Abstract
Inducible Degrader Of the Low-density lipoprotein receptor (IDOL) is an E3 ubiquitin ligase that mediates the ubiquitination and degradation of the low-density lipoprotein receptor (LDLR). IDOL expression is controlled at the transcriptional level by the cholesterol-sensing nuclear receptor LXR. In response to rising cellular sterol levels, activated LXR induces IDOL production, thereby limiting further uptake of exogenous cholesterol through the LDLR pathway. The LXR–IDOL–LDLR mechanism for feedback inhibition of cholesterol uptake is independent of and complementary to the SREBP pathway. Since the initial description of the LXR–IDOL pathway, biochemical studies have helped to define the structural basis for both IDOL target recognition and LDLR ubiquitin transfer. Recent work has also suggested links between IDOL and human lipid metabolism.
Cholesterol is essential for mammalian cells because it plays important roles in a series of biological processes, including the biosynthesis of steroid hormones and the maintenance of membrane integrity.1 However, excessive free cholesterol is toxic to cells. Therefore, cholesterol levels in individual cells as well as in whole animals are tightly regulated through the coordinated action of the LXR and SREBP transcription factors.1, 2 Systemic cholesterol homeostasis reflects a finely tuned balance between dietary uptake, endogenous synthesis, reverse cholesterol transport and removal from the body via biliary and intestinal excretion.1
Elevated levels of cholesterol in the bloodstream are a strong risk factor for cardiovascular disease.3 Diets rich in cholesterol and saturated fat, combined with sedentary lifestyles, present a challenge for the endogenous homeostatic mechanisms designed to maintain optimal cholesterol levels. This has likely been a major contributor to the increased prevalence of cardiovascular disease. Although statin drugs have been beneficial in reducing mortality associated with cardiovascular disease,4 new therapeutic strategies are needed to complement statins, because many individuals fail to reach their target cholesterol levels with statins alone. IDOL (Inducible Degrader Of the Low-density lipoprotein receptor) has recently been identified as an E3 ubiquitin ligase that modulates cholesterol levels by regulating the stability of the low-density lipoprotein receptor (LDLR) via a pathway that is independent of the mechanism of action of statins.5 Therefore, a better understanding of the LXR–IDOL pathway could potentially provide insight into novel approaches to the treatment of cardiovascular disease.
Role of the LDLR in cholesterol homeostasis
The LDLR is a major determinant of plasma cholesterol levels. This cell surface receptor is expressed primarily in liver and removes cholesterol-carrying LDL from plasma by receptor-mediated endocytosis.6 Familial hypercholesterolemia (FH) is a rare human genetic disorder that usually results from a mutation in the coding region of the LDLR that disrupts its function.7 Mice lacking the LDLR are also hypercholesterolemic and provide a widely used model for the study of atherosclerosis.8 The transcription of LDLR is primarily under the control of the transcription factor SREBP-2, the first sterol-regulated transcription factor to be described.9 SREBP-2 is an endoplasmic reticulum (ER) transmembrane protein that exists in a complex with the sterol-sensing proteins SREBP cleavage-activating protein (SCAP) and insulin-induced gene 1 (INSIG1). The SREBP/SCAP complex moves to the Golgi apparatus when the intracellular cholesterol concentration decreases. Proteases in the Golgi cleave SREBP-2 to release the active SREBP-2 transcription factor, which translocates to the nucleus and stimulates transcription of LDLR.1 Like many other plasma membrane receptors, LDLR is transported to the plasma membrane via the ER-Golgi pathway. LDL-bound LDLR is endocytosed through a clathrin-dependent pathway and, after releasing its cargo in the late endosome, the LDLR is either recycled back to the plasma membrane or degraded in the lysosome.10
Subsequent to the description of the SREBP pathway, it was discovered that the LDLR is also regulated by post-translational mechanisms that govern its stability. Proprotein convertase subtilisin/kexin type 9 (PCSK9) binds directly to the extracellular domain of the LDLR and alters its stability and trafficking by interfering with its recycling after endocytosis, thereby promoting its lysosomal degradation.11-13 The physiological importance of PCSK9 in cholesterol metabolism is supported by genetic evidence in both mouse models and in humans. Patients with gain- or loss-of-function PCSK9 mutations show markedly altered plasma cholesterol levels.14, 15 Interestingly, like its target the LDLR, the expression of PCSK9 is also induced by activated SREBP. Although the physiological basis for PCSK9 regulation by SREBPs is not entirely understood, its regulation by SREBP-1 may relate to the metabolism of very low-density lipoprotein (VLDL). The expression of PCSK9 in the setting of SREBP-1 activation in the liver would be expected prevent newly secreted VLDL particles from being reclaimed by hepatocytes, thereby facilitating their delivery to peripheral tissues.16, 17 In this context, it is important to note that the statin drugs increase PCSK9 levels at the same time that they increase LDLR expression and this limits their cholesterol-lowering efficacy. Therefore, targeting the LDLR from different angles using a combination of drugs might improve the treatment of hypercholesterolemia.18
Regulation of IDOL expression by LXR
The nuclear receptors LXRα and LXRβ work in a complementary fashion with SREBPs to maintain cholesterol homeostasis. When cellular cholesterol levels rise, oxysterols are formed and serve as ligands for LXRs.19 Activation of LXR by oxysterols, particularly 24(S),25-epoxycholesterol and 22(R)-hydroxycholesterol,20 induces the expression of genes encoding proteins essential for cholesterol efflux from cells, including ATP-binding cassette transporter A1 (ABCA1) and ABCG1, phospholipid transfer protein (PLTP), ADP-ribosylation factor-like 7 (ARL7) and apolipoprotein (Apo)E.21, 22 The expression of these factors promotes the transfer of excess intracellular cholesterol to extracellular acceptors, such as ApoA-I, ApoE and high-density lipoprotein (HDL), thereby facilitating reverse cholesterol transport from peripheral tissues.23-25
Several years ago, our group made the unexpected observation that in addition to regulating cholesterol efflux, the LXR signaling pathway also exerts a strong influence on the cellular uptake of cholesterol through the LDLR. Treatment of various cell types including macrophages and hepatocytes with the LXR agonists GW3965 and T0901317 markedly inhibits the binding and uptake of LDL by these cells.5 Also surprising was the observation that this effect occurs in the absence of changes in LDLR mRNA levels. Rather, LXR activation was shown to lead to rapid elimination of LDLR protein from the cell surface.5
Through transcription profiling studies and candidate gene analyses, we identified the E3 ubiquitin ligase MYLIP (re-named IDOL) as the mediator of the effects of LXR on the LDLR. IDOL/MYLIP had been studied previously in cultured neurons and proposed to interact with myosin regulatory light chain and to alter neurite outgrowth.26 However, this factor had not been linked to lipid metabolism and its biological targets for ubiquitination had not been definitively established. The IDOL gene promoter is a direct target for binding and regulation by LXR/RXR heterodimers in both mice and humans. Accordingly, the expression of IDOL mRNA is induced by LXR agonists in multiple cell types in vitro and in multiple tissues in vivo, including intestine, macrophages and fat.5 Interestingly, although it is highly induced by LXR agonists in human hepatocyte cell lines, the expression of IDOL does not appear to respond as well to LXR agonists in mouse liver.5 It is possible that there is an as yet unknown mechanism governing IDOL responsiveness to LXR activation in different tissues and organisms.
The IDOL protein contains a FERM (4.1 band, ezrin, radixin and moesin) homology domain at the amino terminus and a RING (“Really Interesting New Gene”) zinc finger putative ubiquitin ligase domain at its carboxyl terminus.26 Other proteins with FERM domains have been shown to interact with the cytoplasmic portions of transmembrane proteins and link these proteins to the cytoskeleton during cell signaling.27 Interestingly, IDOL is the only FERM-containing protein that also possesses a RING ubiquitin ligase motif.
IDOL mediates ubiquitination and degradation of the LDLR
The ability of IDOL to facilitate LDLR protein degradation was initially revealed by a series of in vitro and in vivo studies. Transfection of IDOL cDNA into cultured cells strongly promotes the degradation of LDLR protein. Furthermore, overexpression of IDOL in mouse liver with an adenoviral vector reduces LDLR abundance and increases plasma cholesterol levels in mice.5 Conversely, downregulation of endogenous IDOL levels by RNA silencing increases LDLR protein levels and enhances LDL uptake, suggesting that IDOL plays a role in endogenous LDLR regulation. Evidence for the importance of the LXR–IDOL axis in vivo was provided by the observation that LXR agonists reduce LDLR protein levels in mice in a tissue-specific manner depending on the degree of IDOL induction.5
The presence of a RING domain in IDOL strongly suggested that IDOL could function as an E3 ubiquitin ligase, although it had not been shown previously to ubiquitinate a specific target protein. Given the ability of IDOL to promote LDLR degradation, we hypothesized that the LDLR itself might be a target for IDOL-dependent ubiquitination. Consistent with this mechanism, we showed that the ubiquitination of LDLR is dramatically enhanced by the overexpression of IDOL. Furthermore, an IDOL protein harboring a point mutation (C387A) in the catalytic RING domain that inactivates the E3 ligase activity, has no effect on LDLR protein levels.5 Since IDOL is a cytosolic protein, the cytoplasmic tail is the only portion of LDLR protein that is accessible to IDOL. Therefore, one or more residues in the tail must be the target(s) for ubiquitination. Mutational analysis revealed that either an intact K830 or an intact C839 residue on the LDLR tail is required for ubiquitin anchoring and IDOL-mediated degradation of the LDLR.5 Similar to many other E3 ubiquitin ligases, in addition to inducing ubiquitination and degradation of the LDLR, IDOL also ubiquitinates itself and therefore undergoes autodegradation. However, the degradation of the LDLR and the autodegradation of IDOL are carried out through distinct pathways. Using proteasome and lysosome specific inhibitors, it has been shown that LDLR degradation is dependent on the lysosomal pathway, while IDOL autodegradation appears to be dependent on the proteasomal pathway. 5
VLDLR and ApoER2 are also targets for IDOL-mediated degradation
The LDLR family is a large group of proteins that play diverse roles in physiology, extending from lipid metabolism to Wnt signaling to neuronal development.28, 29 Recent work indicates that, in addition to the LDLR, IDOL also ubiquitinates the two closely related LDLR family members [VLDLR and ApoE receptor 2 (ApoER2)] and promotes their degradation.30 Furthermore, available evidence strongly suggests that LDLR, VLDLR and ApoER2 are the only IDOL target proteins in mammals. 31 Similar to LDLR, pharmacologic activation of LXR with synthetic agonists in mice decreases VLDLR and ApoER2 expression in vivo. Interestingly, VLDLR and ApoER2 have been implicated in neuronal migration in the brain as well as lipid metabolism,29 suggesting a potential role for the LXR–IDOL signaling axis in the central nervous system. In support of this possibility, cell culture studies have shown that LXR activation decreases Reelin binding to VLDLR and reduces downstream Dab1 phosphorylation.30 Further studies will be required to define the role of IDOL in brain development and function.
Complementary regulation of the LDLR by the SREBP and the IDOL pathways
The physiological relevance of the LXR–IDOL pathway in regulating cholesterol homeostasis was further substantiated by the analysis of IDOL-deficient embryonic stem (ES) cells. Cells lacking IDOL expression exhibit higher basal levels of LDLR protein and internalize LDL more readily than wild-type cells.32 Furthermore, synthetic LXR agonists have no effect on protein levels of the LDLR or the uptake of LDL in IDOL-null ES cells, indicating that IDOL is absolutely required for LXR's effect on the LDLR.32
IDOL-null cells also proved useful for dissecting the contribution of the LXR and SREBP pathways to the feedback control of LDL uptake by sterols. We found that the LXR– IDOL pathway was activated more rapidly in response to changing sterol concentrations compared to the SREBP pathway. Oxysterol activation of LXR reduces plasma membrane LDLR protein in WT cells with a half-maximum effect as early as 2 h. The reduction in LDLR protein is slower in IDOL-deficient ES cells, where downregulation of the LDLR can only be achieved by SREBP-mediated suppression of LDLR gene transcription.32 This observation suggests a role for IDOL in the acute responses to changing cholesterol levels.
Interestingly, the effects of IDOL on LDLR protein levels appear to be independent of — and additive with — two currently available pharmacologic pathways for LDLR regulation, the statin (SREBP) and PCSK9 pathways. Statin drugs act by inhibiting cholesterol biosynthesis and triggering SREBP-mediated upregulation of LDLR gene transcription.32, 33 Loss of IDOL expression in cultured ES cells further stabilizes the increased LDLR protein levels induced by statin drugs,32 leading to increased protein levels on the cell surface. As discussed earlier, the SREBP pathway also induces the expression of PCSK9, which reduces membrane LDLR by promoting its endocytosis.12 Analysis of IDOL-deficient ES cells established that the mechanisms of action of PCSK9 and IDOL are distinct. Unlike IDOL, which works intracellularly, PCSK9 induces the internalization of the LDLR primarily by binding to the extracellular domain.11, 13 Furthermore, PCSK9 is still able to stimulate LDLR degradation in cells lacking IDOL, indicating that IDOL and PCSK9 function in complementary but independent pathways.32
The structural basis for substrate recognition by IDOL
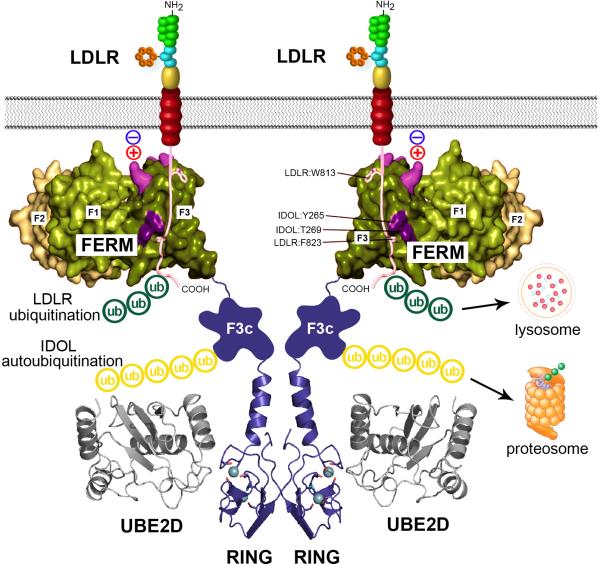
The mechanisms of substrate recognition and ubiquitin transfer in IDOL-dependent protein degradation have been elucidated by several recent studies.31, 34, 35 As mentioned earlier, IDOL is the only protein in the human genome that possesses both a RING E3 ligase domain and a FERM protein–protein interaction motif. It is now clear that these two domains, although both required for IDOL action, are functionally independent. Given that FERM domains have been shown to mediate sequence-specific protein–protein interactions in other proteins, we hypothesized that this domain might be responsibility for IDOL target recognition. Indeed, we showed that the IDOL FERM domain binds directly to a recognition sequence in the cytoplasmic tails of its three known targets–LDLR, VLDLR, and ApoER2 (Figure 1).31 This binding does not require the ubiquitin E3 ligase activity or RING domain, given that target interaction can be observed with isolated FERM domain or with full length IDOL protein in which the structure of the RING domain has been disrupted by mutation.
Figure 1.
Model for the mechanism of recognition and ubiquitination of the LDLR by IDOL. IDOL is an E3 ubiquitin ligase that facilitates the ubiquitination and degradation of the LDLR. IDOL contains an N-terminal FERM domain and a C-terminal RING domain. The FERM domain mediates the recognition between IDOL and the cytoplasmic tail of the LDLR via conserved residues in the F3 subdomain. In addition, residues in the F3 subdomain that are in proximity to the plasma membrane facilitate membrane association and the interaction between IDOL and the LDLR. The RING domain mediates the dimerization of IDOL and the interaction between IDOL and UBE2D, and therefore is essential for the ubiquitin transfer. Ubiquitin can be transferred from UBE2D directly to the lysine and cysteine residues at the cytoplamic tail of the LDLR, leading to its degradation in the lysosome. Alternatively, uniquitin can also be transferred to the lysine residues in the F3c subdomain of IDOL, leading to the autodegradation of IDOL in the proteasome.
Most FERM proteins contain three distinct subdomain folds termed F1, F2 and F3. The IDOL FERM domain is unusual among FERM domains in that it contains a duplication of the phosphotyrosine-binding (PTB)-like fold present in the F3 region. We have referred to these distinct PTB domains as F3a and F3b.31 Mutational studies demonstrated that residues Y265 and T269, which reside in the F3b subdomain, were especially important for IDOL-induced LDLR degradation. An independent study also implicated the neighboring H272 residue.34 Therefore, the F3b subdomain PTB-like fold appears to mediate the recognition between IDOL and the LDLR.31 We further showed that the F3b domain of IDOL binds to a distinct recognition sequence (WxxKNxxSI/MxF) in the cytoplasmic tails of its three target proteins: LDLR, VLDLR, and ApoER2.30 Sequence alignment and structural modeling of these lipoprotein receptors suggested that W813, I821, and F823 in the LDLR tail were key residues mediating the interaction with the IDOL F3b domain, and these predictions were confirmed by mutagenesis.31 Lipoprotein receptors carrying mutations in the IDOL recognition sequence are resistant to IDOL-dependent degradation. Interestingly, the mechanism of IDOL target recognition has been highly conserved in evolution. Defense receptor (DNR)-1, the Drosophila homolog of IDOL, is capable of degrading the human LDLR in mammalian cells,30 and key residues in the DNR-1 F3b subdomain mediating LDLR recognition are functionally conserved.31
Interestingly, cellular membrane appears to be a critical component in the nucleation of the IDOL–LDLR complex. IDOL is unable to promote the degradation of fusion proteins containing the soluble LDLR cytoplasmic domain, but can degrade a heterologous membrane protein carrying the LDLR tail.30 In addition to binding lipoprotein receptor tails, the IDOL FERM domain mediates interaction with negatively charged phospholipids. A combination of biochemical and cell-based assays demonstrated that this tripartite interaction between the FERM domain, membrane phospholipids, and the lipoprotein receptor tail is required for IDOL-dependent degradation (Figure 1).31
Structural basis of IDOL-mediated LDLR ubiquitination
The ubiquitination of proteins involves a cascade of ubiquitin transfer reactions carried out by three categories of enzymes, named E1, E2, and E3. A single E1 can transfer ubiquitin to several species of E2 enzymes, and each E2 acts in concert with either one or several E3 enzymes to induce substrate ubiquitination and the autoubiquitination of E3.36 Although IDOL had been identified as the E3 required for LXR-dependent ubiquitination of the LDLR, the identity of the E2 partner remained unknown until recently. A screen of a panel of E2 enzymes using an in vitro IDOL autoubiquitination assay led to the identification of the UBE2D family (UBE2D1-4) as E2 partners for the IDOL autoubiquitination.35 UBE2Ds are capable of catalyzing the addition of ubiquitin using both K48 and K63 linkages, consistent with the observation that ubiquitin chains formed by IDOL in vitro contain multiple linkages.35 Importantly, UBE2Ds support not only IDOL autoubiquitination, but also IDOL-dependent ubiquitination of the LDLR in a cell-free system. Furthermore, inhibition of UBE2D activity by overexpressing a dominant-negative UBE2D enzyme inhibits the ability of IDOL to degrade the LDLR in cells, strongly suggesting that these are major physiologic IDOL partners.35 However, they may not be the only partners. The UBE2N/UBE2V1 complex has also been shown to mediate ubiquitination of the LDLR using K63-specific ubiquitin linkage.34 The relative importance of these E2 enzymes and of K48 and K63 linkage ubiquitination in IDOL function remain to be clarified.
Structure-guided mutagenesis of the UBE2D-IDOL complex further demonstrated that disruption of the interaction interface between UBE2D and IDOL inhibits the degradation of the LDLR.35 Additionally, the RING domain also mediates the dimerization of IDOL, and dimerization is also required for the ability of IDOL to autoubiquitinate and to ubiquitinate the LDLR (Figure 1). Mutations that disrupt IDOL dimerization also render IDOL unable to degrade the LDLR.35 This finding is consistent with previous reports that dimerization is essential for the function of many other RING-domain containing E3 ligases. 37-39 In-depth structural analysis suggested that the dimerization facilitates ubiquitin transfer by preferentially binding the E2–ubiquitin thioester across the dimer and activating the thioester bond for catalysis.39
Involvement of IDOL in human cholesterol metabolism
IDOL is expressed in a variety of human tissues, but its role in human cholesterol metabolism remains an open question. Until recently, there was no description of any gain- or loss-of-function mutation in the human IDOL gene, and therefore the physiological effect of increased or reduced IDOL activity is still largely unknown in humans. Interestingly, several recent genome-wide association studies have identified SNPs in the region of IDOL gene that are linked to total cholesterol and LDL-cholesterol levels in different cohorts.40, 41 In addition, a recent genome-wide association study found SNPs in the region of IDOL gene to be significantly associated with reduced LDL-cholesterol levels in response to treatment with statin drugs.42
We recently characterized a nonsynonymous single nucleotide polymorphism (SNP) (N342S) in the coding region of the IDOL gene that alters the LDLR degradation activity of the protein.43 Residue 342 is located in the IDOL FERM domain, the region required for the physical interaction between IDOL and the LDLR. The N342 allele is more active in LDLR degradation assays compared to the S342 allele. Furthermore, in a cohort of Mexican hyperlipidemic patients, subjects homozygous for the less active S342 allele exhibited a 7% lower plasma cholesterol levels, a result that would be consistent with increased hepatic LDLR expression.43 These results provide in vivo evidence that modulating IDOL activity can affect the plasma LDL levels in humans. However, the physiologic and pharmacologic relevance of the IDOL pathway for disease pathogenesis or treatment remains to be clarified. There have been no complete loss-of-function or dominant-negative IDOL alleles described to date; therefore, the effects associated with dramatic changes in IDOL activity are unknown. Similarly, it is not known whether pharmacologic inhibition of IDOL-dependent LDLR degradation would substantially affect human lipid levels.
Going forward, it will be important to consider potential functions of the LXR–IDOL pathway in humans beyond the potential to impact hepatic LDLR expression. IDOL is expressed and responsive to LXR activation in other metabolically active tissues, including adipose tissue intestine, as well as in macrophages. The consequence of alteration in LDLR, VLDLR, and ApoER2 expression in these contexts will need to be considered. Moreover, identification of VLDLR and ApoER2 as IDOL targets suggests that IDOL may play as yet unrecognized roles in the central nervous system.30 It will be interesting to determine if genetic variations at the IDOL locus are associated with neurodevelopmental abnormalities in humans.
Acknowledgements
Funding: P.T. is an investigator of the Howard Hughes Medical Institute and was support by HL-066088. P.T., K.R., L.G.F and S.G.Y. are supported by NIH P01 HL090553.
Footnotes
Disclosures: The authors have nothing to disclose.
References
- 1.Brown MS, Goldstein JL. The srebp pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 2.Tontonoz P, Mangelsdorf DJ. Liver x receptor signaling pathways in cardiovascular disease. Molecular endocrinology. 2003;17:985–993. doi: 10.1210/me.2003-0061. [DOI] [PubMed] [Google Scholar]
- 3.Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, Ford E, Furie K, Go A, Greenlund K, Haase N, Hailpern S, Ho M, Howard V, Kissela B, Kittner S, Lackland D, Lisabeth L, Marelli A, McDermott M, Meigs J, Mozaffarian D, Nichol G, O'Donnell C, Roger V, Rosamond W, Sacco R, Sorlie P, Stafford R, Steinberger J, Thom T, Wasserthiel-Smoller S, Wong N, Wylie-Rosett J, Hong Y. Heart disease and stroke statistics--2009 update: A report from the american heart association statistics committee and stroke statistics subcommittee. Circulation. 2009;119:e21–181. doi: 10.1161/CIRCULATIONAHA.108.191261. [DOI] [PubMed] [Google Scholar]
- 4.Pedersen TR, Tobert JA. Simvastatin: A review. Expert opinion on pharmacotherapy. 2004;5:2583–2596. doi: 10.1517/14656566.5.12.2583. [DOI] [PubMed] [Google Scholar]
- 5.Zelcer N, Hong C, Boyadjian R, Tontonoz P. Lxr regulates cholesterol uptake through idol-dependent ubiquitination of the ldl receptor. Science. 2009;325:100–104. doi: 10.1126/science.1168974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown MS, Goldstein JL. A receptor-mediated pathway for cholesterol homeostasis. Science. 1986;232:34–47. doi: 10.1126/science.3513311. [DOI] [PubMed] [Google Scholar]
- 7.Hobbs HH, Russell DW, Brown MS, Goldstein JL. The ldl receptor locus in familial hypercholesterolemia: Mutational analysis of a membrane protein. Annual review of genetics. 1990;24:133–170. doi: 10.1146/annurev.ge.24.120190.001025. [DOI] [PubMed] [Google Scholar]
- 8.Ishibashi S, Brown MS, Goldstein JL, Gerard RD, Hammer RE, Herz J. Hypercholesterolemia in low density lipoprotein receptor knockout mice and its reversal by adenovirus-mediated gene delivery. J Clin Invest. 1993;92:883–893. doi: 10.1172/JCI116663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 10.Brown MS, Goldstein JL. Receptor-mediated endocytosis: Insights from the lipoprotein receptor system. Proc Natl Acad Sci U S A. 1979;76:3330–3337. doi: 10.1073/pnas.76.7.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon HJ, Lagace TA, McNutt MC, Horton JD, Deisenhofer J. Molecular basis for ldl receptor recognition by pcsk9. Proc Natl Acad Sci U S A. 2008;105:1820–1825. doi: 10.1073/pnas.0712064105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang DW, Garuti R, Tang WJ, Cohen JC, Hobbs HH. Structural requirements for pcsk9-mediated degradation of the low-density lipoprotein receptor. Proc Natl Acad Sci U S A. 2008;105:13045–13050. doi: 10.1073/pnas.0806312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang DW, Lagace TA, Garuti R, Zhao Z, McDonald M, Horton JD, Cohen JC, Hobbs HH. Binding of proprotein convertase subtilisin/kexin type 9 to epidermal growth factor-like repeat a of low density lipoprotein receptor decreases receptor recycling and increases degradation. J Biol Chem. 2007;282:18602–18612. doi: 10.1074/jbc.M702027200. [DOI] [PubMed] [Google Scholar]
- 14.Cohen JC, Boerwinkle E, Mosley TH, Jr., Hobbs HH. Sequence variations in pcsk9, low ldl, and protection against coronary heart disease. The New England journal of medicine. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 15.Lagace TA, Curtis DE, Garuti R, McNutt MC, Park SW, Prather HB, Anderson NN, Ho YK, Hammer RE, Horton JD. Secreted pcsk9 decreases the number of ldl receptors in hepatocytes and in livers of parabiotic mice. J Clin Invest. 2006;116:2995–3005. doi: 10.1172/JCI29383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grefhorst A, McNutt MC, Lagace TA, Horton JD. Plasma pcsk9 preferentially reduces liver ldl receptors in mice. Journal of lipid research. 2008;49:1303–1311. doi: 10.1194/jlr.M800027-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams KJ, Brocia RW, Fisher EA. The unstirred water layer as a site of control of apolipoprotein b secretion. J Biol Chem. 1990;265:16741–16744. [PubMed] [Google Scholar]
- 18.Steinberg D, Witztum JL. Inhibition of pcsk9: A powerful weapon for achieving ideal ldl cholesterol levels. Proc Natl Acad Sci U S A. 2009;106:9546–9547. doi: 10.1073/pnas.0904560106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janowski BA, Grogan MJ, Jones SA, Wisely GB, Kliewer SA, Corey EJ, Mangelsdorf DJ. Structural requirements of ligands for the oxysterol liver x receptors lxralpha and lxrbeta. Proc Natl Acad Sci U S A. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor lxr alpha. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 21.Repa JJ, Turley SD, Lobaccaro JA, Medina J, Li L, Lustig K, Shan B, Heyman RA, Dietschy JM, Mangelsdorf DJ. Regulation of absorption and abc1-mediated efflux of cholesterol by rxr heterodimers. Science. 2000;289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- 22.Venkateswaran A, Laffitte BA, Joseph SB, Mak PA, Wilpitz DC, Edwards PA, Tontonoz P. Control of cellular cholesterol efflux by the nuclear oxysterol receptor lxr alpha. Proc Natl Acad Sci U S A. 2000;97:12097–12102. doi: 10.1073/pnas.200367697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bortnick AE, Rothblat GH, Stoudt G, Hoppe KL, Royer LJ, McNeish J, Francone OL. The correlation of atp-binding cassette 1 mrna levels with cholesterol efflux from various cell lines. J Biol Chem. 2000;275:28634–28640. doi: 10.1074/jbc.M003407200. [DOI] [PubMed] [Google Scholar]
- 24.Klucken J, Buchler C, Orso E, Kaminski WE, Porsch-Ozcurumez M, Liebisch G, Kapinsky M, Diederich W, Drobnik W, Dean M, Allikmets R, Schmitz G. Abcg1 (abc8), the human homolog of the drosophila white gene, is a regulator of macrophage cholesterol and phospholipid transport. Proc Natl Acad Sci U S A. 2000;97:817–822. doi: 10.1073/pnas.97.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang N, Silver DL, Costet P, Tall AR. Specific binding of apoa-i, enhanced cholesterol efflux, and altered plasma membrane morphology in cells expressing abc1. J Biol Chem. 2000;275:33053–33058. doi: 10.1074/jbc.M005438200. [DOI] [PubMed] [Google Scholar]
- 26.Olsson PA, Korhonen L, Mercer EA, Lindholm D. Mir is a novel erm-like protein that interacts with myosin regulatory light chain and inhibits neurite outgrowth. J Biol Chem. 1999;274:36288–36292. doi: 10.1074/jbc.274.51.36288. [DOI] [PubMed] [Google Scholar]
- 27.McClatchey AI, Fehon RG. Merlin and the erm proteins--regulators of receptor distribution and signaling at the cell cortex. Trends in cell biology. 2009;19:198–206. doi: 10.1016/j.tcb.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An ldl-receptor-related protein mediates wnt signalling in mice. Nature. 2000;407:535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- 29.Trommsdorff M, Gotthardt M, Hiesberger T, Shelton J, Stockinger W, Nimpf J, Hammer RE, Richardson JA, Herz J. Reeler/disabled-like disruption of neuronal migration in knockout mice lacking the vldl receptor and apoe receptor 2. Cell. 1999;97:689–701. doi: 10.1016/s0092-8674(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 30.Hong C, Duit S, Jalonen P, Out R, Scheer L, Sorrentino V, Boyadjian R, Rodenburg KW, Foley E, Korhonen L, Lindholm D, Nimpf J, van Berkel TJ, Tontonoz P, Zelcer N. The e3 ubiquitin ligase idol induces the degradation of the low density lipoprotein receptor family members vldlr and apoer2. J Biol Chem. 2010;285:19720–19726. doi: 10.1074/jbc.M110.123729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calkin AC, Goult BT, Zhang L, Fairall L, Hong C, Schwabe JW, Tontonoz P. Ferm-dependent e3 ligase recognition is a conserved mechanism for targeted degradation of lipoprotein receptors. Proc Natl Acad Sci U S A. 2011;108:20107–20112. doi: 10.1073/pnas.1111589108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scotti E, Hong C, Yoshinaga Y, Tu Y, Hu Y, Zelcer N, Boyadjian R, de Jong PJ, Young SG, Fong LG, Tontonoz P. Targeted disruption of the idol gene alters cellular regulation of the low-density lipoprotein receptor by sterols and liver x receptor agonists. Molecular and cellular biology. 2011;31:1885–1893. doi: 10.1128/MCB.01469-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao JK, Laufs U. Pleiotropic effects of statins. Annual review of pharmacology and toxicology. 2005;45:89–118. doi: 10.1146/annurev.pharmtox.45.120403.095748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sorrentino V, Scheer L, Santos A, Reits E, Bleijlevens B, Zelcer N. Distinct functional domains contribute to degradation of the low density lipoprotein receptor (ldlr) by the e3 ubiquitin ligase inducible degrader of the ldlr (idol). J Biol Chem. 2011;286:30190–30199. doi: 10.1074/jbc.M111.249557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Fairall L, Goult BT, Calkin AC, Hong C, Millard CJ, Tontonoz P, Schwabe JW. The idol-ube2d complex mediates sterol-dependent degradation of the ldl receptor. Genes Dev. 2011;25:1262–1274. doi: 10.1101/gad.2056211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 37.Johnson AE, Collier SE, Ohi MD, Gould KL. Fission yeast dma1 requires ring domain dimerization for its ubiquitin ligase activity and mitotic checkpoint function. J Biol Chem. 2012;287:25741–25748. doi: 10.1074/jbc.M112.349712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liew CW, Sun H, Hunter T, Day CL. Ring domain dimerization is essential for rnf4 function. The Biochemical journal. 2010;431:23–29. doi: 10.1042/BJ20100957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plechanovova A, Jaffray EG, McMahon SA, Johnson KA, Navratilova I, Naismith JH, Hay RT. Mechanism of ubiquitylation by dimeric ring ligase rnf4. Nature structural & molecular biology. 2011;18:1052–1059. doi: 10.1038/nsmb.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chasman DI, Pare G, Mora S, Hopewell JC, Peloso G, Clarke R, Cupples LA, Hamsten A, Kathiresan S, Malarstig A, Ordovas JM, Ripatti S, Parker AN, Miletich JP, Ridker PM. Forty-three loci associated with plasma lipoprotein size, concentration, and cholesterol content in genome-wide analysis. PLoS genetics. 2009;5:e1000730. doi: 10.1371/journal.pgen.1000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, Johansen CT, Fouchier SW, Isaacs A, Peloso GM, Barbalic M, Ricketts SL, Bis JC, Aulchenko YS, Thorleifsson G, Feitosa MF, Chambers J, Orho-Melander M, Melander O, Johnson T, Li X, Guo X, Li M, Shin Cho Y, Jin Go M, Jin Kim Y, Lee JY, Park T, Kim K, Sim X, Twee-Hee Ong R, Croteau-Chonka DC, Lange LA, Smith JD, Song K, Hua Zhao J, Yuan X, Luan J, Lamina C, Ziegler A, Zhang W, Zee RY, Wright AF, Witteman JC, Wilson JF, Willemsen G, Wichmann HE, Whitfield JB, Waterworth DM, Wareham NJ, Waeber G, Vollenweider P, Voight BF, Vitart V, Uitterlinden AG, Uda M, Tuomilehto J, Thompson JR, Tanaka T, Surakka I, Stringham HM, Spector TD, Soranzo N, Smit JH, Sinisalo J, Silander K, Sijbrands EJ, Scuteri A, Scott J, Schlessinger D, Sanna S, Salomaa V, Saharinen J, Sabatti C, Ruokonen A, Rudan I, Rose LM, Roberts R, Rieder M, Psaty BM, Pramstaller PP, Pichler I, Perola M, Penninx BW, Pedersen NL, Pattaro C, Parker AN, Pare G, Oostra BA, O'Donnell CJ, Nieminen MS, Nickerson DA, Montgomery GW, Meitinger T, McPherson R, McCarthy MI, McArdle W, Masson D, Martin NG, Marroni F, Mangino M, Magnusson PK, Lucas G, Luben R, Loos RJ, Lokki ML, Lettre G, Langenberg C, Launer LJ, Lakatta EG, Laaksonen R, Kyvik KO, Kronenberg F, Konig IR, Khaw KT, Kaprio J, Kaplan LM, Johansson A, Jarvelin MR, Janssens AC, Ingelsson E, Igl W, Kees Hovingh G, Hottenga JJ, Hofman A, Hicks AA, Hengstenberg C, Heid IM, Hayward C, Havulinna AS, Hastie ND, Harris TB, Haritunians T, Hall AS, Gyllensten U, Guiducci C, Groop LC, Gonzalez E, Gieger C, Freimer NB, Ferrucci L, Erdmann J, Elliott P, Ejebe KG, Doring A, Dominiczak AF, Demissie S, Deloukas P, de Geus EJ, de Faire U, Crawford G, Collins FS, Chen YD, Caulfield MJ, Campbell H, Burtt NP, Bonnycastle LL, Boomsma DI, Boekholdt SM, Bergman RN, Barroso I, Bandinelli S, Ballantyne CM, Assimes TL, Quertermous T, Altshuler D, Seielstad M, Wong TY, Tai ES, Feranil AB, Kuzawa CW, Adair LS, Taylor HA, Jr., Borecki IB, Gabriel SB, Wilson JG, Holm H, Thorsteinsdottir U, Gudnason V, Krauss RM, Mohlke KL, Ordovas JM, Munroe PB, Kooner JS, Tall AR, Hegele RA, Kastelein JJ, Schadt EE, Rotter JI, Boerwinkle E, Strachan DP, Mooser V, Stefansson K, Reilly MP, Samani NJ, Schunkert H, Cupples LA, Sandhu MS, Ridker PM, Rader DJ, van Duijn CM, Peltonen L, Abecasis GR, Boehnke M, Kathiresan S. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chasman DI, Giulianini F, Macfadyen J, Barratt BJ, Nyberg F, Ridker PM. Genetic determinants of statin-induced low-density lipoprotein cholesterol reduction: The justification for the use of statins in prevention: An intervention trial evaluating rosuvastatin (jupiter) trial. Circulation. Cardiovascular genetics. 2012;5:257–264. doi: 10.1161/CIRCGENETICS.111.961144. [DOI] [PubMed] [Google Scholar]
- 43.Weissglas-Volkov D, Calkin AC, Tusie-Luna T, Sinsheimer JS, Zelcer N, Riba L, Tino AM, Ordonez-Sanchez ML, Cruz-Bautista I, Aguilar-Salinas CA, Tontonoz P, Pajukanta P. The n342s mylip polymorphism is associated with high total cholesterol and increased ldl receptor degradation in humans. J Clin Invest. 2011;121:3062–3071. doi: 10.1172/JCI45504. [DOI] [PMC free article] [PubMed] [Google Scholar]