Abstract
Purpose
The developing lens expresses at least three different FGF receptor genes (Fgfr1-3) and FGFs have been shown to induce lens epithelial cells to differentiate into fiber cells both in vitro and in vivo. While the loss of Fgfr2 alone does not prevent fiber differentiation and the loss of Fgfr3 alone does not appear to affect lens development the independent role of Fgfr1 in lens development has not been reported. These experiments were conducted to determine if Fgfr1 plays an independent essential role in lens development.
Methods
To address this question, we took two complementary approaches. First, we employed the aphakia (ak) lens complementation system to show that Fgfr1-deficient embryonic stem (ES) cells were able to form a normal embryonic lens that maintains a normal pattern of crystallin gene expression. Second, we employed the Cre-loxP system to achieve lens-specific inactivation of Fgfr1.
Results
Fgfr1-null embryonic stem cells were able to rescue normal embryonic lens development in chimeric combination with aphakia mutant embryos. In addition, conditional deletion of Fgfr1 does not compromise lens development either before or after birth.
Conclusions
The results of both approaches suggest that lens epithelial cell integrity, cell cycle regulation and lens fiber differentiation are intact in the Fgfr1-deficient lens. Overall, our results demonstrate that Fgfr1 is not cell autonomously essential for lens development and suggests functional redundancy among different FGFRs with respect to lens fiber differentiation.
INTRODUCTION
The vertebrate lens arises from head surface ectoderm in response to a series of inductive interactions [reviewed in 1]. Lens development begins with the thickening of surface ectoderm cells overlying the optic vesicle to form a lens placode, which subsequently invaginates to form the lens pit. The lens pit then fuses and detaches from the surface ectoderm to form the initially hollow lens vesicle. Cells lining the posterior part of the lens vesicle withdraw from the cell cycle and elongate to form the primary lens fiber cells that obliterate the vesicle cavity, while the anterior lens vesicle cells remain as the lens epithelium. Secondary lens fiber cells subsequently withdraw from the cell cycle and differentiate from the anterior epithelial cells as they are displaced toward the equatorial region of the lens by continuous lens epithelial proliferation. Both primary and secondary fiber cells lose their nuclei and cellular organelles upon maturation.
Much recent effort has focused on elucidating the molecular mechanisms of lens fiber differentiation, and several pieces of evidence supports the hypothesis that fibroblast growth factor (FGF) signaling plays an important role in this process. Members of the FGF family exhibit dynamic spatio-temporal expression patterns in ocular tissues, suggesting that they may be important for both morphogenesis and maintenance of the lens via both autocrine and paracrine mechanisms [2-9]. Furthermore, FGFs were shown to stimulate explants of lens epithelial cells from neonatal rats to proliferate, migrate, and differentiate into lens fibers in a dose-dependent manner, a process reminiscent of endogenous lens fiber differentiation [10, 11]. FGFs also stimulate these responses in lens epithelial cells from adult animals, though with less potency, suggesting that FGFs may continue to induce lens fiber cell differentiation throughout life [12]. More importantly, in vivo studies using transgenic mice demonstrated that lens-specific expression of several different FGF ligands induced lens epithelial cells to exit from the cell cycle prematurely and undergo ectopic fiber differentiation [13-15].
Different members of FGF receptor family (Fgfr1-3) show distinct spatio-temporal expression patterns during lens development, suggesting that they may play distinct roles in lens fiber differentiation. Fgfr3 is expressed in both epithelial and fiber cells [16]. The Fgfr2 IIIcisoform is expressed throughout the lens epithelium, transitional zone and at a lower level in maturing fibers while the Fgfr2 IIIb isoform is weakly expressed in the lens epithelium but is upregulated in transitional zone where epithelial cells are committed to fiber differentiation [16]. During early lens development, Fgfr1 is first detected in lens vesicle and cornea, but its expression level increases with the onset of primary fiber cell differentiation. Later, Fgfr1 expression is highly upregulated in newly differentiated lens fiber cells [17, 18]. Transgenic mice expressing secreted dimers or truncated (dominant negative) versions of FGFRs show defects in lens growth and differentiation [19-22]. Furthermore, transgenic overexpression of a dominant-negative Fgfr1 in the presumptive lens ectoderm perturbs early lens morphogenesis, suggesting that FGF receptor signaling might be required at the lens induction stage as well [23]. However, the functional specificity of individual FGF receptors during lens development is poorly understood. Fgfr3-null mice are viable and do not exhibit lens defects [24]. Fgfr2 plays an essential role in the proper development of the trophoblast, and as a result, Fgfr2-null embryos die before the onset of lens fiber differentiation [25, 26]. This embryonic lethality can be rescued by making aggregation chimeras with tetraploid wild-type embryos and Fgfr2-deficient diploid embryos. Using this approach, Li et al showed that Fgfr2-deficient mice survive to birth with lenses that clearly undergo morphological aspects of fiber cell differentiation [27]. In addition, the lens is not among the many tissues affected by inactivation of the Fgfr2 IIIb isoform [28]. More recent experiments, utilizing a conditional mutant allele of Fgfr2 [29] and the LeCre mouse strain where Cre expression initiates in the ocular surface ectoderm at the lens placode stage [30] demonstrated that early inactivation of Fgfr2 leads to increased lens cell apoptosis and inefficient cell cycle withdrawal upon fiber differentiation [31]. Mouse embryos homozygous for null mutations in Fgfr1 die shortly after gastrulation as a result of defects in mesoderm patterning [32-34]. Mice harboring hypomorphic Fgfr1 alleles can survive to birth without lens defects, but the presence of ~20% wild-type Fgfr1 transcripts in these mice made it difficult to evaluate the role of Fgfr1 in lens fiber differentiation [35].
To determine whether Fgfr1 is specifically important for lens development, we utilized two complementary approaches. First, we took advantage of the aphakia (ak) lens complementation system that has been used successfully for evaluating the role of Rb1 in lens development [36]. The recessive aphakia (ak) mutation is characterized by an arrest of lens development shortly after lens pit formation as the result of a dramatic reduction in the expression of the Pitx3 transcription factor [37, 38]. The cell-autonomous nature of the lens defect in ak/ak mice, makes cells from these embryos unable to contribute to normal lens development in chimeric mice. Therefore in ak/ak ↔ ES cell chimeras, lens fiber development is entirely dependent on the ES cell contribution [36]. We demonstrated that Fgfr1-null embryonic stem (ES) cells are capable of forming lenses with normal morphology that also maintained normal expression patterns of lens crystallins, thus rescuing the lens deficit in aphakia mice. Second, we employed the Cre-loxP system to inactivate Fgfr1 in a tissue-specific manner using a transgenic mouse line that we generated, MLR10 [39], which expresses Cre recombinase from the lens vesicle stage of lens development, and a conditional mutant allele of Fgfr1 created by the insertion of loxP sites in introns 7 and 15, flanking the exons responsible for the transmembrane domain and the majority of the tyrosine kinase domain of the receptor [40]. Morphological and gene expression analyses of mice with lens-specific inactivation of Fgfr1 showed normal embryonic and postnatal lens development. Therefore, Fgfr1 is not independently essential in the lens developmental lineage for lens fiber differentiation.
METHODS
All animal experiments conformed to the ARVO Statement for Use of Animals in Ophthalmic and Vision Research and were also performed according to protocols approved by the Columbus Children's Hospital Institutional Animal Care and Use Committee.
Generation and analysis of ak/ak ↔ Fgfr1Δtmk/Δtmk chimeras
The Fgfr1Δtmk/+ ES cell clone C16 and the Fgfr1Δtmk/Δtmk ES cell clone C5B were generated as previously described [33]. The lens complementation system developed by Liegeois et al was used to generate ak/ak ↔ Fgfr1Δtmk/Δtmk chimeras [36]. Briefly, ES cells were either injected into homozygous aphakia (ak/ak) blastocysts or aggregated with morula stage ak/ak embryos before transfer back to pseudopregnant females. The embryos were then allowed to develop to embryonic day 14.5 (E14.5) when they were collected for analysis. Pregnant females carrying the ak/ak ↔ Fgfr1 ES cell chimeras were euthanized and embryos were removed, fixed and stained for β-galactosidase activity with X-gal as described [13, 39].
Generation of transgenic mice and genotyping
MLR10 transgenic mice, described previously [39], were made on an FVB/N inbred background and crossed to mice carrying the null (Fgfr1Δtmk) and conditional (Fgfr1flox) alleles of Fgfr1 maintained on genetic background that was a mixture of 129SV, C57BL/6 and FVB/N alleles. The Fgfr1Δtmk allele was detected by PCR as described [32]. The Cre transgene in MLR10 was detected by PCR with primers PR4 (5’-GCATTCCAGCTGCTGACGGT-3’) and CRE-AS (5’-CAGCCCGGACCGACGATGAAG-3’) that generate a DNA fragment of 577bp. The Fgfr1flox allele was constructed as described [40] and amplified with primers Fgfr1 i75’ (5’-AGGTTCCCTCCTCTTGGATGA-3’) and Fgfr1 i73’: (5’-CTGGGTCAGTGTGGACAGTGT-3’) that produce a PCR product of 332bp. The thermal cycling condition: 95°C for 30 seconds, 56°C for 30 seconds, 72°C for 40 seconds, 35 cycles.
Histology, immunofluorescence and in-situ Hybridization
Embryos and adult eyes were fixed in 4% paraformaldehyde overnight at 4°C, processed and embedded in paraffin. Embedded samples were sectioned at 5 μm. For immunofluorescence, paraffin-embedded ocular tissue sections on slides were deparaffinized and rehydrated in an ethanol series ending in PBS. After blocking with 0.5% bovine serum albumin and 1% Triton X-100 for 30 minutes at room temperature, tissue sections were incubated with antibodies to α-crystallins (1:3000 dilution), β-crystallins (1:3000 dilution) or γ-crystallins (1:5000 dilution) (provided by Dr. J. Samuel Zigler Jr., National Eye Institute, Bethesda, MD) at 4°C overnight. After brief washes, the slides were incubated with Cy-2 or Cy-3 labeled secondary anti-rabbit antibodies (Jackson Immunoresearch Laboratories, West Grove, PA) for 1 hour at room temperature. The sections were then counterstained with DAPI (Vector Laboratories, Burlingame, CA).
Radioactive in-situ hybridization on sections of E14.5 embryos with riboprobes specific for FoxE3 (from Dr. Milan Jamrich, Baylor College of Medicine, Houston, TX), Pax6, Six3, Prox1, p57kip2, c-Maf, and Sox1 (kindly provided by Dr. Paul Overbeek, Baylor College of Medicine) was performed as described previously [20]. At least four mutant lenses and four control lenses were hybridized with each probe. After the hybridization was complete, the slides were exposed to Kodak NTB-2 emulsion (Kodak, Rochester, NY) for 4-6 days at 4°C before development.
RNAse protection assay
Total RNA was extracted from 8 to 10 mouse lenses for each genotype at postnatal day 30 using TRIZOL (Invitrogen, Carlsbad, CA). A 415bp fragment of Fgfr1 cDNA containing exonic sequences from the region surrounding the second loxP site in the Fgfr1flox allele (nucleotides 1990-2404 of GenBank accession number NM010206) was used as probe for Fgfr1 expression in an RNase protection assay. A 126bp fragment of hypoxanthineguanine phosphoribosyl transferase (Hprt) cDNA (nucleotides 116-241 of GenBank accession number NM013556) was used as probe for internal control. The RNAse protection assays were performed using RPAIII kit (Ambion, Austin, TX) according to manufacturer's instructions. The alternatively spliced Fgfr1 transcript from the Ffgr1Δtmk allele was amplified with primers Fgfr1e55’ (5’-GAATTGGAGGCTACAAGGTTCGCTATGC-3’) and Fgfr1e163’ (5’-TGTCGGGAAAGCTGGGTGAGTACTGG-3’), and the amplified band was gel purified and sequenced to reveal a splice junction between exons 7 and 15.
Relative Real Time RT-PCR assay
Lenses from wildtype and MLR10 Fgfr1flox/flox mice were isolated at postnatal day 17 and separated into lens fiber and epithelial fractions by peeling off and trimming lens capsules with adherent epithelia. Lens fractions were pooled from 8 lenses from each genotype. Total RNA for each fraction was isolated as described above. cDNA was synthesized from 5μg of lens epithelium or fiber cell total RNA and mixed with 200ng of random hexamers (Promega, Madison, WI) and 10nmol of dNTPs. The mixture was heated to 60°C for 5 minutes before addition of 40 units of RNASEOUT RNAse inhibitor and 15 units of SUPERSCRIPTII reverse transcriptase (Invitrogen). The reaction was carried out in 50mM Tris-HCl (pH 8.3), 75mM KCl, 5mM MgCl2 and 10mM DTT at 25°C for 10 minutes followed by 50 minutes at 42°C. Then the reaction was heated to 70°C for 15 minutes followed by incubation with 20 units of RNase H (Invitrogen) at 37°C for 30minutes. Real-time relative RTPCR reactions included cDNA prepared from isolated lens fiber or lens epithelial RNA, 0.5 uM each of forward and reverse primers in QuantiTect SYBR Green PCR Master Mix (Qiagen Inc.) in a total volume of 25 ul. Quantitative PCR amplification was performed using ABI Prism 7700 Sequence Detection System equipped with Sequence Detector v1.6.3 software (Perkin-Elmer Applied Biosystems Inc., Foster City, CA) for data acquisition and analysis. Wild type Fgfr1 transcripts were amplified with primers Fgfr1e95’ (5’-GAATTGGAGGCTACAAGGTTCGCTATGC-3’) and Fgfr1e103’ (5’-TGTCGGGAAAG CTGGGTGAGTACTGG-3’). Cycling parameters for Fgfr1 transcripts were: 94°C for 1 minute and 20 seconds, 65°C for 1 minutes, 72°C for 1 minute and 30 seconds, 35 cycles. As internal control for RT-PCR, Hprt transcripts were amplified with primers HprtE2S from exon 2 (5’-AGCGATGATGAACCAGGTTA-3’) and HprtE4AS from exon 4 (5’-GTTGAGAGATCATC TCCACC-3’). Hprt transcripts were amplified under following conditions: 95°C for 30 seconds, 57°C for 30 seconds, 72°C for 1 minute, 35 cycles. Amplification of correct PCR products was evaluated by agarose gel electrophoresis and further confirmed by sequencing analysis with dye terminator and ABI 373 Sequencer (Perkin-Elmer Applied Biosystems Inc.). Amplification reactions of serial dilutions of plasmids carrying cDNA fragments encompassing these primers were conducted in parallel to generate standard curves for each primer pair. The relative quantity of transcripts in different cDNA samples was calculated using this standard curve. And the quantity of Fgfr1 transcripts was normalized against the amount of Hprt transcripts in the same sample. The quantitation was carried out in triplicate. To assess the purity of the epithelial RNA fraction, relative real-time RT-PCR was carried out as described above to compare the expression of γD-crystallin in the epithelial and fiber RNA samples using primers GUNIVF (5’-CTGCTGGATGCTCTATGAGC-3’) and GDR (5’-TTCCGTGAACTCTATCACTTGGC -3’) as described [41].
RESULTS
Lens complementation assay with ak/ak ↔ Fgfr1Δtmk/Δtmk chimeras
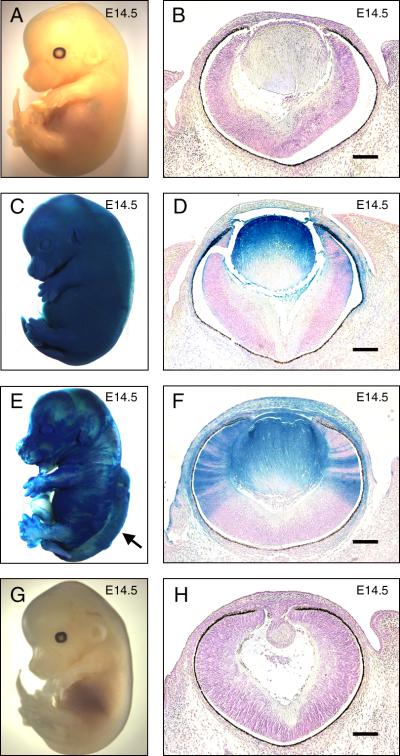
Since Fgfr1Δtmk/Δtmk (homozygous Fgfr1-null) embryos die shortly after gastrulation, ES cell clones carrying the Rosa β-geo26 (Rosa26) transgene [42] and either heterozygous or homozygous for Fgfr1Δtmk allele [33] were used in a chimeric analysis with ak/ak embryos to test the ability of Fgfr1-null cells to contribute to normal lens development. X-gal staining for LacZ activity revealed the ES cell contributions in the chimeras (Figure 1). Fgfr1+/+ ak/ak cells occasionally rescued the perigastrulation lethality of Fgfr1 deficiency in the chimeras and allowed us to evaluate the lens-forming capacity of Fgfr1Δtmk/Δtmk ES cells (Table 1). We subjectively designated anything over 20% overall ES cell contribution to the chimeric embryo as high degree chimeras. Seven ak/ak ↔Fgfr1Δtmk/+ embryos were generated, four of which showed high degree chimerism and complete rescue of the ak/ak lens deficit (Figure 1C). In these embryos, the morphology of lenses derived from ES cells was indistinguishable from wild-type controls, indicating complete rescue of lens defects relative to ak/ak embryos (Figure 1B, D, H). Two high percentage ak/ak↔Fgfr1Δtmk/+ chimeras were born and survived to adulthood, and both displayed, by gross observation, normal development of the lens and other eye structures compared to adult ak/ak mice (data not shown). In contrast, a significant portion of chimeras made with Fgfr1Δtmk/Δtmk ES cells were lost during early embryonic development due to lethality associated with homozygosity for the Fgfr1Δtmk allele. As a result, the majority of surviving ak/ak↔Fgfr1Δtmk/Δtmk exhibited relatively less contribution from Fgfr1Δtmk/Δtmk ES cells (Table 1). Nonetheless, live embryos displaying relatively high contribution from Fgfr1Δtmk/Δtmk ES cells were obtained (Figure 1E) though they were rare and never survived beyond birth (Table 1). Further histological analysis showed that lenses derived from Fgfr1Δtmk/Δtmk ES cells maintained a normal morphology compared to wild-type controls, thus rescuing the ak/ak lens phenotype and restoring normal anterior segment structure (Figure 1B, F, H). Neither wild-type nor ak/ak E14.5 embryos showed evidence of LacZ expression upon X-gal staining (Figure 1A, B, G, H).
Figure 1.
Histological analysis of ak/ak ↔ Fgfr1Δtmk/Δtmk chimeras. ES cell-derived tissues were identified by the β-galactosidase expression. X-gal staining was performed on wild-type (A, B), ak/ak ↔ Fgfr1Δtmk/+ (C, D), ak/ak↔ Fgfr1Δtmk/Δtmk (E, F) and ak/ak (G, H) embryos at E14.5. Both whole mount pictures (A, C, E, G) and embryonic eye sections (B, D, F, H) are shown. The presence of rescued lens that stained positive for X-gal in ak/ak ↔ Fgfr1Δtmk/Δtmk (F) demonstrates that Fgfr1-deficient ES cells are capable of forming lenses with normal morphology. The arrow in (E) indicates a neural tube defect in the ak/ak ↔ Fgfr1Δtmk/Δtmk chimera. Although the lenses in (D) and (F) are completely ES-cell derived, the lack of penetration of the X-gal staining reagents in whole mount embryos prevented the staining of the deep lens fibers. Scale bars: 100μm.
Table 1.
Generation of ak/ak chimeras with Fgfr1Δtmk/Δtmk and Fgfr1Δtmk/+ ES cells
| Chimera | Total Embryos | Non-chimeric | Chimeric | Neural Tube Defect | Lens Rescue | |||
|---|---|---|---|---|---|---|---|---|
| Low | High | None | Partial | Full | ||||
| ak/ak↔Fgfr1Δtmk/+ | 85 | 80 | 1 | 4 | 0 | 1 | 1 | 8 |
| ak/ak↔Fgfr1Δtmk/Δtmk | 56 | 46 | 7 | 3 | 2 | 7 | 11 | 2 |
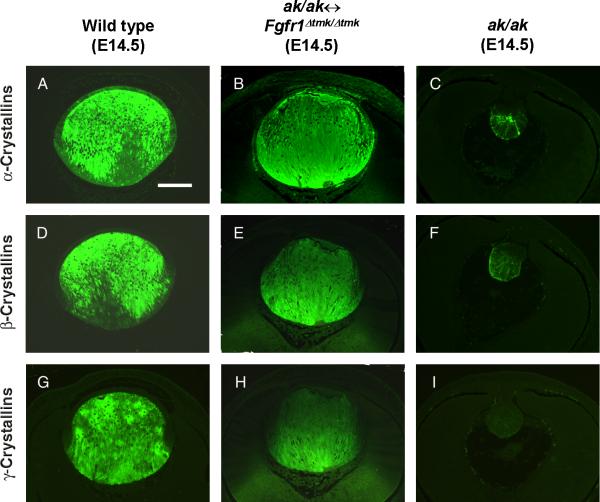
To analyze lens fiber differentiation in the Fgfr1-deficient lenses, expression of lens α-, β- and γ-crystallins were studied by immunofluorescence. In the E14.5 wild-type lens, α-crystallins were expressed in both lens epithelial and fiber cells (Figure 2A). In lenses derived from Fgfr1Δtmk/Δtmk ES cells, a similar expression of α-crystallins was detected, whereas in ak/ak lens, α-crystallins were only weakly expressed in the most posterior regions of the degenerating lens (Figure 2B, C). Expression of differentiation-specific β- and γ-crystallins was restricted to the lens fiber cells of both the wild-type and Fgfr1-deficient lenses (Figure 2D, E, G, H). In contrast, ak/ak lenses expressed β-crystallins at a barely detectable level and no γ-crystallin expression was detected (Figure 2F, I). Therefore, the ak/ak ↔ Fgfr1Δtmk/Δtmk chimeras demonstrated that there is no cell autonomous requirement for Fgfr1 in normal embryonic lens fiber differentiation.
Figure 2.
Analysis of crystallin gene expression in lenses derived from Fgfr1Δtmk/Δtmk ES cells. Expression of α-crystallins (A-C), β-crystallins (D-F) and γ-crystallins (G-I) were analyzed by immunofluorescence with specific antibodies. The expression of these lens structural proteins in rescued lenses from ak/ak ↔ Fgfr1Δtmk/Δtmk chimeras (B, E, H) are shown together with those of wild-type (A, D, G) and ak/ak (C, F, I) control lenses. Lenses derived from Fgfr1Δtmk/Δtmk ES cells expressed these crystallin genes at levels comparable to those of wild-type lenses. Notice α-crystallins are expressed in both the lens epithelial and fiber cells. Whereas β-, and γ-crystallins expression was only detected in the lens fiber cells. In ak/ak lenses, α-, and β-crystallins were expressed at very low level and no γ-crystallin expression was observed. Scale bar: 100μm.
Lens-specific inactivation of Fgfr1 by MLR10 transgenic mice
Although the high degree ak/ak ↔ Fgfr1Δtmk/Δtmk chimeras exhibited complete rescue of the ak/ak lens defects, significant ES cell contributions in these chimeras invariably resulted in neural tube defects inconsistent with postnatal survival. The difficulty of getting enough high degree ak/ak ↔ Fgfr1Δtmk/Δtmk chimeras with lens rescue prevented us from performing further molecular analyses and made it impossible to evaluate the effect of null mutation in Fgfr1 on postnatal lens development. To address this problem, we took a conditional gene targeting approach using the Cre-loxP system to inactivate Fgfr1 in a lens-specific manner. To drive Cre expression to the lens, we used MLR10 transgenic mice, where ocular Cre expression is restricted to the lens, commencing at the lens vesicle stage of development [39]. We also utilized mice carrying a conditional allele of Fgfr1 (Fgfr1flox) which has two loxP sites introduced separately into introns 7 and 15 of the Fgfr1 gene through homologous recombination in ES cells [40]. Cre recombinase recognizes the inserted loxP sites within Fgfr1 and catalyzes the deletion of exons 8 through 15 encoding the transmembrane domain and most of intracellular tyrosine kinase domain. Previous reports using these mice suggest that the loxP sites do not interfere with the expression of Fgfr1 prior to Cre mediated recombination, and several different Cre recombinase transgenes have been shown to efficiently delete exons 8-15 in vivo to generate a loss-of-function mutation in Fgfr1 [35, 40, 43, 44]. The novel allele generated after Cre-mediated recombination, Fgfr1Δflox, does not appear to act in a dominant-negative manner and instead behaves similarly to Fgfr1-null alleles described previously [32, 45].
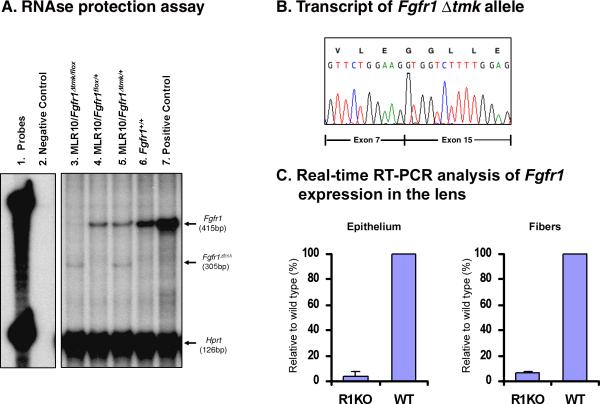
Mice heterozygous for both the Fgfr1Δtmk allele and the Cre transgene (MLR10/Fgfr1Δtmk/+) were crossed with Fgfr1flox mice. In the resulting transgenic compound heterozygotes of both targeted Fgfr1 alleles, MLR10/Fgfr1Δtmk/flox, Cre recombinase needs only to recombine one Fgfr1flox allele in each lens cell to inactivate the Fgfr1 gene. While MLR10 transgenic mice have been shown to efficiently delete a loxP flanked neo gene in both the lens epithelium and lens fiber cells of Cre reporter mice [39], we sought to directly determine the efficiency of Cre-mediated recombination and inactivation of the Fgfr1flox allele. Therefore, we conducted an RNAse protection assay using adult lens total RNA. The 415bp probe used in the assay was derived from the Fgfr1 cDNA sequence containing exons 14 through 16. Since the Fgfr1Δtmk allele has exons 8 to 14 deleted, and the Fgfr1flox allele targets exons 8 to 15, this probe could detect not only wild-type Fgfr1 expression but also potential alternative transcripts generated by the Fgfr1Δtmk and Fgfr1flox alleles respectively. The 415bp-protected fragment, specific for the wild-type Fgfr1 transcript, was detected in total RNA from both the wild-type adult lens and E15.5 whole embryos (Figure 3A, lanes 6 and 7). However, in lens total RNA from the MLR10/Fgfr1Δtmk/+ mice, both a 415bp protected wild-type Fgfr1 transcript and a smaller hybridizing band was observed (Figure 3A, lane 5). The size of the smaller fragment was consistent with an alternatively spliced transcript joining exons flanking the neo cassette from the Fgfr1Δtmk allele. This was confirmed by amplifying MLR10/Fgfr1Δtmk/+ lens total RNA through RT-PCR using PCR primers derived from sequences in exons outside the targeted region. Sequence analysis of this Fgfr1 product showed that exon 7 had been spliced to exon 15 (Figure 3B), leading to frameshift mutation and premature stop codon downstream of this aberrant splice junction. In total lens RNA from MLR10/Fgfr1flox/+ adult mice, only the wild- type Fgfr1 transcript was detected. No alternative transcripts were observed, indicating that the Fgfr1Δflox allele generated after Cre-mediated recombination is a true null allele in that the remaining Fgfr1 sequences did not drive the expression of a detectable transcript (Figure 3A, lane 4). Also, it does not appear, according to the RNAse protection experiment, that the expression of the wild-type allele was upregulated in the presence of a non-functional Fgfr1 allele (Fig. 4A, lanes 4 and 5). However, in lenses from MLR10/Fgfr1Δtmk/flox mice, the transcript from Fgfr1Δtmk allele was consistently detected, but the wild-type Fgfr1 transcript was absent, suggesting that Cre recombinase in MLR10 transgenic mice had recombined the Fgfr1flox allele and inactivated Fgfr1 expression efficiently (Figure 3A, lane 3). Similarly, when a probe derived from exonic sequences targeted by both Fgfr1 mutant alleles was used in an RNAse protection assay, wild-type Fgfr1 expression was absent from adult lens total RNA from MLR10/Fgfr1Δtmk/flox (data not shown). Furthermore, quantitative RT-PCR analysis on whole lens RNA revealed that Fgfr1 expression was almost entirely eliminated from lens of MLR10/Fgfr1Δtmk/flox mice (data not shown).
Figure 3.
Lens-specific inactivation of Fgfr1 by MLR10 transgenic mice. An. RNAse protection assay using an Fgfr1 cDNA probe spanning exons 14 to 16 detected a 415 bp protected fragment indicative of the wild-type Fgfr1 transcript in whole embryo E15.5 total RNA (Lane 7), as well as in adult lens total RNA from wild-type (Lane 6) and heterozygous animals (Lanes 4, 5). In lens RNA from MLR10/Fgfr1Δtmk/+, an additional 305 bp protected fragment was detected, which indicated alternative splicing of the Fgfr1Δtmk allele (Lanes 3 and 5). In lens RNA from MLR10/Fgfr1Δtmk/flox mice, the wild-type transcript was absent indicating the floxed allele of Fgfr1 was recombined efficiently by the Cre recombinase (Lane 3). B. Sequencing of the PCR-amplified smaller transcript revealed an alternative transcript where exon 7 was spliced onto exon 15 because of the deletion of genomic sequence spanning exon 8 through exon 14 in the Fgfr1Δtmk allele. C. Quantitative real time-PCR analysis of Cre-mediated Fgfr1 inactivation in the lens epithelial and fiber cells. In the lens epithelium of MLR10/Fgfr1flox/flox mice, the level of Fgfr1 transcripts was only 3.87% that of the wild type lens epithelium. Similarly, lens fiber cells from MLR10/Fgfr1Δtmk/flox mice only retain 6.11% of Fgfr1 transcripts relative to wild type lens fibers. Error bars represent the standard deviation present in samples analyzed in triplicate.
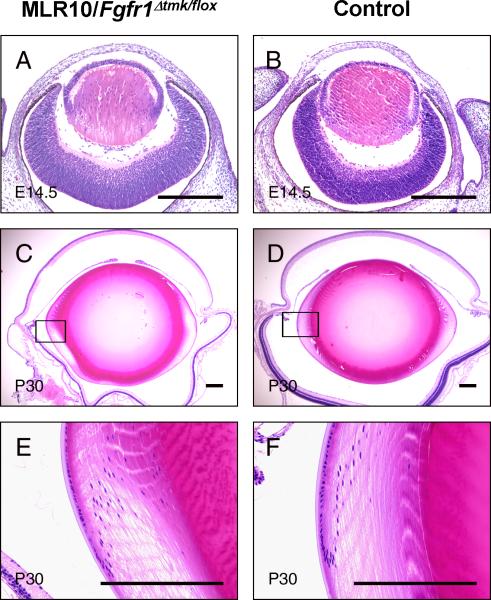
Figure 4.
Morphology of the eye from MLR10/Fgfr1Δtmk/flox mice. E14.5 embryo (A) and 30-day-old adult mice (P30) (C, E) lenses from MLR10/Fgfr1Δtmk/flox mice were compared to wild-type age-matched control lenses (B, D, and F). The equatorial regions of adult lenses (boxed in C, D) were shown under higher magnification (E, F). Notice the size and morphology of Fgfr1-deficient lens was indistinguishable from that of wild-type controls. The retinal degeneration seen in the adult MLR10/Fgfr1Δtmk/flox mouse (C, E) is unrelated to either the Cre transgene or the Fgfr1 mutations and is the result of homozygosity for the rd1 mutation segregating in the mixed genetic background. Scale bar: 200μm.
Given the fact that the bulk of the lens consists of lens fibers, whereas the epithelial cells only constitute a small portion of the lens, it is likely that analysis of RNA extracted from the whole lens may not accurately reflect the efficiency of Cre-mediated Fgfr1 inactivation in the epithelial lineage. To address this possibility, we carried out quantitative RT-PCR analyses of Fgfr1 expression in the epithelial and fiber cells of lenses from MLR10/Fgfr1flox/flox mice collected at weaning age. The results indicated that Fgfr1 transcripts in MLR10 mice homozygous for the conditional Fgfr1 mutation contained only 3.9% the amount of Fgfr1 transcripts present in the wildtype lens epithelial cells. Similarly the lens fiber cells from these mice contained only 6.1% of the transcripts present in wildtype fiber cells (Fig. 3C). Therefore, Cre recombinase expressed by the lens of MLR10 transgenic mice was clearly able to efficiently recombine and inactivate the Fgfr1flox allele in the entire lens. The purity of the lens epithelial cell RNA fraction was assessed by relative expression of γD-crystallin transcripts by a similar real time RT-PCR assay. The lens epithelial RNA sample used produced only 0.17% of the signal generated by the fiber cell RNA fraction for γD-crystallin (data not shown).
Embryonic and postnatal lens development in mice with lens-specific inactivation of Fgfr1: Litter sizes were normal in crosses between MLR10/Fgfr1Δtmk/+ and Fgfr1flox/+ mice, and different genotypes were distributed according to normal Mendelian ratios. No obvious developmental abnormalities were detected in any of the resulting genotypes. Histological analyses showed normal morphology of the MLR10/Fgfr1Δtm/f/lox lenses relative to that of wild-type control lenses at E14.5 and adult stage (Figure 4A-F).
To determine if MLR10/Fgfr1Δtmk/flox lenses were normal at the molecular level, we analyzed lens sections for the expression of genes critical for normal lens development. In-situ hybridization for detection of FoxE3, Pax6 and Six3 mRNA demonstrated an unaltered pattern of expression in MLR10/Fgfr1Δtmk/flox E14.5 embryonic lenses relative to the wild-type control lenses (data not shown). Normal expression of FoxE3, Pax6, and Six3 suggested that proliferation and maintenance of lens epithelial cells were intact in the absence of Fgfr1.
To investigate genes thought to be important for cell cycle regulation during fiber cell differentiation in MLR10/Fgfr1Δtmk/flox mice, the expression pattern of p57kip2 and Prox1 was studied by radioactive in-situ hybridization on E14.5 lens sections. The pattern of expression for these genes was also unchanged in the MLR10/Fgfr1Δtmk/lox mice (data not shown). Therefore, it appeared that the regulation of genes controlling cell cycle withdrawal in the developing lens was not affected by the absence of Fgfr1.
Likewise, in situ hybridization demonstrated that the expression patterns of c-Maf and Sox1, transcription factors thought to be important for lens fiber differentiation, were unchanged in the lens fiber cells of MLR10/Fgfr1Δtmk/flox E14.5 embryos (data not shown). Thus, the expression of these transcription factors suggested that lens fiber differentiation was normal in MLR10/Fgfr1Δtmk/flox mice. Immunofluorescence analyses with antibodies against α- and β-crystallins performed on tissue sections of E14.5 day lenses from both MLR10/Fgfr1Δtmk/flox and wild-type embryos failed to reveal any differences (data not shown), providing further evidence that, within the lens, Fgfr1 is not essential for lens fiber differentiation.
DISCUSSION
Although there is evidence suggesting that FGF signaling is likely to play an important role in lens induction [23], lens cell proliferation and fiber cell differentiation [reviewed in 46], the essential ligands and receptors for these processes remain elusive. The family of FGF receptor tyrosine kinases that bind FGFs with high affinity are at the top of this signaling cascade, and activation of extracellular regulated kinase (ERK) is likely to be a required endpoint of FGF-induced lens cell proliferation and fiber differentiation [47]. Since previous studies demonstrated that lens fiber differentiation is not prevented in mouse embryos carrying null mutations of either Fgfr3 or Fgfr2 genes, we hypothesized that if any single FGF receptor gene is essential for lens fiber differentiation, it was most likely to be Fgfr1. Null mutations of Fgfr1 lead to embryonic lethality prior to ocular induction, therefore the function of Fgfr1 in lens development could not be elucidated in Fgfr1-null mice.
To address whether Fgfr1 plays a role during lens development, we took advantage of aphakia complementation system and a tissue-specific knockout approach. We demonstrated that, although complete rescue was rare, ES cells lacking Fgfr1 expression are capable of forming morphlogically normal lenses that expresses α-, β- and γ-crystallins in a normal pattern. We also demonstrated, by both RNAse protection and relative real-time RT-PCR, that Cre-expressing MLR10 transgenic mice can efficiently recombine the conditional (loxP-flanked) allele of Fgfr1 creating lenses where the majority of cells completely lack Fgfr1 transcripts.
Using these two complementary approaches we have demonstrated that the expression of Fgfr1 is not cell autonomously required for lens fiber cell differentiation. This conclusion is based on several pieces of evidence. First, cells exhibiting normal fiber cell morphology were produced both by cells homozygous null for Fgfr1 in ak/ak ↔ Fgfr1Δtmk/Δtmk chimeras and in cells that were compound heterozygous for the Fgfr1-null and recombined conditional allele. We also examined molecular aspects of fiber cell differentiation in lens cells lacking Fgfr1 expression. FoxE3 is expressed in the lens epithelial cells and plays an important role in the proliferation and survival of the lens epithelium [48]. No change in the expression of FoxE3 was detected in Fgfr1-deficient lens cells. The transcription factors Pax6 and Six3 are expressed in the lens and neural retina and play important roles in regulating ocular development [49-51]. In addition, mutual regulation of these two transcription factors in the developing lens has been suggested previously [52]. Since FGF signaling is likely to play a role in regulating Pax6 expression in developing lens [23], we looked at the expression patterns of both of these genes, and again, no differences were detected between wild-type and Fgfr1 deficient lens cells. During lens development, proliferating lens epithelial cells withdraw from the cell cycle at the equatorial region of the lens and initiate fiber differentiation. Cell cycle regulators such as p57kip2 and Prox1 are essential in mediating this cell cycle exit. p57kip2 blocks cell cycle progression by inhibiting the activity of cyclin-dependent kinases and lens fiber cells in p57kip2-deficient mice exhibit inappropriate fiber cell proliferation [53]. Prox1 may act indirectly in this process by regulating the expression of p57kip2[54]. Both Prox1 and p57kip2 appeared to be expressed normally in Fgfr1 deficient lens cells. The transcription factors c-Maf and Sox1 have been shown to mediate high-level expression of crystallin genes in the lens, and are essential for normal lens fiber differentiation [41, 55-58]. The expression patterns of these transcription factors as well as the expression patterns of α-, β- and γ-crystallins in the Fgfr1-deficient lenses were indistinguishable from those of the wild-type lenses. Therefore, from morphological, cell cycle and differentiation-specific gene expression analyses, Fgfr1-deficient lens cells were not compromised in their ability to undergo fiber cell differentiation.
Neither the chimeric nor the conditional knockout approaches could definitively demonstrate that Fgfr1 was dispensable for the initiation of lens induction. This is because, in the case of the chimeras made between Fgfr1 null and ak/ak embryos, we did not demonstrate that Fgfr1 null cells could independently undergo lens induction in the absence of neighboring ak/ak cells as the aphakia deficiency only becomes evident subsequent to initial lens induction. In the case of the conditional Fgfr1 deletion by the MLR10 Cre transgenic mice, Cre transgene expression does not commence until the lens pit stage, subsequent to lens induction. However, the fact that Fgfr1-null ES cells are capable of differentiating into apparently normal fiber cells demonstrates that there is no cell autonomous requirement of FGFR1 for lens fiber development and normal morphology in mice even in cell lineages that never express Fgfr1.
The notion that no single Fgfr gene is essential for lens fiber differentiation seems to contradict previous evidence suggesting the importance of FGF receptor signaling in lens fiber differentiation or promotion of lens cell proliferation or survival [19-21, 59]. However, it is possible that different Fgfr genes exhibit functional redundancy in mediating lens fiber differentiation. In support of functional redundancy, although Fgfr3-deficient mice do not demonstrate lens defects, lens-specific expression of a secreted version of FGFR3 leads to delayed lens fiber differentiation and reduced ERK phosphorylation levels in vivo, suggesting that Fgfr(s) other than Fgfr3 are present to transduce FGF signals in the lens [22]. The redundant nature of Fgfrs during lens fiber differentiation may also explain the severe lens defect observed in transgenic mice overexpressing dominant-negative FGFR1 that can heterodimerize with other FGFRs and therefore abolish the functions of multiple Fgfrs simultaneously [20, 21, 60]. Moreover, the expression pattern of different Fgfrs during lens development is characterized by their increased expression in regions of the lens where fiber differentiation occurs. Thus, it is possible that these FGFRs all contribute to lens fiber differentiation. Recent evidence, however, demonstrates that Fgfr2 does play a role in lens development that can't be entirely compensated for by the normal expression of other Fgfr genes [31]. Conditional deletion of different Fgfr genes in combination or at different stages of lens development will likely reveal additional requirements for Fgfr signaling in the induction or maturation of lens cells.
ACKNOWLEDGEMENTS
The experiments described in this manuscript were performed at Columbus Children's Research Institute. The authors wish to thank Sadeq Kharzai for technical assistance. Dr. David Cunningham for helpful discussion. Drs. Milan Jamrich, Paul Overbeek, J. Samuel Zigler and Brian Sauer for probes, plasmids and antibodies. Drs. Claudia M. Garcia and David C. Beebe for critical review of the manuscript and helpful suggestions. This work is supported by a grant (EY12995) from The National Eye Institute.
REFERENCES
- 1.Grainger RM. Embryonic lens induction: shedding light on vertebrate tissue determination. Trends Genet. 1992;8(10):349–55. doi: 10.1016/0168-9525(92)90280-h. [DOI] [PubMed] [Google Scholar]
- 2.de Iongh R, McAvoy JW. Distribution of acidic and basic fibroblast growth factors (FGF) in the foetal rat eye: implications for lens development. Growth Factors. 1992;6(2):159–77. [PubMed] [Google Scholar]
- 3.de Iongh R, McAvoy JW. Spatio-temporal distribution of acidic and basic FGF indicates a role for FGF in rat lens morphogenesis. Dev Dyn. 1993;198(3):190–202. doi: 10.1002/aja.1001980305. [DOI] [PubMed] [Google Scholar]
- 4.Lovicu FJ, McAvoy JW. Localization of acidic fibroblast growth factor, basic fibroblast growth factor, and heparan sulphate proteoglycan in rat lens: implications for lens polarity and growth patterns. Invest Ophthalmol Vis Sci. 1993;34(12):3355–65. [PubMed] [Google Scholar]
- 5.Schulz MW, Chamberlain CG, de Iongh RU, McAvoy JW. Acidic and basic FGF in ocular media and lens: implications for lens polarity and growth patterns. Development. 1993;118(1):117–26. doi: 10.1242/dev.118.1.117. [DOI] [PubMed] [Google Scholar]
- 6.Lovicu FJ, de Iongh RU, McAvoy JW. Expression of FGF-1 and FGF-2 mRNA during lens morphogenesis, differentiation and growth. Curr Eye Res. 1997;16(3):222–30. doi: 10.1076/ceyr.16.3.222.15408. [DOI] [PubMed] [Google Scholar]
- 7.Consigli SA, Lyser KM, Joseph-Silverstein J. The temporal and spatial expression of basic fibroblast growth factor during ocular development in the chicken. Invest Ophthalmol Vis Sci. 1993;34(3):559–66. [PubMed] [Google Scholar]
- 8.Bugra K, Oliver L, Jacquemin E, Laurent M, Courtois Y, Hicks D. Acidic fibroblast growth factor is expressed abundantly by photoreceptors within the developing and mature rat retina. Eur J Neurosci. 1993;5(12):1586–95. doi: 10.1111/j.1460-9568.1993.tb00228.x. [DOI] [PubMed] [Google Scholar]
- 9.Caruelle D, Groux-Muscatelli B, Gaudric A, Sestier C, Coscas G, Caruelle JP, et al. Immunological study of acidic fibroblast growth factor (aFGF) distribution in the eye. J Cell Biochem. 1989;39(2):117–28. doi: 10.1002/jcb.240390204. [DOI] [PubMed] [Google Scholar]
- 10.Lovicu FJ, McAvoy JW. Structural analysis of lens epithelial explants induced to differentiate into fibres by fibroblast growth factor (FGF). Exp Eye Res. 1989;49(3):479–94. doi: 10.1016/0014-4835(89)90056-0. [DOI] [PubMed] [Google Scholar]
- 11.McAvoy JW, Chamberlain CG. Fibroblast growth factor (FGF) induces different responses in lens epithelial cells depending on its concentration. Development. 1989;107(2):221–8. doi: 10.1242/dev.107.2.221. [DOI] [PubMed] [Google Scholar]
- 12.Richardson NA, McAvoy JW, Chamberlain CG. Age of rats affects response of lens epithelial explants to fibroblast growth factor. Exp Eye Res. 1992;55(5):649–56. doi: 10.1016/0014-4835(92)90169-s. [DOI] [PubMed] [Google Scholar]
- 13.Robinson ML, Overbeek PA, Verran DJ, Grizzle WE, Stockard CR, Friesel R, et al. Extracellular FGF-1 acts as a lens differentiation factor in transgenic mice. Development. 1995;121(2):505–14. doi: 10.1242/dev.121.2.505. [DOI] [PubMed] [Google Scholar]
- 14.Robinson ML, Ohtaka-Maruyama C, Chan CC, Jamieson S, Dickson C, Overbeek PA, et al. Disregulation of ocular morphogenesis by lens-specific expression of FGF-3/int-2 in transgenic mice. Dev Biol. 1998;198(1):13–31. doi: 10.1006/dbio.1998.8879. [DOI] [PubMed] [Google Scholar]
- 15.Lovicu FJ, Overbeek PA. Overlapping effects of different members of the FGF family on lens fiber differentiation in transgenic mice. Development. 1998;125(17):3365–77. doi: 10.1242/dev.125.17.3365. [DOI] [PubMed] [Google Scholar]
- 16.de Iongh RU, Lovicu FJ, Chamberlain CG, McAvoy JW. Differential expression of fibroblast growth factor receptors during rat lens morphogenesis and growth. Invest Ophthalmol Vis Sci. 1997;38(9):1688–99. [PubMed] [Google Scholar]
- 17.de Iongh RU, Lovicu FJ, Hanneken A, Baird A, McAvoy JW. FGF receptor-1 (flg) expression is correlated with fibre differentiation during rat lens morphogenesis and growth. Dev Dyn. 1996;206(4):412–26. doi: 10.1002/(SICI)1097-0177(199608)206:4<412::AID-AJA7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 18.Ohuchi H, Koyama E, Myokai F, Nohno T, Shiraga F, Matsuo T, et al. Expression patterns of two fibroblast growth factor receptor genes during early chick eye development. Exp Eye Res. 1994;58(6):649–58. doi: 10.1006/exer.1994.1062. [DOI] [PubMed] [Google Scholar]
- 19.Chow RL, Roux GD, Roghani M, Palmer MA, Rifkin DB, Moscatelli DA, et al. FGF suppresses apoptosis and induces differentiation of fibre cells in the mouse lens. Development. 1995;121(12):4383–93. doi: 10.1242/dev.121.12.4383. [DOI] [PubMed] [Google Scholar]
- 20.Robinson ML, MacMillan-Crow LA, Thompson JA, Overbeek PA. Expression of a truncated FGF receptor results in defective lens development in transgenic mice. Development. 1995;121(12):3959–67. doi: 10.1242/dev.121.12.3959. [DOI] [PubMed] [Google Scholar]
- 21.Stolen CM, Griep AE. Disruption of lens fiber cell differentiation and survival at multiple stages by region-specific expression of truncated FGF receptors. Dev Biol. 2000;217(2):205–20. doi: 10.1006/dbio.1999.9557. [DOI] [PubMed] [Google Scholar]
- 22.Govindarajan V, Overbeek PA. Secreted FGFR3, but not FGFR1, inhibits lens fiber differentiation. Development. 2001;128(9):1617–27. doi: 10.1242/dev.128.9.1617. [DOI] [PubMed] [Google Scholar]
- 23.Faber SC, Dimanlig P, Makarenkova HP, Shirke S, Ko K, Lang RA. Fgf receptor signaling plays a role in lens induction. Development. 2001;128(22):4425–38. doi: 10.1242/dev.128.22.4425. [DOI] [PubMed] [Google Scholar]
- 24.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84(6):911–21. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 25.Xu X, Weinstein M, Li C, Naski M, Cohen RI, Ornitz DM, et al. Fibroblast growth factor receptor 2 (FGFR2)-mediated reciprocal regulation loop between FGF8 and FGF10 is essential for limb induction. Development. 1998;125(4):753–65. doi: 10.1242/dev.125.4.753. [DOI] [PubMed] [Google Scholar]
- 26.Arman E, Haffner-Krausz R, Chen Y, Heath JK, Lonai P. Targeted disruption of fibroblast growth factor (FGF) receptor 2 suggests a role for FGF signaling in pregastrulation mammalian development. Proc Natl Acad Sci U S A. 1998;95(9):5082–7. doi: 10.1073/pnas.95.9.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li C, Guo H, Xu X, Weinberg W, Deng CX. Fibroblast growth factor receptor 2 (Fgfr2) plays an important role in eyelid and skin formation and patterning. Dev Dyn. 2001;222(3):471–83. doi: 10.1002/dvdy.1205. [DOI] [PubMed] [Google Scholar]
- 28.Revest JM, Spencer-Dene B, Kerr K, De Moerlooze L, Rosewell I, Dickson C. Fibroblast growth factor receptor 2-IIIb acts upstream of Shh and Fgf4 and is required for limb bud maintenance but not for the induction of Fgf8, Fgf10, Msx1, or Bmp4. Dev Biol. 2001;231(1):47–62. doi: 10.1006/dbio.2000.0144. [DOI] [PubMed] [Google Scholar]
- 29.Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, et al. Conditional inactivation of FGF receptor 2 reveals an essential role for FGF signaling in the regulation of osteoblast function and bone growth. Development. 2003;130(13):3063–74. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- 30.Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000;14(21):2701–11. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia CM, Yu K, Zhao H, Ashery-Padan R, Ornitz DM, Robinson ML, et al. Signaling through FGF receptor-2 is required for lens cell survival and for withdrawal from the cell cycle during lens fiber cell differentiation. Dev Dyn. 2005;233(2):516–27. doi: 10.1002/dvdy.20356. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi TP, Harpal K, Henkemeyer M, Rossant J. fgfr-1 is required for embryonic growth and mesodermal patterning during mouse gastrulation. Genes Dev. 1994;8(24):3032–44. doi: 10.1101/gad.8.24.3032. [DOI] [PubMed] [Google Scholar]
- 33.Ciruna BG, Schwartz L, Harpal K, Yamaguchi TP, Rossant J. Chimeric analysis of fibroblast growth factor receptor-1 (Fgfr1) function: a role for FGFR1 in morphogenetic movement through the primitive streak. Development. 1997;124(14):2829–41. doi: 10.1242/dev.124.14.2829. [DOI] [PubMed] [Google Scholar]
- 34.Ciruna B, Rossant J. FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell. 2001;1(1):37–49. doi: 10.1016/s1534-5807(01)00017-x. [DOI] [PubMed] [Google Scholar]
- 35.Pirvola U, Ylikoski J, Trokovic R, Hebert JM, McConnell SK, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35(4):671–80. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- 36.Liegeois NJ, Horner JW, DePinho RA. Lens complementation system for the genetic analysis of growth, differentiation, and apoptosis in vivo. Proc Natl Acad Sci U S A. 1996;93(3):1303–7. doi: 10.1073/pnas.93.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Semina EV, Murray JC, Reiter R, Hrstka RF, Graw J. Deletion in the promoter region and altered expression of Pitx3 homeobox gene in aphakia mice. Hum Mol Genet. 2000;9(11):1575–85. doi: 10.1093/hmg/9.11.1575. [DOI] [PubMed] [Google Scholar]
- 38.Rieger DK, Reichenberger E, McLean W, Sidow A, Olsen BR. A double-deletion mutation in the Pitx3 gene causes arrested lens development in aphakia mice. Genomics. 2001;72(1):61–72. doi: 10.1006/geno.2000.6464. [DOI] [PubMed] [Google Scholar]
- 39.Zhao H, Yang Y, Rizo CM, Overbeek PA, Robinson ML. Insertion of a Pax6 consensus binding site into the alphaA-crystallin promoter acts as a lens epithelial cell enhancer in transgenic mice. Invest Ophthalmol Vis Sci. 2004;45(6):1930–9. doi: 10.1167/iovs.03-0856. [DOI] [PubMed] [Google Scholar]
- 40.Trokovic R, Trokovic N, Hernesniemi S, Pirvola U, Vogt Weisenhorn DM, Rossant J, et al. FGFR1 is independently required in both developing mid- and hindbrain for sustained response to isthmic signals. Embo J. 2003;22(8):1811–23. doi: 10.1093/emboj/cdg169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishiguchi S, Wood H, Kondoh H, Lovell-Badge R, Episkopou V. Sox1 directly regulates the gamma-crystallin genes and is essential for lens development in mice. Genes Dev. 1998;12(6):776–81. doi: 10.1101/gad.12.6.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5(9):1513–23. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- 43.Hebert JM, Lin M, Partanen J, Rossant J, McConnell SK. FGF signaling through FGFR1 is required for olfactory bulb morphogenesis. Development. 2003;130(6):1101–11. doi: 10.1242/dev.00334. [DOI] [PubMed] [Google Scholar]
- 44.Trokovic N, Trokovic R, Mai P, Partanen J. Fgfr1 regulates patterning of the pharyngeal region. Genes Dev. 2003;17(1):141–53. doi: 10.1101/gad.250703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deng CX, Wynshaw-Boris A, Shen MM, Daugherty C, Ornitz DM, Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 1994;8(24):3045–57. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 46.McAvoy JW, Chamberlain CG, de Iongh RU, Hales AM, Lovicu FJ. Lens development. Eye. 1999;13(Pt 3b):425–37. doi: 10.1038/eye.1999.117. [DOI] [PubMed] [Google Scholar]
- 47.Lovicu FJ, McAvoy JW. FGF-induced lens cell proliferation and differentiation is dependent on MAPK (ERK1/2) signalling. Development. 2001;128(24):5075–84. doi: 10.1242/dev.128.24.5075. [DOI] [PubMed] [Google Scholar]
- 48.Blixt A, Mahlapuu M, Aitola M, Pelto-Huikko M, Enerback S, Carlsson P. A forkhead gene, FoxE3, is essential for lens epithelial proliferation and closure of the lens vesicle. Genes Dev. 2000;14(2):245–54. [PMC free article] [PubMed] [Google Scholar]
- 49.Ashery-Padan R, Gruss P. Pax6 lights-up the way for eye development. Curr Opin Cell Biol. 2001;13(6):706–14. doi: 10.1016/s0955-0674(00)00274-x. [DOI] [PubMed] [Google Scholar]
- 50.Carl M, Loosli F, Wittbrodt J. Six3 inactivation reveals its essential role for the formation and patterning of the vertebrate eye. Development. 2002;129(17):4057–63. doi: 10.1242/dev.129.17.4057. [DOI] [PubMed] [Google Scholar]
- 51.Lengler J, Krausz E, Tomarev S, Prescott A, Quinlan RA, Graw J. Antagonistic action of Six3 and Prox1 at the gamma-crystallin promoter. Nucleic Acids Res. 2001;29(2):515–26. doi: 10.1093/nar/29.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goudreau G, Petrou P, Reneker LW, Graw J, Loster J, Gruss P. Mutually regulated expression of Pax6 and Six3 and its implications for the Pax6 haploinsufficient lens phenotype. Proc Natl Acad Sci U S A. 2002;99(13):8719–24. doi: 10.1073/pnas.132195699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang P, Wong C, DePinho RA, Harper JW, Elledge SJ. Cooperation between the Cdk inhibitors p27(KIP1) and p57(KIP2) in the control of tissue growth and development. Genes Dev. 1998;12(20):3162–7. doi: 10.1101/gad.12.20.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21(3):318–22. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- 55.Kawauchi S, Takahashi S, Nakajima O, Ogino H, Morita M, Nishizawa M, et al. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J Biol Chem. 1999;274(27):19254–60. doi: 10.1074/jbc.274.27.19254. [DOI] [PubMed] [Google Scholar]
- 56.Kim JI, Li T, Ho IC, Grusby MJ, Glimcher LH. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc Natl Acad Sci U S A. 1999;96(7):3781–5. doi: 10.1073/pnas.96.7.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ring BZ, Cordes SP, Overbeek PA, Barsh GS. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development. 2000;127(2):307–17. doi: 10.1242/dev.127.2.307. [DOI] [PubMed] [Google Scholar]
- 58.Kamachi Y, Uchikawa M, Collignon J, Lovell-Badge R, Kondoh H. Involvement of Sox1, 2 and 3 in the early and subsequent molecular events of lens induction. Development. 1998;125(13):2521–32. doi: 10.1242/dev.125.13.2521. [DOI] [PubMed] [Google Scholar]
- 59.Stolen CM, Jackson MW, Griep AE. Overexpression of FGF-2 modulates fiber cell differentiation and survival in the mouse lens. Development. 1997;124(20):4009–17. doi: 10.1242/dev.124.20.4009. [DOI] [PubMed] [Google Scholar]
- 60.Ueno H, Gunn M, Dell K, Tseng A, Jr., Williams L. A truncated form of fibroblast growth factor receptor 1 inhibits signal transduction by multiple types of fibroblast growth factor receptor. J Biol Chem. 1992;267(3):1470–6. [PubMed] [Google Scholar]