Abstract
Objective:
Acute Otitis Media (AOM) is a common bacterial infection in childhood that causes an inflammatory response in the middle ear. Leukocytes produce different inflammatory molecules in-vitro when stimulated with Gram-positive and Gram-negative bacteria. The major causes of AOM are Streptococcus pneumoniae, Nontypeable Haemophilus influenzae andMoraxella catarrhalis. We sought to assess differences in cytokines, chemokines and expression of Toll-like receptors (TLRs) at onset of AOM based on bacterial culture results.
Study Design:
Middle ear fluids (MEF) from 66 children with AOM were studied.
Methods:
Innate immune genes, cytokines (IL-6, IL-8, IL-10, IL1-β, TNF-α), chemokines (CCL2, CCL3, CCL4, CCR5, CXCR3) and Toll-like receptor (TLR2, TLR4 & TLR9) expression was measured using real-time RT-PCR from MEF collected in-vivo by tympanocentesis.
Results:
Culture positive MEFs had higher levels of all cytokines and chemokines (9-300 folds) as compared to MEFs that were culture negative. PCR positive/culture negative MEF for otopathogens showed significant differences (p<0.01) in TLR2, TLR4 and TLR9 expression (6-31 fold), but cytokine and chemokine levels were similar compared to PCR negative/culture negative MEF. No significant differences were found in the cytokine/chemokine/TLR levels among the bacterial otopathogen species. However, higher levels of TLRs, and all the cytokine and chemokines were detected when more than one bacterial species was present compared to single otopathogens.
Conclusion:
Expression levels of pro-inflammatory cytokines/chemokines and TLRs are elevated in AOM children with a bacterial otopathogen, and are dependent on the number of bacterial species identified.
Level of Evidence:
NA
Keywords: Acute otitis media, middle ear fluid, cytokine/chemokine, toll-like receptors
Introduction
Acute otitis media (AOM) in childhood accounts for more pediatric office visits and antibiotics prescription than any other childhood disease. Bacterial infection is the dominant etiology of AOM, most commonly Streptococcus pneumoniae (Spn), Nontypeable Haemophilus influenzae (NTHi) and Moraxella catarrhalis (Mcat). However, bacterial pathogens are not detected by culture from the middle-ear fluid (MEF) in approximately 15-30% of AOM cases.1-4 Preceding viral upper respiratory infection is the most important predisposing factor for the bacterial invasion of middle ear resulting in AOM.5 Some studies have shown that viruses can be found in the middle-ear fluid either alone or together with bacteria and suggest that viruses may be causative agents of AOM in samples that are culture negative for bacteria.5,6 The pathogenesis of AOM involves a complex interplay between viruses, bacteria, and the host's inflammatory response.
The first step in the activation of human innate response against microbes is the recognition of pathogens by antigen presenting cells (APCs which recognize pathogen associated molecular patterns (PAMPs), generating intracellular signals and subsequent production of cytokines and chemokines.7 Toll-like receptors on APCs bind to infecting microbes by recognition of PAMPc and directly induce host-defense responses. TLR9 specifically recognizes the unmethylated cytidine-phosphate-guanosine (CpG) motifs in bacterial DNA. TLR2, TLR4 and TLR9 signaling have been shown to be critical to the regulation of infection in otitis media.8,9 Locally produced cytokines play a major role in the pathogenesis of AOM especially in initiation and maintenance of inflammation.The role of chemokines like The role of chemokines like CCL2, CCL3 and CCR5 contributing to inflammation has also been explored in the pathogenesis of experimental otitis media.10,11,12 TLR genes and signaling molecules are up-regulated in OM in murine models. Deletion of several key innate immune genes has resulted in persistent OM in mice, coupled with an inability to clear bacterial infection from the middle ear.13
A better understanding of the pathogenesis and the inflammatory process associated with AOM would be helpful in developing more effective strategies for the management and prevention of this childhood disease. The aim of the present study was to evaluate the differences and relationships between the expression of cytokines, chemokines and TLRs at onset of AOM based on bacterial culture results. Specifically variations in expression of inflammatory cytokines interleukin-6 (IL-6), IL-8, IL-10, IL1β and tumor necrosis factor alpha (TNF-α); chemokines CCL2 (MCP-1), CCL3, CCL4, CCR5 and CXCR3 and Toll like receptors TLR2, TLR4, TLR9 were compared in middle ear fluids (collected by tympanocentesis) of children with AOM where any of the major otopathogens were detected by culture compared to negative culture MEFs. Comparisons were made in culture negative MEFs in relation to PCR detection of otopathogens (Spn, NTHi and Mcat). Previous studies have suggested that gram-positive and gram-negative bacteria elicit different patterns of pro-inflammatory cytokines and chemokines.14-16 We investigated whether these differences in cytokine, chemokine and TLRs expression levels exist in relation to Gram negative vs Gram positive bacteria in the MEF during AOM.
Methods
Study populations and sample collection
The details of the study design have been previously described.17 Children were enrolled in the study at age 6 months and prospectively followed until 30-36 months of age. Diagnosis of AOM by validated otoscopists was based on signs of inflammation (bulging eardrum) of the tympanic membrane and presence of middle ear fluid (MEF) documented by tympanocentesis. More than one MEF sample was collected by tympanocentesis from children if they have multiple episodes of AOM. Only one MEF sample was included in this study analysis from each child except two children whose occurrence of AOM was more than 6 months apart. MEF was cultured and tested by PCR for otopathogens. Participants received all doses of pneumococcal conjugate vaccine (PCV) according to the U.S. schedule; either PCV7 or PCV13 depending on the date of enrollment. The study was approved by the Rochester General Hospital IRB.
Microbiology and PCR detection of pathogens
Sampling procedures, microbiology processing and identification, and molecular testing for organism identification have been previously described. 3,18,19 Bacterial identification was performed on all MEF samples using standard laboratory culture techniques. Typically 75-80% MEF has common detectable otopathogens isolates such as Spn (~31%), NTHi (33%) and Mcat (14%); ~20% of the MEF during AOM were culture negative as shown in previous studies. 3, 4 Multiplex-PCR targeting 16S gene was used for bacterial detection of Spn, NTHi and Mcat in culture negative samples. The primers information and PCR experiment condition has been described in detail previously.18 It is possible that other relevant bacteria could be present in culture negative MEF samples if tested via PCR but focus was given only to 3 otopathogens of interest.
Isolation of RNA from MEF samples
MEF diluted in PBS was centrifuged for 10 minutes at 4°C. After aliquoting the supernatant, 1 ml TRIZOL reagent was added to the pellet, mixed by pipetting and left at room temperature for 5 minutes. Samples were stored at −80°C. For RNA isolation, 100μl Bromo-chloro phenol/ml was added to thawed samples containing TRIZOL followed by gentle shaking for 5-10 sec to ensure a uniform cloudy solution. The mixture were incubated at room temperature for 5 minutes and then centrifuged at 12,000 g for 10 min at 4°C. After centrifugation, two distinct layers formed. The layers were separated and 5μg/ml RNase-free glycogen was added to the aqueous layer followed by gentle shaking. 500μl of isopropanol was added to the aqueous layer, mixed well by shaking and incubated at room temperature for 5 minutes. After this step the entire contents were transferred to a RNA isolation column from QIAGEN RNeasy kit and manufacturer’s instructions followed. To remove traces of DNA if any, a DNA digestion step was performed using DNase I enzyme from Qiagen. After RNA isolation, RNase inhibitor was added to each sample to avoid degradation. Absence of DNA was confirmed by PCR and the quality and quantity of RNA was determined by spectrophotometry and agarose gel electrophoresis. Criteria for inclusion in downstream applications was based on OD 260/280 of >2.0 and the absence of visible degradation.
cDNA synthesis and realtime PCR
First strand cDNA was generated for each RNA samples isolated from MEF with the Maxima® First Strand cDNA Synthesis Kit (Fermentas) and manufacturer’s instructions followed. For the innate immune response molecules, human primers for each specific cytokine, chemokine and Toll like receptor were purchased from Applied Biosystems. Real-time PCR assays were run for each sample containing 50-100 ng of cDNA as a template and specific primers using SYBER Green dye on an iCycler IQ from Bio-Rad. Both 18S rRNA and GapDH genes expression was used as a control to normalize the data. The standard amplification conditions were comprised of 38 cycles, each consisting of 15 sec of denaturation at 95°C, followed by 30 sec of annealing at the optimal temperature and 30 sec of elongation at 72°C.
Data analysis
We used a semi-quantitative/relative quantitative approach to analyze the real-time PCR data from the clinical samples for relative mRNA concentration. The primary assumption with this approach was that the additive effect of concentration, gene, and replicate can be adjusted by subtracting Ct number of the target gene from that of a reference gene, which will provide ΔCt. Average of 18S rRNA gene and GapDH gene data were used as reference controls. The result presented represent relative expression 2−ΔCt or fold change given by the equation 2−ΔΔCt, a convenient way to analyze the relative changes in gene expression from real-time quantitative PCR experiments.20,21 Mann-Whitney tests were used to assess significant differences.
Results
Bacterial pathogen detection by culture and PCR
Sixty-six children (median age 16 months: range 6 months to 30 months of age and mostly Caucasians) with AOM were included in this study. Middle ear fluids were obtained from each child using tympanocentesis. MEF cultures were positive for a common bacterial otopathogens (NTHi, Spn and Mcat) in 42 cases and 24 MEF samples were culture negative. Among 42 culture positive middle ear fluids, 10 NTHi, 12 Spn, 10 Mcat and 10 mixed otopathogens (NTHi and/or Spn and/or Mcat) from MEF samples were included in this study.
PCR was performed on all the samples. All culture-positive samples were also positive by PCR. In several culture positive samples, the PCR revealed not only bacteria found by culture but also one or two additional otopathogens. Among the 24 culture-negative MEF samples, only 10 were negative by PCR. Thus, additional otopathogen were detected with PCR in the samples negative by culture.
Inflammatory innate molecules response in relation to bacterial culture findings
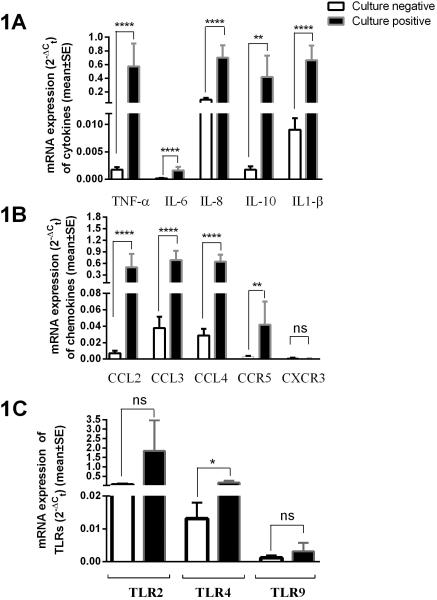
Cytokine (TNF-α, IL-6, IL-8, IL-10 and IL1-β,) and chemokine (CCL2, CCL3, CCL4, CCR5, CXCR3) levels and Toll-like receptor (TLR2, TLR4 & TLR9) responses were measured using real-time RT-PCR from middle-ear fluids (MEF) related to the culture findings. When relative mRNA expression was compared for cytokines and chemokines, culture positive MEFs had significantly higher levels (8-330 fold differences) compared to culture negative MEF samples as shown in figure 1A and 1B indicating higher proinflammatory responses in MEF samples when bacterial otopathogens were detected. The relative mRNA expression of TLRs responses in comparison of culture positive and culture negative samples varied as shown in figure 1C. TLR4 expression was >10 fold higher in culture positive compared to culture negative MEF samples. A similar, but not statistically significant trend was observed for TLR2. No difference in TLR9 responses was observed.
Figure 1.
Comparison of tumor-necrosis factor- α (TNF-α), interleukins (IL-6, IL-8, IL-10 & IL-1β) (shown in 1A) chemokines (CCL2, CCL3, CCL4) and chemokines receptors (CCR5, CXCR3) levels (shown in 1B) and toll-like receptors (TLR2, TLR4 & TLR9) expression (shown in 1C) in MEF of children with AOM. Results shown are relative mRNA expression of culture negative (n=24) vs. culture positive (n=42) MEF samples.
Note: All studied cytokines and chemokines showed significantly higher expression in culture positive compared to culture negative MEFs. **** p <0.0001, **; p <0.001, * p value <0.05, ns: not significant
Since we prospectively followed the children after each AOM event we sought to determine if the clinical course in the subjects had an association of TLR levels and a next episode of AOM in these subjects. There was no association observed in the level of TLR expression with the likelihood of a next AOM.
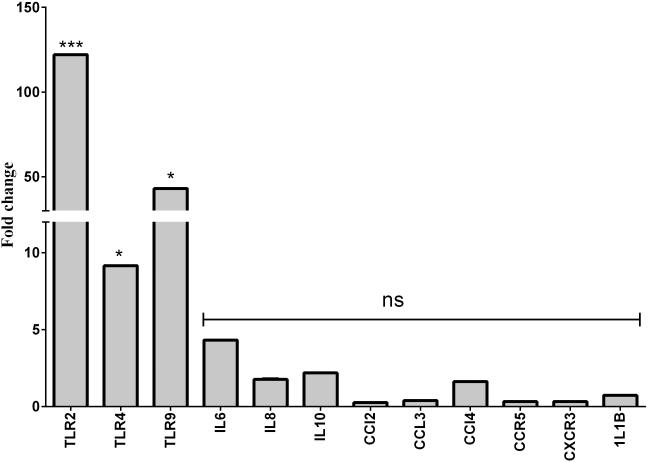
Inflammatory innate molecules response in relation to PCR finding of bacterial otopathogens in culture negative samples
24 culture negative samples were analyzed for Spn, NTHi and Mcat by PCR. 14 culture negative samples showed presence of Spn, NTHi or Mcat by PCR. 10 samples were negative for 3 bacterial otopathogens DNA of interest. Cytokine and chemokine levels and TLRs expression was compared in PCR positive compared to PCR negative samples in culture negative MEFs. Figure 2 shows the fold differences in mRNA for various cytokines, chemokines and TLRs. TLR2, TLR4 and TLR9 expression were found to be significantly higher (p<0.05) in PCR positive samples (6.4-30.9 fold difference) compared to PCR negative MEF samples; No significant difference was observed in the expression of any cytokine and chemokines in relation to PCR detection of bacterial pathogens in culture negative MEFs.
Figure 2.
Fold change mRNA expression of innate gene molecules from PCR negative (n=10) vs PCR positive (n=14) for bacterial (S. pneumoniae, NTHi and M.catarrhalis) DNA from MEF samples that were culture negative
Note: *** mean <0.001, * mean <0.05, ns: not significant.
Inflammatory innate responses in relation to individual bacterial otopathogens
All the culture positive MEF samples were grouped according to whether NTHi, Spn, Mcat or mixed otopathogens were detected and the mRNA expression of various inflammatory innate responses from each group was compared to culture negative samples. Since culture negative samples have very low expression of innate immune molecules, its data was used to compute the relative expression among all groups given as fold change using the formula 2−ΔΔCt . Table 1 shows the fold change in innate immune response levels in MEF of children with AOM caused by different groups. Only TLR9, IL-10 and CXCR3 of the 13 tested molecules didn’t show significant difference from negative culture samples. Although some variations are observed among the groups of single otopathogens, they did not reach statistically significant difference due to individual variability among the different MEF. However, there is a trend of higher expression in MEF containing mix pathogens compared to single otopathogens.
Table 1.
Fold change Innate immune response in MEF of AOM children with different bacterial otopathogens compared to culture negative MEF
| Innate immune molecules |
S. pneumoniae
(n=12) |
NTHi
(n=10) |
M. catarrhalis
(n=10) |
Mix pathogens (Spn +or Mcat +or NTHi+) (n=10) |
|---|---|---|---|---|
| TLR2 | 10.8 | 36.5 | 6.1 | 53.5 |
| TLR4 | 4.7 | 16.0 | 7.8 | 30.5 |
| TLR9 | 1.0 | 1.4 | 2.1 | 222.1* |
| TNF-α | 16.8 | 55.4 | 45.2 | 126.2 |
| IL-6 | 10.4 | 17.1 | 34.5 | 18.0 |
| IL-8 | 3.7 | 13.4 | 9.3 | 10.4 |
| IL-10 | 2.0 | 1.3 | 3.7 | 20.9 |
| IL-1-β | 20.6 | 63.1 | 55.1 | 131.1 |
| CCL2 | 5.4 | 10.5 | 12.6 | 21.4 |
| CCL3 | 6.0 | 28.1 | 20.3 | 34.8 |
| CCL4 | 11.6 | 39.3 | 29.2 | 21.7 |
| CXCR3 | 0.1 | 0.3 | 0.3 | 5.4 |
| CCR5 | 16.8 | 8.3 | 19.1 | 49.7 |
Note: Almost all the culture positive samples with any bacteria have significant higher expression (p <0.05) of all the innate molecules compared to culture negative MEFs samples except the values shown in bold.
Expression is significantly different compared to single pathogens
Discussion
In this study, we show that detection of otopathogenic bacteria by culture is highly associated with significantly higher inflammatory cytokine and chemokine levels and higher TLR expression compared to otopathogen culture negative but PCR-positive MEF samples and compared to culture/PCR negative samples. The result suggests that differences in inflammatory cytokine and chemokine levels and TLRs expression varies with the presence of otopathogenic bacteria and with bacterial diversity (more than one bacterial species) in the MEF.
Previously, Brazilaia et al 22 found that IL-6 was lower in MEF when bacteria were eliminated from the MEF after antibiotic treatment an observation that is consistent with our observation. Similarly, we observed low levels of IL8, TNF-α and IL1-β in MEF when bacterial cultures were negative from MEF, again consistent with previous studies.23-26 But here for the first time low levels of IL-10, CCL2, CCL3, CCL4 and CCR5 were also measured in culture negative MEF samples. Most prior studies were focusing on individual cytokines. Our data extend those studies by extending the measurements to several additional pro-inflammatory cytokines and chemokines.
IL-10 plays an important role in down-regulating the inflammatory response. We have shown in a previous study that higher levels of IL-10 in serum are detectable at the onset of AOM caused by Spn compared to NTHi and Mcat.27 CCL2 has been shown to be a key molecule contributing to inflammation through CCR2-mediated recruitment of monocytes.12 The role of chemokine CCL3 in experimental otitis media has also been explored and shown to be a downstream effector of TNF-mediated inflammation.10 Others have observed higher expression of CCR5 and CXCR3 receptors during AOM. CXCR3 and CCR5 were preferentially expressed on Th1 cells.11 Therefore the increase expression levels of CXCR3 and CCR5 suggests an active and ongoing immune response against AOM infection mediated by the Th1 arm of the adaptive immune response. Neutrophils are the main cells found in MEF at onset of AOM and a prior study has found that encounter of neutrophils with bacterial pathogens modulates chemokine receptor expression patterns.28
When we compared TLR expression in relation to culture results of MEF, TLR4 showed significantly higher expression and TLR2 a trend for higher expression in culture positive MEF. Using an otitis media model, Leichtle et al has shown that TLR4 signaling probably induces TLR2 expression 8 and TLR2 activation is critical for bacterial clearance and timely resolution of otitis media. Although we did not detect differences in TLR9 expression in culture positive vs. culture negative MEFs, the role of DNA sensing via TLR9 has also been shown in otitis media pathogenesis and recovery.9 Based on these previous studies and our own, similar expression of TLR2 and TLR9 in culture positive vs. culture negative samples suggests that expression of these receptors is involved in the resolution of otitis media.
Negative MEF cultures might occur due to technical inadequacies, therefore we compared innate immune response differences in culture negative MEF by applying PCR techniques for the detection of bacterial otopathogens. We didn’t find a significant difference in the studied cytokine or chemokines when comparing culture negative/PCR positive MEFs with culture/ PCR negative MEFS; the result is consistent with a previous study. 26 However we did find significantly higher expression of TLR2, TLR4 and TLR9 in culture negative/ PCR positive MEF compared to culture and PCR negative MEF suggesting that TLR signaling can be up-regulated with traces of bacterial DNA. Culture negative/PCR positive data suggest that bacteria are not alive or the level of bacteria in MEF is below detection limits. Intact innate immune signaling system has been shown to be critical to recovery from bacterial OM in an animal model.13 TLR2, TLR4 and TLR9 play important role in bacterial defense.
We did not seek to detect respiratory viruses in the MEFs we studied. A viral etiology has been proposed for AOM especially if MEF is culture negative for otopathogens.5,6,29 However, we can conclude that if respiratory viruses were present in MEF that they induce low pro-inflammatory responses. Consistent with our conclusion, Stol K et al showed no association of viruses and changes in cytokines levels in chronic otitis media.30
We compared innate immunity molecule expression in culture positive MEFs that contain gram positive (Spn), gram negative (NTHi or Mcat) and mixtures of bacteria (NTHi or Spn or Mcat). No significant difference was observed in the innate response whether AOM was caused by gram positive compared to gram negative bacteria. However, for the first time we demonstrated increased levels of innate immune response molecules when more than one pathogen was causing AOM.
Our observations provide a clinical and immunologic correlation for AOM. At onset of infection there is a brisk rise in pro-inflammatory cytokines, and chemokines and up-regulation of TLRs, most likely expressed predominantly by immune cells present in MEF. This immunologic phenomenon is reflected clinically by the signs and symptoms of inflammation in the middle ear. After onset of the pro-inflammatory response the time course of recovery from inflammation involves fairly rapid disappearance of elevated cytokines and chemokines and somewhat slower disappearance of immune response cells expressing higher levels of TLR expression. Thus our results suggest that when a culture of MEF is negative but PCR detection is positive for an otopathogen the bacterial infection was either not of very recent onset or was already controlled and perhaps resolving (eradication of the bacteria) when the MEF sample was obtained. Thirdly, combinations of otopathogens during a single AOM event have been shown to occur much more frequently in recurrent AOM, chronic AOM, and in Bedoins vs. Jews living in Israel (associated with epidemiologic and other risk factors).3,31,32 The finding of higher cytokine, chemokine and TLR levels in multiple otopathogen infections compared to single otopathogen infections supports the notion that multiple otopathogen infections results in more damaging inflammatory responses to the middle ear as a bystander negative effect of the more robust immune response.
Conclusion
We found cytokine and chemokine levels and TLR expression differs when otopathogens are present in sufficient numbers to be detected by culture as compared to when bacterial load is below culture detection but within the limit of detection by molecular techniques (PCR). Moreover, in culture negative but PCR positive MEFs, pro-inflammatory responses measured in levels of cytokines and chemokines are low but TLR stimulation occurs.
Acknowledgements
The authors thank all children and parents who participated in this study. The authors are grateful to the staff of Legacy Pediatrics for securing samples and Paridhi Malik for help in some experiments.
Financial Disclosure: This study was funded by the Hearing Health Foundation (Dr. Kaur, PI) and NIH NIDCD RO1 08671 (Dr. Pichichero).
Footnotes
Conflicts of Interest: Authors have no conflicts to declare.
References
- 1.Bluestone CDK, Microbiology JO. In: Otitis media in infants and children. Klein BaJO., editor. W. B. Saunders; Philadelphia, Pa.: 2001. pp. 79–101. [Google Scholar]
- 2.Casey JR, Pichichero ME. Changes in frequency and pathogens causing acute otitis media in 1995-2003. The Pediatric infectious disease journal. 2004;23:824–828. doi: 10.1097/01.inf.0000136871.51792.19. [DOI] [PubMed] [Google Scholar]
- 3.Casey JR, Adlowitz DG, Pichichero ME. New patterns in the otopathogens causing acute otitis media six to eight years after introduction of pneumococcal conjugate vaccine. The Pediatric infectious disease journal. 2010;29:304–309. doi: 10.1097/INF.0b013e3181c1bc48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casey JR, Kaur R, Friedel VC, Pichichero ME. Acute otitis media otopathogens during 2008 to 2010 in Rochester, New York. The Pediatric infectious disease journal. 2013;32:805–809. doi: 10.1097/INF.0b013e31828d9acc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heikkinen T, Chonmaitree T. Importance of respiratory viruses in acute otitis media. Clinical microbiology reviews. 2003;16:230–241. doi: 10.1128/CMR.16.2.230-241.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Patel JA, Nair S, Grady J, et al. Systemic cytokine response profiles associated with respiratory virus-induced acute otitis media. The Pediatric infectious disease journal. 2009;28:407–411. doi: 10.1097/INF.0b013e318194b7c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nature immunology. 2004;5:975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- 8.Leichtle A, Hernandez M, Pak K, et al. TLR4-mediated induction of TLR2 signaling is critical in the pathogenesis and resolution of otitis media. Innate immunity. 2009;15:205–215. doi: 10.1177/1753425909103170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leichtle A, Hernandez M, Lee J, et al. The role of DNA sensing and innate immune receptor TLR9 in otitis media. Innate immunity. 2012;18:3–13. doi: 10.1177/1753425910393539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leichtle A, Hernandez M, Ebmeyer J, et al. CC chemokine ligand 3 overcomes the bacteriocidal and phagocytic defect of macrophages and hastens recovery from experimental otitis media in TNF−/− mice. Journal of immunology. 2010;184:3087–3097. doi: 10.4049/jimmunol.0901167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qin S, Rottman JB, Myers P, et al. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. The Journal of clinical investigation. 1998;101:746–754. doi: 10.1172/JCI1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo JI, Pan H, Oh S, Lim DJ, Moon SK. Spiral ligament fibrocyte-derived MCP-1/CCL2 contributes to inner ear inflammation secondary to nontypeable H. influenzae-induced otitis media. BMC infectious diseases. 2010:10–314. doi: 10.1186/1471-2334-10-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leichtle A, Lai Y, Wollenberg B, Wasserman SI, Ryan AF. Innate signaling in otitis media: pathogenesis and recovery. Current allergy and asthma reports. 2011;11:78–84. doi: 10.1007/s11882-010-0158-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hessle C, Andersson B, Wold AE. Gram-positive bacteria are potent inducers of monocytic interleukin-12 (IL-12) while gram-negative bacteria preferentially stimulate IL-10 production. Infection and immunity. 2000;68:3581–3586. doi: 10.1128/iai.68.6.3581-3586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hessle CC, Andersson B, Wold AE. Gram-negative, but not Gram-positive, bacteria elicit strong PGE2 production in human monocytes. Inflammation. 2003;27:329–332. doi: 10.1023/b:ifla.0000006700.41614.21. [DOI] [PubMed] [Google Scholar]
- 16.Hessle CC, Andersson B, Wold AE. Gram-positive and Gram-negative bacteria elicit different patterns of pro-inflammatory cytokines in human monocytes. Cytokine. 2005;30:311–318. doi: 10.1016/j.cyto.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Friedel V, Zilora S, Bogaard D, Casey JR, Pichichero ME. Five-year prospective study of paediatric acute otitis media in Rochester, NY: modelling analysis of the risk of pneumococcal colonization in the nasopharynx and infection. Epidemiology and Infection. 2013 doi: 10.1017/S0950268813003178. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaur R, Adlowitz DG, Casey JR, Zeng M, Pichichero ME. Simultaneous assay for four bacterial species including Alloiococcus otitidis using multiplex-PCR in children with culture negative acute otitis media. The Pediatric infectious disease journal. 2010;29:741–745. doi: 10.1097/INF.0b013e3181d9e639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaur R, Chang A, Xu Q, Casey JR, Pichichero ME. Phylogenetic relatedness and diversity of non-typable Haemophilus influenzae in the nasopharynx and middle ear fluid of children with acute otitis media. Journal of medical microbiology. 2011;60:1841–1848. doi: 10.1099/jmm.0.034041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nature protocols. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 21.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 22.Barzilaia A, Dekel B, Dagan R, Leibovitz E. Middle ear effusion IL-6 concentration in bacterial and non-bacterial acute otitis media. Acta paediatrica. 2000;89:1068–1071. doi: 10.1080/713794564. [DOI] [PubMed] [Google Scholar]
- 23.Barzilai A, Dekel B, Dagan R, Passwell JH, Leibovitz E. Cytokine analysis of middle ear effusions during acute otitis media: significant reduction in tumor necrosis factor alpha concentrations correlates with bacterial eradication. The Pediatric infectious disease journal. 1999;18:301–303. doi: 10.1097/00006454-199903000-00022. [DOI] [PubMed] [Google Scholar]
- 24.Barzilai A, Leibovitz E, Laver JH, et al. Dynamics of interleukin-1 production in middle ear fluid during acute otitis media treated with antibiotics. Infection. 1999;27:173–176. doi: 10.1007/BF02561523. [DOI] [PubMed] [Google Scholar]
- 25.Leibovitz E, Dagan R, Laver JH, et al. Interleukin 8 in middle ear fluid during acute otitis media: correlation with aetiology and bacterial eradication. Archives of disease in childhood. 2000;82:165–168. doi: 10.1136/adc.82.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skovbjerg S, Roos K, Nowrouzian F, et al. High cytokine levels in perforated acute otitis media exudates containing live bacteria. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2010;16:1382–1388. doi: 10.1111/j.1469-0691.2010.03083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu K, Kaur R, Almudevar A, Pichichero ME. Higher serum levels of interleukin 10 occur at onset of acute otitis media caused by Streptococcus pneumoniae compared to Haemophilus influenzae and Moraxella catarrhalis. The Laryngoscope. 2013;123:1500–1505. doi: 10.1002/lary.23973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartl D, Krauss-Etschmann S, Koller B, et al. Infiltrated neutrophils acquire novel chemokine receptor expression and chemokine responsiveness in chronic inflammatory lung diseases. Journal of immunology. 2008;181:8053–8067. doi: 10.4049/jimmunol.181.11.8053. [DOI] [PubMed] [Google Scholar]
- 29.Chonmaitree T, Heikkinen T. Viruses and acute otitis media. The Pediatric infectious disease journal. 2000;19:1005–1007. doi: 10.1097/00006454-200010000-00013. [DOI] [PubMed] [Google Scholar]
- 30.Stol K, Diavatopoulos DA, Graamans K, et al. Inflammation in the middle ear of children with recurrent or chronic otitis media is associated with bacterial load. The Pediatric infectious disease journal. 2012;31:1128–1134. doi: 10.1097/INF.0b013e3182611d6b. [DOI] [PubMed] [Google Scholar]
- 31.Falup-Pecurariu O, Leibovitz E, Mercas A, et al. Pneumococcal acute otitis media in infants and children in central Romania, 2009-2011: microbiological characteristics and potential coverage by pneumococcal conjugate vaccines. International journal of infectious diseases : IJID : official publication of the International Society for Infectious Diseases. 2013;17:e702–706. doi: 10.1016/j.ijid.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 32.Pichichero ME, Casey JR, Hoberman A, Schwartz R. Pathogens causing recurrent and difficult-to-treat acute otitis media, 2003-2006. Clinical pediatrics. 2008;47:901–906. doi: 10.1177/0009922808319966. [DOI] [PubMed] [Google Scholar]