Abstract
Background
Control of classical swine fever (CSF) by vaccination ideally requires that field strain infection can be detected irrespective of the vaccination status of the herd. To inform on the usefulness of molecular tests compatible with genetic Differentiation of Infected from Vaccinated Animals (DIVA) principles when using live-attenuated vaccines, tonsil homogenates from a vaccination-challenge experiment were analyzed using a differential real-time qRT-PCR for the C-strain vaccine or real-time qRT-PCR assays developed to specifically detect the challenge strains used.
Results
In animals with high or moderate levels of blood viraemia, which were not, or not fully, protected by vaccination, challenge virus RNA was readily detected in tonsil homogenates. In three out of the seven vaccinated animals that had high or moderate viraemia, the vaccine strain RNA also could be detected but at lower levels. Lower but varying levels of challenge and/or vaccine virus RNA were detected in tonsil homogenate samples from animals with no or low-level viraemia, and in groups solely consisting of such animals, no transmission of infection to naïve in-contact animals occurred. In one group of animals that were vaccinated 3 days prior to challenge, viraemia levels varied from high to absent and transmission of challenge virus to naïve in-contact animals occurred. The DIVA assay revealed challenge virus in all tonsil homogenates from this group, even in those animals that did not have viraemia and were protected from clinical disease by vaccination. Such animals, particularly in a low biosecurity/informal farm setting, could constitute a risk for disease control in the field.
Conclusions
Genetic DIVA testing is useful for detecting the presence of field virus infection especially in non-viraemic animals without overt clinical signs but which are incompletely protected by vaccination. Such tests could particularly be useful to inform decisions prior to and during cessation of a control strategy that employs vaccination.
Keywords: Classical swine fever virus, DIVA PCR test, Infection, Vaccination, C-strain
Background
Classical swine fever virus (CSFV), a Pestivirus in the family Flaviviridae, causes a severe, often fatal, hemorrhagic disease of pigs and remains a serious threat to pig welfare and pork production worldwide [1]. The live, attenuated Riemser C-strain is a safe and effective vaccine for CSFV [2,3] that provides robust immunity to infection and prevents virus transmission within 5 days of vaccination [4]. C-strain vaccines are widely used in regions where classical swine fever (CSF) remains endemic, such as Asia and Central-South America [2]. Within the EU, use of C-strain vaccination in domestic pig herds is only authorized during emergency situations as the vaccine does not allow for DIVA analysis of samples using serological assays. However, deployment of C-strain vaccine in oral baits is permitted to control CSF in wild boar [5,6] and has been used to assist in the eradication of CSF from low biosecurity/informal swine holdings in Romania [7]. Whilst development of an efficacious DIVA vaccine is progressing [8-10] and the corresponding real-time RT-PCR [11] and serological DIVA tests have been developed [12], until such time as a DIVA capability is widely available, pre-existing C-strain vaccines remain important tools for CSF control.
CSFV vaccine viral RNA is not always readily detected post vaccination. Blood and tissue homogenate samples obtained following intramuscular vaccination may contain detectable levels of vaccine viral RNA when monitored by CSFV-specific, real-time RT-PCR assays. Commonly, post-vaccination positive results in blood are only obtained following inoculation with high doses of vaccine and, if detectable, are transient and occur for a maximum of 16 days post-vaccination (dpv) [8,13,14]. Low levels of vaccine RNA are detectable in tonsil or oropharyngeal fluid samples for extended periods of time [14,15], particularly in tonsil samples, with 98dpv being the longest duration reported [8]. These findings indicate that vaccination elicits a transient low-level viraemia which is hard to detect and does not result in high viral loads in vivo. There is no evidence that vaccinated pigs shed or horizontally transmit the C-strain vaccine ([14,16] and data not shown).
Leifer and colleagues developed a real-time RT-PCR assay to specifically detect the C-strain for use as a genetic DIVA test in circumstances where this vaccine might be used [17]. This assay was subsequently modified to accommodate nucleotide variations found in different C-strain vaccine batches and now also detects all genotype 1.1 CSFV strains, including the C-strain [18]. This modified genotype 1.1 assay has been successfully used, in combination with a real-time RT-PCR assay that detects all known CSFV strains, to distinguish C-strain vaccine from the circulating field strains which do not belong to genotype 1. For example, following a campaign to vaccinate wild boar in Germany using oral baits, C-strain vaccine could be distinguished from the genotype 2.3 field virus in CSFV RNA positive samples obtained from hunted wild boar [19]. Another differential assay has been developed that can distinguish the genetically similar Riemser C-strain, HCLV and LPC vaccine strains from most field strain genotypes except some of the genotype 3 strains [20,21]. Other differential real-time RT-PCR tests have been developed to specifically detect diverse CSFV vaccine strains [22-24]. However, because of sequence variation, they are not specific for the Riemser C-strain vaccine licensed for use in the EU.
Little is known regarding the likelihood of detecting vaccine or field CSFV strains in vaccinated and then challenged domestic pigs. We therefore assessed wild-type and vaccine CSFV RNA levels in pigs that were vaccinated with the Riemser C-strain and then challenged intranasally with one of two moderately virulent field strains [4], either the genotype 2.1 UK2000/7.1 strain [25], or the genotype 3.3 Thai CBR/93 strain [26]. Challenge strains with distinct genotypes to the vaccine strain were chosen to enable assessment of vaccine efficacy. The genetic diversity of CSFV strains has impeded efforts to develop a differential assay that will specifically detect all virulent field strains but exclude all vaccine strains. In this study, we therefore developed differential real-time, qRT-PCR assays that were specifically designed to detect the individual challenge strains. The RNA levels of the vaccine or challenge viruses in tonsil homogenates from pigs with varied status of protection or disease were then assessed.
Results
In order to assess the specificity of each differential PCR design, a template mixture containing 104 copies of C-strain, CBR/93 and UK2000/7.1 RNA was analyzed using the differential genotype 1.1 real-time RT-PCR developed for the Riemser C-strain [17,18] together with quantification standards or differential real-time qRT-PCR assays developed specifically to detect the challenge strains. The specificity of the genotype 1.1 assay depends on a single nucleotide at the 3’end of each primer, and, as expected, only produced an amplicon from the C-strain template and not from the other viral strains (Figure 1A). Additional amplicons were consistently produced in lower concentrations when using this primer pair, possibly owing to the requirement for a redundant primer. In contrast, primers used in the assays for detection of challenge strains CBR/93 or UK2000/7.1 (HE5 and HE4.1/4.2) amplified all strains equivalently, therefore the discrimination by these assays depended entirely on the specificity of the cognate locked nucleic acid (LNA) probe. Each real-time qRT-PCR assay was specific for the intended target strain and did not detect the non-cognate strain (Figure 1B to D). The sensitivity of the differential assays for the challenge strains were evaluated using a standard curve prepared from a serial 10-fold dilution of viral RNA from the corresponding viral strain that had been quantified using a pan-CSFV PCR [27]. Although having a less sensitive detection limit than the 10 copies/assay reported for the genotype 1.1 assay [17], the challenge strain differential assays allowed reliable quantification of template RNA down to 102 copies/assay (Table 1).
Figure 1.
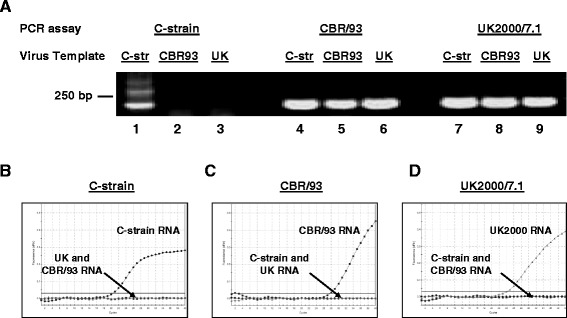
Specificity of differential PCR assays for detection of C-strain (C-str), CBR/93 and UK2000/7.1 (UK) RNA templates. Panel A: Whereas the genotype 1 specific real-time RT-PCR assay previously described [17,18] only produced a C-strain vaccine amplicon (Lane 1–3), the primer mix HE5 and HE4.1/4.2 amplified the C-strain (C-str), CBR/93 and UK2000/7.1 (UK) strains equally (Lanes 4–9). Panels B to D: Differential real-time RT-PCR analysis of a mixture of 104 copies of the three viral RNA templates revealed that the C-strain (B), CBR/93 (C) or UK2000/7.1 (D) assays were specific for their respective templates.
Table 1.
Detection limits of the challenge strain-specific probes assessed using a dilution series of standard concentrations of their cognate templates
| DIVA assay Ct | ||
|---|---|---|
| CBR/93 | UK2000/7.1 | |
| Standard copy number | ||
| 5 × 10 5 | 27.3 | 30.7 |
| 5 × 10 4 | 31.3 | 34.2 |
| 5 × 10 3 | 34.9 | 37.6 |
| 5 × 10 2 | 38.5 | 42.0 |
| 5 × 10 1 | None | None |
Vaccination with the C-strain produces very low or undetectable levels of viraemia in blood ([8,13,14] and unpublished observations). To identify the most appropriate tissue sample for detection of C-strain, homogenates of tonsil, spleen and lateral retropharyngeal, ileocecal, mandibular and pre-scapular lymph nodes from animals euthanized between 11 and 33 days post-challenge were assessed. Tonsil samples allowed most consistent detection of C-strain RNA in vaccinated animals (data not shown) and this was in agreement with previous studies [8,14,15,28]. The amounts of total CSFV, C-strain, CBR/93 and UK2000/7.1 RNA were determined in tonsil tissue homogenates prepared from animals that had been vaccinated 5, 3 or 1 days prior to challenge with either the CBR/93 or UK2000/7.1 (Table 2). Control homogenates were prepared from mock-vaccinated, infected animals [4]. As expected, the total viral load increased as the level of protection afforded by vaccination decreased. When high or moderate blood viraemia occurred, this correlated with a high level of total CSFV in tonsil which was mainly comprised of challenge virus. The animals in this category (all animals in Groups A and E and some animals in Groups D and H) had clinical signs of CSF and such animals would be expected to be identified as infected. Herds harboring such animals would also be correctly designated CSFV positive and indeed, naïve animals co-housed with these experimental groups also developed CSF.
Table 2.
Detection of CSFV in tonsil extracts
| (A) CBR/93 | (B) UK2000/7.1 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pig group and number | Blood viraemia | Tonsil RNA levels (log 10 copies/ng) | Clinical outcome | End day (dpi) | Pig group and number | Blood viraemia | Tonsil RNA levels (log 10 copies/ng) | Clinical outcome | End day (dpi) | ||||
| CBR/93 | C-strain | Total CSFV | UK2000/7.1 | C-strain | Total CSFV | ||||||||
| Mock vaccination* | Mock vaccination* | ||||||||||||
| A.1 | High | 7.10 | 0 | 8.62 | Euthanized | 17 | E.1 | High | 9.39 | 0 | 9.43 | Euthanized | 17 |
| A.2 | Moderate | 4.99 | 0 | 8.21 | Euthanized | 21 | E.2 | Moderate | 8.50 | 0 | 8.87 | Euthanized | 15 |
| A.3 | High | 6.23 | 0 | 8.18 | Euthanized | 21 | E.3 | High | 8.35 | 0 | 8.56 | Euthanized | 15 |
| A.4 | Moderate | 6.02 | 0 | 8.06 | Euthanized | 17 | E.4 | High | 7.98 | 0 | 8.48 | Euthanized | 17 |
| Vaccination -5 dpi | Vaccination -5 dpi | ||||||||||||
| B.1 | Low | < assay | 4.61 | 5.19 | Healthy | 25 | F.1 | Low | 0 | 3.64 | 4.38 | Healthy | 29 |
| B.2 | ⊗ | 0 | 2.76 | 3.62 | Healthy | 25 | F.2 | ⊗ | 0 | 3.23 | 3.61 | Healthy | 29 |
| B.3 | ⊗ | 0 | 1.61 | 3.76 | Healthy | 30 | F.3 | ⊗ | 0 | 2.93 | 3.42 | Healthy | 30 |
| B.4 | ⊗ | 0 | 1.92 | 3.25 | Healthy | 30 | F.4 | ⊗ | 0 | 2.19 | 3.41 | Healthy | 30 |
| B.5 | ⊗ | 0 | 1.66 | 2.96 | Healthy | 30 | F.5 | ⊗ | 0 | 1.81 | 0.87 | Healthy | 29 |
| B.6 | ⊗ | 0 | 2.21 | 2.87 | Healthy | 25 | F.6 | Low | 0 | 1.83 | 0.11 | Healthy | 30 |
| Vaccination -3 dpi | Vaccination -3 dpi* | ||||||||||||
| C.1 | Low | < assay | 0 | 4.17 | Healthy | 33 | G.1 | High | 7.79 | 2.75 | 8.33 | unrelated inf. | 22 |
| C.2 | Low | 0 | 1.65 | 3.74 | Healthy | 32 | G.2 | Low | 8.01 | 0 | 8.23 | Healthy | 28 |
| C.3 | ⊗ | 0 | 1.24 | 3.45 | Healthy | 33 | G.3 | Low | 2.75 | 4.29 | 5.13 | Healthy | 28 |
| C.4 | Low | 0.5 | 1.45 | 3.43 | Healthy | 32 | G.4 | ⊗ | 1.06 | 3.80 | 4.78 | Healthy | 24 |
| C.5 | Low | < assay | 1.35 | 3.16 | Healthy | 32 | G.5 | Low | 2.63 | 1.74 | 4.08 | Healthy | 24 |
| C.6 | ⊗ | 0 | 0 | 2.51 | Healthy | 33 | G.6 | ⊗ | 1.98 | 1.12 | 3.35 | Healthy | 24 |
| Vaccination -1 dpi* | Vaccination -1 dpi* | ||||||||||||
| D.1 | High | 7.28 | 0 | 8.69 | Euthanized | 19 | H.1 | High | 7.51 | 0 | 8.69 | Euthanized | 19 |
| D.2 | Moderate | 7.75 | 4.26 | 8.68 | Euthanized | 12 | H.2 | High | 7.98 | 0 | 8.21 | Euthanized | 12 |
| D.3 | Moderate | 3.03 | < assay | 5.62 | Healthy | 23 | H.3 | Moderate | 7.99 | 4.16 | 8.16 | Euthanized | 11 |
| D.4 | Low | 2.28 | 1.02 | 5.10 | unrelated inf. | 19 | H.4 | Low | 4.65 | 0 | 5.43 | Healthy | 23 |
| D.5 | Low | 0.76 | 2.53 | 4.15 | Healthy | 23 | H.5 | Low | 2.59 | 1.00 | 4.41 | Healthy | 23 |
| D.6 | Low | < assay | < assay | 2.77 | Healthy | 23 | H.6 | Low | 2.67 | 1.58 | 4.16 | Healthy | 23 |
*Naïve animals co-housed together with these groups became CSFV infected.
Pigs were mock-vaccinated or vaccinated at 1, 3, or 5 days before inoculation with (A) the CBR/93 or (B) the UK2000/7.1 field strains. Blood viraemia is recorded as not detectable (⊗), low, moderate or high. The amount of CBR/93, UK20007.1 or C-strain vaccine RNA determined using differential real-time qRT-PCR or of total CSFV RNA quantified using a routine real-time qRT-PCR assay is shown as log10 viral RNA copy per ng 18S RNA. Bold type indicates high copy number. Samples with high Ct values below the quantifiable limit of an assay (< assay) or no Ct value (0) are indicated. The clinical outcome shows those animals that reached the humane endpoint and were euthanized. Animals indicated as healthy had clinical scores that did not exceed 4. Tonsil samples were obtained on the indicated days post inoculation (dpi).
The situation was more complex in Groups B, C and F (Table 2) where the animals developed low or no viraemia. These animals were protected by vaccination, appeared clinically healthy and did not transmit CSFV to naïve animals placed in contact. In one viraemic animal (C4), a mixture of challenge and vaccine strain could be detected in tonsil. However, in other animals, the source of viraemia was probably the C-strain (CBR/93 challenged animals B.1, C.2, and C.5 and UK2000/7.1 challenged animals F.1 and F.6) or could not be distinguished (CBR93 challenged animal C.1). In the absence of blood viraemia, if CSFV RNA was detected in tonsil samples, this was due to C-strain vaccine and challenge virus was not detectable in tonsil tissue.
The results obtained for Group G, challenged with the UK2000/7.1 strain three days post-vaccination, were varied. This group included one animal (G.1) that developed clinical signs unrelated to CSFV infection and subsequent elevated CSFV viraemia. All other challenged animals in the group remained clinically healthy, despite low levels of viraemia in some (G2, G3 and G5) but naïve in-contact animals became infected. Interestingly, two animals (G.4 and G.6) did not develop viraemia and low levels of challenge virus could be detected in tonsil. It is possible that virus shed by the viraemic animal G.1 was taken up by animals G.4 and G.6 and was retained in tonsil but vaccine-mediated protection limited infection.
Discussion
In this study, the C-strain vaccine, CBR/93 and UK2000/7.1 CSFV strains could clearly be individually distinguished in clinical samples using a differential qRT-PCR approach. In high or moderately viraemic animals, the challenge strain contributed the majority of total viral RNA in tonsil extracts. In the field, such infected animals would likely have clinical signs and would be correctly identified as CSFV positive by routine CSFV molecular tests that do not discriminate vaccine from challenge virus. Consequently, a herd including such animals would be identified as infected. In animals with low-level viraemia, tonsil extracts contained detectable levels of either vaccine and/or challenge virus. In a vaccination strategy these animals would be difficult to identify based on clinical criteria and testing with non-discriminatory tests could either miss a field infection or lead to incorrect declarations of disease in a herd.
This study has shown that the situation is even more complex in those animals which do not develop viraemia but have variable protection status and may be infected with field virus. In the non-viraemic animals that had been challenged with the CBR/93 strain 3 days after vaccination, the vaccine strain could be detected in tonsil samples from one animal (Table 2, C.3) but the identity of the strain could not be determined in the second animal (C.6). Although the four other challenged animals exhibited a transient viraemia, a very low level of challenge virus was only found in one animal and in-contact animals remained healthy, indicating there was no transmission of challenge virus [4]. A converse outcome was observed when animals were challenged with the UK2000/7.1 strain at 3 days post-vaccination (Group G). The presence of one animal with a high level of viraemia in this group (G.1), which most likely resulted in infection of in-contact animals [4], did not allow clear conclusions to be made. However, it is noteworthy that the challenge strain RNA could be detected in tonsil samples from two non-viraemic, challenged animals that were apparently protected by vaccination (G.4 and G.6). Other studies have shown that detection of challenge virus does not always indicate infection. Presence of wild-type virus RNA in clinically healthy, vaccinated animals has been reported [15,29] and retention of challenge virus RNA in oropharyngeal fluid of a vaccinated animal was reported not to cause infection of naïve in-contact animals [15] possibly as a result of virus being sequestered in a non-infectious state within immune complexes or phagocytic cells. The likelihood of infectious virus being transmitted from such non-viraemic, vaccinated, animals to initiate infection remains unclear, but could compromise a vaccination campaign for CSFV eradication, particularly in low biosecurity farming sectors.
These risks can be better understood by applying DIVA assays for monitoring a vaccination strategy. Ideally, a DIVA assay would detect field strain infection, irrespective of whether animals had been vaccinated. Field-strain specific RT-PCR assays have been developed [22,23], however they rely upon pre-existing sequence information about circulating field strains and are susceptible to viral genome changes as such assays often rely on one or two nucleotide differences between the field strain and vaccine. An alternative approach is to test samples for the presence of all CSFV strains and then apply a vaccine-specific test [17,18,20]. This strategy assumes that an animal is not infected if vaccine-specific RNA is detected in tonsil tissue. Whilst this may be an acceptable risk in a wild boar vaccination campaign, a potential failure to detect field virus in the domestic pig sector would be of concern. This issue can be overcome by use of a DIVA vaccine, such as the candidate CP7_E2alf vaccine, that has the potential for allowing serological and genetic DIVA assays to be applied [8-10].
Conclusions
Our DIVA study has shown that non-immune animals (mock-vaccinated or vaccinated 1 day prior to challenge) which develop a high or moderate level of blood viraemia, harbor challenge virus that can clearly be detected in tonsil tissue. Completely protected animals (vaccinated 5 days or more before challenge) develop low, if any viraemia. Only vaccine virus was detected in the tonsil of the protected animals, therefore applying a DIVA strategy based on the detection of total CSFV to exclude infection based on the detection of vaccine would represent little risk of missing infection in an individual animal. However, for animals that are not completely protected, for example due to poor pre-existing health status or exposure to field virus before or shortly after vaccination, there is a greater risk of individual animals harboring a mixture of field and vaccine viruses. The potential consequence of this risk to the particular vaccination and testing strategy must therefore be considered. Vaccinated domestic pigs that also carry infectious virus, while probably not common, are difficult to identify within a herd. Such animals could potentially serve as a reservoir of infectious virus and initiate re-emergence of disease after the end of a vaccination campaign. This risk could be mitigated by imposing movement restrictions for two weeks after vaccination and monitoring CSFV in unvaccinated sentinel animals within a herd. These control efforts could be supported by applying genetic DIVA tests to assess herds or slaughter animals during and before ending a vaccination campaign.
Methods
Tissue samples were obtained at necroscopy during a previously described study [4] and stored at −80°C. Briefly, in this study conducted following AHVLA Ethics committee approval and in accordance with the UK Animals (Scientific Procedures) Act 1986, ten-week old domestic pigs, were vaccinated by intramuscular injection of 100PD50 of the Riemser C-strain vaccine (Riemser Arzneimittel) according to the manufacturer’s protocol at intervals of 1, 3, or 5 days prior to intranasal inoculation with either the UK2000/7.1 [25] or CBR/93 field strain [26]. Control pigs were mock vaccinated prior to inoculation. Pigs were euthanized on completion of the study at 30–33 days post inoculation (dpi) or on the dpi indicated in Table 2 as determined by the score assigned to their clinical signs [30]. Blood viraemia results were reported previously [4] and are indicated in Table 2 as positive if total CSFV viral RNA could be detected in blood from an animal on two or more consecutive sampling days. Viraemia was designated low if log10 viral RNA copy number/μl blood did not exceed 3.99, moderate if between 4.00 and 5.99 and high if in excess of 6.00.
Tissue samples between approximately 0.2 to 0.5 g were homogenized in 1.5 ml PBS using an Omni-TH Tissue Homogenizer (Omni International). Homogenates were extracted using the QIAamp viral RNA mini kit (Qiagen). The total CSFV genomic RNA copy number was determined by real-time RT-PCR [27,31] together with quantification standards. The C-strain was detected using a differential real-time qRT-PCR assay [17] with a modified 5’ primer [18] as the C-strain batch used to vaccinate pigs contained a mixed population of virus genotypes with a purine base in the 1649 position corresponding to the 3’ terminal nucleotide of the 5’ primer used in this discriminatory assay. The PCR strategy to differentiate the UK2000/7.1 and Thai CBR/93 strains from the vaccine virus was designed to equally amplify a 233 nucleotide region within the Npro gene using the 5’ primer HE5 (TGGGAGTGGAGGAACCG) and a 2:1 molar ratio mixture of the 3’ primers HE4.1/4.2 (GCCCATGTAGTCCTGATAGWAGAC) and (GCCCATGTAGTCCTGATAAAAGAC). Locked nucleic acid (LNA) probes were used to discriminate the wild-type challenge strains from the vaccine. The probes used to detect the two challenge strains were UK2000/7.1: Fam-CCACT(+A)AC(+T)GG(+T)CCTAG(+G)TG(+G)TTT-BHQ1 and CBR/93: Fam-TAG(+G)TTCAG(+C)GTTG(+A)(+C)(+T)GTG-BHQ1. Reaction mixes were prepared using the qRT-PCR one-step SuperScript III kit (Life Technologies) and contained 12.5 μl 2× RT-PCR reaction mix, 0.5 μl Superscript III/Taq polymerase enzyme mix, 600 nM of the 5’ primer and the 3’ primer mix, 100 nM probe, 50 nM ROX and 5 μl template RNA in a total reaction volume of 25 μl. The thermocycling program used for the differential PCR reactions was: 50°C for 30 min and 95°C for 2 min followed by 45 cycles of 95°C for 15 s and 60°C for 1 min. In order to obtain RNA quantification standards, RNA was extracted from stocks of each virus strain and the concentration of total viral genomic RNA copies was quantified using an in vitro transcribed RNA standard and the CSFV-specific qRT-PCR [27]. These stocks were then used to prepare ten-fold dilution series that allowed generation of standard curves specific for each strain and quantification of the respective viral RNA copy numbers in each sample. Samples that had a Ct value higher than the quantifiable detection limit of an assay are indicated (<assay). The amount of 18S RNA in the tonsil tissue extracts was quantified using a QuantumRNA Universal 18S kit (Ambion) and was used to normalize the viral copy number result according to total RNA input. Viral copy number amounts in tonsil homogenates measured as log10 viral RNA copy number/ng 18S RNA were similarly classified as low if below 3.99, intermediate if between 4.00 and 5.99 and high if in excess of 6.00.
Acknowledgements
The authors would like to thank colleagues who participated in the vaccination-challenge studies that provided the samples used in this study and Dr Falko Steinbach for critical review of the manuscript. This work was funded by Defra Project SE0778.
Footnotes
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
HE designed the protocol and discriminatory qRT-PCR assays, interpreted the data and wrote the manuscript. BC acquired, collated and analyzed the data. OS developed and optimized the experimental methods used. HC consulted on the experimental design and critically reviewed the manuscript. All authors have approved the final version of the manuscript.
Contributor Information
Helen E Everett, Email: Helen.Everett@apha.gsi.gov.uk.
Bentley S Crudgington, Email: bentley.crudgington@apha.gsi.gov.uk.
Olubukola Sosan-Soulé, Email: bukisoule@gmail.com.
Helen R Crooke, Email: helen.crooke@apha.gsi.gov.uk.
References
- 1.Edwards S, Fukusho A, Lefevre PC, Lipowski A, Pejsak Z, Roehe P, Westergaard J. Classical swine fever: the global situation. Vet Microbiol. 2000;73(2–3):103–119. doi: 10.1016/S0378-1135(00)00138-3. [DOI] [PubMed] [Google Scholar]
- 2.Suradhat S, Damrongwatanapokin S, Thanawongnuwech R. Factors critical for successful vaccination against classical swine fever in endemic areas. Vet Microbiol. 2007;119(1):1–9. doi: 10.1016/j.vetmic.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Huang YL, Deng MC, Wang FI, Huang CC, Chang CY. The challenges of classical swine fever control: modified live and E2 subunit vaccines. Virus Res. 2014;179:1–11. doi: 10.1016/j.virusres.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 4.Graham SP, Everett HE, Haines FJ, Johns HL, Sosan OA, Salguero FJ, Clifford DJ, Steinbach F, Drew TW, Crooke HR. Challenge of pigs with classical swine fever viruses after C-strain vaccination reveals remarkably rapid protection and insights into early immunity. PLoS One. 2012;7(1):e29310. doi: 10.1371/journal.pone.0029310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Oirschot JT. Emergency vaccination against classical swine fever. Dev Biol (Basel) 2003;114:259–267. [PubMed] [Google Scholar]
- 6.von Ruden S, Staubach C, Kaden V, Hess RG, Blicke J, Kuhne S, Sonnenburg J, Frohlich A, Teuffert J, Moennig V. Retrospective analysis of the oral immunisation of wild boar populations against classical swine fever virus (CSFV) in region Eifel of Rhineland-Palatinate. Vet Microbiol. 2008;132(1–2):29–38. doi: 10.1016/j.vetmic.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 7.Postel A, Moennig V, Becher P. Classical swine fever in Europe–the current situation. Berl Munch Tierarztl Wochenschr. 2013;126(11–12):468–475. [PubMed] [Google Scholar]
- 8.Tignon M, Kulcsar G, Haegeman A, Barna T, Fabian K, Levai R, Van der Stede Y, Farsang A, Vrancken R, Belak K, Koenen F. Classical swine fever: comparison of oronasal immunisation with CP7E2alf marker and C-strain vaccines in domestic pigs. Vet Microbiol. 2010;142(1–2):59–68. doi: 10.1016/j.vetmic.2009.09.044. [DOI] [PubMed] [Google Scholar]
- 9.Blome S, Gabriel C, Schmeiser S, Meyer D, Meindl-Bohmer A, Koenen F, Beer M. Efficacy of marker vaccine candidate CP7_E2alf against challenge with classical swine fever virus isolates of different genotypes. Vet Microbiol. 2014;169(1–2):8–17. doi: 10.1016/j.vetmic.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Uttenthal A, Parida S, Rasmussen TB, Paton DJ, Haas B, Dundon WG. Strategies for differentiating infection in vaccinated animals (DIVA) for foot-and-mouth disease, classical swine fever and avian influenza. Expert Rev Vaccines. 2010;9(1):73–87. doi: 10.1586/erv.09.130. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Hoffmann B, Baule C, Beer M, Belak S, Widen F. Two real-time RT-PCR assays of classical swine fever virus, developed for the genetic differentiation of naturally infected from vaccinated wild boars. J Virol Methods. 2009;159(1):131–133. doi: 10.1016/j.jviromet.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Aebischer A, Muller M, Hofmann MA. Two newly developed E(rns)-based ELISAs allow the differentiation of Classical Swine Fever virus-infected from marker-vaccinated animals and the discrimination of pestivirus antibodies. Vet Microbiol. 2013;161(3–4):274–285. doi: 10.1016/j.vetmic.2012.07.046. [DOI] [PubMed] [Google Scholar]
- 13.Lorena J, Barlic-Maganja D, Lojkic M, Madic J, Grom J, Cac Z, Roic B, Terzic S, Lojkic I, Polancec D, Cajavec S. Classical swine fever virus (C strain) distribution in organ samples of inoculated piglets. Vet Microbiol. 2001;81(1):1–8. doi: 10.1016/S0378-1135(01)00321-2. [DOI] [PubMed] [Google Scholar]
- 14.Koenig P, Hoffmann B, Depner KR, Reimann I, Teifke JP, Beer M. Detection of classical swine fever vaccine virus in blood and tissue samples of pigs vaccinated either with a conventional C-strain vaccine or a modified live marker vaccine. Vet Microbiol. 2007;120(3–4):343–351. doi: 10.1016/j.vetmic.2006.10.034. [DOI] [PubMed] [Google Scholar]
- 15.Eble PL, Geurts Y, Quak S, Moonen-Leusen HW, Blome S, Hofmann MA, Koenen F, Beer M, Loeffen WL. Efficacy of chimeric Pestivirus vaccine candidates against classical swine fever: protection and DIVA characteristics. Vet Microbiol. 2013;162(2–4):437–446. doi: 10.1016/j.vetmic.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 16.Kaden V, Lange E, Kuster H, Muller T, Lange B. An update on safety studies on the attenuated "RIEMSER Schweinepestoralvakzine" for vaccination of wild boar against classical swine fever. Vet Microbiol. 2010;143(2–4):133–138. doi: 10.1016/j.vetmic.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 17.Leifer I, Depner K, Blome S, Le Potier MF, Le Dimna M, Beer M, Hoffmann B. Differentiation of C-strain "Riems" or CP7_E2alf vaccinated animals from animals infected by classical swine fever virus field strains using real-time RT-PCR. J Virol Methods. 2009;158(1–2):114–122. doi: 10.1016/j.jviromet.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Leifer I, Everett H, Hoffmann B, Sosan O, Crooke H, Beer M, Blome S. Escape of classical swine fever C-strain vaccine virus from detection by C-strain specific real-time RT-PCR caused by a point mutation in the primer-binding site. J Virol Methods. 2010;166(1–2):98–100. doi: 10.1016/j.jviromet.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Blome S, Gabriel C, Staubach C, Leifer I, Strebelow G, Beer M. Genetic differentiation of infected from vaccinated animals after implementation of an emergency vaccination strategy against classical swine fever in wild boar. Vet Microbiol. 2011;153(3-4):373–376. doi: 10.1016/j.vetmic.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 20.Liu L, Xia H, Everett H, Sosan O, Crooke H, Meindl-Bohmer A, Qiu HJ, Moennig V, Belak S, Widen F. A generic real-time TaqMan assay for specific detection of lapinized Chinese vaccines against classical swine fever. J Virol Methods. 2011;175(2):170–174. doi: 10.1016/j.jviromet.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 21.Widen F, Everett H, Blome S, Fernandez Pinero J, Uttenthal A, Cortey M, von Rosen T, Tignon M, Liu L. Comparison of two real-time RT-PCR assays for differentiation of C-strain vaccinated from classical swine fever infected pigs and wild boars. Res Vet Sci. 2014;97(2):455–457. doi: 10.1016/j.rvsc.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Zhao JJ, Cheng D, Li N, Sun Y, Shi Z, Zhu QH, Tu C, Tong GZ, Qiu HJ. Evaluation of a multiplex real-time RT-PCR for quantitative and differential detection of wild-type viruses and C-strain vaccine of Classical swine fever virus. Vet Microbiol. 2008;126(1–3):1–10. doi: 10.1016/j.vetmic.2007.04.046. [DOI] [PubMed] [Google Scholar]
- 23.Cho HS, Park SJ, Park NY. Development of a reverse-transcription polymerase chain reaction assay with fluorogenic probes to discriminate Korean wild-type and vaccine isolates of Classical swine fever virus. Can J Vet Res. 2006;70(3):226–229. [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang XJ, Han QY, Sun Y, Belak S, Liu L, Qiu HJ. Development of a loop-mediated isothermal amplification for visual detection of the HCLV vaccine against classical swine fever in China. J Virol Methods. 2011;171(1):200–205. doi: 10.1016/j.jviromet.2010.10.025. [DOI] [PubMed] [Google Scholar]
- 25.Sandvik T, Drew T, Paton D. CSF virus in East Anglia: where from? Vet Rec. 2000;147(9):251. [PubMed] [Google Scholar]
- 26.Parchariyanon S, Inui K, Damrongwatanapokin S, Pinyochon W, Lowings P, Paton D. Sequence analysis of E2 glycoprotein genes of classical swine fever viruses: identification of a novel genogroup in Thailand. Dtsch Tierarztl Wochenschr. 2000;107(6):236–238. [PubMed] [Google Scholar]
- 27.Hoffmann B, Beer M, Schelp C, Schirrmeier H, Depner K. Validation of a real-time RT-PCR assay for sensitive and specific detection of classical swine fever. J Virol Methods. 2005;130(1–2):36–44. doi: 10.1016/j.jviromet.2005.05.030. [DOI] [PubMed] [Google Scholar]
- 28.Kaden V, Lange E, Riebe R, Lange B. Classical swine fever virus Strain 'C': how long is it detectable after oral vaccination? J Vet Med B Infect Dis Vet Public Health. 2004;51(6):260–262. doi: 10.1111/j.1439-0450.2004.00766.x. [DOI] [PubMed] [Google Scholar]
- 29.Huang YL, Pang VF, Pan CH, Chen TH, Jong MH, Huang TS, Jeng CR. Development of a reverse transcription multiplex real-time PCR for the detection and genotyping of classical swine fever virus. J Virol Methods. 2009;160(1–2):111–118. doi: 10.1016/j.jviromet.2009.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Everett H, Salguero FJ, Graham SP, Haines F, Johns H, Clifford D, Nunez A, La Rocca SA, Parchariyanon S, Steinbach F, Drew T, Crooke H. Characterisation of experimental infections of domestic pigs with genotype 2.1 and 3.3 isolates of classical swine fever virus. Vet Microbiol. 2010;142(1–2):26–33. doi: 10.1016/j.vetmic.2009.09.039. [DOI] [PubMed] [Google Scholar]
- 31.Graham SP, Everett HE, Johns HL, Haines FJ, La Rocca SA, Khatri M, Wright IK, Drew T, Crooke HR. Characterisation of virus-specific peripheral blood cell cytokine responses following vaccination or infection with classical swine fever viruses. Vet Microbiol. 2010;142(1–2):34–40. doi: 10.1016/j.vetmic.2009.09.040. [DOI] [PubMed] [Google Scholar]


