Abstract
Objectives
Recent clinical trial data cast doubt on the utility of genotype-guided warfarin dosing, specifically showing worse dosing with a pharmacogenetic versus clinical dosing algorithm in African Americans. However, many genotypes important in African Americans were not accounted for. We aimed to determine if omission of the CYP2C9*5, *6, *8, *11 alleles and rs12777823 G>A genotype affects performance of dosing algorithms in African Americans.
Methods
In a cohort of 274 warfarin-treated African Americans, we examined the association between the CYP2C9*5, *6, *8, *11 alleles and rs12777823 G>A genotype and warfarin dose prediction error with pharmacogenetic algorithms used in clinical trials.
Results
The warfarindosing.org algorithm over-estimated doses by a median (IQR) of 1.2 (0.02 to 2.6) mg/day in rs12777823 heterozygotes (p<0.001 for predicted versus observed dose), 2.0 (0.6 to 2.8) mg/day in rs12777823 variant homozygotes (p=0.004), and 2.2 (0.5 to 2.9) mg/day in carriers of a CYP2C9 variant (p<0.001). The International Warfarin Pharmacogenetics Consortium (IWPC) algorithm under-dosed warfarin by 0.8 (−2.3 to 0.4) mg/day for patients with the rs12777823 GG genotype (p<0.001) and over-dosed warfarin by 0.7 (−0.4 to 1.9) mg/day in carriers of a variant CYP2C9 allele (p=0.04). Modifying the warfarindosing.org algorithm to adjust for variants important in African Americans led to better dose prediction than either the original warfarindosing.org (p<0.01) or IWPC (p<0.01) algorithm.
Conclusions
These data suggest that, when providing genotype-guided warfarin dosing, failure to account for variants important in African Americans leads to significant dosing error in this population.
Keywords: Pharmacogenomics, warfarin, genotype, CYP2C9, rs12777823
Introduction
While warfarin remains the most commonly prescribed oral anticoagulant[1], its use is complicated by a narrow therapeutic index and wide inter-individual variability in dose requirements.[2] Numerous studies have consistently and unequivocally shown that genotype affects warfarin dose requirements.[3] Accordingly, Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines recommend dosing warfarin based on genotype when genetic information is available and using either the International Warfarin Pharmacogenetics Consortium (IWPC) or www.warfarindosing.org (WD) algorithm to assist with dosing.[4-6]
Two randomized, controlled trials evaluating the clinical utility of genotype-guided warfarin dosing were published in late 2013. The European Pharmacogenetics of Anticoagulation Therapy (EU-PACT) trial found that dosing warfarin with a modified version of the IWPC algorithm was superior to conventional dosing in the homogenous European population.[7] The Clarification of Optimal Anticoagulation through Genetics (COAG) trial compared warfarin dosing with the WD pharmacogenetic algorithm and found no benefit with this approach compared to dosing based on clinical factors alone in the ethnically diverse population.[8] Moreover, African Americans were more likely to be over-dosed with the pharmacogenetic algorithm. Investigators have questioned why African Americans had worse outcomes with genotype-guided dosing, and we and others have proposed that this might be due to ethnic differences in genotype frequencies.[9-12] Specifically, pharmacogenetic algorithms used in the EU-PACT and COAG trials only included the vitamin K epoxide reductase complex 1 (VKORC1) −1639 G>A (rs9923231), cytochrome P450 2C9 (CYP2C9) *2 and CYP2C9*3 polymorphisms.[5, 6, 13] While these are the major genetic determinants of warfarin dose variability in Europeans,[14, 15] additional variants are important in African Americans.[16]
The CYP2C9*5, *6, *8. and *11 alleles occur almost exclusively in persons of African descent and significantly reduce warfarin clearance and dose requirements.[17-22] In a recent genome-wide association study (GWAS), the rs12777823 G>A variant emerged as an additional predictor of lower dose requirements in African Americans, and pharmacokinetic analysis showed reduced S-warfarin clearance in rs12777823A carriers.[23] A recent study demonstrates improved dose prediction in African Americans with an algorithm that includes variants important in this population.[24] Based on these data, we hypothesized that significant dosing error would occur from use of recommended warfarin pharmacogenetic dosing algorithms, such as that used in the COAG trial, in African Americans who have the CYP2C9*5, *6, *8, *11 or rs12777823 A allele. The objective of this study was to determine the effect of CYP2C9*5, *6, *8, and *11 alleles and rs12777823 G>A genotype on warfarin dose prediction with the WD and IWPC algorithms in African Americans.
Methods
Study population
Adult, warfarin-treated African Americans (by self-report) on a stable warfarin dose, defined as the dose that produced a therapeutic INR for ≥2 consecutive clinic visits ≥14 days apart, were enrolled. After obtaining written informed consent, a genetic sample (buccal cell or venous blood) was obtained and clinical data were collected from the electronic health record. The study protocol was approved by the University of Illinois at Chicago Institutional Review Board.
Genotyping
Genomic DNA was isolated from buccal cells or whole blood using a Puregene kit (Qiagen, Valencia, CA). The CYP2C9*2 (p.R144C, rs1799853), *3 (p.I359L, rs1057910), *5 (p.D360E, rs28371686), *6 (c.818delA, rs9332131), *8 (p.R150H, rs7900194), *11 (p.R335W, rs28371685), rs12777823 G>A, and VKORC1-1639 G>A genotypes were determined by PCR and pyrosequencing, as previously described.[22] The −1766 C>T (rs9332096) genotype was used to detect the CYP2C9*8 allele because it is in strong linkage disequilibrium with R150H (r2=0.89, D’=0.95), and can be detected via pyrosequencing, whereas R150H cannot.[25] In addition, the −1766 C>T polymorphism decreases gene expression thereby contributing to the functional effects of the CYP2C9*8 allele.[25] Individual genetic ancestry was determined in the majority of patients using 105 autosomal DNA ancestry informative markers, as previously described.[26]
Statistical analysis
The predicted warfarin maintenance dose was calculated for each patient using the IWPC and WD algorithms. Dose prediction error was defined as the predicted dose minus the observed (actual) dose.[27] The correlation between dose prediction error and percent West African ancestry was assessed by Spearman correlation. Predicted doses and dose prediction error were compared between carriers and non-carriers of a CYP2C9*5, *6, *8, and *11 allele and between rs12777823 G>A genotype groups by the Kruskal-Wallis or Wilcoxon-Mann-Whitney U Test. Dose prediction error was also examined with CYP2C9*8 alone, given how frequently it occurs in African Americans (reported allele frequency of 0.047),[28] and with the rs12777823 G>A genotype alone after excluding patients with a variant CYP2C9*2, *3, *5, *6, *8, or *11 allele. Additionally, Bland-Altman plots were created to visualize the amount of disagreement between predicted and actual doses. Finally, we tested whether adjusting doses predicted by the WD algorithm to account for the CYP2C9*5, *6, *8, *11, and rs12777823 variants or use of newer algorithms that include African-specific variants would provide a more accurate means of dosing warfarin for African Americans. Power calculations were based on identifying differences in dose prediction error between carriers and non-carriers of a CYP2C9 variant. Assuming an alpha of 0.05 and a standard deviation of 1.3 mg/day in dose prediction error based on previous data[6] including at least 30 patients in each genotype group was estimated to provide 80% power to detect a difference of 1.0 mg/day in dose prediction error between groups. All statistical analyses were performed with the SAS software package, version 9.2 (SAS Institute, Cary, NC).
Results
A total of 274 African Americans on a stable warfarin dose were included, with their characteristics and genotype frequencies summarized in Table 1. Genotypes frequencies were similar to those previously reported in persons of African descent.[21, 29] The mean observed warfarin maintenance dose was 6.6 ± 2.5 mg/day in the study population. The IWPC algorithm predicted lower doses (6.1 ± 1.3 mg/day; p<0.0001), and the WD algorithm predicted higher doses (7.2 ± 1.8 mg/day; p<0.0001) than observed.
Table 1. Baseline characteristics and genotype frequencies.
| Variable or genotype | n = 274 |
|---|---|
| Age (years) | 55 ± 16 |
| Male sex | 76 (28) |
| BSA (m2) | 2.1 ± 0.3 |
| Current smoker | 47 (17) |
| Primary warfarin indication | |
| DVT or PE | 150 (55) |
| Atrial fibrillation or flutter | 38 (14) |
| Secondary stroke prevention | 36 (13) |
| PVD | 14 (5) |
| Heart valve replacement | 17 (6) |
| Other* | 19 (7) |
| Amiodarone use | 7 (2.6) |
| CYP2C9 inducer† use | 10 (3.7) |
| Genotype | |
| VKORC1-1639 G>A | |
| GG | 227 (82.9) |
| GA | 45 (16.4) |
| AA | 2 (0.7) |
| rs12777823 G>A | |
| GG | 157 (57.3) |
| GA | 97 (35.4) |
| AA | 20 (7.3) |
| CYP2C9 | |
| *1/*1 | 214 (78.1) |
| *1/*2 | 11 (4.0) |
| *1/*3 | 3 (1.1) |
| *1/*5 | 2 (0.7) |
| *1/*6 | 6 (2.2) |
| *1/*8 | 23 (8.4) |
| *1/*11 | 4 (1.5) |
| *2/*3 | 1 (0.4) |
| *2/*5 | 1 (0.4) |
| *2/*11 | 1 (0.4) |
| *5/*8 | 2 (0.7) |
| *8/*8 | 5 (1.8) |
| *8/*11 | 1 (0.4) |
Mean ± SD or No. (%)
left ventricular or right atrial thrombus, sinus thrombosis, cortical vein thrombosis
phenytoin or carbamazepine
BSA, body surface area; DVT, deep vein thrombosis; PE, pulmonary embolism; TIA, transient ischemic attack; PVD, peripheral vascular disease
For 220 patients for whom individual genetic ancestry estimates were determined, the mean percent of West African ancestry was 82 ± 13%. Percent West African ancestry was weakly but positively correlated with dose prediction error with the WD (r=0.148, p=0.028), but not IWPC (r=0.088, p=0.194) algorithm.
Observed and predicted doses, stratified by CYP2C9 and rs12777823 genotypes, are shown in Tables 2 and 3. As previously reported, individuals with a CYP2C9*5, *6, *8, *11 or rs12777823 A allele required significantly lower doses compared to those without a variant.[22, 23] The IWPC and WD algorithms predicted similar doses regardless of rs12777823 genotype. The IWPC algorithm also predicted similar doses regardless of CYP2C9 genotype; however, the WD algorithm tended to predict higher doses in CYP2C9 variant carriers versus non-carriers.
Table 2. Dosing error with pharmacogenomic algorithms based on CYP2C9*5, *6, *8, or *11 genotype.
|
CYP2C9*5, *6, *8, or *11 |
|||
|---|---|---|---|
| Non-carrier (n=229) |
Carrier (n=45) |
p value* | |
| Dose (mg/day) | |||
| Observed | 6.4 (5.0 to 8.2) | 5.0 (4.0 to 7.5) | 0.0063 |
| IWPC | 6.0 (5.1 to 6.9)† | 6.3 (5.4 to 6.9)§ | 0.2535 |
| WD | 7.0 (6.0 to 8.4)‡ | 7.7 (6.4 to 8.6)# | 0.0559 |
| Prediction error (mg/day) | |||
| IWPC | −0.5 (−1.9 to 0.6) | 0.7 (−0.4 to 1.9) | <0.0001 |
| WD | 0.5 (−0.9 to 1.6) | 2.2 (0.5 to 2.9) | <0.0001 |
Median (IQR)
Comparison between genotype groups
p=0.005 for IWPC predicted versus observed dose in non-carriers
p=0.009 for WD predicted versus observed dose in non-carriers
p=0.04 for IWPC predicted versus observed dose in carriers
p<0.001 for WD predicted versus observed dose in carriers
Table 3. Dosing error with pharmacogenomic algorithms based on rs12777823 genotype.
| rs12777823 genotype |
||||
|---|---|---|---|---|
| GG (n=157) |
GA (n=97) |
AA (n=20) |
p value* | |
| Dose (mg/day) | ||||
| Observed | 6.8 (5.0 to 8.6) | 5.7 (4.3 to 7.5) | 5.2 (4.3 to 7.3) | 0.0047 |
| IWPC | 6.0 (5.1 to 6.9)† | 6.0 (5.3 to 7.0) | 6.3 (4.9 to 6.8) | 0.5803 |
| WD | 7.0 (5.9 to 8.3) | 7.2 (6.3 to 8.6)‡ | 7.7 (6.2 to 8.5)§ | 0.1665 |
| Prediction error (mg/day) | ||||
| IWPC | −0.8 (−2.3 to 0.4) | 0.1 (−1.2 to 1.6) | 0.7 (−0.4 to 1.5 ) | <0.0001 |
| WD | 0.3 (−1.4 to 1.4) | 1.2 (0.02 to 2.6) | 2.0 (0.6 to 2.8) | <0.0001 |
Median (IQR)
Comparison between genotype groups
p<0.001 for IWPC predicted versus observed dose with the GG genotype
p<0.001 for WD predicted versus observed dose with the GA genotype
p=0.0036 for WD predicted versus observed dose with the AA genotype
To examine the performance of the IWPC and WD algorithms by genotype, we compared observed and predicted doses within each genotype strata. There were significant dose prediction errors by the CYP2C9 (Table 2) and rs12777823 (Table 3) genotypes with both algorithms. The WD algorithm predicted higher doses than the IWPC algorithm across genotypes (p<0.01 for all CYP2C9 and rs12777823 genotype groups). Both algorithms over-estimated doses for patients with a CYP2C9*5, *6, *8, or *11 variant. When limiting our comparison to carriers and non-carriers of the CYP2C9*8, one of the most common CYP2C9 variants in African Americans, the WD algorithm over-estimated doses in carriers by a median (IQR) of 2.3 (0.4 to 3.0) mg/day (p<0.001, for the predicted versus observed dose), while the IWPC algorithm had a dose prediction error of 1.0 (−1.3 to 1.9) mg/day (p=0.13).
For the rs12777823 genotype, the IWPC algorithm under-estimated doses for patients with the GG genotype by 0.8 mg/day, while the WD algorithm over-estimated doses by approximately 1 mg/day for heterozygotes and 2 mg/day for variant allele homozygotes. After excluding patients with a CYP2C9 variant, the associations between the rs12777823 genotype and dose prediction error remained. Specifically, among 229 patients without a CYP2C9*5, *6, *8, or *11 allele, the IWPC algorithm under-dosed patients with the rs12777823 GG genotype (n=147) by 0.9 mg/day (p<0.01), and the WD algorithm over-dosed those with the rs12777823 GA or AA genotype (n=82) by 1.1 mg/day (p<0.01). Interestingly, when removing CYP2C9 variant carriers, we noticed that disproportionately more patients with rs12777823 A allele versus without an rs12777823 A allele also had a CYP2C9*5, *6, *8, or *11 allele (78% versus 22%, p<0.0001), suggesting linkage disequilibrium among SNPs. We took this into account when constructing the WD-African American (WD-AA) model below.
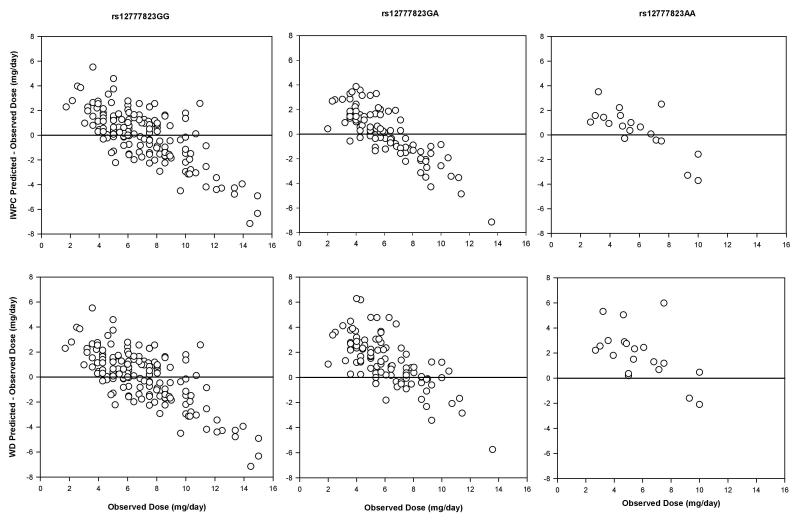
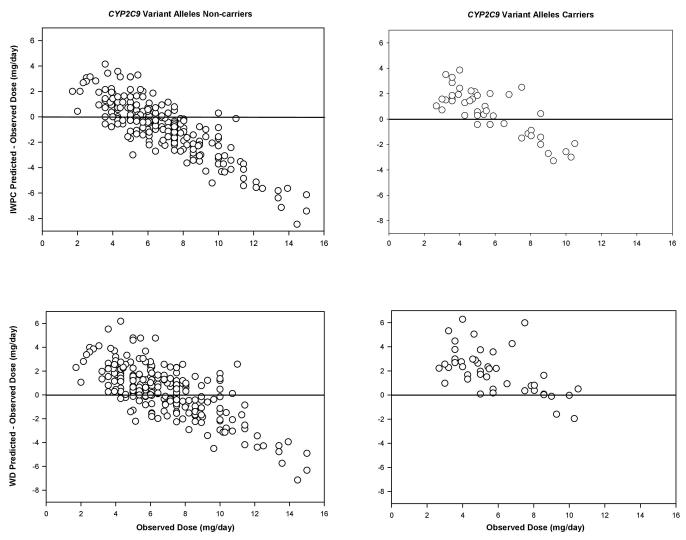
Figures 1 and 2 show the level of disagreement between the IWPC and WD predicted doses and observed doses, stratified by the CYP2C9 and rs12777823 genotypes. The visual presentation of the individual data points further illustrates the extent to which the algorithms over- or under-estimated doses based on genotype. Specifically, the WD algorithm over-dosed warfarin by >1 mg/day in 64% of those with a CYP2C9 variant and 70% of patients with the rs12777823 AA genotype. The IWPC over-dosed warfarin by >1 mg/day in 47% of CYP2C9 variant allele carriers and under-estimated doses by >1 mg/day in 42% of those with the rs12777823 GG genotype.
Figure 1.
Bland-Altman plots of the dose prediction error with the International Warfarin Pharmacogenetics Consortium (IWPC) and www.warfarindosing.org (WD) algorithms stratified by rs12777823 G>A genotype.
Figure 2.
Bland-Altman plots of the dose prediction error with the International Warfarin Pharmacogenetics Consortium (IWPC) and www.warfarindosing.org (WD) algorithms stratified by CYP2C9*5, *6, *8, or *11 carrier status.
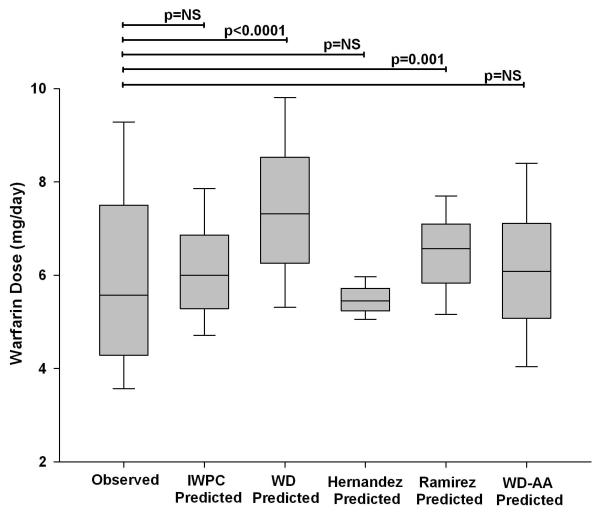
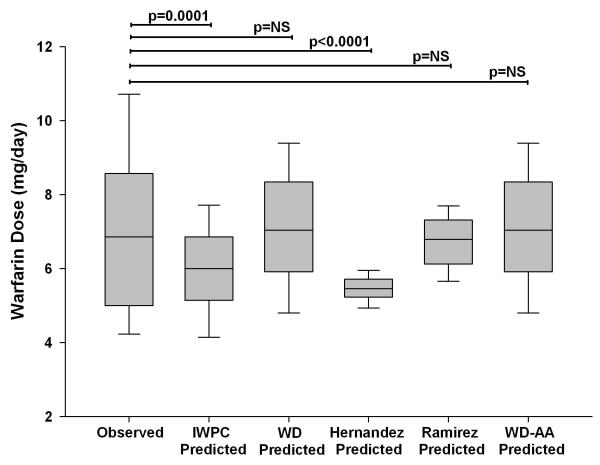
Next, we examined performance of the WD algorithm, which was used in the COAG trial, after making adjustments to account for the CYP2C9 and rs12777823 variants. The WD algorithm available through http://www.warfarindosing.org contains an option for inputting CYP2C9*5 and *6, although this was not done in the COAG trial. We entered these alleles when present and then reduced the resulting predicted dose by 20% for patients with a CYP2C9*8 or *11 allele and by 7 mg/week for the rs12777823 GA genotype and 9 mg/week for the rs12777823 AA genotype. These reductions were based on previous pharmacokinetic data for CYP2C9 variants and specific suggestions in the GWAS paper describing the association between the rs12777823 SNP and warfarin dose in African Americans.[19, 23, 30] Since the rs12777823 A and CYP2C9*5, *6, *8, and *11 allele tended to occur together, as described above, we only reduced doses for rs12777823 G>A variant for patients without a CYP2C9*5, *6, *8, or *11 allele to avoid over-adjustment. The dose predicted by the new WD-African American algorithm was 6.6 ± 1.8 mg/day for the population overall, which was similar to the observed dose (p=0.97), lower than the WD predicted dose (p=0.001), and higher than the IWPC predicted dose (p=0.002). The new algorithm performed better than the WD algorithm in CYP2C9*5, *6, *8, *11 or rs12777823 A allele carriers (p<0.0001) and better than the IWPC algorithm in patients without a variant allele (p=0.0001), as shown in Figures 3 and 4.
Figure 3.
Observed and predicted doses with the International Warfarin Pharmacogenetics Consortium (IWPC), www.warfarindosing.org (WD), WD-African American (WD-AA), Hernandez et al[24] and Ramirez et al[31] algorithms in carriers of the CYP2C9*5, *6, *8, *11 or rs12777823A alleles carriers.
Lines within boxes represent medians; lower and upper boundaries represent IQRs; and whiskers represent tenth and 90th percentiles. The Hernandez algorithm includes the VKORC1 - 1639 G>A, CYP2C9*2, *3, *5, *8, and *11; rs12777823; VKORC1 rs61162043 and CYP2C9 rs7089580 variants. The Ramirez algorithm includes VKORC1-1639 G>A, CYP2C9*2, *3, *6, *8, and *11, and CYP4F2 V433M. All variants were genotyped as previously described[22, 26] to determine algorithm-predicted doses.
Figure 4.
Observed and predicted doses with the International Warfarin Pharmacogenetics Consortium (IWPC), www.warfarindosing.org (WD), WD-African American (WD-AA), Hernandez et al[24] and Ramirez et al[31] algorithms in non-carriers of the CYP2C9*5, *6, *8, *11 or rs12777823A alleles.
Lines within boxes represent medians; lower and upper boundaries represent IQRs; and whiskers represent tenth and 90th percentiles. The Hernandez algorithm includes the VKORC1 - 1639 G>A, CYP2C9*2, *3, *5, *8, and *11; rs12777823; VKORC1 rs61162043 and CYP2C9 rs7089580 variants. The Ramirez algorithm includes VKORC1-1639 G>A, CYP2C9*2, *3, *6, *8, and *11, and CYP4F2 V433M. All variants were genotyped as previously described[22, 26] to determine algorithm-predicted doses.
Finally, we evaluated the performance of two existing algorithms. An algorithm by Hernandez et al,[24] which includes the CYP2C9*5, *8, *11, and rs12777823 variants, performed well in variant allele carriers but under-dosed non-carriers (Figures 3 and 4). In contrast, an algorithm by Ramirez et al,[31] which includes the CYP2C9*6, *8, and *11 (but not CYP2C9*5 or rs12777823), performed well in those without a variant but over-dosed variant allele carriers. Further analysis showed that this was due to over-dosing patients with a rs12777823 variant (data not shown).
Discussion
We found that warfarin pharmacogenetic dosing algorithms used in recent clinical trials perform poorly in African Americans with the CYP2C9*5, *6, *8, *11 or rs12777823 variant. Specifically, when accounting for only the VKORC1, CYP2C9*2, and CYP2C9*3 genotypes, as done in the COAG trial, the WD algorithm significantly overdoses CYP2C9*5, *6, *8,*11, and rs12777823 A allele carriers, sometimes by 2 mg/day or more. The IWPC algorithm over-dosed CYP2C9 variant carriers, while it under-dosed rs12777823 G allele homozygotes. These algorithms were derived from different patient cohorts, which led to differing coefficients included in the algorithms and likely contributed to their varying performance. Although weak, there was a significant correlation between West African ancestry and dose prediction error with the WD, but not IWPC algorithm, suggesting that the WD algorithm may especially perform poorly in African Americans.
The VKORC1-1639 G>A and CYP2C9*2 and *3 genotypes are widely believed to be the major genetic determinants of warfarin dose requirements across populations, as evident by dose recommendations based exclusively on these genotypes in the FDA-approved warfarin label [32] However, data on additional variants important in African Americans have emerged since the warfarin label update. In particular, the rs12777823 polymorphism, located on chromosome 10 near the CYP2C18 gene, is common (e.g. allele frequency of approximately 25%) and associated with reduced S-warfarin clearance and lower dose requirements in African Americans.[23] Interestingly, the allele is also common in other populations, but not associated with warfarin response. A proposed explanation is that rs12777823 G>A is in linkage disequilibrium with one of more functional variants in African Americans, but not other groups.[23] Our finding that, compared to the rs12777823 GG genotype, the rs12777823 A allele occurred disproportionately more often with a CYP2C9*5, *6, *8, or *11 allele supports this. However, many African Americans have the rs12777823 variant in absence of a CYP2C9*5, *6, *8, or *11 allele, and they too require lower warfarin doses. In addition, we showed that the dosing error with the rs12777823 remains after controlling for CYP2C9 variants. Therefore, the mechanism underlying associations with the rs12777823 genotype in African Americans is not solely due to linkage disequilibrium with CYP2C9*5, *6, *8, and *11, and remains to be further elucidated.
Additional data have also emerged with the CYP2C9 variants. Specifically, in-vitro and in-vivo studies have demonstrate lower S-warfarin clearance with the CYP2C9*8 allele, which occurs in approximately 12% of African Americans.[19] Recent data also provide further evidence of reduced warfarin clearance and dose requirements with the *5, *6, and *11 alleles.[30, 33]
The CYP2C9*5, *6, *8, *11, or rs12777823 variants were not included in dosing algorithms used for either of the recent clinical trials. This was probably of no consequence in the EU-PACT trial, in which 99% of participants were European.[7] The EU-PACT trial demonstrated a significant improvement in time spent with the therapeutic INR range during the initial 12 weeks of warfarin therapy with genotype-guided versus conventional dosing. In contrast, the COAG trial, which was conducted in a much more diverse population, showed no difference in the time in range during the first 4 weeks of therapy with a pharmacogenetic versus clinical algorithm.[8] African Americans, who comprised approximately one-third of the COAG trial population, did worse with pharmacogenetic dosing, with a higher likelihood of supratherapeutic INR values with pharmacogenetic versus clinically based dosing. Our data suggest that omission of the CYP2C9*5, *6, *8, *11 and rs12777823 genotypes was a major reason genotype-guided dosing performed poorly in African Americans. Specifically, the higher incidence of supratherapeutic anticoagulation with genotype-guided dosing in COAG is consistent with the over-estimation of doses we observed with the WD algorithm in CYP2C9*5, *6, *8, *11 and rs12777823 carriers, who likely comprised 50% of the African Americans COAG trial based on allele frequency data.
There are a couple of ways to account for the African-specific genotypes when dosing warfarin. One is to use an algorithm derived from an African American population that contains these variants, such as that recently described by Hernandez et al.[24] Another algorithm by Ramirez et al[31] accounts for many of the African-specific CYP2C9 variants, but not rs12777823. We found that the Hernandez algorithm, but not the Ramirez algorithm, performed well in variant allele carriers. This is not surprising since the Ramirez algorithm does not include the rs12777823 allele, and it reinforces the importance of this variant in dosing warfarin for African Americans. Many of the patients in the current study were included in the derivation cohort for Hernandez et al algorithm, preventing an unbiased evaluation. However, our findings are consistent with previous examination of the algorithm in an independent African American cohort, which showed that the algorithm better predicted the therapeutic warfarin dose (r=0.51) than the IWPC algorithm (r=0.38).
Another approach would be to use a universal algorithm, derived from a multi-ethnic population, such as either the IWPC or WD algorithm, and then further adjust dose based on additional CYP2C9 and rs12777823 genotypes. This approach would allow the use of the same algorithm in all patients, regardless of ethnicity. This approach proved to more accurately predict warfarin dose in our patients than the “unadjusted” WD algorithm, with no difference in observed and predicted doses with the WD-AA algorithm.
While not examined in the current study since data were collected after the warfarin initiation period and after stable dosing was achieved, there are previous data demonstrating an increased risk for over-anticoagulation and bleeding with variants that reduce S-warfarin clearance, and these data extend to African American patients.[34-36] Most relevant to this study, in a diverse population consisting of 50% African Americans, Limdi et al[35] reported a significantly higher risk for major hemorrhage with warfarin among carriers versus non-carriers of a CYP2C9*2, *3, *5, *6, or *11 allele. Similar to these alleles, the rs12777823 A and CYP2C9*8 alleles are associated with reduced S-warfarin clearance, and thus, would be expected to have similar effects on bleeding risk.[19, 23, 30] A recent meta-analysis of nine studies showed that genotype-guided dosing of vitamin K antagonists reduces the risk for serious bleeding events compared to clinically-guided approaches.[37] However, most of the patients included in these studies were European, and genotyping focused on the VKORC1 and CYP2C9*2 and *3 variants. Our data suggest that the benefit of genotype-guided dosing on bleeding risk in African Americans will not likely be realized unless variants that reduce warfarin clearance in this population are accounted for.
We focused on variants with pharmacokinetic data supporting their association with warfarin dose requirements in the current study. All of the variants tested are associated with reduced dose requirements, and our data provide an explanation for the higher risk for over-anticoagulation with use of recommended pharmacogenetic algorithms in African Americans.[8] However, it is important to note that there are also variants that have been associated with higher warfarin dose requirements in African Americans.[26, 38] Ultimately, identifying and accounting for both low- and high-dose variants could further improve dosing.
Conclusions
In summary, our data indicate that, when dosing warfarin based on genotype, it is important to account for variants that are either common or specifically influence warfarin response in African Americans and that not doing so can lead to significant over-dosing in a large portion of the African American population. Genotype-guided dosing has been shown to be of benefit in a European population for whom important variants are well defined.[7] The clinical utility of genotype-guided warfarin dosing in African Americans, when accounting for variants important in this population, remains to be determined. Our data help explain why African Americans fared worse with genotype-guided dosing in the COAG trial. While it had been suggested that the limited number of genotypes considered in the trial contributed to poor outcomes in African Americans,[10, 12] our data are the first to support this and to show the degree to which failure to account for variants important in African Americans affects dosing error. Another clinical trial of genotype-guided warfarin dosing is underway, and unlike previous trials, it will be powered to assess clinical outcomes.[39] However, similar to previous studies, genotyping will be limited to variants important in Europeans. Whether funding agencies will support additional clinical trials to examine the utility of genotype-guided warfarin dosing in minority populations is questionable. However, genomic data are becoming more readily available to guide therapy, which makes the question of whether or not to genotype at the time of warfarin initiation less relevant. A more important question as preemptive genotyping becomes more widespread will be how to apply genetic information available at the time of warfarin prescribing to appropriately inform dosing across ethnic groups.
Supplementary Material
Acknowledgments
Founding Sources
This work was supported an American Heart Association Midwest Affiliate Spring 2010 Grant-In-Aid (10GRNT3750024) and a University of Illinois at Chicago College of Pharmacy Hans Vahlteich Research Award. Work by EAN was supported by NIH NHLBI (K23HL112908). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Disclosures
KD, SW, SRP, APB, RAK and EAN declared no conflict of interest. LHC is a co-investigator for U.S. Utility Patent Application No. 12/572,908, titled “CYP2C9*8 alleles correlate with decreased warfarin metabolism and increased warfarin sensitivity”. Published: May 27, 2010; Pub. No. US 2010/0130599.
References
- 1.Kirley K, Qato DM, Kornfield R, Stafford RS, Alexander GC. National trends in oral anticoagulant use in the United States, 2007 to 2011. Circ Cardiovasc Qual Outcomes. 2012;5(5):615–621. doi: 10.1161/CIRCOUTCOMES.112.967299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wadelius M, Chen LY, Lindh JD, Eriksson N, Ghori MJ, Bumpstead S, et al. The largest prospective warfarin-treated cohort supports genetic forecasting. Blood. 2009;113(4):784–792. doi: 10.1182/blood-2008-04-149070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wadelius M, Pirmohamed M. Pharmacogenetics of warfarin: current status and future challenges. Pharmacogenomics J. 2007;7(2):99–111. doi: 10.1038/sj.tpj.6500417. [DOI] [PubMed] [Google Scholar]
- 4.Johnson JA, Gong L, Whirl-Carrillo M, Gage BF, Scott SA, Stein CM, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. 2011;90(4):625–629. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360(8):753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gage BF, Eby C, Johnson JA, Deych E, Rieder MJ, Ridker PM, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84(3):326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pirmohamed M, Burnside G, Eriksson N, Jorgensen AL, Toh CH, Nicholson T, et al. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med. 2013;369(24):2294–2303. doi: 10.1056/NEJMoa1311386. [DOI] [PubMed] [Google Scholar]
- 8.Kimmel SE, French B, Kasner SE, Johnson JA, Anderson JL, Gage BF, et al. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med. 2013;369(24):2283–2293. doi: 10.1056/NEJMoa1310669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maitland-van der Zee AH, Daly AK, Kamali F, Manolopoulous VG, Verhoef TI, Wadelius M, et al. Patients benefit from genetics-guided coumarin anticoagulant therapy. Clin Pharmacol Ther. 2014;96(1):15–17. doi: 10.1038/clpt.2014.44. [DOI] [PubMed] [Google Scholar]
- 10.Scott SA, Lubitz SA. Warfarin pharmacogenetic trials: is there a future for pharmacogenetic-guided dosing? Pharmacogenomics. 2014;15(6):719–722. doi: 10.2217/pgs.14.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavallari LH, Kittles RA, Perera MA. Genotype-guided dosing of vitamin K antagonists. N Engl J Med. 2014;370(18):1763. doi: 10.1056/NEJMc1402521. [DOI] [PubMed] [Google Scholar]
- 12.Daneshjou R, Klein TE, Altman RB. Genotype-guided dosing of vitamin K antagonists. N Engl J Med. 2014;370(18):1762–1763. doi: 10.1056/NEJMc1402521#SA4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avery PJ, Jorgensen A, Hamberg AK, Wadelius M, Pirmohamed M, Kamali F, et al. A proposal for an individualized pharmacogenetics-based warfarin initiation dose regimen for patients commencing anticoagulation therapy. Clin Pharmacol Ther. 2011;90(5):701–706. doi: 10.1038/clpt.2011.186. [DOI] [PubMed] [Google Scholar]
- 14.Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112(4):1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeuchi F, McGinnis R, Bourgeois S, Barnes C, Eriksson N, Soranzo N, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genetics. 2009;5(3):e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavallari LH, Perera MA. The future of warfarin pharmacogenetics in under-represented minority groups. Future Cardiol. 2012;8(4):563–576. doi: 10.2217/fca.12.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickmann LJ, Rettie AE, Kneller MB, Kim RB, Wood AJ, Stein CM, et al. Identification and functional characterization of a new CYP2C9 variant (CYP2C9*5) expressed among African Americans. Mol Pharmacol. 2001;60(2):382–387. doi: 10.1124/mol.60.2.382. [DOI] [PubMed] [Google Scholar]
- 18.Redman AR, Dickmann LJ, Kidd RS, Goldstein JA, Ritchie DM, Hon YY. CYP2C9 genetic polymorphisms and warfarin. Clin Appl Thromb Hemost. 2004;10(2):149–154. doi: 10.1177/107602960401000205. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Jeong H, Takahashi H, Drozda K, Patel SR, Shapiro NL, et al. Decreased warfarin clearance associated with the CYP2C9 R150H (*8) polymorphism. Clin Pharmacol Ther. 2012;91(4):660–665. doi: 10.1038/clpt.2011.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tai G, Farin F, Rieder MJ, Dreisbach AW, Veenstra DL, Verlinde CL, et al. In-vitro and in-vivo effects of the CYP2C9*11 polymorphism on warfarin metabolism and dose. Pharmacogenet Genomics. 2005;15(7):475–481. doi: 10.1097/01.fpc.0000162005.80857.98. [DOI] [PubMed] [Google Scholar]
- 21.Limdi NA, Arnett DK, Goldstein JA, Beasley TM, McGwin G, Adler BK, et al. Influence of CYP2C9 and VKORC1 on warfarin dose, anticoagulation attainment and maintenance among European-Americans and African-Americans. Pharmacogenomics. 2008;9(5):511–526. doi: 10.2217/14622416.9.5.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavallari LH, Langaee TY, Momary KM, Shapiro NL, Nutescu EA, Coty WA, et al. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87(4):459–464. doi: 10.1038/clpt.2009.223. [DOI] [PubMed] [Google Scholar]
- 23.Perera MA, Cavallari LH, Limdi NA, Gamazon ER, Konkashbaev A, Daneshjou R, et al. Genetic variants associated with warfarin dose in African-American individuals: a genome-wide association study. Lancet. 2013;382(9894):790–796. doi: 10.1016/S0140-6736(13)60681-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez W, Gamazon ER, Aquino-Michaels K, Patel S, O’Brien TJ, Harralson AF, et al. Ethnicity-specific pharmacogenetics: the case of warfarin in African Americans. Pharmacogenomics J. 2014;14(3):223–228. doi: 10.1038/tpj.2013.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cavallari LH, Vaynshteyn D, Freeman KM, Wang D, Perera MA, Takahashi H, et al. CYP2C9 promoter region single-nucleotide polymorphisms linked to the R150H polymorphism are functional suggesting their role in CYP2C9*8-mediated effects. Pharmacogenet Genomics. 2013;23(4):228–231. doi: 10.1097/FPC.0b013e32835e95c7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perera MA, Gamazon E, Cavallari LH, Patel SR, Poindexter S, Kittles RA, et al. The missing association: sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African Americans. Clin Pharmacol Ther. 2011;89(3):408–415. doi: 10.1038/clpt.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roper N, Storer B, Bona R, Fang M. Validation and comparison of pharmacogenetics-based warfarin dosing algorithms for application of pharmacogenetic testing. J Mol Diagn. 2010;12(3):283–291. doi: 10.2353/jmoldx.2010.090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott SA, Jaremko M, Lubitz SA, Kornreich R, Halperin JL, Desnick RJ. CYP2C9*8 is prevalent among African-Americans: implications for pharmacogenetic dosing. Pharmacogenomics. 2009;10(8):1243–1255. doi: 10.2217/pgs.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martis S, Peter I, Hulot JS, Kornreich R, Desnick RJ, Scott SA. Multi-ethnic distribution of clinically relevant CYP2C genotypes and haplotypes. Pharmacogenomics J. 2013;13(4):369–377. doi: 10.1038/tpj.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niinuma Y, Saito T, Takahashi M, Tsukada C, Ito M, Hirasawa N, et al. Functional characterization of 32 CYP2C9 allelic variants. Pharmacogenomics J. 2013;23(4):228–231. doi: 10.1038/tpj.2013.22. [DOI] [PubMed] [Google Scholar]
- 31.Ramirez AH, Shi Y, Schildcrout JS, Delaney JT, Xu H, Oetjens MT, et al. Predicting warfarin dosage in European-Americans and African-Americans using DNA samples linked to an electronic health record. Pharmacogenomics. 2012;13(4):407–418. doi: 10.2217/pgs.11.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coumadin (warfarin sodium) package insert. Bristol-Myers Squibb; Princeton, NJ: Jan, 2010. [Google Scholar]
- 33.Mitchell C, Gregersen N, Krause A. Novel CYP2C9 and VKORC1 gene variants associated with warfarin dosage variability in the South African black population. Pharmacogenomics. 2011;12(7):953–963. doi: 10.2217/pgs.11.36. [DOI] [PubMed] [Google Scholar]
- 34.Higashi MK, Veenstra DL, Kondo LM, Wittkowsky AK, Srinouanprachanh SL, Farin FM, et al. Association between CYP2C9 genetic variants and anticoagulation-related outcomes during warfarin therapy. JAMA. 2002;287(13):1690–1698. doi: 10.1001/jama.287.13.1690. [DOI] [PubMed] [Google Scholar]
- 35.Limdi NA, McGwin G, Goldstein JA, Beasley TM, Arnett DK, Adler BK, et al. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther. 2008;83(2):312–321. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biss TT, Avery PJ, Williams MD, Brandao LR, Grainger JD, Kamali F. The VKORC1 and CYP2C9 genotypes are associated with over-anticoagulation during initiation of warfarin therapy in children. J Thromb Haemost. 2013;11(2):373–375. doi: 10.1111/jth.12072. [DOI] [PubMed] [Google Scholar]
- 37.Franchini M, Mengoli C, Cruciani M, Bonfanti C, Mannucci PM. Effects on bleeding complications of pharmacogenetic testing for initial dosing of vitamin K antagonists: a systematic review and meta-analysis. J Thromb Haemost. 2014;12(9):1480–1487. doi: 10.1111/jth.12647. [DOI] [PubMed] [Google Scholar]
- 38.Voora D, Koboldt DC, King CR, Lenzini PA, Eby CS, Porche-Sorbet R, et al. A polymorphism in the VKORC1 regulator calumenin predicts higher warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87(4):445–451. doi: 10.1038/clpt.2009.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Do EJ, Lenzini P, Eby CS, Bass AR, McMillin GA, Stevens SM, et al. Genetics informatics trial (GIFT) of warfarin to prevent deep vein thrombosis (DVT): rationale and study design. Pharmacogenomics J. 2012;12(5):417–424. doi: 10.1038/tpj.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.