Abstract
The extracellular matrix (ECM) is a complex, spatially inhomogeneous environment that is host to myriad cell–receptor interactions that promote changes in cell behavior. These biological systems can be probed and simulated with engineered surfaces,but doing so demands careful control over the arrangement of ligands. Here, we describe how such surfaces can be fabricated by utilizing polymer pen lithography (PPL), which is a cantilever-free scanning probe lithographic method that utilizes polymeric pen arrays to generate patterns over large areas. With the advent of PPL, fundamental questions in cell biology can be answered by recapitulating cell–ECM interactions to explore how these interactions lead to changes in cell behavior. Here, we describe an approach for the combinatorial screening of cell adhesion behavior to gain understanding of how ECM protein feature size dictates osteogenic differentiation of mesenchymal stem cells. The technique outlined here is generalizable to other biological systems and can be paired with quantitative analytical methods to probe important processes such as cell polarization, proliferation, signaling, and differentiation.
INTRODUCTION
Probing the extracellular microenvironment to obtain information about how cell–receptor interactions occur and trigger downstream signaling cascades is a rapidly growing field of multidisciplinary research. However, understanding the effect of cell–receptor events requires careful control at the molecular level over a variety of parameters, such as the arrangement, density, and position of extracellular factors. These parameters can be controlled by molecular patterning techniques, such as dippen nanolithography (DPN) or polymer pen lithography (PPL), thus allowing one to answer fundamental questions pertaining to cellular function and the response of cells to external stimuli. These tools are ideally suited for creating models of the extracellular matrix (ECM) at the length scales of focal adhesions in a deliberate, programmable, and systematic fashion; therefore, they are well suited for studying the fundamental underpinnings of cell adhesion, motility, stem cell differentiation, and many other biological processes.
In 1999, Mirkin and co-workers invented DPN, a scanning probe technique that allows one to deliver molecules onto a surface in a direct-write manner with sub-50 nm resolution (Piner, Zhu, Xu, Hong, & Mirkin, 1999). In this technique, an atomic force microscope (AFM) probe, which consists of a cantilever with a sharp tip at its end, is coated with a solution containing a molecule of interest or “ink.” This coated probe is then used to deposit molecules onto a substrate, much as a pen deposits ink onto paper. The DPN process is mediated by the formation of a water meniscus between the tip and the substrate which allows the ink material to diffuse from the tip to the surface, as was initially demonstrated by the deposition and formation of self-assembled monolayers (SAMs) composed of alkanethiols on Au-coated substrates. Since then, this technique has been used to deposit many other biologically relevant materials such as proteins (Lee, Park, Mirkin, Smith, & Mrksich, 2002), DNA (Demers et al., 2002), polymers (Su, Aslam, Fu, Wu, & Dravid, 2004), and a variety of small organic and inorganic molecules on many different substrates (Ginger, Zhang, & Mirkin, 2004; Salaita et al., 2005; Salaita, Wang, & Mirkin, 2007; Su, Liu, Li, Dravid, & Mirkin, 2002). Once patterned, nanoscale arrangements of biomolecules can then be used as arrays for combinatorial screening and to address fundamental questions in cell biology (Giam et al., 2012). The functionality of such arrays demonstrates the relevance of DPN as a method of choice for conducting studies inthe biological sciences. Forexample,Niemeyer and co-workersshowedthatDPN can be used to generate protein arrays and probe protein–receptor interactions inside living cells and that this method can be coupled with traditional biological techniques for the study of biological processes (Gandor et al., 2013).
Following extensive research into methods of patterning diverse materials by DPN, it was recognized that throughput was the main limiting factor for many studies, as it will take prohibitively long to pattern cm2 areas with a single probe. To address this, 1D and 2D arrays of cantilevers were developed to transition this technique from a serial writing process to a parallel one (Salaita et al., 2005); however, reliance on these fragile and expensive cantilever arrays limited wide-spread adoption of these approaches. To circumvent these limitations, a new architecture that did not rely on cantilevers for mechanical compliance of the probes was developed to generate arbitrary patterns in a high-throughput fashion (Huo et al., 2008). This cantilever-free method, termed PPL, utilizes a 2D array of elastomeric pyramidal probes that rest on an elastomeric film on a rigid planar substrate to deliver molecules to a surface with nanoscale control over large areas. In contrast to microcontact printing, which relies on a lithographically defined master (Qin, Xia, & Whitesides, 2010), PPL is a mask-free technique, allowing a user to pattern arbitrary arrangements of materials on a surface. Furthermore, pen arrays with as many as 11 million tips can be easily and cheaply fabricated, thus significantly increasing the patterning throughput without compromising robustness and cost.
In this chapter, we detail a canonical PPL-based cellular biology experiment that involves the patterning of the ECM protein fibronectin. In addition to outlining the procedure for generating patterns and observing cell differentiation, we elaborate on a unique feature of PPL that enables patterning of combinatorial libraries. This ability arises from the deformable nature of the elastomeric pens, which gives rise to a force-dependent feature size. Because of this, PPL can be used to generate patterns with a gradient of feature sizes by intentionally tilting the pen array with respect to the sample. Of course, uniform features on a sample can also be generated if the pen array is leveled with respect to surface. This powerful technique ultimately allows for the screening of conditions that dictate cell adhesion in a combinatorial fashion and was recently shown to be useful for the study of osteogenic induction of mesenchymal stem cells (MSCs) by culturing MSCs on fibronectin-patterned substrates with a range of feature sizes from the nano- to microscale (Giam et al., 2012). The questions addressed in this chapter, that is, the role of nanoscale variations of ECM proteins on MSC differentiation, are subtle given that many mechanical and chemical factors are known to play a role in differentiation. For example, Discher and co-workers initially observed that MSCs cultured on substrates with different stiffnesses underwent either neurogenesis, myogenesis, or osteogenesis, showing that material properties directed the expression of lineage-specific markers and promoted downstream differentiation (Engler, Sen, Sweeney, & Discher, 2006). Additionally, a recent paper by (Trappmann et al., 2012) showed that the porosity of the substrate also contributes to cell differentiation. These studies are representative of the growing trend of exploring the role of physical cues on stem cell differentiation (Lutolf, Gilbert, & Blau, 2009). Despite the growing body of work on these topics, there are still many questions to be answered about how the interactions between the cell membrane and the surface trigger signaling pathways that promote downstream changes in the cellular machinery (Dalby et al., 2007; McMurray et al., 2011). Interestingly, the observations that physical and chemical features of the surface direct differentiation hint at the opportunity for developing tunable substrates with ligand gradients that can be used to rapidly study cell adhesion, polarization, differentiation of cells, or other relevant biological processes.
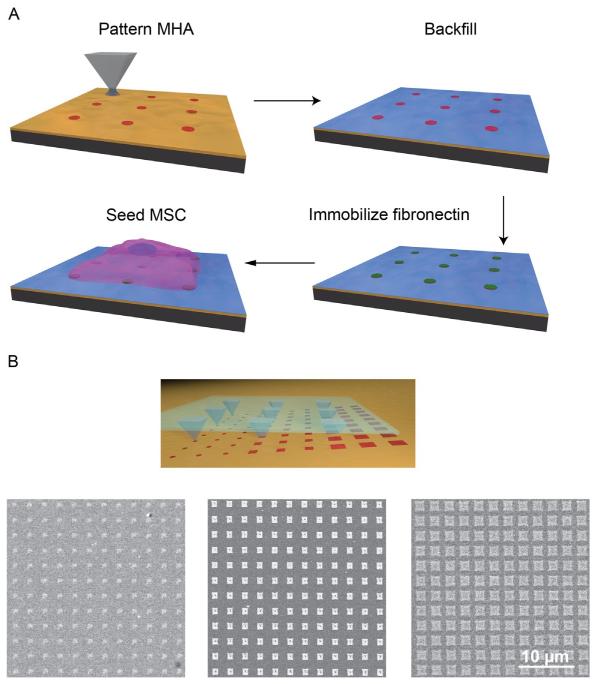
Herein, we outline the steps required for performing studies to address the questions in this field. The general process of fabricating nano- to micrometer scale protein patterns with PPL and using them to study cellular behavior is (Fig. 14.1A):
FIGURE 14.1.
Polymer pen lithography (PPL) process used to generate extracellular matrix (ECM) protein patterns. (A) Schematic shows the major steps of this cantilever-free scanning probe technique which include: deposition of a template SAM followed by addition of a passivation layer, protein attachment to the patterned areas, seeding cells on the molecular patterns. (B) Scheme for generating combinatorial libraries by tilting the polymer pen array. A range of feature sizes (475 nm to 1.2 μm) were obtained by controlling the tilt angle across one substrate. The resulting patterns were arranged in a 60 μm × 60 μm area, where each pen wrote a 15 ×15 array with 4 μm pitch. The resulting patterns were imaged by scanning electron microscopy after etching.
Fabrication of polymer pen arrays
Patterning of 16-mercaptohexadecanoic acid (MHA) SAMs
Fibronectin immobilization on MHA patterns
Combinatorial screening of MSC attachment
MSC differentiation and analysis
14.1 MATERIALS AND INSTRUMENTATION
Reverse osmosis purified >18 MΩ • cm H2O (e.g., NANOpure) for rinsing and cleaning steps.
Ethanol (≥99.5%, Sigma-Aldrich, cat. no. 459844) for cleaning glass slides and preparation of solution.
Acetone (≥99.9%, Sigma-Aldrich, cat. no. 270725) for cleaning and photoresist lift-off.
2-Propanol (99.9%, Sigma-Aldrich, cat. no. 650447) for cleaning and silicon etching.
Shipley S1805 (MicroChem, cat. no. 10018321) photoresist.
Developer MF-319 (MicroChem, cat. no. 10018042) for developing the photoresist after the exposure.
4″ {10 0} Si wafer (Nova Electronic Materials, cat. no. STK8414) with 5000Å thermal or wet oxide for photolithography.
Buffered HF improved (Transene Etchants, pH 5.0) for SiO2.
Si etchant was prepared by mixing 750 g of potassium hydroxide (99.995% (metals basis) Sigma-Aldrich, cat. no. 306568) and 500 mL of 2-propanol in 2 L NANOpure H2O.
Heptadecafluoro-1,1,2,2-tetra(hydrodecyl)trichlorosilane (Gelest, cat. no. SIH5841.0) for SiF coating.
Toluene (≥99.5% Sigma-Aldrich, cat. no. 155004) for SiF coating.
Hard polydimethylsiloxane (h-PDMS) prepolymer was prepared by mixing 500 g divinylmethylsiloxane (Gelest, product code VDT-731) with 20 μL Pt divinyltetramethyldisiloxane (Gelest, product code SIP6831.2) and 344 μL 1,3,5,7-tetramethyl-1,3,5,7-tetravinylcyclotetrasiloxane (Gelest, product code SIT7900.0). The h-PDMS prepolymer was stirred for 5 days before use.
25–35% (methylhydrosiloxane) 65–70% (dimethylsiloxane) copolymer (Gelest, product code HMS-301) for making h-PDMS.
MHA (90%, Sigma-Aldrich, cat. no. 674435) for forming SAMS on Au-coated surfaces.
(1-Mercapto-11-undecyl)hexa(ethylene glycol) 1 mM in ethanol (99%, Asemblon, cat. no. 231043-011) for backfilling the surface after patterning to prevent nonspecific adsorption of proteins and cells.
Auetchingsolutionwaspreparedbymixinganaqueous20 mMsolutionofthiourea (≥99.0% Sigma-Aldrich, cat. no. T8656), with a 13.3 mM solution of iron(III) nitrate nonahydrate (99.99%, Sigma-Aldrich, cat. no. 254223) and hydrochloric acid solution (pH 1) (diluted from 37% stock, Sigma-Aldrich, cat. no. 320331) in a 1:1:1 ratio. We recommend that this solution is freshly prepared prior to use. The etchant can be used for ~1 day, and then should be disposed of and remade fresh.
Phosphate-buffered saline (PBS) 1 ×, pH 7.4 (GIBCO, cat. no. 10010-023) for rinsing steps after protein immobilization and cell adhesion.
Human plasma fibronectin (1 mg/mL, Millipore, cat. no. FC010) for protein immobilization. 10 mM Co(NO3)2 (99.9%, Sigma-Aldrich, cat. no. 230375) solution in ethanol for chelating fibronectin to the MHA SAM.
Anti-human fibronectin produced in rabbit (Sigma-Aldrich, cat. no. F3648-5 ML) for immunofluorescence.
Human alkaline phosphatase (H-300) (Santa Cruz Biotechnology, cat. no. SC30203) for immunofluorescence.
Goat anti-rabbit IgG conjugated to AF 568 (Invitrogen, cat. no. A11036) for immunofluorescence.
Au substrate, prepared by physical vapor deposition (i.e., e-beam or thermal evaporation) of 30 nm Au thin film onto a Si wafer or glass slide. Use of a Cr or Ti 5 nm adhesion layer is highly recommended. When possible, substrate should be used within 1 week of deposition.
Confocal microscope (Nikon C-Si inverted laser confocal microscope).
Optical microscope (Zeiss Axiovert).
Plasma cleaner (PDC-001, Harrick Plasma).
Scanning probe lithography platform (Park XE-150, Park Systems).
E-beam evaporator (PVD 75, Lesker).
Human MSCs (Lonza, cat. no. PT-2501).
MSC growth medium (MSGCM, Lonza, cat. no. PT-3001).
14.2 METHODS
14.2.1 Fabrication of master arrays as molds for polymer pen arrays
Masters for molding polymer pen arrays are fabricated using conventional microfabrication techniques. The photolithography mask used for defining arrays consists of an array of circular holes arranged in a square lattice, where the hole diameter corresponds to the tip base width and the hole-to-hole pitch determines the pen-to-pen pitch. The square arrays of holes are grouped into 2 cm × 2 cm arrays; this size was chosen because it easily fits onto standard glass slides.
To begin, spin-coat a 500 nm oxide-coated 4″ Si {10 0} wafer with S1805 by spinning for 5 s at 500 rpm, followed immediately by 40 s at 4000 rpm, both with 500 rpm/s ramping rates. Soft bake the resist-coated wafer at 115 °C for 60 s, allow it to cool to room temperature, and then expose it in a mask aligner with a dose of ~ 75 mJ/cm2. Note that overexposure is not typically problematic as it will translate to slightly taller pyramids while underexposure will cause the pattern transfer process to fail. Therefore, it is best to err on the side of overexposure. After exposure, develop the patterns in MF-319 for 60 s, rinse with DI H2O for 10 s, and then blow dry with flowing N2. It is important to note that the photolithography protocol outlined here with S8015 is just an example; the patterns can be defined using any photoresist and developer. If a different resist is to be used, consult the manufacturer’s data sheets and follow their recommended procedure with regard to exposure and development.
Once photolithography is complete, plasma clean the resist-coated wafer in an air plasma for 2 min at 10 W to remove residual photoresist from the hole regions. After plasma cleaning, submerse the wafer in a buffered oxide etch (BOE) for 6 min to transfer the resist patterns into the underlying oxide. Remove the wafer from the BOE solution, rinse with DI H2O, blow dry with N2, and then submerse the wafer in acetone for 5 min to remove the resist. After resist removal, sequentially rinse the wafer with acetone, DI H2O, isopropanol (IPA), and then blow dry with N2.
Upon resist removal, place the wafer in an anisotropic KOH etchant solution. This solution consists of 30% w/w KOH in DI H2O, stirring at 75 °C, with IPA added once the KOH reaches temperature in a 1:4 ratio of IPA:H2O. Once the temperature of the etchant has stabilized, suspend the wafer in the etchant solution, below the IPA layer and facing the bottom of the container. Etching will take 50–70 min; the extent of etching can be checked by removing the wafer, rinsing with DI H2O, blowing dry with N2, and then imaging the tips in an optical microscope. By changing the focal plane, it should be possible to see individual pens converge to a point; if they converge to a square, the wafer should be returned to the etchant until this is no longer the case. Note that the orientation and level of the wafer in the etchant is important and care must be taken to keep the wafer consistently placed within the etchant solution.
Once the pyramidal pits have been etched into the Si, place the master in BOE for 5 min to strip the remaining oxide, and then rinse with DI H2O and blow dry with N2. Subject the masters to an air plasma for 2 min at 30 W to regrow a uniform oxide layer, and then place the masters in a desiccator with a solution consisting of 8–12 drops of heptadecafluoro-1,1,2,2-tetra(hydrodecyl)trichlorosilane in 2 mL of toluene and immediately pull vacuum on the desiccator, taking care to mix the fluorosilane and start the vacuum as quickly as possible. Once the toluene begins to boil, turn off the vacuum and leave the masters exposed to the fluorosilane solution for 12–24 h. After silanization, immerse the masters in toluene for 5 min to remove any multilayers, and then blow dry with N2. This step should render the masters hydrophobic, thus facilitating the removal of PDMS in later steps.
14.2.2 Fabrication of polymer pen arrays
Masters of a defined pen-to-pen pitch and array dimension will be used to generate their complementary polymer pen arrays. After thoroughly mixing 3.4 g of h-PDMS prepolymer with 1.0 g copolymer, place the h-PDMS mixture in a desiccator under vacuum for ~15 min to remove trapped air bubbles. During degassing, clean a glass slide by rinsing with IPA and then plasma clean in air for 5 min. Once the PDMS precursor is degassed and a glass slide is prepared, pour a few drops of the PDMS mixture on the master pen array and carefully place the glass slide on the PDMS drop. Let the PDMS cure in an oven at 80 °C for 24–48 h. After this baking step, the PDMS pen array should be cured and no longer flow in response to pressure. Peel off the glass slide from the surface of the master by carefully wedging a razor blade between the glass slide and the master and slowly lifting the glass slide to separate it from the master array. Using a razor blade, remove residual PDMS on the periphery of the PDMS tips and clean the surface with a N2 stream.
14.2.3 Patterning of MHA SAMs
Prior to inking the tip array, place the array in an air plasma for 2 min at 10 W to render the surface hydrophilic. Next, drop cast 100 μL of a solution of 5 mM MHA in ethanol and let the solution evaporate for at least 10 min. Next, for integration into the scanning probe lithography system, mount the tip array on a magnetic holder using double-sided carbon tape and load it onto the AFM head. Place an Au-coated no. 2 glass slide on the stage and move the x–y control stage so that the pen array is directly below the area of the substrate that will be patterned.
Bring the z-stage slowly into approach with the substrate, being careful not to crush the tips against the substrate. Once the pen array is about 100 μm away from the substrate, move the z-stage in small increments (i.e., ~10 μm). Contact between the pen array and the substrate can be visually identified by the extent of deformation of the polymeric tips upon contact with the surface; this is easily seen by a transition in color at the center of the tip from light (not in contact) to dark (just in contact) and then back to light (hard contact). For small features, gentle contact and small tip deformation is desired. Conversely, if large features are desired, greater tip deformation and harder contact with the substrate is required.
To level the polymer pen array optically, check the four corners of the tip array for contact with the substrate and make sure that all four corners experience the same amount of deformation. If the corners are in different degrees of contact, tilt the sample stage until the sample is level. In order to not damage the probes and ensure rapid leveling, it is ideal to only move one direction at a time (i.e., level the stage in the x-direction first), and make sure to raise the probes from the surface before tilting the stage up in either direction. To create uniform patterns, all four corners of the pen array should be equidistant from the substrate, that is, the amount of tip deformation should be uniform across all corners. A gradient of different feature size patterns can be generated by tilting the stage across the x- or y-axis as desired. The tilting angle will define the size of the features that can be generated, with areas that are in closer proximity to the substrate giving rise to larger features (Fig. 14.1B).
After leveling, the desired pattern should be designed and executed by the patterning software. After printing, the sample will be patterned with SAMs of the alkanethiol, however, these patterns are only directly visible through AFM measurements. In order to rapidly visualize the patterns and ensure successful pattern transfer, the sample can be broken in half and a portion of the patterned substrate can be etched to visualize the patterned features. This procedure works because SAMs form an etch mask where the unpatterned regions will be rapidly removed. The residual Au features can be easily seen in an optical microscope or scanning electron microscope (SEM). To etch the Au on the sacrificial section of the sample, incubate the sacrificial area in a solution of gold etchant for ~7 min. The incubation time in the etching solution will vary according to the thickness of the Au film, with thicker films requiring longer etching times. In general, the rate of etching corresponds to approximately 4 nm/min. To stop the etching, rinse copiously with NANOpure water and blow dry with a N2 stream. Following etching, the patterns will appear light against a dark background under optical microscopy or SEM. If the patterns are satisfactory, take the portion of the substrate that was not etched and proceed with the protein immobilization steps.
14.2.4 Fibronectin immobilization on MHA patterns
Following MHA deposition, backfill the unpatterned Au areas by submersing the substrate in a 1 mM ethanolic solution of (1-mercapto-11-undecyl)hexa(ethylene glycol) for 1 h to prevent nonspecific attachment of biomolecules or cells. Next, remove the substrate and rinse copiously with ethanol and dry with a N2 stream. Place the substrate in an aqueous solution of 10 mM Co(NO3)2 for 5 min. During this step, the cobalt ions chelate to the carboxylic acid groups on MHA, thus directing the attachment of fibronectin without compromising binding site availability and protein functionality. Rinse the substrate with ethanol followed by water to remove any residual Co(NO3)2 solution, and then dry with flowing N2. Finally, place the substrate in an aqueous solution of 50 μg/mL fibronectin in 1 × PBS and incubate the sample overnight on a shaker at 100 rpm and 4 °C.
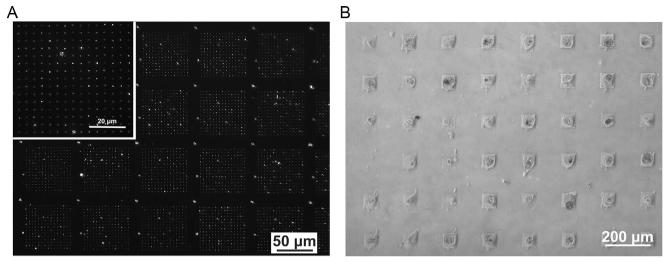
After fibronectin immobilization, rinse with copious amounts of 1 × PBS and dry with a N2 stream. At this point, the substrate should present fibronectin patterns and can be subsequently used for cell adhesion studies. To verify protein immobilization, proceed to immunostaining by incubating the substrate in a 1:100 solution of human antifibronectin antibody (produced in rabbit) in 1 PBS on a shaker stirring (100 rpm) overnight at 4 °C. Remove the substrate from the primary antibody solution, rinse with 1 × PBS, and add a 1:250 solution of fluorophore-labeled secondary antibody (goat anti-rabbit AF 568) diluted in 1 × PBS for 1 h. Rinse with 1 × PBS, NANOpure H2O, dry under flowing N2, and then image with fluorescence microscopy (Fig. 14.2A). The fluorescence micrograph in Fig. 14.2A shows fluorescently labeled fibronectin patterns arranged in a 15 ×15 array over a × 60 μm 60 μm area. These patterns were generated using an array with a pen-to-pen spacing of 80 μm.
FIGURE 14.2.
(A) The resulting fibronectin patterns can be visualized by immunofluorescence staining with AF 468 conjugated antibody. The pitch of the master array used to generate these features corresponded to 80 μm. (B) Optical micrograph of mesenchymal stem cells (MSCs) seeded on fibronectin adhesion sites displays only one cell per 60 μm × 60 μm patterned square region. A master array having a pitch of 180 μm was used to generate these features.
14.2.5 Combinatorial screening of MSC attachment
Combinatorial screening of MSC adhesion is necessary to first understand how fibronectin adhesive sites affect cell spreading, adhesion, and subsequent differentiation. Prior to the start of experiments, culture human MSCs under standard growth conditions in normal growth medium at 37 °C with 5% CO2. We recommend the use of MSCs between passages 2 and 5. Proceed to seed MSCs on patterned substrates presenting a gradient of fibronectin feature sizes at a density of 3000 cells/cm2; this cell density is recommended to ensure that a single cell can be attached to each patterned area (Fig. 14.2B). At this point, we recommend handling the substrate with care to prevent cell detachment.
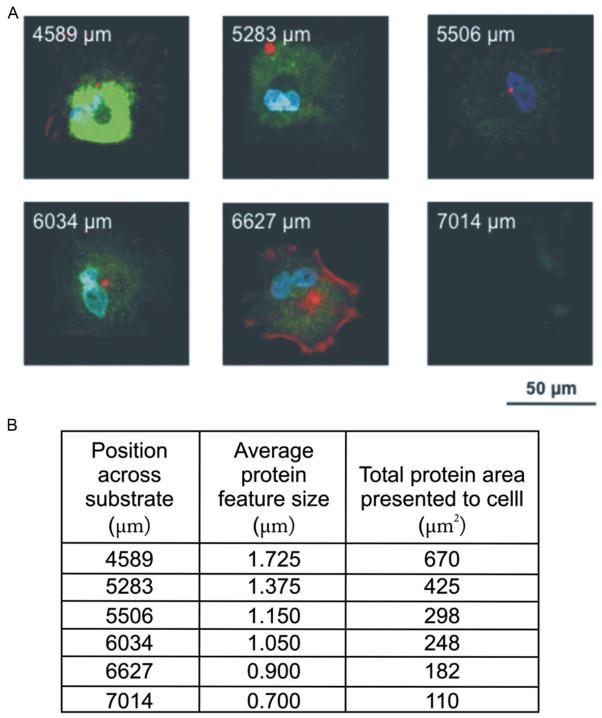
The total area of fibronectin presented to the cell within a patterned region can be modified by defining the dimensions of the array prior to patterning the MHA template. In the case of MSCs, we have previously shown that a total projected area of 3600 μm2 (60 μm × 60 μm) is sufficient to induce MSC attachment without compromising cell viability. Within this patterned region, one can easily probe how MSC spreading changes as a function of total amount of fibronectin immobilized. At this point, it is evident that combinatorial screening is an efficient approach to determine the total area of fibronectin necessary to obtain fully spread MSCs over a patterned region, because it allows a wide variety of parameters to be explored in a single experiment. After 1 week of cell growth on the patterns, the extent of cell attachment and spreading can be monitored by immunofluorescence using standard protocols for cytoskeleton staining, such as actin staining with phalloidin AF568, and immunofluorescence using primary and fluorescently labeled secondary antibodies. As shown in Fig. 14.3, cells cultured on patterns presenting 225 μm2 of total fibronectin lead to complete attachment of MSCs.
FIGURE 14.3.
Stem cell adhesion study using combinatorial fibronectin patterns to evaluate cell attachment and spreading. (A) A tilted polymer pen array (180 μm pen spacing) was used to generate 15 ×15 array of fibronectin features spaced by 4 μm. MSCs were cultured on these patterns for 1 week and subsequently stained for alkaline phosphatase (ALP) (green), actin (red), and the nucleus (blue). The labels on the resulting patterns shown in (B) indicate the x-position of patterns of a certain feature size across a substrate. The total area presented to the cell can be obtained by squaring the average protein feature size and multiplying by 225 for a 15 ×15 array of dot features.
14.2.6 MSC differentiation and analysis
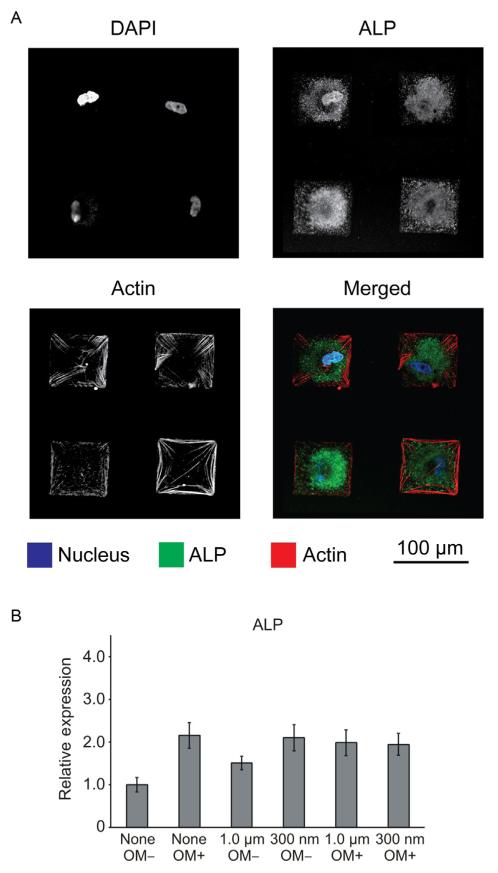
To investigate the effect of fibronectin feature size on MSC differentiation, generate 1 μm and 300 nm fibronectin homogeneous size patterns by leveling the polymer pen array. Proceed to seed MSCs as described earlier and culture the MSCs in the absence of osteogenic differentiation factors for 1 week. Harvest the cells and collect mRNA to quantify expression levels of osteogenic biomarkers, such as alkaline phosphatase (ALP), osteocalcin (OCN), and osteopontin (OPN), using RT-PCR and Western Blotting. In general, the primer sequences for these targets can be found in the literature or obtained commercially. Qualitative expression of osteogenic markers can be monitored by immunofluorescence (Fig. 14.4A). Depending on the application, certain cytoskeletal components, such as actin, can be easily labeled by commercially available kits, without the need for specific antibodies. Specifically, 300 nm diameter fibronectin features were found to be more effective at inducing osteogenesis and expression of related biomarkers, such as ALP, than 1 μm diameter fibronectin patterns, as measured by RT-PCR (Fig. 14.4B) and Western Blotting. It is important to point out that elevated expression of osteogenic markers occurred in the absence of differentiation medium, solely as a response to nanoscale structures, even when the total amount of fibronectin presented to MSCs was held constant. These results further demonstrate the importance of designing substrates that present micro- and nanoscale cues to program-specific cell processes.
FIGURE 14.4.
Immunofluorescence micrographs show the expression of osteogenic marker alkaline phosphatase (ALP) in MSCs cultured on homogeneous (300 nm diameter) fibronectin patterns. (A) The confocal microscopy images in the merged channel display samples stained for actin in red, ALP in green, and the nucleus in blue. (B) Quantitative RT-PCR results for ALP were normalized to GAPDH levels. The bar graph also displays results that were normalized to the negative control: no pattern in absence of osteogenic media (OM –).
14.3 DISCUSSION
The work by Giam et al. provides a compelling example of what can be learned in a biological context by using PPL. Key to the success of this study was the ability to first generate combinatorial libraries of features for high-throughput screening and then select experimental conditions and generate statistically significant homogenous patterns (Giam et al., 2012). By using these methods, the authors were able to address the question of how fibronectin feature size and the formation of focal adhesions mediate osteogenic differentiation. Specifically, 300 nm diameter fibronectin features were found to be more effective at inducing osteogenesis and expression of related biomarkers than 1 μm diameter fibronectin patterns, as measured by RT-PCR and Western Blotting. It follows then that the cell–substrate binding signaling events that occur in the ECM are transduced to biochemical cues that trigger intracellular differentiation events. To better understand this phenomenon, focal adhesion kinase (FAK) phosphorylation was studied, an event known to precede expression of osteogenic markers, which led the authors to discover that levels of phosphorylated FAK indeed increased when MSCs were cultured on 300 nm fibronectin patterns, even when the total amount of fibronectin beneath a given cell was conserved. These results further demonstrate the importance of designing substrates that present micro- and nanoscale cues to program specific cell processes.
More generally, the outlined protocol presents PPL as a highly versatile and practical approach for generating a range of protein patterns in a high-throughput fashion. This cantilever-free scanning probe technique, which relies on elastomeric pen arrays to deposit molecules, offers an attractive method for generating bioactive surfaces that present gradients of ligand density in addition to protein patterns of uniform size by simply tilting the pen array or holding it level, respectively. An important advantage of this technique over other microcontact printing approaches is the ability to generate thousands of arbitrary patterns rapidly over large-scale areas without the need for fabricating multiple masks, thus enabling rapid prototyping. Patterning over large areas is particularly important for experiments in cell biology, where a statistically significant sample size is necessary for both mRNA and protein quantification. More importantly, tilting the polymer pen array allows one to generate combinatorial libraries where both size and density of the patterns are controlled with high precision and registration on the substrate. For the life sciences, this feature is of particular interest as the spatial profiling of several cell–substrate interactions opens an attractive avenue to elucidate changes in the biochemical signaling pathways of the cellular machinery. In addition, PPL provides a highly robust method that, in principle, can be coupled with microscopy techniques to obtain spatiotemporal profiles of cell–receptor interactions that would yield quantitative information for understanding the kinetics of events at the single cell level.
While we have described the methodology to template features of MHA on Au-coated substrates, this approach can be extended to deliver different inks to generate substrates presenting multiple functionalities. Multiplexed printing can be achieved with the assistance of an inkjet-printing platform to first deliver the inks to microfabricated reservoirs (Zheng et al., 2009). By choosing the periodicity of the reservoirs to match the pen-to-pen spacing of the pen, it was possible to achieve perfect registration and eliminate cross-contamination. Inking different pens in the array with different inks allows one to print features of varying composition, adding another experimental handle to the capabilities of PPL. In particular, having combinatorial libraries of multiple ligands would allow scientists to design systematic studies ranging from fundamental questions, such as how ECM factors affect cell polarization, to application-driven questions related to high-throughput drug screening or tissue engineering.
SUMMARY
PPL allows one to construct molecularly patterned surfaces of sizes large enough to be applicable in biologically relevant studies that contain features defined in high resolution. In particular, we have described an approach that utilized patterned small molecules that form SAMs as templates for an ECM protein (here, fibronectin) in order to define adhesion sites for cellular attachment. After protein immobilization, the surfaces were incubated with cells, and their behavior was studied using standard cell biology techniques, such as immunofluorescence, RT-PCR, and Western Blotting. Critical to the success of this work was the ability to first create a gradient of feature sizes that allowed for the screening of reaction conditions of interest. This powerful paradigm of utilizing combinatorial screening in conjunction with uniform samples composed of region of interest conditions can potentially be applied to many cell systems beyond MSCs.
Acknowledgments
This material is based upon work supported by the Air Force Office of Scientific Research (AFOSR), under award number FA9550-08-1-0124, SPAWAR Systems Center Pacific Code 22550 under award number N66001-08-1-2044, the National Cancer Institute under award number U54CA151880, the National Science Foundation (NSF) Nanoscale Sciences and Engineering Initiative under award number EEC-0647560, and the Defense Advanced Research Projects Agency (DARPA). D.J.E. is supported by a DoD, Air Force Office of Scientific Research, National Defense Science and Engineering Graduate (NDSEG) Fellowship, 32 CFR 168a. K.A.B. gratefully acknowledges support from Northwestern University’s International Institute for Nanotechnology.
References
- Dalby MJ, Gadegaard N, Tare R, Andar A, Riehle MO, Herzyk P, et al. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nature Materials. 2007;6(12):997–1003. doi: 10.1038/nmat2013. http://dx.doi.org/10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- Demers LM, Ginger DS, Park SJ, Li Z, Chung SW, Mirkin CA. Direct patterning of modified oligonucleotides on metals and insulators by dip-pen nanolithography. Science. 2002;296(5574):1836–1838. doi: 10.1126/science.1071480. http://dx.doi.org/10.1126/science.1071480. [DOI] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126(4):677–689. doi: 10.1016/j.cell.2006.06.044. http://dx.doi.org/10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Gandor S, Reisewitz S, Venkatachalapathy M, Arrabito G, Reibner M, Schroder H, et al. A protein-interaction array inside a living cell. Angewandte Chemie, International Edition in English. 2013;52(18):4790–4794. doi: 10.1002/anie.201209127. http://dx.doi.org/10.1002/anie.201209127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giam LR, Massich MD, Hao L, Shin Wong L, Mader CC, Mirkin CA. Scanning probe-enabled nanocombinatorics define the relationship between fibronectin feature size and stem cell fate. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(12):4377–4382. doi: 10.1073/pnas.1201086109. http://dx.doi.org/10.1073/pnas.1201086109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginger DS, Zhang H, Mirkin CA. The evolution of dip-pen nanolithography. Angewandte Chemie, International Edition in English. 2004;43(1):30–45. doi: 10.1002/anie.200300608. http://dx.doi.org/10.1002/anie.200300608. [DOI] [PubMed] [Google Scholar]
- Huo F, Zheng Z, Zheng G, Giam LR, Zhang H, Mirkin CA. Polymer pen lithography. Science. 2008;321(5896):1658–1660. doi: 10.1126/science.1162193. http://dx.doi.org/10.1126/science.1162193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KB, Park SJ, Mirkin CA, Smith JC, Mrksich M. Protein nanoarrays generated by dip-pen nanolithography. Science. 2002;295(5560):1702–1705. doi: 10.1126/science.1067172. http://dx.doi.org/10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- Lutolf MP, Gilbert PM, Blau HM. Designing materials to direct stem-cell fate. Nature. 2009;462(7272):433–441. doi: 10.1038/nature08602. http://dx.doi.org/10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray RJ, Gadegaard N, Tsimbouri PM, Burgess KV, McNamara LE, Tare R, et al. Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nature Materials. 2011;10(8):637–644. doi: 10.1038/nmat3058. http://dx.doi.org/10.1038/nmat3058. [DOI] [PubMed] [Google Scholar]
- Piner RD, Zhu J, Xu F, Hong S, Mirkin CA. “Dip-Pen” nanolithography. Science. 1999;283(5402):661–663. doi: 10.1126/science.283.5402.661. [DOI] [PubMed] [Google Scholar]
- Qin D, Xia Y, Whitesides GM. Soft lithography for micro- and nanoscale patterning. Nature Protocols. 2010;5(3):491–502. doi: 10.1038/nprot.2009.234. http://dx.doi.org/10.1038/nprot.2009.234. [DOI] [PubMed] [Google Scholar]
- Salaita K, Lee SW, Wang X, Huang L, Dellinger TM, Liu C, et al. Sub-100 nm, centimeter-scale, parallel dip-pen nanolithography. Small. 2005;1(10):940–945. doi: 10.1002/smll.200500202. http://dx.doi.org/10.1002/smll.200500202. [DOI] [PubMed] [Google Scholar]
- Salaita K, Wang Y, Mirkin CA. Applications of dip-pen nanolithography. Nature Nanotechnology. 2007;2(3):145–155. doi: 10.1038/nnano.2007.39. http://dx.doi.org/10.1038/nnano.2007.39. [DOI] [PubMed] [Google Scholar]
- Su M, Aslam M, Fu L, Wu NQ, Dravid VP. Dip-pen nanopatterning of photosensitive conducting polymer using a monomer ink. Applied Physics Letters. 2004;84(21):4200–4202. http://dx.doi.org/10.1063/1.1737469. [Google Scholar]
- Su M, Liu X, Li SY, Dravid VP, Mirkin CA. Moving beyond molecules: patterning solid-state features via dip-pen nanolithography with sol-based inks. Journal of the American Chemical Society. 2002;124(8):1560–1561. doi: 10.1021/ja012502y. [DOI] [PubMed] [Google Scholar]
- Trappmann B, Gautrot JE, Connelly JT, Strange DG, Li Y, Oyen ML, et al. Extracellular-matrix tethering regulates stem-cell fate. Nature Materials. 2012;11(7):642–649. doi: 10.1038/nmat3339. http://dx.doi.org/10.1038/nmat3339. [DOI] [PubMed] [Google Scholar]
- Zheng ZJ, Daniel WL, Giam LR, Huo FW, Senesi AJ, Zheng GF, et al. Multiplexed protein arrays enabled by polymer pen lithography: Addressing the inking challenge. Angewandte Chemie, International Edition in English. 2009;48(41):7626–7629. doi: 10.1002/anie.200902649. http://dx.doi.org/10.1002/Anie.200902649. [DOI] [PMC free article] [PubMed] [Google Scholar]