Abstract
Visible/near-infrared (Vis/NIR) hyperspectral imaging was employed to determine the spatial distribution of total nitrogen in pepper plant. Hyperspectral images of samples (leaves, stems, and roots of pepper plants) were acquired and their total nitrogen contents (TNCs) were measured using Dumas combustion method. Mean spectra of all samples were extracted from regions of interest (ROIs) in hyperspectral images. Random frog (RF) algorithm was implemented to select important wavelengths which carried effective information for predicting the TNCs in leaf, stem, root, and whole-plant (leaf-stem-root), respectively. Based on full spectra and the selected important wavelengths, the quantitative relationships between spectral data and the corresponding TNCs in organs (leaf, stem, and root) and whole-plant (leaf-stem-root) were separately developed using partial least-squares regression (PLSR). As a result, the PLSR model built by the important wavelengths for predicting TNCs in whole-plant (leaf-stem-root) offered a promising result of correlation coefficient (R) for prediction (RP = 0.876) and root mean square error (RMSE) for prediction (RMSEP = 0.426%). Finally, the TNC of each pixel within ROI of the sample was estimated to generate the spatial distribution map of TNC in pepper plant. The achievements of the research indicated that hyperspectral imaging is promising and presents a powerful potential to determine nitrogen contents spatial distribution in pepper plant.
Introduction
Pepper (Capsicum annuum L) is one of the popular international spices in cooking [1], [2]. At present, world production of pepper is about 19 million tons fresh fruit from 1.5 million ha [3]. Nitrogen (N) is an essential element of vegetative growth, flowering, and fruit bearing of pepper plants [4]–[8], as well as in all metabolic processes of cellular structure and genetic coding [9]. In order to strengthen and stabilize plant growth and yield production, farmers most of the case will over-apply N fertilizer to ensure enough N for crop requirements. The excessive N fertilizer could result in a series of problems, such as lodging, delayed maturity, diseases, and weed [10], [11]. Therefore, it is crucial to investigate the N status and obtain the real-time spatial distribution information of N content in crops at different growth stages, so as to improve field management efficiency and economic benefit from agricultural production, as well as contribute to sustainable agriculture [12], [13].
Conventionally, measurement of total nitrogen content (TNC) in crops based on chemical analysis is a destructive measurement, which is high-cost and time-consuming [14]. Researchers demonstrated that TNC in plant was directly related to the formation of chlorophyll [15], [16]. Therefore, chlorophyll content in leaves can theoretically be considered as an indicator of the N status [17], [18]. The soil and plant analyzer development (SPAD) reading, an indirect value representing chlorophyll content, is measured by the SPAD meter with a “point-based” way [19]. This method merely provides information on chlorophyll content in leaves at one point, which is not sufficient to obtain the accurate chlorophyll concentration of the whole leaf. Moreover, SPAD meter cannot provide the chlorophyll information of other parts of the plant (e.g. buds, stems). In recent years, spectroscopic technology, particularly visible and near-infrared (Vis/NIR) spectroscopy, has been used for detection of the agricultural and sideline products. For plants, several researchers [20]–[24] employed spectroscopy technique to study and diagnose the nutrient status, N contents, and yield of pepper. Unfortunately, spectroscopic techniques fail to provide information on spatial distribution of quality parameters, which is essential to the analysis of products’ features [25]. Remote sensing technique can provide chlorophyll information of a large number of plants in an area with satellite or airplane sensors scanning. Airborne visible/infrared imaging spectrometer (AVIRIS) in NASA was used to determine forest canopy lignin, nitrogen, and ecosystem [26]. However, this approach is not suitable for providing chlorophyll information about individual plants [27].
Hyperspectral imaging combines conventional spectroscopy with imaging techniques to acquire both spectral and spatial information simultaneously for detecting physical, chemical, and biological attributes of the samples [28]–[30]. Relying on this capability, hyperspectral imaging has witnessed tremendous growth and been widely applied in agriculture [31]. Examples of nitrogen analyzing in plant included: mapping of foliar nitrogen of mangroves and explanation of spatial nitrogen variation in relation to local environmental variables [32], determination of nitrogen (N), phosphorus (P), and potassium (K) content and visualization of chemical distribution in oilseed rape leaves [33], estimation of leaf nitrogen accumulation in wheat [34], diagnostics of N deficiency by virtue of chlorophyll content in cucumber leaf [19], assessment of the leaf N content in wheat [35], estimation of canopy N concentration in temperate forest [36]. A majority of studies have investigated the chemical components in certain organ tissue (e.g. leaves) of crops. However, few studies on TNCs spatial distribution of plants using hyperspectral imaging have been reported.
In the current study, hyperspectral imaging technique was employed to map the spatial distribution of total nitrogen in pepper plant. The specific objectives were as follows: (1) to acquire hyperspectral images and measure TNCs of samples (leaves, stems, and roots) using Dumas combustion method; (2) to extract the spectral data and employ Random frog (RF) to select important wavelengths; (3) to build multivariate calibration models for predicting TNCs in organs (leaf, stem, and root) and whole-plant (leaf-stem-root) by partial least-squares regression (PLSR) based on full spectra and the selected important wavelengths; (4) to apply the optimal PLSR model to predict TNC of each pixel in samples and generate spatial distribution of TNCs in whole pepper plant.
Materials and Methods
Samples preparation
A total of 40 pepper plants (Capsicum frutescens L. conoides (Mill.) Bailey) were taken for this research. According to the crop management of National Farmers Information Service (NAFIS) [37], pepper plants were planted in the same environmental conditions and field management in greenhouses at Zhejiang University, Hangzhou (120°09′E, 30°14′N), China.
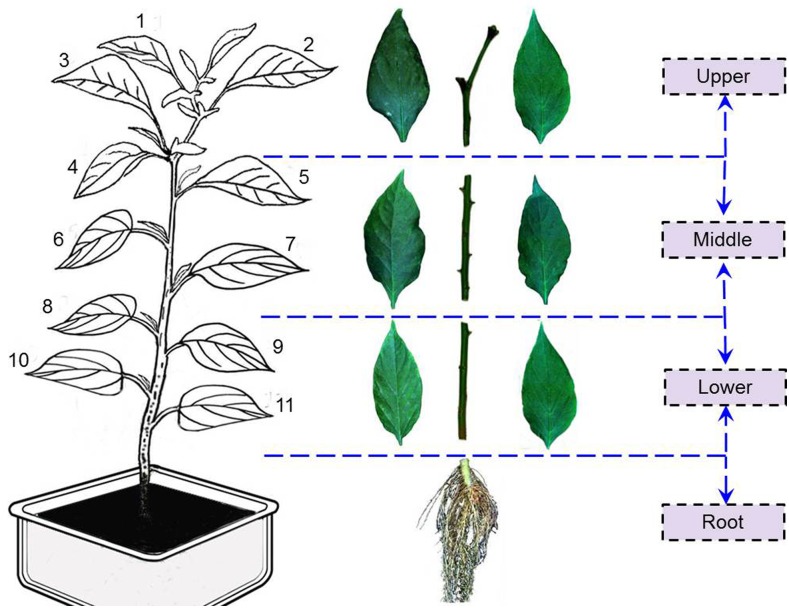
After transplanting about 6 weeks, all pepper plants began to send forth flower buds during early blooming stage. According to the growth status of the pepper plants, several leaves (few yellow leaves in lower position and some small fresh leaves in upper position) were excluded, and then the remaining 11 true leaves with similar size were numbered from upper to lower (Fig. 1). In each pepper plant, 2 leaves (petioles were removed) were randomly sampled from the upper (1st–4th), middle (5th–8th), and lower (9th–11th) positions [38]. Each 2 leaves in the sampled position were viewed as a sample for subsequent data analysis, because the dry matter of one leaf failed to satisfy the demands of total nitrogen measurement. Meanwhile, the whole stem was divided into 3 parts according to positions (upper, middle, and lower) of petiole node. Lastly, root of each pepper plant was dug out from the soil and impurities were cleaned up. The position of sampled leaf, stem, and root is marked in Fig. 1. According to the preceding process, a total of 280 samples containing 120 leaf samples, 120 stems, and 40 roots were collected from 40 pepper plants. These samples were then used for hyperspectral data acquisition and TNCs measurement.
Figure 1. Positions of sampled leaf, stem, and root samples in one exemplary pepper plant.
Acquisition and calibration of hyperspectral images
Three-dimensional hyperspectral data of all samples were acquired by a laboratorial hyperspectral imaging device with the reflectance mode. The hyperspectral imaging device consists of: a mobile platform operated by a stepper motor (IRCP0076, Isuzu Optics Crop, Taiwan, China); an assembled illumination source with two 150-W quartz tungsten halogen lamps (Fiber-Lite DC950 Illuminator, Dolan Jenner Industries Inc., USA); an imaging spectrograph (ImSpectorV10E, Spectral Imaging Ltd., Finland) covering the spectral range of 380–1,030 nm with 512 wavebands; a CCD camera (C8484-05, Hamamatsu city, Japan) coupled with a zoom lens (OLES23, Specim, Spectral Imaging Ltd., Oulu, Finland); and a computer with the spectral imaging system V10E software (Isuzu Optics Corp, Taiwan, China), which is used to set and adjust the parameters of the device, including exposure time, motor speed, imaging acquisition, wavelength range, image correction. In all, the whole system (except the computer) was assembled in a dark chamber to minimize the effects of ambient light during the sample scanning.
As Yu et al. [39] described, the hyperspectral imaging device was calibrated before samples were scanned. Parameters of the device for image acquisition are summarized in Table 1. Subsequently, a hyperspectral image which was named “hypercube” with dimension of (x, y, λ) was generated with line scanning [39]. In detail, a hypercube contained n-pixels in y-direction (depending on the size of the sample), 672 pixels in x-direction, and 512 wavebands in λ-direction. Then, the raw hyperspectral images were calibrated with white and dark references using the following (1) and this process was implemented by the hyperspectral imaging analyzer software.
| (1) |
where
Table 1. Parameters of hyperspectral imaging device for acquisition of images.
| Parameters | Corresponding values |
| Spectral range | 380–1,030 nm |
| Spectral resolution | 2.8 nm |
| Moving speed of the mobile platform | 2.1 mm/s |
| Spatial dimension of images | 672×512 (spatial × spectral) pixels |
| Exposure time | 0.008 s |
| The distance from lens to sample | 295 mm |
Icalibrated is the calibrated hyperspectral image of the sample;
Iraw is the raw hyperspectral image of the sample;
Iwhite is the white reference image with 99% reflectance acquired from a white reference ceramic tile;
Idark is the dark reference image with 0% reflectance obtained by the camera lens thoroughly covered with its opaque cap.
Measurement of TNCs using Dumas combustion method
The referenced method for TNC measurement was Dumas combustion using Nitrogen Analyzer Rapid N cube (Elementar Analysensysteme GmbH, Hanau, Germany). After hyperspectral images acquisition, all samples were dried completely to the constant weight [40]. Then, the samples were grinded by a Tissuelyser-48 (Jingxin Experimental Technology, Shanghai, China) operating in 70 Hz, 150 s. 50±1 mg dry matter of each sample was used to determine the TNC. After a series of processing (combustion, reduction, purification, detection, etc.), TNCs (%) of samples were obtained through the Rapid N software. All the measurements were carried out in a room at an approximate constant temperature of 25°C and a relative humidity of 20–30%.
Hyperspectral data processing and analyzing
Hyperspectral data extraction
By implementing the mask function in the ENVI software, the target region of each sample was separated from hyperspectral image. The separated region was identified as the region of interest (ROI) of the corresponding sample. The spectra of each pixel in ROIs were then averaged and the average spectrum was considered as the spectrum of a sample. The spectra of other samples were obtained in the same way as mentioned above. Because of the low signal-to-noise ratio (SNR), the reflectance in two regions (380–420 nm and 1,000–1,030 nm) was rather low. Therefore, hyperspectral data were resized to the spectral range of 420–1,000 nm with 460 wavebands.
Variable selection
Selection of important wavelength is of critical significance for removing the redundant information from high-dimensional data and optimizing calibration models for producing reliable results [41], [42]. Thus, identification of important variables carrying the most valuable and authentic information is a challenging task in the current hyperspectral data analysis [43].
In the present study, Random frog (RF) methodology was carried out to select important wavelengths. RF is a novel and efficient technique for variable selection, which borrows the framework of reversible jump Markov Chain Monte Carlo (RJMCMC) methods [44], [45]. It is employed to perform feature extraction for selecting a series of variables which describe the correlation between the predictor variables and the response variables [46]. In interior of RF algorithm, partial least squares regression (PLSR) is viewed as a modeling method. X (n × p) stands for the spectral matrix consisting of n samples in rows and p variables in columns. And Y (n×1) denotes the property of interest.
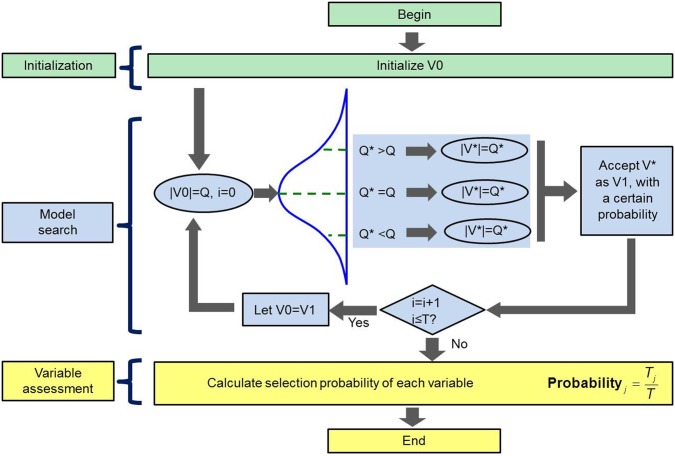
Before running RF algorithm, five parameters (T, Q, θ, ω, and η, details were listed in Table 2) should be assigned with proper values. As shown in Fig. 2, random frog works in three steps [47]:
Table 2. Parameters for running random frog.
| Parameters | Function | Value |
| T | number of iterations | 10,000 |
| Q | number of variables in the initialized variable set | 50 |
| θ | control variance of a normal distribution | 0.3 |
| ω | a coefficient explained in step 2 | 3 |
| η | represent the upper bound of the probability | 0.1 |
Figure 2. The flowchart of the Random frog algorithm from Li et al. [45].
A variable subset V0 consisting of Q variables is initialized randomly;
A candidate variable subset V* is proposed based on Q* variables, accept V* with a certain probability as V1 and replace V0 using V1;
Selection probability of each variable after N iterations is computed. The details of RF algorithm could be found in literature [44], [45].
As Monte Carlo strategy is embedded in RF algorithm, the selection probability of variables is unable to be reproduced exactly [44], [45]. Generally, RF is implemented a couple of times (depending on the data) to minimize the influence of this random factor [44], [45].
Multivariate data analyses
Partial least-squares regression (PLSR), one of the reliable analytical tools for modeling [48]–[51], has a wide application in multivariable data analysis and regression. It possesses an advantage to solve the situation when the number of variables is larger than that of samples, and when there is colinearity among variables [52]. The PLSR model employs several latent variables (LVs) instead of real variables and develops the calibration model applying leave-one-out cross-validation (LOOCV) method [53], [54]. The LOOCV method is applied to validate the performance and evaluate over-fitting of the calibration models. In this study, the quantitative relationships between spectral reflectance values and TNCs of samples were established by employing PLSR.
Evaluation of models
The performances of models were evaluated by correlation coefficient (R) and root mean square error (RMSE) across the calibration (RC, RMSEC), cross-validation (RCV, RMSECV), and prediction (RP, RMSEP) sets of samples. Generally, an optimal model should provide high R values, low RMSE values, and a small difference between calibration and prediction [40].
Chemical imaging
Chemical imaging is a technique for building visual color images to display the spatial distribution of chemical components in heterogeneity [55]. The chemical value of each pixel can be predicted by inputting its corresponding spectral reflectance values into the established quantitative model [19]. Spatial distribution of chemical components could be generated with the established quantitative model combined with image processing. To observe the variance of TNCs in whole pepper plant, distribution maps of TNCs were required. In particular, an optimal model built using the mean spectra of important wavelengths was applied to predict the TNC in each pixel. Subsequently, the spatial position of each pixel along with its TNC was used to form the spatial distribution maps [52].
Software tools
The measured TNCs statistics and analysis of variance (ANOVA) was finished using Microsoft Office Excel 2010 and IBM SPSS Statistics (version 20.0, IBM Corporation, Armonk, New York, USA). Hyperspectral data was extracted with ENVI software (Version 4.6, ITT Visual Information Solutions, Boulder, USA). The processes of spectral data analyses were carried out by “The Unscrambler X10.1” (CAMO PROCESS AS, Oslo, Norway). In addition, Random frog (version 2.0) toolbox and the developed program for building TNC spatial distribution maps were worked in MATLAB 7.8 (R2009a) software (The MathWorks, Inc., Natick, MA, USA). OriginPro 8.0 SR0 (Origin Lab Corporation, Northampton, MA, USA) software was employed to design the spectral curves. All processes were run on a PC (CPU: Intel Core i3-3220 @3.30 GHz, RAM: 4.00 GB) operating with Windows 7.
Results and Discussion
Statistics of the TNCs measured by Dumas combustion (TNCs-DC)
The statistical results of TNCs in pepper plant samples measured by Dumas combustion are presented in Table 3. The results were similar to previous study of Zhang et al. [33], Nelson et al. [56], and Stevenson [57]. The mean TNCs-DC in leaves (3.295%), roots (1.186%), and stems (0.992%) displayed a descending tendency. Meanwhile, the TNCs-DC in groups of leaf and stem conveyed an ascending trend in lower/middle/upper positions, ranging from 2.703% to 3.129%, then to 4.053% in leaves, and from 0.736% to 0.849%, then to 1.390% in stems. Also in Table 3, results of analysis of variance (ANOVA) analyses of the TNCs-DC in different samples revealed an apparent difference among leaf/stem/root groups, as well as the leaf and stem groups in upper/middle/lower positions.
Table 3. Statistic results of all samples’ TNCs-DC.
| Samples | Positions | N.[1] | Mean (%) ± S. D.[2] |
| Leaf | Upper | 40 | 4.053±0.498a |
| Middle | 40 | 3.129±0.321b | |
| Lower | 40 | 2.703±0.325c | |
| Stem | Upper | 40 | 1.390±0.366a |
| Middle | 40 | 0.849±0.175b | |
| Lower | 40 | 0.736±0.102c | |
| Leaf | All | 120 | 3.295±0.686a |
| Stem | All | 120 | 0.992±0.374c |
| Root | All | 40 | 1.186±0.143b |
| Leaf-root-stem | All | 280 | 2.007±1.231 |
Note: Different letters (a, b, c) in the same column indicate statistical significance at the 5% level by Tamhane’s T2 test.
N.: Number of samples;
S. D.: Standard deviation of the group.
One-way analysis of variance (ANOVA) was used to generate these results, which were obtained using IBM SPSS Statistics (Version 20.0, IBM Corporation, Armonk, New York, USA). The results of TNCs-DC exhibited significant differences between groups (leaf, stem, and root) and within groups (leaves/stems in different positions).
In this research, the SPXY (sample set partitioning based on joint x–y distances) method proposed by Galvao et al. [58] was implemented to divide all samples into calibration sets with 210 samples (90 leaves, 90 stems, and 30 roots) and prediction sets with 70 samples (30 leaves, 30 stems, and 10 roots) for subsequent hyperspectral TNC analyses. Table 4 shows a summary of statistical analysis on the TNCs-DC of all samples in calibration and prediction sets. There was an evident variation in TNCs-DC in the calibration set, ranging from 2.264% to 4.871% for leaf, 0.847% to 1.470% for root, 0.556% to 2.135% for stem, and 0.556% to 4.871% for whole-plant samples. Samples with a wide compositional distribution of the TNCs-DC were collected, which was of great importance to build stable calibration models.
Table 4. Summary of statistical analyses of TNCs-DC in calibration and prediction sets.
| Samples | Calibration set | Prediction set | ||||||
| N.[1] | Max.[3] (%) | Min.[4] (%) | Mean (%) ± S.D.[2] | N.[1] | Max.[3] (%) | Min.[4] (%) | Mean (%) ± S.D.[2] | |
| Leaf | 90 | 4.871 | 2.264 | 3.357±0.701 | 30 | 4.731 | 2.398 | 3.108±0.612 |
| Root | 30 | 1.470 | 0.847 | 1.183±0.156 | 10 | 1.301 | 1.035 | 1.194±0.103 |
| Stem | 90 | 2.135 | 0.556 | 1.040±0.403 | 30 | 1.636 | 0.599 | 0.849±0.218 |
| All | 210 | 4.871 | 0.556 | 2.114±1.292 | 70 | 4.731 | 0.599 | 1.927±1.218 |
Note:
N.: Number of samples;
S. D.: Standard deviation of the group;
Max.: Maximum;
Min.: Minimum;
Calibrations set with wide variation range of TNCs-DC could benefit for building robust models. Cross-validation set had the same results with the calibration set, which were not motioned in this table. In this study, leave-one-out cross-validation (one sample randomly chosen from calibration set was retained at a time and the rest of samples in calibration set were used to build the model) was used to verify the reproducibility and robustness of models.
Overview of spectral features
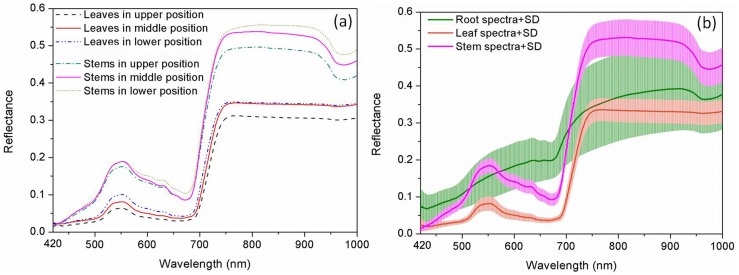
Fig. 3 demonstrates the mean spectral reflectance curves of samples covering the range of 420–1,000 nm. In Fig. 3(a), there were similar profiles of curves of leaves and stems in upper/middle/lower positions, however, the discrepancy of two groups only appeared on the different spectral reflectance values. Moreover, spectral reflectance of leaves and stems in lower/middle/upper positions displayed an ascending gradient. In detail, molecular vibration around 550 nm is caused by chlorophyll, which related to N concentrations [20], [32]. Small valley of the curves around 960 nm is attributed to the O-H second overtone, which is related to water in plants [19], [39], [41], [55], [59]. The region of 680–750 nm with high reflectance values generally refers to the “red edge” [60].
Figure 3. Spectral curves of all pepper plant samples covering the range of 420–1,000 nm.
(a) mean spectral reflectance curves of all leaves and stems in upper, middle, and lower positions; (b) mean spectral reflectance and standard deviation (SD) of leaves, stems, and roots across all samples.
The mean spectral reflectance and standard deviation (SD) of leaf/stem/root groups are illustrated in Fig. 3(b). The mean reflectance and SD of the stem was consistently higher than that of the leaf across the whole tested range. There were several intersections of means for each three groups (roots, stems, and leaves), which was attributed to their different tissue structures. The spectra of organs (leaf, stem, and root) resulted from a complex combination of scattering processes and overlapping absorptions arising from water and biochemical components [61]. In addition, the trend of the root curve was relatively flat and different from those of leaf and stem, both of which were similar to the study reported by San Juan Martínez [21]. Actually, some soil particles adhered to the surface of root, resulting in spectrum similar to the soil where the plant grew [21], [62].
Selection of important wavelengths using Random frog algorithm
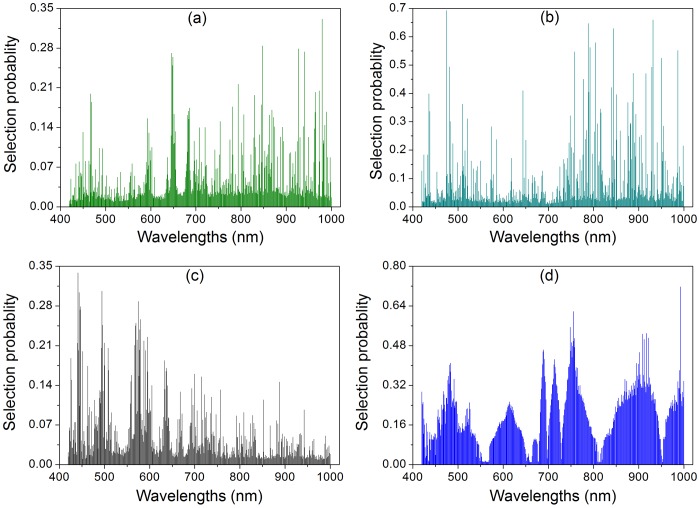
In this research, RF was executed 50 times and the average value over these 50 runs was taken as the criterion for estimating the importance of each variable. The selection probability (SP) of wavelengths is shown in Fig. 4. From these SP curves, it could be found that a small number of wavelengths exhibited an extremely high SP, whereas the SP of most wavelengths was relatively low. This indicated that the majority of wavelengths provided a weak relevance to the TNCs predicted by hyperspectral imaging (TNCs-HSI). All the variables were ranked in descending order, according to the SP. To investigate the influence of the number of wavelengths in the model, as well as to seek an optimal number of variables [45], different wavelengths for benefiting the TNCs-HSI in organs (leaves, stems, and roots) and the whole-plant (leaf-stem-root) are collected in Table 5.
Figure 4. Selection probability (SP) of each wavelength averaged over 50 runs of Random frog for TNCs-HSI prediction of (a) leaves, (b) stems, (c) roots, and (d) whole-plant samples (leaf-stem-root).
Table 5. Important wavelengths for predicting TNCs in leaves, stems, roots, and whole-plant (leaf-stem-root) based on Random frog (RF).
| Samples | Important wavelength (nm) |
| Leaves | 980, 848, 928, 941, 647, 649, 648, 794, 973, 965 |
| Stems | 475, 932, 788, 844, 804, 792, 987, 759, 951 |
| Roots | 445, 495, 444, 575, 448, 446, 580 |
| Whole-plant | 992, 756, 749, 918, 909, 921, 759, 912 |
The selected important wavelengths of leaves were mainly scattered in three regions (around 650, 790, and 970 nm), in agreement with previous researches [22], [23], [33], [31]. The important wavelengths for TNCs-HSI in stems were principally clustered in 790 and 980 nm. However, the important wavelengths of root were largely distributed around the regions of 445 and 580 nm, which were similarly related to the wavelengths characteristic for soil [21], [63]. For whole-plant (leaf-stem-root), the selected important wavelengths at 756, 749, and 759 nm were assigned to the third overtones of N-H stretching around 785 nm [31], [64], [65]. The selected important wavelength at 992 nm was close to the second overtones O-H stretching near 970 nm [39], and the remaining selected important wavelengths at 918, 909, 921, and 912 nm were ascribed to the third overtones of free monomer C-H stretching (934 nm) in carboxylic acid [31].
Establishment of the PLSR models
The PLSR predictive models of the TNCs-HSI in organs (leaf, stem, and root) and whole-plant (leaf-stem-root) were separately established based on the full spectra and the selected important wavelengths. The results of these models are enumerated in Table 6. Organ prediction methods provided more accurate TNC estimates, compared with the whole-plant model. Meanwhile, PLSR models based on both full spectra and the selected important wavelengths showed indeed only minute changes between the calibration and the respective cross-validation values. However, organ prediction methods needed three models to accrue TNCs-HSI in different organs, which enhanced the complexity of determining the TNCs-HSI of whole-plant. Furthermore, the pepper plant should be taken as a whole experimental subject. Hence, the whole-plant models with more samples (the sum of roots, stems, and leaves) were introduced to obtain TNCs-HSI in pepper plant and obtain good performance.
Table 6. Results of PLSR models for TNCs-HSI analysis based on full spectra (F-PLSR) and the selected important wavelengths (RF-PLSR).
| Samples | Models | N.[1] | LVs | Calibration | Cross-Validation | Prediction | |||
| RC | RMSEC (%) | RCV | RMSECV (%) | RP | RMSEP (%) | ||||
| Leaf | F-PLSR[5] | 460 | 17 | 0.974 | 0.156 | 0.924 | 0.269 | 0.934 | 0.223 |
| RF-PLSR[6] | 10 | 3 | 0.817 | 0.401 | 0.798 | 0.420 | 0.828 | 0.356 | |
| Stem | F-PLSR | 460 | 16 | 0.987 | 0.063 | 0.917 | 0.168 | 0.930 | 0.084 |
| RF-PLSR | 9 | 5 | 0.820 | 0.229 | 0.773 | 0.254 | 0.724 | 0.157 | |
| Root | F-PLSR | 460 | 11 | 0.992 | 0.020 | 0.931 | 0.064 | 0.915 | 0.045 |
| RF-PLSR | 7 | 7 | 0.773 | 0.097 | 0.592 | 0.130 | 0.797 | 0.068 | |
| Whole-plant | F-PLSR | 460 | 6 | 0.904 | 0.388 | 0.893 | 0.409 | 0.908 | 0.351 |
| RF-PLSR | 8 | 4 | 0.878 | 0.451 | 0.874 | 0.468 | 0.876 | 0.426 | |
Note:
N.: Number of wavelengths used for analysis;
F-PLSR models meant the PLSR models established by using full spectra;
RF-PLSR models represented the PLSR models built by important wavelengths selected by RF algorithm. LVs, RC, RMSEC, RCV, RMSECV, RP, and RMSEP could be found in the text.
From the results of whole-plant models (Table 6), the PLSR model based on full spectra (F-PLSR) with 6 latent variables (LVs) provided relatively robust results to benefit the TNCs-HIS, compared with PLSR model built using important wavelengths selected by RF (RF-PLSR). When switching from F-PLSR model (406 variables) to RF-PLSR model (8 variables), 98.26% of variables were eliminated. Meanwhile, the correlation coefficients of calibration, cross-validation, and prediction (RC, RCV, and RP) only showed a slight reduction of 0.026, 0.019, and 0.032, while their respective errors RMSEC, RMSECV, and RMSEP exhibited small changes of 0.063, 0.059, and 0.075. It meant that the predictive power of the RF-PLSR model would slightly drop for the whole-plant TNCs-HSI prediction in comparison with the F-PLSR model. However, the F-PLSR model with 460 wavelengths (varieties) would bring about a time-consuming data process [51], [53], and failed to offer a simple and effective linear for predicting the TNCs-HSI. In addition, several models were less over-fitted, which might be caused by the improper values of LVs. The above results revealed that the RF-PLSR model of whole-plant proved best-suited for gaining the TNCs-HSI in pepper plants.
The promising quantitative relationship between the spectral reflectance values and the TNCs-DC of samples was established through the RF-PLSR model. A multi-linear function for the TNCs-HSI prediction of the whole-plant (leaf-stem-root) was obtained:
| (2) |
where
λi nm is the spectral reflectance value at the wavelength of i nm;
YTNC is the TNCs-HSI estimate of whole pepper plant.
Spatial distribution maps of TNCs in pepper plant
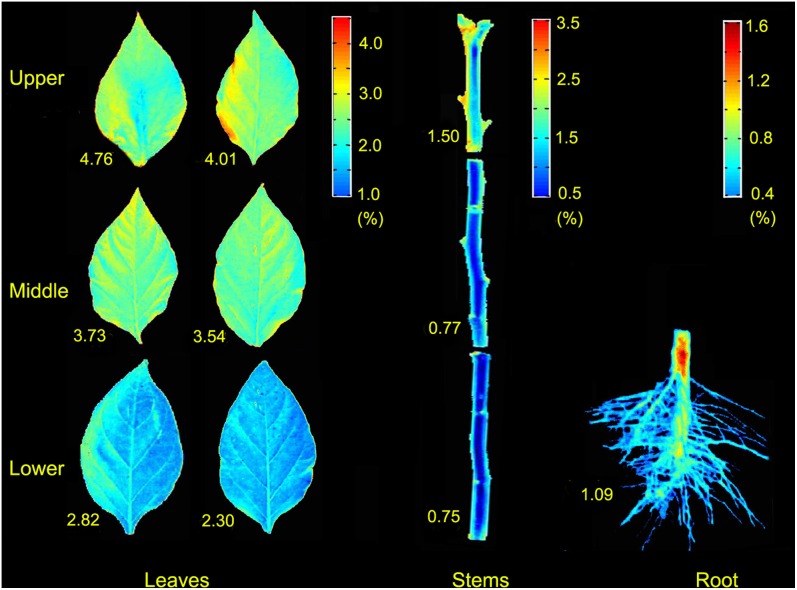
The linear function (2) obtained from the RF-PLSR model was employed to predict the TNC of each pixel within the leaf, stem, and root images. Pixels with similar spectral patterns in raw hyperspectral images would produce similar predicted values of the TNCs-HSI, and then would appear in similar colors in the resulting chemical images [55]. As a result, the spatial distribution maps of the TNCs-HSI in 10 samples (6 leaves and 3 stems in upper, middle, and lower positions and a root) of pepper plant were generated in Fig. 5. The TNCs-DC covered a broad range from 0.75% (stem in lower) to 4.76% (leaf in upper). Pixels providing similar spectral information in original hyperspectral images would result in similar results of the TNCs-HSI [51], [55], thus producing similar colors in the resultant chemical images. Indeed some samples (stems in middle/lower and root) with the relatively low TNCs-DC might show similar color, which was hard to distinguish based on the pixels of high or low TNCs. Hence, in the predictive map, three color scales described the ranges of the TNCs-HSI in each spot of the leaves, stems, and root samples, respectively. Compared with the images of original samples, the difference of the TNCs-HSI coloring within a sample could be easily identified by the naked eyes.
Figure 5. Spatial distribution maps of TNCs-HSI in samples of an exemplary tested pepper plant included six leaves, three stems, and one root, respectively.
TNCs-HSI of samples in hyperspectral images were computed based on linear function (2) and TNCs-HSI distribution was achieved in MATLAB software. The numbers accompanying each sample map denote the respective TNC-DC value.
As shown in Fig. 5, three linear color scales in different colors from red to blue represented the different TNCs-HSI from high to low, respectively. The 10 resulting images of samples revealed the changing spatial tendency of the TNCs-HSI. Nitrogen status showed a decreasing trend from upper to lower positions in leaves and stems, which was in alignment with the measured nitrogen variation shown in Table 3 and the respective TNC-DC values of those samples. This phenomenon could be explained as follows: N is a removable and active element and exists in the form of organic matter in a living plant. It can be reabsorbed from older leaves to young leaves [19]. Meanwhile, N and other nutrients in plant are transported from roots to growth center (fresh leaves) with the xylem sap in stems under the transpiration pull [66], [67], resulting in the high TNCs in fresh leaves. In addition, TNC distribution is related to seasons, plant age, and photon flux density of leaves in different positions [68].
According to plant physiology [69]–[72], N element is absorbed on the root surface in the form of ammonium (NH4+) or nitrate (NO3−) by the movement of water in the soil [73]. Then, N ions enter the conduit of xylem in the root tissues through the apoplast pathway. The sap ascending in the xylem of nitrogen-fixing legumes carries nitrogen compounds originating from inorganic soil nitrogen (mainly NO3− absorbed by the roots). Early growth of the leaf depends largely on phloem for intake of nitrogen and other nutrients that transported with xylem. At last, nutriments are transported from roots to various organs and tissues through the xylem sap under the physiological activities.
It is worth noticing that the maps of the upper and middle leaves displayed a relatively even color within the mesophyll part of individual images. In the images of the lower leaves (Fig. 5), pixels in the midrib part with light blue implied the relatively high TNCs-HSI. For stems, petiole nodes with orange showed the relatively low TNCs-HSI. In the map of root, hair parts colored blue revealed the extremely low TNCs-HSI, as well as in stems and leaves.
Furthermore, colors in the contour of stem samples were distinctly different in other regions, which might be caused by its tridimensional structure. Samples with the planar or tridimensional structure might result in different spectral characteristics [74]. This issue would be deliberated in further work.
Conclusions
In this research, RF method was employed to select eight important wavelengths (992, 756, 749, 918, 909, 921, 758, and 912 nm) for successful prediction of whole-plant (leaf-stem-root) total nitrogen contents. After that, PLSR was employed to build the quantitative relationship between the spectral reflectance and TNCs-DC of samples based on full spectra and important wavelengths. The RF-PLSR model of whole-plant with results of RP = 0.876 and RMSEP = 0.426% was considered the optimal model for the TNCs-HSI prediction in pepper plants. Lastly, the TNCs-HSI of all pixels in samples were calculated by applying the optimal PLSR model. Meanwhile, spatial distribution maps of the TNCs-HSI in pepper plant samples were constructed by using a developed image processing procedure. It could be inferred that the differences in TNCs within pepper plant could provide important information for monitoring nutrient distribution.
In further research, more varieties of pepper plant with different geographical locations, ages, time, and broader chemical values should be taken into account for developing more adequate and accurate TNC prediction models, which could provide a theoretical guidance for crop nutrient diagnosis and field management.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.
Funding Statement
This study was supported by 863 National High-Tech Research and Development Plan (Project No. 2013AA102301). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Johnson CD, Decoteau DR (1996) Nitrogen and potassium fertility affects jalapeño pepper plant growth, pod yield, and pungency. HortScience 31:1119–1123. [Google Scholar]
- 2. Bosland PW (1992) Chiles: a diverse crop. HortTechnology 2:6–10. [Google Scholar]
- 3.FAOSTAT (2001) Crop water information: pepper. FAO Water Development and Management Unit. Available: http://www.fao.org/nr/water/cropinfo_pepper. html.
- 4. Feng L, Fang H, Zhou WJ, He Y (2006) Nitrogen stress measurement of canola based on multi-spectral charged coupled device imaging sensor. Spectroscopy and Spectral Analysis 26:1749–1752. [PubMed] [Google Scholar]
- 5. Van Hezewijk BH, De Clerck-Floate RA, Moyer JR (2008) Effect of nitrogen on the preference and performance of a biological control agent for an invasive plant. Biological Control 46:332–340. [Google Scholar]
- 6. Kumagai E, Araki T, Ueno O (2009) Effect of nitrogen-deficiency on midday photoinhibition in flag leaves of different rice (Oryza sativa L.) cultivars. Photosynthetica 47:241–246. [Google Scholar]
- 7. Evans RD (2001) Physiological mechanisms influencing plant nitrogen isotope composition. Trends in Plant Science 6:121–126. [DOI] [PubMed] [Google Scholar]
- 8. Bondada BR, Oosterhuis DM (2001) Canopy photosynthesis, specific leaf weight, and yield components of cotton under varying nitrogen supply. Journal of Plant Nutrition 24:469–477. [Google Scholar]
- 9.Mattson WJ (1980) Herbivory in relation to plant nitrogen content. Annual Review of Ecology and Systematics: 119–161.
- 10. Diacono M, Rubino P, Montemurro F (2012) Precision nitrogen management of wheat. A review. Agronomy for Sustainable Development 33:219–241. [Google Scholar]
- 11.Skjødt P, Hansen PM, Jørgensen RN (2003) Sensor based nitrogen fertilization increasing grain protein yield in winter wheat. Risø Report.
- 12. Blackmer TM, Schepers JS, Varvel GE, Walter-Shea EA (1996) Nitrogen deficiency detection using shortwave radiation from irrigated corn canopies. Agronomy Journal 88(1):1–5. [Google Scholar]
- 13. Cassman KG, Dobermann A, Walters DT (2002) Agroecosystems, nitrogen-use efficiency, and nitrogen management. AMBIO: A Journal of the Human Environment 31:132–140. [DOI] [PubMed] [Google Scholar]
- 14. Barbarino E, Lourenço SO (2009) Comparison of CHN analysis and hach acid digestion to quantify total nitrogen in marine organisms. Limnology and Oceanography: Methods 7:751–760. [Google Scholar]
- 15. Boussadia O, Steppe K, Zgallai H, Ben El Hadj S, Braham M, et al. (2010) Effects of nitrogen deficiency on leaf photosynthesis, carbohydrate status and biomass production in two olive cultivars ‘Meski’ and ‘Koroneiki’. Scientia Horticulturae 123:336–342. [Google Scholar]
- 16. Kamnev AA, Sadovnikova YN, Antonyuk LP (2008) Effects of nitrogen deficiency and wheat lectin on the composition and structure of some biopolymers of azospirillum brasilense Sp245. Microbiology 77:240–242. [PubMed] [Google Scholar]
- 17. Li J, Yang J, Fei P, Song J, Li D, et al. (2009) Responses of rice leaf thickness, SPAD readings and chlorophyll a/b ratios to different nitrogen supply rates in paddy field. Field Crops Research 114:426–432. [Google Scholar]
- 18. Shaahan MM, El-Sayed AA, Abou El-Nour EAA (1999) Predicting nitrogen, magnesium and iron nutritional status in some perennial crops using a portable chlorophyll meter. Scientia Horticulturae 82:339–348. [Google Scholar]
- 19. Shi J-Y, Zou X-B, Zhao J-W, Wang K-L, Chen Z-W, et al. (2012) Nondestructive diagnostics of nitrogen deficiency by cucumber leaf chlorophyll distribution map based on near infrared hyperspectral imaging. Scientia Horticulturae 138:190–197. [Google Scholar]
- 20. de la Cruz-Durán JA, Sánchez-García P, Galvis-Spínola A, Carrillo-Salazar JA (2011) Spectral indexes in sweet pepper for diagnosis of nitrogen nutrient status. Terra Latinoamericana 29(3):259–265. [Google Scholar]
- 21.San Juan Martínez E (2014) Diagnosis de nitrógeno en chile de agua identificada por reflectancia. Instituto de ensenanza e investigacion en ciencias agricolas. Available: http://hdl.handle.net/10521/2285.
- 22.Durán C (2013) Reflectancia en hojas de pimiento y fresa para el diagnóstico nutrimental. Instituto de ensenanza e investigacion en ciencias agricolas. Available: http://hdl.handle.net/10521/1902.
- 23.Gasga Peña R (2011) Reflectancia en hojas de chile manzano (Capsicum pubescens R. y P.) para estimar contenido de nitrógeno. Institucion de ensenanza e investigacion en ciencias agricolas. Available: http://hdl.handle.net/10521/468.
- 24. Demirel K, Genc L, Bahar E, Inalpulat M, Smith S, Kizil U (2014) Yield estimate using spectral indices in eggplant and bell pepper grown under deficit irrigation. Fresenius Environmental Bulletin 23(5):1232–1237. [Google Scholar]
- 25.Yu K-Q, Zhao Y-R, Liu Z-Y, Li X-L, Liu F, et al. (2014) Application of visible and near-infrared hyperspectral imaging for detection of defective features in loquat. Food and Bioprocess Technology DOI: 10.1007/s11947-014-1357-z, in press.
- 26. Martin ME, Aber JD (1997) High spectral resolution remote sensing of forest canopy lignin, nitrogen, and ecosystem processes. Ecological Applications 7(2):431–443. [Google Scholar]
- 27. Bronson KF, Booker JD, Bordovsky JP, Keeling JW, Wheeler TA, et al. (2006) Site-specific irrigation and nitrogen management for cotton production in the southern high plains. Agronomy Journal 98(1):212–219. [Google Scholar]
- 28. Jackman P, Sun D-W, Du CJ, Allen P, Downey G (2008) Prediction of beef eating quality from colour, marbling and wavelet texture features. Meat Science 80:1273–1281. [DOI] [PubMed] [Google Scholar]
- 29. Jackman P, Sun D-W, Du C-J, Allen P (2009) Prediction of beef eating qualities from colour, marbling and wavelet surface texture features using homogenous carcass treatment. Pattern Recognition 42:751–763. [Google Scholar]
- 30. Valous NA, Mendoza F, Sun D-W, Allen P (2009) Colour calibration of a laboratory computer vision system for quality evaluation of pre-sliced hams. Meat Science 81:132–141. [DOI] [PubMed] [Google Scholar]
- 31. Liu D, Sun D-W, Zeng X-A (2013) Recent advances in wavelength selection techniques for hyperspectral image processing in the food industry. Food and Bioprocess Technology 7:307–323. [Google Scholar]
- 32. Fauzi A, Skidmore AK, Gils HV, Schlerf M, Heitkönig I (2013) Shrimp pond effluent dominates foliar nitrogen in disturbed mangroves as mapped using hyperspectral imagery. Marine Pollution Bulletin 76(1):42–51. [DOI] [PubMed] [Google Scholar]
- 33. Zhang X, Liu F, He Y, Gong X (2013) Detecting macronutrients content and distribution in oilseed rape leaves based on hyperspectral imaging. Biosystems Engineering 115:56–65. [Google Scholar]
- 34. Yao X, Zhu Y, Tian Y, Feng W, Cao W (2010) Exploring hyperspectral bands and estimation indices for leaf nitrogen accumulation in wheat. International Journal of Applied Earth Observation and Geoinformation 12:89–100. [Google Scholar]
- 35. Vigneau N, Ecarnot M, Rabatel G, Roumet P (2011) Potential of field hyperspectral imaging as a non destructive method to assess leaf nitrogen content in wheat. Field Crops Research 122:25–31. [Google Scholar]
- 36. Smith ML, Martin ME, Plourde L, Ollinger SV (2003) Analysis of hyperspectral data for estimation of temperate forest canopy nitrogen concentration: comparison between an airborne (AVIRIS) and a spaceborne (Hyperion) sensor. IEEE Transactions on Geoscience and Remote Sensing 41:1332–1337. [Google Scholar]
- 37.National Farmers Information Service (6 Jan 2014). Crop Management in the Greenhouse: Nursery Establishment. NAFIS. Available: http://www.nafis.go.ke/vegetables/tomatoes/crop-management-in-the-greenhouse/.
- 38. Zhao C, Wang J, Huang W, Zhou Q (2009) Spectral indices sensitively discriminating wheat genotypes of different canopy architectures. Precision Agriculture 11:557–567. [Google Scholar]
- 39. Yu K, Zhao Y, Li X, Shao Y, Zhu F, et al. (2014) Identification of crack features in fresh jujube using Vis/NIR hyperspectral imaging combined with image processing. Computers and Electronics in Agriculture 103:1–10. [Google Scholar]
- 40. Li X, Luo L, He Y, Xu N (2013) Determination of dry matter content of tea by near and middle infrared spectroscopy coupled with wavelet-based data mining algorithms. Computers and Electronics in Agriculture 98:46–53. [Google Scholar]
- 41. Zou X, Zhao J, Mel H, Mao H, Shi J, et al. (2010) Independent component analysis in information extraction from visible/near-infrared hyperspectral imaging data of cucumber leaves. Chemometrics and Intelligent Laboratory Systems 104:265–270. [Google Scholar]
- 42. Wu D, He Y, Nie P, Cao F, Bao Y (2010) Hybrid variable selection in visible and near-infrared spectral analysis for non-invasive quality determination of grape juice. Analytica Chimica Acta 659:229–237. [DOI] [PubMed] [Google Scholar]
- 43. Cheng JH, Qu JH, Sun D-W, Zeng X-A (2014) Visible/near-infrared hyperspectral imaging prediction of textural firmness of grass carp (Ctenopharungodon idella) as affected by frozen storage. Food Research International 56:190–198. [Google Scholar]
- 44. Yun Y-H, Li H-D, Wood LRE, Fan W, Wang J-J, et al. (2013) An efficient method of wavelength interval selection based on Random frog for multivariate spectral calibration. Spectrochimica Acta Part A: Molecular and Biomolecular Spectroscopy 111:31–36. [DOI] [PubMed] [Google Scholar]
- 45. Li H-D, Xu Q-S, Liang Y-Z (2012) Random frog: an efficient reversible jump Markov Chain Monte Carlo-like approach for variable selection with applications to gene selection and disease classification. Analytica Chimica Acta 740:20–26. [DOI] [PubMed] [Google Scholar]
- 46. Moscetti R, Haff RP, Saranwong S, Monarca D, Cecchini M, et al. (2014) Nondestructive detection of insect infested chestnuts based on NIR spectroscopy. Postharvest Biology and Technology 87:88–94. [Google Scholar]
- 47. Yan J, Zhu W, Kong B, Lu B, Yun H, et al. (2014) A combinational strategy of model distribution and outlier comparison to define application domain in quantitative activity relationship. Molecular Informatics 33(8):503–513. [DOI] [PubMed] [Google Scholar]
- 48. Li F, Mistele B, Hu Y, Chen X, Schmidhalter U (2014) Reflectance estimation of canopy nitrogen content in winter wheat using optimised hyperspectral spectral indices and partial least-squares regression. European Journal of Agronomy 52:198–209. [Google Scholar]
- 49. Wu D, He Y, Feng S, Sun D-W (2008) Study on infrared spectroscopy technique for fast measurement of protein content in milk powder based on LS-SVM. Journal of Food Engineering 84:124–131. [Google Scholar]
- 50. Wu D, Nie P, Cuello J, He Y, Wang Z, et al. (2011) Application of visible and near infrared spectroscopy for rapid and non-invasive quantification of common adulterants in Spirulina powder. Journal of Food Engineering 102:278–286. [Google Scholar]
- 51. He H-J, Wu D, Sun D-W (2013) Non-destructive and rapid analysis of moisture distribution in farmed Atlantic salmon (Salmo salar) fillets using visible and near-infrared hyperspectral imaging. Innovative Food Science & Emerging Technologies 18:237–245. [Google Scholar]
- 52. He H-J, Wu D, Sun D-W (2014) Potential of hyperspectral imaging combined with chemometric analysis for assessing and visualising tenderness distribution in raw farmed salmon fillets. Journal of Food Engineering 126:156–164. [Google Scholar]
- 53. Barbin DF, Sun D-W, Su C (2013) NIR hyperspectral imaging as non-destructive evaluation tool for the recognition of fresh and frozen-thawed porcine longissimus dorsi muscles. Innovative Food Science & Emerging Technologies 18:226–236. [Google Scholar]
- 54. Wold S, Sjöström M, Eriksson L (2001) PLS-regression: a basic tool of chemometrics. Chemometrics and Intelligent Laboratory Systems 58:109–130. [Google Scholar]
- 55. Zhu F, Zhang H, Shao Y, He Y, Ngadi M (2013) Mapping of fat and moisture distribution in atlantic salmon using near-infrared hyperspectral imaging. Food and Bioprocess Technology 7:1208–1214. [Google Scholar]
- 56. Nelson DW, Sommers LE (1973) Determination of total nitrogen in plant material. Agronomy Journal 65(1):109–112. [Google Scholar]
- 57.Stevenson FJ (1982) Nitrogen in agricultural soils. In: Olson RA, Kurtz LT, editors. Crop nitrogen requirements, utilization, and fertilization. American Society of Agronomy, Crop Science Society of America, Soil Science Society of America. pp. 567–604.
- 58. Galvao RK, Araujo MC, Jose GE, Pontes MJ, Silva EC, et al. (2005) A method for calibration and validation subset partitioning. Talanta 67:736–740. [DOI] [PubMed] [Google Scholar]
- 59. Penuelas J, Filella I, Biel C, Serrano L, Save R (1993) The reflectance at the 950–970 nm region as an indicator of plant water status. International Journal Remote Sensing 14:1887–1905. [Google Scholar]
- 60. Cho MA, Skidmore AK (2006) A new technique for extracting the red edge position from hyperspectral data: the linear extrapolation method. Remote Sensing of Environment 101:181–193. [Google Scholar]
- 61. Kokaly RF (2001) Investigating a physical basis for spectroscopic estimates of leaf nitrogen concentration. Remote Sensing of Environment 75:153–161. [Google Scholar]
- 62. Li D, Chen X, Peng Z, Chen S, Chen W, et al. (2012) Prediction of soil organic matter content in a litchi orchard of South China using spectral indices. Soil and Tillage Research 123:78–86. [Google Scholar]
- 63. Rossel RAV, Behrens T (2010) Using data mining to model and interpret soil diffuse reflectance spectra. Geoderma 158:46–54. [Google Scholar]
- 64. Lee YJ, Yang CM, Chang KW, Shen Y (2008) A simple spectral index using reflectance of 735 nm to assess nitrogen status of rice canopy. Agronomy Journal 100:205–212. [Google Scholar]
- 65. Inoue Y, Sakaiya E, Zhu Y, Takahashi W (2012) Diagnostic mapping of canopy nitrogen content in rice based on hyperspectral measurements. Remote Sensing of Environment 126:210–221. [Google Scholar]
- 66. Elmasry G, Kamruzzaman M, Sun D-W, Allen P (2012) Principles and applications of hyperspectral imaging in quality evaluation of agro-food products: a review. Critical Reviews in Food Science and Nutrition 52:999–1023. [DOI] [PubMed] [Google Scholar]
- 67. Hörtensteiner S, Feller U (2002) Nitrogen metabolism and remobilization during senescence. Journal of Experimental Botany 53:927–937. [DOI] [PubMed] [Google Scholar]
- 68. Anten NPR, Miyazawa K, Hikosaka K, Nagashima H, Hirose T (1998) Leaf nitrogen distribution in relation to leaf age and photon flux density in dominant and subordinate plants in dense stands of a dicotyledonous herb. Oecologia 113:314–324. [DOI] [PubMed] [Google Scholar]
- 69.Nobel PS (1983) Biophysical plant physiology and ecology. W.H. Freeman and Company.
- 70.Hopkins WG, Hüner NP (1995) Introduction to plant physiology (fourth edition). New York: J Wiley.
- 71. Pate JS (1973) Uptake, assimilation and transport of nitrogen compounds by plants. Soil Biology and Biochemistry 5(1):109–119. [Google Scholar]
- 72. Pate JS (1980) Transport and partitioning of nitrogenous solutes. Annual Review of Plant Physiology 31:313–340. [Google Scholar]
- 73. Tylova-Munzarova E, Lorenzen B, Brix H, Votrubova O (2005) The effects of NH4+ and NO3− on growth, resource allocation and nitrogen uptake kinetics of phragmites australis and glyceria maxima. Aquatic Botany 81:326–342. [Google Scholar]
- 74. Geladi P, Burger J, Lestander T (2004) Hyperspectral imaging: calibration problems and solutions. Chemometrics and Intelligent Laboratory Systems 72(2):209–217. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper.