Abstract
Background
Fevers of unknown origin constitute a substantial disease burden in Southeast Asia. In majority of the cases, the cause of acute febrile illness is not identified.
Methods
We used MassTag PCR, a multiplex assay platform, to test for the presence of 15 viral respiratory agents from 85 patients with unexplained respiratory illness representing six disease clusters that occurred in Cambodia between 2009 and 2012.
Results
We detected a virus in 37 (44%) of the cases. Human rhinovirus, the virus detected most frequently, was found in both children and adults. The viruses most frequently detected in children and adults, respectively, were respiratory syncytial virus and enterovirus 68. Sequence analysis indicated that two distinct clades of enterovirus 68 were circulating during this time period.
Conclusions
This is the first report of enterovirus 68 in Cambodia and contributes to the appreciation of this virus as an important respiratory pathogen.
Keywords: Enterovirus 68, Respiratory disease, RSV
Introduction
Acute respiratory infections (ARI) are among the leading causes of morbidity and mortality in both developed and developing nations [1-3]. In Cambodia, pneumonia is the third highest cause of death in children under five [4,5]. In December 2006, a passive hospital-based surveillance study was initiated to identify the causes of acute undifferentiated fever in patients seeking healthcare in Cambodia. Between 2006 and 2009, 9,997 patients from peri-urban and rural Cambodian health centers were enrolled in the study. The most frequent diagnoses included influenza, dengue, malaria and leptospirosis [6]. No diagnosis was obtained in 62% of cases [6]. In a separate analysis using samples from December 2006 to December 2008, 4,233 patients with respiratory disease were screened for influenza virus by real-time reverse-transcriptase polymerase chain reaction (rRT–PCR). Of these patients, 1,151 (27.2%) were positive for influenza [7]. In a separate study, Sreng et al. analyzed 2,805 patients with influenza-like illness in Cambodia between 2006 and 2008 and showed that only 9.6% tested positive for influenza [8]. To address the high rate of undiagnosed ARI cases in Cambodia, we selected six clusters of previously unexplained respiratory illness from 2009 to 2012 for analysis by MassTag PCR, a high throughput multiplex screening platform. We identified a potential viral pathogen in 44% (37/85) of the samples, confirming the utility of multiplex diagnostic assays in clinical microbiology and providing insights into ARI in Cambodia.
Results
From 2009 to 2012, 17,363 samples were collected from 18 health clinics in six provinces of Cambodia and screened for influenza virus, dengue virus and Plasmodium spp (Figure 1). From amongst the samples that tested negative for these agents, we selected 6 clusters representing 85 patients with unexplained acute respiratory illness for further analysis. 88% (75/85) of the patients exhibited influenza-like symptoms (Table 1). The archived specimens from the 6 clusters were screened by MassTag PCR for viral respiratory pathogens [9,10]. At least one virus was detected in 44% (37/85) samples (Figure 2A). Infection with two viruses was detected in 8 samples. Rhinovirus (HRV) was the most frequently detected virus (n = 17), followed by respiratory syncytial virus (RSV; n = 15), and enterovirus (HEV; n = 9) (Figure 2B).
Figure 1.
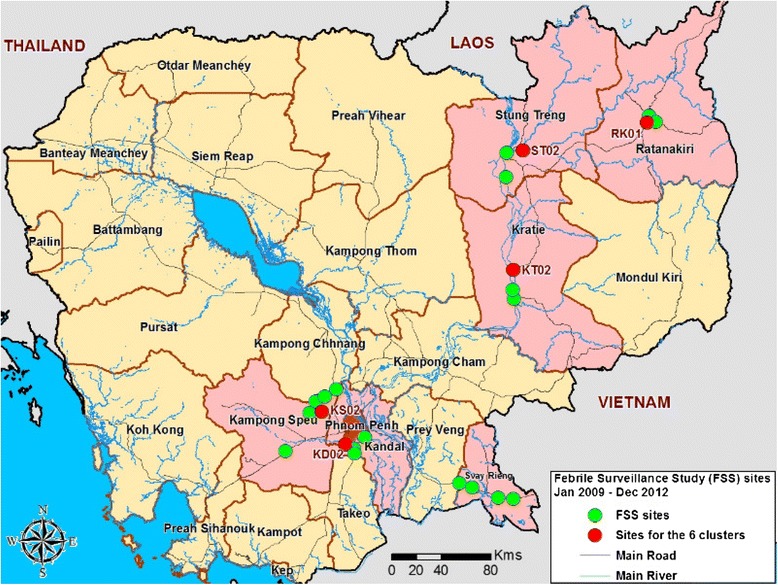
Map of NAMRU-2PP Febrile Surveillance Study health center sites from January 2009 to December 2012. The locations of the 6 clusters are indicated in red. Pink represents the 6 provinces where the study was conducted.
Table 1.
Demographics and clinical characteristics of the 6 analyzed clusters
| Year | 2009 | 2010 | 2011 | 2012 | ||
|---|---|---|---|---|---|---|
| province (site code) | Kandal (KD02) | Kampong Speu (KS02) | Kandal (KD02) | Ratanakiri (RK01) | SteungTreng (ST02) | Kratie (KT02) |
| enrollment period, (day duration) | 24-31 Aug (8 d) | 12-16 Oct (7 d) | 05-18 Mar (14 d) | 25 Jul-12 Sep (50 d) | 13-16 Feb (4 d) | 4 Apr (1 d) |
| season | Rainy | Rainy | Dry | Rainy | Dry | Dry |
| number of cases | 20 | 22 | 11 | 11 | 11 | 10 |
| median age [range] | 36.5 [4–71] | 11 [2–60] | 26 [6–45] | 4 [2–5] | 19 [18–49] | 23 [2–43] |
| female/male, n (%) | 9 (45)/11 (55) | 12 (55)/10 (45) | 8 (73)/3 (27) | 4 (36)/7 (64) | 4 (36)/7 (64) | 8 (80)/2 (20) |
| fever present, median (duration day) | 4 [2–4] | 4 [2–6] | 3 [2–4] | 4 [2–6] | 3 [2–3] | 3 [2–5] |
| virus type detected (n) | EV-D68 (6); HRV-C (4); HRV-A; RSV-B | RSV-B (5); EV-D68; HRV-A; HRV-B; HRV-C | ADV; HRV-B | RSV-A (9); CV-B4; EV-D68; HCoV-OC43; HRV-C | HRV-B (3); HCoV-OC43 | HRV-A (3); HRV-C; HCoV-229E |
| Symptoms | ||||||
| headache, n (%) | 18 (90) | 21 (95.5) | 11 (100) | 7 (63.5), 4 NA‡ | 11 (100) | 10 (100) |
| cough, n (%) | 18 (90) | 22 (100) | 11 (100) | 11 (100) | 8 (73) | 5 (50) |
| malaise, n (%) | 15 (75) | 21 (95.5) | 4 (36.5) | 11 (100) | 0 | 0 |
| chills, n (%) | 0 | 21 (95.5) | 5 (45.5) | 3 (27.5) | 7 (63.5) | 2 (20) |
| muscle ache n (%) | 3 (15) | 0 | 1 (9) | 0, 4 NA | 0 | 7 (70) |
| joint pain, n (%) | 8 (40) | 0 | 1 (9) | 0, 6 NA | 2 (18) | 1 (10) |
| seizure, n (%) | 0 | 0 | 0 | 1 (9) | 0 | 0 |
| sore throat, n (%) | 12 (60) | 4 (18) | 11 (100) | 3 (27.5), 8 NA | 1 (9) | 6 (60) |
| shortness of breath, n (%) | 0 | 0 | 0 | 10 (91) | 0 | 0 |
| nausea, n (%) | 0 | 0 | 0 | 9 (82) | 0 | 0 |
| vomiting, n (%) | 1 (5) | 0 | 10 (91) | 8 (73) | 0 | 0 |
| abdominal cramps, n (%) | 1 (5) | 0 | 2 (18) | 1 (9), 4 NA | 0 | 0 |
| Site Diagnosis* | ||||||
| pharyngitis, n (%) | 11 (55) | 3 (13.5) | 10 (91) | 0 | 0 | 0 |
| influenza, n (%) | 7 (35) | 5 (23) | 1 (9) | 0 | 0 | 7 (70) |
| pneumonia, n (%) | 1 (5) | 0 | 0 | 11 (100) | 0 | 0 |
| bronchitis, n (%) | 0 | 1 (4.5) | 0 | 0 | 0 | 0 |
| upper respiratory infection, n (%) | 0 | 13 (59) | 0 | 0 | 0 | 0 |
| malaria, n (%) | 0 | 0 | 0 | 0 | 11 (100) | 3 (30) |
| other, n (%) | 1 typhoid fever | 0 | 0 | 0 | 0 | 0 |
‡NA = Not available; HRV-A/B/C = human rhinovirus type A/ B/C; EV-D68 = enterovirus D68; HCoV = human coronavirus; ADV = adenovirus; RSV-A/B = respiratory syncytial virus A/B; CV-B4 = coxsackievirus B4.
*all samples tested negative for influenza virus, dengue virus and Plasmodium spp.in laboratory assays.
Figure 2.
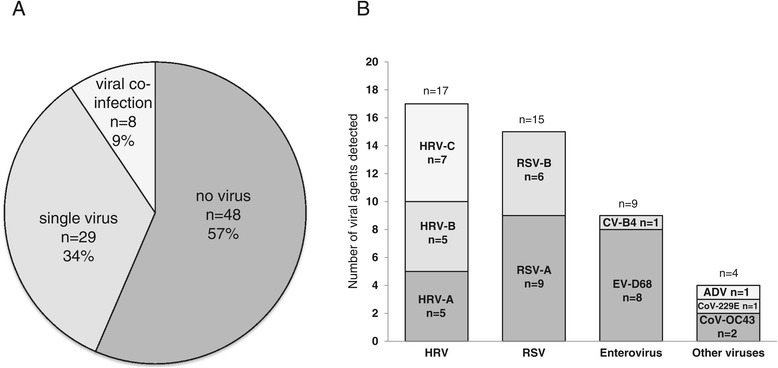
Summary of agents detected by MassTag PCR. A) Percentage of viral-positive samples B) Distribution of viruses detected in all samples. Each bar represents the number of all detected viruses.
KD02 cluster
The KD02 cluster samples were collected from 20 patients in Kandal who presented for medical attention over an eight-day period in August, 2009. Eighteen had influenza-like illness (ILI), one had a presumptive diagnosis of typhoid fever and one was diagnosed with pneumonia (Table 1). Analysis of these samples by MassTag PCR detected a viral agent in 11 samples (Table 2). HEV was detected in six samples; one of them also contained RSV-B. To determine the HEV serotype, we amplified and sequenced a 600 nucleotide (nt) VP4-VP2 genome fragment from each sample. Sequence analysis indicated that all six HEV-positive samples contained a single strain of enterovirus 68 (EV-D68) (100% nt identity in VP4-VP2). Unlike most HEVs, EV-D68 is predominately a respiratory pathogen. Recent genetic analysis of EV-D68 indicated three main clades of the virus circulating worldwide [11]. To determine the clades circulating in Cambodia, we amplified and sequenced a 339 nt fragment of VP1 from all six samples. The Cambodian EV-D68 strain was most similar to EV-D68 strains originating from Asia between 2006 and 2010 (99% identity to strains from Japan, Philippines and China) and clustered with strains belonging to clade A, which has been implicated in recent EV-D68 outbreaks worldwide [11-14] (Figure 3). Comparison of amino acids within the VP1 relative to the EV-D68 Fermon reference strain revealed the majority of the substitutions were in the putative surface-exposed BC and DE loops (Figure 4). All six EV-D68 positive samples originated from adults (four men, two women) ranging between 24 to 71 years of age. Five of the six patients presented with pharyngitis, headache, cough and malaise. The exception, a 36-year-old male, presented with abdominal cramps, and joint pain, and was initially diagnosed with typhoid fever. Since both symptoms are atypical of EV-D68 infection, they may have been caused by a co-infecting agent.
Table 2.
Clinical description of virus positive cases in the KD02, KS02 and RK01 clusters
| Site | Date enrolled | Virus detected | Age | Gender | Sore throat | Cough | Temperature | Site diagnosis |
|---|---|---|---|---|---|---|---|---|
| KD02 | 25-Aug-2009 | EV-D68 | 46 | male | No | Yes | 38.4 | influenza |
| KD02 | 26-Aug-2009 | EV-D68 | 71 | female | Yes | Yes | 38.4 | pharyngitis |
| KD02 | 26-Aug-2009 | EV-D68 | 36 | male | No | No | 38.8 | typhoid fever |
| KD02 | 27-Aug-2009 | EV-D68 | 24 | female | Yes | Yes | 38.3 | pharyngitis |
| KD02 | 27-Aug-2009 | EV-D68 | 38 | male | No | Yes | 38.7 | pharyngitis |
| KD02 | 31-Aug-2009 | EV-D68, RSV-B | 51 | male | Yes | Yes | 38.7 | pharyngitis |
| KD02 | 24-Aug-2009 | HRV-C | 4 | male | No | Yes | 39.0 | pharyngitis |
| KD02 | 26-Aug-2009 | HRV-C | 11 | female | Yes | Yes | 38.6 | pharyngitis |
| KD02 | 26-Aug-2009 | HRV-A | 59 | female | Yes | Yes | 38.5 | pneumonia |
| KD02 | 26-Aug-2009 | HRV-C | 40 | male | No | Yes | 38.8 | influenza |
| KD02 | 31-Aug-2009 | HRV-C | 37 | male | Yes | Yes | 39.2 | pharyngitis |
| KS02 | 12-Oct-2009 | RSV-B | 8 | male | No | Yes | 39.0 | influenza |
| KS02 | 13-Oct-2009 | RSV-B | 3 | female | No | Yes | 38.8 | URI^ |
| KS02 | 13-Oct-2009 | RSV-B, HRV-C | 3 | female | No | Yes | 38.5 | URI |
| KS02 | 13-Oct-2009 | RSV-B | 12 | male | Yes | Yes | 39.5 | pharyngitis |
| KS02 | 15-Oct-2009 | HRV-A | 2 | female | No | Yes | 39.0 | URI |
| KS02 | 16-Oct-2009 | EV-D68 | 60 | female | No | Yes | 38.6 | influenza |
| KS02 | 16-Oct-2009 | RSV-B | 4 | female | No | Yes | 38.6 | influenza |
| RK01 | 25-Jul-2011 | RSV-A | 3 | female | Yes | Yes | 38.5 | pneumonia |
| RK01 | 1-Aug-2011 | RSV-A | 2 | male | NA* | Yes | 40.0 | pneumonia |
| RK01 | 15-Aug-2011 | RSV-A | 5 | female | NA | Yes | 39.0 | pneumonia |
| RK01 | 15-Aug-2011 | RSV-A | 3 | male | NA | Yes | 40.0 | pneumonia |
| RK01 | 18-Aug-2011 | CV-B4, RSV-A | 2 | male | NA | Yes | 39.0 | pneumonia |
| RK01 | 18-Aug-2011 | EV-D68, RSV-A | 4 | male | NA | Yes | 38.5 | pneumonia |
| RK01 | 18-Aug-2011 | HRV-C, RSV-A | 4 | male | NA | Yes | 39.8 | pneumonia |
| RK01 | 25-Aug-2011 | HCOV-OC43, RSV-A | 5 | female | NA | Yes | 38.5 | pneumonia |
| RK01 | 12-Sep-2011 | RSV-A | 4 | male | Yes | Yes | 38.3 | pneumonia |
^URI: Upper Respiratory Infection; NA* = Data not available.
Figure 3.
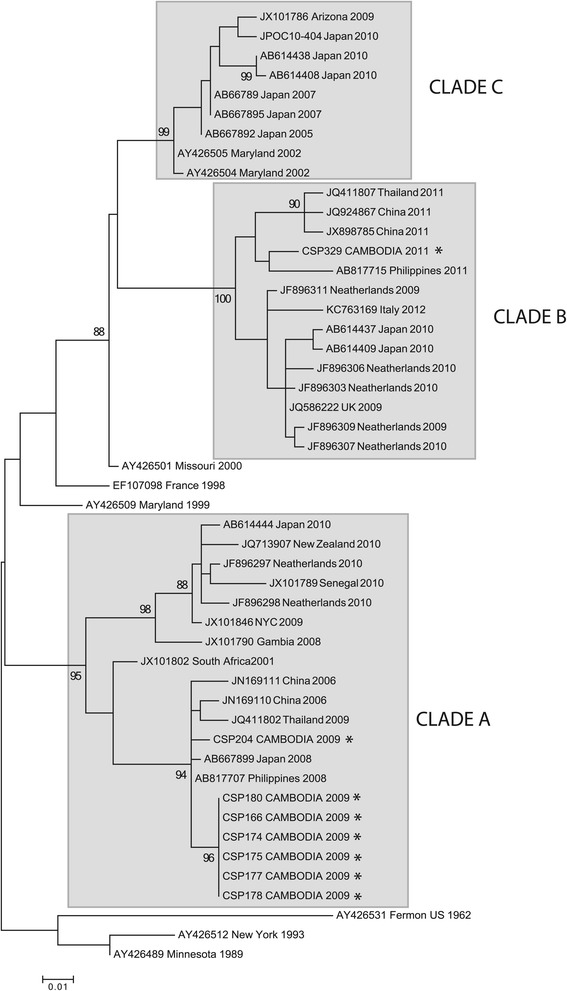
Maximum-likelihood phylogenetic tree of EV-D68 based on a 339 base pair fragment of the VP1 gene. For clarity, only select sequences were included in the tree. The three main clades are shown in gray. The sequences from the current study are indicated by *.
Figure 4.

Comparison of 112 amino acids of VP-1 from the EV-D68 Fermon strain to the eight Cambodian sequences generated in this study. Coordinates are provided relative to the Fermon VP1 sequence (accession number AY426531). The putative antigen binding loops BC and DE are indicted in gray. The amino acid differences in the Cambodian strains relative to Fermon are indicated by *.
Five other samples from the cluster contained a HRV; amplification and sequencing of the VP4-VP2 genome fragment revealed that four of these were HRV-C and one was HRV-A (Table 2). Three HRV-positive samples originated from adults and two from children.
KS02 cluster
The cluster of 22 cases from Kampong Speu province, collected within 7 days in October 2009, contained 16 patients <15 years of age. Upper respiratory tract infection was the most common diagnosis. 21 of 22 patients had complaints of headache (95%), malaise (95%), chills (95%), and all 22 had cough (100%) (Table 1).
Testing revealed that five cases were positive for RSV-B, all in patients 12 years old and under. One sample was also co-infected with HRV-C (Table 2). A single sample contained HRV-A. Of the six adult cases, one contained EV-D68. Sequence analysis indicates this virus (CSP-204) was similar to the strain from the Kandal cluster with 98.7% and 98.6% nt identity in the VP4-VP2 and VP1, respectively (100% amino acid (aa) identity in both proteins) (Figures 3 and 4).
RK01 cluster
Between July 25th and September 12th of 2011 a cluster of 11 children less than five years of age from Ratanakiri received a diagnosis of pneumonia. Ten of them had shortness of breath; eight had nausea and vomiting. MassTag PCR detected RSV-A in nine cases (82%). Four RSV-A positive samples were co-infected with other viruses, identified as, coxsackie virus B4 (CV-B4), human coronavirus (hCoV-OC43), HRV-C, or EV-D68 (Table 1). Sequence analysis of a VP1 fragment amplified from this EV-D68 strain indicated that this strain was only 86% identical (92% aa identity) to the Cambodian strains from 2009, and clustered with EV-D68 sequences from clade B (Figures 3 and 4). The highest similarity was to strains reported from the Philippines in 2008 (99% nt identity in the VP1) [11,15,16].
KD02 (2010), ST02, and KT02 clusters
The remaining three clusters comprising 32 cases all originated during the dry season in Cambodia (November through May). In cluster KD02, 10 of 11 patients were initially diagnosed with pharyngitis. All 11 patients in this cluster reported headache, cough and sore throat. ADV and HRV-B represented the lone viral agents detected in this cluster. In cluster ST02 all 11 cases were initially diagnosed with malaria and complained of fever (100%), cough (73%) and chills (63.5%). HRV-B was detected in three samples, and one sample contained CoV-OC43. In cluster KT02, 70% of the cases were diagnosed with influenza; the remaining 30% of cases were diagnosed with malaria (Table 1). HRV was detected in four of these samples (3 HRV-A and 1 HRV-C); CoV-229E was present in one sample.
Discussion
Our analysis of 6 clusters of previously undiagnosed ARI from Cambodia revealed the presence of potential viral pathogens in 44% of cases. The agents most commonly detected were HRV, RSV, and HEV. Our results are similar to published data from a Cambodian children cohort study wherein the predominant agents found were HRV and RSV [17]; but differ in that we also report findings in adults wherein the predominant organisms found were HRV and EV-D68.
RSV is a frequent cause of respiratory disease in children and was the predominant agent detected in pediatric clusters in Ratanakiri (RK01; RSV-A) and Kampong Speu (KS02; RSV-B) respectively. The two clusters of RSV occurred during the rainy season suggesting that RSV infections may represent a significant cause of viral pneumonia in Cambodian children during this period [17-19].
EV-D68 was the primary agent detected in a cluster initially diagnosed as pharyngitis/influenza and was also detected in individual cases from two other clusters. Our study represents the initial report linking EV-D68 with respiratory disease in Cambodia. First isolated in the 1960s, EV-D68 is atypical among EVs in respiratory tract tropism [20,21]. Since the late 2000′s, there has been a global increase of respiratory disease outbreaks associated with EV-D68. EV-D68 has been reported in outbreaks of respiratory disease in Asia (China, Thailand, Japan, the Philippines), Europe (Italy, Finland, the Netherlands, the UK, France), Oceania (New Zealand), Africa (the Gambia, Senegal, and South Africa) and the United States [11-13,15,16,22-24]. The clinical presentation of EV-D68 infections in these outbreaks ranged from acute, mild illness to severe pneumonia, and, in rare instances, death. Retrospective studies have shown epidemic emergence of EV-D68 in some countries, leading to the hypothesis that evolution of the virus may have increased its fitness, enhancing its global expansion [11,13]. In our study we found that multiple clades of EV-D68 were circulating in Cambodia between 2009–2011, similar to reports from other countries [12,13,16]. We detected representatives of two distinct EV-D68 clades, both with variations in the putative antigenic sites relative to the EV-D68 reference strain. It has been suggested that changes within the antigenic sites of the capsid may have enabled the recent worldwide emergence of EV-D68 [25].
The majority of EV-D68 positive cases in our study originated from adults. Although EV-D68 was originally isolated from children, recent reports link EV-D68 to ARI in both children and adults, though the severity of symptoms may differ [13,26].
During the preparation of this manuscript, a large outbreak of respiratory disease in children linked to EV-D68 has occurred in the US [27]. This outbreak, which has resulted in large number of hospitalized cases, represents the first documented large-scale outbreak of disease due to EV-D68.
Conclusions
Our findings demonstrate the utility of multiplex diagnostic assays for differential diagnosis of respiratory tract infections and underscore the increasing importance of EV-68 in respiratory disease.
Materials and methods
Sample collection
Beginning in December 2006, hospital-based surveillance of acute febrile illness was conducted in six provinces of eastern Cambodia: Kandal, Kampong Speu, Kratie, Ratanakiri, Stung Treng and Svay Rieng (Figure 1) [6]. Over a six-year period subjects were enrolled at 18 health clinics within these provinces. Ten were located in Kandal and Kampong Speu, approximately 40 to 75 Km from the capital Phnom Penh, respectively, whereas the others were located in more remote areas with difficult access during rainy seasons.
Between 2009 and 2012, 17,363 patients were enrolled and tested by PCR for influenza virus and dengue virus (upper respiratory swab (URS) and serum, respectively) and by Giemsa stain of whole blood for Plasmodium spp. Inclusion/Exclusion criteria, testing algorithm, specimen processing and testing were as described by Kasper et al. However the catchment area of the study changed as per Figure 1 [6]. Samples were collected under the auspices of study protocols #NAMRU2.2005.0004 and #037 NECHR approved by the U.S. Naval Medical Research Center Institutional Review Board and The Cambodian Ethical Committee for Health and Research, respectively. All research was in compliance with applicable federal regulations governing the protection of human subjects. All enrolled patients provided written consent. For patients <18 years of age, consent was obtained from a parent or legal guardian.
Cluster definition
Patients enrolled at the same site within a two-week span with similar clinical symptoms were included in individual clusters. We selected five such case clusters for further analysis. Two clusters originated from Kandal, and one each from Kampong Speu, Steung Treng, and Kratie provinces (Figure 1). Additionally, we selected a cluster of children under five years of age who were diagnosed at the Ratanakiri (RK01) field site with pneumonia. Patients in this cluster were enrolled within a 50-day period. Together the six clusters represented 85 patients.
MassTag PCR
Total nucleic acid was extracted from 250 μl of URS using the EasyMag automated extraction platform (BioMérieux). Total nucleic acid (TNA) was eluted in 40 μl. cDNA was generated using 10 μl of TNA and SuperScript II Reverse Transcriptase kit (Life Technologies). All samples were screened by MassTag PCR using respiratory panels composed of 15 known respiratory viruses [9,10]. The panels included primer sets for detection of 14 RNA viruses (influenza A and B, human respiratory syncytial virus A and B (RSV-A, RSV-B), human parainfluenza virus 1, 2, 3, 4A and 4B, human metapneumovirus, human coronavirus (hCoV) OC43 and 229E, human enterovirus (HEV)/rhinovirus (HRV)), and 1 DNA virus (adenovirus (ADV). All samples positive by MassTag PCR were confirmed by single agent PCR and dideoxy sequencing of the resulting amplification products.
Data analysis
Mass Tag PCR data were analyzed using non-parametric statistics. The confidence score was calculated using fold inter-quartile range distance of the positive control or the test sample from the 95th percentile cut-point of the negative control distribution for a tag. The score is similar to parametric z-score but is distribution free.
PCR and dideoxy sequencing
The list of primers can be found in Table 3. For each reaction, quantified positive control standards, samples, and non-template controls were used. In HEV-positive samples the serotype was identified using a consensus nested PCR assay that amplifies a 600-nucleotide region within the VP4-2 gene. The cycling conditions used for MassTag PCR were also used in individual PCR assays with the exception of the HEV VP4/2 nested assay (annealing temperature of 55°C).
Table 3.
Sequences of primers used in the study
| FLUA-F | GTCTTCTAACCGAGGTCGAAACG |
|---|---|
| FLUA-R | GCATTTTGGACAAAGCGTCTACG |
| FLUB-F | CACAGCAAAAACAATGAATGGA |
| FLUB-R | AGCACTTCCATTACATCCTTTGC |
| RSVA-F | AGATCAACTTCTGTCATCCAGCAA |
| RSVA-R | GCACATCATAATTAGGAGTATCAAT |
| RSVB-F | AAGATGCAAATCATAAATTCACAGGA |
| RSVB-R | TGATATCCAGCATCTTTAAGTATCTTTATAGTG |
| Coronavirus229E-F | GGCGCAAGAATTCAGAACCA |
| Coronavirus229E-R | TAAGAGCCGCAGCAACTGC |
| CoronavirusOC43-F | TGTGCCTATTGCACCAGGAGT |
| CoronavirusOC43-R | CCCGATCGACAATGTCAGC |
| HPIV1-F | TACTTTTGACACATTTAGTTCCAGGAG |
| HPIV1-R | CGGTACTTCTTTGACCAGGTATAATTG |
| HPIV2-F | GGACTTGGAACAAGATGGCCT |
| HPIV2-R | AGCATGAGAGCYTTTAATTTCTGGA |
| HPIV3-F | GCTTTCAGACAAGATGGAACAGTG |
| HPIV3-R | GCATKATTGACCCAATCTGATCC |
| HPIV4A-F | AACAGAAGGAAATGATGGTGGAAC |
| HPIV4A-R | TGCTGTGGATGTATGGGCAG |
| HPIV4B-F | AGAAGAAAACAACGATGAGACAAGG |
| HPIV4B-R | GTTTCCCTGGTTCACTCTCTTCA |
| EV/RV-F | TCCTCCGGCCCCTGAATGYGGCTAAT |
| EV/RV-R | GGAAACACGGWCACCCAAAGTA |
| MPV-F | GCGAGARATGGGYCCHGAATCTG |
| MPV-R | CCTGARGCATTDCCRAGAACAACAC |
| ADV-F | CCCMTTYAACCACCACCG |
| ADV-R | ACATCCTTBCKGAAGTTCCA |
| EV/RV VP4/2 round 1-F* | TCIGGIARYTTCCACCACCAICC |
| EV/RV VP4/2 round 1-R* | CTCCGGCCCCTGAATRYGGCTAA |
| EV/RV VP4/2 round 2-F* | ACCRASTACTTTGGGTGTCCGTG |
| EV/RV VP4/2 round 2-R* | CCGG YAAYTTCCASCACCA |
| EV-D68 VP1 round 1-F* | AACGCCGAACTTGGYGTG |
| EV-D68 VP1 round 1-R* | GGTAAGRGCACCAGTKGGT |
| EV-D68 VP1 round 2-F* | TCCCTAGCTTAAAYGCAGTTG |
| EV-D68 VP1 round 2-R* | CCAGTGGGTACRAACATTGC |
| Coronavirus-F* | GGTTGGGAYTAYCCTAARTGTGA |
| Coronavirus-R* | CCATCATCAGAWARAATCATCAT |
| RSVA-F* | GGTGCAGGGCAAGTGATGTTA |
| RSVA-R* | GCCAGCAGCATTGCCTAATAC |
| RSVB-F* | ATGGTTCAGGGCAAGTAATGCT |
| RSVB-R* | TCTCCTCCCAACTTCTGTGCA |
* indicates primers used in individual PCR assays.
All EV-D68 sequences were deposited in GenBank under accession numbers KJ556320–KJ556335. All base pair coordinates are provided relative to the original 1962 EV-D68 isolate, the Fermon strain (GenBank accession no. AY426531). A Maximum-likelihood phylogenetic tree of EV-D68 based on a 339 nucleotide fragment of the VP1 gene was constructed using Mega 5.2 software.
Acknowledgements
This work was supported by grants from the National Institutes of Health AI057158 (Northeast Biodefense Center-Lipkin), USAID PREDICT and the Defense Threat Reduction Agency. This work was funded in part by grants from the U.S. Department of Defense, Armed Forces Health Surveillance Center, Global Emerging Infection Surveillance and Response Systems (AFHSC-GEIS).
The authors are thankful to the patients who volunteered to participate in our study, to the Cambodian Ministry of Health, and to the health care workers at the field sites for helping to enroll the patients. We would like to acknowledge NAMRU-2 PP staff. We would like to thank Diane McFadden for her input on this manuscript. We would also like to thank NAMRU-2 PP staff, especially NAMRU-2 Scientific Director Capt. Patrick Blair for his critical reading of, and input for this manuscript.
Disclaimer
The opinions or assertions expressed herein are the private views of the authors and are not to be construed as representing those of the Department of Defense or Department of the Navy. For some authors this work was prepared as part of their official duties as U.S. Government employees. Title 17 U.S.C. §105 provides that ‘Copyright protection under this title is not available for any work of the United States Government.’ Title 17 U.S.C. §101 defines a U.S. Government work as a work prepared by a military service member or employee of the U.S. Government as part of that person’s official duties.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NL and WIL conceived the study. AR, USA, and SN participated in sample collection. NL, NM, and SS performed all molecular assays. KJ performed all bioinformatics analysis. NL, RT, DH, and WIL analyzed data and wrote the manuscript. All authors approved the final manuscript.
Contributor Information
Nary Ly, Email: naryly@gmail.com.
Rafal Tokarz, Email: rt2249@cumc.columbia.edu.
Nischay Mishra, Email: nm2641@cumc.columbia.edu.
Stephen Sameroff, Email: scs2178@columbia.edu.
Komal Jain, Email: kj2230@cumc.columbia.edu.
Agus Rachmat, Email: Agus.Rachmat@namru2.org.kh.
Ung Sam An, Email: usa@camnet.com.kh.
Steven Newell, Email: Newell@med.navy.mil.
Dustin J Harrison, Email: Dustin.Harrison@namru2.org.kh.
W Ian Lipkin, Email: wil2001@columbia.edu.
References
- 1.Luksic I, Kearns PK, Scott F, Rudan I, Campbell H, Nair H. Viral etiology of hospitalized acute lower respiratory infections in children under 5 years of age – a systematic review and meta-analysis. Croat Med J. 2013;54(2):122–134. doi: 10.3325/cmj.2013.54.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudan I, O'Brien KL, Nair H, Liu L, Theodoratou E, Qazi S, Luksic I, Fischer Walker CL, Black RE, Campbell H. Epidemiology and etiology of childhood pneumonia in 2010: estimates of incidence, severe morbidity, mortality, underlying risk factors and causative pathogens for 192 countries. J Glob Health. 2013;3(1):010401. doi: 10.7189/jogh.03.010101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudan I, Tomaskovic L, Boschi-Pinto C, Campbell H. Global estimate of the incidence of clinical pneumonia among children under five years of age. Bull World Health Organ. 2004;82(12):895–903. [PMC free article] [PubMed] [Google Scholar]
- 4.Profiles W-CCH: Cambodia pneumonia under 5. Geneva, Switzerland: World Health Organization Western Pacific Region, Country Health Information Profiles; 2010:45-65
- 5.Profiles W-CCH: Acute Respiratory Infection Cambodia. Geneva, Switzerland: World Health Organization Western Pacific Region, Country Health Information Profiles; 2011:43-61.
- 6.Kasper MR, Blair PJ, Touch S, Sokhal B, Yasuda CY, Williams M, Richards AL, Burgess TH, Wierzba TF, Putnam SD. Infectious etiologies of acute febrile illness among patients seeking health care in south-central Cambodia. Am J Trop Med Hyg. 2012;86(2):246–253. doi: 10.4269/ajtmh.2012.11-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blair PJ, Wierzba TF, Touch S, Vonthanak S, Xu X, Garten RJ, Okomo-Adhiambo MA, Klimov AI, Kasper MR, Putnam SD. Influenza epidemiology and characterization of influenza viruses in patients seeking treatment for acute fever in Cambodia. Epidemiol Infect. 2010;138(2):199–209. doi: 10.1017/S095026880999063X. [DOI] [PubMed] [Google Scholar]
- 8.Sreng B, Touch S, Sovann L, Heng S, Rathmony H, Huch C, Chea N, Kosal S, Siriarayaporn P, Pathanapornpandh N, Rehmet S, Cavailler P, Vong S, Bushy P. A description of influenza-like illness (ILI) sentinel surveillance in Cambodia, 2006–2008. Southeast Asian J Trop Med Public Health. 2010;41(1):97–104. [PubMed] [Google Scholar]
- 9.Briese T, Palacios G, Kokoris M, Jabado O, Liu Z, Renwick N, Kapoor V, Casas I, Pozo F, Limberger R, Perez-Brena P, Ju J, Lipkin WI. Diagnostic system for rapid and sensitive differential detection of pathogens. Emerg Infect Dis. 2005;11(2):310–313. doi: 10.3201/eid1102.040492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamson D, Renwick N, Kapoor V, Liu Z, Palacios G, Ju J, Dean A, St George K, Briese T, Lipkin WI. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J Infect Dis. 2006;194(10):1398–1402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tokarz R, Firth C, Madhi SA, Howie SR, Wu W, Sall AA, Haq S, Briese T, Lipkin WI. Worldwide emergence of multiple clades of enterovirus 68. J Gen Virol. 2012;93(Pt 9):1952–1958. doi: 10.1099/vir.0.043935-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeda T, Mizuta K, Abiko C, Aoki Y, Itagaki T, Katsushima F, Katsushima Y, Matsuzaki Y, Fuji N, Imamura T, Oshitani H, Noda M, Kimura H, Ahiko T. Acute respiratory infections due to enterovirus 68 in Yamagata, Japan between 2005 and 2010. Microbiol Immunol. 2012;56(2):139–143. doi: 10.1111/j.1348-0421.2012.00411.x. [DOI] [PubMed] [Google Scholar]
- 13.Meijer A, van der Sanden S, Snijders BE, Jaramillo-Gutierrez G, Bont L, van der Ent CK, Overduin P, Jenny SL, Jusic E, van der Avoort HG, Smith GJ, Donker GA, Koopmans MP. Emergence and epidemic occurrence of enterovirus 68 respiratory infections in The Netherlands in 2010. Virology. 2012;423(1):49–57. doi: 10.1016/j.virol.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 14.Todd AK, Hall RJ, Wang J, Peacey M, McTavish S, Rand CJ, Stanton JA, Taylor S, Huang QS. Detection and whole genome sequence analysis of an enterovirus 68 cluster. Virol J. 2013;10:103. doi: 10.1186/1743-422X-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imamura T, Fuji N, Suzuki A, Tamaki R, Saito M, Aniceto R, Galang H, Sombrero L, Lupisan S, Oshitani H. Enterovirus 68 among children with severe acute respiratory infection, the Philippines. Emerg Infect Dis. 2011;17(8):1430–1435. doi: 10.3201/eid1708.101328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piralla A, Girello A, Grignani M, Gozalo-Marguello M, Marchi A, Marseglia G, Baldanti F: Phylogenetic characterization of enterovirus 68 strains in patients with respiratory syndromes in Italy.J Med Virol 2013, 86(9):1590-1593. [DOI] [PMC free article] [PubMed]
- 17.Guerrier G, Goyet S, Chheng ET, Rammaert B, Borand L, Te V, Try PL, Sareth R, Cavailler P, Mayaud C, Guillard B, Vong S, Buchy P, Tarantola A. Acute viral lower respiratory tract infections in Cambodian children: clinical and epidemiologic characteristics. Pediatr Infect Dis J. 2013;32(1):e8–e13. doi: 10.1097/INF.0b013e31826fd40d. [DOI] [PubMed] [Google Scholar]
- 18.Arnott A, Vong S, Mardy S, Chu S, Naughtin M, Sovann L, Buecher C, Beaute J, Rith S, Borand L, Asgari N, Frutos R, Guillard B, Touch S, Deubel V, Buchy P. A study of the genetic variability of human respiratory syncytial virus (HRSV) in Cambodia reveals the existence of a new HRSV group B genotype. J Clin Microbiol. 2011;49(10):3504–3513. doi: 10.1128/JCM.01131-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vong S, Guillard B, Borand L, Rammaert B, Goyet S, Te V, Lorn Try P, Hem S, Rith S, Ly S, Cavailler P, Mayaud C, Buchy P. Acute lower respiratory infections in >/= 5 year -old hospitalized patients in Cambodia, a low-income tropical country: clinical characteristics and pathogenic etiology. BMC Infect Dis. 2013;13:97. doi: 10.1186/1471-2334-13-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schieble JH, Fox VL, Lennette EH. A probable new human picornavirus associated with respiratory diseases. Am J Epidemiol. 1967;85(2):297–310. doi: 10.1093/oxfordjournals.aje.a120693. [DOI] [PubMed] [Google Scholar]
- 21.Khetsuriani N, Lamonte-Fowlkes A, Oberst S, Pallansch MA. Enterovirus surveillance–United States, 1970–2005. MMWR Surveill Summ. 2006;55(8):1–20. [PubMed] [Google Scholar]
- 22.Centers for Disease Control and Prevention Clusters of acute respiratory illness associated with human enterovirus 68--Asia, Europe, and United States, 2008–2010. MMWR Morb Mortal Wkly Rep. 2011;60(38):1301–1304. [PubMed] [Google Scholar]
- 23.Linsuwanon P, Puenpa J, Suwannakarn K, Auksornkitti V, Vichiwattana P, Korkong S, Theamboonlers A, Poovorawan Y. Molecular epidemiology and evolution of human enterovirus serotype 68 in Thailand, 2006–2011. PLoS One. 2012;7(5):e35190. doi: 10.1371/journal.pone.0035190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiang Z, Gonzalez R, Wang Z, Ren L, Xiao Y, Li J, Li Y, Vernet G, Paranhos-Baccala G, Jin Q, Wang J. Coxsackievirus A21, enterovirus 68, and acute respiratory tract infection, China. Emerg Infect Dis. 2012;18(5):821–824. doi: 10.3201/eid1805.111376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imamura T, Okamoto M, Nakakita S, Suzuki A, Saito M, Tamaki R, Lupisan S, Roy CN, Hiramatsu H, Sugawara KE, Mizuta K, Matsuzaki Y, Suzuki Y, Oshitani H. Antigenic and receptor binding properties of enterovirus 68. J Virol. 2014;88(5):2374–2384. doi: 10.1128/JVI.03070-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tokarz R, Kapoor V, Wu W, Lurio J, Jain K, Mostashari F, Briese T, Lipkin WI. Longitudinal molecular microbial analysis of influenza-like illness in New York City, May 2009 through May 2010. Virol J. 2011;8:288. doi: 10.1186/1743-422X-8-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Centers for Disease Control and Prevention, Severe Respiratory Illness Associated with Enterovirus D68-Missouri and Illinois, 2014. Morbidity and Mortality Weekly Report, 2014. 63(36):798-799. [PMC free article] [PubMed]


