Abstract
While available evidence supports the role of genetics in the pathogenesis of placental abruption (PA), PA-related placental genome variations and maternal-placental genetic interactions have not been investigated. Maternal blood and placental samples collected from participants in the Peruvian Abruptio Placentae Epidemiology study were genotyped using Illumina’s Cardio-Metabochip platform. We examined 118,782 genome-wide SNPs and 333 SNPs in 32 candidate genes from mitochondrial biogenesis and oxidative phosphorylation pathways in placental DNA from 280 PA cases and 244 controls. We assessed maternal-placental interactions in the candidate gene SNPS and two imprinted regions (IGF2/H19 and C19MC). Univariate and penalized logistic regression models were fit to estimate odds ratios. We examined the combined effect of multiple SNPs on PA risk using weighted genetic risk scores (WGRS) with repeated ten-fold cross-validations. A multinomial model was used to investigate maternal-placental genetic interactions. In placental genome-wide and candidate gene analyses, no SNP was significant after false discovery rate correction. The top genome-wide association study (GWAS) hits were rs544201, rs1484464 (CTNNA2), rs4149570 (TNFRSF1A) and rs13055470 (ZNRF3) (p-values: 1.11e-05 to 3.54e-05). The top 200 SNPs of the GWAS overrepresented genes involved in cell cycle, growth and proliferation. The top candidate gene hits were rs16949118 (COX10) and rs7609948 (THRB) (p-values: 6.00e-03 and 8.19e-03). Participants in the highest quartile of WGRS based on cross-validations using SNPs selected from the GWAS and candidate gene analyses had a 8.40-fold (95% CI: 5.8–12.56) and a 4.46-fold (95% CI: 2.94–6.72) higher odds of PA compared to participants in the lowest quartile. We found maternal-placental genetic interactions on PA risk for two SNPs in PPARG (chr3∶12313450 and chr3∶12412978) and maternal imprinting effects for multiple SNPs in the C19MC and IGF2/H19 regions. Variations in the placental genome and interactions between maternal-placental genetic variations may contribute to PA risk. Larger studies may help advance our understanding of PA pathogenesis.
Introduction
Placental abruption (PA), the premature separation of the placenta from the uterine wall prior to delivery of the fetus, complicates about 1% of pregnancies and is an important cause of maternal and neonatal morbidity and mortality [1]–[7]. Evidence from studies conducted during the last three decades suggests that hypertensive disorders, advanced maternal age, grand-multiparity, thrombophilia, cigarette smoking, illicit drug use and external trauma to the abdomen are associated with an increased risk of PA [8]–[13]. In addition, other putative risk factors have also been recently described for PA, including maternal iron deficiency anemia, hyperhomocystinemia, mood and anxiety disorders, migraine and headache disorders, maternal infection and/or inflammation [4], [14]–[21]. Pathophysiologic mechanisms involved in PA include uteroplacental underperfusion, chronic hypoxemia, uteroplacental ischemia and infarctions, and thrombosis [5], [13], [22]–[23].
As a multi-factorial disorder of complex origin, PA aggregates in families of women with the condition [24], suggesting a strong role for genetic predisposition, a thesis supported by a number of candidate gene studies [25]–[29]. Findings from recent PA-related genome-wide association studies (GWAS) and candidate gene association studies (mitochondrial biogenesis and oxidative phosphorylation pathway genes) in the maternal genome by our group provided suggestive evidence supporting associations of variation in maternal cardiometabolic genes with risk of PA [30], [31]. Given these findings indicative of the importance of genetic susceptibility factors in PA and the evidence highlighting the central role of placental pathology in PA [5], [28], [32], we hypothesized that genetic variations in the placental genome, particularly those variants in mitochondrial biogenesis (MB) and oxidative phosphorylation (OP) pathways, are associated with PA risk. Further, using data from both maternal and placental genomes, we examined interactions between placental and maternal genetic variations (in MB-OP pathway genes and imprinted regions) on risk of PA.
Materials and Methods
Study Setting and Study Population
The current study was conducted in the setting of the Peruvian Abruptio Placentae Epidemiology (PAPE) study that has been described before [30], [31]. Briefly, PAPE study participants were recruited and enrolled among patients admitted for obstetrical services to the Hospital Nacional Dos de Mayo, Instituto Especializado Materno Perinatal, and Hospital Madre-Niño San Bartolomé in Lima, Peru, between August 2002 and May 2004 and between September 2006 and September 2008. Hospital admission and delivery logs were monitored daily to identify PA cases among new admissions to antepartum, emergency room, and labor and delivery wards of participating hospitals. PA was diagnosed based on evidence of retroplacental bleeding (fresh blood) entrapped between the decidua and the placenta or blood clots behind the placenta and any two of the following: (i) vaginal bleeding in late pregnancy not due to placenta previa or cervical lesions; (ii) uterine tenderness and/or abdominal pain; and, (iii) non-reassuring fetal status or death. Controls were selected from among pregnant women who delivered at participating hospitals during the study period and did not have a diagnosis of PA in the current pregnancy. For the current study, investigating associations of placental genome with risk of PA, 280 PA cases and 244 controls who provided placental samples at delivery were included. A subset of these cases and controls that also provided blood samples (222 PA cases and 198 controls) were also included in the maternal-placental genetic interaction investigations.
Ethical approval for the study was granted by the Institutional Review Boards (IRB) of Hospital Nacional Dos de Mayo, Instituto Especializado Materno Perinatal, Hospital Madre-Niño San Bartolomé in Lima, Peru and the IRB of Swedish Medical Center, Seattle, WA. All participants provided written informed consent in accordance with the principles of the declaration of Helsinki.
Data collection
Standardized structured questionnaires administered by trained research personnel were used to collect information on socio-demographic characteristics (including maternal age, marital status, employment status during pregnancy, and smoking and alcohol consumption before and during pregnancy), and medical history. A brief physical examination was conducted to measure maternal height, weight, and mid-arm circumference. Medical records were reviewed to abstract information on course and outcomes of the pregnancy. At delivery, placental samples were collected for DNA extraction and genotyping as described below.
Placental sample collection, DNA extraction, and genome-wide genotyping
Placentas were collected immediately after delivery. Placentas were weighed, double bagged and transported in coolers. The chorionic plate and overlying membranes were stripped and tissue biopsies (approximately 0.5 cm3 each) were obtained from 8 sites (4 maternal and 4 fetal). For this study, biopsy samples taken from the fetal side were sampled for genomic DNA extraction. Biopsies were placed in cryotubes, snap frozen in liquid nitrogen, and stored at −80°C until analysis. The Gentra PureGene Cell kit for DNA preparations (Qiagen, Hilden, Germany) was used to extract DNA from placental samples. Genotyping was conducted using the Illumina Cardio-Metabochip (Illumina Inc, San Diego, CA) platform [30], a high-density custom array designed to include 217,697 SNPs that represent DNA variations at regions previously related to diseases and traits relevant to metabolic and atherosclerotic-cardiovascular endpoints [33], [34]. During the assay manufacturing process 20,972 SNPs (9.6%) failed, resulting in 196,725 SNPs available for genotyping, downstream quality control and statistical analyses [34], [35].
Candidate gene/SNP selection
For the candidate association study, 32 genes that were involved in mitochondrial biogenesis and oxidative phosphorylation were selected based on literature [31] and a total of 333 SNPs belonging to these genes and found in the Cardio-MetaboChip were included in the candidate gene association analyses. For the maternal-placental genetic interaction study, 325 of these SNPs that also passed quality control in maternal blood genomes, as well as SNPs in imprinted regions (5 SNPs in IGF2/H19 and 33 SNPs in C19MC) included in the Cardio-MetaboChip were analyzed.
Data quality control
Quality control and preprocessing were performed on the genotype data. Individuals with genotyping failure in more than 10% of SNPs were removed (n = 2). SNPs with minor allele frequency (MAF) less than 1% or that failed to be genotyped in more than 10% of the study samples were removed (n = 77, 276), as well as SNPs not in Hardy-Weinberg equilibrium (HWE) among controls (n = 667). After these quality control procedures, a total of 118,782 genome-wide SNPs were examined among 280 PA cases and 244 controls. Similar quality control procedures were performed on the maternal blood genotype data, resulting in 222 PA case and 198 controls among maternal-placental pairs.
Statistical analyses
Univariate logistic regression model was used to estimate odds ratio (OR) and 95% confidence interval (95% CI) relating each SNP with risk of PA, in the genome-wide and candidate gene analyses. For multiple testing correction, a false discovery rate (FDR) procedure was used [36]. Functions and functional relationships of genes represented by the top 200 genome-wide SNPs were obtained by pathway analysis using the Ingenuity Pathway Analysis (IPA, Ingenuity Systems, www.ingenuity.com) software. Gene-enrichment network score based on a modified Fisher's exact test were calculated to rank biological significance of networks in relation to PA.
In multivariable analyses, we applied penalized logistic regression models to identify sets of SNPs that are jointly associated with the risk of PA. These penalized approaches have previously been applied in the context of GWAS and have shown promising results [37]–[38]. These methods allow the selection of relevant variables or groups of variables and the estimation of their regression coefficients [39]. The number of selected variables is guided by a penalty parameter: the larger the parameter, the smaller the selected subset. A 20-fold cross-validation approach was performed to select the penalty parameter and the value yielding the smallest prediction error was used. For the genome-wide SNP analysis, we applied a lasso regression [39]. One characteristic of lasso regression is that it selects a single variable among a set of correlated variables. To circumvent this, SNPs in high linkage disequilibrium with a selected SNP were also considered using an r2 threshold of 0.8 within 500 kb. For SNPs in the candidate gene analyses, a group penalty approach was used to account for the membership in a gene [40]. Furthermore, we considered a bi-level selection approach that uses a composite minimax concave penalty [41], [42] to select candidate genes associated with PA as well as relevant SNPs within those genes. These penalized regression methods do not accommodate missing values and the software BEAGLE version 3.3.2 [43] was used to impute missing genotypes.
For weighted genetic risk score (WGRS) analyses [44], a 10-fold cross-validation procedure was implemented to protect against model over-fitting, which arises from using the same data to estimate the regression parameters used in computing WGRS and to evaluate the association between PA risk and WGRS [45]. The procedure consisted of randomly partitioning the data into 10 equal size subsamples, using nine of the subsamples as training set and the left-out one as validation set, with each subsample being used in turn as a test set. For each fold, a multivariate logistic regression model was fit on the training set using the SNPs selected from multivariate analyses. A weighted approach was then used to compute Genetic Risk Scores (GRS) in the validation set by multiplying the number of risk allele for each locus by its associated effect size estimated from the training set. Once the WGRS were obtained for all individuals, the subjects were categorized into four groups defined by the quartiles in the control. A logistic regression model was then fit to examine the association of the WGRS with PA risk using the lowest quartile (Group 1) as a reference and adjusting for infant sex and population admixture. This 10-fold cross-validation procedure was repeated 1000 times to account for the variability in randomly partitioning the data into subsamples. The receiver operating characteristics (ROC) curve for each of the replicates was evaluated. The estimated effect sizes and AUCs over the 1000 replicates were used to obtain the respective point estimates and confidence intervals.
Maternal-placental interaction analyses (for candidate genes and imprinted regions) were performed using a multinomial model proposed by [46] and implemented in the EMIM and PREMIM software tools [47]. The method requires some biological assumptions, such as Hardy-Weinberg equilibrium (HWE), random mating, and rare disease. For each SNP, four models were considered and a model selection procedure based on the Bayesian information criterion (BIC) was applied. The four models correspond to allele effects operating only at the fetal level (Model F), allele effects operating only at the maternal level (Model M), an additive effect of maternal and fetal effects (Model M+F), and a model that includes a maternal-placental interaction effect (Model I). For the latter, we used a parametrization that introduces two interaction terms capturing incompatibility between maternal and placental genotypes; the interaction effects operate when the infant has one copy and the mother has either zero or two copies of the risk allele [46], [48]. Maternal imprinting effect, which corresponds to the factor multiplying the disease risk if the infant inherits a risk allele from the mother, was tested using a likelihood ratio test [46], [49].
Adjustment for the first four principal components was done for all univariate and multivariable logistic regression models to take into account population stratification. The various statistical analyses were conducted using a combination of software tools: PLINK, PREMIM, EMIM, Haploview, and R. As for multivariable approaches the R packages ncvreg and grpreg were used [41], [50]. The pathway analyses were conducted using the Ingenuity Pathway Analysis (IPA) software.
Results
Table 1 shows selected characteristics of PA cases and controls. Average maternal age in both groups was around 27 years. Alcohol use during pregnancy and preeclampsia/eclampsia were more common among PA cases than controls. As expected, infant birthweight and gestational age at delivery were lower for PA cases compared with controls.
Table 1. Socio-demographic and reproductive characteristics and infant outcomes in the study sample, Lima, Peru.
| Maternal Characteristics | Placental Abruption | ||
| Cases | Controls | ||
| (n = 280) | (n = 244) | p-value** | |
| Maternal age at delivery, years * | 27.03 (6.5) | 27.3 (6.6) | 0.517 |
| <35 | 239 (85%) | 200 (82%) | 0.337 |
| ≥35 | 40 (14%) | 43 (18%) | |
| Nulliparous | 115 (41%) | 95 (39%) | 0.655 |
| Less than High school education | 220 (79%) | 186 (76%) | 0.598 |
| Employed during pregnancy | 126 (45%) | 108 (44%) | 0.935 |
| Planned pregnancy | 114 (41%) | 99 (.41%) | 0.929 |
| No prenatal vitamin | 82 (29%) | 65 (27%) | 0.559 |
| Smoked during pregnancy | 12 (4%) | 5 (2%) | 0.216 |
| Alcohol use during pregnancy | 9 (3%) | 0 (0) | 0.004 |
| Pre-pregnancy BMI, kg/m2 * | 23.5 (3.5) | 23.9 (3.9) | 0.228 |
| <18.5 | 14 (5%) | 8 (3%) | |
| 18.5–24.9 | 179 (64%) | 149 (61%) | |
| 25.0–29.9 | 57 (2%) | 56 (23%) | |
| ≥30.0 | 13 (5%) | 18 (7%) | |
| Chronic hypertension | 8 (3%) | 4 (2%) | 0.391 |
| Preeclampsia or Eclampsia | 58 (21%) | 29 (12%) | 0.005 |
| History of placental abruption | 2 (7%) | 0 (0) | 0.498 |
| Infant birthweight, grams * | 2357 (888) | 3058 (825) | 2.20E-16 |
| Gestational age at delivery, weeks * | 35 (4.3) | 37.8 (3.5) | 2.68E-15 |
*Mean (standard deviation), otherwise count (%).
**t-test and chi-square test respectively used for continuous and categorical variables.
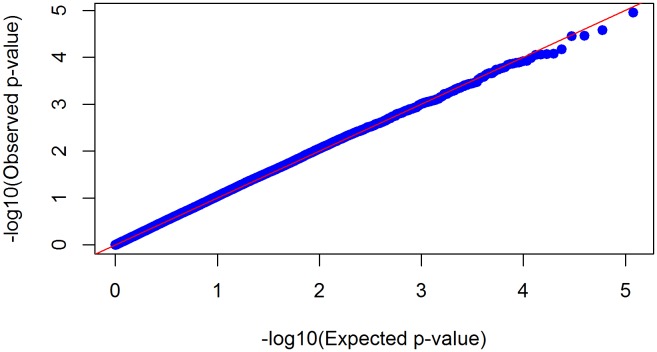
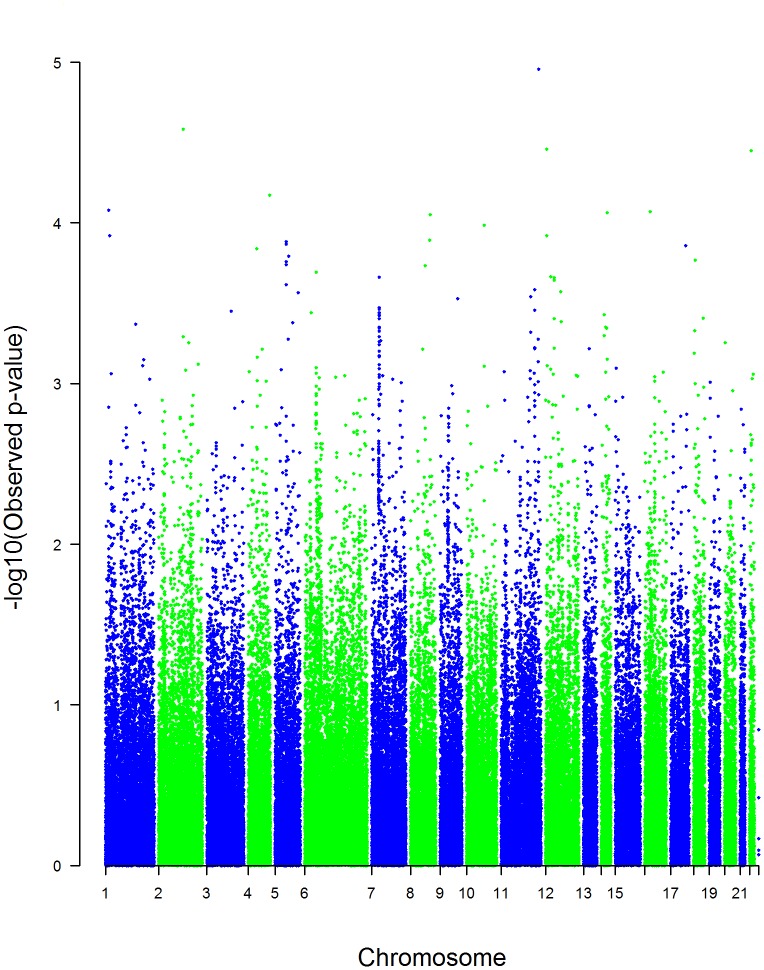
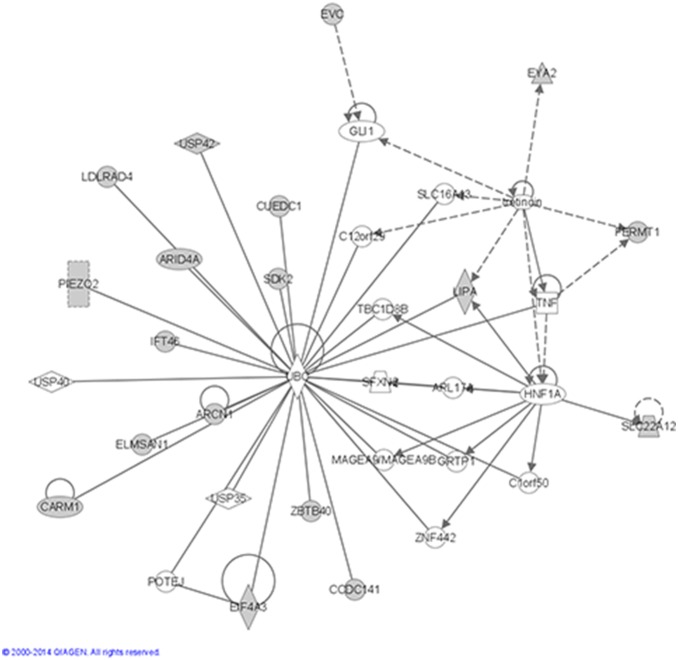
In the GWAS analyses, we did not observe significant genomic inflation (λ = 1.17) (Fig. 1). None of the FDR-corrected p-values were lower than a 0.05 threshold (Table 2). The top GWAS hits were rs544201, rs1484464 (CTNNA2), rs4149570 (TNFRSF1A) and rs13055470 (ZNRF3) (nominal p-values: 1.11e-05 to 3.54e-05) (Fig. 2 and Table 2). Functions of 56 genes represented by the top 200 SNPs from univariate GWAS analyses were examined using IPA. The top five networks from these analyses with p-values less than 0.05 are shown in Table 3. The top network enriched by these genes was a network of cell cycle, growth and proliferation (score = 43, p-value = 2.12e-19) (Fig. 3). In the candidate gene analyses, none of the FDR-corrected p-values were lower than a 0.05 threshold. The top hits in these analyses were rs16949118 (COX10) and rs7609948 (THRB) (nominal p-values: 6.00e-03 and 8.19e-03, respectively) (Table 4). In addition, several SNPs in the PPARG gene were among the top hits in the candidate gene analyses.
Figure 1. Quantile-Quantile plot.
Quantile-quantile plot (QQ-plot) of raw p-values from univariate GWAS analysis adjusting for population stratification (genomic inflation factor λ = 1.168).
Table 2. Top 20 hits from univariate analyses examining genome-wide genetic variations related to placental abruption risk.
| NCBIGene Name | SCANGene Name | SNP | MinorAllele | MAF | OR (95% CI) | Empiricalp-value |
| rs544201 | T | 0.13 | 0.33 (0.21–0.54) | 1.11E-05 | ||
| CTNNA2 | CTNNA2 | rs1484464 | G | 0.3 | 1.80 (1.37–2.38) | 2.62E-05 |
| TNFRSF1A | TNFRSF1A | rs4149570 | A | 0.21 | 1.88 (1.40–2.53) | 3.46E-05 |
| ZNRF3 | ZNRF3 | rs13055470 | A | 0.31 | 0.52 (0.39–0.71) | 3.54E-05 |
| ACSL1 | LOC11394 | rs9997745 | A | 0.03 | 3.78 (1.97–7.26) | 6.73E-05 |
| rs10754855 | A | 0.26 | 0.52 (0.38–0.72) | 8.34E-05 | ||
| rs3096425 | G | 0.47 | 0.58 (0.44–0.76) | 8.50E-05 | ||
| rs12896434 | A | 0.19 | 1.84 (1.36–2.49) | 8.61E-05 | ||
| rs2436893 | A | 0.3 | 1.72 (1.31–2.25) | 8.86E-05 | ||
| LIPA | LIPA | rs7922269 | A | 0.44 | 0.58 (0.43–0.76) | 1.03E-04 |
| SCNN1A | SCNN1A | rs2228576 | A | 0.19 | 1.83 (1.34–2.48) | 1.20E-04 |
| ZBTB40 | ZBTB4 | rs12725956 | G | 0.08 | 2.24 (1.49–3.38) | 1.20E-04 |
| rs3133572 | A | 0.15 | 0.44 (0.29–0.67) | 1.28E-04 | ||
| ZBED3-AS1 | rs4457053 | G | 0.45 | 0.59 (0.46–0.78) | 1.31E-04 | |
| ZBED3-AS1 | rs7708285 | G | 0.45 | 0.59 (0.46–0.78) | 1.36E-04 | |
| SDK2 | rs9913193 | G | 0.25 | 0.54 (0.39–0.74) | 1.38E-04 | |
| ADAMTS3 | ADAMTS3 | rs4694121 | A | 0.55 | 0.60 (0.46–0.78) | 1.45E-04 |
| rs6871240 | A | 0.18 | 1.8 (1.33–2.46) | 1.62E-04 | ||
| PIEZO2 | rs9964303 | A | 0.3 | 0.57 (0.42–0.76) | 1.70E-04 | |
| chr5∶76460816 | C | 0.45 | 0.60 (0.46–0.78) | 0.000174 |
Abbreviations: MAF = Minor Allele Frequency in Controls.
Figure 2. Manhattan plot.
Manhattan plot of raw p-values from univariate GWAS analysis adjusting for population stratification.
Table 3. Significant networks represented by top GWAS hits.
| Genes from GWAS analysis with adjustment for the first four components | |||||
| ID | Molecules inNetwork | Score | FocusMolecules | Top Diseasesand Functions | P-value |
| 1 | ARCN1,ARID4A,ARL17A,C12orf29,C1orf50,CARM1,CCDC141,CUEDC1,EIF4A3,ELMSAN1,EVC,EYA2,FERMT1,GLI1,GRTP1,HNF1A,IFT46,LDLRAD4,LIPA,MAGEA9/MAGEA9B,PIEZO2,POTEJ,SDK2,SFXN2,SLC16A13,SLC22A12,TBC1D8B,TNF,tretinoin,UBC,USP35,USP40,USP42,ZBTB40,ZNF442 | 43 | 18 | Cell Cycle,Cellular Growthand Proliferation,Gene Expression | 2.12E-19 |
| 2 | ACSL1,Akt,ANGPTL4,ARAP3,ARHGAP26,Cyp2j9,Focal adhesion kinase,ganglioside GD2,GLIS3,GYS2,IL22R1-IL10R2,Immunoglobulin,INSRR,Insulin,IPO8,LPP,miR-491–5p (and other miRNAs w/seed GUGGGGA),NCOA7,NFkB (complex),NRG4,NTN4,P38 MAPK,PID1,PKN3,PTPRB,RAB31,RGS5,SCNN1A,SLC2A5,SLC30A7,SLC35B2,SLC3A1,TNFRSF1A,TRAF1-TRAF2-TRAF3,WISP3 | 28 | 13 | Endocrine SystemDevelopment andFunction, TissueMorphology,Cellular Development | 6.91E-13 |
| 3 | ADAMDEC1,ADAMTS3,AKT2,ART3,ATP10A,beta-estradiol,Ca2+,Calmodulin-Camk4a2+,CCDC82,Cetn4,COA4,CPA2,CTNNA2,DGKB,DOK6,ERBB2,FURIN,GDPD3,HDAC4,HNF4A,Interferon alpha,INTS4,KCNIP1,MAPK1,MITF,NUDT6,PDE11A,PRR15L,RHOBTB2,SLC24A3,SPAG16,Timd2,TMEM258,TRPM1,ZNRF3 | 28 | 13 | Energy Production,Molecular Transport,Nucleic AcidMetabolism | 6.91E-13 |
| 4 | SYT14,SYT16 | 3 | 1 | Hereditary Disorder,NeurologicalDisease, Cancer | 4.98E-02 |
| 5 | KIAA1524,SLC12A8 | 3 | 1 | Cancer, CellularMovement,GastrointestinalDisease | 4.98E-02 |
The networks were generated using Ingenuity Pathways Analysis (Ingenuity Systems, www.ingenuity.com). Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base (IPKB) and overlaid onto a global molecular network developed from information contained in the IPKB. Scores, corresponding to degree of enrichment, are negative log of p-values from Fisher’s exact test. Genes in bold (focus molecules) are genes that correspond to top hit SNPs in our genome-wide association study of placental abruption.
Figure 3. Molecules in the top network.
Representation of molecules in the top network enriched by genes corresponding to the top 200 SNPs from univariate GWAS analyses.
Table 4. Top 20 SNPs in univariate analyses of candidate genes in relation to risk of placental abruption.
| SCAN Gene Name | SNP | MinorAllele | MAF | OR (95% CI) | Empirical p-value |
| COX10 | rs16949118 | A | 0.09 | 1.74 (1.17–2.59) | 0.006003 |
| THRB | rs7609948 | A | 0.19 | 1.48 (1.11–1.99) | 0.008191 |
| COX5A | chr15∶73015771 | G | 0.01 | 2.80 (1.09–7.21) | 0.03232 |
| THRB | rs17787283 | A | 0.12 | 0.64 (0.42–0.97) | 0.03637 |
| PRKCA | rs3848426 | A | 0.29 | 1.33 (1.02–1.74) | 0.03651 |
| PPARG | chr3∶12388339 | C | 0.23 | 1.37 (1.015–1.848) | 0.03935 |
| NDUFS4 | rs1388111 | A | 0.51 | 0.78 (0.61–1.01) | 0.05589 |
| CAMK2B | chr7∶44224020 | A | 0.33 | 1.31 (0.99–1.72) | 0.05679 |
| CAMK2D | rs4834348 | A | 0.18 | 0.70 (0.49–1.01) | 0.05689 |
| PPARG | rs11709077 | A | 0.26 | 1.31 (0.98–1.75) | 0.06663 |
| NDUFC2-KCTD14 | rs627297 | C | 0.18 | 0.74 (0.53–1.03) | 0.07345 |
| CAMK2B | chr7∶44224468 | A | 0.33 | 1.28 (0.97–1.69) | 0.07898 |
| THRB | rs12639293 | A | 0.29 | 1.28 (0.97–1.69) | 0.07978 |
| PPARG | chr3∶12326521 | C | 0.26 | 1.29 (0.96–1.72) | 0.08784 |
| PPARG | rs4135275 | G | 0.28 | 0.78 (0.58–1.05) | 0.0958 |
| NDUFS4 | rs2168662 | G | 0.45 | 0.80 (0.62–1.04) | 0.09672 |
| TUFM | chr16∶28763228 | A | 0.24 | 0.76 (0.55–1.06) | 0.1012 |
| PPARG | chr3∶12344401 | A | 0.26 | 1.27 (0.95–1.69) | 0.1048 |
| PPARG | chr3∶12352344 | A | 0.3 | 0.79 (0.59–1.05) | 0.1053 |
| PPARG | chr3∶12340308 | A | 0.26 | 1.27 (0.95–1.69) | 0.1059 |
Abbreviations: MAF = Minor Allele Frequency in Controls.
Among 118,782 SNPs included in the GWAS analyses, six SNPs were selected using lasso regression (Table 5). All six SNPs were among the top 200 hits, including the top three hits, identified using the univariate logistic regression analyses. When fitting a multiple logistic regression model with the selected SNPs, all 6 SNPs had empirical p-values lower than 0.05. Using a group penalty and bi-level selection approach, we identified 22 SNPs (in 14 genes) among the >300 SNPs included in the candidate gene analyses (Table 6). In multiple logistic regression analysis that included these SNPs, 11 SNPs had empirical p-values less than 0.05 (Table 6). WGRS were computed using SNPs selected from the respective GWAS and candidate gene multivariable analyses (Table 7). Both WGRSs were significantly associated with risk of PA (p-values<0.001). Participants in the highest quartiles for cross-validated GWAS-based WGRS and candidate gene-based WGRS had a 8.4 (95% CI: 5.8–12.56) and a 4.46 (95% CI: 2.94–6.72) fold higher odds of PA compared with participants in the respective referent quartiles (quartile 1) adjusting for infant sex and population admixture. The cross-validated AUCs for the ROC curves were estimated to be 0.71 (95% CI: 0.69–0.73) for the GWAS-based WGRS and 0.67 (95% CI: 0.65–0.7) for the candidate gene-based WGRS, confirming that the WGRS models have good predictive ability (S1 Fig. and S2 Fig.).
Table 5. Multiple logistic regression using SNPs selected in lasso regression.
| Gene | SNP | SNPs in high LD* | Minor Allele | MAF | Odds Ratio | Empirical P-value | rank in univariate GWAS |
| rs544201 | T | 0,13 | 0.33 (0.26–0.56) | 4,19E-05 | 1 | ||
| CTNNA2 | rs1484464 | G | 0,30 | 1.69 (1.46–2.26) | 4,24E-04 | 2 | |
| TNFRSF1A | rs4149570 | rs2228576 | A | 0,21 | 1.77 (1.53–2.41) | 2,58E-04 | 3 |
| rs10754855 | A | 0,26 | 0.58 (0.49–0.81) | 1,35E-03 | 6 | ||
| ARHGAP26 | rs17287593 | G | 0,01 | 4.55 (2.99–10.67) | 5,04E-04 | 47 | |
| IFT46 | rs2277292 | rs2277295; rs17122278 | A | 0,05 | 0.23 (0.14–0.6) | 2,55E-03 | 60 |
*R2>0.8 within 500 kb.
Table 6. Multiple logistic regression based on SNPs selected from candidate genes using a bi-level selection approach.
| Gene | SNP | Minor Allele | MAF | Odds Ratio | Empirical P-value |
| PPARG | chr3∶12388339 | C | 0.23 | 1.68 (1.21–2.34) | 2.19E-03 |
| THRB | rs7609948 | A | 0.19 | 1.6 (1.17–2.18) | 3.31E-03 |
| NDUFA10 | rs6437237 | A | 0.48 | 0.63 (0.45–0.87) | 5.73E-03 |
| NDUFA10 | rs4149549 | A | 0.19 | 1.7 (1.15–2.51) | 8.12E-03 |
| COX10 | rs16949118 | A | 0.09 | 1.77 (1.15–2.7) | 8.72E-03 |
| THRB | rs17787283 | A | 0.12 | 0.56 (0.35–0.88) | 1.15E-02 |
| NDUFS4 | rs1388111 | A | 0.51 | 0.71 (0.55–0.93) | 1.31E-02 |
| NDUFA12 | rs11107847 | A | 0.45 | 0.71 (0.54–0.94) | 1.57E-02 |
| PPARG | chr3∶12363563 | C | 0.14 | 1.59 (1.05–2.41) | 2.75E-02 |
| NR1H3 | chr11∶47226512 | A | 0.3 | 1.39 (1.03–1.85) | 2.86E-02 |
| COX5A | chr15∶73015771 | G | 0.01 | 2.55 (1.05–6.22) | 3.87E-02 |
| CAMK2D | rs4834348 | A | 0.18 | 0.7 (0.47–1.02) | 6.48E-02 |
| NDUFC2-KCTD14 | rs627297 | C | 0.18 | 0.73 (0.52–1.03) | 7.41E-02 |
| PPA2 | rs2298733 | C | 0.17 | 0.72 (0.5–1.03) | 7.58E-02 |
| PRKCA | rs3848426 | A | 0.29 | 1.27 (0.96–1.69) | 9.95E-02 |
| PPARG | chr3∶12412978 | A | 0.03 | 1.85 (0.87–3.93) | 1.07E-01 |
| COX5A | chr15∶73011246 | A | 0.17 | 0.74 (0.51–1.09) | 1.31E-01 |
| PRKCA | chr17∶61760907 | G | 0.34 | 1.23 (0.92–1.64) | 1.56E-01 |
| THRB | rs17014418 | G | 0.02 | 0.5 (0.17–1.42) | 1.94E-01 |
| COX7B2 | rs17598636 | G | 0.02 | 0.54 (0.19–1.53) | 2.45E-01 |
| NR1H3 | chr11∶47233666 | A | 0.07 | 0.76 (0.44–1.32) | 3.33E-01 |
| CAMK2B | chr7∶44227306 | A | 0.02 | 1.36 (0.5–3.68) | 5.47E-01 |
Table 7. Association between risk of placental abruption and weighted genetic risk score (WGRS) computed from SNPs selected in multivariable analyses using repeated 10-fold cross-validations.
| Weighted Genetic Risk Score (GRS) | |||||
| 1st Quartile | 2nd Quartile | 3rd Quartile | 4th Quartile | p-value | |
| Genome-wide Association Analysis* | |||||
| OR ** | 1 | 2.53 | 3.78 | 8.40 | <0.001 |
| (95% CI) | (Ref.) | (1.47–4.71) | (2.2–6.4) | (5.8–12.56) | |
*Cross-validated WGRS computed from SNPs selected from multivariable analyses.
**Cross-validated odds ratios (and 95% confidence intervals) from logistic regression models adjusted for sex, and population admixture, p-values associated to chi-square global test.
We observed evidence for maternal-placental genetic interaction (on PA risk) for 23 candidate SNPs (Table 8). In particular, maternal-placental genetic interaction on PA risk was found for two SNPs in PPARG (chr3∶12313450 and chr3∶12412978). The model selection procedure based on the BIC for the SNP chr3∶12412978, is shown in Table 9. The smallest BICcorresponds to Model I meaning that Model I fits the data best. In the imprinting effect analyses, we found that six SNPs in the C19MC region (of the 33 that were examined) and two SNPs in the IGF2-H19 region (of the five that were examined) showed evidence for maternal imprinting effect (empirical p-value <0.05) (Table 10 and Table 11). In addition, borderline imprinting effects were detected for a number of other SNPs in these regions.
Table 8. SNPs selected with maternal-placental interaction as best fitting model.
| Gene | SNP |
| PPARG | chr3∶12412978 |
| PPARG | chr3∶12313450 |
| PRKCA | chr17∶61743445 |
| CAMK2B | chr7∶44226231 |
| THRB | rs12639293 |
| LRPPRC | rs11899538 |
| COX5A | chr15∶73008298 |
| COX5A | chr15∶73012861 |
| COX5A | chr15∶73001842 |
| COX5A | chr15∶73008918 |
| PRKCA | rs9896575 |
| THRB | rs9809150 |
| PRKCA | chr17∶61732949 |
| NR1H3 | chr11∶47233666 |
| PRKCA | chr17∶61736374 |
| CAMK2B | rs2075076 |
| CAMK2B | rs1127065 |
| THRB | rs2362186 |
| PRKCA | chr17∶61735430 |
| PRKCA | chr17∶61735623 |
| COX10 | rs16949118 |
| TRNT1 | rs7629889 |
| PPARGC1A | rs12650562 |
Table 9. Sample model selection procedure for SNP chr312412978.
| chr3∶12412978 Models compared | Loglikelihood of first model | Likelihood ratio test | BIC of the first model |
| Model I vs Model M+F | −349.57 | 13.9504 | 726.0741 |
| Model M+F vs Model F | −356.546 | 22.9475 | 726.5577 |
| Model M+F vs Model M | −356.546 | 19.13 | 726.5577 |
| Model M+F vs Null Model | −356.546 | 34.423 | 726.5577 |
| Model M vs Model NULL | −366.111 | 15.293 | 738.9543 |
| Model F vs Model NULL | −368.019 | 11.4755 | 742.7718 |
| Model Null | −373.757 |
Table 10. Results of imprinting analysis for SNP mapping to C19MC.
| C19MC site | |||
| SNP | Position | Empirical p-value | Gene |
| rs12608629 | 57617596 | 0.101 | |
| rs12985487 | 57757473 | 0.973 | |
| rs12327640 | 57758705 | 0.089 | |
| rs179320 | 57780245 | 0.293 | ZNF701 |
| rs12974834 | 57855966 | 0.027 | ZNF83 |
| rs12976870 | 57859746 | 0.1 | ZNF83 |
| rs4802981 | 57887238 | 0.564 | ZNF83 |
| rs4802987 | 57925085 | 0.059 | ZNF611 |
| rs4801931 | 57928058 | 0.351 | ZNF611 |
| rs10407762 | 57979913 | 0.543 | ZNF600 |
| rs12461390 | 57997054 | 0.809 | ZNF600 |
| rs12982980 | 58041662 | 0.032 | ZNF468 |
| rs8112177 | 58108780 | 0.004 | ZNF888 |
| rs7251313 | 58116136 | 0.118 | ZNF888 |
| rs12972202 | 58161816 | 0.926 | |
| rs12610001 | 58162603 | 0.917 | |
| rs1650966 | 58166272 | 0.899 | ZNF702P |
| rs7258746 | 58246131 | 0.3 | ERVV-2 |
| rs10405102 | 58262679 | 0.715 | ZNF160 |
| rs7254015 | 58272963 | 0.655 | ZNF160 |
| rs17300167 | 58296859 | 0.537 | ZNF160 |
| rs10423215 | 58340885 | 0.02 | ZNF347 |
| rs4803058 | 58386953 | 0.786 | ZNF665 |
| rs11669754 | 58389180 | 0.45 | ZNF665 |
| rs6509732 | 58394320 | 0.628 | |
| rs11084227 | 58419094 | 0.593 | |
| rs8100275 | 58442327 | 0.004 | ZNF677 |
| rs2965261 | 58467498 | 0.676 | |
| rs4263048 | 58469142 | 0.068 | |
| rs7250240 | 58614327 | 0.002 | ZNF765 |
| rs7258566 | 58704092 | 0.128 | |
| rs12982082 | 58717730 | 0.898 | ZNF331 |
| rs4994351 | 58724856 | 0.468 | ZNF331 |
Rows in bold correspond to p-value<0.05.
Table 11. Results of imprinting analysis for SNP mapping to IGF2-H19.
| IGF2-H19 site | |||
| SNP | position | Empirical p-value | Gene |
| rs965912 | 1900778 | 0.061 | TNNT3 |
| rs6578974 | 2052309 | 0.199 | |
| rs7924768 | 2064648 | 0.038 | |
| rs7926624 | 2085606 | 0.03 | |
| rs1004446 | 2126719 | 0.05 | IGF2 |
Rows in bold correspond to p-value<0.05.
Discussion
In this placental GWAS and candidate gene study of PA, no SNP had a significant association with PA risk following correction for false discovery. The top GWAS hits were rs544201, rs1484464 (CTNNA2), rs4149570 (TNFRSF1A), and rs13055470 (ZNRF3). The top 200 SNPs of the GWAS were in genes of pathways involved in cell cycle, growth and proliferation. The top candidate gene hits were rs16949118 (COX10) and rs7609948 (THRB). Using repeated ten-fold cross-validations, participants in the highest quartiles of WGRS based on SNPs selected from GWAS and candidate gene analyses had 8.40-fold (95% CI: 5.8–12.56) and 4.46-fold (95% CI: 2.94–6.72) higher odds of PA compared to participants in the respective lowest quartiles. We also found evidence of maternal-placental genetic interaction on PA risk for two SNPs in PPARG (chr3∶12313450 and chr3∶12412978) and maternal imprinting effects for multiple SNPs in the C19MC and IGF2/H19 regions.
A number of studies have investigated genetic risk factors of PA. Most of these prior studies were candidate gene studies including investigations of thrombophilia and rennin-angiotensin system pathways, folate metabolism pathways, and interleukin receptor related pathways [24], [28], [29], [51]. Besides inconsistencies between reports of previous associations, few studies evaluated genome-wide variations and PA risk [24], [28], [29], [30], [31], [51]. Given the multi-factorial nature of PA pathogenesis, GWAS studies can potentially provide important information concerning possible genetic susceptibility factors and related novel pathways that play a role in occurrence of PA. To our knowledge, no prior study investigated PA risk and genetic variation in the placenta, where abnormal vasculature, thrombosis, lesions, and reduced perfusion, all culminate in the eventual occurrence of PA [5], [52].
In the current study, we did not have top hits of the GWAS that passed statistical significance after correcting for multiple testing. However, a number of SNPs among the top hits deserve mention. Of note, the top 10 SNPs represented five known genes including CTNNA2, TNFRSF1A, ZNRF3, ACSL1, and LIPA. The CTNNA2 gene codes for a protein linking cadherin adhesion receptors with the cytoskeleton. The SNP we identified in this gene, rs1484464, has been investigated in relation to a number of phenotypes, including tobacco use disorder, smoking cessation, coronary artery disease, lipid metabolism (HDL, total cholesterol, triglycerides) disorders, waist-hip ratio, CRP, glucose levels, insulin resistance, body mass index, blood pressure, and interleukin levels; however, significant associations have not been reported [53]–[62]. The TNFRSF1A is a gene member of the TNFR superfamily that activates the transcription factor NF-KB, mediates apoptosis, and functions as regulators of inflammation. The SNP we identified in the current study has been associated with susceptibility to inflammatory bowel disease, as well as response to anti-TNF treatment, in a large Danish cohort [63], [64]. It has also been associated with reduced expression of TNF alpha receptor [65].
While the ZNRF3 gene is encoding for an E3 ubiquitin-protein ligase that acts as a negative regulator of the Wnt signaling pathway and a tumor suppressor, no previous report, to our knowledge, exists on the ZNRF3 SNP we identified in the current study. The ACSL1 gene codes for an isozyme of the long chain fatty acid coenzyme A ligase family that converts free long-chain fatty acids into fatty acyl-CoA esters, a nuclear-encoded mitochondrial protein, and plays key role in lipid biosynthesis and fatty acid degradation. The rs9997745 SNP in ACSL1 we identified has been previously associated with metabolic syndrome, fasting glucose, insulin levels, and insulin resistance [66]. Similarly, the LIPA gene encodes for lipase A, also known as cholesterol ester hydrolase, an enzyme that functions in the lysosome to catalyze the hydrolysis of cholesteryl esters and triglycerides. The SNP in LIPA, rs792269, is associated with total cholesterol, LDL cholesterol, and triglycerides [67]. None of the other top ten hits in our list had any previous associations reported. Our pathway analyses revealed that genes participating in cell cycle, cell growth and proliferation, and gene expression were overly represented by SNPs that were among the top hit of our GWAS. It is well known that disruptions in underlying normal placental growth and development are key underlying pathways that may later lead to the occurrence of PA [68].
The top hits in the candidate gene association analyses included SNPs in the COX10 (rs16949118) and THRB gene (rs7609948). COX10 encodes for the cytochrome C oxidase protein, the terminal component of the mitochondrial respiratory chain that catalyzes the electron transfer from reduced cytochrome C to oxygen. Genetic variations in this gene have been related to several diseases with underlying mitochondrial dysfunction including Alzheimers’ disease, neurodegenerative diseases and other childhood disorders [69]–[71]. The THRB gene encodes a protein that is a receptor for triidothyronine. Several studies have reported associations between disorders in thyroid (particularly low thyroid levels) and placental disorders including PA [72]–[74]. The SNPs identified in our study, however, have not been associated with phenotypes or clinical outcomes. In addition to these SNPs, several other SNPs in the PPARG gene, belonging to the PPAR-family of genes that have been well-described in relation to placental growth and development, were among the top hits in the candidate gene analyses [75], [76].
We examined associations between GRS, calculated from top hits of the GWAS and candidate gene analyses, respectively, and risk of PA and demonstrated strong associations between GRS and PA risk in both instances. While we did not use separate training and testing samples, we have used a cross-validation approach to protect against overfitting. These preliminary analyses are helpful to summarize identified effects of genetic variations, and could help in the construction of predictive models in future studies [77], [78]. In particular, such genetic prediction have advantages, over non-genetic prediction models, as they are highly stable over time and are more suited for assessment of lifetime risk. In fact, their utility improves over time [78]. In addition, the decreasing genotyping cost and minimal invasiveness associated with obtaining samples highlight the potential importance of genetic prediction scores. On the other hand, the need for large study populations, that comprise of training and testing sets, and identification of genetic variants that individually account for large effects, are potential challenges in this area of research.
In the current study, for two SNPs in PPARG (chr3∶12313450 and chr3∶12412978), models with maternal-genetic interaction on PA risk were found to fit the data best. A number of studies have previously reported interactions between maternal and fetal metabolic genes on maternal and fetal outcomes [79]–[84]. Liang et al. have previously reported significant interaction between maternal and fetal genetic variations at the G308A SNP of TNF-alpha on risk of preterm delivery [85]. Similarly, other investigators have also reported significant maternal-fetal genotype interactions in IL-1beta -511C/T genotype, 4845GG genotype of IL-1alpha, along with the G308A SNP of TNF-alpha on preterm delivery risk [84]. Sinsheimer et al. have particularly stressed that the complex interplay of maternal and fetal genetics can be important for phenotypes originating with the placenta, given the importance of both maternal and fetal related (paternal) risk factors to placenta-based diseases and demonstrated gene expression differences in placenta-related pathologies (e.g. preeclampsia) [83]. Interestingly, the gene that has been highlighted in relation to maternal genetic variation and fetal sex interactions on risk of gestational diabetes is the PPARG gene [81] More specifically, Hocher et al. have demonstrated that mothers carrying G alleles of the Pro12Ala polymorphism delivering a girl had a higher total glycated hemoglobin (6.81) versus mother carrying the same alleles but delivering boys (5.85) (p-value = 0.0015) [81]. However, to our knowledge, no prior study investigated maternal-placental genotype interactions in relation to PA risk. Similarly, placental growth and development is primarily under the control of fetal genes inherited from the father [5], [86]. Imprinted paternal alleles regulate formation of placenta and membranes surrounding the embryo, whereas the development of the embryo itself requires contribution from the maternally derived alleles [5], [86]. Our findings suggest maternal imprinting effects for multiple SNPs in the C19MC and IGF2/H19 regions. While these imprinting sites are well described for several conditions including placental growth and development, our findings are novel in terms of linking imprinting at these sites to PA risk [87], [88].
Given our sample size and related concerns regarding limited available statistical power, in exploratory analyses, we conducted maternal-placental interaction analyses of the PPARG gene using a haplotype-based approach. Tag SNPs from this gene were identified and haplotype blocks were defined using the Haploview software version 4.2 [89], [90]. A total of four haplotype blocks tagged by 29 SNPs were identified. For each haplotype block, three possible diplotypes HH, HH0, H0H0 as described in [80] were defined with the haplotype “H” denoting the “relevant” haplotype and “H0” denoting all other haplotypes. Each haplotype with frequency greater than 0.05 was considered as a potential “relevant” haplotype. For each “relevant” haplotype a procedure similar to the SNP interaction analysis was performed through the EMIM and PREMIM software tools. The “relevant” haplotype with the smallest BIC after evaluating all potential “relevant” haplotypes in each haplotype block was selected. This haplotype was called the optimal “relevant” haplotype. Findings from the exploratory interaction analyses are presented in Table 12. For haplotype block 1 a maternal-only effect was selected whereas for haplotype block 2 there was no evidence of haplotype effect. For haplotype block 3 a placental-only effect was selected. For both haplotype blocks (1 and 3), there was evidence that group 3 had a significantly lower risk of PA compared to the reference group (RR = 0.21, 95% CI: 0.066–0.68 and RR = 0.51, 95% CI: 0.293–0.878, respectively). For haplotype block 4, there was evidence of maternal-placental interaction effect. The group 3 showed a significantly higher risk of PA (RR = 5.71 95% CI: 3.398–9.604) whereas group 5 presented a significantly lower risk (RR = 0.18, 95% CI: 0.037–0.884) compared to the reference group. While findings from these haplotype based interaction analyses are encouraging, caution is warranted in interpreting these results due to uncertainties in the haplotype estimation process and the simplified model that considers diplotypes rather than evaluating all haplotypes using a reference.
Table 12. Results of haplotype-haplotype interaction analysis for haplotype blocks in PPARG gene.
| Haplotype block 1; optimal “relevant” haplotype denoted H | ||
| Maternal Model | Relative Risk | CI (95%) |
| gpRef {H0H0_H0H0; H0H0_H0 H} | 1 | |
| gp2 {H0 H_H0H0; H0 H_H0 H; H0 H_HH} | 0.6912 | (0.386–1.237) |
| gp3 {HH_H0 H; HH_HH} | 0.2123 | (0.066–0.68) |
Diplotypes shown as maternal_placental.
This study expands the literature concerning the genetics of PA in a number of respects. We evaluated placental genetic variations, assessed interactions between maternal and placental genetic variations and examined placental imprinting effects on PA risk. However, several study limitations should be considered when interpreting our findings. First, our GWAS study has limited power to examine associations between genetic variations and disease risk, particularly for SNPs that have low to very low minor allele frequencies. Similarly, our statistical power to detect SNP-SNP interactions on PA risk was limited. To the extent possible we have tried to employ approaches that involve genetic risk scores to assess placental genetic variations contributions to PA risk. Second, many of the identified top hits of our analyses, both in the main effect and interaction models, have not been previously described either in genetic epidemiology or basic science investigations. Therefore, an important next step, along with replication efforts, should involve characterization of functional effects of these variations. Third, our imprinting effect assessment was based on mother-offspring dyad data and could benefit from triad data that also has information on fathers. Fourth, our GRS-based analyses, because of limited statistical power, did not involve independent training and testing sets, which would have been ideal for evaluating their predictive power, but rather relied on repeated ten-fold cross-validations. The WGRS will provide a summary estimate of effects of multiple SNPs, and can provide specific hypotheses that can be tested in future studies. Finally, findings from our study need to be cautiously generalized to other populations with different genetic make-up, population admixture, and environmental risk factors.
In sum, we found that variations in the placental genome and interactions between maternal-placental genetic variations may contribute to PA risk. We reported several novel loci where placental genetic variations may be associated with PA risk as well as several novel loci for maternal-placental genetic interactions on PA risk. Future larger investigations may help advance our understanding of PA pathogenesis.
Supporting Information
ROC curves for 1000 cross-validation replicates of the GWAS-based WGRS model with associated average AUC and 95% CI.
(TIF)
ROC curves for 1000 cross-validation replicates of the candidate gene-based WGRS with associated average AUC and 95% CI.
(TIF)
Data Availability
The authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. Data cannot be made available as they contain identifying information. To access the individual level data, please contact Professor Michelle A. Williams at mawilliams@hsph.harvard.edu.
Funding Statement
This study was supported by grants from the National Institutes of Health, the Eunice Kennedy Shriver National Institute of Child Health and Human Development (R01HD059827) and the National Heart Lung and Blood Institute (K01HL10374). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Leunen K, Hall DR, Odendaal HJ, Grove D (2003) The profile and complications of women with placental abruption and intrauterine death. J Trop Pediatr 49(4):231–234. [DOI] [PubMed] [Google Scholar]
- 2. Odendaal HJ, Hall DR, Grove D (2000) Risk factors for and perinatal mortality of abruptio placentae in patients hospitalised for early onset severe pre-eclampsia - a case controlled study. J Obstet Gynaecol 20(4):358–364. [DOI] [PubMed] [Google Scholar]
- 3. Naeye RL, Harkness WL, Utts J (1977) Abruptio placentae and perinatal death: a prospective study. Am J Obstet Gynecol 128(7):740–746. [DOI] [PubMed] [Google Scholar]
- 4. Naeye RL (1980) Abruptio placentae and placenta previa: frequency, perinatal mortality, and cigarette smoking. Obstet Gynecol 55(6):701–704. [PubMed] [Google Scholar]
- 5. Ananth CV, Wilcox AJ (2001) Placental abruption and perinatal mortality in the United States. Am J Epidemiol 153(4):332–337. [DOI] [PubMed] [Google Scholar]
- 6. Salafia CM, Minior VK, Pezzullo JC, Popek EJ, Rosenkrantz TS, et al. (1995) Intrauterine growth restriction in infants of less than thirty-two weeks' gestation: associated placental pathologic features. Am J Obstet Gynecol 173(4):1049–1057. [DOI] [PubMed] [Google Scholar]
- 7. Spinillo A, Fazzi E, Stronati M, Ometto A, Iasci A, et al. (1993) Severity of abruptio placentae and neurodevelopmental outcome in low birth weight infants. Early Hum Dev 35(1):45–54. [DOI] [PubMed] [Google Scholar]
- 8. Williams MA, Lieberman E, Mittendorf R, Monson RR, Schoenbaum SC (1991) Risk factors for abruptio placentae. Am J Epidemiol 134(9):965–972. [DOI] [PubMed] [Google Scholar]
- 9. Ananth CV, Savitz DA, Luther ER (1996) Maternal cigarette smoking as a risk factor for placental abruption, placenta previa, and uterine bleeding in pregnancy. Am J Epidemiol 144(9):881–889. [DOI] [PubMed] [Google Scholar]
- 10. Younis JS, Samueloff A (2003) Gestational vascular complications. Best Pract Res Clin Haematol 16(2):135–151. [DOI] [PubMed] [Google Scholar]
- 11. Sanchez SE, Pacora PN, Farfan JH, Fernandez A, Qiu C, et al. (2006) Risk factors of abruptio placentae among Peruvian women. Am J Obstet Gynecol 194(1):225–230. [DOI] [PubMed] [Google Scholar]
- 12. Oyelese Y, Ananth CV (2006) Placental abruption. Obstet Gynecol 108(4):1005–1016. [DOI] [PubMed] [Google Scholar]
- 13. Ananth CV, Peltier MR, Chavez MR, Kirby RS, Getahun D, et al. (2007) Recurrence of ischemic placental disease. Obstet Gynecol 110(1):128–33. [DOI] [PubMed] [Google Scholar]
- 14. Duthie SJ, King PA, To WK, Lopes A, Ma HK (1991) A case controlled study of pregnancy complicated by severe maternal anaemia. Aust N Z J Obstet Gynaecol 31(2):125–137. [DOI] [PubMed] [Google Scholar]
- 15. Arnold DL, Williams MA, Miller RS, Qiu C, Sorensen TK (2009) Iron deficiency anemia, cigarette smoking and risk of abruptio placentae. J Obstet Gynaecol Res. 35(3):446–452. [DOI] [PubMed] [Google Scholar]
- 16. Ray JG, Laskin CA (1999) Folic acid and homocyst(e)ine metabolic defects and the risk of placental abruption, pre-eclampsia and spontaneous pregnancy loss: A systematic review. Placenta 20(7):519–529. [DOI] [PubMed] [Google Scholar]
- 17. de Paz NC, Sanchez SE, Huaman LE, Chang GD, Pacora PN, et al. (2011) Risk of placental abruption in relation to maternal depressive, anxiety and stress symptoms. J Affect Disord. 130(1–2):280–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez SE, Williams MA, Pacora PN, Ananth CV, Qiu C, et al. (2010) Risk of placental abruption in relation to migraines and headaches. BMC Womens Health doi: 10.1186/1472-6874-10-30. [DOI] [PMC free article] [PubMed]
- 19. Ananth CV, Oyelese Y, Srinivas N, Yeo L, Vintzileos AM (2004) Preterm premature rupture of membranes, intrauterine infection, and oligohydramnios: risk factors for placental abruption. Obstet Gynecol 104(1):71–7. [DOI] [PubMed] [Google Scholar]
- 20. Williams MA, Sanchez SE, Ananth CV, Hevner K, Qiu C, et al. (2013) Maternal blood mitochondrial DNA copy number and placental abruption risk: results from a preliminary study. Int J Mol Epidemiol Genet 4(2):120–127. [PMC free article] [PubMed] [Google Scholar]
- 21. Signore C, Mills JL, Qian C, Yu K, Lam C, et al. (2006) Circulating angiogenic factors and placental abruption. Obstet Gynecol 108(2):338–344. [DOI] [PubMed] [Google Scholar]
- 22. Ananth CV, Peltier MR, Kinzler WL, Smulian JC, Vintzileos AM (2007) Chronic hypertension and risk of placental abruption: is the association modified by ischemic placental disease. Am J Obstet Gynecol e197(3) 273:e1–7. [DOI] [PubMed] [Google Scholar]
- 23. Iams J (1998) Prevention of preterm birth. N Engl J Med 338(1):54–66. [DOI] [PubMed] [Google Scholar]
- 24. Toivonen S, Keski-Nisula L, Saarikoski S, Heinonen S (2004) Risk of placental abruption in first-degree relatives of index patients. Clin Genet 66(3):244–246. [DOI] [PubMed] [Google Scholar]
- 25. Dizon-Townson D, Miller C, Sibai B, Spong CY, Thom E, et al. (2005) The relationship of the factor V Leiden mutation and pregnancy outcomes for mother and fetus. Obstet Gynecol 106(3):517–524. [DOI] [PubMed] [Google Scholar]
- 26. Jaaskelainen E, Toivonen S, Romppanen EL, Helisalmi S, Keski-Nisula L, et al. (2004) M385T polymorphism in the factor V gene, but not Leiden mutation, is associated with placental abruption in Finnish women. Placenta 25(8–9):730–734. [DOI] [PubMed] [Google Scholar]
- 27. Jarvenpaa J, Pakkila M, Savolainen ER, Perheentupa A, Jarvela I, et al. (2006) Evaluation of Factor V Leiden, Prothrombin and Methylenetetrahydrofolate Reductase Gene Mutations in Patients with Severe Pregnancy Complications in Northern Finland. Gynecol Obstet Invest 62(1):28–32. [DOI] [PubMed] [Google Scholar]
- 28. Gargano JW, Holzman CB, Senagore PK, Reuss ML, Pathak DR, et al. (2009) Polymorphisms in thrombophilia and renin-angiotensin system pathways, preterm delivery, and evidence of placental hemorrhage. Am J Obstet Gynecol 201(3) 317:e1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zdoukopoulos N, Zintzaras E (2008) Genetic risk factors for placental abruption: a HuGE review and meta-analysis. Epidemiology 19(2):309–323. [DOI] [PubMed] [Google Scholar]
- 30. Moore A, Enquobahrie DA, Sanchez SE, Ananth CV, Pacora PN, et al. (2012) A genome-wide association study of variations in maternal cardiometabolic genes and risk of placental abruption. Int J Mol Epidemiol Genet 3:305–313. [PMC free article] [PubMed] [Google Scholar]
- 31. Workalemahu T, Enquobahrie DA, Moore A, Sanchez SE, Ananth CV, et al. (2013) Genome-wide and candidate gene association studies of placental abruption. Int J Mol Epidemiol Genet 4:128–139. [PMC free article] [PubMed] [Google Scholar]
- 32. Gargano JW, Holzman CB, Senagore PK, Reuss ML, Pathak DR, et al. (2010) Evidence of placental haemorrhage and preterm delivery. BJOG 117(4):445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lyssenko V, Laakso M (2013) Genetic screening for the risk of type 2 diabetes: worthless or valuable? Diabetes Care 36(2):S120–S126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, et al. (2012) The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. PLoS Genet 8(8): e1002793+. [DOI] [PMC free article] [PubMed]
- 35.Crawford DC, Goodloe R, Brown-Gentry K, Wilson S, Roberson J, et al. (2013) Characterization of the Metabochip in diverse populations from the International HapMap Project in the Epidemiologic Architecture for Genes Linked to Environment (EAGLE) project. PacSympBiocomput 188–199. [PMC free article] [PubMed]
- 36. Benjamini Y, Yekutieli D (2001) The Control of the False Discovery Rate in Multiple Testing Under Dependency. Ann Statist 29:1165–1188. [Google Scholar]
- 37. Ayers KL, Cordell HJ (2010) SNP selection in genome-wide and candidate gene studies via penalized logistic regression. Genetic epidemiology 34:879–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoggart CJ, Whittaker JC, Iorio MD, Balding DJ (2008) Simultaneous analysis of all SNPs in genome-wide and re-sequencing studies. PLoS Genet doi: 10.1371/journal.pgen.1000130. [DOI] [PMC free article] [PubMed]
- 39. Tibshirani R (1996) Regression shrinkage and selection via the lasso. J Roy Stat Soc B 58:267–288 10.1111/j.1467-9868.2011.00771.x [DOI] [Google Scholar]
- 40. Zhou H, Sehl ME, Sinsheimer JS, Lange K (2010) Association screening of common and rare genetic variants by penalized regression. Bioinformatics 26(19):2375–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Breheny P, Huang J (2009) Penalized methods for bi-level variable selection. Statistics and its interface 2:369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang J, Breheny P, Ma S (2012) A selective review of group selection in high dimensional models. Statistical Science 27:481–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing data inference for whole genome association studies using localized haplotype clustering. Am J Hum Genet 81:1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kundu S, Aulchenko YS, van Duijn CM, Janssens ACJW (2011) PredictABEL: an R package for the assessment of risk prediction models. Eur J Epidemiol 26:261–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burgess S, Thompson SG (2013) Use of allele scores as instrumental variables for Mendelian randomization. Int J Epidemiol 42(4):1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ainsworth HF, Unwin J, Jamison DL, Cordell HJ (2011) Investigation of maternal effects, maternal-fetal interactions and parent-of-origin effects (imprinting), using mothers and their offspring. Genetic Epidemiology 35:19–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howey R, Cordell H (2012) PREMIM and EMIM: Tools for estimation of maternal, imprinting and interaction effects using multinomial modeling. BMC Bioinformatics doi: 10.1186/1471-2105-13-149. [DOI] [PMC free article] [PubMed]
- 48. Sinsheimer JS, Palmer CGS, Woodward JA (2003) Detecting genotype combinations that increase risk for disease: maternal-fetal genotype incompatibility test. Genet Epidemiol 24:1–13. [DOI] [PubMed] [Google Scholar]
- 49. Weinberg CR, Wilcox AJ, Lie RT (1998) A log-linear approach to case-parent-triad data: assessing effects of disease genes that act either directly or through maternal effects and that may be subject to parental imprinting. Am J Hum Genet 62:969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Breheny P, Huang J (2011) Coordinate descent algorithms for nonconvex penalized regression, with applications to biological feature selection. Ann Appl Statist 5:232–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ananth CV, Peltier MR, Moore DF, Kinzler WL, Leclerc D, et al. (2008) Reduced folate carrier 80A–>G polymorphism, plasma folate, and risk of placental abruption. Hum Genet 124(2):137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ananth CV, Oyelese Y, Prasad V, Getahun D, Smulian JC (2006) Evidence of placental abruption as a chronic process: associations with vaginal bleeding early in pregnancy and placental lesions. Eur J Obstet Gynecol Reprod Biol 128(1–2):15–21. [DOI] [PubMed] [Google Scholar]
- 53. Uhl GR, Liu QR, Drgon T, Johnson C, Walther D, et al. (2008) Molecular genetics of successful smoking cessation: convergent genome-wide association study results. Archives of general psychiatry 65(6):683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Uhl GR, Drgon T, Johnson C, Walther D, David SP, et al. (2010) Genome-wide association for smoking cessation success: participants in the Patch in Practice trial of nicotine replacement. Pharmacogenomics 11(3):357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bailey SD, Xie C, Do R, Montpetit A, Diaz R, et al. (2010) Variation at the NFATC2 locus increases the risk of thiazolidinedione-induced edema in the Diabetes REduction Assessment with ramipril and rosiglitazone Medication (DREAM) study. Diabetes Care 33(10):2250–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Benjamin EJ1, Dupuis J, Larson MG, Lunetta KL, Booth SL et al. (2007) Genome-wide association with select biomarker traits in the Framingham Heart Study. BMC Med Genet 19 8 Suppl 1: S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Meigs JB, Manning AK, Fox CS, Florez JC, Liu C, et al. (2007) Genome-wide association with diabetes-related traits in the Framingham Heart Study. BMC Med Genet 19 8 Suppl 1: S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kathiresan S, Manning AK, Demissie S, D'Agostino RB, Surti A, et al. (2007) A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet 19 8 Suppl 1: S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fox CS, Heard-Costa N, Cupples LA, Dupuis J, Vasan RS, et al. (2007) Genome-wide association to body mass index and waist circumference: the Framingham Heart Study 100K project. BMC Med Genet 19 8 Suppl 1: S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Levy D, Larson MG, Benjamin EJ, Newton-Cheh C, Wang TJ, et al. (2007) Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet 19 8 Suppl 1: S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wilk JB, Walter RE, Laramie JM, Gottlieb DJ, O'Connor GT (2007) Framingham Heart Study genome-wide association: results for pulmonary function measures. BMC Med Genet 19 8 Suppl 1: S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Melzer D, Perry JR, Hernandez D, Corsi AM, Stevens K, et al. (2008) A genome-wide association study identifies protein quantitative trait loci (pQTLs). PLoS Genet doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed]
- 63.Bank S, Skytt Andersen P, Burisch J, Pedersen N, Roug S, et al. (2014) Polymorphisms in the inflammatory pathway genes TLR2, TLR4, TLR9, LY96, NFKBIA, NFKB1, TNFA, TNFRSF1A, IL6R, IL10, IL23R, PTPN22, and PPARG are associated with susceptibility of inflammatory bowel disease in a Danish cohort. PLoS One doi: 10.1371/journal.pone.0098815. [DOI] [PMC free article] [PubMed]
- 64.Bank S, Andersen PS, Burisch J, Pedersen N, Roug S, et al. (2014) Associations between functional polymorphisms in the NFκB signaling pathway and response to anti-TNF treatment in Danish patients with inflammatory bowel disease. Pharmacogenomics J:doi: 10.1038/tpj.2014.19. [DOI] [PubMed]
- 65.Sennikov SV, Vasilyev FF, Lopatnikova JA, Shkaruba NS, Silkov AN (2014) Polymorphisms in the tumor necrosis factor receptor genes affect the expression levels of membrane-bound type I and type II receptors. Mediators Inflamm doi: 10.1155/2014/745909. [DOI] [PMC free article] [PubMed]
- 66. Phillips CM, Goumidi L, Bertrais S, Field MR, Cupples LA, et al. (2010) Gene-nutrient interactions with dietary fat modulate the association between genetic variation of the ACSL1 gene and metabolic syndrome. J Lipid Res 51(7):1793–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, et al. (2013) Discovery and refinement of loci associated with lipid levels. Nature Genetics 45(11):1274–1283 Available: http://www.sph.umich.edu/csg/abecasis/public/lipids2013/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Di Simone N, Maggiano N, Caliandro D, Riccardi P, Evangelista A, et al. (2003) Homocysteine induces trophoblast cell death with apoptotic features. Biol Reprod 69(4):1129–1134. [DOI] [PubMed] [Google Scholar]
- 69. Pitceathly RD, Taanman JW, Rahman S, Meunier B, Sadowski M, et al. (2013) COX10 mutations resulting in complex multisystem mitochondrial disease that remains stable into adulthood. JAMA Neurol 70(12):1556–1561. [DOI] [PubMed] [Google Scholar]
- 70. Fukui H, Diaz F, Garcia S, Moraes CT (2007) Cytochrome c oxidase deficiency in neurons decreases both oxidative stress and amyloid formation in a mouse model of Alzheimer's disease. Proc Natl Acad Sci USA 104(35):14163–14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Darin N, Moslemi AR, Lebon S, Rustin P, Holme E, et al. (2003) Genotypes and clinical phenotypes in children with cytochrome-c oxidase deficiency. Neuropediatrics 34(6):311–317. [DOI] [PubMed] [Google Scholar]
- 72. Breathnach FM, Donnelly J, Cooley SM, Geary M, Malone FD (2013) Subclinical hypothyroidism as a risk factor for placental abruption: evidence from a low-risk primigravid population. Aust N Z J Obstet Gynaecol 53(6):553–560. [DOI] [PubMed] [Google Scholar]
- 73. Männistö T, Mendola P, Grewal J, Xie Y, Chen Z, et al. (2013) Thyroid diseases and adverse pregnancy outcomes in a contemporary US cohort. J Clin Endocrinol Meta 98(7):2725–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Haddow JE, McClain MR, Palomaki GE, Neveux LM, Lambert-Messerlian G, et al. (2011) Thyroperoxidase and thyroglobulin antibodies in early pregnancy and placental abruption. Obstet Gynecol 117(2 Pt 1):287–292. [DOI] [PubMed] [Google Scholar]
- 75. Wieser F, Waite L, Depoix C, Taylor RN (2008) PPAR Action in Human Placental Development and Pregnancy and Its Complications. PPAR Res 2008:527048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Borel V, Gallot D, Marceau G, Sapin V, Blanchon L (2008) Placental implications of peroxisome proliferator-activated receptors in gestation and parturition. PPAR Res 2008:758562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dudbridge F (2013) Power and predictive accuracy of polygenic risk scores. PLoS Genet doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed]
- 78.Jostins L, Barrett JC (2011) Genetic risk prediction in complex disease. Hum Mol Genet doi: 10.1093/hmg/ddr378. [DOI] [PMC free article] [PubMed]
- 79. Lupo PJ, Mitchell LE, Canfield MA, Shaw GM, Olshan AF, et al. (2014) Maternal-fetal metabolic gene-gene interactions and risk of neural tube defects. Mol Genet Metab 111(1):46–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li M, Erickson SW, Hobbs CA, Li J, Tang X, et al. (2014) Detecting maternal-fetal genotype interactions associated with conotruncal heart defects: a haplotype-based analysis with penalized logistic regression. Genet Epidemiol 38(3):198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Hocher B, Schlemm L, Haumann H, Poralla C, Chen YP, et al. (2010) Interaction of maternal peroxisome proliferator-activated receptor gamma2 Pro12Ala polymorphism with fetal sex affects maternal glycemic control during pregnancy. Pharmacogenet Genomics 20(2):139–142. [DOI] [PubMed] [Google Scholar]
- 82.Liang M, Wang X, Li J, Yang F, Fang Z, et al. (2010) Association of combined maternal-fetal TNF-alpha gene G308A genotypes with preterm delivery: a gene-gene interaction study. J Biomed Biotechnol doi: 10.1155/2010/396184. [DOI] [PMC free article] [PubMed]
- 83. Sinsheimer JS, Elston RC, Fu WJ (2010) Gene-Gene Interaction in Maternal and Perinatal Research. Journal of Biomedicine and Biotechnology 2010:4597–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yılmaz Y, Verdi H, Taneri A, Yazıcı AC, Ecevit AN, et al. (2012) Maternal-fetal proinflammatory cytokine gene polymorphism and preterm birth. DNA Cell Biol 31(1):92–97. [DOI] [PubMed] [Google Scholar]
- 85.Liang M, Wang X, Li J, Yang F, Fang Z, et al. (2010) Association of combined maternal-fetal TNF-alpha gene G308A genotypes with preterm delivery: a gene-gene interaction study. J Biomed Biotechnol doi: 10.1155/2010/396184. [DOI] [PMC free article] [PubMed]
- 86. Bartolomei MS, Tilghman SM (1997) Genomic imprinting in mammals. Annu Rev Genet 31:493–525. [DOI] [PubMed] [Google Scholar]
- 87. Noguer-Dance M, Abu-Amero S, Al-Khtib M, Lefèvre A, Coullin P, et al. (2010) The primate-specific microRNA gene cluster (C19MC) is imprinted in the placenta. Hum Mol Genet 19:3566–3582. [DOI] [PubMed] [Google Scholar]
- 88. Ishida M, Moore GE (2013) The role of imprinted genes in humans. Mol Aspects Med 34:826–840. [DOI] [PubMed] [Google Scholar]
- 89. Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. [DOI] [PubMed] [Google Scholar]
- 90. De Bakker PIW, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, et al. (2005) Efficiency and power in genetic association studies. Nature Genetics 37:1217–1223. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ROC curves for 1000 cross-validation replicates of the GWAS-based WGRS model with associated average AUC and 95% CI.
(TIF)
ROC curves for 1000 cross-validation replicates of the candidate gene-based WGRS with associated average AUC and 95% CI.
(TIF)
Data Availability Statement
The authors confirm that, for approved reasons, some access restrictions apply to the data underlying the findings. Data cannot be made available as they contain identifying information. To access the individual level data, please contact Professor Michelle A. Williams at mawilliams@hsph.harvard.edu.