Abstract
Fatty Acid Desaturase (FADS) genes and their variants have been associated with multiple metabolic phenotypes including liver enzymes and hepatic fat accumulation but the detailed mechanism remains unclear. We aimed to delineate the role of FADSs in modulating lipid composition in human liver. We performed a targeted lipidomic analysis of a variety of phospholipids, sphingolipids and ceramides among 154 human liver tissue samples. The associations between previously Genome-wide Association Studies (GWAS)-identified six FADS single nucleotide polymorphisms (SNPs) and these lipid levels as well as total hepatic fat content (HFC) were tested. The potential function of these SNPs in regulating transcription of 3 FADS genes (FADS1, FADS2 and FADS3) in the locus was also investigated. We found that while these SNPs were in high linkage disequilibrium (r2 >0.8), the rare alleles of these SNPs were consistently and significantly associated with the accumulation of multiple very-long-chain fatty acids (VLCFAs), with C47H85O13P (C36:4), a phosphatidylinositol (PI) and C43H80O8PN (C38:3), a phosphatidylethanolamine (PE) reached the Bonferroni corrected significance (p<3×10−4). Meanwhile, these SNPs were significantly associated with increased ratios between the more saturated and relatively less saturated forms of VLCFAs, especially between PEs, PIs and phosphatidylcholines (PCs) (p≤3.5×10−6). These alleles were also associated with increased total HFC (p<0.05). Further analyses revealed that these alleles were associated with decreased hepatic expression of FADS1 (p=0.0018 for rs174556), but not FADS2 or FADS3 (p>0.05).
Conclusion
Our findings revealed critical insight into the mechanism underlying FADS1 and its polymorphisms in modulating hepatic lipid deposition by altering gene transcription and controlling lipid composition in human livers.
Keywords: FADS1, polymorphism, hepatic fat content, hepatic lipid accumulation, lipidomics
INTRODUCTION
It is widely recognized that status of polyunsaturated fatty acid (PUFA), especially long-chain PUFA (LC-PUFA) has a major impact on human health and has been demonstrated to be associated with clinical outcomes in various physiological processes including cardiovascular, immunology, peripheral and central nervous systems (1). Encoded respectively by the fatty acid desaturase 1 (FADS1) and 2 (FADS2) genes, the delta-5 desaturase (D5D) and delta-6 desaturase (D6D) are membrane-bound enzymes that catalyze the rate-limiting formation of endogenous long-chain PUFA (1–5). The FADS1 and FADS2 genes are clustered on human chromosome 11 (11q12-q13.1), which also includes the FADS3 gene with unclear function (6, 7). Previous candidate gene-based association studies identified correlation between FADS polymorphisms and complex diseases such as coronary artery disease (CAD) (8, 9), metabolic syndrome (10), allergic rhinitis and atopic eczema (2, 11), attention deficit/hyperactivity disorder (ADHD) (12) and intelligence development (13, 14). Significant associations were also reported in several recent genome-wide associations studies (GWAS) that revealed multiple polymorphisms at the FADS locus associated with various metabolic phenotypes especially serum lipid compositions and metabolic perturbations (See Table S1 for summary). These findings strongly suggest a functional linkage between genetic variations in FADS locus and multiple human diseases via altering the LC-PUFA metabolism (4, 5). However, a few important questions remain to be addressed. First, given the high linkage disequilibrium (LD) level across the FADS gene cluster (please see supplemental Fig S1), there thus far lacks a clear validation which FADS gene(s) mediates these genotype-phenotype correlations. Second, the regulation of lipid metabolism is a complex systematic process involving a large number of genes in multiple tissues/organs throughout the organism. However, a mechanistic linkage between genetic variations within FADS genes, PUFA metabolism and functional implications in critical tissues/organs has not been established.
The liver is the most important organ involved in lipid synthesis and metabolism. Homeostasis of circulating PUFA levels is significantly controlled by the dietary intake and hepatic metabolism (15). Therefore, the variability in lipid perturbations as well as the underlying disorders may be largely attributed to the variation in the function of FADS genes in the liver. Unfortunately, a complete delineation of the role of polymorphisms in modulating FADS genes as well as lipid profile in liver tissue has not been achieved to date. Collectively, filling this knowledge gap will not only reveal the mechanism that FADS genes involved in hepatic fat deposition, which is highly relevant to certain chronic liver disorders [e.g. nonalcoholic fatty liver disease (NAFLD)]; but will also help establish the causal pathways underlying the dysregulation of lipid metabolism in other human diseases mentioned above. We therefore conducted this study to explore the mechanistic pathway linking genetic polymorphisms at the FADS locus, FADS gene expression, lipid profile and metabolism as well as total fat accumulation in human livers, using an integrated multi-omics approach including genomic, transcriptomic and lipidomic data obtained from human liver samples.
MATERIALS AND METHODS
Samples and Associated Genomic and Transcriptomic Data
Liver tissue samples (n=206) collected from liver transplantation donors were used in this study. The sample procurement procedure and related information have been described in our previous publication (16). The use of these livers and the performance of this study were approved by the Institutional Review Board (IRB) of Purdue University. These samples have been previously analyzed for SNP genotypes and mRNA expression using Illumina quad 610 SNP array and Agilent expression arrays (Agilent-014850 4×44k arrays, GPL4133). After removing samples with incomplete demographic and co-factors (age, gender, race and BMI) as well as samples with record of heavy drinking, 154 remaining samples were used to conduct the subsequent analyses.
Measurement of Hepatic Fat Content (HFC)
Total lipid fraction was extracted from ~50mg liver tissue and measured using the method described previously (17). An aliquot of liver homogenate (20µl) was set aside for subsequent measurement of total protein concentration using the standard Bradford method. The extracted total lipid fraction was then used to perform lipidomic analysis. The hepatic contents of total fat and individual lipids were normalized to the total protein concentration for further analysis.
Lipidomic Analysis
Targeted lipidomic analysis of 357 lipids (Table S2) was conducted using an automated electrospray ionization (ESI)-tandem mass spectrometry approach according to a protocol described previously (18). More technical details were included in the supplemental materials.
Western Blotting
The protein expression of FADS1 was examined with Western blotting. Detailed protocol was included in the supplemental materials.
Genotyping
We genotyped rs738409 of the Patatin-like phospholipase domain-containing protein 3 (PNPLA3) gene using a Taqman assay according to the manufacturer instructions (Life Technologies, Forster City, CA, USA).
Data Processing and Statistical Analysis
Six SNPs (rs174576, rs1535, rs174546, rs102275, rs174537 and rs174556) from the FADS gene cluster were investigated in this study. These SNPs were available on the Illumina quad 610 SNP array platform, and were also either identified in previous GWAS and/or in LD (r2≥0.8) with them (Table S1 listed all previous studies identifying the associations with these SNPs). SNP genotype and gene expression information were extracted from the previously published data (16). The analysis of the HapMap data revealed that these 6 SNPs are in strong LD LD (r2≥0.8) (Fig S1).
We performed an integrated analysis to understand the role of FADS polymorphisms as well as FADS genes in regulating hepatic lipid composition, using various omics data. The detailed analysis plan was included in the supplemental materials. A flowchart describing study design along with key observations was also shown in Fig S2.
RESULTS
Measurement of Hepatic Fat Content and Lipid Profile
Among the liver samples, the median total hepatic fat content (mg fat/mg total protein) was 0.093 (range 0.04–0.58). The frequency distribution of the hepatic fat content among all samples was plotted in Fig S3. The targeted lipidomic profiling was conducted focusing on 357 individual lipid species. After profiling, 169 lipids with detectable levels in the liver were included in the data analyses and each individual lipid level was normalized to total protein content.
Association between FADS SNPs and Hepatic Content of Individual Lipids, Lipid Ratios and Total HFC
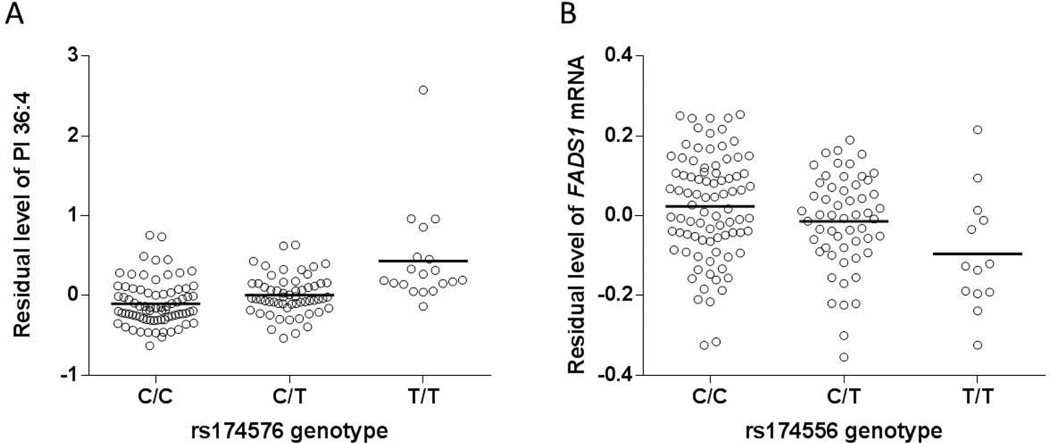
In order to test whether 6 FADS SNPs were associated with the hepatic lipid profile, these SNPs were correlated with the hepatic content of 169 individual lipids. We found that the hepatic content of two phospholipids, C47H85O13P (C36:4), a phosphatidylinositol (PI) and C43H80O8PN (C38:3), a phosphatidylethanolamine (PE) were significantly associated with genotype of most SNPs (5 out of all 6 SNPs reached the Bonferroni corrected significance p<3×10−4) (Table 1). Compared to the PE, the PI lipid level was more strongly associated with all the SNPs. SNP rs174576 possessed the highest correlation with the two lipids levels (p=5.3×10−8 and 1.9×10−5, respectively), with its minor allele associated with increased hepatic lipid level (Fig 1A).
Table 1.
Association between FADS SNPs and individual lipid levels.
| SNP | Physical Position |
C47H85O13P PI(36:4) | C43H80O8PN PE(38:3) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Beta | R2 | P-value | Adjusted P* | Beta | R2 | P-value | Adjusted P | ||
| rs174537 | 61309256 | 0.218 | 0.175 | 6.6E-08 | 2.9E-08 | 0.111 | 0.109 | 3.0E-05 | 3.3E-05 |
| rs102275 | 61314379 | 0.215 | 0.174 | 7.7E-08 | 3.3E-08 | 0.112 | 0.114 | 1.9E-05 | 1.8E-05 |
| rs174546 | 61326406 | 0.218 | 0.175 | 6.6E-08 | 2.9E-08 | 0.111 | 0.109 | 3.0E-05 | 3.3E-05 |
| rs174556 | 61337211 | 0.235 | 0.175 | 6.6E-08 | 2.7E-08 | 0.102 | 0.079 | 4.1E-04 | 5.1E-04 |
| rs1535 | 61354548 | 0.218 | 0.175 | 6.6E-08 | 2.9E-08 | 0.111 | 0.109 | 3.0E-05 | 3.3E-05 |
| rs174576 | 61360086 | 0.221 | 0.178 | 5.3E-08 | 2.3E-08 | 0.114 | 0.114 | 1.9E-05 | 2.1E-05 |
Adjusted p refers to the p values after adjusting for rs738409 genotype.
Fig 1.
Association between SNP genotype (rs174556 as an example) and hepatic lipid level and FADS1 mRNA expression. A) Association between rs174576 and the hepatic level of PI 36:4. B) Association between FADS1 polymorphism rs174556 as an example and FADS1 mRNA expression. Plots were demonstrated as the correlation between the genotypes and the residual hepatic FADS1 mRNA expression or PI 36:4 levels after adjusted for covariates. Horizontal bars indicate mean values.
Previous studies have suggested that the use of metabolite concentration ratios as a surrogate measurement of enzymatic activity helps reduce non-genetic variation and yielded stronger statistical associations between genetic alleles and metabolite profile (19, 20). We therefore conducted the association between these 6 SNPs and the 14,196 pair-wise ratios of all 169 lipids. We observed that these SNPs were significantly associated with a number of lipid ratios even after Bonferroni correction (p<3.5×10−6). These lipids were particularly very long chain fatty acids (VLCFA) especially PEs, PIs and phosphatidylcholines (PCs). Interestingly, the results consistently suggested that the minor allele of each SNP was associated with higher ratios of more saturated over less saturated lipid forms, reflecting the functional variability in fatty acid desaturase (Table S3). The strongest association observed was between rs102275 and the ratio of a pair of PI C47H83O13P (C38:4) and (C47H85O13P, C38:3) (p=4.3×10−10) (Table S3).
To test whether this modulation also alter total fat accumulation in the liver, SNPs were associated with the total HFC measured by biochemical extraction method. We found that all SNPs were consistently associated with the total HFC (p<0.04 for all tests) (Table 2). Consistently, the minor alleles were associated with increased hepatic fat deposition.
Table 2.
Association between FADS polymorphisms and hepatic fat content.
| SNP | Physical Position |
HFC | |||
|---|---|---|---|---|---|
| Beta | R2 | P-value | Adjusted P* | ||
| rs174537 | 61309256 | 0.116 | 0.034 | 0.023 | 0.021 |
| rs102275 | 61314379 | 0.095 | 0.023 | 0.061 | 0.052 |
| rs174546 | 61326406 | 0.116 | 0.034 | 0.023 | 0.021 |
| rs174556 | 61337211 | 0.115 | 0.028 | 0.037 | 0.037 |
| rs1535 | 61354548 | 0.116 | 0.034 | 0.023 | 0.021 |
| rs174576 | 61360086 | 0.112 | 0.031 | 0.029 | 0.027 |
Adjusted p refers to the p values after adjusting for rs738409 genotype.
Potential Effect of the PNPLA3 Polymorphism rs738409 on the Association between FADS SNPs and Lipids Phenotypes
The PNPLA3 polymorphism rs738409 is thus far the single strongest genetic risk factor for hepatic lipid accumulation and NAFLD (21). To evaluate whether this polymorphism may confound the associations observed between FADS polymorphisms and hepatic lipid traits, we conducted a conditional analysis by controlling the rs738409 genotype. After this, associations between the FADS SNPs and HFC or individual lipids remained significant (Table 1 and 2). In our dataset, rs738409 was marginally associated with total HFC (p=0.08, data not shown) but not with individual lipids (p>0.1 for both lipids, data not shown).
Genome-wide Association Analyses of Hepatic Contents of Lipids and Lipid Ratios Associated with the FADS SNPs
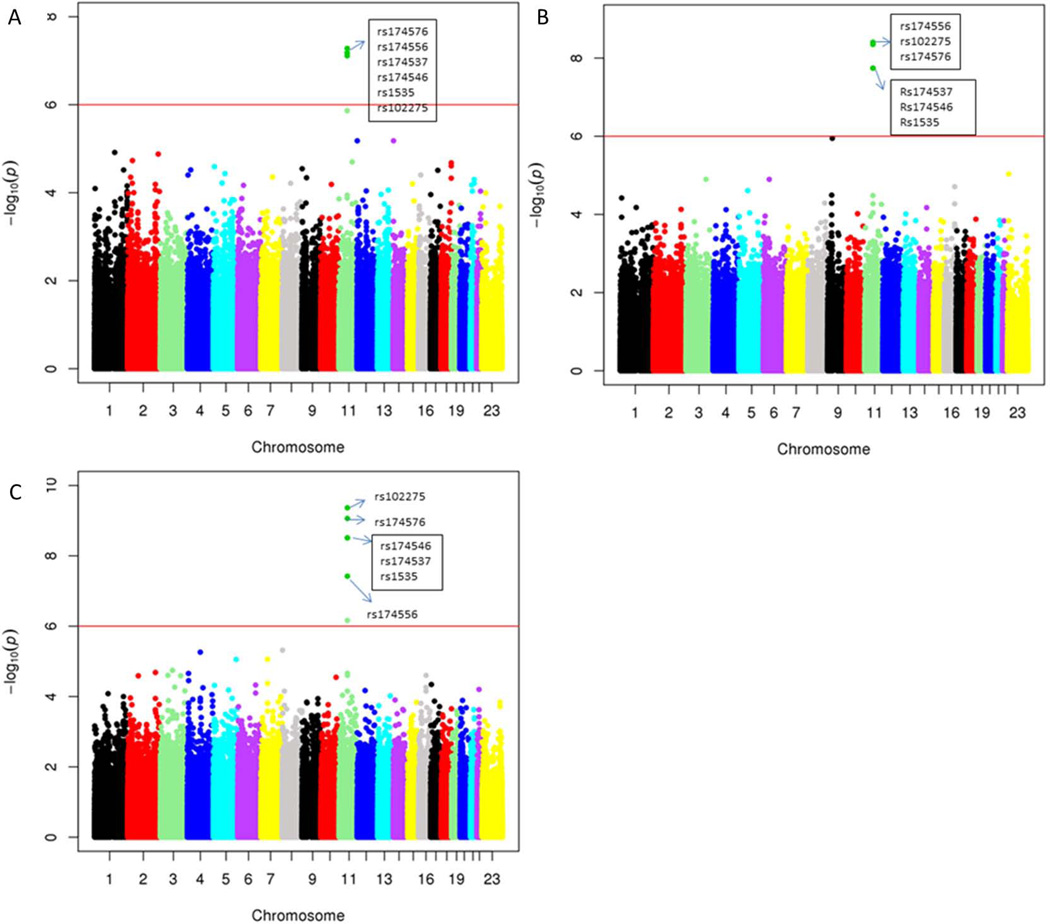
We next analyzed to see whether the FADS1 SNPs were the only polymorphisms affecting the hepatic contents of the PI C36:4 as well as the lipid ratios identified above. After the GWAS analyses, the 6 FADS polymorphisms were still the most significant genome-wide associated SNPs with the level of PI C36:4 (Fig 2A). Similarly, the FADS polymorphisms were also the most significant SNPs genome-wide associated with most of the lipid ratios identified above. (See Fig 2B–C with C47H85013P/C49H85013P and C47H83O13P/C47H85O13P as examples).
Fig 2.
Manhattan plots of quantitative traits loci (QTL) mapping. A) PI (36:4) (C47H85O13P); B) lipid ratio C47H85O13P/C49H85O13P and C) lipid ratio C47H83O13P/C47H85O13P. Each dot represented the –log(p) value for one SNP. The significance level (10−6) based on Bonferroni correction was indicated in each plot.
Association between FADS SNPs and Hepatic mRNA levels of FADS Genes
We further set out to understand the mechanism underlying this genotype-lipid association. Given the multiple FADS gene clustered in the region as well as their overlapping functions, we first focused on the question which FADS gene (s) was potentially involved in the genotype-phenotype correlations that were identified in the previous studies (Table S1). We found that all 6 SNPs were consistently and significantly associated with FADS1 gene expression, but not with FADS2 or FADS3 (Table 3). The rs174556 exerted the highest correlation with FADS1 gene expression (p=1.8×10−3), which remains significant after Bonferroni correction (p<0.017) (Fig 1B).
Table 3.
Association between SNPs and FADS mRNA expression.
| SNP | MAF | Physical Position |
FADS1 | FADS2 | FADS3 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta | R2 | P-value | Beta | R2 | P-value | Beta | R2 | P-value | |||
| rs174537 | 0.31 | 61309256 | −0.035 | 0.035 | 0.021 | 0.017 | 0.009 | 0.2468 | −3.8E-16 | 5.5E-03 | 0.36 |
| rs102275 | 0.35 | 61314379 | −0.032 | 0.030 | 0.032 | 0.020 | 0.013 | 0.1660 | −3.7E-16 | 5.5E-03 | 0.36 |
| rs174546 | 0.31 | 61326406 | −0.035 | 0.035 | 0.021 | 0.017 | 0.009 | 0.2468 | −3.8E-16 | 5.5E-03 | 0.36 |
| rs174556 | 0.26 | 61337211 | −0.051 | 0.062 | 0.0018 | 0.016 | 0.007 | 0.2880 | −4.1E-16 | 5.7E-03 | 0.35 |
| rs1535 | 0.31 | 61354548 | −0.035 | 0.035 | 0.021 | 0.017 | 0.009 | 0.2468 | −3.8E-16 | 5.5E-03 | 0.36 |
| rs174576 | 0.31 | 61360086 | −0.033 | 0.031 | 0.029 | 0.016 | 0.008 | 0.2719 | −3.9E-16 | 5.9E-03 | 0.34 |
To further screen whether additional SNPs located in the locus were also associated with FADS gene expression, we performed a locus-wide cis-eQTL mapping for the three FADS genes. We included all genotyped and imputed SNPs [k=2,177 after quality control (QC) filtering] in the region spanning from 500kb upstream and downstream of the FADS cluster (hg19, Chr11: 61,071,142–62,159,006), which is far beyond the LD block harboring the 6 SNPs (Fig S1). We found that the SNPs in the LD block were consistently associated with FADS1 mRNA expression (most significant SNPs 10−3<p<10−2) but not with FADS2 or FADS3 (p>0.05 for both), albeit the results did not reach statistical significance after Bonferroni correction (p<2.3×10−5). Meanwhile, although a few other SNPs were associated with each of the three FADS genes at the level of p=0.01, they were in weak LD (r2≤0.2) with the SNPs in the LD block (Fig S4A–C). In case that nonsynonymous or splicing variants in FADS2 and FADS3 that may be in LD with the 6 SNPs but do not alter gene expression, we manually screened the nonsynonymous SNPs and the SNPs located within ±50bp region of all exon-intron boundaries in these two genes in 1000 Genome data. No such a SNP with significant LD (r2≥0.5) with the 6 core SNPs was found.
Given the non-significant association after Bonferroni correction for the locus-wide expression quantitative traits loci (eQTL) mapping, we further validated the results in an additional hepatic eQTL dataset published recently (22). Again, imputed and genotyped SNP data (k=1,619 after QC) in the same region were used. We found that in this population, the SNPs in the LD block were strongly associated with the expression of FADS1 (10−8<p<10−6) but much weakly with FADS2 (10−4<p<10−2) and no association with FADS3 (p>0.05) (Fig S5A–E). After Bonferroni correction, only the results for FADS1 remained significant (corrected significance p=3×10−5). It should be noted that FADS1 and FADS2 mRNA levels in this population were highly correlated (p=1.2×10−48) (Fig S5F), possibly explained the weak but consistent associations between these SNPs and FADS2 expression. Collectively, these data strongly suggested that the SNPs in the LD block are cis-regulators for FADS1 gene transcription.
Association between FADS SNPs and FADS1 Protein Expression
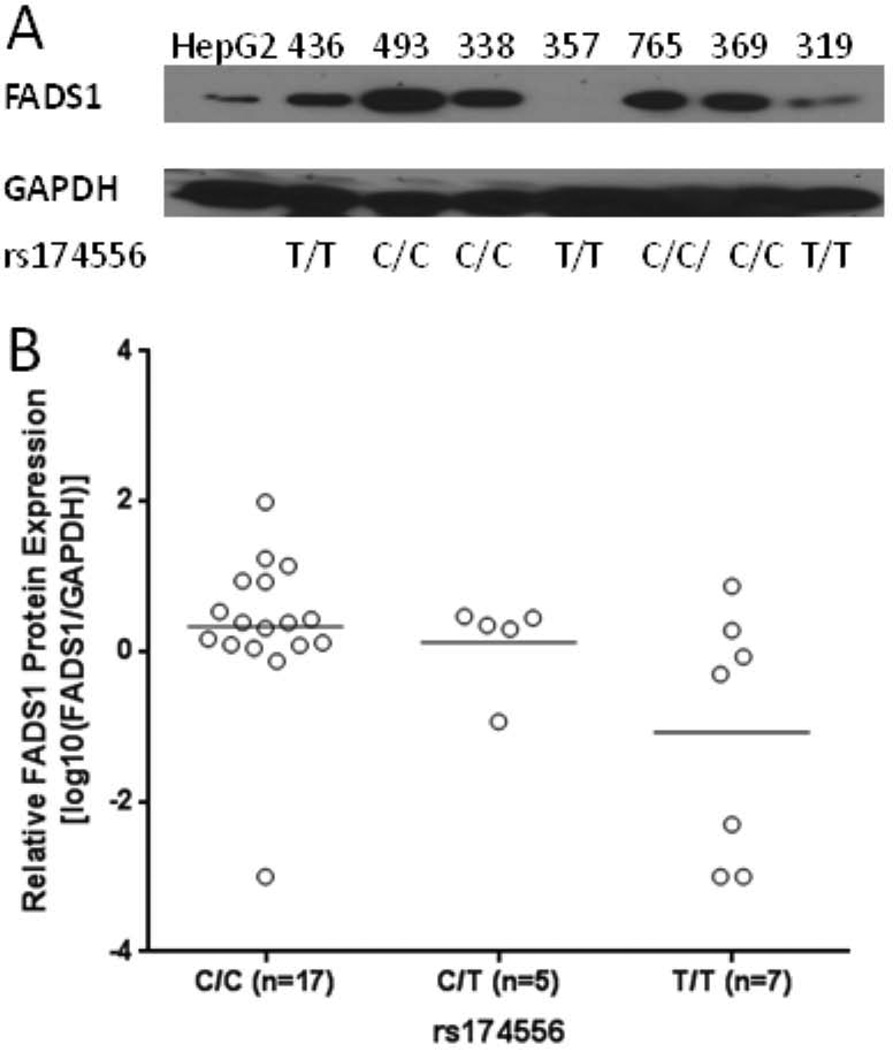
We also set out to test whether the FADS SNPs determine FADS1 protein level in the livers. We performed immunoblotting of the FADS1 protein in the livers with enough protein samples available (n=29). We found that these SNPs (represented by rs174556) were significantly associated with FADS1 protein level (R2=0.22, p=0.01) (Fig 3A–B).
Fig 3.
Association between FADS polymorphisms (rs174556 as an example) and FADS1 protein level. A) The protein levels of FADS1 and GAPDH (as a loading control) determined with Western blotting. B) Correlation between rs174556 and FADS1/GAPDH ratios (+log10 transformed) (R2=0.22, p=0.01). Samples with non-detectable FADS1 but normal GAPDH level were assigned a ratio of 0.001 to facilitate the log transformation.
Interaction between FADS SNPs and Transcription Factors (TFs)
To further elucidate the potential mechanism underlying the regulation of FADS1 gene transcription by the SNPs, we hypothesized that the 6 SNPs and/or their LD proxies (r2≥0.8) may be more likely to interact with trans-regulatory factors, e.g. TFs, than other SNPs in the locus. To test this, we investigated the distribution of these SNPs and their LD proxies in the TF binding sites (TFBS) identified by the Encyclopedia of DNA Elements (ENCODE) project, and compared this data to that of the SNPs that are not in the LD block but with similar minor allele frequency (MAF) (≥0.25). We found that among 42 highly linked SNPs, 29 (69%) are located in TFBS. This ratio was significantly higher than that of the SNPs out of the LD block [132 (43%) out of 309 SNPs; Chi-square test, p=0.001], suggesting that indeed the 6 SNPs and their LD proxies are more likely to interact with TFs.
DISCUSSION
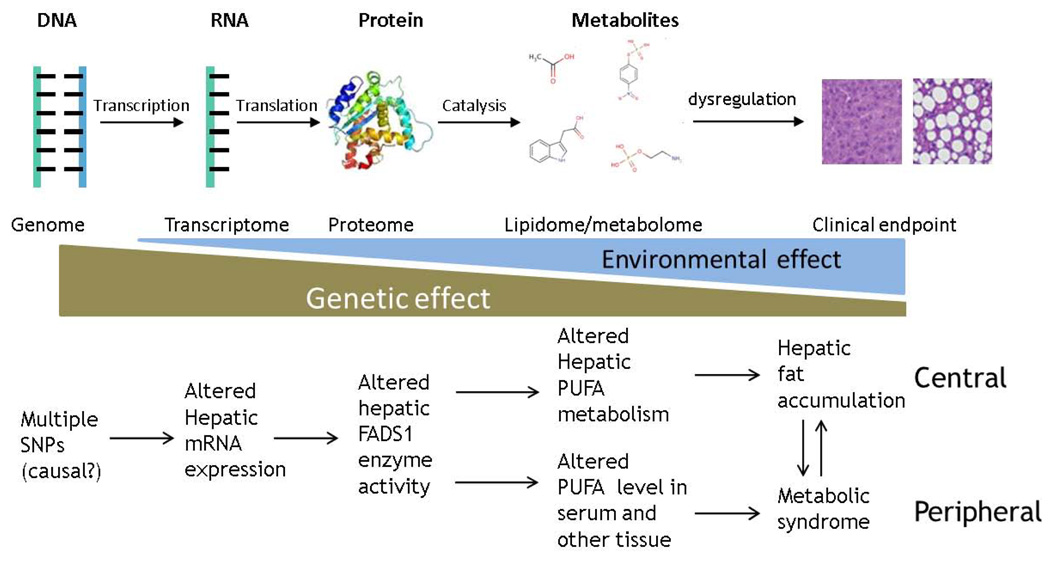
SNPs located at the FADS locus have been consistently associated with multiple phenotypes related to lipid metabolism, especially serum lipids composition. Our study for the first time observed that these SNPs are also significantly associated with lipid composition and deposition in the liver tissue. While the detailed intermediate mechanism underlying these FADS1 genotype-phenotype associations remains unexplored thus far, our study demonstrated that 1) specific hepatic lipids are affected by altered FADS1 gene expression/function, and 2) FADS1 but not FADS2 or FADS3 is the gene mediating these associations. Our study established the role of FADS1 in lipid composition in the liver as a central organ, and revealed a causal pathway encompassing DNA variation to gene/protein expression/function, followed by intermediate lipid metabolism, and eventually leading to hepatic fat accumulation. Combined with previous observations between these polymorphisms and metabolism of specific lipids e.g. in blood and breast milk (see Table S1 for summary) as peripheral tissues, our findings completed the picture showing the central role of FADS1 in lipid homeostasis in human body and underlying health and disease (Fig 4).
Fig 4.
Schematic illustration of the findings in our and previous studies. The upper panel demonstrated the general intermediate molecular mechanism underlying the genotype-phenotype association. The effect size of genetic factors on various intermediate phenotypes decreases in general along with the increased distance from DNA, while environmental effect may increase accordingly, underscoring the power of the integration of intermediate phenotypes with different omics approaches in understanding complex endpoint phenotypes. The lower panel showed the intermediate mechanism underlying the association between FADS1 alleles and various metabolic phenotypes. The genetic effect of these polymorphisms on FADS1 mRNA expression and enzyme function is conserved in both central (liver) and peripheral tissues/organs (breast, blood, etc.) in humans, which together composes the complete role of the FADS1 biology in metabolic perturbations. The importance of systems approach was therefore highlighted.
Our findings suggested a role of FADS1 and its polymorphisms involved in hepatic total fat accumulation by modulating the accumulation of VLCFAs, especially PEs, PCs and PIs. The associations between the SNPs and HFC were weaker compared to their associations with individual lipids and lipid ratios. This further reflects the greater effect size of genetic alleles on intermediate phenotypes compared to the endpoint phenotypes as pointed out previously (19) (Fig 4), especially when the sample size is limited. Our results were consistent with our previous study where a strong negative association between FADS1 mRNA expression and total hepatic fat content in human livers was demonstrated (23). The involvement of FADS1 in hepatic fat accumulation is also supported by the strong association between the same set of SNPs and liver enzyme levels in a previous GWAS (24). Liver function has been negatively correlated with fat accumulation, and the variability in hepatic fat content and hepatic function share similar genetic basis, e.g. genetic variations in PNPLA3 gene (21, 25–27), although our data suggest that the influence of the FADS1 polymorphism on hepatic lipids accumulation is likely independent of PNPLA3 variant. In addition, fat accumulation among hepatocytes is highly correlated with various metabolic perturbations including NAFLD, insulin resistance, hyperlipidemia, obesity and cardiovascular diseases. Our findings thus are also in concert with the associations between FADS SNPs and these perturbations in previous studies (8–10, 28–33). For examples, a study by Araya et al showed significant reductions in the activities of D5 and D6 desaturases in the liver of obese NAFLD samples which was postulated to play a role in altering lipid homeostasis and causing hepatic lipid accumulation (32). Others also showed association between decreased D5 desaturase activity with adverse metabolic risk profiles and in the pathogenesis of atherosclerosis (29, 30).
Our study also identified specific hepatic lipids significantly affected by FADS1 and its polymorphism. First, among 169 hepatic lipids, we identified two phospholipids whose content in the liver was significantly associated with FADS1 SNPs. Furthermore, this association stayed to be the most significant association in the subsequent genome-wide association study (lipid-QTL mapping), suggesting a highly specific linkage between these SNPs, FADS1 mRNA/protein expression and these two individual lipids. This linkage was further confirmed by the strong correlation between FADS1 SNPs and pair-wise lipid ratios. In particular, the minor allele carriers demonstrated to be associated with higher ratios of more saturated over less saturated lipids, suggesting that these SNPs are associated with reduced D5 desaturase enzyme activity. Indeed, the minor allele was also associated with reduced FADS1 protein level. This finding is consistent with previous studies showing that FADS polymorphisms are associated with increased desaturation precursor while decreased desaturation products of fatty acid, PUFA and LC-PUFA in serum, plasma, erythrocyte and breast milk in both candidate gene based and genome-wide studies (2, 19, 20, 28, 34–38). In particular, several lipids [e.g. PC (36:4), PC (36:5), PC (38:4) and PI (38:4)] in the liver which were significantly associated with these SNPs in our study were the same serum lipids showing significant association with the same SNPs in previous GWAS (19, 37), suggesting a highly specific role of FADS1 on the metabolism of these lipids. It is possible that hepatic FADS1 may determine the levels of these lipids in the liver, and these lipids are subsequently secreted into blood.
Given the clustering of three FADS genes and the high LD level across the locus, as well as the similarity in the function of FADS1/2, it remains to be unclear which gene is exactly causal to these phenotypes. Our study confirmed that FADS1 is the causal gene involved in the previously observed genotype-phenotype associations. This notion is supported by 1) the correlation between the core SNPs and transcription level of FADS1 but not FADS2 or FADS3, even in different populations; 2) the lack of other functional variants (missense or splicing) in FADS2 or FADS3 that are in LD with the core SNPs but do not alter gene transcription; and 3) the enrichment of the SNPs in the LD block at the TFBS. Our data thus warrants further investigation of the role of FADS1 in regulating PUFA metabolism and affecting susceptibility to various metabolic perturbations.
Our study fostered strong rationale for new hypotheses to be tested in the next step. First, although we observed a strong correlation between those SNPs, FADS1 gene/protein expression and hepatic lipid metabolism, the SNPs studied here are in high LD. It is unclear which SNP(s) is causal for the altered FADS1 gene function, since at least 29 SNPs in the LD block demonstrated to interfere with TFs (Table S4). How these TFs interplay with the polymorphisms and lead to the altered gene transcription remain to be investigated. Second, how altered modulation of the specific hepatic lipids identified in our study is involved in total hepatic fat accumulation represents another interesting but unaddressed question, especially many of these lipids were also associated with the same SNPs in other tissue and fluids. Given the intrinsic linkage between hepatic lipid accumulation and metabolic disorders, it is very possible that these lipids may be critical bioactive molecules involved in energy metabolism and homeostasis. It is thus warranted to further explore their roles in lipids homeostasis, e.g. whether blocking their synthesis or increase the intake of its desaturated substrates may improve/prevent hepatic fat accumulation and/or other metabolic comorbidities. Recent studies have demonstrated encouraging results in taking LC-PUFA such as prolonged n-3 polyunsaturated fatty acid for treating NAFLD (39–43). One study has also shown the potential protective effect of dietary PIs supplement on the development of NAFLD, although a detailed composition of the dietary PIs was unknown (44).
In conclusion, our study for the first time delineated the detailed mechanism in human livers that FADS polymorphisms significantly control the hepatic lipid composition, which is further involved in hepatic fat accumulation. The findings provided clues in understanding the general role of FADS1 gene in conferring susceptibility to other metabolic perturbations as well. Our work highlights the power of multi-level integrated omics (systems biology) approach to better understand causal mechanisms behind the genotype-phenotype associations noted by GWAS.
Supplementary Material
ACKNOWLEDGEMENT
We thank Dr. Xiaoliang Wang who kindly provided all imputed data for the Asian population eQTL mapping for our analyses.
FINANCIAL SUPPORT
This study was supported in part by the NIH/NIDDK grant (R21 DK090437) (WL) and the start-up fund (WL) by the Department of Medicinal Chemistry and Molecular Pharmacology, Purdue University.
LIST OF ABBREVIATIONS
- FADS
Fatty Acid Desaturase
- GWAS
Genome-wide Association Study
- SNP
Single Nucleotide Polymorphism
- HFC
Hepatic Fat Content
- VLCFA
Very Long Chain Fatty Acid
- PI
Phosphatidylinositol
- PE
Phosphatidylethanolamine
- PC
Phosphatidylcholines
- PUFA
Polyunsaturated Fatty Acid
- LC-PUFA
Long Chain Polyunsaturated Fatty Acid
- D5D
Delta-5 desaturase
- D6D
Delta-6 desaturase
- CAD
Coronary Artery Disease
- ADHD
Attention Deficit/Hyperactivity Disorder
- LD
Linkage Disequilibrium
- NAFLD
Nonalcoholic Fatty Liver Disease
- BMI
Body Mass Index
- ESI
Electrospray Ionization
- PNPLA3
Patatin-like Phospholipase Domain-containing Protein 3 gene
- VLCFA
Very Long Chain Fatty Acids
- eQTL
Expression Quantitative Traits Loci
- QC
Quality Control
- TF
Transcription Factor
- TFBS
Transcription Factor Binding Site
- ENCODE
The Encyclopedia of DNA Elements
- MAF
minor allele frequency (MAF)
Footnotes
CONFLICT OF INTEREST
The authors declared no conflict of interest.
Contributor Information
Libo Wang, Email: wang220@purdue.edu.
Shaminie Athinarayanan, Email: sathinar@purdue.edu.
Guanglong Jiang, Email: ggjiang@iu.edu.
Naga Chalasani, Email: nchalasa@iu.edu.
Min Zhang, Email: minzhang@purdue.edu.
Wanqing Liu, Email: liu781@purdue.edu.
REFERENCES
- 1.Glaser C, Heinrich J, Koletzko B. Role of FADS1 and FADS2 polymorphisms in polyunsaturated fatty acid metabolism. Metabolism. 2010;59:993–999. doi: 10.1016/j.metabol.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 2.Schaeffer L, Gohlke H, Muller M, Heid IM, Palmer LJ, Kompauer I, Demmelmair H, et al. Common genetic variants of the FADS1 FADS2 gene cluster and their reconstructed haplotypes are associated with the fatty acid composition in phospholipids. Hum Mol Genet. 2006;15:1745–1756. doi: 10.1093/hmg/ddl117. [DOI] [PubMed] [Google Scholar]
- 3.Lattka E, Illig T, Koletzko B, Heinrich J. Genetic variants of the FADS1 FADS2 gene cluster as related to essential fatty acid metabolism. Curr Opin Lipidol. 2010;21:64–69. doi: 10.1097/MOL.0b013e3283327ca8. [DOI] [PubMed] [Google Scholar]
- 4.Lattka E, Illig T, Heinrich J, Koletzko B. Do FADS genotypes enhance our knowledge about fatty acid related phenotypes? Clin Nutr. 2010;29:277–287. doi: 10.1016/j.clnu.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Merino DM, Ma DW, Mutch DM. Genetic variation in lipid desaturases and its impact on the development of human disease. Lipids Health Dis. 2010;9:63. doi: 10.1186/1476-511X-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura MT, Nara TY. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu Rev Nutr. 2004;24:345–376. doi: 10.1146/annurev.nutr.24.121803.063211. [DOI] [PubMed] [Google Scholar]
- 7.Marquardt A, Stohr H, White K, Weber BH. cDNA cloning, genomic structure, and chromosomal localization of three members of the human fatty acid desaturase family. Genomics. 2000;66:175–183. doi: 10.1006/geno.2000.6196. [DOI] [PubMed] [Google Scholar]
- 8.Martinelli N, Girelli D, Malerba G, Guarini P, Illig T, Trabetti E, Sandri M, et al. FADS genotypes and desaturase activity estimated by the ratio of arachidonic acid to linoleic acid are associated with inflammation and coronary artery disease. Am J Clin Nutr. 2008;88:941–949. doi: 10.1093/ajcn/88.4.941. [DOI] [PubMed] [Google Scholar]
- 9.Song Z, Cao H, Qin L, Jiang Y. A case-control study between gene polymorphisms of polyunsaturated fatty acid metabolic rate-limiting enzymes and acute coronary syndrome in Chinese Han population. Biomed Res Int. 2013;2013:928178. doi: 10.1155/2013/928178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Truong H, DiBello JR, Ruiz-Narvaez E, Kraft P, Campos H, Baylin A. Does genetic variation in the Delta6-desaturase promoter modify the association between alpha-linolenic acid and the prevalence of metabolic syndrome? Am J Clin Nutr. 2009;89:920–925. doi: 10.3945/ajcn.2008.27107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rzehak P, Thijs C, Standl M, Mommers M, Glaser C, Jansen E, Klopp N, et al. Variants of the FADS1 FADS2 gene cluster, blood levels of polyunsaturated fatty acids and eczema in children within the first 2 years of life. PLoS One. 2010;5:e13261. doi: 10.1371/journal.pone.0013261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brookes KJ, Chen W, Xu X, Taylor E, Asherson P. Association of fatty acid desaturase genes with attention-deficit/hyperactivity disorder. Biol Psychiatry. 2006;60:1053–1061. doi: 10.1016/j.biopsych.2006.04.025. [DOI] [PubMed] [Google Scholar]
- 13.Caspi A, Williams B, Kim-Cohen J, Craig IW, Milne BJ, Poulton R, Schalkwyk LC, et al. Moderation of breastfeeding effects on the IQ by genetic variation in fatty acid metabolism. Proc Natl Acad Sci U S A. 2007;104:18860–18865. doi: 10.1073/pnas.0704292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steer CD, Davey Smith G, Emmett PM, Hibbeln JR, Golding J. FADS2 polymorphisms modify the effect of breastfeeding on child IQ. PLoS One. 2010;5:e11570. doi: 10.1371/journal.pone.0011570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rapoport SI, Igarashi M, Gao F. Quantitative contributions of diet and liver synthesis to docosahexaenoic acid homeostasis. Prostaglandins Leukot Essent Fatty Acids. 2010;82:273–276. doi: 10.1016/j.plefa.2010.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Innocenti F, Cooper GM, Stanaway IB, Gamazon ER, Smith JD, Mirkov S, Ramirez J, et al. Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet. 2011;7:e1002078. doi: 10.1371/journal.pgen.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, Song J, Mirkov S, Xiao SY, Hart J, Liu W. Comparing morphometric, biochemical, and visual measurements of macrovesicular steatosis of liver. Hum Pathol. 2011;42:356–360. doi: 10.1016/j.humpath.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilaru A, Isaac G, Tamura P, Baxter D, Duncan SR, Venables BJ, Welti R, et al. Lipid profiling reveals tissue-specific differences for ethanolamide lipids in mice lacking fatty acid amide hydrolase. Lipids. 2010;45:863–875. doi: 10.1007/s11745-010-3457-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gieger C, Geistlinger L, Altmaier E, Hrabe de Angelis M, Kronenberg F, Meitinger T, Mewes HW, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Illig T, Gieger C, Zhai G, Romisch-Margl W, Wang-Sattler R, Prehn C, Altmaier E, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42:137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang X, Tang H, Teng M, Li Z, Li J, Fan J, Zhong L, et al. Mapping of hepatic expression quantitative trait loci (eQTLs) in a Han Chinese population. J Med Genet. 2014;51:319–326. doi: 10.1136/jmedgenet-2013-102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mirkov S, Myers JL, Ramirez J, Liu W. SNPs affecting serum metabolomic traits may regulate gene transcription and lipid accumulation in the liver. Metabolism. 2012;61:1523–1527. doi: 10.1016/j.metabol.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chambers JC, Zhang WH, Sehmi J, Li XZ, Wass MN, Van der Harst P, Holm H, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43 doi: 10.1038/ng.970. 1131-U1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanyal AJ. NASH: A global health problem. Hepatol Res. 2011;41:670–674. doi: 10.1111/j.1872-034X.2011.00824.x. [DOI] [PubMed] [Google Scholar]
- 26.Guichelaar MM, Gawrieh S, Olivier M, Viker K, Krishnan A, Sanderson S, Malinchoc M, et al. Interactions of allelic variance of PNPLA3 with nongenetic factors in predicting nonalcoholic steatohepatitis and nonhepatic complications of severe obesity. Obesity (Silver Spring) 2013;21:1935–1941. doi: 10.1002/oby.20327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Verrijken A, Beckers S, Francque S, Hilden H, Caron S, Zegers D, Ruppert M, et al. A gene variant of PNPLA3, but not of APOC3, is associated with histological parameters of NAFLD in an obese population. Obesity (Silver Spring) 2013;21:2138–2145. doi: 10.1002/oby.20366. [DOI] [PubMed] [Google Scholar]
- 28.Malerba G, Schaeffer L, Xumerle L, Klopp N, Trabetti E, Biscuola M, Cavallari U, et al. SNPs of the FADS gene cluster are associated with polyunsaturated fatty acids in a cohort of patients with cardiovascular disease. Lipids. 2008;43:289–299. doi: 10.1007/s11745-008-3158-5. [DOI] [PubMed] [Google Scholar]
- 29.Das UN. A defect in the activity of Delta(6) and Delta(5) desaturases may be a factor in the initiation and progression of atherosclerosis. Prostaglandins Leukotrienes and Essential Fatty Acids. 2007;76:251–268. doi: 10.1016/j.plefa.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Murakami K, Sasaki S, Takahashi Y, Uenishi K, Watanabe T, Kohri T, Yarnasaki M, et al. Lower estimates of delta-5 desaturase and elongase activity are related to adverse profiles for several metabolic risk factors in young Japanese women. Nutrition Research. 2008;28:816–824. doi: 10.1016/j.nutres.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Allard JP, Aghdassi E, Mohammed S, Raman M, Avand G, Arendt BM, Jalali P, et al. Nutritional assessment and hepatic fatty acid composition in non-alcoholic fatty liver disease (NAFLD): a cross-sectional study. J Hepatol. 2008;48:300–307. doi: 10.1016/j.jhep.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 32.Araya J, Rodrigo R, Pettinelli P, Araya AV, Poniachik J, Videla LA. Decreased liver fatty acid delta-6 and delta-5 desaturase activity in obese patients. Obesity (Silver Spring) 2010;18:1460–1463. doi: 10.1038/oby.2009.379. [DOI] [PubMed] [Google Scholar]
- 33.Simopoulos AP. Genetic variants in the metabolism of omega-6 and omega-3 fatty acids: their role in the determination of nutritional requirements and chronic disease risk. Exp Biol Med (Maywood) 2010;235:785–795. doi: 10.1258/ebm.2010.009298. [DOI] [PubMed] [Google Scholar]
- 34.Xie L, Innis SM. Genetic variants of the FADS1 FADS2 gene cluster are associated with altered (n-6) and (n-3) essential fatty acids in plasma and erythrocyte phospholipids in women during pregnancy and in breast milk during lactation. J Nutr. 2008;138:2222–2228. doi: 10.3945/jn.108.096156. [DOI] [PubMed] [Google Scholar]
- 35.Molto-Puigmarti C, Plat J, Mensink RP, Muller A, Jansen E, Zeegers MP, Thijs C. FADS1 FADS2 gene variants modify the association between fish intake and the docosahexaenoic acid proportions in human milk. Am J Clin Nutr. 2010;91:1368–1376. doi: 10.3945/ajcn.2009.28789. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka T, Shen J, Abecasis GR, Kisialiou A, Ordovas JM, Guralnik JM, Singleton A, et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 2009;5:e1000338. doi: 10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demirkan A, van Duijn CM, Ugocsai P, Isaacs A, Pramstaller PP, Liebisch G, Wilson JF, et al. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet. 2012;8:e1002490. doi: 10.1371/journal.pgen.1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemaitre RN, Tanaka T, Tang W, Manichaikul A, Foy M, Kabagambe EK, Nettleton JA, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet. 2011;7:e1002193. doi: 10.1371/journal.pgen.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Capanni M, Calella F, Biagini MR, Genise S, Raimondi L, Bedogni G, Svegliati-Baroni G, et al. Prolonged n-3 polyunsaturated fatty acid supplementation ameliorates hepatic steatosis in patients with non-alcoholic fatty liver disease: a pilot study. Aliment Pharmacol Ther. 2006;23:1143–1151. doi: 10.1111/j.1365-2036.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- 40.Spadaro L, Magliocco O, Spampinato D, Piro S, Oliveri C, Alagona C, Papa G, et al. Effects of n-3 polyunsaturated fatty acids in subjects with nonalcoholic fatty liver disease. Dig Liver Dis. 2008;40:194–199. doi: 10.1016/j.dld.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 41.Nobili V, Bedogni G, Alisi A, Pietrobattista A, Rise P, Galli C, Agostoni C. Docosahexaenoic acid supplementation decreases liver fat content in children with non-alcoholic fatty liver disease: double-blind randomised controlled clinical trial. Arch Dis Child. 2011;96:350–353. doi: 10.1136/adc.2010.192401. [DOI] [PubMed] [Google Scholar]
- 42.Janczyk W, Socha P, Lebensztejn D, Wierzbicka A, Mazur A, Neuhoff-Murawska J, Matusik P. Omega-3 fatty acids for treatment of non-alcoholic fatty liver disease: design and rationale of randomized controlled trial. BMC Pediatr. 2013;13:85. doi: 10.1186/1471-2431-13-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabatti C, Service SK, Hartikainen AL, Pouta A, Ripatti S, Brodsky J, Jones CG, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirouchi B, Nagao K, Inoue N, Furuya K, Koga S, Matsumoto H, Yanagita T. Dietary phosphatidylinositol prevents the development of nonalcoholic fatty liver disease in Zucker (fa/fa) rats. J Agric Food Chem. 2008;56:2375–2379. doi: 10.1021/jf703578d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.