Abstract
β-catenin is a key signaling molecule in the canonical Wnt pathway, which is involved in animal development. However, little information has been reported for β-catenin in bivalves. In the present study, we cloned a homolog of β-catenin from the scallop Chlamys farreri and determined its expression characteristics. The full-length cDNA of β-catenin was 3,353 bp, including a 2,511 bp open reading frame that encoded a predicted 836 amino acid protein. Level of the β-catenin mRNA increased significantly (P<0.05) with C. farreri gonadal development and presented a sexually dimorphic expression pattern in the gonads, which was significantly high in ovaries detected by quantitative real-time polymerase chain reaction (qRT-PCR). Immunohistochemical analysis revealed that the β-catenin was mainly located in germ cells of the gonads, with obvious positive immune signals in the oogonia and oocytes of ovaries as well as in the spermatogonia and spermatocytes of testes, implying β-catenin might be involved in the gametogenesis of C. farreri. Furthermore, when 0.1 µg/mL and 0.2 µg/mL DKK-1 (an inhibitor of the canonical Wnt pathway) were added in vitro to culture medium containing testis cells of C. farreri, the expression of β-catenin decreased significantly detected by qRT-PCR (P<0.05), suggesting the canonical Wnt signal pathway exists in the scallop testis. Similarly, when 50 µM and 100 µM quercetin (an inhibitor of β-catenin) were added in vitro to the culture system, Dax1 expression was significantly down-regulated compared with controls (P<0.05), implying the β-catenin is an upstream gene of Dax1 and is involved in the regulation of C. farreri spermatogenesis.
Introduction
The Wnt signaling transduction pathway plays an important role in many developmental processes, including vertebrate limb regeneration, nervous system development, body axis formation, and adrenocortical development [1], [2], [3]. Current studies in invertebrates such as Drosophila melanogaster, Caenorhabditis elegans, and Hemicentrotus pulcherrimus, and vertebrates such as Danio rerio, Xenopus laevis, Mus musculus, and Homo sapiens have provided abundant information regarding Wnt signaling in signaling transduction and function [4]–[10].
To date, at least three Wnt intracellular signaling pathways have been identified: the Wnt/β-catenin pathway, the Wnt/planar polarity pathway, and the Wnt/calcium pathway [11], [12], [13]. The Wnt/β-catenin pathway, also named the “canonical” Wnt pathway is the best understood, and controls various developmental processes including cell fate determination, differentiation, and survival, by stabilizing β-catenin [14]. In the absence of a Wnt ligand, cytoplasmic β-catenin is degraded by interactions with a destruction complex formed by three proteins, APC (adenomatous polyposis coli), Axin, and GSK3β (glycogen synthase kinase-3β). The presence of Wnt prevents the degradation of β-catenin, thus the stabilized β-catenin is transported to the nucleus and activates gene transcriptions through direct interactions with T-cell factor/lymphoid enhancer factor (Tcf/Lef) [15]. The Wnt/planar polarity pathway and Wnt/calcium pathway are two major noncanonical Wnt pathways. The Wnt/planar polarity pathway (also called the Wnt/JNK pathway) is characterized by the activation of jun-N-terminal kinase (JNK) involving small GTPases of the Rho family such as RhoA, Rac, or Cdc42 [16]. The Wnt/calcium pathway employs the second-messenger systems of G-proteins to mobilize intracellular calcium stores and activate atypical protein kinase C (PKC) and other calcium-responsive pathways [17], [18]. The noncanonical Wnt pathway is required for the formation of vertebrate tissues, maintenance of adult stem cells, and the suppression of tumors [19].
β-catenin is a key transcriptional effector of the canonical Wnt signal transduction pathway but also functions as a cell adhesion molecule at the plasma membrane by linking cadherins to α-catenin [20]. Structurally, β-catenin consists of an N-terminal region, a central region, and a C-terminal region [21]. The N-terminal region contains a consensus site required for the phosphorylation of GSK-β. The C-terminal region functions as a transactivator required for the activation of target genes [22]. The central region contains 12 imperfect armadillo repeats (ARM). Each repeat forms three alpha-helixes that are arranged in a compact superhelix [23], which is required for interactions with proteins such as cadherins, Axins, APC, and Tcf/Lef [24], [25], [26], [27], [28]. Recently, increasing numbers of studies have found that β-catenin functions in mammalian sex determination and differentiation. In mice, the activation of β-catenin in somatic cells of XY gonads effectively blocks testis development, including disruption of testis cord formation, and down-regulates the expression of testis marker genes, finally leading to male-to-female sex reversal. This implies that mouse β-catenin is a key anti-testis molecule [29]. Furthermore, loss of β-catenin in the SF1-positive population of fetal somatic cells causes morphological defects in ovaries such as the appearance of testis-specific coelomic vessels and the loss of female germ cells, while morphogenesis, Sertoli cell differentiation, or masculinization in testes are not affected, suggesting β-catenin is necessary only for ovarian differentiation but is dispensable for testis development [30]. Moreover, it is well documented in human embryo kidney 293 (HEK293) cells that β-catenin activates the expression of Dax1 through the canonical Wnt pathway. Therefore, the Wnt-β-catenin-Dax1 signaling pathway exists in these cells [31]. Similar results are obtained in mouse ovaries during sexual differentiation, in which the disruption of Wnt4 (an upstream activator of β-catenin) significantly decreased the expression of Dax1, implying that this signal pathway may commonly exist in mammals [31].
Shellfish comprise a group of bivalves with rich diversity in species and biological characteristics. Sex is an important characteristic, and sexual variability induced usually by interactions between environmental factors and genes is common in bivalves. However, the molecular mechanisms involved in sex variability are not clear because of a deficiency of data regarding sex-related genes. The scallop Chlamys farreri (Jones and Preston 1904) is an important commercial shellfish in China and is characterized by its relatively stable sex composition [32], [33]. Therefore, it is a good experimental species to identify sex-related genes and their functions. In the previous studies, C. farreri Dax1 is suggested to involve in both oogenesis and spermatogenesis while DAX1 expressed much higher in testis than in ovary [34]. To understand the upstream genes of Dax1 during C. farreri spermatogenesis, we cloned β-catenin cDNA in the present study using the Rapid Amplification of cDNA Ends (RACE) technique, and determined its expression pattern in gonads during the reproductive cycle of C. farreri. Furthermore, the type of Wnt pathway and correlations between the β-catenin and Dax1 in C. farreri spermatogenesis were investigated using an in vitro cultured testis cells treated with DKK-1 and quercetin. Our aim was to reveal the β-catenin expression characteristics in the developing gonads of C. farreri, and provide evidence for the existence of the Wnt-β-catenin-Dax1 pathway in the C. farreri spermatogenesis.
Materials and Methods
Ethical statement
The collection and handling of these animals was approved by the Animal Care and Use Committee at the Ocean University of China.
Animals and sampling
Healthy male and female scallops C. farreri with mean shell height 6.27±0.32 cm were purchased from the Aquatic Product Market (Qingdao, China). Gonads were dissected and weighed. Parts of the gonads were immediately frozen in liquid nitrogen, then stored at −80°C until RNA extraction. Parts of the gonads were fixed with Bouin's solution (Picric acid:formaldehyde:acetic acid = 15∶5∶1) for 24 h, embedded in paraffin wax, sliced at a thickness of 5 µm, and stained by hematoxylin-eosin (H&E) for histological analysis to determine developmental stages of the gonads. Parts of the gonads were fixed in 4% paraformaldehyde (pH 7.4) at 4°C for 24 h, and then dehydrated through a methanol series (25, 50, 75, and 100%) and stored in 100% methanol at −20°C for immunohistochemistry analysis.
According to the morphologic characteristics described by Liao et al. [32], the gonads (ovaries and testes) were grouped into four stages based on histological structure and the gonadosomatic index (GSI = gonad weight/soft tissue weight ×100): proliferative stage (GSI = 3.18±0.008 for ovaries and GSI = 3.89±0.008 for testes), growth stage (GSI = 4.20±0.013 for ovaries and GSI = 3.93±0.012 for testes), mature stage (GSI = 4.41±0.004 for ovaries and GSI = 4.53±0.009 for testes), and resting stage (GSI = 2.68±0.006 for ovaries and GSI = 2.46±0.009 for testes).
Isolation of scallop β-catenin full-length cDNA
Total RNA was isolated from C. farreri ovaries at the proliferative stage with Trizol reagents (Takara Bio Inc., Otsu, Japan) according to the manufacturer's instructions. The total RNA was then treated with DNase (Takara Bio Inc.) and purified using the RNeasy mini kit (TransGen Bio Inc., Beijing, China). RACE-Ready First-strand cDNA was synthesized, and 5′ and 3′ RACE PCR was conducted using the SMART-RACE cDNA Amplification kit (Invitrogen, Carlsbad, CA, USA). The specific primers (5′-CATTCAGCGTGTAGCAGCAGGAGTCCTC-3′ for 3′ RACE and 5′- CTTCGGACATGCGGAAAAGGACTGCTGC-3′ for 5′ RACE) were designed according to a 325 bp expressed sequence tag (EST) sequence (GenBank accession no. DT718886) with BLAST search from the C. farreri EST collection (http://www.ncbi.nlm.nih.gov/nucest/DT718886.1). RACE cDNAs were denatured at 94°C for 5 min, followed by 35 cycles at 94°C for 30 s, 68°C for 30 s, and 72°C for 3 min, ending with a 5-min extension at 72°C. PCR products were separated on an agarose gel (1.2%) and purified with a DNA purification kit (Takara Bio Inc.). Then, the purified 5′ and 3′ RACE products were subcloned into a PMD-18T vector (Takara Bio Inc.) and sequenced.
Bioinformatics analysis
5′ and 3′ RACE fragments were assembled using DNAstar software (DNAStar, WI, USA) to get the full-length cDNA of β-catenin. Sequence identity and similarity of the C. farreri β-catenin with other known β-catenins were analyzed using the online BLAST suite of programs at the National Center for Biotechnology Information. Phylogenetic analysis was conducted using MEGA software (version 4.0) with the neighbor-joining method.
Quantitative real-time polymerase chain reaction (qRT-PCR) analysis
qRT-PCR for analyzing expression levels of the β-catenin in gonads of C. farreri during the reproductive cycle was conducted as described previously [35]. Total RNA was isolated from the gonads from different stages and was transcribed to cDNA using the PrimeScript RT reagent Kit (Takara Bio Inc.) as the initial templates for qRT-PCR. Specific primers for amplifying a 196 bp fragment were designed according to the non-conservative domain of C. farreri β-catenin; forward primer: 5′-CAATCAGCAGCAAGGGTGGA-3′ and reverse primer: 5′-TCTGGGAACATGGCGTCTCG-3′. The β-actin gene was used as reference gene following previous studies in C. farreri [36], [37], [38], [39] and a 129 bp fragment of β-actin (GenBank accession no. AY335441) was amplified using the forward primer: 5′-TTCTTGGGAATGGAATCTGC-3′ and reverse primer: 5′-ATTGTGCTACCACCGGAAAG-3′. Primer specificity during the qRT-PCR was verified by a single distinct peak obtained by melting curve analysis. Quantification of target and reference genes was conducted simultaneously using the ABI 7500 detection system (Applied Biosystems) with SYBR Green Master Mix (Takara Bio Inc.). The qRT-PCR reaction consisted of 5 min at 94°C, followed by 40 cycles of 94°C for 15 s, and 60°C for 1 min. Gonads from six individuals at the same stage were sampled, and duplicate assays for each gonad sample were conducted. Data were analyzed using the ABI 7500 system SDS software version 1.4 (Applied Biosystems) with automatically set baseline and cycle threshold values. The 2−ΔΔct method was used to analyze the relative expression levels of the β-catenin [40].
All data were presented as means ± SEM of six samples with two parallel repetitions. Differences between means were tested using one-way analysis of variance (ANOVA) followed by Duncan's post-hoc test (SPSS software version 18.0; SPSS Inc., Chicago, IL, USA), and the significant level was set at P<0.05. All assays in the qRT-PCR were validated in compliance with “the MIQE guidelines” [41].
Immunohistochemistry
Based on the analysis of antigen clusters (DNAstar software), the partial ORF fragment (encoded from 378 amino acids to 836 amino acids) of the β-catenin was predicated to have a relative high antigenicity, and was amplified using the forward primer: 5′-GGATCCTCCAGTAACAAGCCAGCTGT-3′ (BamHI site underlined) and reverse primer: 5′-CTCGAGCAGATCAGTGTCATACCAGTTG-3′ (XhoI site underlined). After subcloning into a PMD-18T vector (Takara Bio Inc.), the recombinant plasmid was extracted and digested by BamHI and XhoI. The digested fragment was then ligated into a BamHI/XhoI site of the bacterial expression vector pet28a (Invitrogen) and verified by sequencing. The recombinant plasmid was then transformed into Escherichia coli BL21 (DE3) (TransGen Bio Inc.). Glutathione-S-transferase (GST)-β-catenin fusion protein was expressed and affinity purified on an anti-GST Ni-NTA His Bind Resins (Invitrogen), and was injected into New Zealand white rabbits for the production of polyclonal antibodies. Antibody specificity was detected by Western blotting using the gonads of C. farreri at the growth stage, as previously described [42]. The sera antibody titer was determined by indirect enzyme-linked immunoassay and the antisera were aliquoted and stored at −80°C. For immunohistochemical analysis, the stored gonads were dehydrated with ethanol, embedded in paraffin, and 5-µm sections were cut using a Histostart 820 Rotary microtome (Reichert, Inc., New York, USA). Sections were deparaffinized in xylene and hydrated in descending ethanol, followed by antigen retrieval in citrate buffer (0.01 mol/L, pH 6.0). To assess the specificity of immunoreactivity, sections of testes and ovaries from different stages were blocked in bovine serum albumin (3%) for 1 h and incubated with pre-immune serum (for the negative control) and anti-β-catenin primary antibody (1∶200 dilution) for 1 h at room temperature, respectively. Specific goat-anti-rabbit IgG (conjugated to horseradish peroxidase, 1∶1,000 dilution) was used as a secondary antibody and incubated for 1 h at room temperature. After washing with PBST (phosphate-buffered saline +0.1% Tween-20), sections were color developed with DAB (3, 3′-diaminobenzidine) and counterstained with hematoxylin. Tissue sections were then observed and images were captured using a Nikon E80i microscope.
Expression of β-catenin and Dax1 in in vitro cultured testis cells treated with DKK-1 and quercetin
Healthy male scallop C. farreri with mean shell height 5.67±0.24 cm and mean GSI of 3.95±0.016 from the growth stage were purchased from the Aquatic Product Market (Qingdao, China). The scallops were maintained in seawater for 1 week at room temperature before sampling. The testis tissues were dissected and cut into pieces (mean size = 1 mm3) in PBS (pH 7.4) containing penicillin (1,000 IU/mL) and streptomycin (800 µg/mL). These tissue pieces were cultivated in vitro at 23°C in primary medium that consisted of L15 medium (pH 7.2–7.4) plus 5% fetal bovine serum, 20.2 g/L NaCl, 0.54 g/L KCl, 0.60 g/L CaCl2, 1 g/L MgSO4, 3.9 g/L MgCl2, 10 ng/mL EGF, 2 ng/mL bFGF, 10 ng/mL LIF, 100 IU/mL penicillin, and 100 µg/mL streptomycin. After approximately 5 h of adherent culture, most somatic cells and germ cells migrated out from these tissue pieces. According to the differential adherent ability, we made the germ cells apart from bottom of the culture dish and suspend in the medium by shaking slightly the dish, meanwhile the somatic cells and tissue pieces were still attached to the bottom. Then the culture medium containing germ cells was collected carefully and transferred to six-well plates in which new primary medium was added. After cultivating at 23°C for 10 h when the germ cells were completely attached to the bottom of the plate, the culture medium was replaced by new medium containing the new primary medium plus human recombinant DKK-1 (Life Technologies Co., Carlsbad, USA) at the concentrations of 0.1 µg/mL or 0.2 µg/mL, and quercetin (National Pharmacy Inc., Beijing, China) at the concentrations of 50 µM or 100 µM for the treated groups, respectively; for the control, it was added only with new primary medium. The germ cells of the treatment group and control were cultivated for 48 h, and then harvested by centrifugation at 100 g for 10 min. After washing with PBS twice, the germ cells were stored in liquid nitrogen until RNA extraction. Three replicates were conducted for each group. Total RNAs from the cells of each sample were extracted with the MicroElute Total RNA kit (Omega Inc., New York, USA). The RNAs were digested with DNase (Takara Bio Inc.), and transcribed to cDNA as described before. The expressions of β-catenin and Dax1 were detected using qRT-PCR. The primers for C. farreri β-catenin were the same as the gonadal expression described above by qRT-PCR detection, and the primers for C. farreri Dax1 (GenBank accession no. JQ071986) were as follows: forward primer 5′-TCTTCCTCGCCTCATTGTCG-3′ and reverse primer 5′-CGTCGGTATTGGAGCCTTTG-3′. The qRT-PCR data of both treated groups and controls were analyzed by the methods described above.
Results
Sequence and evolutionary analysis of β-catenin in C. farreri
The 3′ and 5′ RACE fragments of target cDNA from C. farreri ovary at the proliferative stage were 1,241 bp and 2,279 bp in length, respectively. A full-length sequence of 3,353 bp was assembled (Fig. 1), which included an open reading frame (ORF) of 2,511 bp encoding 836 amino acids, and a 3′ and 5′ untranslated region (UTR) of 643 bp and 199 bp, respectively (GenBank accession number JQ071985). The putative protein was 91.8 kDa with an isoelectric point of 5.76. Structural analysis showed it contained three putative regions of β-catenin: an N-terminal region, a C-terminal region, and a central region. The central region contained a 42-aa ARM in 12 repeats, with a smaller insertion between repeat 10 and 11 (Fig. 1). The N-terminal region had a 21-aa GSK-β consensus phosphorylation site, and the C-terminal region had a 72-aa transactivator domain required for the activation of target gene (Fig. 1). The target protein sequence had high identity with that of other species β-catenin, especially the ARM repeat region that shared 96%, 91%, 89%, 83%, 84%, 83%, 84%, and 84% identities with Crassostrea gigas, Platynereis dumerilii, Branchiostoma floridae, Strongylocentrotus purpuratus, Gallus gallus, Ciona intestinalis, H. sapiens, and X. laevis, respectively. In contrast, the N- and C-terminal regions were more divergent compared with the ARM repeat region. Characteristics described above indicated the target sequence in this study was C. farreri β-catenin.
Figure 1. Full-length cDNA sequence and deduced amino acids of C. farreri β-catenin.
Lower case text indicates the 5′ and 3′ UTR sequences of β-catenin; upper case text indicates the encoding sequence. The start codon (ATG) and stop codon (TGA) are double underlined. The putative ARM repeat regions (1–12) are boxed. The N-terminal putative GSK-β consensus phosphorylation site and the C-terminal transactivator region are underlined.
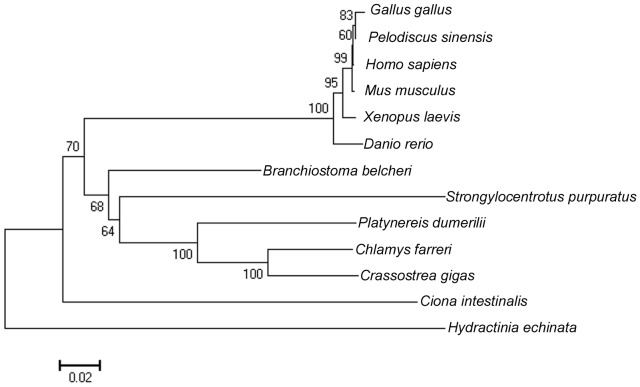
Phylogenic analysis indicated that C. farreri β-catenin clustered primarily with that of C. gigas, and together formed a subcluster with that of P. dumerilii, which then clustered with that of S. purpuratus and Branchiostoma belcheri successively, finally clustering with the branch formed by vertebrates (Fig. 2).
Figure 2. Phylogenic analysis of β-catenin among different species.
GenBank accession numbers: Gallus gallus: AAB80856; Pelodiscus sinensis: BAD74125; Homo sapiens: CAA61107; Mus musculus: NP_001159374; Xenopus laevis: AAA49670; Danio rerio: NP_571134; Branchiostoma belcheri: BAD12593; Strongylocentrotus purpuratus: NP_001027543; Platynereis dumerilii: ABQ85061; Chlamys farreri: JQ071985; Crassostrea gigas: AFL93714; Ciona intestinalis: NP_001027779; Hydractinia echinata: ACZ56248.
Quantitative expression of β-catenin mRNA in the gonads during the reproductive cycle
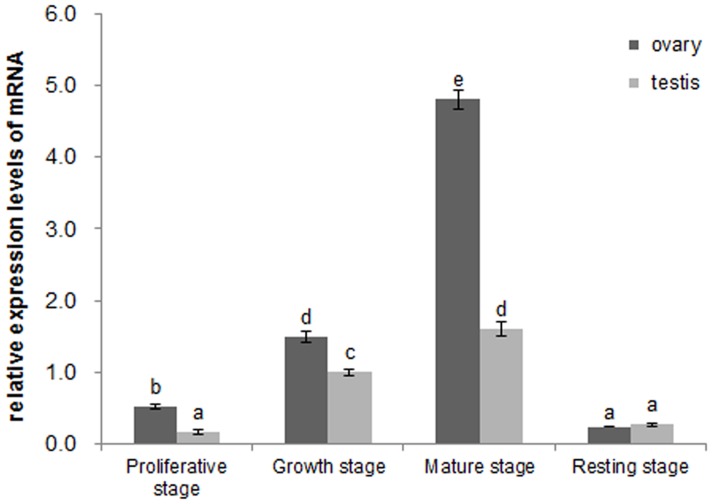
qRT-PCR results (Fig. 3) indicated that the expression of β-catenin increased significantly from the proliferative stage to mature stage (P<0.05) in C. farreri gonads during the reproductive cycle. Then, the level markedly decreased to a minimal level at the resting stage (P<0.05). Significant differences at expression levels (P<0.05) were also found between testes and ovaries at the same stage except the resting stage, which was approximately two times higher in the ovary than testis at the proliferative stage, 0.5 times higher at the growth stage, and two times higher at the mature stage.
Figure 3. Expression levels of β-catenin mRNA in C. farreri gonads by qRT-PCR.
The expression level of β-catenin mRNA in testis at the growth stage is set as 1.00 to calibrate the relative levels in gonads at different stages. Values are the means ± SEM; n = 6. The different letters indicate statistically significant differences (P<0.05).
Cyto-location of β-catenin in the gonads during gametogenesis
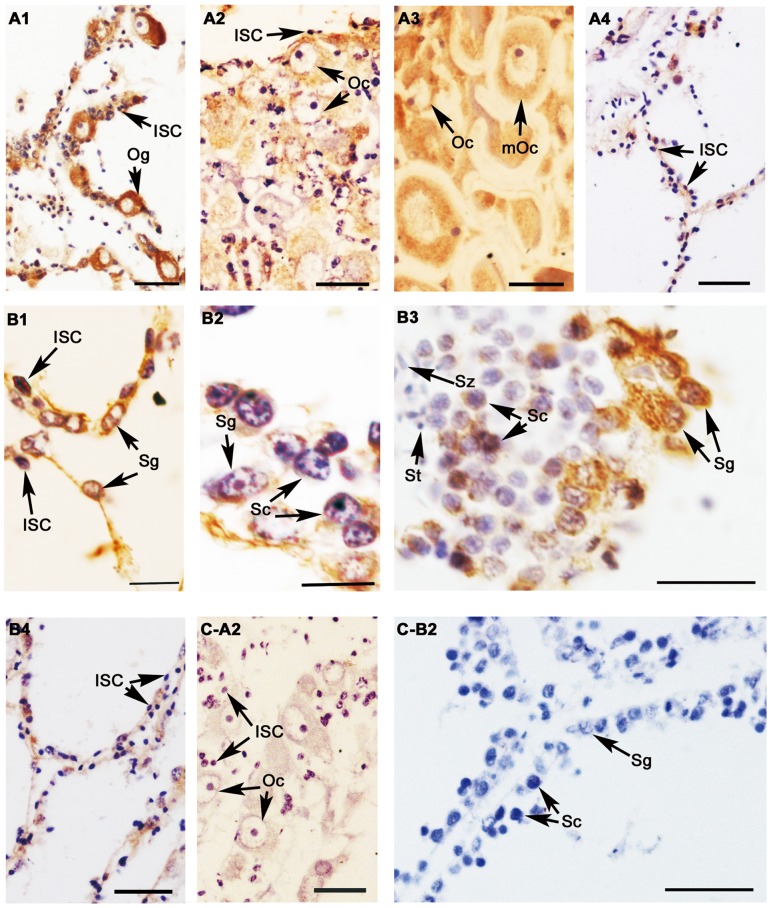
The titer of β-catenin polyclonal antibody was detected to be 1∶512,000, and a single band with a molecular mass of approximately 90 kDa in gonads at the growth stage was detected by Western blotting (Fig. 4), indicating the β-catenin polyclonal antibody was specificity. In the testis, β-catenin immunoreactivity was observed in spermatogonia and spermatocytes, however no visible signal was observed in spermatid and spermatozoon during spermatogenesis (Fig. 5 B1–B4). In the ovary, β-catenin was detected in the oogonia and oocytes at all developmental stages (Fig. 5 A1–A4). In testis and ovary of the resting stage, the immunoreactive signals declined to a very low level. Moreover, weak signals were also found in the intragonadal somatic cells (ISCs) which are characterized by small size, extremely heterogenous and pleomorphic nucleus.
Figure 4. Expression of β-catenin detected by Western blotting in C. farreri gonads at the growth stage.
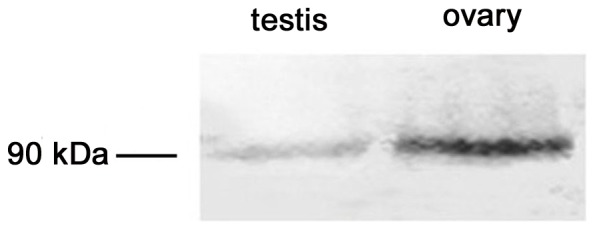
Figure 5. Location of β-catenin protein in C. farreri gonads detected by immunohistochemistry.
A, B: Positive signals are brown and represent anti-β-catenin in ovaries (A) and testes (B). C: Negative control with preimmune serum; 1: proliferative stage; 2: growth stage; 3: mature stage; 4: resting stage; ISC: intragonadal somatic cell; Og: oogonium; Oc: oocyte; mOc: mature oocyte; Sg: spermatogonium; Sc: spermatocyte; St: spermatid; Sz: spermatozoon; Scale bars: B1, B2, and B3 are 10 µm, others are 30 µm.
DKK-1 down-regulates β-catenin and Dax1 expression in cultured C. farrei testis cells
qRT-PCR analysis demonstrated transcriptional levels of β-catenin and Dax1 decreased significantly (P<0.05) in C. farreri testis cells cultured in vitro, by a concentration-dependent mechanism when DKK-1 was added for 48 h. The expression levels of β-catenin decreased about 15% in the 0.1 µg/mL DKK-1 group and 26% in the 0.2 µg/mL DKK-1 group, compared with the control group (Fig. 6). Dax1 expression levels decreased about 16% and 55%, respectively (Fig. 7).
Figure 6. Relative mRNA expression of β-catenin in cultured testis cells in vitro treated with DKK-1 for 48 h.
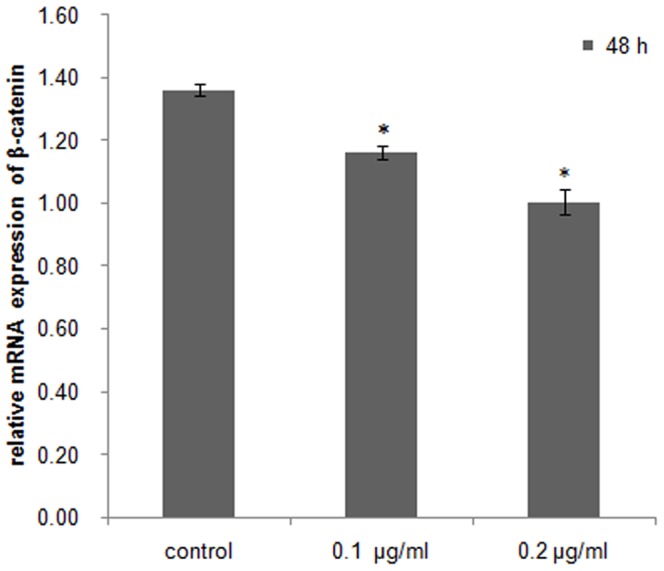
Asterisks indicate significant differences (P<0.05) between the treated group and the control.
Figure 7. Relative mRNA expression of Dax1 in cultured testis cells in vitro treated with DKK-1 for 48 h.
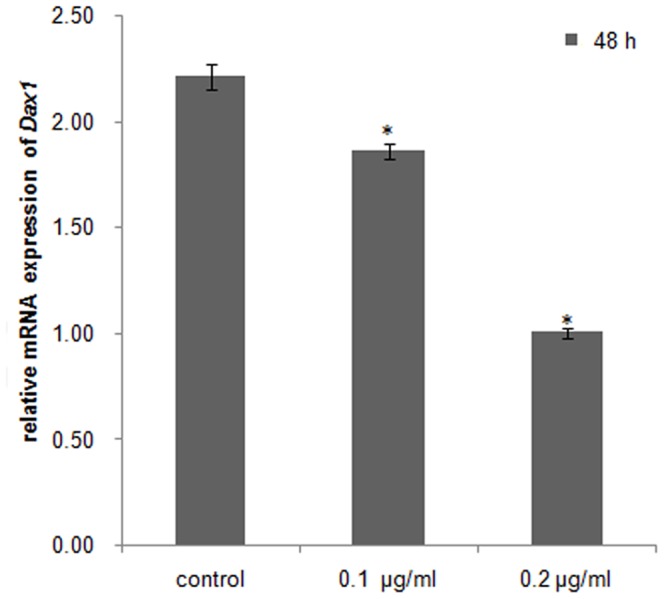
Asterisks indicate significant differences (P<0.05) between the treated group and the control.
Quercetin down-regulates β-catenin and Dax1 expression in cultured C. farrei testis cells
Quercetin suppression of β-catenin and Dax1 transcriptional activity in scallop testis cells was concentration dependent. In the cells treated for 48 h, β-catenin mRNA levels were reduced by approximately 44% in the 50 µM quercetin group and 56% in the 100 µM quercetin group compared with controls (Fig. 8), and Dax1 mRNA levels decreased by about 12% and 25%, respectively (Fig. 9).
Figure 8. Expression of β-catenin in cultured testis cells in vitro treated with quercetin for 48 h.
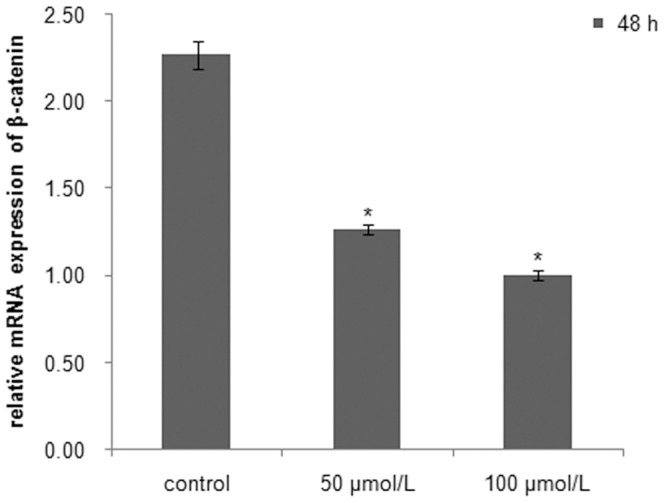
Asterisks indicate significant differences (P<0.05) between the treated group and the control.
Figure 9. Expression of Dax1 in cultured testis cells in vitro treated with quercetin for 48 h.
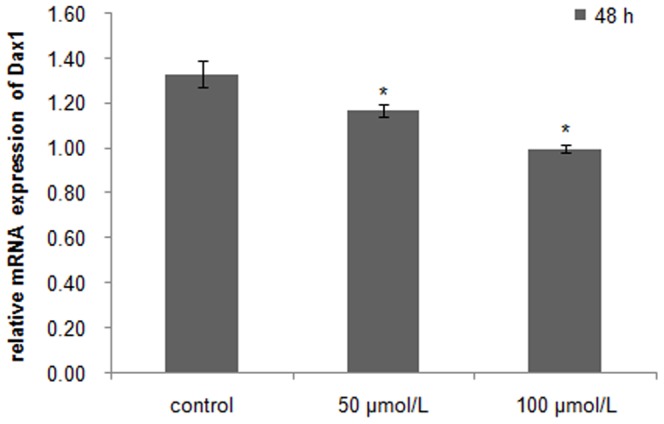
Asterisks indicate significant differences (P<0.05) between the treated group and the control.
Discussion
β-catenin is an evolutionary conserved protein in C. farreri
In the present study, we cloned and characterized a homolog of β-catenin from the scallop C. farreri. Like most known β-catenins, C. farreri β-catenin has three putative regions, an N-terminal, a C-terminal, and a central region that contains a 42-aa ARM repeating domain that repeats 12 times (Fig. 1). The deduced amino acid sequence of the target protein is highly conservative, particularly in the ARM repeat region shared 96%, 91%, and 89% identities with C. gigas, P. dumerilii, and B. floridae β-catenin, respectively. This result was consistent with previous studies reported in Hydractinia echinata, G. gallus, M. musculus, and D. rerio [23], [43], [44], [45]. The highly conserved region is thought to be required for β-catenin binding to Axin, a negative regulator of the Wnt signaling pathway, and forming a complex with GSK3β, and interacting with E-cadherin, APC and members of the LEF/TCF family [21]. Most of these interactions are involved in the regulation of the Wnt signaling pathway [22], [46].
Potential involvement of β-catenin in gametogenesis of C. farreri
Generally, the structure and developmental status of the gonads are non-permanent and change as an annual reproductive cycle in most marine animals. In the adult scallop, gametogenesis is initiated annually through mitosis and the differentiation of spermatogonia/oogonia at the beginning of the proliferative stage. This progress is gone on through meiosis and differentiation of germ cells to form mature gametes in the germinal scini of the growth and mature gonads [32]. In the present study, C. farreri β-catenin was located in the germ cells (oogonia and oocytes of ovaries, spermatogonia and spermatocytes of testes) and its expression levels increased significantly (P<0.05) in both testes and ovaries during the development, implying the β-catenin might be involved in gametogenesis and gonadal development. Furthermore, we observed the expression of C. farreri β-catenin presented a sexually dimorphic expression pattern in gonads, with significantly (P<0.05) higher levels in ovaries than in testes during the reproductive cycle, except for gonads of the resting stage. Immunohistochemical analysis demonstrated strong β-catenin staining in ovaries while a relatively weak signal was observed in testes, indicating its sex dimorphic expression at the protein level. A similar result has also been observed in mice, where β-catenin is predominately expressed in ovaries [30]. Furthermore, in mouse ovaries, β-catenin is activated by Rspo1, and Rspo1 knockout mice shows masculinized gonads, suggesting β-catenin is required for the development of ovaries [47]. Based on the similarity of expression, we hypothesized β-catenin might be involved in the gametogenesis of C. farreri. Further study is required to reveal the precise function of β-catenin in bivalve gonads.
The canonical Wnt signaling pathway exists in scallop gonads
Wnt regulates a variety of cellular events via canonical or noncanonical pathways [14], [2]. Wnt antagonists can be divided into two functional classes, the sFRP (secreted Frizzled-related protein) class and the DKK (Dickkopf) class. Members of sFRP include the sFRP family, WIF-1 (Wnt inhibitory factor 1), and Cerberus. The sFRP members can directly bind to Wnts [48], [49] to repress both the canonical and non-canonical Wnt signaling pathways. The DKK class is composed of four members, DKK-1 to DKK-4, which binds directly to the LRP5/LRP6 component of the canonical Wnt receptor complex to specifically inhibit the canonical Wnt pathway [49], [50], [51]. This down-regulates the expression of β-catenin, which is a downstream and key mediator molecule in the canonical Wnt pathway [20]. In the present study, the addition of DKK-1 at a concentration of 0.1 or 0.2 µg/mL to cultured testis cells of C. farreri significantly (P<0.05) down-regulated the expression of β-catenin compared with the control. This result is coincident with that of adult human mesenchymal stem cells from bone marrow stroma (hMSCs). In hMSCs, the canonical Wnt signaling pathway has been confirmed to exist, and DKK-1 at a concentration of 0.1 µg/mL can down-regulate the expression of β-catenin both in the cytoplasm and nucleus of cultured hMSCs [49]. Thus, our findings in the present study implied that the canonical Wnt signal pathway exists in C. farreri gonads.
β-catenin is the upstream gene of Dax1
β-catenin is a major component of the canonical Wnt pathway that participates in the regulation of gene expression. In HEK293 cells, β-catenin activated the transcription of Dax1 by interacting with Ad4BP/SF-1 in vitro, thus β-catenin is upstream of Dax1 [31]. The same conclusion is also obtained by an in vivo study in mice, where mutation of Wnt4, an upstream activator of β-catenin, causes the down-regulated expression of Dax1, suggesting the conservation of this signaling pathway in mammals [31]. To identify whether this relationship between β-catenin and Dax1 also exists in C. farreri, we examined the expressions of β-catenin and Dax1 in cultured testis cells in vitro treated with quercetin, an inhibitor against β-catenin/Tcf signaling that down-regulates the expression of β-catenin or its downstream elements in SW480 colon cancer cells and HEK293 cells [52], [53]. As expected, it was clearly shown in C. farreri testis cells cultured in vitro that the transcription of β-catenin was significantly down-regulated after treatment with quercetin for 48 h, particularly at concentrations of 100 µM (Fig. 8), which down-regulated level by approximately 56% compared with the control. This indicated quercetin inhibited β-catenin expression in C. farreri testis cells. Furthermore, the expression levels of Dax1 were also down-regulated when β-catenin expression decreased in the cultured testis cells treated with quercetin (Figs. 8 and 9) or DKK-1 (Figs. 6 and 7), suggesting Dax1 is probably the downstream gene of β-catenin.
Conclusions
In the present study, we cloned and characterized a 3,353 bp full-length cDNA sequence of β-catenin from C. farreri. The β-catenin might play a role in C. farreri gametogenesis, and a dimorphic pattern of β-catenin expression has been revealed, which was significantly higher in ovaries than testes at the same developmental stage. In addition, we showed the canonical Wnt signaling pathway exists in C. farreri spermatogenesis and the β-catenin is the upstream gene of Dax1 in this pathway. The β-catenin in C. farreri should be functionally conserved as in mammals.
Supporting Information
Expression levels of β-catenin mRNA in C. farreri gonads by qRT-PCR.
(XLSX)
Expression of β-catenin and Dax1 in in vitro cultured testis cells treated with DKK-1 and quercetin. A: Relative mRNA expression of β-catenin in cultured testis cells in vitro treated with DKK-1 for 48 h; B: Relative mRNA expression of Dax1 in cultured testis cells in vitro treated with DKK-1 for 48 h; C: Expression of β-catenin in cultured testis cells in vitro treated with quercetin for 48 h; D: Expression of Dax1 in cultured testis cells in vitro treated with quercetin for 48 h.
(XLSX)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. All the accession numbers of genes can be found in the National Center for Biotechnology Information (NCBI).
Funding Statement
This work was supported by the National High Technology Research and Development Program of China (2012AA10A402). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kawakami Y, Esteban CR, Raya M (2006) Wnt/β-catenin signaling regulates vertebrate limb regeneration. Genes Dev 20:3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Freese JL, Pino D, Pleasure SJ (2010) Wnt signaling in development and disease. Neurobiol Dis 38:148–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. El Wakil A, Lalli E (2011) The Wnt/β-catenin pathway in adrenocortical development and cancer. Mol Cell Endocrinol 332:32–37. [DOI] [PubMed] [Google Scholar]
- 4. Woll PS, Morris JK, Painschab MS, Marcus RK, Kohn AD, et al. (2007) Wnt signaling promotes hemato-endothelial cell development from human embryonic stem cells. Blood 111(1):122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lin G, Slack JM (2008) Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev Biol 316:323–335. [DOI] [PubMed] [Google Scholar]
- 6. McFarland KA, Topczewska JM, Weidinger G, Dorsky RI, Appel B (2008) Hh and Wnt signaling regulate formation of olig2+ neurons in the zebrafish cerebellum. Dev Biol 318:162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zinovyeva AY, Yamamoto Y, Sawa H, Forrester WC (2008) Complex network of Wnt signaling regulates neuronal migrations during Caenorhabditis elegans development. Genetics 179:1357–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Byrum CA, Xu R, Bince JM, McClay DR, Wikramanayake AH (2009) Blocking Dishevelled signaling in the noncanonical Wnt pathway in sea urchins disrupts endoderm formation and spiculogenesis, but not secondary mesoderm formation. Dev Dyn 238:1649–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mani P, Jarrell A, Myers J, Atit R (2010) Visualizing canonical Wnt signaling during mouse craniofacial development. Dev Dyn 239:354–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Strutt D, Madder D, Chaudhary V, Artymiuk PJ (2012) Structure function dissection of the frizzled receptor in Drosophila melanogaster suggests different mechanisms of action in planar polarity and canonical Wnt signaling. Genetics 192:1295–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuhl M, Sheldahl LC, Park M, Miller JR, Moon RT (2000) The Wnt/Ca2+ pathway: a new vertebrate Wnt signaling pathway takes shape. Trends Genet 16:279–283. [DOI] [PubMed] [Google Scholar]
- 12. Mlodzik M (2002) Planar cell polarization: do the same mechanisms regulate drosophila tissue polarity and vertebrate gastrulation? Trends Genet18:564–571. [DOI] [PubMed] [Google Scholar]
- 13. Logan CY, Nusse R (2004) The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20:781–810. [DOI] [PubMed] [Google Scholar]
- 14. MacDonald BT, Tamai K, He X (2009) Wnt/β-catenin Signaling: Components, Mechanisms, and Diseases. Dev Cell 17:9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gottardi CJ, Gumbiner BM (2004) Distinct molecular forms of β-catenin are targeted to adhesive or transcriptional complexes. J Cell Biol 167(2):339–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Habas R, Dawid IB, He X (2003) Coactivation of Rac and Rho by Wnt/Frizzled signaling is required for vertebrate gastrulation. Genes Dev 17:295–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Slusarski DC, Yang-Snyder J, Busa WB, Moon RT (1997) Modulation of embryonic intracellular Ca2+ signaling by Wnt-5A. Dev Biol 182:114–120. [DOI] [PubMed] [Google Scholar]
- 18. Wang HY, Malbon CC (2003) Wnt signaling, Ca2+, and cyclic GMP: visualizing Frizzled functions. Science 300:1529–1530. [DOI] [PubMed] [Google Scholar]
- 19. Sugimura R, Li L (2010) Noncanonical Wnt signaling in vertebrate development, stem cells, and diseases. Birth Defects Res 90:243–256. [DOI] [PubMed] [Google Scholar]
- 20. Willert K, Nusse R (1998) β-catenin: a key mediator of Wnt signaling. Curr Opin Genet Dev 8:95–102. [DOI] [PubMed] [Google Scholar]
- 21. Schneider SQ, Finnerty JR, Martindale MQ (2003) Protein evolution: structure–function relationships of the oncogene β-catenin in the evolution of multicellular animals. J Exp Zool B 295:25–44. [DOI] [PubMed] [Google Scholar]
- 22. Akiyama T (2000) Wnt/β-catenin signaling. Cytokine Growth Factor Rev 11:273–282. [DOI] [PubMed] [Google Scholar]
- 23. Huber AH, Nelson WJ, Weis WI (1997) Three-dimensional structure of the armadillo repeat region of β-catenin. Cell 90:871–882. [DOI] [PubMed] [Google Scholar]
- 24. Rubinfeld B, Albert I, Porri E, Munemitsu S, Polakis P (1997) Loss of β-catenin regulation by the APC tumor suppressor protein correlates with loss of structure due to common somatic mutations of the gene. Cancer Res 57:4624–30. [PubMed] [Google Scholar]
- 25. Nakamura T, Hamada F, Ishidate T, Anai K, Kawahara K, et al. (1998) Axin, an inhibitor of the Wnt signalling pathway, interacts with β-catenin,GSK-3β and APC and reduces the β-catenin level. Genes Cells 3:395–403. [DOI] [PubMed] [Google Scholar]
- 26. Von Kries JP, Winbeck G, Asbrand C, Schwarz-Romond T, Sochnikova N, et al. (2000) Hot spots in β-catenin for interactions with LEF-1, conductin and APC. Nat Struct Biol 7:800–807. [DOI] [PubMed] [Google Scholar]
- 27. Graham TA, Weaver C, Mao F, Kimelman D, Xu W (2000) Crystal structure of a β-catenin/Tcf complex. Cell 103:885–896. [DOI] [PubMed] [Google Scholar]
- 28. Huber AH, Weis WI (2001) The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell 105:391–402. [DOI] [PubMed] [Google Scholar]
- 29. Maatouk DM, DiNapoli L, Alvers A, Parker KL, Taketo MM, et al. (2008) Stabilization of β-catenin in XY gonads causes male-to-female sex-reversal. Hum Mol Genet 17:2949–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu CF, Bingham N, Parker K, Yao HH (2009) Sex-specific roles of β-catenin in mouse gonadal development. Hum Mol Genet 18:405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mizusaki H, Kawabe K, Mukai T, Ariyoshi E, Kasahara M, et al. (2003) Dax-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1) gene transcription is regulated by wnt4 in the female developing gonad. Mol Endocrinol 17:507–519. [DOI] [PubMed] [Google Scholar]
- 32. Liao CY, Xu YF, Wang YL (1983) Reproductive cycles of scallop Chlamys farreri (Jones and Preston 1904) at QingDao. J Fish China (in Chinese) 1:1–13. [Google Scholar]
- 33. Yang FY, Zhang B (2009) Comparative nutrition components in various populations of Chlamys farreri . J Anhui Agri Sci (in Chinese) 37(9):4073–4075. [Google Scholar]
- 34. Li H, Liu J, Huang X, Wang D, Zhang Z (2014) Characterization, expression and function analysis of DAX1 gene of Scallop (Chlamys farreri Jones and Preston 1904) during its gametogenesis. J Ocean Univ China 13(4):696–704. [Google Scholar]
- 35. Zhou QR, Shao MY, Qin ZK, Kang KH, Zhang ZF (2010) Cloning, characterization, and expression analysis of the DEAD-box family genes, Fc-vasa and Fc-PL10a, in Chinese shrimp (Fenneropenaeus chinensis). Chin J Oceanol Limnol 28:37–45. [Google Scholar]
- 36. Zhang H, Pan L, Zhang L (2012) Molecular cloning and characterization of estrogen receptor gene in the Scallop Chlamys farreri: Expression profiles in response to endocrine disrupting chemicals. Comp Biochem Physiol part C 156:51–57. [DOI] [PubMed] [Google Scholar]
- 37. Yang J, Wang L, Zhang H, Qiu L, Wang H, et al. (2011) C-type lectin in Chlamys farreri (CfLec-1) mediating immune recognition and opsonization. PLoS ONE 6(2):e17089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo H, Bao Z, Li J, Lian S, Wang S, et al. (2012) Molecular characterization of TGF-beta type I receptor gene (Tgfbr1) in Chlamys farreri, and the association of allelic variants with growth traits. PLoS ONE 7(11):e51005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liu J, Zhang Z, Ma X, Liang S, Yang D (2014) Characteristics of 17β-hydroxysteroid dehydrogenase 8 and its potential role in gonad of Zhikong scallop Chlamys farreri . J Steroid Biochem Mol Biol 141:77–86. [DOI] [PubMed] [Google Scholar]
- 40. Kenneth JL, Thomas DS (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔct method. Methods 25:402–408. [DOI] [PubMed] [Google Scholar]
- 41. Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, et al. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55(4):611–622. [DOI] [PubMed] [Google Scholar]
- 42. Hu X, Bao Z, Hu J, Shao M, Zhang L, et al. (2006) Cloning and characterization of tryptophan 2, 3-dioxygenase gene of Zhikong scallop Chlamys farreri (Jones and Preston 1904). Aquac Res 37:1187–1194. [Google Scholar]
- 43. Hobmayer E, Hatta M, Fischer R, Fujisawa T, Holstein TW, et al. (1996) Identification of a hydra homologue of the β-catenin/plakoglobin/armadillo gene family. Gene 172:155–159. [DOI] [PubMed] [Google Scholar]
- 44. Lu J, Chuong CM, Widelitz RB (1997) Isolation and characterization of chicken β-catenin. Gene 196:201–207. [DOI] [PubMed] [Google Scholar]
- 45. Xing Y, Takemaru K, Liu J, Berndt JD, Zheng JJ, et al. (2008) Crystal structure of a full-length β-catenin. Structure 16:478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hulsken J, Birchmeier W, Behrens J (1994) E-cadherin and APC compete for the interaction with β-catenin and the cytoskeleton. J Cell Biol 127:2061–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chassot AA, Ranc F, Gregoire EP, Roepers-Gajadien HL, Taketo MM, et al. (2008) Activation of β-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum Mol Genet 17:1264–1277. [DOI] [PubMed] [Google Scholar]
- 48. Kawano Y, Kypta R (2003) Secreted antagonists of the Wnt signaling pathway. J Cell Sci 116:2627–2634. [DOI] [PubMed] [Google Scholar]
- 49. Gregory CA, Singh H, Perry AS, Prockop DJ (2003) The Wnt signaling inhibitor dickkopf-1 is required for reentry into the cell cycle of human adult stem cells from bone marrow. J Biol Chem 278:28067–28078. [DOI] [PubMed] [Google Scholar]
- 50. Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA (2001) Novel mechanism of Wnt signaling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat Cell Biol 3:683–686. [DOI] [PubMed] [Google Scholar]
- 51. Mao B, Niehrs C (2003) Kremen2 modulates Dickkopf2 activity during Wnt/lRP6 signaling. Gene 302:179–183. [DOI] [PubMed] [Google Scholar]
- 52. Park CH, Chang JY, Hahm ER, Park S, Kim HK, et al. (2005) Quercetin, a potent inhibitor against β-catenin / Tcf signaling in SW480 colon cancer cells. Biochem Biophys Res Commun 328:227–234. [DOI] [PubMed] [Google Scholar]
- 53. Shan BE, Wang MX, Li RQ (2009) Quercetin inhibit human SW480 colon cancer growth in association with inhibition of cyclin D1 and surviving expression through Wnt/β-catenin signaling pathway. Cancer Invest 27:604–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression levels of β-catenin mRNA in C. farreri gonads by qRT-PCR.
(XLSX)
Expression of β-catenin and Dax1 in in vitro cultured testis cells treated with DKK-1 and quercetin. A: Relative mRNA expression of β-catenin in cultured testis cells in vitro treated with DKK-1 for 48 h; B: Relative mRNA expression of Dax1 in cultured testis cells in vitro treated with DKK-1 for 48 h; C: Expression of β-catenin in cultured testis cells in vitro treated with quercetin for 48 h; D: Expression of Dax1 in cultured testis cells in vitro treated with quercetin for 48 h.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. All the accession numbers of genes can be found in the National Center for Biotechnology Information (NCBI).