Abstract
Scope
Established epithelial cell lines equipped with pattern recognition receptors such as the Toll-like receptor (TLR)-2 are common tools for immune response studies on invading pathogens, e.g. the obligate intracellular species of Chlamydia. Moreover, such models are widely used to elucidate fatty acid-mediated immune effects. In several transformed cell lines, however, unusual loss of metabolic functions was described. The cell lines A549 and HeLa are poorly characterized in this respect. Therefore, we comparatively assessed the metabolic capacity of A549 and HeLa prior to proposed application as in vitro model for fatty acid effects on chlamydial infection.
Methodology/Principal Findings
We incubated both cell lines either with substrates (C18∶2n−6 or C18∶3n−3) or products (C18∶3n−6, C18∶4n−3) of fatty acid desaturase-2 (FADS2), and analysed the fatty acid profiles after 24 h and 72 h by gas chromatography. Based on these data, we suspected that the complete discontinuation of normal biosynthesis of long-chain polyunsaturated fatty acids (LC-PUFA) in HeLa was due to loss of FADS2 function. Consequently, prostaglandin E2 (PGE2) formation was less inducible by TLR2 stimulation in HeLa, likely as a result of not only insufficient supply of precursors but also weak cyclooxygenase-2 (COX-2) response. In accordance, Chlamydia infection rates were consistently lower in HeLa than in A549. Sequence analysis revealed no alteration within the FADS2 gene in HeLa. The FADS2 expression level, however, was significantly lower and, in contrast to A549, not regulated by C18∶2n−6. A549 exhibited regular fatty acid metabolism and enzyme functionality.
Conclusions/Significance
Our data show that HeLa cells considerably differ from A549 at several stages of fatty acid metabolism. The poor metabolic potential of HeLa, mainly concerning FADS2 upstream of COX-2 function, calls into question whether these cells represent a good model to unveil fatty acid or downstream eicosanoid effects in the course of intracellular bacterial infection.
Introduction
The cell lines A549, a human airway epithelial cell line, and HeLa, which originate from the human cervix epithelium, are widely used to represent the epithelium as the first barrier to encounter and defend microbial infection [1], [2]. Invading pathogens, such as the obligate intracellular species of Chlamydia, are recognized by prominent host defence receptors referred to as Toll-like receptors (TLR) [3]. TLR2, for instance, predominantly recognizes the chlamydial major outer membrane protein (MOMP), and is therefore considered to be critical for Chlamydia-mediated host cell activation and pathology [4]–[6]. Engagement of TLR2 immediately initiates downstream signaling towards COX-2-catalysed formation and release of n−6 LC-PUFA-derived eicosanoids to propagate a specific immune response [2], [7], [8]. An altered PUFA biosynthesis pathway entailing an insufficient supply of eicosanoid and downstream docosanoid precursors would therefore have serious consequences for adequate orchestration of the local immune response. Since in vitro models can only provide insights into a limited part of the in vivo situation, it is highly important to ensure appropriate functionality regarding the part that should be displayed. Immortalized cell lines with tendency for continuous growth have been recognized to be prone to loss of function [9], and little is known concerning the suitability of A549 and HeLa to serve as reliable in vitro models for such studies on immune responses involving fatty acids or their metabolites.
Most mammalian cells synthesize n−6 or n−3 LC-PUFA in a sophisticated anabolic cascade with alternating desaturation and elongation steps from dietary supplied C18∶2n−6 and C18∶3n−3 [10]. The initial and rate limiting step is catalysed by the FADS2-encoded Δ-6 desaturase that introduces a defined double bond into the acyl chain of its main substrates C18∶2n−6 and C18∶3n−3 to produce C18∶3n−6 and C18∶4n−3, respectively. Moreover, an alternative Δ-8 desaturase activity via C20∶2n−6 and C20∶3n−3 has also been linked to FADS2 [11]. Following chain elongation, the next desaturation reaction undertaken by FADS1 with Δ-5 activity is required to synthesize C20∶4n−6 and C20∶5n−3, which are further metabolized to docosaenoics, again with the help of FADS2. Finally, and catalysed by COX-2 activity, C20- and C22-PUFA give rise to highly bioactive derivatives such as eicosanoids and docosanoids, which play crucial roles, e.g., in inflammation and its resolution [12], [13].
Herein, we show that the loss of FADS2 function entailed a complete discontinuation of normal PUFA biosynthesis in HeLa. Consequently, PGE2 formation was less inducible in HeLa, likely as a result of not only insufficient supply of precursors but also weak COX-2 response. These results may serve as an explanation for the observation that Chlamydia infection rates were consistently lower and less stable in HeLa than in A549.
Materials and Methods
Cell culture
HeLa (cervix adenocarcinoma epithelial cell line; ATCC number: CCL-2) or A549 (type II alveolar adenocarcinoma epithelial cell line; ATCC number: CCL-185) were seeded in 25 cm2 tissue culture flasks at a density of 1.5×105/mL. Cells were cultured for 24 h in a total volume of 6 mL DMEM/10% FBS alone or supplemented with either 33 µM C18∶2n−6, C18∶3n−3, C18∶3n−6, or C18∶4n−3 (all from Sigma-Aldrich, Taufkirchen, Germany). Free fatty acids were dissolved in sterile DMSO (Roth, Karlsruhe, Germany) to produce a 100 mM stock solution and stored under nitrogen in aliquots at −20°C. A corresponding volume of DMSO accounting for maximum 0.03% was added to the fatty acid free cultures. After 24 h (and 72 h for some experiments), cells were harvested and either processed for viability determination, fatty acid analysis, assessment of cell surface CD36, or gene expression and sequence analysis. For stimulation of COX-2-mediated PGE2 formation, 1.5×105 cells/mL were seeded into 6-well plates and cultured for 20 h in the presence of either 33 µM C18∶2n−6 or C18∶3n−6. Subsequently, 0.1 µg/mL PAM2CSK4 (acts as TLR2 agonist [14], Cayla-Invivogen, Toulouse, France) or 1 ng/mL recombinant TNF-α (Life Technologies, Darmstadt, Germany) was added. Corresponding wells were 1 h preincubated with 2 µM of a selective COX-2 inhibitor (sc-58125) or 200 nM of a selective COX-1 inhibitor (sc-560; both Biomol, Hamburg, Germany) before stimulation. Control cultures contained DMSO vehicle. After 8 h, cells were detached, centrifuged and processed for flow cytometry or protein quantification. Supernatants were collected and stored at −20°C until PGE2 analysis. Before they were used in the experiments, cells were subcultivated at least six times after thawing. Cultures were held under standard conditions at 37°C, 5% CO2, 95% humidity. Finally, in both A549 and HeLa a DNA profiling using 8 different and highly polymorphic short tandem repeat (STR) loci was carried out in order to confirm authenticity of each cell line (Authentication service, Leibniz-Institute DSMZ, Braunschweig, Germany).
Viability and absolute cell count determination
Viability was flow cytometrically measured by annexin-V/propidium iodide exclusion staining as described previously [15]. Absolute cell count was quantified by flow cytometry using counting beads (eBioscience, Frankfurt/Main, Germany).
Fatty acid analysis
Total lipids were extracted from PBS-washed cell pellets using a methanol/chloroform mixture according to Bligh and Dyer [16]. Subsequently, a transesterification was performed by incubating samples with 0.5 N methanolic sodium hydroxide at 100°C for 10 min followed by methanolic boron trifluoride (10% w/w; Supelco, Bellefonte, PA, USA) treatment at 100°C for 5 min. Subsequently, fatty acid methyl esters (FAME) were extracted with n-hexan, separated in a gas chromatograph (GC 17a V3, Shimadzu, Kyoto, Japan) using a fused-silica capillary column with medium polarity (DB 225 MS, 60 m×0.25 mm i.d., 0.25 µm film thickness; Agilent Technologies, Santa Clara, USA) and detected downstream by a flame ionization detector (FID). GC conditions were as previously described [17]. Peak area integration was accomplished using GC solution software version 2.3 in comparison to previously measured reference standards (BR2, BR4, and ME93 from Larodan/CPS-Chemie, Aachen, Germany; Menhaden from Sigma-Aldrich, Taufkirchen, Germany; 463 and 674 from Nu-Chek-Prep, Elysian, USA).
FADS2 and FADS1 expression analysis
First strand cDNA was synthesized from 2 µg extracted total RNA (Total RNA and protein isolation kit, Macherey-Nagel, Düren, Germany) using random oligo(dT) primers and reagents according to the conditions supplied in the Transcriptor First Strand cDNA Synthesis Kit from Roche Diagnostics (Mannheim, Germany). Samples were stored at −20°C until further use. Target primer sequences and probes were retrieved from the Universal Probe Library (UPL, Roche) and are listed in Table 1. Primers were purchased from Biomers (Ulm, Germany). PCR efficiencies for each primer pair were determined by serial cDNA dilutions. The 20 µl reaction mixture contained 5 µL cDNA, 3 µL water, 0.5 µL (0.5 µM final concentration) of each primer, 1 µL (0.1 µM) probe and 10 µL 1×Light Cycler480 Probe Master Mix (Roche). All reactions were performed in technical triplicates in the Light Cycler480 Instrument (Roche) with a PCR profile including an initial denaturation step of 10 min at 95°C followed by 45 amplification cycles each at 95°C for 10 s, 60°C for 30 s and 72°C for 1 s, and a terminal cooling period of 10 s at 40°C. Analysis of the PCR results was carried out with the Light Cycler480 Software (Roche). Expression levels were determined by calculating relative quantifications using the ΔΔCT-method with TUBA1A (α1-tubulin) as the reference gene.
Table 1. Primers and UPL probes used for real-time gene expression analysis.
| Gene name | GenBank accession# | Nucleotide sequence (5′→3′) | UPL probes | Amplicon | PCR efficiency |
| FADS2 * | NM_004265.3 | FW: ttaacttccagattgagcacca | #60 | nt 1286–nt 1351 | 1.87 |
| RV: gggcgatcttgtgtaagttgt | |||||
| FADS1 | NM_013402.4 | FW: cattttgtgattggccacct | #11 | nt 891–nt 986 | 2.05 |
| RV: gtctttgcggaagcagttg | |||||
| TUBA1A | NM_006009.3 | FW: cttcgtctccgccatcag | #58 | nt 304–nt 398 | 1.94 |
| RV: ttgccaatctggacacca |
*Products on intended targets: transcript variants 1–3.
Sequence analysis of FADS2 in HeLa
For full-length sequencing encompassing the entire FADS2 coding region in HeLa cells, DNA was extracted using the Cristal DNA extraction Kit (Bio Lab Products, Hamburg, Germany) according to the instructions. PCR was performed with Pfu polymerases (Fermentas, St. Leon Roth, Germany) with 0.3 µM of each primer and conditions as recommended by the manufacturer. After initial denaturation at 94°C for 2 min, 35 cycles each of 94°C for 30 s, 60°C for 20 s, and 72°C for 90 s were performed. Primers (TIB Molbiol, Berlin, Germany) used for the amplification are listed in Table 2. Subsequently, the PCR products were cloned into pJet1.2 (Fermentas) and sequenced using the sequencing primers pJet1.2-F or pJet1.2-R at LCG sequencing service (Berlin, Germany).
Table 2. Primers used for FADS2 encoding sequence analysis.
| Coding exon no. | Primer sequence (5′→3′) | Position in AP002380 |
| 1, tr. var. 2 and 3 | FW: aatgggctcctttcctgctc | 137,054-73 |
| RV: ggctcagggacctacccc | 137,713-30 | |
| 1, tr. var. 1 | FW: ccgctggccttcgaaagat | 148,918-36 |
| RV: agatgcactttccttccctacag | 149,713-35 | |
| 2 | FW: gatcacaggagaccccgttt | 158,653-72 |
| RV: cagttactggtcatggggcaag | 158,966-87 | |
| 3/4 | FW: gcattggctgttgaaatggc | 161,249-68 |
| RV: accctgtagcctcagagcac | 161,807-26 | |
| 5 | FW: cctgaagtgctatccccgag | 169,042-61 |
| RV: tgtcagctctctaggcttcct | 169,477-97 | |
| 6/7 | FW: ggtgtgccctgagcagatag | 177,906-25 |
| RV: ccagctgtgtccccaactc | 178,537-55 | |
| 8/9/10 | FW: agagggcaggttgcacctaa | 183,938-57 |
| RV: gcccccacattctgcattct | 184,860-79 | |
| 11/12 | FW: atagagcacaaacgccctcc | 185,935-54 |
| RV: catacaccagcctctcggac | 186,777-96 |
tr. var. - transcript variants.
Flow cytometry for determination of cyclooxygenases and CD36 expression
For intracellular quantification of cyclooxygenase protein expression, cells were permeabilized by washing with PBS/0.1% BSA/0.1% saponine and stained with Multicolor anti-human COX-1-FITC/anti-human COX-2-PE monoclonal antibodies (mAb; clones AS70/AS57, BD, Heidelberg, Germany) as described previously [18]. To assess surface expression of CD36, cells were stained with anti-human CD36-APC mAb (clone TR9, Abcam, Cambridge, UK). Non-specific fluorescence was controlled by incubation with isotype-matched antibodies. Samples were measured on a FACSCalibur flow cytometer and analysed using CELLQUEST software (BD).
PGE2 formation
PGE2 formation was quantified in supernatants using a competitive enzyme immunoassay (Cayman, Ann Arbor, USA) with indicated sensitivity of 50 pg/mL and limit of detection at 15 pg/mL. Data are related to the protein content of the corresponding cells which was determined by employing the Lowry method using bovine gamma-globulin as standard [19]. The colorimetric assay was purchased from Bio-Rad (München, Germany).
Assessment of Chlamydia infection rates
HeLa and A549 were seeded in 12-well plates at a density of 1.5×105 cells/mL and cultured for 24 h. Monolayers were infected with Chlamydia psittaci strain DC15 (DSM27008) at a multiplicity of infection (MOI) of 4 as described [20]. The strain was obtained from the collection of the National Reference Laboratory for Chlamydiosis at Friedrich-Loeffler-Institut Jena, Germany (Head: Dr. K. Sachse). 48 h post infection (p.i.), cells were trypsinized, fixed with methanol and processed for flow cytometric quantification of chlamydial infection as previously described [21]. For immunofluorescence staining, cells were inoculated with strain DC15 at indicated MOI of 4 in shell vials on coverslips. 48 h p.i., methanol-fixed coverslips with monolayers were mounted on slides, stained with fluorochrome-labeled antibody against Chlamydiaceae-specific lipopolysaccharide (IMAGEN Chlamydia kit, Oxoid Limited, Wesel, Germany) and visualized using fluorescence microscopy, according to Goellner et al. [20].
Statistics
Based on likelihood ratio test (LRT), a linear model with generalized least squares was used to determine the relationship between the outcomes FAME and FADS2 mRNA expression, respectively, and the two factors cell line (HeLa and A549) and fatty acid (FA) treatment (without and with) or the interaction of these factors. This model was considered appropriate because of variance inhomogeneity within the factor FA treatment. For comparison of data on COX-2 expression and PGE2 formation, the stimulation condition was entered as third independent factor. To avoid heteroscedasticity, data were log-transformed. Post hoc test was conducted using Tukey-Kramer test and p values were adjusted for multiple comparisons. All calculations were carried out using the gls-function from R package nlme version 3.1–103. Data on FADS ratios and infection rates were evaluated using 2-tailed Student’s t-test (SPSS software version 19.0, SPSS Inc., Illinois USA). To indicate differences of desaturation activity between HeLa and A549, the following product to substrate ratios were estimated and compared: (C18∶3n−6+C20∶3n−6)/C18∶2n−6 and (C18∶4n−3+C20∶4n−3)/C18∶3n−3 for FADS2 function (after 24 h incubation with C18∶2n−6 and C18∶3n−3, respectively), and C20∶4n−6/C20∶3n−6 and C20∶5n−3/C20∶4n−3 for FADS1 activity (after 24 h incubation with C18∶3n−6 and C18∶4n−3, respectively). Unless otherwise indicated, data are expressed as means ± SEM of four independent experiments that were performed based on different cryopreserve lots. Significance of difference was set at p≤.05.
Results and Discussion
Effect of fatty acid incubation on viability and cell growth
In both HeLa and A549 cell lines, there was no negative effect of any fatty acid treatment on viability and cell growth over an incubation period of 24 h to 72 h compared to the DMSO-control (data not shown).
PUFA biosynthesis
Following a 24-h incubation with 33 µM C18∶2n−6 or C18∶3n−3, the intracellular contents of these fatty acids increased in A549 from 2.0±0.3% and 0.1±0.1% to 14.6±0.7% and 10.2±0.3% of total FAME, respectively (Table 3). Concomitantly, all following intermediates of the n−6 and n−3 LC-PUFA biosynthesis were detected in abundance up to and including C22-PUFA (n−3 PUFA observed after 72 h; Fig. 1B), whose production is essentially dependent on efficient enzyme activity for desaturation (FADS2 and FADS1) and acyl chain elongation (fatty acid elongases ELOVL5 and ELOVL2; Fig. 2) [22]. Ratios reflecting direct FADS2 activity were calculated in A549 at 24 h as 0.24±0.06 for n−6 and 0.46±0.1 for n−3 (DMSO-ctrl.-corrected p≤.001 vs. HeLa, Fig. 3). Taking into account that FADS2 products are themselves substrates for subsequent enzymes of the progressional LC-PUFA biosynthesis, then consideration of the allover product (additional increase in all detected eicosaenoics and docosaenoics in sum) to substrate ratios allows a more comprehensive assessment of the metabolic capacity, particularly as FADS2 catalyzes a second step in the cascade of LC-PUFA synthesis. Thus, n−6- and n−3-related desaturation ratios increased to 0.65±0.15 for n−6 and to 1.17±0.11 for n−3 (data not shown) whilst retaining the ratio of n−6 to n−3 products at approximately 1∶2. These data indicate a substrate preference of FADS2 for n−3 over n−6 fatty acids in A549 cells, provided that both substrates are initially present in equimolar concentrations, and corroborate previous results obtained from HepG2 (human hepatocellular carcinoma cells) and cardiomyocytes [23], [24].
Table 3. Metabolic capacity of HeLa compared to A549.
| FADS2required | HeLa | A549 | FA treat-ment | Cell line | FA treat-ment×cell line | FADS2passedover | HeLa | A549 | FA treat-ment | Cell line | FA treat-ment×cell line | ||||
| C18∶2 n −6 a | − | + | − | + | p = | p = | p = | C18∶3 n −6 b | − | + | − | + | p = | p = | p = |
| C18∶2n−6 | 2.1±0.3 | 17.2±1.0 | 2.0±0.3 | 14.6±0.7 | <.001 | .442 | .014 | 2.1±0.3 | 2.3±0.2 | 2.0±0.3 | 1.4±0.2 | .438 | .402 | .024 | |
| C18∶3n−6 | 0.0±0.0 | 0.0±0.0 | 0.3±0.2 | 0.8±0.1 | .810 | .006 | .002 | 0.0±0.0 | 2.2±0.5 | 0.3±0.2 | 5.4±0.8 | <.001 | .468 | <.001 | |
| C20∶2n−6c | 0.0±0.1 | 1.4±0.2 | 0.1±0.1 | 1.0±0.1 | <.001 | .601 | .008 | 0.0±0.1 | 0.0±0.0 | 0.1±0.1 | 0.0±0.1 | .379 | .554 | .921 | |
| C20∶3n−6 | 0.3±0.0 | 0.2±0.2 | 1.0±0.0 | 3.5±0.6 | .634 | <.001 | <.001 | 0.3±0.0 | 7.8±0.8 | 1.0±0.0 | 13.9±1.3 | <.001 | <.001 | <.001 | |
| C20∶4n−6 | 1.8±0.3 | 1.9±0.1 | 3.9±0.4 | 8.9±1.3 | .824 | <.001 | <.001 | 1.8±0.3 | 8.0±0.8 | 3.9±0.4 | 11.7±1.2 | <.001 | <.001 | .148 | |
| C22∶4n−6 | 0.1±0.1 | 0.2±0.0 | 0.1±0.1 | 1.8±0.5 | .883 | .826 | <.001 | 0.1±0.1 | 1.2±0.1 | 0.1±0.1 | 2.1±0.4 | <.001 | .550 | .004 | |
| C22∶5n−6d | 0.0±0.0 | 0.0±0.0 | 0.0±0.1 | 0.3±0.1 | 1.00 | .319 | <.001 | 0.0±0.0 | 0.0±0.0 | 0.0±0.1 | 0.3±0.1 | 1.00 | .472 | .003 | |
| C18∶3 n −3 a | − | + | − | + | C18∶4 n −3 b | − | + | − | + | ||||||
| C18∶3n−3 | 0.1±0.1 | 16.2±2.1 | 0.1±0.1 | 10.2±0.3 | <.001 | .236 | .001 | 0.1±0.1 | 0.1±0.1 | 0.1±0.1 | 0.0±0.1 | .602 | .150 | .813 | |
| C18∶4n−3 | 0.1±0.1 | 0.0±0.0 | 0.0±0.1 | 0.9±0.2 | <.001 | <.001 | <.001 | 0.1±0.1 | 2.3±1.0 | 0.0±0.1 | 4.7±1.3 | <.001 | .721 | .041 | |
| C20∶3n−3c | 0.0±0.0 | 2.3±0.2 | 0.0±0.1 | 1.0±0.1 | <.001 | .685 | <.001 | 0.0±0.0 | 0.2±0.2 | 0.0±0.1 | 0.0±0.0 | .501 | .366 | .308 | |
| C20∶4n−3 | 0.0±0.0 | 0.0±0.0 | 0.0±0.0 | 3.8±0.2 | <.001 | <.001 | <.001 | 0.0±0.0 | 9.2±0.2 | 0.0±0.0 | 10.1±1.3 | <.001 | .577 | .296 | |
| C20∶5n−3 | 0.4±0.1 | 0.4±0.0 | 0.5±0.0 | 6.4±0.4 | <.001 | <.001 | <.001 | 0.4±0.1 | 7.3±1.3 | 0.5±0.0 | 6.5±1.7 | <.001 | .006 | .509 | |
| C22∶5n−3 | 0.8±0.1 | 0.7±0.2 | 0.8±0.1 | 3.1±0.2 | <.001 | <.001 | <.001 | 0.8±0.1 | 3.5±0.9 | 0.8±0.1 | 2.4±0.8 | <.001 | .769 | .199 | |
| C22∶6n−3d | 1.0±0.2 | 1.0±0.1 | 1.7±0.1 | 1.6±0.0e | .808 | <.001 | .364 | 1.0±0.2 | 0.9±0.1 | 1.7±0.1 | 1.9±0.0e | .036 | <.001 | .233 | |
Fatty acid profiles were determined by GC-FID analysis of cellular lipid extracts and are expressed as % of total FAME. Data represent means ± SD.
Cells were incubated without (−) or with (+) 33 µM of C18∶2n−6 and C18∶3n−3, respectively, for 24 h. Both are substrates for FADS2.
Cells were incubated without (−) or with (+) 33 µM of the FADS2 products C18∶3n−6 and C18∶4n−3 for 24 h.
Considered Δ8-desaturation products.
Synthesis requires upstream a second step of FADS2-encoded activity.
After 72 h, significant increase in A459 to 2.7±0.2 when incubated with C18∶3n−3 (FA treatment×cell line: p = .002; see also Figure 1B, peak 14) and to 2.8±0.1 when incubated with C18∶4n−3 (FA treatment×cell line: p = .011).
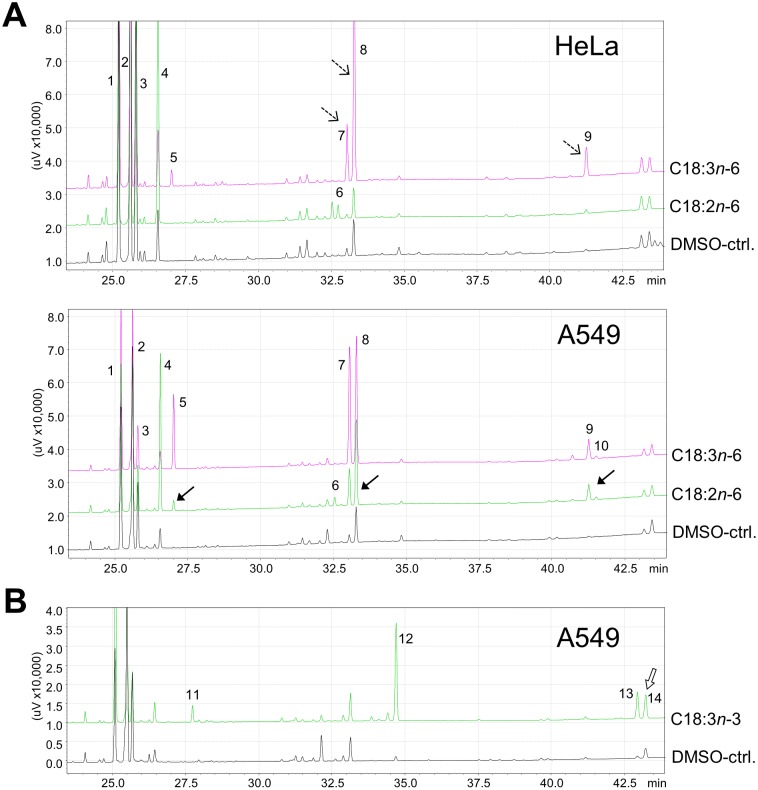
Figure 1. Partial chromatograms of cellular lipids of HeLa and A549.
Cells were incubated without (DMSO-treated control) or with the indicated fatty acid (33 µM) for (A) 24 h or (B) for 72 h. 1–C18∶0, 2–C18∶1c9, 3–C18∶1c11, 4–C18∶2n−6, 5–C18∶3n−6, 6–C20∶2n−6, 7–C20∶3n−6, 8–C20∶4n−6, 9–C22∶4n−6, 10–C22∶5n−6, 11–C18∶3n−3, 12–C20∶5n−3, 13–C22∶5n−3, 14–C22∶6n−3. A: C20∶2n−6 (peak 6) was the only metabolite identified in HeLa following incubation with C18∶2n−6. In contrast, all detectable elongated and desaturated products of C18∶2n−6 increased in A549 (filled-headed arrows). When C18∶3n−6 was the supplement, the respective metabolites increased in HeLa, too (broken-lined arrows). B: As a product of C18∶3n−3, C22∶6n−3 (peak 14) increased in A549 after 72 h.
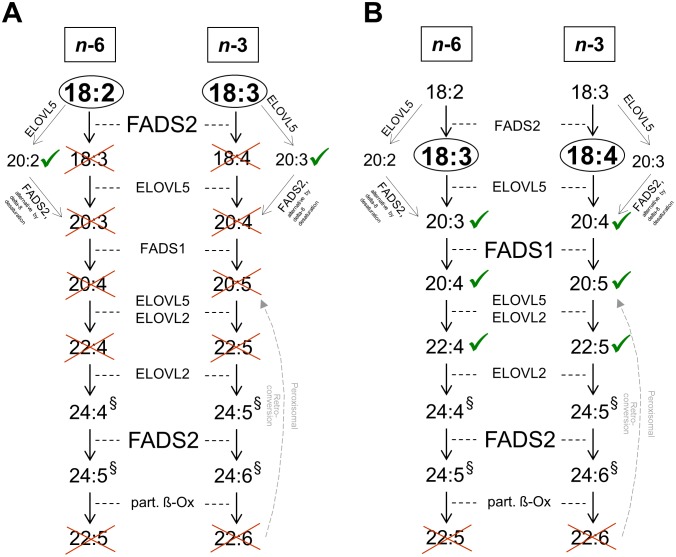
Figure 2. HeLa lack FADS2 but not FADS1 function.
A: HeLa failed to metabolize C18∶2n−6 and C18∶3n−3 to their respective Δ-6 desaturation products. However, the respective elongation products increased. B: Bypassing the (first) FADS2-dependent step by incubation with C18∶3n−6 and C18∶4n−3, respectively, led to synthesis of all subsequent products, including those involving FADS1, up the second step that required FADS2 function. §C24-LC-PUFA were not detectable due to methodological limitations. Ticked: product emerged, crossed: product not generated. ELOVL – fatty acid elongase, part. ß-Ox – partial ß-oxidation.
Figure 3. Product-to-substrate ratios as estimates of desaturase activity in HeLa and A549.
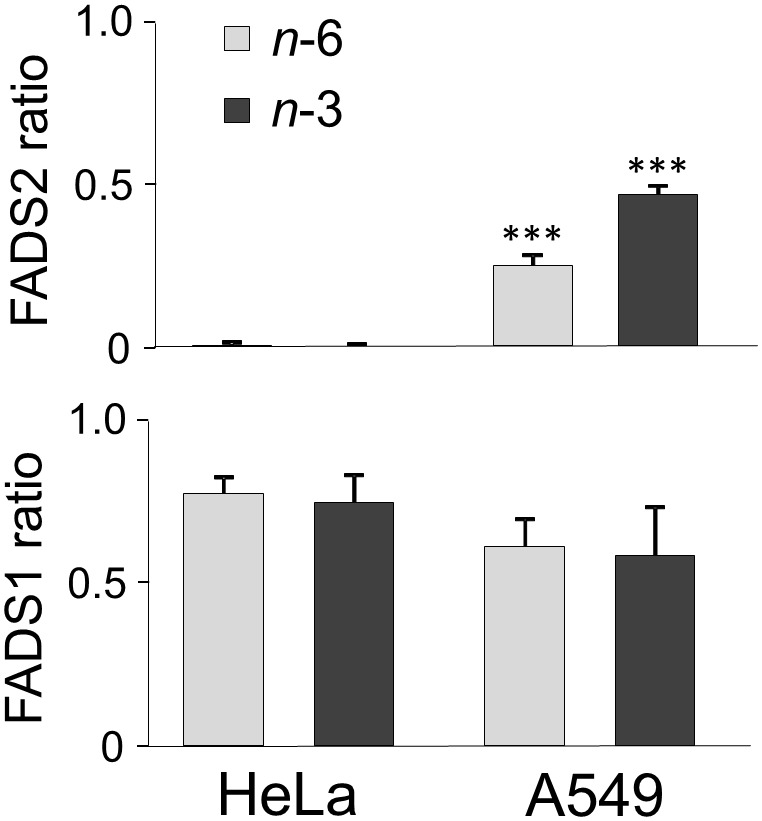
FADS2 ratio: (C18∶3n−6+C20∶3n−6)/C18∶2n−6 and (C18∶4n−3+C20∶4n−3)/C18∶3n−3 after 24 h (with the second FADS2-dependent step not considered), FADS1 ratio: C20∶4n−6/C20∶3n−6 and C20∶5n−3/C20∶4n−3 after 24 h. Increase in FADS2 ratios is significantly different from HeLa (***p≤.001).
In HeLa, after 24 h incubation with 33 µM of the respective fatty acid, the percentages of C18∶2n−6 or C18∶3n−3 also increased from 2.1±0.3% and 0.1±0.1% to 17.2±1.0% and 16.2±2.1% of total FAME, respectively (Table 3). However, products directly involving FADS2 activity, such as C18∶3n−6 or C18∶4n−3, were not detected at the expected concentrations (Fig. 1A), reflected by FADS2 ratios of 0±0 for both n−6 and n−3 (Fig. 3). In contrast, there was an increase in the respective elongation products C20∶2n−6 and C20∶3n−3, considered as substrates for the alternative FADS2-catalysed Δ-8 desaturation [11] (Table 3, Figs. 1, 2A). In several cancer cell lines with FADS2 deficiency, such as K562 and MCF7, it has been shown that FADS1 operates in a compensatory manner to produce unusual C20∶3 and C20∶4 of unclear physiological relevance with butylene- instead of the methylene-interrupted double bonds [25], [26]. However, since we detected neither the usual form nor unusual C20∶3 or C20∶4, both C20∶2n−6 and C20∶3n−3 denoted dead-end products in HeLa. Interestingly, when HeLa were incubated with the Δ-6 desaturation products C18∶3n−6 and C18∶4n−3 with an aim to passing over this rate limiting step of PUFA biosynthesis, concentrations of all the following metabolites increased within the cellular lipid fraction up to the second step that involves FADS2 activity (Table 3, Fig. 2B). Moreover, FADS1 ratios for both n−6 and n−3 were similar for A549 and HeLa (Fig. 3).
FADS2 expression and sequence analyses
Since firstly, several single nucleotide polymorphisms (SNPs) are associated with decreased FADS2 activity [27], [28] and altered transcriptional regulation [29], and secondly, its gene cluster locus on human chromosome 11q12–13.1 is susceptible to carcinogenic alterations [rev. in 26], we hypothesized that the lacking capacity for Δ-6 desaturation of HeLa resides at the transcriptional level. Therefore, we pursued the following two strategies: first, we compared HeLa with A549 regarding FADS2 mRNA expression, and second, we proceeded to a FADS2-specific sequence analysis of genomic DNA in HeLa. Because previous observations suggest that FADS2 is regulated by both its substrates and products [30], we performed gene expression analysis in cultures after 24 h incubation with either C18∶2n−6 (FADS2 substrate) or C18∶3n−6 (FADS2 product). In A549, the amplicon of FADS2 was detected in relative abundance. Presumably as a negative feedback response, FADS2 mRNA was down-regulated upon 24 h incubation with C18∶2n−6 (p≤.005 vs. DMSO-ctrl.) and, to a lesser and non-significant extent, also in response to C18∶3n−6 (Fig. 4). In HeLa, the expression level of FADS2 mRNA was only about a quarter of that in A549 and neither altered by C18∶2n−6 nor by C18∶3n−6 (Fig. 4). This outcome supports our observation that HeLa cells failed to metabolize C18∶2n−6 and C18∶3n−3 to their respective Δ-6 desaturation products (Table 3) what indicates lacking FADS2 functionality.
Figure 4. Real-time quantitative expression of FADS2 mRNA in HeLa and A549.
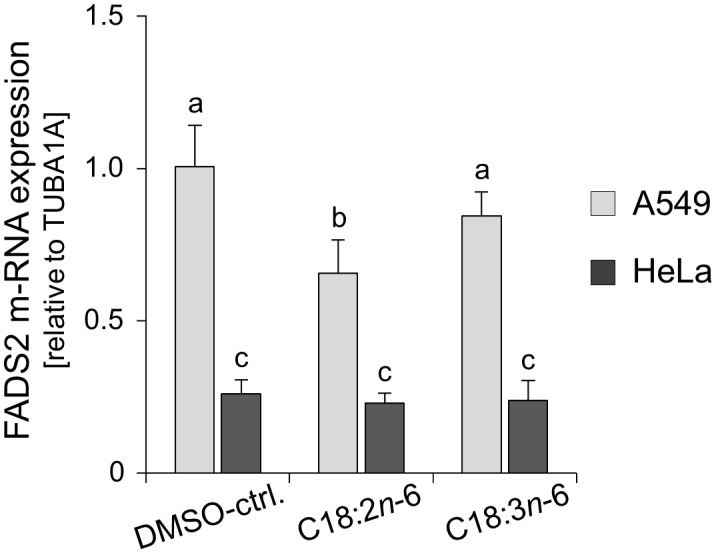
Cells were incubated without (DMSO-treated control) or with 33 µM C18∶2n−6 or C18∶3n−6 for 24 h. Different letters indicate significant differences: a vs. b, and b vs. c, p≤.005.
Many transformed cells that are not capable of Δ-6 desaturation have been shown to possess Δ-5 desaturating ability at functional and transcriptional levels [9]. In accordance, the FADS1 expression levels in HeLa were not different from those detected in A549 (data not shown), what fits with the data obtained from GC-FID analysis (Table 3).
To verify the DNA sequence of FADS2 in HeLa, all coding exon fragments were cloned, sequenced and compared with the published sequence (GenBank: AP002380). Since we did not find any sequence alteration within the coding region of FADS2, we can only speculate about the conditions that led to a loss of FADS2 function in HeLa. Maybe this deficit is the consequence of alterations within the promoter region of FADS2, as suggested [31], or post transcriptionally, of unusual protein folding that, as yet, remains elusive.
Cyclooxygenase-mediated PGE2 formation and expression of CD36
Using the example of PGE2, we further looked at eicosanoid formation, which crucially depends on an efficient upstream biosynthesis of PUFA. We hypothesized that the lacking activity of FADS2 in HeLa consequently resulted in an altered pattern of PGE2 formation from upstream C18∶2n−6 as compared with C18∶3n−6 as well as compared with A549. In both HeLa [32] and A549 [8], TLR2 stimulation rapidly induces COX-2 through p38 mitogen activated protein kinase (MAPK) and nuclear factor kappa B (NFκB) pathways. Moreover, both HeLa and A549 highly express PGE synthase in response to cytokine stimulation, the enzyme acting downstream from COX-2 to convert the prostaglandin endoperoxide H2 (PGH2), deriving from C20∶4n−6, into PGE2 [33]. For induction of cyclooxygenase activity, we mimicked chlamydial infection by TLR2 engagement. Both HeLa and A549 up-regulate TLR2 upon stimulation with microbial components [34]. TLR2 signaling is dependent on the expression of co-receptors. As HeLa lack TLR1 expression and are non-responsive to TLR2/TLR1 receptor pair ligands [6], we used PAM2CSK4 to induce TLR2/TLR6 signaling with subsequent activation of the COX-2 pathway. Stimulation with TNF-α as a TLR-independent COX-2 activator served as positive control.
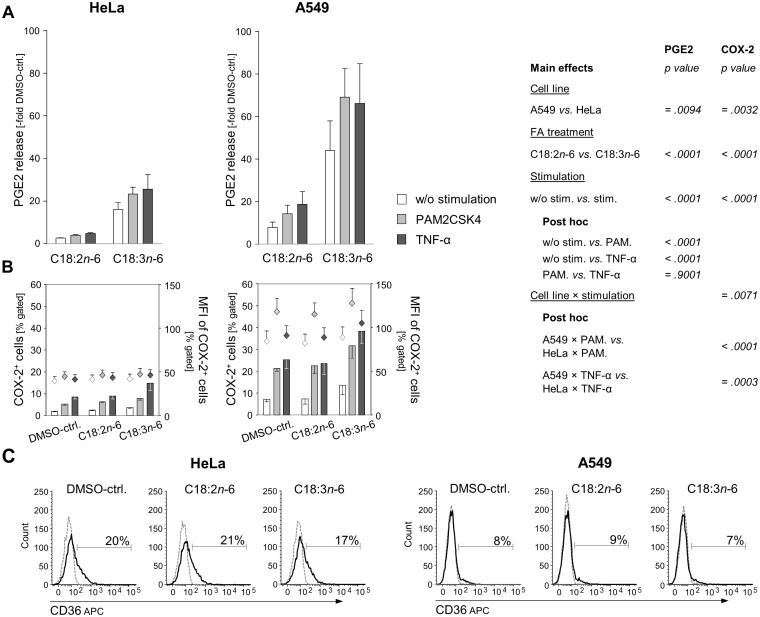
In general and in agreement with the literature, PGE2 was well detectable in both cell lines following stimulation procedures. In order to make both cell lines comparable, we related the changes in PGE2 to the respective DMSO-controls. In HeLa, C18∶2n−6 elevated the PGE2 release 3-fold compared to the DMSO-ctrl. After preincubation with C18∶2n−6, there was only a weak additional increase in PGE2 in response to PAM2CSK4 (4-fold compared to the DMSO-ctrl.) or TNF-α (5-fold compared to the DMSO-ctrl.; Fig. 5A). In contrast, when HeLa were preincubated with C18∶3n−6, PGE2 release was enhanced by the factor of 16 and augmented further following either stimulation procedure (23-fold and 26-fold, respectively, over the DMSO-ctrl.; main effect FA treatment: p<.0001). This outcome is in line with our observation that the cascade of LC-PUFA synthesis stops at the level of FADS2 in HeLa (Fig. 2) with consequences for downstream eicosanoid formation. In A549, in both C18∶2n−6 and C18∶3n−6 pretreatments, PGE2 formation increased in response to PAM2CSK4 and TNF-α, respectively. The increase in PGE2 over the DMSO-control was generally stronger in A549 than in HeLa treatments (main effect cell line: p = .0094; Fig. 5A) and fits with the expression patterns of COX-2 (main effect cell line: p = .0032; Fig. 5B).
Figure 5. Induction of PGE2 formation and expression of COX-2 and CD36 in HeLa and A549.
Cells were incubated without (DMSO-treated control) or with 33 µM C18∶2n−6 or C18∶3n−6 for 20 h. Subsequently, cells were stimulated with the TLR2 agonist PAM2CSK4 or TNF-α for further 8 h. A: PGE2 in supernatants was quantified by EIA technique and is expressed as -fold increase over the unstimulated DMSO-treated control. Means ± SEM. B: Percentage of COX-2 positive cells were determined by intracellular staining and flow cytometric analysis. C: Representative histograms of HeLa and A549 incubated without (DMSO-ctrl.) or with 33 µM C18∶2n−6 or C18∶3n−6 for 4 h and stained for CD36. Markers were set in reference to the isotype controls (dotted lines).
After 8 h stimulation, the percentage of A549 cells positive for COX-2 raised approximately 4-fold to 25% (Fig. 5B). The stimulation effect of PAM2CSK4 was comparable with that of TNF-α. In the presence of C18∶3n−6, but not C18∶2n−6, the percentage of COX-2 positive cells further increased to 30% (PAM2CSK4) and 38% (TNF-α), respectively. The mean fluorescence intensity (MFI) reflecting the COX-2 expression on a per cell basis was highest in response to PAM2CSK4. In contrast to C18∶2n−6, C18∶3n−6 exerted a stimulatory effect on COX-2 expression itself (main effect FA treatment: p<.0001). We have recently shown that the presence of its substrates sufficed to induce COX-2 [18], what is in line with previous findings [35], [36].
Aiming at elucidating how C18∶3n−6 exerts this stimulatory effect itself we measured the expression of the transmembrane scavenger receptor CD36. TLR2/6 signaling is closely linked to CD36 that acts as an accessory co-receptor, sensing and clustering TLR2 ligands, to facilitate TLR2-mediated NFκB activation [37]. But CD36 has also been reported to initiate a downstream cascade upon ligand binding TLR-independently [38]. A main function of CD36, therefore alternatively named fatty acid translocase (FAT), is the transmembrane transport of long-chain fatty acids (LCFA). The degree of unsaturation has previously been suggested to be a determinant for CD36 regulation [39]. Nevertheless, measuring the surface expression of CD36 on both A549 and HeLa revealed no regulatory effect of either C18∶2n−6 or C18∶3n−6 treatment for 4 h (Fig. 5C) or 24 h (not shown), whereby the expression level of CD36 was generally higher in HeLa than in A549 (Fig. 5C). Hence, of special interest is the finding of Chêne et al., that the number of double bonds of the LCFA the cell is exposed to appears to be critical for the mobilization of C20∶4n−6 from membrane phospholipids via phospholipase A2 [35].
In HeLa, the basal level of COX-2 was approximately half as much as in A549 (Fig. 5B), what accords to the basal levels of C20∶4n−6 (Table 3). Consequently, the percentage of COX-2 positive HeLa cells increased hardly over 10% upon either fatty acid or stimulus treatment (interaction effect cell line×stimulation: p≤.0071; Fig. 5B). It is likely that COX-1 contributed to PGE2 synthesis, because specific inhibition of COX-1 prior to stimulation abrogated the stimulation effect and PGE2 remained on the level detected in the unstimulated fatty acid treatments (not shown). Previous data suggest that inefficiency of COX-2 can partly be compensated by COX-1 activity in HeLa [32]. Nevertheless, COX-1 expression was detected in less than 2% of the cells, was not different between the two cell lines, and remained unaltered upon either fatty acid or stimulus treatment (data not shown).
Rates of chlamydial infection
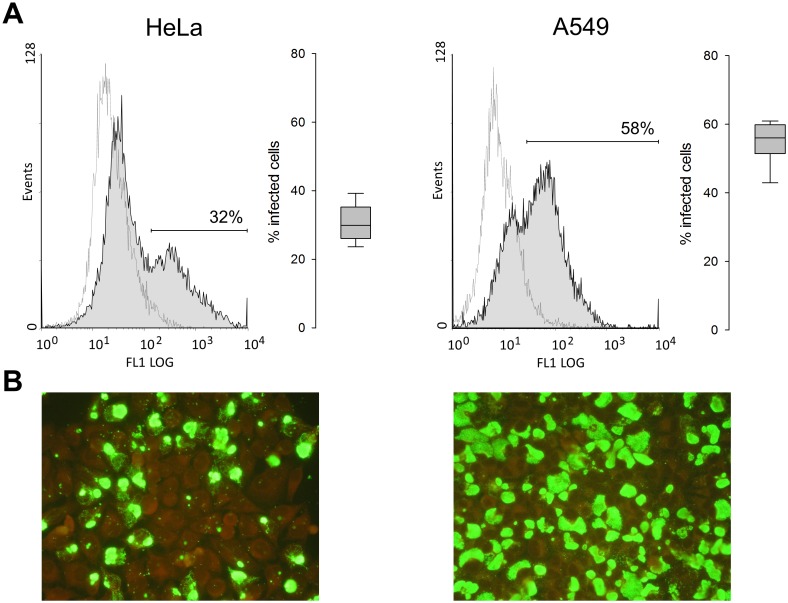
As a strategy for long-term survival within the host, Chlamydia ssp. promote target cell survival in the early phase of infection by inducing upregulation of COX-2 und subsequent PGE2 production. It was shown that blocking of COX-2 reduces the infection rate in vitro and diminishes the infectious load in vivo, likely by compromising the completion of the developmental cycle [40]. Fig. 6 shows that infection rates 48 h p.i. with strain DC15 at MOI of 4 were significantly lower in HeLa than in A549 (31±5% vs. 55±6% positive cells, as determined by flow cytometry; p = <.001), which is in accordance with weak COX-2 response following stimulation in HeLa (Fig. 5B).
Figure 6. Flow cytometric and fluorescence microscopic assessment of chlamydial infection of HeLa and A549.
Cells were inoculated with strain DC15 at MOI of 4 and analysed 48 h p.i. A: Representative histograms of fluorescence intensities of infected cells (filled peaks) and uninfected controls (dotted lines) stained with FITC-labeled mAb (BDI168) against Chlamydia-specific lipopolysaccharide. Boxplots represent five independent experiments, each done in duplicate. B: Cells were stained using the IMAGEN kit. Infection was visualized using fluorescence microscopy at a magnification of ×40.
Conclusion
In conclusion, our data show that HeLa cells differ considerably from A549 at several stages of fatty acid metabolism. The poor metabolic potential of HeLa, mainly concerning FADS2 upstream of COX-2 function, calls into question whether these cells represent a good model to unveil fatty acid or downstream eicosanoid effects in the course of intracellular bacterial infection. Moreover, our findings implicate that it is of basic interest to characterize the metabolic capacity of epithelial cell lines prior to use.
Acknowledgments
The authors thank Lars Uhlmann, Gisela Schultheiss, Doris Haase, Susanne Merkel, and Marina Giorgi from the Department of Physiology and Biochemistry of Nutrition, Federal Research Institute of Nutrition and Food, Karlsruhe, Germany, and Sabine Scharf from the Institute of Molecular Pathogenesis, Federal Research Institute for Animal Health, Jena, Germany, for their excellent technical assistance.
Funding Statement
This study was financially supported by a grant from the German Research Council DFG Ja 893/5) and a grant provided by the North Rhine-Westphalian Ministry of Environment, Conservation, Agriculture and Consumers Protection (MUNLV) within the Inter-Departmental Center of Sustainable Agriculture (USL). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Birchler T, Seibl R, Büchner K, Loeliger S, Seger R, et al. (2001) Human Toll-like receptor 2 mediates the induction of the antimicrobial peptide human beta-defensin 2 in response to bacterial lipoprotein. Eur J Immunol 31:3131–7. [DOI] [PubMed] [Google Scholar]
- 2. Fukuda EY, Lad SP, Mikolon DP, Iacobelli-Martinez M, Li E (2005) Activation of lipid metabolism contributes to interleukin-8 production during Chlamydia trachomatis infection of cervical epithelial cells. Infect Immun 73:4017–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Janeway CA Jr, Medzhitov R (2002) Innate immune recognition. Annu Rev Immunol 20:197–216. [DOI] [PubMed] [Google Scholar]
- 4. Joyee AG, Yang X (2008) Role of toll-like receptors in immune responses to chlamydial infections. Curr Pharm Des 14:593–600. [DOI] [PubMed] [Google Scholar]
- 5. Beckett EL, Phipps S, Starkey MR, Horvat JC, Beagley KW, et al. (2012) TLR2, but not TLR4, is required for effective host defence against Chlamydia respiratory tract infection in early life. PLoS ONE 7:e39460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Massari P, Toussi DN, Tifrea DF, de la Maza LM (2013) Toll-like receptor 2-dependent activity of native major outer membrane protein proteosomes of Chlamydia trachomatis. Infect Immun 81:303–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bernard JJ, Gallo RL (2010) Cyclooxygenase-2 enhances antimicrobial peptide expression and killing of Staphylococcus aureus. J Immunol 185:6535–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu F, Xu Z, Zhang R, Wu Z, Lim JH, et al. (2008) Nontypeable Haemophilus influenzae induces COX-2 and PGE2 expression in lung epithelial cells via activation of p38 MAPK and NF-kappa B. Respir Res. 9:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grammatikos SI, Subbaiah PV, Victor TA, Miller WM (1994) Diversity in the ability to elongate and desaturate essential (n-6 and n-3) fatty acids. Ann N Y Acad Sci 745:92–105. [DOI] [PubMed] [Google Scholar]
- 10. Sprecher H, Luthria DL, Mohammed BS, Baykousheva SP (1995) Reevaluation of the pathways for the biosynthesis of polyunsaturated fatty acids. J Lipid Res 36:2471–7. [PubMed] [Google Scholar]
- 11. Park WJ, Kothapalli KS, Lawrence P, Tyburczy C, Brenna JT (2009) An alternate pathway to long-chain polyunsaturates: the FADS2 gene product Delta8-desaturates 20: 2n−6 and 20: 3n−3. J Lipid Res 50:1195–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Calder PC (2009) Polyunsaturated fatty acids and inflammatory processes: New twists in an old tale. Biochimie 91:791–5. [DOI] [PubMed] [Google Scholar]
- 13. Serhan CN (2005) Novel eicosanoid and docosanoid mediators: resolvins, docosatrienes, and neuroprotectins. Curr Opin Clin Nutr Metab Care 8:115–21. [DOI] [PubMed] [Google Scholar]
- 14. Buwitt-Beckmann U, Heine H, Wiesmüller KH, Jung G, Brock R, et al. (2005) Toll-like receptor 6-independent signaling by diacylated lipopeptides. Eur J Immunol 35:282–9. [DOI] [PubMed] [Google Scholar]
- 15. Jaudszus A, Jahreis G, Schlörmann W, Fischer J, Kramer R, et al. (2012) Vaccenic acid-mediated reduction in cytokine production is independent of c9, t11-CLA in peripheral blood mononuclear cells. Biochim Biophys Acta 1821:1316–22. [DOI] [PubMed] [Google Scholar]
- 16. Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37:911–7. [DOI] [PubMed] [Google Scholar]
- 17. Jaudszus A, Möckel P, Hamelmann E, Jahreis G (2010) Trans-10, cis-12-CLA-caused lipodystrophy is associated with profound changes of fatty acid profiles of liver, white adipose tissue and erythrocytes in mice: possible link to tissue-specific alterations of fatty acid desaturation. Ann Nutr Metab 57:103–11. [DOI] [PubMed] [Google Scholar]
- 18. Jaudszus A, Gruen M, Watzl B, Ness C, Roth A, et al. (2013) Evaluation of suppressive and pro-resolving effects of EPA and DHA in human primary monocytes and T-helper cells. J Lipid Res 54:923–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–75. [PubMed] [Google Scholar]
- 20. Goellner S, Schubert E, Liebler-Tenorio E, Hotzel H, Saluz HP, et al. (2006) Transcriptional response patterns of Chlamydophila psittaci in different in vitro models of persistent infection. Infect Immun 47:4801–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grün M, Liebisch M, Sauerwein H, Jahreis G, Sachse K (2009) Flow cytometric quantification of chlamydial infection in cell culture. J Microbiol Methods 78:360–2. [DOI] [PubMed] [Google Scholar]
- 22. Guillou H, Zadravec D, Martin PG, Jacobsson A (2010) The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog Lipid Res 49:186–99. [DOI] [PubMed] [Google Scholar]
- 23. Angeletti C, de Alaniz MJ (1995) Fatty acid uptake and metabolism in HepG2 human-hepatoma cells. Mol Cell Biochem 143:99–105. [DOI] [PubMed] [Google Scholar]
- 24. Hrelia S, Lopez Jimenez JA, Bordoni A, Nvarro SZ, Horrobin DF, et al. (1995) Essential fatty acid metabolism in cultured rat cardiomyocytes in response to either N-6 or N-3 fatty acid supplementation. Biochem Biophys Res Commun 216:11–9. [DOI] [PubMed] [Google Scholar]
- 26. Park WJ, Kothapalli KS, Lawrence P, Brenna JT (2011) FADS2 function loss at the cancer hotspot 11q13 locus diverts lipid signaling precursor synthesis to unusual eicosanoid fatty acids. PLoS ONE 6:e28186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schaeffer L, Gohlke H, Muller M, Heid IM, Palmer LJ, et al. (2006) Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet 15:1745–56. [DOI] [PubMed] [Google Scholar]
- 28. Bokor S, Dumont J, Spinneker A, Gonzalez-Gross M, Nova E, et al. (2010) Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J Lipid Res 51:2325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nwankwo JO, Spector AA, Domann FF (2003) A nucleotide insertion in the transcriptional regulatory region of FADS2 gives rise to human delta-6-desaturase deficiency. J Lipid Res 44:2311–9. [DOI] [PubMed] [Google Scholar]
- 30. Portolesi R, Powell BC, Gibson RA (2008) Delta6 desaturase mRNA abundance in HepG2 is suppressed by unsaturated fatty acids. Lipids 43:91–5. [DOI] [PubMed] [Google Scholar]
- 31. Lattka E, Eggers S, Moeller G, Heim K, Weber M, et al. (2010) A common FADS2 promoter polymorphism increases promoter activity and facilitates binding of transcription factor ELK1. J Lipid Res 51:182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Deva R, Shankaranarayanan P, Ciccoli R, Nigam S (2003) Candida albicans induces selectively transcriptional activation of cyclooxygenase-2 in HeLa cells: pivotal roles of Toll-like receptors, p38 mitogen-activated protein kinase, and NF-kappa B. J Immunol. 171:3047–55. [DOI] [PubMed] [Google Scholar]
- 33. Jakobsson P, Thorén S, Morgenstern R, Samuelsson B (1999) Identification of human prostaglandin E synthase: A microsomal, glutathione-dependent, inducible enzyme, constituting a potential novel drug target. Proc Natl Acad Sci USA 96:7220–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shimasaki S, Koga T, Shuto T, Suico MA, Sato T, et al. (2010) Endoplasmatic reticulum stress increases the expression and function of toll-like receptor-2 in epithelial cells. Biochem Biophys Res Commun 402:235–40. [DOI] [PubMed] [Google Scholar]
- 35. Chêne G, Dubourdeau M, Balard P, Escoubet-Lozach L, Orphila C, et al. (2007) n-3 and n-6 polyunsaturated fatty acids induce the expression of COX-2 via PPARgamma activation in human keratinocyte HaCaT cells. Biochim Biophys Acta 1771:576–89. [DOI] [PubMed] [Google Scholar]
- 36. Derecka K, Sheldrick EL, Wathes DC, Abayasekara DR, Flint AP (2008) A PPAR-independent pathway to PUFA-induced COX-2 expression. Mol Cell Endocrinol 287:65–71. [DOI] [PubMed] [Google Scholar]
- 37. Triantafilou M, Gamper FG, Haston RM, Mouratis MA, Morath S, et al. (2006) Membrane sorting of Toll-like receptor (TLR)-2/6 and TLR2/1 heterodimers at the cell surface determines heterotypic associations with CD36 and intracellular targeting. J Biol Chem 281:31002–11. [DOI] [PubMed] [Google Scholar]
- 38. Baranova IN, Kurlander R, Bocharov AV, Vishnyakova TG, Chen Z, et al. (2008) Role of human CD36 in bacterial recognition, phagocytosis, and pathogen-induced JNK-mediated signaling. J Immunol 181:7147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xie P, Zhang AT, Wang C, Azzam MM, Zou XT (2012) Molecular cloning, characterization, and expression analysis of fatty acid translocase (FAT/CD36) in the pigeon (Columba livia domestica). Poult Sci 91:1670–9. [DOI] [PubMed] [Google Scholar]
- 40. Liu W, Dubinett S, Patterson SL, Kelly KA (2006) COX-2 inhibition affects growth rate of Chlamydia muridarum within epithelial cells. Microbes Infect 8:478–86. [DOI] [PubMed] [Google Scholar]