Abstract
Acinus-S’ is a co-repressor for retinoic acid receptor (RAR)-dependent gene transcription and has been suggested to be involved in RNA processing. In this study the role of Acinus isoforms in regulating pre-mRNA splicing was explored using in vivo splicing assays. Both Acinus-L and Acinus-S’, with the activity of Acinus-L higher than that of Acinus-S’, increase the splicing of a retinoic acid (RA)-responsive minigene containing a weak 5′ splice site but not a RA-responsive minigene containing a strong 5′ splice site. RA treatment further enhances the splicing of the weak 5′ splice site by Acinus in a dose- and time-dependent manner, suggesting a RA-dependent activity in addition to a RA-independent activity of Acinus. The RA-independent effect of Acinus occurs to varying degrees using minigene constructs containing several different promoters while the RA-dependent splicing activity of Acinus is specific for transcripts derived from the minigene driven by a RA response element (RARE)-containing promoter. This suggests that the ligand-dependent splicing activity of Acinus is related to the RA-activated RAR bound to the RARE. The RRM domain is necessary for the RA-dependent splicing activity of Acinus and the RA-independent splicing activity of Acinus is repressed by RNPS1. Importantly, measurement of the splicing of endogenous human RARβ and Bcl-x in vivo demonstrates that Acinus stimulates the use of the weaker alternative 5′ splice site of these two genes in a RA-dependent manner for RARβ and a RA-independent manner for Bcl-x. Taken together, these studies demonstrate that Acinus functions in both RAR-dependent splicing and RAR-dependent transcription.
Keywords: Acinus, RAR, retinoic acid, alternative splicing
Retinoic acid (RA) acts as a ligand for the RA-receptor (RAR)/retinoid X receptor (RXR) heterodimer to activate target gene expression (for review, see Chambon, 1996; Evans and Mangelsdorf, 2014). RARs and RXRs display the typical conserved modular structure, which includes five or six structurally and functionally distinct domains termed A-F, characteristic of all members of the nuclear hormone receptor superfamily. Conformational changes within the AF-2 region (the ligand-dependent transcription activation function 2) located in the E domain that are induced by ligand binding leads to the release of corepressor complexes (such as HDAC-containing NCoR and SMRT complexes) and the recruitment of coactivator complexes (such as SRC-p160 and CBP/p300 family members). This acts as the ‘switch’ between transcriptional repression and transcriptional activation.
The structurally flexible N-terminal AF-1 region (the ligand-independent transcription activation function 1) in the A/B domain has a propensity to adopt a more structured conformation through intra- and/or intermolecular protein-protein interactions upon ligand binding. AF-1 plays an important role in cooperation with AF-2 in ligand-activated transcription by nuclear receptors. The AF-1 region of different members of the nuclear receptor superfamily has been shown to associate with the general transcription machinery, co-repressor proteins, coactivator proteins and other transcription factors or coregulatory proteins (for review, see Lavery and McEwan, 2005). Acinus-S’ is one of the proteins identified in our lab to interact with A/B region (AF-1) of RARs and regulate the transcription of RAR target genes (Vucetic et al., 2008).
Apoptotic Chromatin Condensation Inducer in the Nucleus (Acinus) has three isoforms, termed Acinus-L, Acinus-S and Acinus-S’ (Figure 1A), which are most likely generated by alternative splicing and/or alternative promoter usage (Sahara et al., 1999). These three isoforms share a common C-terminus containing a RRM domain and a region rich in RS dipeptides (two typical structural features of SR/SR-related splicing factors), and differ in their N-termini. To date, no distinct function(s) for any of these three isoforms has been identified.
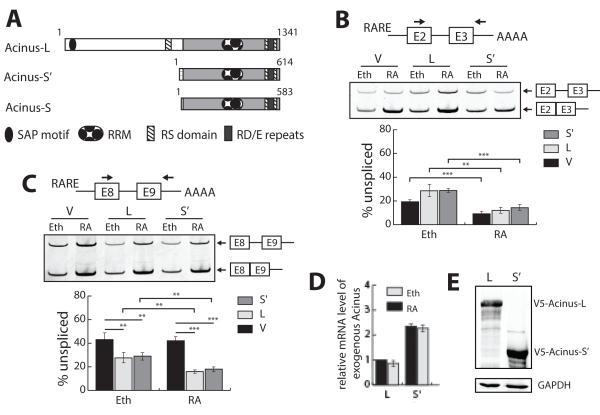
Figure 1. Both Acinus-S’ and Acinus-L cooperate with RA to activate the splicing of a RA-responsive reporter minigene containing a weak 5′ splice site.
A. Functional domains of the three human Acinus isoforms. B. Acinus does not facilitate the splicing of RARE-tuba1bE2-E3 pre-mRNA containing a strong 5′splice site. C. Acinus and RA cooperatively activate the splicing of RARE-tubg1E8-E9 pre-RNA containing a weak 5′splice site. D. Relative level of exogenous Acinus-L and Acinus-S’ mRNA in transfected cells. E. Relative level of exogenous Acinus-L and Acinus-S’ in transfected cells. 293A cells were transfected with pRARE-tuba1bE2-E3 (B) or pRARE-tubg1E8-E9 (C-E) along with the expression vector DNA for RARβ (B–E) and the expression vector DNA for either V5-Acinus-L or V5-Acinus-S’ (B-E). The V5 empty vector DNA (pcDNA3.1nV5DEST) was transfected as a control for pV5-Acinus-L or pV5-Acinus-S’ (B-E). In addition, the internal control Renilla vector DNA (pRL-CMV) was transfected as a normalizer for transfection efficiency (B-E). Twenty-four hr following transfection, cells were treated with ethanol (B-E) or RA (10−6 M) (B-D) for 24 hr. RNA was prepared and used to analyze splicing in Panels B-C or relative mRNA levels in Panel D. In Panels B-C, RT-PCR was performed to analyze splicing of the minigene pre-mRNAs using the indicated forward and reverse primers (see diagram for location). Spliced and unspliced PCR products were resolved on polyacrylamide gels, stained using SYTO 60 and quantitated using LI-COR Odyssey Infrared Imaging System. Percentage of unspliced was calculated as the ratio of the intensity of PCR products of the unspliced transcripts to that of the sum of the spliced and unspliced transcripts. In Panel D, RT-qPCR was performed to quantitate the relative mRNA levels of exogenous Acinus-L and Acinus-S’. The mRNA levels were normalized to Renilla mRNA levels. The normalized relative mRNA level of exogenous Acinus-L coexpressed with RARE-tubg1E8-E9 in cells treated with ethanol was set to 1. The forward primer is located within the C-terminal common region of Acinus isoforms and the reverse primer is located within the bovine growth hormone polyadenylation region on the V5 vector. In Panel E, whole protein extracts were isolated and western blot was performed using anti-V5 or anti-GAPDH antibodies. V, V5 empty vector; L, V5-Acinus L; S’ V5-Acinus-S’; Eth, ethanol; RA, retinoic acid. Values represent mean ± SD from 3 independent experiments. ** p < 0.01, *** p < 0.001, unpaired t test.
The potential role of Acinus in RNA processing has been suggested by identification of Acinus as a component of the spliceosome (Rappsilber et al., 2002; Zhou et al., 2002), nuclear speckles (Saitoh et al., 2004) and the exon junction complex (EJC) (Tange et al., 2005). However, the role of Acinus in RNA processing has not been described. Several SR-related proteins such as PGC-1 and CAPER function as both hormone-dependent transcriptional coregulators and splicing cofactors for nuclear receptors (Dowhan et al., 2005; Jung et al., 2002; Monsalve et al., 2000). Since Acinus-S’ acts as a co-repressor for RAR-dependent gene transcription and is suggested to be involved in RNA processing, it is likely that Acinus has dual activities in both RAR-dependent transcription and splicing.
In the present work, both Acinus-L and Acinus-S’ are shown to be splicing cofactors (with the activity of Acinus-L higher than that of Acinus-S’) that facilitate constitutive splicing of pre-mRNAs containing a weak 5′ splice site and regulate alternative splicing in favor of the isoform generated from the weaker alternative 5′ splice site. In addition, both Acinus-L and Acinus-S’ have a RA-dependent splicing activity specific for RA-responsive genes containing a retinoic acid response element (RARE), which suggests that Acinus has an additional function in RAR-dependent splicing.
MATERIALS AND METHODS
Cells
293A cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 2mM glutamine, 100 μg/ml penicillin, and 100 units/ml streptomycin. The cells were maintained in a humidified 5% CO2 incubator at 37°C. All transfections were performed using GenJet™ DNA In Vitro Transfection Reagent (SignaGen Laboratories, Rockville, MD) according to the manufacturer’s protocol. The ratio of GenJet Reagent (μl):DNA (μg) was 3:1. Six hr after transfection, the medium was changed to complete medium containing 10% charcoal/dextran-treated fetal bovine serum. Twenty-four hr after transfection, cells were treated with RA prepared in ethanol (generous gift from Hoffman-LaRoche) or rosiglitazone (Cayman Chemicals, Ann Arbor, MI) prepared in DMSO at the indicated concentrations for up to an additional 24hr. Control cells were treated with ethanol or DMSO.
Plasmids for in vivo splicing assay
pRARE-tuba1bE2-E3 was constructed by replacing the luciferase fragment of pRAR-Luc (Panomics, Fremont, CA) with the intact TUBA1B exon 2-intron 2-exon 3 from the minigene pcDNA3.1(+)-tuba1bE2-E3. pRARE-tubg1E8-E9, pSp1RE-tubg1E8-E9 and pPPRE-tubg1E8-E9 were constructed by replacing the luciferase fragment of pRAR-Luc, pSp1-Luc and pPPAR-Luc (Panomics), respectively, with the intact TUBG1 exon 8-intron 8- exon 9 from the minigene pcDNA3.1(+)-tubg1E8-E9 (CMV-tubg1E8-E9). pcDNA3.1(+)-tuba1bE2-E3 and pcDNA3.1(+)-tubg1E8-E9 were kind gifts from Dr. Dong-Er Zhang (University of California, San Diego, CA) (Ahn et al., 2011). pV5-Acinus-L and pV5-Acinus-S’ were constructed using Invitrogen Gateway® (Life Technologies, Grand Island, NY) cloning technology to clone the full length coding sequence of human Acinus-L and Acinus-S’, respectively, into pcDNA3.1/nV5-DEST destination vector. pV5-Acinus-S’ (ΔRRM) was constructed using QuickChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Inc., Wilmington, DE) to delete RRM (amino acids 301-360) of Acinus-S’. pV5-Acinus-S’ (ΔC) was constructed using QuickChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Inc) for introducing an in-frame premature stop codon by replacing G at nucleotide 1228 of the coding sequence of Acinus-S’ with T and an out-of-frame premature stop codon by replacing G at nucleotide 1233 of the coding sequence of Acinus-S’ with T to generate the C-terminal 205 amino acid truncated Acinus-S’. Other DNA expression constructs used were pOPRSVICAT-RARβ (Soprano et al., 2000); pCMX-PPARγ (kind gift from Dr. Ronald Evans, Salk Institute for Biological Studies, La Jolla, CA) (Kliewer et al., 1992) and pCMV-3XFLAG-RNPS1 (kind gift from Dr. Akila Mayeda, Fujita Health University, Japan and Dr. Eiji Sakashita, Jichi Medical University, Japan) (Sakashita et al., 2004) and pRL-CMV (Promega, Madison, WI).
Splicing minigene reporter assays
293A cells were transfected with the indicated combinations of DNA: one of the splicing reporter minigene plasmid DNAs (0.8 μg), pOPRSVICAT-RARβ or pCMX-PPARγ expression vector DNA (0.3 μg), V5-Acinus-L, V5-Acinus-S’ or empty pcDNA3/nV5-DEST expression vector DNA (3 μg), and pRL-CMV DNA (6 ng). Twenty-four hr following transfection cells were treated for an additional 24 hr with 10−6 M RA, 50 μM rosiglitazone or carrier (ethanol or DMSO). RNA was isolated using RNA-Bee™ reagent (Tel Test Inc, Gainesville, FL) following the manufacturer’s protocol. To remove any contaminating DNA, RNA samples were treated with RQ1 RNase-Free DNase (Promega) followed by clean up with E.Z.N.A.™ MicroElute RNA Clean Up Kit (Omega Bio-tek, Inc, Norcross, GA) following the manufacturers’ protocol. Following purification, RNA was reverse transcribed using High Capacity cDNA Reverse Transcription Kit from Applied Biosystems (Foster City, CA). PCR was performed using Go-Taq Flexi DNA Polymerase (Promega). As a control for DNA contamination, equal amounts of each purified RNA sample were amplified by PCR without reverse transcription. The forward and reverse primers were designed to target the first exon of the minigene and the transcribed sequence from the plasmid vector downstream of the last exon (Integrated DNA Technologies, Coralville, IA) allowing for the detection of both the spliced and unspliced RNA products. Specifically, primers to detect RARE-E2-E3 were forward 5′-CCGGGCTGTGTTTGTAGACT-3′ and reverse 5′-ATCATGTCTGCTCGAAGC-3′; CMV-E8-E9 were forward 5′-CGGCTACACCCCTCTCACTA-3′ and reverse 5′-TAGAAGGCACAGTCGAGG-3′; and RARE-E8-E9, Sp1-E8-E9 and PPRE-E8-E9 were forward 5′-CGGCTACACCCCTCTCACTA-3′ and reverse 5′-ATCATGTCTGCTCGAAGC-3′. Spliced and unspliced products of the minigenes were resolved on a polyacrylamide gel and stained using SYTO 60 red fluorescent nucleic acid stain (Life Technologies, Grand Island, NY) (dilution factor: 1:10,000) for 1 hr at room temperature. Stained gels were scanned and quantitated using a LI-COR Odyssey instrument using the 700 nm channel with a 0.25 mm focus offset.
Endogenous Gene Alternative Splicing
Following transfection of 293A cells with pV5-Acinus-L, pV5-Acinus-S’or empty pcDNA3/nV5-DEST expression vector DNA (4μg) and treatment for 24 hr with 10−6 M RA or ethanol, RNA was isolated, treated with RQ1 RNase-Free DNase, reverse transcribed, amplified by PCR and stained with SYTO 60 red fluorescent dye as described for the splicing minigene reporter assay. Primers (Integrated DNA Technologies) to detect alternatively spliced products of human RARβ were forward 5′-ATCCGAGCAGGGTTTGTCTG-3′ and reverse 5′-TACACTCGAGGGGGAGGAAG-3′ and for human Bcl-x were forward 5′-GGGCATTCAGTGACCCTGACA-3′ and reverse 5′-GGTGGGAGGTAGAGTGGAT’3′.
Western blot
Western blot analysis of total cell protein extracts was performed essentially as previously described (Vucetic et al., 2008; Zhao et al., 2009). Primary antibodies used were goat anti-GAPDH (sc-48176, Santa Cruz, Santa Cruz, CA), mouse anti-V5 (R960-25, Life Technologies), and mouse anti-FLAG (F1804, Sigma-Aldrich, St. Louis, MI). Secondary antibodies used were donkey anti-mouse IRDye 800CW and donkey anti-goat IRDye 680CW purchased from LI-COR, Lincoln, NE. Images were captured and quantitated using the LI-COR Odyssey instrument and software. GAPDH levels were used as the loading control.
Quantitative RT-PCR
Total RNA was isolated, treated with RQ1 RNase-Free DNase and reverse transcribed as described for the splicing minigene reporter assay. Quantitative real-time PCR was performed using SYBR Green PCR Master mix (Fermentas,Thermo Fisher Scientific, Rockford, IL) essentially as previously described (Vucetic et al., 2008; Zhao et al., 2009). The primers (Integrated DNA Technologies) included: exogenous human Acinus (both L or S’): forward, 5′-GACCGCCGCCGCAAGGAACGTG-3′ and reverse, 5′-TAGAAGGCACAGTCGAGG-3′ (bovine growth hormone polyadenylation sequence transcribed from the plasmid vector); RARE-tubg1E8-E9 or PPRE-tubg1E8-E9 total RNA: forward, 5′-GCCAGCGTGAGGAAGACC-3′ and reverse, 5′-ATCATGTCTGCTGAAGC-3′; and Renilla: forward, 5′-GATAACTGGTCCGCAGTGGT-3′ and reverse, 5′-TTGCCTGATTTGCCCATACC-3′. Changes in gene expression were calculated using the ddCT method for relative quantification of each target gene normalized to the internal transfection control Renilla. All primers used yielded a dissociation curve with a single peak and a single PCR product of the appropriate size.
RESULTS
Effect of Acinus on Splicing of Pre-mRNAs Transcribed from Reporter Minigenes Driven by a RARE-TATA Promoter
In order to examine the role of Acinus-S’ and Acinus-L in pre-mRNA splicing of RAR-regulated genes, we utilized two splicing reporter minigenes driven by RARE-TATA promoter constructed from the TUBA1B minigene (pCMV-tuba1bE2-E3) and the TUBG1 minigene (pCMV-tubg1E8-E9), and named pRARE-tuba1bE2-E3 and pRARE-tubg1E8-E9, respectively (Figure 1B and 1C). The E2-E3 (intact exon 2-intron 2-exon 3) region of the TUBA1B gene contains a strong (canonical) 5′ splice site (5′ss), and the E8-E9 (intact exon 8-intron 8-exon 9) region of the TUBG1gene contains a weak 5′ss (Ahn et al., 2011). To test the effect of Acinus on the splicing of pre-mRNAs transcribed from pRARE-tuba1bE2-E3 and pRARE-tubg1E8-E9, 293A cells were transfected with these two minigenes individually along with the expression vector DNAs for RARβ and either Acinus-L or Acinus-S’. Cells were treated with ethanol (control) or RA for 24 hr. RT-PCR was performed to analyze splicing efficiency using primer pairs that span the intron of either RARE-tuba1b-E2-E3 pre-mRNA or RARE-tubg1E8-E9 pre-mRNA to allow detection of both the unspliced and spliced RNAs produced by each of these reporter minigenes.
As expected for a strong 5′ss, less than 20% of total RARE-tub1bE2-E3 pre-mRNA is unspliced in empty vector control cells treated with ethanol (Figure 1B). Treatment with RA significantly enhances the splicing of RARE-tub1bE2-E3 pre-mRNA resulting in only 10% unspliced pre-mRNA in the empty vector cells. Overexpression of either Acinus-L or Acinus-S’ slightly represses the splicing of RARE-tub1bE2-E3 pre-mRNA both in the presence and absence of RA. Therefore the splicing of RARE-tuba1bE2-E3 pre-mRNA is enhanced by RA independent of Acinus overexpression, suggesting that RA facilitates the splicing of RARE-tuba1bE2-E3 pre-mRNA by a mechanism that does not involve Acinus.
On the other hand, approximately 43% of RNA transcribed from the pRARE-tubg1E8-E9 minigene containing a weak 5′ss is unspliced both in the absence and presence of RA (Figure 1C). Overexpression of Acinus (either Acinus-L or Acinus-S’) significantly increases the splicing of RARE-tubg1E8-E9 pre-mRNA with this effect strongly enhanced by RA treatment. Approximately 30% of the total mRNA is unspliced in the absence of RA and 17% is unspliced in the presence of RA in the cells overexpressing either Acinus-L or Acinus-S’ (Figure 1C). These data demonstrate that Acinus increases the splicing of RARE-tubg1E8-E9 pre-mRNA and RA further enhances this effect of Acinus.
In order to determine the expression levels of exogenous Acinus-L and Acinus-S’ in the transfected cells to allow a comparison of their effect on the splicing of RARE-tubg1E8-E9 pre-mRNA, the relative mRNA level of exogenous Acinus-L or Acinus-S’ in each sample was measured by real-time PCR (qPCR) and normalized for transfection efficiency to the internal control Renilla. The relative mRNA level of exogenous Acinus-L was less than half of that of exogenous Acinus-S’ (Figure 1D). Furthermore the level of exogenous Acinus-S’ protein was also more than 2 times that of exogenous Acinus-L protein (Figure 1E). Since Acinus-S’ and Acinus-L activate the splicing of RARE-tubg1E8-E9 pre-mRNA to a similar extent but Acinus-S’ levels are at least 2 times higher than that of Acinus-L (Figure 1C, D and E), we infer that this splicing activity of Acinus-L is higher than that of Acinus-S’ by at least 2-fold.
Acinus has Promoter Specificity in Activating Ligand-dependent Splicing of Pre-mRNAs from Exon 8-9 Region of TUBG1
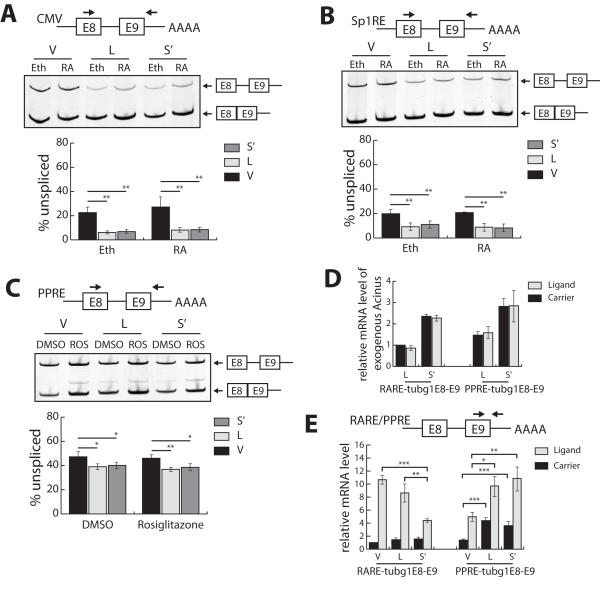
Since Acinus and RA cooperate in the splicing of RARE-tubg1E8-E9 pre-mRNA, we next wished to determine if this effect is due to the presence of the RARE in the promoter of this minigene by examining the splicing of the same TUBG1 exon 8-9 pre-mRNA under the regulation of two constitutive promoters: CMV (pCMV-tubg1E8-E9) and Sp1 (pSp1RE-tubg1E8-E9). Experiments were performed as described in Figure 1C except that pRARE-tubg1E8-E9 DNA was replaced with either pCMV-tubg1E8-E9 or pSp1RE-tubg1E8-E9 DNA. RT-PCR analysis demonstrated that approximately 25% of RNA transcribed from pCMV-tubg1E8-E9 and 20% of RNA transcribed from pSp1RE-tubg1E8-E9 is unspliced both in the presence and absence of RA (Figures 2A and 2B, respectively). Expression of Acinus-L or Acinus-S’ increased the splicing of pre-mRNAs transcribed from these two minigenes with approximately 8% (CMV-tubg1-E8-E9) and 10% (Sp1RE-tubg1-E8-E9) of the total RNA remaining unspliced both in the presence and absence of RA (Figures 2A and 2B, respectively). Therefore, unlike the splicing of RARE-tubg1E8-E9 pre-mRNA, RA does not cooperate with Acinus in activating the splicing of TUBG1 exons 8-9 transcribed from minigenes driven by two non-RA responsive promoters.
Figure 2. The cooperation between RA and Acinus isoforms in the splicing of a weak 5′ splice site is promoter specific.
A-B. Acinus activates the splicing of CMV-tubg1E8-E9 pre-mRNA (A) and Sp1RE-tubg1E8-E9 pre-mRNA (B) containing a weak 5′splice site without cooperation with RA. C. Acinus activates the splicing of PPRE-tubg1E8-E9 pre-mRNA containing a weak 5′ splice site without cooperation with rosiglitazone. D. The relative level of exogenous Acinus-L mRNA and Acinus-S’ mRNA in cells transfected with pRARE-tubg1E8-E9 and pPPRE-tub1E8-E9. E. The relative level of RARE-tubg1E8-E9 mRNA and PPRE-tubg1E8-E9 mRNA in transfected cells. 293A cells were transfected with pCMV-tubg1E8-E9 (A), pSp1RE- tubg1E8-E9 DNA (B), pPPRE- tubg1E8-E9 (C-E) or pRARE-tubg1E8-E9 (D-E) splicing reporter DNA along with the expression vector DNA for RARβ (A, B, D and E) or PPARγ (C-E) and the expression vector DNA for either V5-Acinus-L or V5-Acinus-S’ (A-E). The V5 empty vector DNA (pcDNA3.1nV5DEST) was transfected as a control for pV5-Acinus-L or pV5-Acinus-S’ (A-E). In addition, the internal control Renilla vector DNA (pRL-CMV) was transfected as a normalizer for transfection efficiency (A-E). Twenty-four hr following transfection, cells in Panels A, B, D, and E were treated with ethanol (Eth) or RA (10−6 M) for 24 hr, and cells in Panels C-E were treated with DMSO or 50 μM Rosiglitazone for 24 hr. Total RNA was prepared and used to analyze splicing as described in the legend to Figure 1. In Panel D, RT-qPCR was performed to quantitate the relative mRNA levels of exogenous Acinus-L and Acinus-S’ as described in Figure 1. In Panel E, schematic diagram of RARE-tubg1E8-E9/PPRE-tubg1E8-E9 is shown in the upper panel. Arrows indicate the positions of the primer pairs for RT-qPCR analysis. All mRNA levels are normalized to Renilla mRNA levels. The normalized relative mRNA level of RARE-tubg1E8-E9 in the empty vector control cells treated with ethanol was set to 1. V, V5 empty vector; L, V5-Acinus-L; S’, V5-Acinus-S’; Eth, ethanol; RA, retinoic acid; DMSO, dimethyl sulfoxide; ROS, rosiglitazone. Values represent mean ± SD from 3 independent experiments. * p < 0.05, ** p < 0.01, *** p < 0.001, unpaired t test.
We also examined the effect of Acinus on the splicing of the TUBG1 exon8-exon9 pre-mRNA under the regulation of another hormone responsive promoter, PPARγ (pPPRE-tubg1E8-E9). Figure 2C shows that approximately 47% of RNA transcribed from pPPRE-tubg1E8-E9 is unspliced in empty vector control cells in the absence and presence of the PPARγ ligand, rosiglitazone. In addition, overexpression of either Acinus-L or Acinus-S’ slightly increases the splicing of PPRE-tubg1E8-E9 pre-mRNA reducing the amount of unspliced pre-mRNA to approximately 40% (Figure 2C). More importantly, unlike the cooperation of Acinus and RA in activating the splicing of RARE-tubg1E8-E9 pre-mRNA, treatment with rosiglitazone has no effect on the activity of Acinus in the splicing of PPRE-tubg1E8-E9 pre-mRNA. This effect is not due to differences in the expression of Acinus isoforms in the RARE-tubg1E8-E9 mRNA expressing cells compared to that in the PPRE-tubg1E8-E9 mRNA expressing cells since the relative mRNA levels of both exogenous Acinus-L and Acinus-S’ are similar in these two experiments (Figure 2D). Furthermore, the lack of cooperation between rosiglitazone and Acinus in the splicing of PPRE-tubg1-E8-E9 pre-mRNA is not likely to be related to the fold activation of transcription by rosiglitazone compared with RA since both rosiglitazone and RA increased the level of TUBG1 exon8-exon9 total RNA to a relatively similar extent (Figure 2E). Note, as we have previously reported, Acinus-S’ repressed RA-dependent transcription (Vucetic et al., 2008). Since pRARE-tubg1E8-E9 and pPPRE-tubg1E8-E9 only differ in one cis-acting DNA sequence (DR5 RARE vs DR1 PPRE) in their promoter regions, which are bound by their cognitive transcriptional activators (RAR or PPAR, respectively), we infer that Acinus has promoter specificity in activating ligand-dependent splicing of pre-mRNAs from exon8-9 region of TUBG1.
RA Enhances the Stimulatory Effect of Acinus on Splicing of RARE-tubg1E8-E9 Pre-mRNA in a Dose-dependent Manner
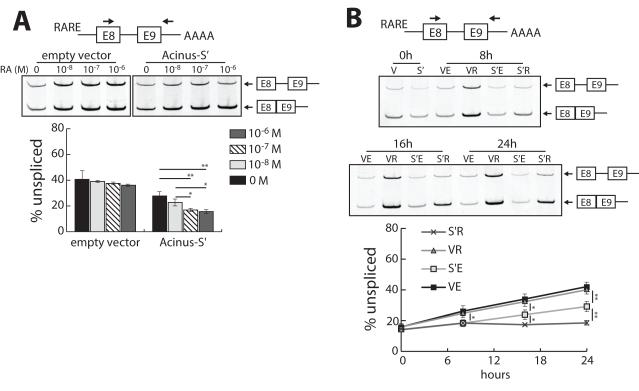
RA treatment enhances the effect of both Acinus-L and Acinus-S’ in activating splicing of RARE-tubg1E8-E9 pre-mRNA (Figure 1C). In order to determine if this effect of RA is dose-dependent, 293A cells were transfected with the minigene pRARE-tubg1E8-E9 along with the expression vectors for RARβ and Acinus-S’. Transfected cells were treated with either ethanol or different concentrations of RA for 24 hr. RT-PCR analysis showed that with increasing concentrations of RA, the splicing of RARE-tubg1E8-E9 pre-mRNA increases only when Acinus is overexpressed (Figure 3A). Maximal splicing of RARE-tubg1E8-E9 pre-mRNA was observed in cells overexpressing Acinus-S’ and treated with 10−7 M RA. These data indicate that RA enhances the effect of Acinus in activating splicing of RARE-tubg1E8-E9 pre-mRNA in a dose-dependent manner, which further supports the idea that RA cooperates with Acinus in facilitating pre-mRNA splicing of RARE-tubg1E8-E9.
Figure 3. The synergistic effect of RA and Acinus on the splicing of a RA-responsive reporter minigene containing a weak 5′ splice site is RA dose-dependent and time-dependent.
A. RA enhances the stimulatory effect of Acinus on the splicing RARE-tubg1E8-E9 pre-mRNA in a dose-dependent manner. B. The synergism of RA and Acinus in promoting the splicing of RARE-tubg1E8-E9 pre-mRNA is time-dependent. In Panels A and B, 293A cells were transfected with pRARE-tubg1E8-E9 splicing reporter DNA, RARβ expression vector DNA, and either V5-Acinus-S’ (S’) expression vector DNA or V5 empty vector DNA (V). Twenty-four hr after transfection cells were treated with ethanol or RA at the indicated concentrations (A) or 10−6 M (B) for 24 hr (A) or times ranging from 0 to 24 hr (B). Total RNA was prepared and used to analyze splicing as described in the legend to Figure 1. Values represent mean ± SD from 3 independent experiments. V, V5 empty vector; S’, V5-Acinus-S’; E, ethanol; R, retinoic acid. * p < 0.05, ** p < 0.01, *** p < 0.001, unpaired t test.
The Cooperation of RA with Acinus in Promoting Splicing of RARE-tubg1E8-E9 Pre-mRNA is Time-dependent
In order to determine how quickly RA treatment can facilitate Acinus in promoting splicing of RARE-tubg1E8-E9 pre-mRNA, a time-course experiment was performed. 293A cells were transfected with the minigene pRARE-tubg1E8-E9 along with the expression vector DNA for RARβ and Acinus-S’. Twenty-four hr after transfection, cells were treated with ethanol or RA. RNA was isolated at 0 hr, 8 hr, 16 hr and 24 hr following RA treatment. Unexpectedly, without overexpression of Acinus, the splicing efficiency of RARE-tubg1E8-E9 pre-mRNA decreases dramatically over time both in the presence and absence of RA (Figure 3B). Interestingly, when Acinus is overexpressed, this time-dependent reduction in splicing of RARE-tubg1E8-E9 pre-mRNA is reduced by approximately 50% in the absence of RA. However, in the presence of both Acinus and RA, the splicing efficiency of RARE-tubg1E8-E9 pre-mRNA is maintained at the level observed at time 0 over the 24 hr period of time (Figure 3B). The effect of Acinus alone (RA-independent) is observed between 0 and 8 hr while the cooperation between RA and Acinus (RA-dependent) in pre-mRNA splicing is observed between 8 and 16 hr of RA treatment. The reduction of splicing efficiency of RARE-tubg1E8-E9 pre-mRNA over time without overexpression of Acinus is likely due to the insufficiency of endogenous Acinus. However, overexpression of Acinus only partly alleviates the reduction of splicing efficiency in the absence of RA. In the presence of RA, Acinus can rescue the reduction of splicing efficiency to the full extent.
The RRM Domain is Critical for the Cooperation of Acinus with RA in Activating the Splicing of RARE-tubg1E8-E9 Pre-mRNA
Acinus contains a RRM domain and a C-terminal region rich in alternating RS and RD/E dipeptides, which are typical domains involved in pre-mRNA splicing. To determine the domain necessary for Acinus- and RA-mediated pre-mRNA splicing, two mammalian DNA expression vectors encoding mutant Acinus-S’ lacking either the C-terminal RS- and RD/E-rich region or the RRM domain were constructed and named pV5-Acinus-S’ (ΔC) (V5-tagged C-terminal truncated Acinus-S’) and pV5-Acinus-S’ (ΔRRM) (V5-tagged RRM domain deletion mutant of Acinus-S’), respectively (Figure 4A).
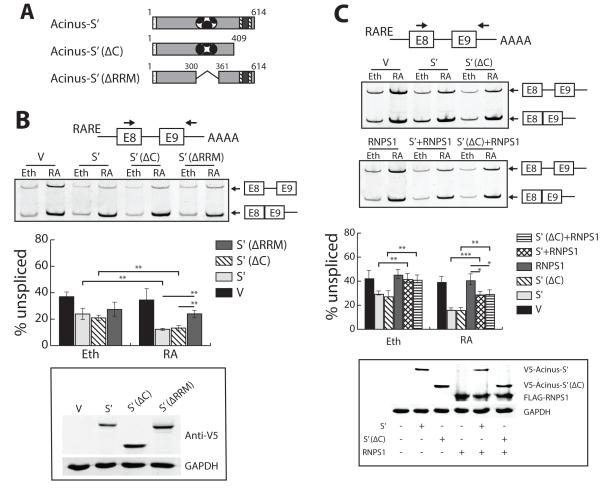
Figure 4. The synergistic effect of RA and Acinus on the splicing of a RA-responsive reporter minigene containing a weak 5′ splice site requires the RRM domain of Acinus, and is independent of RNPS1.
A. Schematic diagram of wild type and Acinus-S’ mutants. B. The RRM domain is critical for the RA-dependent activity of Acinus in the splicing of RARE-tubg1E8-E9 pre-mRNA. C. Only the RA-independent effect of Acinus on the splicing of RARE-tubg1E8-E9 pre-mRNA is repressed by RNPS1. In Panels B and C, 293A cells were transfected with RARE-tubg1E8-E9 splicing reporter DNA, RARβ expression vector DNA, and one of the following V5-tagged expression vector DNAs [wild type V5-Acinus-S’, V5-Acinus-S’ (ΔC), V5-Acinus-S’ (δRRM) or V5-empty expression vector DNA (see schematic diagram in Panel A)], and 3XFlag-tagged RNPS1 expression vector DNA (Panel C only). Twenty-four hrs after transfection cell were treated with ethanol or 10−6 M RA for an additional 24 hr. Total RNA was prepared and used to analyze splicing as described in the legend to Figure 1 and whole cell protein extracts were analyzed by Western blot using V-5 antibody to detect wild type and mutant Acinus-S’, Flag antibody to detect RNPS1 and GAPDH antibody (C). Values represent mean ± SD from 3 independent experiments. V, V5 empty vector; S’, V5-Acinus-S’; S’(δC), V5-Acinus-S’ (ΔC); S’(ΔRRM), V-Acinus-S’ (ΔRRM); Eth, ethanol; RA, retinoic acid. * p < 0.05, ** p < 0.01, unpaired t test.
To test the ability of these mutant Acinus-S’ proteins to regulate the splicing of RARE-tubg1E8-E9 pre-mRNA compared to wild type Acinus-S’, 293A cells were transfected with expression vector DNA for wild type or each mutant Acinus-S’ along with the minigene pRARE-tubg1E8-E9 DNA and the expression vector DNA for RARβ. Twenty-four hr after transfection, cells were treated with ethanol or RA for 24 hr. The expression level of each of these mutant Acinus-S’ proteins in 293A cells is similar (Figure 4B). RT-PCR analysis demonstrated that Acinus-S’ (ΔC) has a similar activity to that of wild type Acinus-S’ in activating the splicing of RARE-tubg1E8-E9 pre-mRNA both in the absence and presence of RA (Figure 4B). Acinus-S’ (ΔRRM), in the absence of RA, had a similar activity to that of wild type Acinus-S’ in activating the splicing of RARE-tubg1E8-E9 pre-mRNA. However, in the presence of RA, deletion of the RRM domain significantly reduced the activity of Acinus-S’ in pre-mRNA splicing of this minigene to a level comparable to that of the ethanol treated sample (Figure 4B). These data demonstrate that the RRM domain of Acinus is critical for the RA-dependent effect of Acinus on the splicing of RARE-tubg1E8-E9 pre-mRNA. However, other region(s) besides the RRM domain alone or the C-terminal RS- and RD/E-rich region alone are responsible for the RA-independent effect of Acinus on the splicing of RARE-tugb1E8-E9 pre-mRNA.
Activity of Acinus in Pre-mRNA Splicing is Repressed by RNPS1
Acinus through its C-terminal region interacts with RNPS1 and SAP18 to form the ASAP complex, which is part of the EJC (Schwerk et al., 2003; Tange et al., 2005). In addition, the sub-nuclear localization of RNPS1 is affected by Acinus (Wang et al., 2014). RNPS1 is a SR-related protein and functions in regulating constitutive and alternative splicing. To address whether Acinus’s activity in pre-mRNA splicing is regulated by its binding partner RNPS1, the splicing of RARE-tubg1E8-E9 pre-mRNA was examined with overexpression of both RNPS1 and wild type Acinus-S’ or C-terminal truncated Acinus-S’. Coexpression of 3xFLAG-tagged RNPS1 and either V5-tagged wild type Acinus-S’ or C-terminal truncated Acinus-S’ in 293A cells was confirmed by western blot (Figure 4C). Splicing of RARE-tubg1E8-E9 pre-mRNA was examined by RT-PCR. As shown in Figure 4C, RNPS1 alone has no effect on the splicing of RARE-tubg1E8-E9 pre-mRNA either in the presence or absence of RA. When RNPS1 is coexpressed with either wild type or C-terminal truncated Acinus, the stimulatory effect of Acinus on splicing of RARE-tubg1E8-E9 pre-mRNA is completely repressed in the absence of RA and partly repressed in the presence of RA. Thus, RNPS1 expression appears to abolish the RA-independent effect of Acinus but not the RA-dependent effect of Acinus on the splicing of RARE-tubg1E8-E9 pre-mRNA. Furthermore, the repressive effect of RNPS1 on the RA-independent activity of Acinus in facilitating the splicing of RARE-tubg1E8-E9 pre-mRNA is not mediated by interaction between RNPS1and the C-terminal region of Acinus.
Acinus Stimulates the Use of the Weaker Alternative 5′ Splice Site of Endogenous RARβ in a RA-dependent Manner and Endogenous Bcl-x in a RA-independent Manner in 293A Cells
In metazoans, alternative splicing plays an important role in generating different protein isoforms that function in diverse cellular processes. Studies have shown that alternative splice sites tend to be weak and to depend on exonic splicing enhancers (ESEs) or intronic splicing enhancers (ISEs) for their recognition. SR and SR-related proteins have important roles in facilitating splice site recognition by binding to nearby ESEs and interacting with the splicing machinery. Acinus, as described above, facilitates the splicing of reporter minigene pre-mRNAs containing a weak 5′ss but not a strong 5′ss with a RA-independent activity for all minigenes examined, and both a RA-independent and RA-dependent effect for only the RA-responsive minigene. This suggests a potential role of Acinus in regulating alternative splicing of endogenous genes by promoting the recognition of the weaker alternative 5′ss.
To test if Acinus regulates the selection of alternative 5′ss, we chose to examine two endogenous genes, human RARβ and human Bcl-x, the former involved in the RA signaling pathway and the latter involved in the apoptosis pathway. Acinus has been reported to be involved in both of these two pathways (Chan et al., 2007; Hu et al., 2007; Hu et al., 2005; Joselin et al., 2006; Sahara et al., 1999; Vucetic et al., 2008). Both of these genes have isoforms generated by alternative usage of 5′ss. RARβ, a RAR isotype, regulates transcription of RA responsive genes critical for embryonic development, cell growth and differentiation. Two isoforms of human RARβ, RARβ2 and RARβ4, are generated from the RARE-containing P2 promoter by alternative 5′ ss selection (Figure 5A) (Nagpal et al., 1992; Shen et al., 1991; Sommer et al., 1999). RARβ2 is the most abundant isoform and the major RA-inducible isoform, and RARβ4 is the minor RA-inducible isoform in the cell. It has been suggested that RARβ2 functions as a tumor suppressor that limits cell growth by regulating gene expression, while RARβ4 functions as a dominant-negative repressor of RARβ2 (Berard et al., 1994; Hayashi et al., 2003; Sommer et al., 1999). On the other hand, Bcl-x belongs to the Bcl-2 family of proteins which are key regulators of apoptosis. Alternative usage of two competing 5′ss generates the antiapoptotic Bcl-xL (usage of the relatively stronger 5′ss) and the shorter proapoptotic Bcl-xS isoform (usage of the weaker 5′ss) (Figure 5B) (Zhou et al., 2008). Bcl-xL is highly expressed in cancer tissues, and its overexpression confers resistance to apoptotic stimuli and favors metastasis (Boise and Thompson, 1997; Du et al., 2007; Olopade et al., 1997). In contrast, Bcl-xS can induce apoptosis and alleviate multidrug resistance (Clarke et al., 1995; Mercatante et al., 2002).
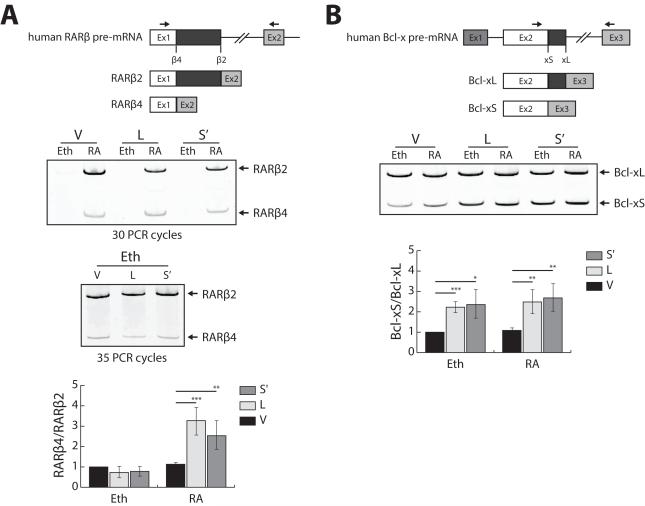
Figure 5. Acinus stimulates the use of the weaker alternative 5′ splice site of endogenous RARβ in a RA-dependent manner (A) and endogenous Bcl-x in a RA-independent manner.
293A cells were transfected with V5-Acinus-L, V-5Acinus-S’ or V5 empty expression vector DNA. Twenty-four hr after transfection, cells were treated with either ethanol or 10−6 M RA. After an additional 24 hr, RNA was isolated and RT-PCR was performed using primers that detect both spliced forms as indicated by the arrows in the schematic diagram of RARβ (A) or Bcl-x (B). The products were separated by gel electrophoresis, stained with SYTO 60 and quantitated using LI-COR Odyssey Infrared Imaging System. Note that for RARβ in Panel A, visualization of the PCR products in the ethanol treated samples required 35 PCR cycles while in the RA treated samples 30 PCR cycles were sufficient. The isoform ratio of the empty vector control treated with ethanol was set to 1. Values represent mean ± SD from 3 independent experiments. V, V5 empty vector; L, V5-Acinus L; S’ V5-Acinus-S’; Eth, ethanol; RA, retinoic acid. * p < 0.05, ** p < 0.01, *** p < 0.001, unpaired t test.
To investigate if Acinus stimulates the use of the weaker alternative 5′ss of endogenous RARβ and Bcl-x, in vivo splicing assays were performed in 293A cells with transient overexpression of Acinus-L or S’. Transfected cells were treated with ethanol or RA for 24 hr. RT-PCR was performed to analyze the relative abundance of alternative splice isoforms.
Overexpression of Acinus-L or Acinus-S’ significantly shifts the ratio of endogenous RARβ4 to RARβ2 in favor of RARβ4 only in the presence of RA, but has no significant effect in the absence of RA (Figure 5A). Interestingly, the total mRNA level of RARβ2/β4 is reduced with overexpression of Acinus (with the effect of Acinus-S’ slightly stronger than that of Acinus-L) in the presence of RA (Figure 5A), which is consistent with the previous results using the reporter minigene (Figures 1C and 2E) and Vucetic et al., 2008. These data indicate that RA is required for the activity of Acinus in stimulating the use of the endogenous RARβ4 5′ss, which further supports the idea that RAR-dependent pre-mRNA splicing requires the cooperation between Acinus and RA for at least some RARE containing genes.
Overexpression of Acinus-L or Acinus-S’ significantly promotes the usage of endogenous Bcl-xS 5′ss both in the absence and presence of RA to the same extent (Figure 5B). This indicates that the activity of Acinus in regulating alternative 5′ss selection of non-RAR-dependent gene pre-mRNA is independent of RA.
DISCUSSION
Acinus has been identified as a component of the human spliceosome and the EJC, suggesting its involvement in RNA processing (Rappsilber et al., 2002; Zhou et al., 2002). However Acinus was shown to have little activity in regulating pre-mRNA splicing (Schwerk et al., 2003; Singh et al., 2010), which raises the question about the function of Acinus found in spliceosomes and the EJC. In addition, whether Acinus-L and Acinus-S’ have distinct functions in regulating pre-mRNA splicing due to their distinct sub-nuclear localizations (Wang et al., 2014) is unclear. Previous studies from our laboratory have identified Acinus-S’ as a corepressor of RAR-dependent gene expression (Vucetic et al., 2008). Transcription and splicing are thought to be physically coupled (for review, see Kornblihtt et al., 2004). Several SR-related proteins have been shown to couple transcription and splicing of target genes, such as PGC-1, CAPER and Pinin (Alpatov et al., 2004; Alpatov et al., 2008; Dowhan et al., 2005; Monsalve et al., 2000). In order to determine if Acinus also functions as a cofactor that couples transcription and splicing of target genes, we explored the role of Acinus-S’ and Acinus-L in regulating pre-mRNA splicing of RAR-dependent genes using in vivo splicing assays.
Using splicing reporter minigene experiments, both Acinus-L and Acinus-S’ were found to facilitate the splicing of pre-mRNAs transcribed from a minigene derived from TUBG1 exon8-9 region which contains a weak 5′ss but not a minigene derived from TUBA1B exon2-3 region which contains a strong 5′ss. Acinus shows a general effect increasing the splicing to varying degrees depending on the promoter driving the transcription of the pre-mRNAs containing the weak 5′ss. Importantly, RA cooperates with Acinus in a dose- and time-dependent manner to further increase the splicing of the tubg1E8-E9 pre-mRNA specifically transcribed by the RARE-containing promoter but not by non-RARE containing promoters, suggesting that the cooperation of RA with Acinus in regulating pre-mRNA splicing is RARE/RAR-dependent and not due to an increase in the expression of a splicing factor by RA. Furthermore, unlike the cooperation of RA and Acinus in regulating the pre-mRNA splicing of RARE-tubg1E8-E9, no cooperation between Acinus and rosiglitazone is observed in regulating the splicing of PPRE-tubg1E8-E9 pre-mRNA. Taken together, this suggests that in addition to a general ligand-independent effect on pre-mRNA splicing, Acinus has a ligand-dependent splicing activity, which is specific to a RARE-containing promoter.
It has been suggested that splicing factors can be recruited by transcription activators, coactivators, carboxy-terminal domain (CTD) of RNA Pol II and general transcriptional factors (Ge et al., 1998; Lai et al., 1999; Monsalve et al., 2000; Rosonina et al., 2003; Yoshida et al., 1999). Specific transcription activators have been shown to recruit distinct combinations of coregulators (Hermanson et al., 2002; Li et al., 2003; Willy et al., 1995), suggesting a promoter specificity for some coregulators. Thus, the minigenes driven by different promoters recruit distinct combinations of coativators when the promoter is active and hence recruit distinct combinations of splicing factors through these coactivators. It is possible that the general activity of Acinus in regulating pre-mRNA splicing of RARE-tubg1E8-E9 and PPRE-tubg1E8-E9 in the absence of ligand (basal transcription) is due to its interaction with specific splicing factors recruited by Pol II and/or other general transcription factors. In the presence of ligand (activated transcription), it is likely that the combination of additional splicing factors recruited through promoter specific coactivators is different between the RARE promoter and the PPRE promoter This results in the enhanced activity of Acinus in regulating the splicing of RARE-tubg1E8-E9 pre-mRNA but not that of PPRE-tubg1E8-E9 pre-mRNA. The splicing factors recruited by the PPRE-specific coactivators possibly either lacks the specific splicing factors necessary for the splicing activity of Acinus or impedes the interaction of the specific splicing factors with Acinus. Hence, no cooperation of rosiglitazone with Acinus in regulating the splicing of PPRE-tubg1E8-E9 pre-mRNA is observed. The constitutively active promoters, such as CMV and Sp1RE, are more like the activated RARE promoter, to which splicing factors are recruited through both promoter-specific activators, coactivators and promoter-independent Pol II and/or general transcriptional factors. Acinus has been shown to interact with A/B domain of RARs through its C-terminal region using in vitro GST pull-down assay (Vucetic et al., 2008). Our data demonstrate that Acinus-S’ (ΔC) has the same activity as the wild type in facilitating the splicing of RARE-tubg1E8-E9. It is likely that Acinus can be recruited by members of transcription machinery or other RAR binding proteins besides RAR.
RA is observed to cooperate with Acinus in facilitating the splicing of the minigene RARE-tubg1E8-E9 pre-mRNA in a dose- and time-dependent manner. The reduction in the splicing efficiency of RARE-tubg1E8-E9 pre-mRNA over time without overexpression of Acinus is likely due to the insufficiency of Acinus since overexpression of Acinus in the absence of RA can alleviate the reduction of the splicing efficiency of this minigene pre-mRNA. The reason responsible for this insufficiency of Acinus over time is unclear but maybe due to an accumulation of minigene pre-mRNA with time of expression after transfection. The maintenance of the splicing efficiency of RARE-tubg1E8-E9 over time requires both Acinus and RA, suggesting that RA treatment increases the recruitment of coactivators with the accompanying recruitment of splicing factors necessary for the splicing activity of Acinus. This idea is also supported by the dose response experiment where the splicing activity of Acinus increases with the increasing concentrations of RA.
It has been thought that the RS domain of SR/SR-related proteins is required for their splicing activity. However, the RS domain of SRSF1has been shown to be dispensable for splicing of some but not all pre-mRNA substrates; and that the requirement for the RS domain in splicing correlates with the strength of the 3′ss (Zhu and Krainer, 2000). This raises the possibility that a strong 3′ss can be recognized by the splicing machinery without the assistance of the RS domain of SR/SR-related splicing factors. In agreement with this, the C-terminal RS- and RD/E-rich region is dispensable for the activity of Acinus in facilitating the splicing of RARE-tubg1E8-E9 pre-mRNA, which has a strong 3′ss (ESE finder 3.0, http://rulai.cshl.edu/tools/ESE) (Ahn et al., 2011; Cartegni et et al., 2003). Therefore it is possible that the splicing of other pre-mRNAs that have a weak 5′ss and a weak 3′ss will require the C-terminal RS- and RD/E-rich region of Acinus.
The RRM domain has been identified as a region required for the RA-dependent activity but not RA-independent activity of Acinus in pre-mRNA splicing. The RRM domain was first identified as an RNA-binding motif, but it has been shown to interact with proteins as well. The interaction between the RRMs of SRSF1 and U1-70K promotes the formation of the spliceosome by recruiting the U1 snRNP to the 5′ss (Cho et al., 2011). The activity of PTB (a regulatory splicing repressor) is modulated by interacting through its RRM2 domain with its corepressor Raver1 (Rideau et al., 2006). Since Acinus-S’ (ΔRRM) retains its RA-independent activity and loses RA-dependent activity, it is likely that the RRM domain of Acinus interacts with specific splicing factors recruited via coactivators upon RA treatment to modulate the splicing activity of Acinus.
Acinus through its C-terminal region interacts with RNPS1 and SAP18 to form a ternary complex termed ASAP (Schwerk et al., 2003). RNPS1 is a splicing cofactor functioning in both constitutive and alternative splicing (Mayeda et al., 1999; Sakashita et al., 2004). We found that unlike Acinus, RNPS1 has no activity in regulating the splicing of RARE-tubg1E8-E9 pre-mRNA, suggesting that RNPS1 might regulate the recognition of different types of splice sites from those of Acinus. In addition, RNPS1 abolishes the RA-independent activity of both Acinus and Acinus (ΔC) but not the RA-dependent activity in facilitating the splicing of RARE-tubg1E8-E9 pre-mRNA. Because RNPS1 also represses the splicing activity of Acinus (δC), which presumably lacks the ability to bind RNPS1, the repression is not likely due to their interaction, instead is more likely due to their competition for specific splicing factors, such as SR proteins. It is possible that in the absence of RA, RNPS1 competes with Acinus for the limited splicing factors recruited by Pol II and general transcriptional factors, with RNPS1 more potent than Acinus, which abolishes the splicing activity of Acinus. In the presence of RA, splicing factors are recruited not only by members of the transcription machinery but also by coactivators. Therefore, the competition between RNPS1and Acinus is alleviated and Acinus displays partial splicing activity due to its RA-dependent activity. This repressive effect of RNPS1 on the splicing activity of Acinus is reminiscent of the repressive effect of the ASAP complex on the splicing activity of RNPS1 (Schwerk et al., 2003). Interestingly, SAP18 has been shown to regulate pre-mRNA splicing with an activity distinct from RNPS1 (Singh et al., 2010). These observations suggest that the three components of the ASAP complex have distinct splicing regulatory activities.
Notably, although both Acinus-L and Acinus-S’ function in facilitating the splicing of the minigene RARE-tubg1E8-E9, the activity of Acinus-L in pre-mRNA splicing is higher than that of Acinus-S’ based on their relative expression levels. It is possible that the unique N-terminal region of Acinus-L is responsible for its higher splicing activity either because Acinus-L is more available than Acinus-S’ to the transcription site of the minigene due to its diffuse localization in the nucleoplasm (Wang et al., 2014) or the large N-terminal unique region of Acinus-L associates with additional splicing factors conferring Acinus-L higher splicing activity.
Our studies further demonstrate that Acinus isoforms (Acinus-L and Acinus-S’) regulate alternative splicing of endogenous RA induced RARβ2/β4 (from the RARE-containing P2 promoter) and endogenous Bcl-x (from a non-RARE-containing promoter) in favor of the isoform generated from a weaker 5′ss. In the absence of RA, Acinus has no effect on the ratio of RARβ4 mRNA to RARβ2 mRNA; while in the presence of RA, Acinus strongly increases the ratio of RARβ4 mRNA to RARβ2 mRNA. The reason that, unlike the minigene driven by a RARE-containing promoter, the regulation of endogenous RARβ alternative splicing by Acinus is completely RA-dependent is probably due to the chromatin structure of endogenous genes. Unlike the transiently transfected minigene DNAs which are naked in the cells, endogenous genes are packaged into nucleosomes. In the absence of RA, the unliganded RAR/RXR heterodimer associated with the RARE of endogenous genes is bound by corepressor complexes, which leads to a compacted chromatin. As described above, splicing factors could be recruited by transcriptional activators, coactivators as well as RNA Pol II and other members of transcription pre-initiation complex. It is likely that the corepressor complexes bound to the RAR/RXR heterodimer and the compact chromatin structure of RARβ not only strongly impairs the recruitment of the transcription machinery but also splicing factors. In agreement with this, it has been shown that no SR splicing factors were detected at the promoter of the uninduced FOS gene (Sapra et al., 2009). Thus, the lack of the activity of Acinus in regulating alternative splicing of endogenous RARβ2/β4 in the absence of RA is likely due to the lack of specific splicing factors (necessary for the splicing activity of Acinus) and/or the inability to recruit Acinus directly to the promoter of RARβ2/β4. On the other hand, consistent with the minigenes driven by non-RARE promoters, Acinus significantly increases the ratio of Bcl-xS to Bcl-xL independent of RA. These findings indicate that the regulatory effect of Acinus on pre-mRNA splicing of endogenous RAR-regulated genes is strictly controlled by RA, while for endogenous non-RAR-regulated genes, RA has no effect on the splicing activity of Acinus. This suggests that RA in conjunction with Acinus not only controls transcription activation but also pre-mRNA splicing of specific endogenous RAR-regulated genes such as RARβ2/β4.
The SR-related proteins involved in coupling transcription and splicing of target genes, such as PGC-1, CAPER and Pinin, function in a promoter-dependent manner (Alpatov et al., 2004; Alpatov et al., 2008; Dowhan et al., 2005; Monsalve et al., 2000). It is most likely that these bifunctional coregulators bridge transcription and splicing by associating with both transcriptional activators, coactivators or Pol II and splicing factors in a promoter-dependent manner. Interestingly, like Acinus, Pinin is also a component of the EJC (Li et al., 2003) and interacts with both RNPS1 and SAP18 to form a complex termed PSAP (Murachelli et al., 2012). In addition, opposite to that of Acinus, Pinin regulates alternative splicing of Bcl-x in favor of Bcl-xL (Leu et al., 2012). These findings suggest that Acinus might have a similar role in coupling transcription and splicing in a promoter-specific manner.
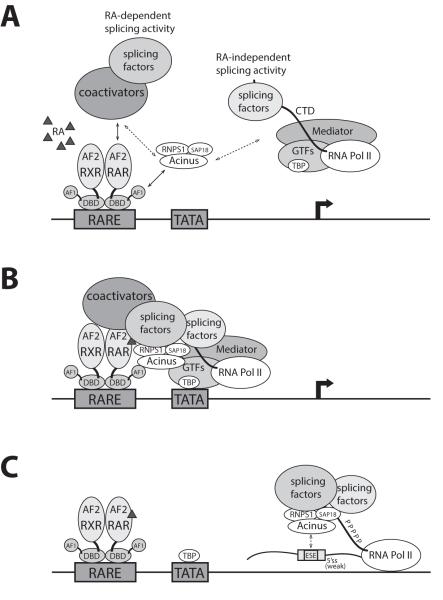
A proposed model for the mechanism of Acinus in coupling transcription and splicing of RAR-dependent gene containing a weak 5′ss is illustrated in Figure 6. It is likely that Acinus together with RNPS1 and SAP18 are recruited to RARE-containing promoters. This may occur through interaction with RAR, RAR-binding proteins and/or the general transcription machinery which includes RNA Pol II associated with splicing factors responsible for the RA-independent activity of Acinus, general transcription factors and mediator. In the presence of RA, the recruitment of coactivators brings more splicing factors required for the RA-dependent splicing activity of Acinus. After transcription initiation, the polymerase transcribes away from the promoter with a heavily phosphorylated CTD and dissociates from the other general transcription factors. The elongating Pol II is associated with a new set of proteins including Acinus and other splicing factors recruited both by Pol II and by coactivators. Acinus travels with Pol II elongation complex to facilitate the recognition of weak 5′ss of pre-mRNA by the splicing machinery. Acinus might directly binds the ESE near the weak 5′ss or indirectly associate with the ESE through interactions with other SR/SR-related splicing factors.
Figure 6. Proposed model for the coupling of RA-dependent transcription and splicing by Acinus.
(A) Recruitment of coactivators and the accompanying recruitment of splicing factors responsible for RA-dependent splicing activity of Acinus in the presence of RA to RARE containing promoter. (B) The pre-initiation complex formed at the RARE-containing promoter. (C) Acinus travels with the elongation complex to facilitate the recognition of weak 5′ss. Solid arrow, direct protein-protein interactions; Dashed arrow, direct or indirect interactions.
ACKNOWLEDGEMENTS
We thank Mr. Zhenping Zhang and Ms. Dorret Lynch for their expert technical assistance. This work was supported by a grant to D.R.S. from the National Institutes of Health (DK067558).
Contract Grant Sponsor: National Institutes of Health
Contract Grant Number: DK067558
REFERENCES
- Ahn EY, DeKelver RC, Lo MC, Nguyen TA, Matsuura S, Boyapati A, Pandit S, Fu XD, Zhang DE. SON controls cell-cycle progression by coordinated regulation of RNA splicing. Mol. Cell. 2011;42:185–198. doi: 10.1016/j.molcel.2011.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpatov R, Munguba GC, Caton P, Joo JH, Shi Y, Hunt ME, Sugrue SP. Nuclear speckle-associated protein Pnn/DRS binds to the transcriptional corepressor CtBP and relieves CtBP-mediated repression of the E-cadherin gene. Mol. Cell. Biol. 2004;24:10223–10235. doi: 10.1128/MCB.24.23.10223-10235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpatov R, Shi Y, Munguba GC, Moghimi B, Joo JH, Bungert J, Sugrue SP. Corepressor CtBP and nuclear speckle protein Pnn/DRS differentially modulate transcription and splicing of the E-cadherin gene. Mol. Cell. Biol. 2008;28:1584–1595. doi: 10.1128/MCB.00421-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berard J, Gaboury L, Landers M, De Repentigny Y, Houle B, Kothary R, Bradley WE. Hyperplasia and tumours in lung, breast and other tissues in mice carrying a RAR beta4-like transgene. EMBO J. 1994;13:5570–5580. doi: 10.1002/j.1460-2075.1994.tb06894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boise LH, Thompson CB. Bcl-x(L) can inhibit apoptosis in cells that have undergone Fas-induced protease activation. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3759–3764. doi: 10.1073/pnas.94.8.3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR. ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 2003;31:3568–3571. doi: 10.1093/nar/gkg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- Chan CB, Liu X, Tang X, Fu H, Ye K. Akt phosphorylation of zyxin mediates its interaction with acinus-S and prevents acinus-triggered chromatin condensation. Cell Death Differ. 2007;14:1688–1699. doi: 10.1038/sj.cdd.4402179. [DOI] [PubMed] [Google Scholar]
- Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. U. S. A. 2011;108:8233–8238. doi: 10.1073/pnas.1017700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MF, Apel IJ, Benedict MA, Eipers PG, Sumantran V, Gonzalez-Garcia M, Doedens M, Fukunaga N, Davidson B, Dick JE, Minn AJ, Boise LH, Thompson CB, Wicha M, Nunez G. A recombinant bcl-x s adenovirus selectively induces apoptosis in cancer cells but not in normal bone marrow cells. Proc. Natl. Acad. Sci. U. S. A. 1995;92:11024–11028. doi: 10.1073/pnas.92.24.11024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O’Malley BW. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol. Cell. 2005;17:429–439. doi: 10.1016/j.molcel.2004.12.025. [DOI] [PubMed] [Google Scholar]
- Du YC, Lewis BC, Hanahan D, Varmus H. Assessing tumor progression factors by somatic gene transfer into a mouse model: Bcl-xL promotes islet tumor cell invasion. PLoS Biol. 2007;5:e276. doi: 10.1371/journal.pbio.0050276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R, Mangelsdorf D. Nuclear receptors, RXR and the big bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Si Y, Wolffe AP. A novel transcriptional coactivator, p52, functionally interacts with the essential splicing factor ASF/SF2. Mol. Cell. 1998;2:751–759. doi: 10.1016/s1097-2765(00)80290-7. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Goodison S, Urquidi V, Tarin D, Lotan D, Lotan R, Tahara E. Differential effects of retinoic acid on the growth of isogenic metastatic and non-metastatic breast cancer cell lines and their association with distinct expression of retinoic acid receptor beta isoforms 2 and 4. Int. J. Oncol. 2003;22:623–629. [PubMed] [Google Scholar]
- Hermanson O, Glass CK, Rosenfeld MG. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol Metab. 2002;13:55–60. doi: 10.1016/s1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- Hu Y, Liu Z, Yang SJ, Ye K. Acinus-provoked protein kinase C delta isoform activation is essential for apoptotic chromatin condensation. Cell Death Differ. 2007;14:2035–2046. doi: 10.1038/sj.cdd.4402214. [DOI] [PubMed] [Google Scholar]
- Hu Y, Yao J, Liu Z, Liu X, Fu H, Ye K. Akt phosphorylates acinus and inhibits its proteolytic cleavage, preventing chromatin condensation. EMBO J. 2005;24:3543–3554. doi: 10.1038/sj.emboj.7600823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joselin AP, Schulze-Osthoff K, Schwerk C. Loss of Acinus inhibits oligonucleosomal DNA fragmentation but not chromatin condensation during apoptosis. J. Biol. Chem. 2006;281:12475–12484. doi: 10.1074/jbc.M509859200. [DOI] [PubMed] [Google Scholar]
- Jung DJ, Na SY, Na DS, Lee JW. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J. Biol. Chem. 2002;277:1229–1234. doi: 10.1074/jbc.M110417200. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Umesono K, Noonan DJ, Heyman RA, Evans RM. Convergence of 9-cis retinoic acid and peroxisome proliferator signalling pathways through heterodimer formation of their receptors. Nature. 1992;358:771–774. doi: 10.1038/358771a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Teh BH, Tarn WY. A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J. Biol. Chem. 1999;274:11832–11841. doi: 10.1074/jbc.274.17.11832. [DOI] [PubMed] [Google Scholar]
- Lavery DN, McEwan IJ. Structure and function of steroid receptor AF1 transactivation domains: induction of active conformations. Biochem. J. 2005;391:449–464. doi: 10.1042/BJ20050872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leu S, Lin YM, Wu CH, Ouyang P. Loss of Pnn expression results in mouse early embryonic lethality and cellular apoptosis through SRSF1-mediated alternative expression of Bcl-xS and ICAD. J. Cell Sci. 2012;125:3164–3172. doi: 10.1242/jcs.100859. [DOI] [PubMed] [Google Scholar]
- Li X, Wong J, Tsai SY, Tsai MJ, O’Malley BW. Progesterone and glucocorticoid receptors recruit distinct coactivator complexes and promote distinct patterns of local chromatin modification. Mol. Cell. Biol. 2003;23:3763–3773. doi: 10.1128/MCB.23.11.3763-3773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda A, Badolato J, Kobayashi R, Zhang MQ, Gardiner EM, Krainer AR. Purification and characterization of human RNPS1: a general activator of pre-mRNA splicing. EMBO J. 1999;18:4560–4570. doi: 10.1093/emboj/18.16.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatante DR, Mohler JL, Kole R. Cellular response to an antisense-mediated shift of Bcl-x pre-mRNA splicing and antineoplastic agents. J. Biol. Chem. 2002;277:49374–49382. doi: 10.1074/jbc.M209236200. [DOI] [PubMed] [Google Scholar]
- Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- Murachelli AG, Ebert J, Basquin C, Le Hir H, Conti E. The structure of the ASAP core complex reveals the existence of a Pinin-containing PSAP complex. Nat Struct Mol Biol. 2012;19:378–386. doi: 10.1038/nsmb.2242. [DOI] [PubMed] [Google Scholar]
- Nagpal S, Zelent A, Chambon P. RAR-beta 4, a retinoic acid receptor isoform is generated from RAR-beta 2 by alternative splicing and usage of a CUG initiator codon. Proc. Natl. Acad. Sci. U. S. A. 1992;89:2718–2722. doi: 10.1073/pnas.89.7.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olopade OI, Adeyanju MO, Safa AR, Hagos F, Mick R, Thompson CB, Recant WM. Overexpression of BCL-x protein in primary breast cancer is associated with high tumor grade and nodal metastases. Cancer J. Sci. Am. 1997;3:230–237. [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideau AP, Gooding C, Simpson PJ, Monie TP, Lorenz M, Huttelmaier S, Singer RH, Matthews S, Curry S, Smith CW. A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nat Struct Mol Biol. 2006;13:839–848. doi: 10.1038/nsmb1137. [DOI] [PubMed] [Google Scholar]
- Rosonina E, Bakowski MA, McCracken S, Blencowe BJ. Transcriptional activators control splicing and 3′-end cleavage levels. J. Biol. Chem. 2003;278:43034–43040. doi: 10.1074/jbc.M307289200. [DOI] [PubMed] [Google Scholar]
- Sahara S, Aoto M, Eguchi MY, Imamoto N, Yoneda Y, Tsujimoto Y. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature. 1999;401:168–173. doi: 10.1038/43678. [DOI] [PubMed] [Google Scholar]
- Saitoh N, Spahr CS, Patterson SD, Bubulya P, Neuwald AF, Spector DL. Proteomic analysis of interchromatin granule clusters. Mol. Biol. Cell. 2004;15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakashita E, Tatsumi S, Werner D, Endo H, Mayeda A. Human RNPS1 and its associated factors: a versatile alternative pre-mRNA splicing regulator in vivo. Mol. Cell. Biol. 2004;24:1174–1187. doi: 10.1128/MCB.24.3.1174-1187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra AK, Anko ML, Grishina I, Lorenz M, Pabis M, Poser I, Rollins J, Weiland EM, Neugebauer KM. SR protein family members display diverse activities in the formation of nascent and mature mRNPs in vivo. Mol. Cell. 2009;34:179–190. doi: 10.1016/j.molcel.2009.02.031. [DOI] [PubMed] [Google Scholar]
- Schwerk C, Prasad J, Degenhardt K, Erdjument-Bromage H, White E, Tempst P, Kidd VJ, Manley JL, Lahti JM, Reinberg D. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol. Cell. Biol. 2003;23:2981–2990. doi: 10.1128/MCB.23.8.2981-2990.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Kruyt FA, den Hertog J, van der Saag PT, Kruijer W. Mouse and human retinoic acid receptor beta 2 promoters: sequence comparison and localization of retinoic acid responsiveness. DNA Seq. 1991;2:111–119. doi: 10.3109/10425179109039679. [DOI] [PubMed] [Google Scholar]
- Singh KK, Erkelenz S, Rattay S, Dehof AK, Hildebrandt A, Schulze-Osthoff K, Schaal H, Schwerk C. Human SAP18 mediates assembly of a splicing regulatory multiprotein complex via its ubiquitin-like fold. RNA. 2010;16:2442–2454. doi: 10.1261/rna.2304410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer KM, Chen LI, Treuting PM, Smith LT, Swisshelm K. Elevated retinoic acid receptor beta(4) protein in human breast tumor cells with nuclear and cytoplasmic localization. Proc. Natl. Acad. Sci. U. S. A. 1999;96:8651–8656. doi: 10.1073/pnas.96.15.8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano DR, Scanlon E, Shukri M, Zhang ZP, Soprano KJ. Murine RARbeta4 displays reduced transactivation activity, lower affinity for retinoic acid, and no anti-AP1 activity. J.Cell. Biochem. 2000;77:604–614. [PubMed] [Google Scholar]
- Tange TO, Shibuya T, Jurica MS, Moore MJ. Biochemical analysis of the EJC reveals two new factors and a stable tetrameric protein core. RNA. 2005;11:1869–1883. doi: 10.1261/rna.2155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucetic Z, Zhang Z, Zhao J, Wang F, Soprano KJ, Soprano DR. Acinus-S’ represses retinoic acid receptor (RAR)-regulated gene expression through interaction with the B domains of RARs. Mol. Cell. Biol. 2008;28:2549–2558. doi: 10.1128/MCB.01199-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Wendling KS, Soprano KJ, Soprano DR. Role of the SAP Motif and the C-terminal RS- and RD/E-rich region in the sub-nuclear localization of Acinus isoforms. J. Cell. Biochem. 2014 doi: 10.1002/jcb.24893. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Makino Y, Tamura T. Association of the rat heterogeneous nuclear RNA-ribonucleoprotein F with TATA-binding protein. FEBS Lett. 1999;457:251–254. doi: 10.1016/s0014-5793(99)01048-0. [DOI] [PubMed] [Google Scholar]
- Zhao J, Zhang Z, Vucetic Z, Soprano KJ, Soprano DR. HACE1: A novel repressor of RAR transcriptional activity. J. Cell. Biochem. 2009;107:482–493. doi: 10.1002/jcb.22146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Ou AC, Cho A, Benz EJ, Jr, Huang SC. Novel splicing factor RBM25 modulates Bcl-x pre-mRNA 5′ splice site selection. Mol. Cell. Biol. 2008;28:5924–5936. doi: 10.1128/MCB.00560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- Zhu J, Kraine AR. Pre-mRNA splicing in the absence of an SR protein RS domain. Genes Dev. 2000;14:3166–3178. doi: 10.1101/gad.189500. [DOI] [PMC free article] [PubMed] [Google Scholar]