is important to recognize that variable ABPP probe reaction rates can skew observed selectively profiles.
In order to validate target occupancy in vivo, each lead inhibitor injected into mice51. After some time, an alkynyl triazole-urea probe was injected to profile residual active enzymes. This probe is poorly reactive with APT enzymes, providing an appropriate kinetic window to profile target occupancy in vivo. Since the rate of the triazole urea probe reaction with APT enzymes is slow, it was then possible to harvest tissues and analyze the results from the in vivo competition. The alkynyl triazole urea probe demonstrated isoform selective APT inhibition in the brain, kidney, lung, and heart. No inhibition was observed in the liver, which accumulates the triazole urea probe and likely shifts the reaction stoichiometry to out-compete the reversible inhibitors. Reduced inhibition was observed in the brain, suggesting these compounds are less efficient at crossing the blood-brain barrier. Overall, piperazine amide APT inhibitors are isoform selective, potent, and in vivo active inhibitors of APT1 and APT2 with no identified off-targets, making them the most promising, drug-like inhibitors to date.
Conclusions
The recent development of APT inhibitors has accelerated our understanding of the cellular regulation of palmitoylation, but has also provided a platform to advance new methodologies in inhibitor identification, selectivity profiling, and cellular validation. APT enzymes are conserved in eukaryotic organisms, suggesting a common biological function, which in mammals includes protein de-palmitoylation of certain target proteins. Surprisingly, there are no reported phenotypes for APTs in standard invertebrate model organism, including yeast, drosophila, and C.elegans. This raises the possibility that for these invertebrates (1) enzymatic depalmitoylation may not be essential, (2) functionally redundant hydrolases compensate for APT deficiency, or (3) APT enzymes are not primarily de-palmitoylases. Future studies demand the generation of vertebrate knockout systems to profile APT mutant phenotypes in higher organisms. Current data suggest APT enzymes are responsible for regulating dynamic palmitoylation, but many of the reported experiments rely on over-expression studies and report subtle changes, leaving room for additional interpretations. With the development of new potent and selective inhibitors and bioorthogonal labeling methods, it is now possible to directly answer these questions on endogenous palmitoylated proteins. Ongoing efforts to optimize APT1 and APT2 inhibitors will require a structural understanding of inhibitor binding, which promises to add structure-guided design principles to focus new medicinal chemistry. Improved competitive inhibitors are the next logical path towards developing selective pharmacological tools for in vivo functional analysis of protein de-palmitoylation.
Acknowledgements
We would like to thank members of the Martin lab for helpful discussions. Funding is provided by the University of Michigan Rackham Merit Fellowship (D.D.), the National Institutes of Health R00CA151460, and the University of Michigan.
References
- 1.Tom CT, Martin BR. ACS Chem Biol. 2013;8:46–57. doi: 10.1021/cb300607e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin BR, Cravatt BF. Nat Methods. 2009;6:135–138. doi: 10.1038/nmeth.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivaldi C, Martin BR, Kieffer-Jaquinod S, Chapel A, Levade T, Garin J, Journet A. PLoS One. 2012;7:e37187. doi: 10.1371/journal.pone.0037187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Martin BR, Cravatt BF, Hofmann SL. J Biol Chem. 2012;287:523–530. doi: 10.1074/jbc.M111.306183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin BR, Wang C, Adibekian A, Tully SE, Cravatt BF. Nat Methods. 2012;9:84–89. doi: 10.1038/nmeth.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang R, Wan J, Arstikaitis P, Takahashi H, Huang K, Bailey AO, Thompson JX, Roth AF, Drisdel RC, Mastro R, Green WN, Yates JR, III, Davis NG, El-Husseini A. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang W, Di Vizio D, Kirchner M, Steen H, Freeman MR. Molecular & Cellular Proteomics. 2010;9:54–70. doi: 10.1074/mcp.M800448-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones Matthew L, Collins Mark O, Goulding D, Choudhary Jyoti S, Rayner Julian C. Cell Host & Microbe. 2012;12:246–258. doi: 10.1016/j.chom.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willumsen BM, Cox AD, Solski PA, Der CJ, Buss JE. Oncogene. 1996;13:1901–1909. [PubMed] [Google Scholar]
- 10.Wedegaertner PB, Bourne HR. Cell. 1994;77:1063–1070. doi: 10.1016/0092-8674(94)90445-6. [DOI] [PubMed] [Google Scholar]
- 11.Baker TL, Zheng H, Walker J, Coloff JL, Buss JE. J. Biol. Chem. 2003;278:19292–19300. doi: 10.1074/jbc.M206956200. [DOI] [PubMed] [Google Scholar]
- 12.Rocks O, Peyker A, Kahms M, Verveer PJ, Koerner C, Lumbierres M, Kuhlmann J, Waldmann H, Wittinghofer A, Bastiaens PIH. Science. 2005;307:1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 13.Duncan JA, Gilman AG. J. Biol. Chem. 1998;273:15830–15837. doi: 10.1074/jbc.273.25.15830. [DOI] [PubMed] [Google Scholar]
- 14.Sugimoto H, Hayashi H, Yamashita S. J Biol Chem. 1996;271:7705–7711. doi: 10.1074/jbc.271.13.7705. [DOI] [PubMed] [Google Scholar]
- 15.Hirano T, Kishi M, Sugimoto H, Taguchi R, Obinata H, Ohshima N, Tatei K, Izumi T. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 2009;1791:797–805. doi: 10.1016/j.bbalip.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 16.Duncan JA, Gilman AG. J. Biol. Chem. 2002;277:31740–31752. doi: 10.1074/jbc.M202505200. [DOI] [PubMed] [Google Scholar]
- 17.Toyoda T, Sugimoto H, Yamashita S. Biochimica et Biophysica Acta (BBA) - Molecular and Cell Biology of Lipids. 1999;1437:182–193. doi: 10.1016/s1388-1981(99)00007-4. [DOI] [PubMed] [Google Scholar]
- 18.Rusch M, Zimmermann TJ, Burger M, Dekker FJ, Gormer K, Triola G, Brockmeyer A, Janning P, Bottcher T, Sieber SA, Vetter IR, Hedberg C, Waldmann H. Angew Chem Int Ed Engl. 2011 doi: 10.1002/anie.201102967. [DOI] [PubMed] [Google Scholar]
- 19.Tomatis VM, Trenchi A, Gomez GA, Daniotti JL. PLoS One. 2010;5:e15045. doi: 10.1371/journal.pone.0015045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong E, Peng S, Chandra G, Sarkar C, Zhang Z, Bagh MB, Mukherjee AB. Journal of Biological Chemistry. 2013;288:9112–9125. doi: 10.1074/jbc.M112.421073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, Hubel K, Dekker F, Hedberg C, Rengarajan B, Drepper C, Waldmann H, Kauppinen S, Greenberg ME, Draguhn A, Rehmsmeier M, Martinez J, Schratt GM. Nat Cell Biol. 2009;11:705–716. doi: 10.1038/ncb1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banerjee S, Neveu P, Kosik KS. Neuron. 2009;64:871–884. doi: 10.1016/j.neuron.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 23.Beronja S, Janki P, Heller E, Lien WH, Keyes BE, Oshimori N, Fuchs E. Nature. 2013;501:185–190. doi: 10.1038/nature12464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burger M, Zimmermann TJ, Kondoh Y, Stege P, Watanabe N, Osada H, Waldmann H, Vetter IR. J Lipid Res. 2012;53:43–50. doi: 10.1194/jlr.M019851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox CS, Liu Y, White CC, Feitosa M, Smith AV, Heard-Costa N, Lohman K, Johnson AD, Foster MC, Greenawalt DM, Griffin P, Ding J, Newman AB, Tylavsky F, Miljkovic I, Kritchevsky SB, Launer L, Garcia M, Eiriksdottir G, Carr JJ, Gudnason V, Harris TB, Cupples LA, Borecki IB. PLoS Genet. 2012;8:e1002695. [Google Scholar]
- 26.Hernandez JL, Majmudar JD, Martin BR. Curr Opin Chem Biol. 2013;17:20–26. doi: 10.1016/j.cbpa.2012.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long JZ, Cravatt BF. Chem Rev. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dekker FJ, Rocks O, Vartak N, Menninger S, Hedberg C, Balamurugan R, Wetzel S, Renner S, Gerauer M, Scholermann B, Rusch M, Kramer JW, Rauh D, Coates GW, Brunsveld L, Bastiaens PI, Waldmann H. Nat Chem Biol. 2010;6:449–456. doi: 10.1038/nchembio.362. [DOI] [PubMed] [Google Scholar]
- 29.Xu J, Hedberg C, Dekker FJ, Li Q, Haigis KM, Hwang E, Waldmann H, Shannon K. Blood. 2012;119:1032–1035. doi: 10.1182/blood-2011-06-358960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tabatadze N, Smejkalova T, Woolley CS. Endocrinology. 2013;154:819–830. doi: 10.1210/en.2012-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemp LE, Rusch M, Adibekian A, Bullen HE, Graindorge A, Freymond C, Rottmann M, Braun-Breton C, Baumeister S, Porfetye AT, Vetter IR, Hedberg C, Soldati-Favre D. J Biol Chem. 2013 doi: 10.1074/jbc.M113.460709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Child MA, Hall CI, Beck JR, Ofori LO, Albrow VE, Garland M, Bowyer PW, Bradley PJ, Powers JC, Boothroyd JC, Weerapana E, Bogyo M. Nat Chem Biol. 2013;9:651–656. doi: 10.1038/nchembio.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cravatt BF, Wright AT, Kozarich JW. Annu Rev Biochem. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 34.Deck P, Pendzialek D, Biel M, Wagner M, Popkirova B, Ludolph B, Kragol G, Kuhlmann J, Giannis A, Waldmann H. Angew Chem Int Ed Engl. 2005;44:4975–4980. doi: 10.1002/anie.200462625. [DOI] [PubMed] [Google Scholar]
- 35.Shi Y, Burn P. Nat Rev Drug Discov. 2004;3:695–710. doi: 10.1038/nrd1469. [DOI] [PubMed] [Google Scholar]
- 36.Weibel EK, Hadvary P, Hochuli E, Kupfer E, Lengsfeld H. J Antibiot (Tokyo) 1987;40:1081–1085. doi: 10.7164/antibiotics.40.1081. [DOI] [PubMed] [Google Scholar]
- 37.Hadvary P, Sidler W, Meister W, Vetter W, Wolfer H. J Biol Chem. 1991;266:2021–2027. [PubMed] [Google Scholar]
- 38.Hedberg C, Dekker FJ, Rusch M, Renner S, Wetzel S, Vartak N, Gerding-Reimers C, Bon RS, Bastiaens PI, Waldmann H. Angew Chem Int Ed Engl. 2011 doi: 10.1002/anie.201102965. [DOI] [PubMed] [Google Scholar]
- 39.Zimmermann TJ, Burger M, Tashiro E, Kondoh Y, Martinez NE, Gormer K, Rosin-Steiner S, Shimizu T, Ozaki S, Mikoshiba K, Watanabe N, Hall D, Vetter IR, Osada H, Hedberg C, Waldmann H. Chembiochem. 2013;14:115–122. doi: 10.1002/cbic.201200571. [DOI] [PubMed] [Google Scholar]
- 40.Smoum R, Rubinstein A, Dembitsky VM, Srebnik M. Chem Rev. 2012;112:4156–4220. doi: 10.1021/cr608202m. [DOI] [PubMed] [Google Scholar]
- 41.Bachovchin DA, Cravatt BF. Nat Rev Drug Discov. 2012;11:52–68. doi: 10.1038/nrd3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Proc Natl Acad Sci U S A. 2010;107:20941–20946. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adibekian A, Martin BR, Wang C, Hsu KL, Bachovchin DA, Niessen S, Hoover H, Cravatt BF. Nat Chem Biol. 2011;7:469–478. doi: 10.1038/nchembio.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsu KL, Tsuboi K, Adibekian A, Pugh H, Masuda K, Cravatt BF. Nat Chem Biol. 2012;8:999–1007. doi: 10.1038/nchembio.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Patricelli MP, Cravatt BF. Proc Natl Acad Sci U S A. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adibekian A, Martin BR, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, Ferguson J, Cravatt BF, Hodder P, Rosen H. Probe Reports from the NIH Molecular Libraries Program. Bethesda (MD): 2010. [Google Scholar]
- 47.Jones ML, Collins MO, Goulding D, Choudhary JS, Rayner JC. Cell Host Microbe. 2012;12:246–258. doi: 10.1016/j.chom.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hall CI, Reese ML, Weerapana E, Child MA, Bowyer PW, Albrow VE, Haraldsen JD, Phillips MR, Sandoval ED, Ward GE, Cravatt BF, Boothroyd JC, Bogyo M. Proc Natl Acad Sci U S A. 2011;108:10568–10573. doi: 10.1073/pnas.1105622108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powers JC, Asgian JL, Ekici OD, James KE. Chem Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- 50.Kerrigan JE, Oleksyszyn J, Kam CM, Selzler J, Powers JC. J Med Chem. 1995;38:544–552. doi: 10.1021/jm00003a017. [DOI] [PubMed] [Google Scholar]
- 51.Adibekian A, Martin BR, Chang JW, Hsu KL, Tsuboi K, Bachovchin DA, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, Ferguson J, Hodder PS, Rosen H, Cravatt BF. J Am Chem Soc. 2012;134:10345–10348. doi: 10.1021/ja303400u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bachovchin DA, Brown SJ, Rosen H, Cravatt BF. Nat Biotechnol. 2009;27:387–394. doi: 10.1038/nbt.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lu H, Tonge PJ. Current Opinion in Chemical Biology. 2010;14:467–474. doi: 10.1016/j.cbpa.2010.06.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsuboi K, Bachovchin DA, Speers AE, Spicer TP, Fernandez-Vega V, Hodder P, Rosen H, Cravatt BF. J Am Chem Soc. 2011;133:16605–16616. doi: 10.1021/ja2066972. [DOI] [PMC free article] [PubMed] [Google Scholar]