Abstract
Isovaleryl-CoA dehydrogenase (IVD) catalyzes the conversion of isovaleryl-CoA to 3-methylcrotonyl-CoA and the transfer of electrons to the electron transfer flavoprotein (ETF). Recombinant human IVD purifies with bound CoA-persulfide. A modified purification protocol was developed to isolate IVD without bound CoA-persulfide and to protect the protein thiols from oxidation. The CoA-persulfide-free IVD specific activity was 112.5 µmol porcine ETF•min−1•mg−1, which was ~20-fold higher than that of its CoA-persulfide bound form. The Km and catalytic efficiency (kcat/Km) for isovaleryl-CoA were 1.0 µM and 4.3 × 106•M−1•sec−1 per monomer, respectively, and its Km for ETF was 2.0 µM. Anaerobic titration of isovaleryl-CoA into an IVD solution resulted in a stable blue complex with increased absorbance at 310 nm, decreased absorbance at 373 and 447 nm, and the appearance of the charge transfer complex band at 584 nm. The apparent dissociation constant (KD app) determined spectrally for isovaleryl-CoA was 0.54 µM. Isovaleryl-CoA, acetoacetyl-CoA, methylenecyclopropylacetyl-CoA, and ETF induced CD spectral changes at the 250–500 nm region while isobutyryl-CoA did not, suggesting conformational changes occur at the flavin ring that are ligand specific. Replacement of the IVD Trp166 with a Phe did not block IVD interaction with ETF, indicating that its indole ring is not essential for electron transfer to ETF. A twelve amino acid synthetic peptide that matches the sequence of the ETF docking peptide competitively inhibited the enzyme reaction when ETF was used as the electron acceptor with a Ki of 1.5 mM.
Keywords: acyl-CoA dehydrogenase, electron-transferring flavoprotein
Isovaleryl-CoA dehydrogenase (IVD;1 EC 1.3.99.10) is an intramitochondrial homotetrameric flavoenzyme in the leucine catabolism pathway that catalyzes the α,β-dehydrogenation of isovaleryl-CoA and transfers the reducing equivalents to the electron transfer flavoprotein (ETF). Deficiency of IVD activity in humans causes the inborn error of metabolism isovaleric acidemia [1, 2]. IVD shares amino acid sequence homology and properties with the growing family of the acyl-CoA dehydrogenases (ACADs). This gene family includes nine members of known function that can be divided into three functional groups. Five of the ACADs are involved in β-oxidation of fatty acids with various chain lengths, three function in branched acyl-CoA amino acid catabolism, and one plays a dual role in lysine metabolism, catalyzing both the α,β-dehydrogenation and decarboxylation of its substrate [3–10]. In addition to these mitochondrial proteins, two predicted large proteins, ACAD10 and ACAD11 have recently been identified in genome databases having “structural fingerprints” of ACADs only at their respective C-terminus part [11].
The reaction catalyzed by these enzymes is initiated upon acyl-CoA substrate binding. In the ensuing reductive half-reaction, while formation of a hydrogen bond between the thioester carbonyl oxygen and the 2′–hydroxyl hydrogen of the FAD ribityl moiety is crucial for activation of the acyl moiety [12–16], a substrate induced strengthening of the hydrogen bond between the flavin C-4 oxygen and the hydroxyl group of a Ser or Thr residue play an important role in activating the flavin [17]. The Cα and Cβ hydrogens of the acyl moiety are abstracted in concert as a proton and a hydride, respectively, forming a stable charge-transfer complex (CTC). The electron transferring flavoprotein (ETF) accepts reducing equivalents from the CTC in the oxidative half-reaction, during which electrons are transferred to the latter and the enoyl-CoA product is released. Mechanistic studies have also concluded that ACADs follow a common ordered Bi Bi type kinetic mechanism [18–22]. The co-crystallization of human medium chain acyl-CoA dehydrogenase (MCAD) with human ETF, has provided additional evidence for the ability of ETF to bind its ACAD partners in the absence of their acyl-CoA substrates [23]. Important insight into the molecular mechanism of electron transfer in the oxidative half-reaction has also been gained with the crystallization of MCAD and the ETF βE165A complex [24].
Many of the genetic defects causing deficiency in IVD activity are point mutations leading to an amino acid substitution in the mutant protein [2, 23]. Some of these have been shown to cause only a mild alteration in enzyme function, correlating to some extent with a mild clinical phenotype in the patient [25, 26]. This correlation has been imperfect, however, and a more thorough understanding of the functional effects of IVD mutants at the enzymatic level requires a well-characterized set of benchmarks for the wild type enzyme. Previous studies of the biophysical and biochemical properties of recombinant IVD and other ACADs have been hampered by two major impediments: difficulty removing tightly bound CoA persulfide and a presumed in vitro formation of a disulfide bond between the solvent accessible γ-thiol groups of C318 and C323 located at the loop connecting α-Helices H and I, which is involved in substrate binding [27]. In this study, a modified protocol was used for purifying recombinant IVD leading to significantly higher specific activity. Kinetic and spectral properties of the purified recombinant human IVD and its interaction with various ligands are presented.
EXPERIMENTAL
Purification of recombinant human ACADs
To isolate IVD, cell growth conditions and purification protocols followed earlier published reports with modifications [28]. E. coli JM105 cells (Amersham Biosciences Corp; Piscataway, NJ) containing the human IVD high expression vector pKKmHIVD [26], and additionally a GroEL/ES expression plasmid with chloramphenicol resistance, were grown overnight in a 200-ml LB broth pre-culture and were used to inoculate 6×1-L cultures in LB broth. (Using richer media, e.g. 2YT supplemented with Mg and phosphate, doubled total cell production, but resulted in poorer IVD production.) The cells were left to grow in the large culture for about 7 hours and IVD expression was induced using IPTG at a final concentration of 0.5 mM. The cultures were then incubated for an additional 20 hrs at 30°C. Cells were harvested by centrifugation and resuspended at 4°C in 2:1 weight to volume of 100 mM Tris, pH 8.0 containing 500 U of DNase. The cells were ruptured using sonication, and EDTA was added to a final concentration of 50 mM. Cellular debris was removed by centrifugation first at 28,000 × g, then 250,000 × g for 60 minutes each. The final supernatant was dialyzed for 4 hours with vigorous stirring in 50 mM Tris pH 8.0, at 4°C. The sample was then loaded on a 16×40 mm DEAE Sepharose FF column pre-equilibrated in 50 mM Tris, pH 8.0 using an ÄKTA FPLC system (Amersham Biosciences Corp; Piscataway, NJ). After washing with 300 ml of 10 mM Tris, pH 8.0, containing 80 mM NaCl, IVD was eluted with a 320 ml linear gradient from 80 to 400 mM NaCl in 10 mM Tris, pH 8.0. The light green fractions containing IVD with a 270/434 nm ratio <12 were pooled and treated with sodium dithionite to remove the bound CoA persulfide (“de-greening”) as below.
To de-green the sample, one molar Tris buffer, pH 8.0 at room temperature, was added to the pooled IVD sample from the anion exchange step to give a final concentration of 200 mM. The sample was degassed and layered with argon using ~10 alternating cycles of vacuum and oxygen-free argon. A pre-weighed amount of recently purchased sodium dithionite, stored away from light and under nitrogen, calculated to give a final concentration of 20 mM was added and dissolved into the partially purified IVD solution. The sample was left under argon for an hour at room temperature, then poured quickly into a dialysis bag and dialyzed under argon for 4 hours in 50 mM Tris, pH 8.5, 10 mM sodium dithionite, at 4°C. The colorless sample was injected onto a 20 µm ceramic hydroxyapatite column (1.5×20 mm) pre-equilibrated with an anaerobic solution of 50 mM Tris buffer, pH 8.0, 10 mM sodium dithionite, and washed with the same buffer at a rate of 1.0 ml/min. Bound proteins were eluted as described earlier [26]. Fractions with 270/434 nm ratio < 5.4 were combined and concentrated. EDTA was added to the sample for a final concentration of 50 mM, frozen with liquid nitrogen, and stored at –80°C. To produce IVD with bound CoA persulfide, the de-greening step was eliminated and the sample containing salt from the anion exchange was directly loaded onto the ceramic hydroxyapatite column. Fractions with a 270/434 nm ratio < 6.5 were combined, concentrated, EDTA added, and stored as above.
Human cDNA sequence coding for the mature form of isobutyryl-CoA dehydrogenase (IBD) cloned into pET21 expression vector, pEThIBD [29], was introduced into an E. coli strain BL21 containing and expressing in addition GroES/EL chaperonins. IBD was purified using a modification of the published protocols [29,30]. The cells producing IBD, following IPTG induction for 3 hrs, was subjected to sonication in 100 mM Tris buffer containing 150 mM EDTA, pH 8. The cell-free extract was subjected to ammonium sulfate precipitation and then dialyzed using two 50 mM phosphate buffer, pH 8. The sample was then loaded on a DEAE-Sepharose and washed with 50 ml of deaerated and argonpurged 50 mM phosphate buffer, pH 8 and 10 mM sodium dithionite. IBD was eluted with a linear phosphate gradient up to 500 mM phosphate, pH 8 and 2 mM dithiothreitol (DTT). Fractions with lowest A270 nm/A448 nm were pooled and dialyzed for 4 hrs. The pooled fractions were re-loaded on the same column and eluted with the same gradient but without the DTT. Fractions with lowest A270 nm/A448 nm were pooled, dialyzed for 4 hrs, 50 mM EDTA included, and stored at −80°C. To produce IBD and SCAD with bound CoA-persulfide, a culture was induced overnight with 0.5 mM IPTG and IBD was purified as above without the dithionite. Recombinant human short/branched chain acyl-CoA dehydrogenase (SBCAD) was purified as previously published [31].
Replacement of the IVD Trp166 with a Phe
Site directed mutagenesis to replace the Trp at position 166 with a Phe was carried out using the QuickChange from Aligent Technologies, Santa Clara, CA, and complimentary primers containing the desired codon modification. The altered plasmid, pKKmhIVD_W166F, was introduced into the E. coli JM105 strain containing the GroEL/ES expressing plasmid. Expression and purification conditions were carried out as above.
ETF Purification
Porcine ETF was purified as previously published [32], except that the dialysis buffer after the 40–60% ammonium sulfate fractionation and the DE-52 running buffer consisted of unbuffered 15 mM dibasic potassium phosphate and 5% glycerol.
Enzyme Assays
The ETF reduction assay was performed using a Perkin Elmer LS50B (Norwalk, CT) fluorescence spectrophotometer with a heated cuvette block set to 30°C at indicated substrate concentrations as described [32,33]. To maintain enzyme stability during the assay, it was necessary to dilute it to ~15 µg IVD/ml of 50 mM Tris, pH 8.0, and 5 mM EDTA in 50% glycerol. Another assayed used was the phenazine methosulfate/2,6-dichlorpheno-indophenol (PMS/DCIP) reduction assay where DCIP decrease in absorbance at 600 nm is monitored as has been described previously [9].
For monitoring the pH effect on ETF fluorescence reduction and IVD activity, a stock buffer cocktail consisting of 80 mM of each of BICINE (N,N–bis[2–hydroxymethyl]glycine), CHES (2–[N–cyclohexylamino]ethanesulfonic acid), HEPES (N–2–hydroxyethylpiperazine–N′–2–ethanesulfonic acid), MES (2–[N–Morpholino] ethanesulfonic acid), and PIPES (piperazine-N,N′–bis[2–ethanesulfonic acid) was prepared and pH adjusted at 6.0, 6.5, 7.0, 7.5, 8.0, 8.5, 9.0, and 9.5. The buffer concentration in the reaction mixture was 10 mM for each.
Binding of ETF Docking Peptide to IVD Site
Twelve-mer peptide representing the ETF docking peptide, ETF βArg191-βLys202, +NH3-RYATLPNIMKAK-CO2−, and a control peptide with βLeu195 replaced with an Asp were synthesized at the University of Pittsburgh Genomics and Proteomics Core Laboratories and dissolved in 50 mM Tris, pH 8.0, at 20 mM final concentration. The peptide concentration was estimated using the extinction coefficient of Tyr (1280 M−1•cm−1) at 280 nm. Final concentrations for the ETF docking peptide in the reaction mixture were 0.434, 0.868, and 1.736 mM measured at ETF concentrations varied at 0.5, 1.0, 1.5, and 2.0 µM. IVD activity was monitored using the ETF fluorescence reduction assay as mentioned above. Data were analyzed using EnzymeKinetics v1.4.1 software from Trinity Software (Campton, NH).
Monitoring the Formation of the Charge Transfer Complex and Determination of the KD app for Isovaleryl-CoA
Formation of the charge transfer complex was monitored by observing the increase in absorbance at the 584 nm region. Spectral scans were performed using a Beckman DU7400 spectrophotometer (Palo Alto, CA). A “de-greened” IVD sample, freshly thawed, was applied to a PD-10 column, Pharmacia, to exchange buffer to a 50 mM Tris, pH 8.0. Quartz cuvette with a round top containing 0.4 ml of 29.86 µM IVD solution was sealed with a rubber plug, and using a needle ten alternating cycles of vacuum and oxygen-free argon were applied to remove oxygen. One µl at a time of a 1.08 mM isovaleryl-CoA freshly dissolved in water was then added to the cuvette using a 50 µl Hamilton syringe attached to an automatic dispenser. Ten seconds of equilibration time were allowed after mixing and the sample was scanned for UV/Visible light absorbance at 250 to 800 nm. Final substrate concentrations varied from 2.2 to 53.4 µM. All data were adjusted for the dilution resulting from substrate addition. Other IVD substrate analogues were titrated similarly, but with different final concentrations as indicated in figures’ legends. The “apparent” dissociation constant (KD app) was calculated as published earlier [34,35]. The titration assays were done similar to the above with similar enzyme (~35 µM) and substrate concentrations each time.
Fluorescence and CD Spectral Measurements
Fluorescence scans were carried out using a Perkin Elmer LS50B fluorescence spectrophotometer using protein samples in 10 mM phosphate, pH 8.0, that were de-aerated, at ambient temperature.
CD measurements were carried out using a Jasco 810 Spectropolarimeter (Easton, MD). All scans were performed in 50 mM Tris, pH 8, at ambient temperature. Baseline scans, with and without substrate, were subtracted from enzyme sample with and without substrate, respectively.
Molecular Modeling
Computer modeling of SBCAD was performed using a Silicon Graphics Fuel workstation (Mountain View, CA) and the Insight II 2005 software package, which included Homology/Modeler, and Discover modules (Accelyrs, San Diego, CA), and the atomic coordinates of monomers A and B of human IVD (PDB: 1IVH, [27]) and rat SCAD (PDB: 1JQI), in the dimer form, as reference molecule [27, 36]. Using Homology, the known protein structures were aligned using the “Superimpose” option for matching their secondary structures. The SBCAD sequence was then aligned to the known structures and the Modeler module was used to build the initial model, where FAD and the acetoacetyl-CoA, from the rat SCAD structure, were included as fixed molecules for each SBCAD monomer in the generated models. Including the ligands was essential to prevent encroachment of residues into the binding site pockets or the collapse of the otherwise unfilled cavities. In each case, several models were generated. Models were inspected manually for anomalous areas at the active site and other regions, and models with significant abnormalities were rejected. When necessary, amino acid residues were examined for flexibility in adopting other energetically similar conformations. In the initial chosen model the acetoacetyl-CoA was manually modified to an S-2-methylbutyryl-CoA to observe proper alignment of various groups at the active site pocket. Finally, the modified initial model was subjected to further refinements using other Insight II modules.
RESULTS
Expression levels and purification efficiency
IVD expressed to about 10% of cellular protein has been purified previously to essential homogeneity. Including the GroEL/ES chaperonins did not improve the level of production. IBD expression and production, however, was significantly increased to ~25–30% of cellular protein by co-production with the GroEL/ES chaperonin. More than 70 mg of IBD protein could be purified from 2 L of culture. This is highest level among all recombinant ACADs we have produced from E. coli, including MCAD, SCAD, VLCAD and ACAD9 (Mohsen and Vockley, unpublished data). The IVD Trp166Phe was not as stable as the wild type and was only partially purified, using the DEAE-Sepharose column, and was not suitable for spectral assays. The mutant enzyme was, however, tested for activity using the ETF fluorescence reduction assay. Activity was comparable to that of wild type.
Evaluation of Recombinant IVD Kinetic Parameters
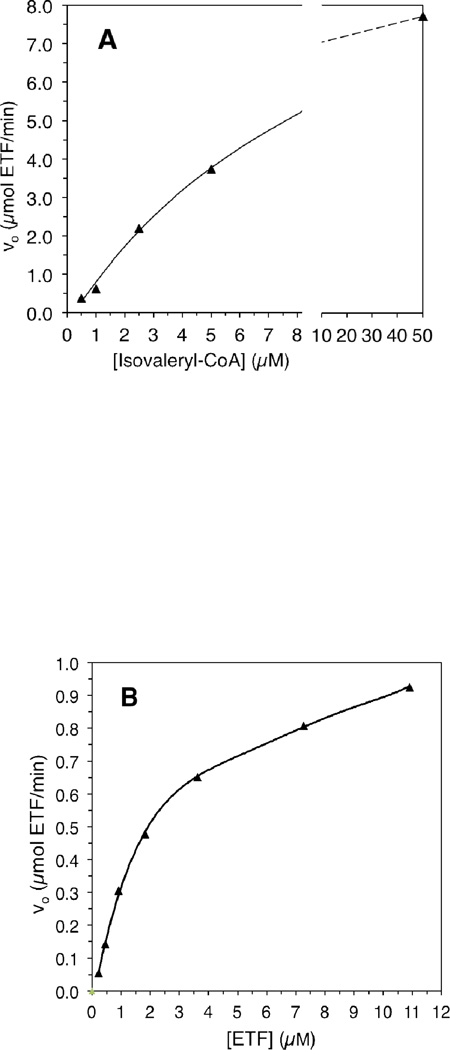
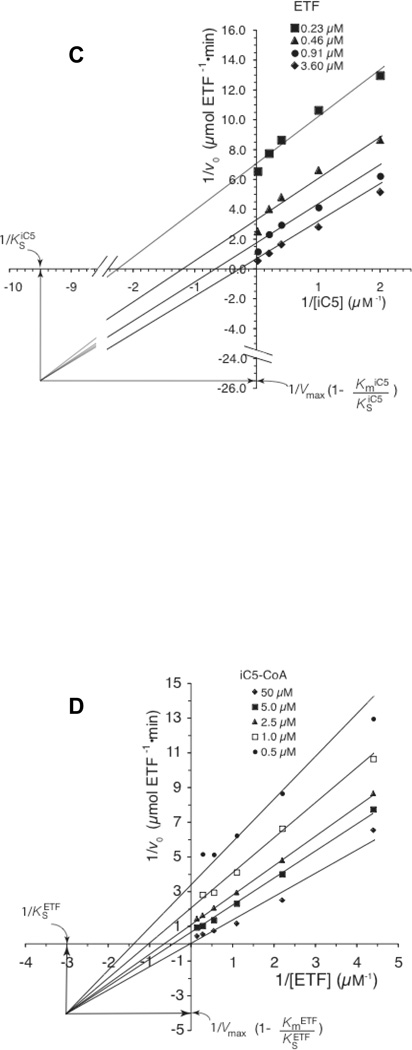
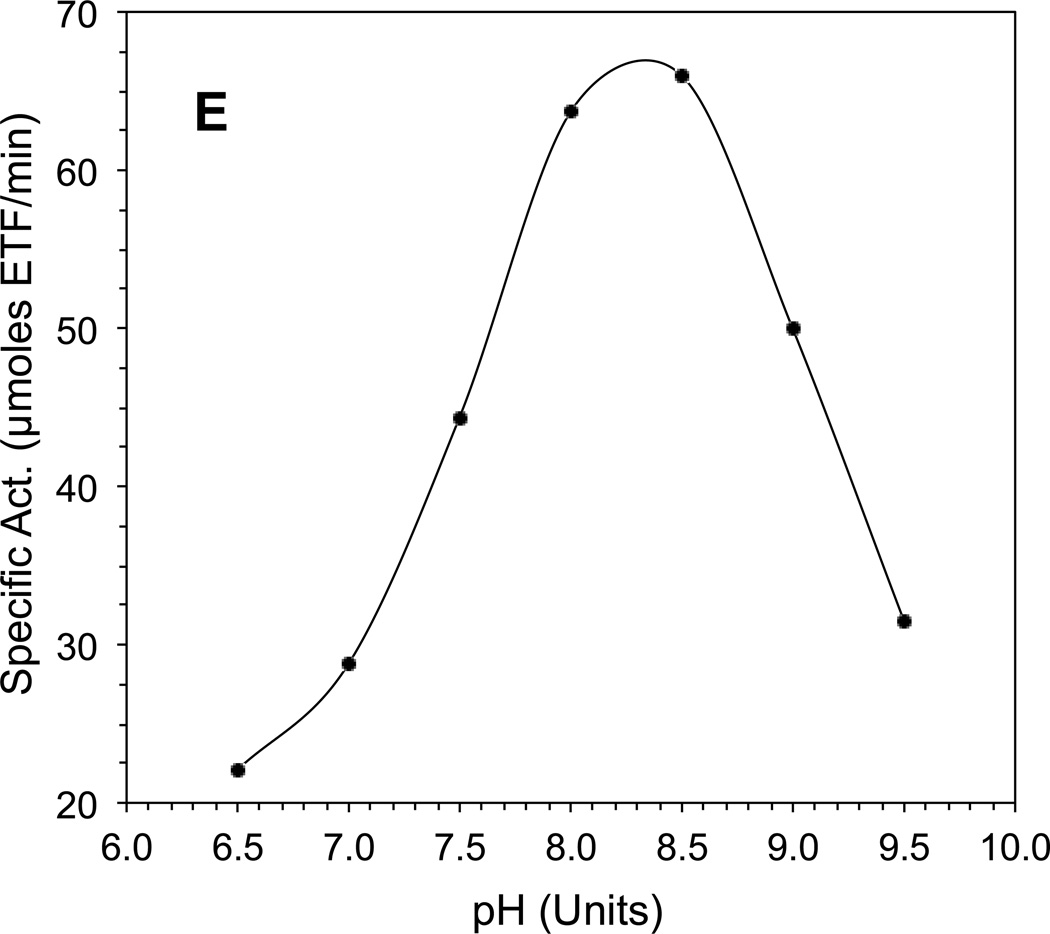
The CoA-persulfide-free IVD had a specific activity of 112.5 µmol porcine ETF•min−1 • mg−1 as measured with the ETF fluorescence reduction assay. Increasing either isovaleryl-CoA or ETF concentration significantly improves the initial velocity (v0) of IVD reduction (Fig. 1A and 1B.) Because of limitations with fluorescence detection, the ETF concentration could not be further increased beyond the highest concentration used (Fig. 1B). The CoApersulfide-free enzyme catalytic efficiency (kcat/Km) was 4.3×106 • M−1 • sec−1 per monomer at 50 µM isovaleryl-CoA when varying ETF concentration. Vmax was higher in the presence of higher amounts of isovaleryl-CoA substrate and higher ETF concentrations, but the Km for either substrate was higher as well. The Km was 0.5 µM or 8.1 µM for isovaleryl-CoA at constant 0.2 µM or 10.9 µM ETF, respectively. The Km for porcine ETF was 0.5 µM, at a constant 0.5 µM isovaleryl-CoA concentration but was 2.37 µM at 50 µM isovaleryl-CoA. pH dependence assays were conducted at a relatively lower ionic strength with total buffer concentration maintained at 50 mM, since high ionic strength has been reported to inhibit ACAD-ETF interaction [37]. IVD showed maximum activity at ~pH 8.3 (Fig. 1D).
Figure 1.
Evaluation of kinetic parameters of recombinant human IVD. (A) Increase in initial velocity with increasing amounts of isovaleryl-CoA. (B) Increase in initial velocity with increasing amounts of ETF. (C) Double reciprocal plot of IVD activity and isovaleryl-CoA concentration in the presence of various amounts of ETF. (D) Double reciprocal plot of IVD activity and ETF concentration in the presence of various amounts of isovaleryl-CoA. The amount of IVD in the reaction was 69 ng IVD (2.6 nM). (E) The pH dependence of the IVD reductive and oxidative half-reactions in the presence of excess ETF as the electron acceptor: Activity was determined by monitoring the reduction in ETF fluorescence under anaerobic conditions, (see text for details), in 50 mM Tris, pH 8.0, at 30°C.
Spectral Properties of the CoA-Persulfide-Bound IVD
The tight binding of the CoA-persulfide (CoAS–S−) by a bacterial butyryl-CoA dehydrogenase (BCAD) has been reported [38], among others. The presence of a CoA persulfide causes a solution of purified recombinant human IVD to appear green as has reported for other ACADs. However, not all recombinant ACADs have been reported to contain a bound CoA-persulfide following purification. The green form of the recombinant IVD stored in buffer containing 50 mM EDTA at –80°C was stable at high concentrations (~12 mg/ml) for several months as determined by measuring enzyme activity following de-greening of diluted (~2 mg/ml) enzyme solution, as outlined in Materials and Methods. When stored at +4°C, purified IVD remained in solution in the presence of the high concentration of EDTA. In the absence of EDTA the enzyme precipitated in more than a day depending on its concentration, leaving a green granular sediments, and clear solution in the case of the CoA-persulfide-bound form or a yellow precipitate for the de-greened enzyme. Adding DTT or β-mercaptoethanol was not effective in preventing precipitation, but the precipitation was more suspended cloudiness rather than a granular sediment.
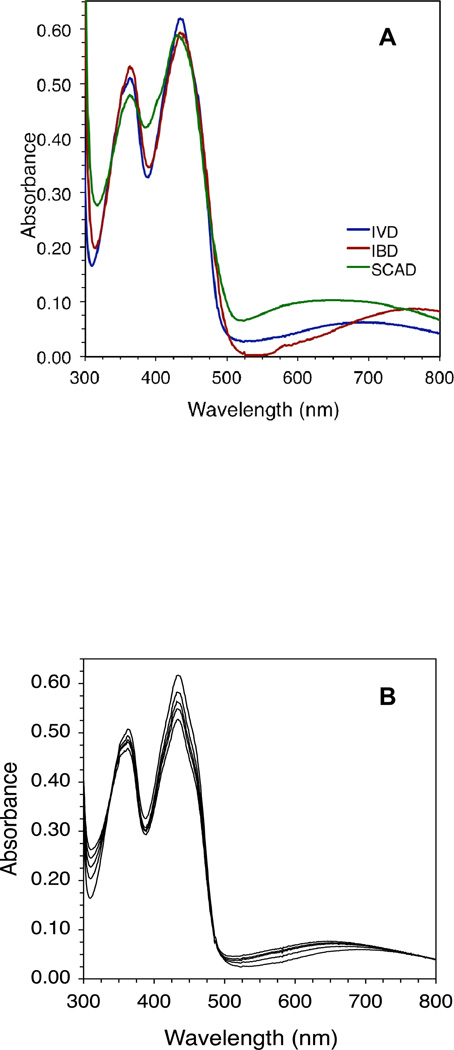
The presence of the CoA-persulfide in the purified green enzyme is demonstrated spectrally by the broad absorbance band extending from ~513 nm to beyond 800 nm and centered around 690 nm (Fig. 2A). The magnitude of this absorbance band is low for IVD, with the A434 nm/A690 nm ratio usually ~10. It is less pronounced than its counterpart in recombinant human IBD which shows a spectrum of CoA-persulfide bound enzyme that has a broad band centered at 760 nm and an A434 nm/A760 nm ratio of 6.9, or SCAD which has a broad band centered at 650 nm and a A434 nm/A650 nm ratio of 5.8 (Fig. 2A). The A434 nm/~A600 nm ratio corresponds well with the intensity of the green color of the enzyme, with IVD having the lightest green color, IBD with a medium green, and SCAD having the darker lime green color. Binding of residual CoA-persulfide to purified IVD is more reliably indicated by a blue shift of the absorbance maximum at 447 nm that appears to correlate with the amount of bound CoA persulfide and could be eliminated by de-greening. Green IVD in an essentially pure state had an A270 nm/A434 nm ratio of ~6.7, compared to an A270 nm/A447 nm ratio of ~5.4 for the de-greened form. The higher A270 nm/A434 nm ratio can be attributed partly to the contribution of the CoA persulfide adenine moiety to an increased absorbance at 270 nm and a decrease in absorbance at the 434 nm region. Other spectral clues to the presence of the CoA persulfide include a shift in absorbance at 373–363 nm, higher absorbance at the 313 nm valley, and lower A434 nm/A313 nm and A434 nm/A363 nm ratios. For de-greened IVD, the A447 nm/A313 nm and A447 nm/A373 nm the ratios are 4.7 and 1.4, respectively. Higher ratios indicate a lower percentage of CoA persulfide-bound subunits.
Figure 2.
Human IVD UV/Visible spectra with bound CoA persulfide: (A) Spectrum of 44 µM IVD and a near equivalent amount of IBD and SCAD with bound CoA-persulfide. (B) IVD with bound CoA persulfide in the presence of increasing amounts (13, 51, 75, and 103 µM) of isovaleryl-CoA to CoA persulfide-bound IVD. Scans were performed under anaerobic conditions in 50 mM Tris, pH 8.0, at ambient temperature.
A 12-fold molar excess of isovaleryl-CoA over enzyme only decreased absorbance in the 434 nm region by 30% even with prolonged incubation time under anaerobic conditioned (Fig. 2B). The long wavelength band characteristic of the CTC shifted to center at 650 nm rather than 584 nm or 760 nm characteristic of the IVD:isovaleryl-CoA CTC and the IVD:CoA-persulfide, respectively.
UV/Visible Spectral Analysis of the Interaction of IVD with Isovaleryl-CoA
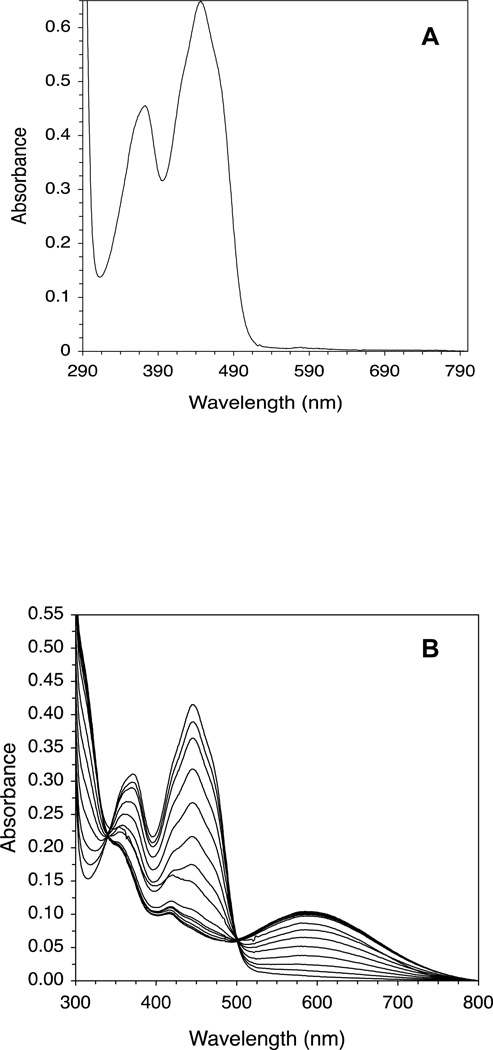
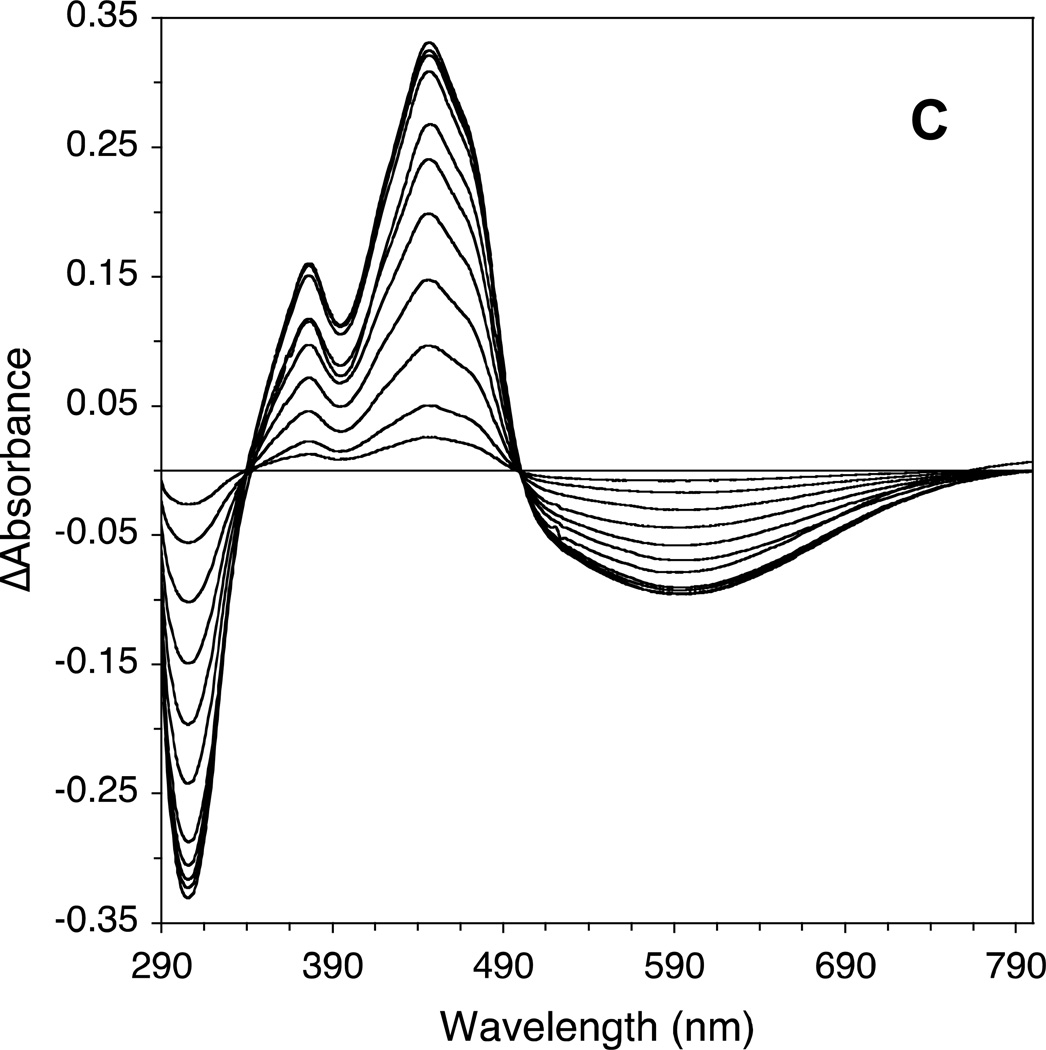
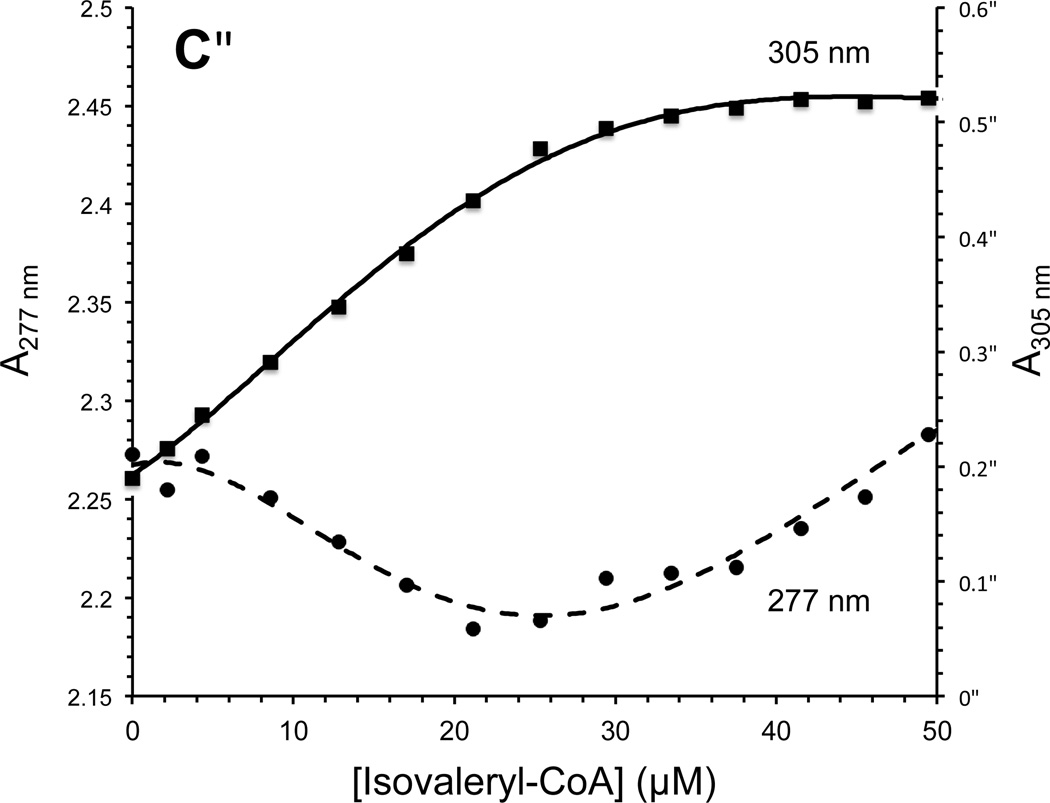
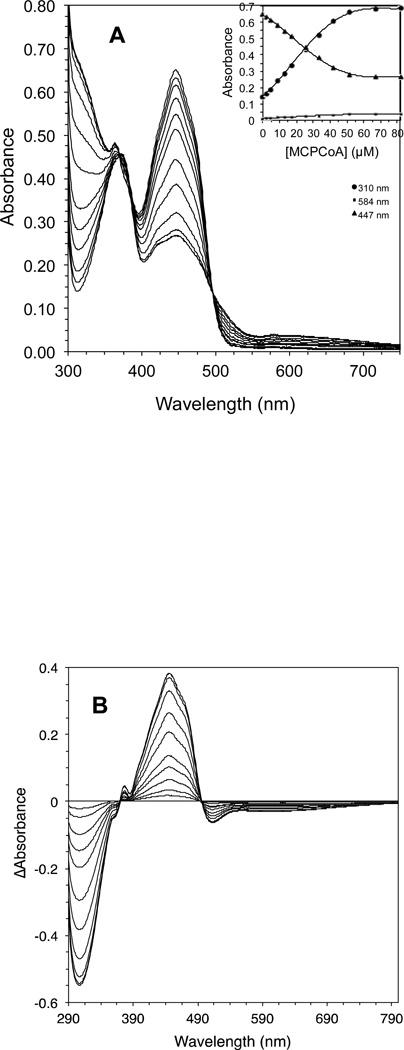
The effect of adding increasing amounts of isovaleryl-CoA on the CoA persulfide-free IVD spectrum is shown (Fig. 3A and C.) The most notable spectral changes include increasing absorbance centered at 310 nm (see Fig 3B), decreasing absorbance at 373 and 447 nm, the appearance of a blue band at 584 nm, and formation of two isosbestic points at 340 and 500 nm. The increase of absorbance at 310 nm and the decrease in absorbance at 373 nm appear to overlap. The increase of absorbance at 310 nm and the decrease at 447 nm are almost a mirror image, and appear to start to plateau near 30 µM of substrate essentially staying constant with increasing amounts of substrate. Fig. 4A plots the increase in absorbance at the 310 nm and 584 nm regions and decrease at the 447 nm versus the increase in isovaleryl-CoA concentrations.
Figure 3.
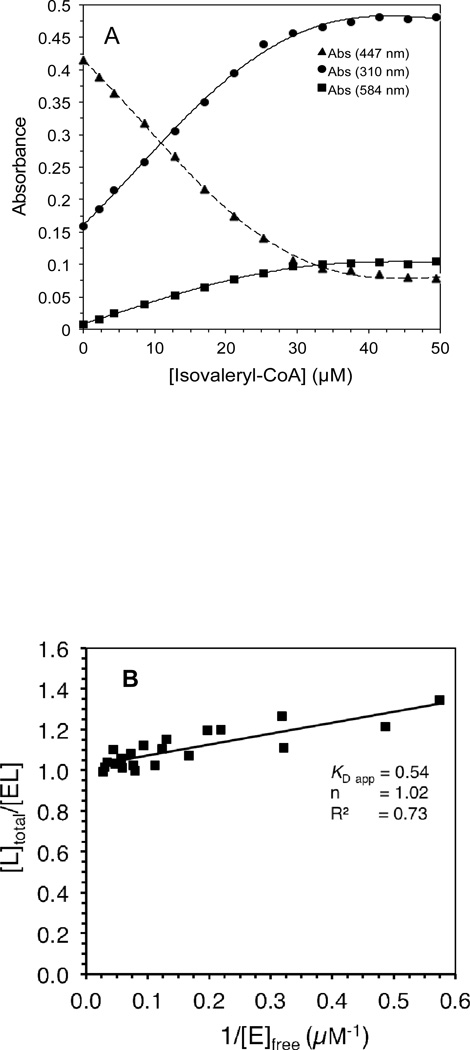
Human IVD UV/Visible spectra: (A) the region at 300–800 nm of the IVD oxidized form. Amount of enzyme was 29.86 µM in 50 mM Tris pH 8 at ambient temperature. Absorbance maxima at this region are at 373 and 447 nm. Shoulders are also observed at 420 and 470 nm, the former is more obvious as a small peak in the substrate-reduced form of the enzyme, see B. (B) Effect of adding increasing amounts of isovaleryl-CoA on IVD spectrum into freshly de-greened IVD under anaerobic conditions. The starting amount of the enzyme was 29.6 µM. The substrate concentrations for the spectra shown, from top to bottom spectra, were 2.2, 4.3, 8.6, 12.8, 17.0, 21.2, 25.3, 29.1, 33.5, 37.5, 45.5, and 49.5 µM. (C) The difference spectra where the spectra generated by adding the increasing amounts of isovaleryl-CoA, in (B), are subtracted from the IVD spectrum in the absence of the ligand.
Figure 4.
Effect of adding increasing amounts of isovaleryl-CoA on IVD absorbance at specific wavelengths: (A) decrease of absorbance at 447 nm, solid triangle, and increase of absorbance at 584 nm, solid square. (B) Stockell plot derivative of the absorbance data, at 447 nm, from three separate titrations. The extrapolated Y intercept represents the number of active sites per monomer, and the slope represents the KD app, the apparent binding constant, see Materials and Methods. E represents one subunit and L is the ligand, isovaleryl–CoA. (C) Changes in absorbance at indicated wavelengths induced by addition of 4.3, 8.6, 12.8, 17.0, 21.2, 25.3, 29.4, 33.5, 37.5, 45.5, and 49.5 µM isovaleryl-CoA.
The band at 584 nm of the substrate-bound IVDred can be attributed to formation of the CTC, as reported for other ACADs. The highest CTC absorbance at 584 nm, presumably with the enzyme essentially totally reduced, gives a ratio of absorbance at 584 nm to that at 447 and 373 nm of 1.56 and 0.71, respectively. The latter ratio (A584 nm/A373 nm) we propose could be used as a substrate productive binding factor (PBF), which would be of use in comparing the binding efficiency of isovaleryl-CoA to wild type and IVD mutants since the CTC band does not always appear in the same intensity with IVD mutants when titrated with the substrate compared to wild type.
It is easy to overlook in the titration spectra, a reproducible decrease in absorbance of IVD centered at 277 nm of an amplitude of 0.1 reaching minimum at a isovaleryl-CoA:IVD ratio of about 1:1, with subsequent increase upon further addition of isovaleryl-CoA (Fig. 4C). This contrasts with apparently exponential increase at 270 nm that parallels the expected increase in presumed free isovaleryl-CoA concentration, which is expected to mask such change. To investigate this, an IVD Trp166Phe was expressed in E. coli, but it was unstable to purify enough for such detailed spectral and kinetic analyses as wild type. Nevertheless, the mutant was active in transferring electrons to ETF with activity being comparable to wild type.
The mathematical model used to calculate the apparent dissociation constant (KD app) is similar as published previously (34). Fig. 4B shows the Stockell plot of combined values of [Ltotal]/[EL] versus reciprocal of [Efree] calculated from spectral data of three separate titrations with isovaleryl-CoA and enzyme differing slightly in each measurement. From these combined substrate titration assay data, the calculated KD app was 0.54 µM, with the calculated number of available monomers (n) being 1.0.
UV/Visible Spectral Analysis of the Interaction of IVD with acetoacetyl-CoA and methylenecyclopropylacetyl-CoA (MCPA-CoA)
Acetoacetyl-CoA is a naturally occurring product of lysine and lipid metabolism. Acetoacetate, an end product of leucine metabolism can be converted to acetoacetyl-CoA by succinyl-CoA:acetoacetate transferase. We examined the affinity of CoA persulfide-free IVD to bind acetoacetyl-CoA using the UV/visible spectral assay (Figures 5A and B). The increase in absorbance at 310 nm and decrease in absorbance at the 373 nm and 447 nm region was similar to that induced by isovaleryl-CoA but less extensive. The peak absorbance at 447 nm was only decreased by ~25% of the highest possible decrease achieved with isovaleryl-CoA. The acetoacetyl-CoA concentration that induced half the maximal spectral changes at the various wavelengths (Kf) was ~18 µM. Concomitant with the decrease in absorbance at 447 nm, there was an increase of absorbance at 565 nm attributed to the formation of a charge transfer complex.
Figure 5.
Effect of adding increasing amounts of acetoacetyl-CoA to IVD: (A) Spectra of 33.7 µM IVD in the presence of 0, 8.9, 17.8, 26.6, 35.3, 44.1, 52.7, 61.4, 70.0, 78.5, and 120.7 µM acetoacetyl-CoA, from upper line to lower one. (B) Difference spectra of IVD calculated from data in (A).
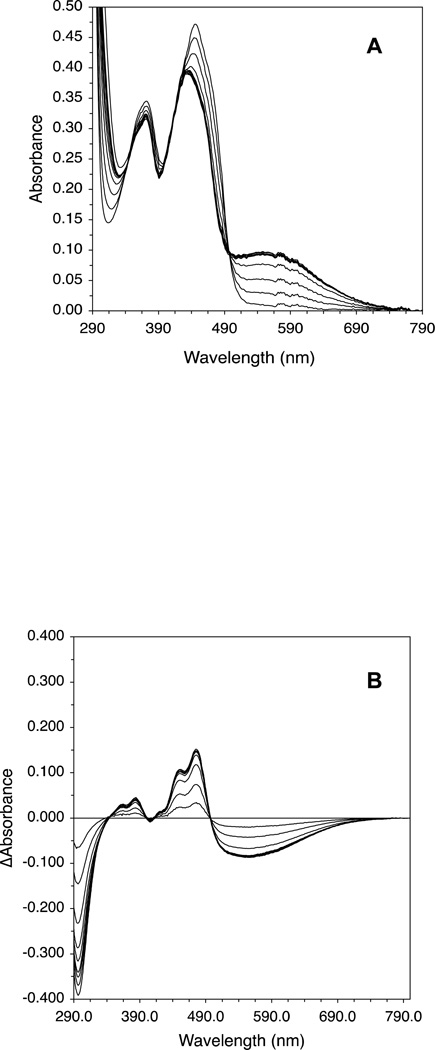
The effect of adding increasing amounts of (±) MCPA-CoA on the IVD absorption spectrum at the 310 and 447 nm regions is shown in Fig. 6A and B. The magnitude of the increase in absorbance at 310 nm was larger than seen with isovaleryl-CoA binding. This can be attributed to the lack of decrease of absorbance at 373 nm related to reduction of the flavin by bona fide substrate (Fig. 3B). The decrease in absorbance at the 447 nm is attributable to the disruption of resonance of the flavin ring resulting from the covalent bond formed involving N-5 of the flavin. The concentration of MCPA-CoA that induced half the maximal spectral changes (Kf) was ~22.5 µM.
Figure 6.
Effect of adding increasing amounts of methylenecyclopropyl-CoA (MCPCoA) to IVD: (A) IVD concentration was 47 µM, MCPCoA concentrations were 0, 2.2, 4.3, 8.6, 12.8, 17.0, 25.3, 33.5, 41.5, 51.4, 66.9, and 81.8 µM, from upper line to lower one. Insets are the increase in absorbance at 310 nm, solid circles; decrease of absorbance at 447 nm, solid triangles; and increase of absorbance at 584 nm, solid squares. (B) Difference spectra of IVD calculated from data in (A).
Circular Dichroism Spectroscopy of Recombinant Human IVD in the Presence of CoA Esters
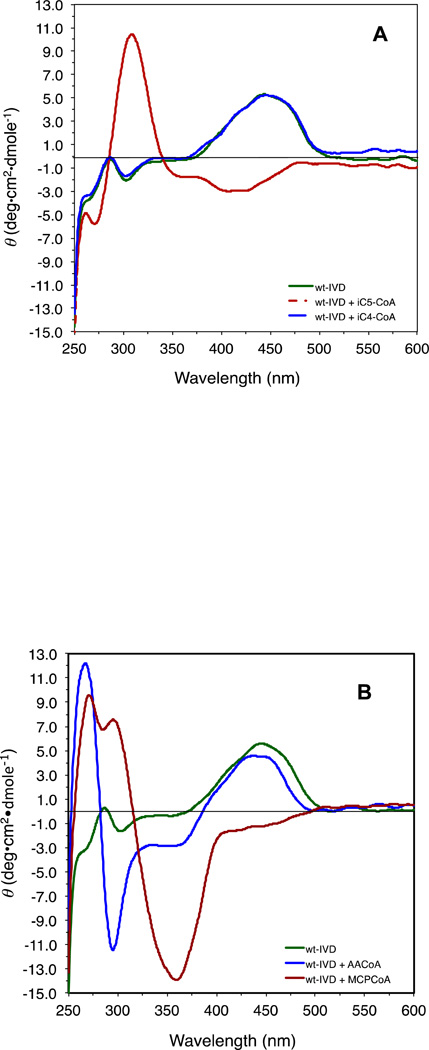
Isovaleryl-CoA induced an inversion of the peak in the 450 nm region, a minor peak at 300 nm region was inverted and intensified (Fig. 7A). The change in the 300 nm region corresponds to the changes in absorbance in the 310 nm region of the UV spectrum. Acetoacetyl-CoA induced markedly different changes in the CD spectrum of IVD compared to bona fide substrate (Fig. 7A and B). The region between 250 to 480 nm showed an inversion from the negative to the positive side at 265 nm almost equal in magnitude to a decrease seen at 295 nm. The peak at the 435 nm region did not change significantly indicating no reduction of the flavin ring in response to acetoacetyl-CoA binding despite formation of a charge transfer complex band at 565 nm as shown above. MCPA-CoA induces yet a different CD signature in the IVD, indicative of a change in conformation of the flavin ring that distinguishes this interaction from that seen with isovaleryl-CoA or acetoacetyl-CoA, Fig 7B.
Figure 7.
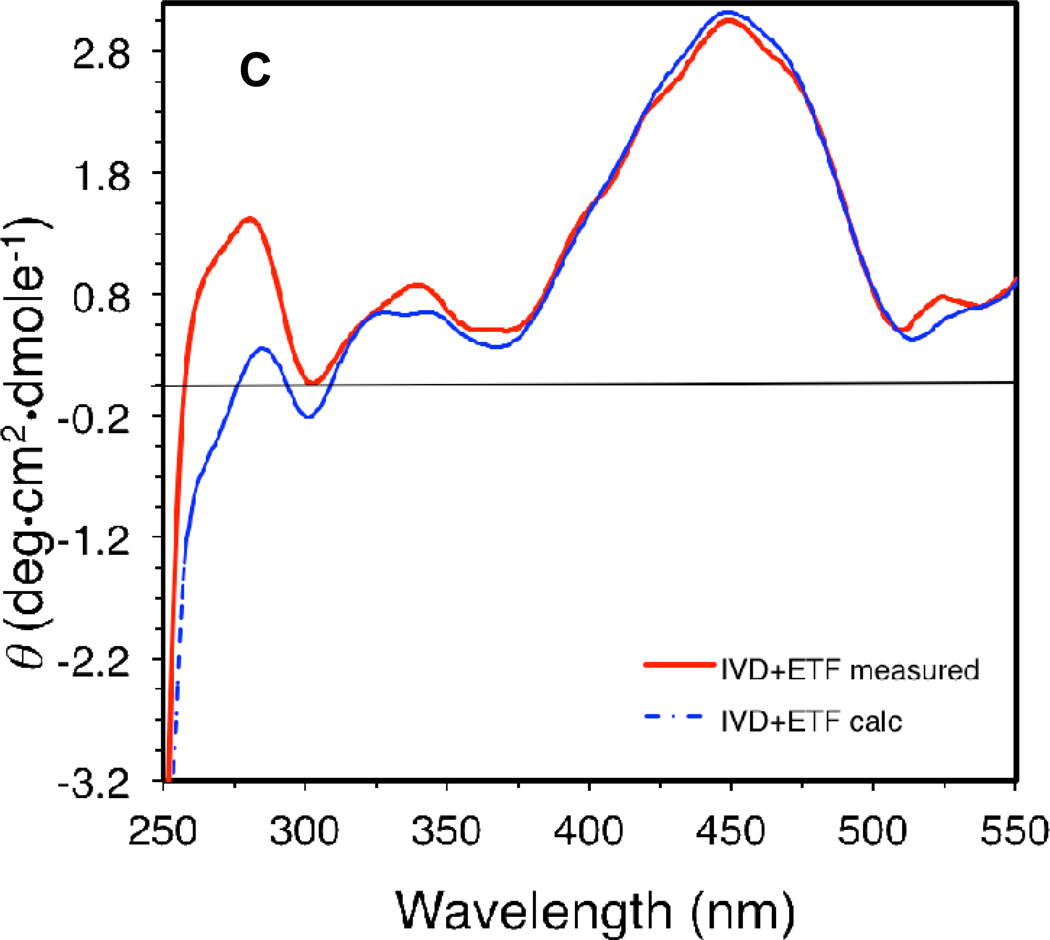
Effect of the presence of ligands on the CD spectrum of IVD: (A) CD scan of recombinant human IVD (10 µM) in the absence of ligands (green line) and presence of ten molar fold isovaleryl-CoA (red line), or isobutyryl-CoA (blue line), under anaerobic conditions. (B) CD spectrum scans of recombinant human IVD in the absence of ligands (green line) and presence of ten molar fold acetoacetyl-CoA (blue line), and MCPCoA (red line). (C) Difference CD scan of a 2:1 molar ratio mixture of recombinant human IVD monomer (11 µM) and porcine ETF heterodimer, respectively; blue line is for the spectrum of the mixture, and the red line is the calculated sum of IVD and ETF scanned separately and adjusted for dilution. All CD scans were performed in 50 mM potassium phosphate, pH 8.0, at ambient temperature.
The effect of the presence of ETF on IVD was examined by monitoring changes in the CD spectrum upon the addition of either protein to the other (Fig. 7C), as well as tryptophan fluorescence of both proteins in solution (see below). A subtle change was observed at the 265 nm area, where a shoulder is inversed to the positive side of the spectrum.
Inhibition of IVD activity in the presence of the ETF docking peptide
While the ETF docking peptide was observed to have an inhibitory effect on IVD activity when measured using the ETF fluorescence assay with Ki estimated to be 1.5 mM peptide, the βLeu195Asp version of the peptide did not show any inhibitory effect. Furthermore, in the presence of the wild type enzyme no change was observed in reduction of PMS/DCIP when used as the electron acceptor.
DISCUSSION
Expression and protein quality
Expression and purification of IVD and IBD were significantly improved using the methodology outlined. While expression of the human IVD cDNA was dramatically enhanced by adjusting for codon bias usage in E. coli as reported earlier [28], co-expression with GroEL/ES did not impact expression as it dramatically did for IBD. Such high expression for IBD coupled with using EDTA proved valuable for improving protein quality as protein manipulation and oxidative damage during purification seemed diminished significantly. The advantage of high concentration of EDTA manifested in higher specific activity can be explained in terms of EDTA being an electron donor protecting groups labile to oxidation against oxygen radicals generated by sonication [39]. While Cys residues are the most suspected to be affected by oxygen radicals [39], other residues, e.g., Met, and Tyr, are also vulnerable to oxidative modification. Use of EDTA at such high concentration has since been a standard for purifying all other ACAD enzymes and has also resulted in higher specific activity of other enzymes (Mohsen, data unpublished).
Another concern for producing ACADs in E. coli is tight binding of CoA persulfide to the substrate binding site. A procedure to remove bound CoA-persulfide from bacterial sources has been reported for several recombinant ACADs [40]. Two approaches followed were successful in eliminating CoA persulfide, induction for a shorter time to minimize E. coli production of CoA persulfide and a novel de-greening method. The advantage of de-greening the sample while bound to column resins is that it is more efficient than either two step anaerobic dialysis in dithionite buffer or aerobic dialysis [40], and an increased yield as the protein elutes from the column in a single and peak rather more than one peak because of a more homogeneous sample. Tight CoA persulfide binding to IVD inhibits reduction of the enzyme upon addition of its bona fide substrate, isovaleryl-CoA [41]. The spectrum of the CoA-persulfide-bound IVD in the presence of excess isovaleryl-CoA may reflect the presence of a mixture of enzyme subunits with two ligands bound, some binding the CoA-persulfide and others the isovaleryl-CoA. This may explain the peak of the long wavelength band characteristic of the CTC shifting to center in between the peak of the characteristic of the IVD:isovaleryl-CoA CTC and the IVD:CoA-persulfide, respectively. While both human recombinant IVD and IBD have been crystallized with bound CoApersulfide, in the former case all of the tetramer active sites were occupied with ligand, while in the latter only two subunits were observed to contain bound ligand [27, 42]. This may imply the presence of communication between neighboring monomers induced by binding of CoA ligands when one, or more, active site is occupied with inhibitors or CoA esters other than the bona fide substrate.
Not all ACADs induce CoA persulfide generation and binding in E. coli. While IVD binds CoA persulfide tightly, recombinant wild type human MCAD does not seem to co-purify with CoA persulfide, though its E376Q mutant does [43]. It has been suggested that the presence of a glutamate at position 99 in MCAD wild type (the equivalent position in IVD is a glycine) hinders binding of the CoA persulfide [44]. This is consistent with undetectable CoA persulfide in purified recombinant human SBCAD [31], which has a glutamate at the equivalent position, while the ligand co-purifies with recombinant bacterial BACD, human IBD, and human and rat SCAD that have a serine at the corresponding position. The glycine at the equivalent position in IVD seems to render binding of the CoA persulfide tighter than for recombinant IBD or SCAD, since de-greening of these latter two proteins is not as difficult (data not shown). Furthermore, the elimination of the negative charge provided by the IVD catalytic base makes removal of the CoA persulfide from purified IVD E254G extremely difficult compared to wild type enzyme (Mohsen and Vockley, unpublished data.) The apparent differential binding affinity of CoA persulfide to these ACADs implies that if it has any relevant modulatory role in vivo [40], such a hypothetical role may affect IVD the most, followed by SCAD and IBD, and that its binding would be affected by pH and the presence heavy metals.
The specific activity of IVD expressed and purified was 14-fold higher from our earlier reported value [33], and was >20-fold higher than that measured for its CoA-persulfide bound form. The kinetic parameters have been significantly improved compared with earlier published data of native IVD purified from other species [33], the longer purification protocol is probably the cause of deterioration in protein integrity. The IVD pH-dependence data is consistent with studies of substrate specificity profiles of SCAD, MCAD, and LCAD using the ferricinium assay, where optimum substrates induced the highest activity at pH values approaching 8.5 [37]. Physiologically higher activity for ACADs at relatively higher pH value is not surprising as this correlates with a higher demand for ATP production. This is in agreement with conclusions by Ghisla et al. who proposed that the ACADs pH-dependence reflect a mechanism for ACADs activity modulation [45].
UV/Visible Spectral Analysis
The spectral changes monitored upon addition of isovaleryl-CoA to IVD solution are consistent with earlier studies. The source of the increase in absorbance at 310 nm has not been determined, but it is likely related to changes at the active site. The indole ring of Trp166 in IVD, which lies parallel to the si face of the dimethylbenzene ring of the flavin, does not seem to be the source since a similar increase in absorbance at 304 nm is observed upon addition of isobutyryl-CoA to IBD (Mohsen, unpublished), which has phenylalanine at the equivalent position. The indole of Trp216 in IBD, which lies perpendicular to the flavin dimethylbenzene ring on its re face side [43], may substitute for a Trp166 structural function. It’s unlikely that the changes at 310 nm are related to the free or bound substrate adenine moiety since further increase in 310 nm absorbance was not observed with further substrate additions, and other CoA ligands, e.g., the CoA-persulfide, does not induce similar changes at the same region.
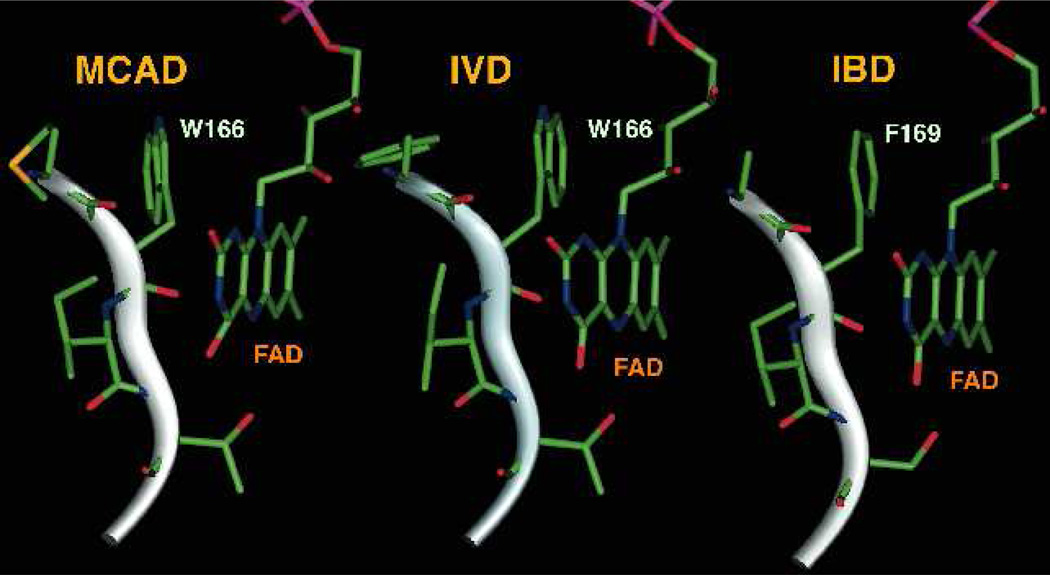
While the source of the absorbance at 277 nm could be the FAD or the substrate, the indole moieties of the tryptophan residues also absorb at this region. Free tryptophan and tyrosine amino acids absorb highest at 277–279 nm and 274–275 nm, respectively, with the former having >4 times the extinction coefficient of the latter at 277 nm. In IVD there are total of 2 tryptophan and 15 tyrosine residues per monomer. Of these two tryptophan residues, Trp166 is the closest of the two to the flavin ring with its indole moiety π-stacked against the flavin dimethylbenzene ring (Fig. 8), as mentioned above and so perhaps is the source of such a subtle change. Reduction of the flavin upon substrate binding is expected to affect this π-π interaction. It has been implied in the MACD:ETF co-crystal structure study that the indole of this residue plays an undefined role in electron transfer between ACADs and ETF [24, 27]. However, a role in the mechanism of electron transfer seems dispensable since replacing IVD Trp166 with a Phe residue did not abolish transfer of electrons to ETF. This is consistent as well with the equivalent position in IBD being a Phe indicating a Trp at this position is dispensable, albeit it is possible that IBD uses a different mechanism for electron transfer to ETF compared to MCAD.
Figure 8.
Stick representation of IVD Trp166 (PDB: 1IVH, [27]) and the equivalent residue in MCAD (PDB: 2A1T, [24]) and IBD (PDB: 1RXO, [42]).
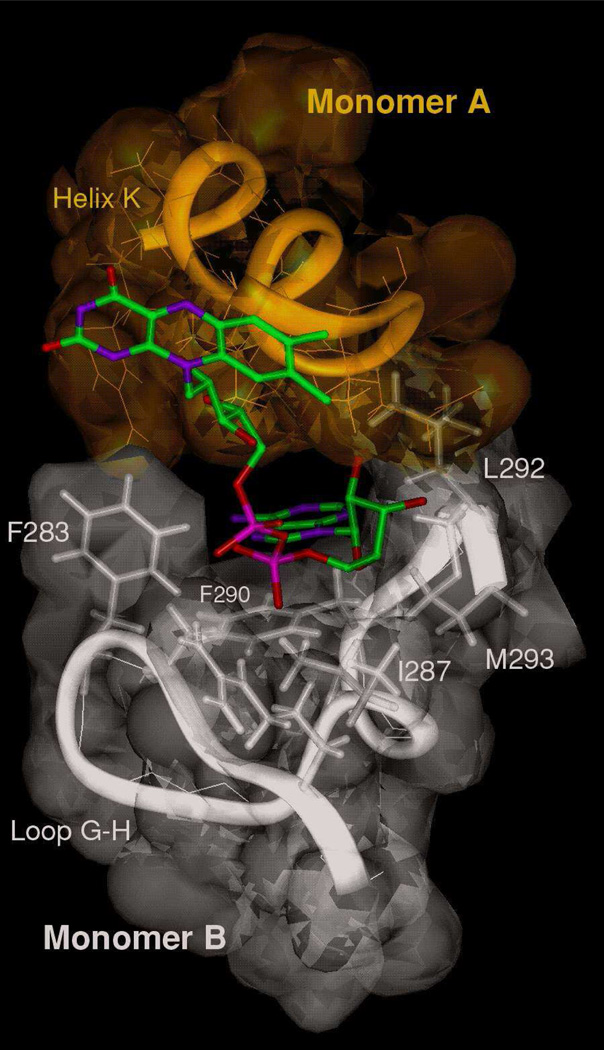
Interest has grown in selective ACADs inhibitors, including MCPA-CoA-like compounds, because of potential therapeutic value in heart disease [46, 47]. MCPA-CoA is the physiological metabolite of hypoglycin A, a naturally occurring toxin in the Ackee plant fruit that peaks just prior to maturity. Toxic levels of hypoglycin A cause Jamaican vomiting sickness, characterized by severe hypoglycemia [48, 49]. MCPA-CoA is a “suicide” inhibitor that targets IVD, as well as other ACADs, binding covalently to the flavin ring of the essential cofactor [47–50]. Despite the irreversible covalent interaction, MCPA-CoA was unable to effect a change in the absorbance at 447 nm to the same extent as isovaleryl-CoA (only ~72% of the latter) even at very large excess of inhibitor to enzyme ratio. This is consistent with the isovaleryl-CoA spectral assay results of the IVD-CoA persulfide bound form, reported above, implying negative cooperativity between subunits may be induced by binding of the inhibitor. While the spectral results with the bona fide substrate, isovaleryl-CoA, suggest that binding at a nearly 1:1 ratio is inconsistent with the hypothesis of communication between subunit exist, the results may not be comparable since the mode of interaction of the inhibitors at the active sites, as supported by from the CD data, is different. Examination of FAD binding in all ACADs with determined crystal structure reveals that several residues from the second subunit (Fig 9) and one from the fourth subunit contribute to binding of the FAD adenine moiety from the first subunit. It is possible that subtle conformational changes induced by ligand binding are transmitted to the second subunit via changes in conformation of the FAD adenine moiety. Physiologically, non-productive ligand binding that does not result in proper alignment of groups at the FAD adenine binding site could be a mechanism for the ACADs to limit accessibility of their active sites and preferentially accept their bona fide substrates.
Figure 9.
Ribbon and stick representation of IVD FAD binding site showing the contribution of the second monomer (B) of the dimer in the binding of the FAD adenine base of the first subunit (A), (PDB: 1IVH, [27]).
Circular Dichroism Spectroscopy of IVD in the Presence of CoA Esters
The effect of the addition of CoA esters on the CD spectrum of IVD was dramatically different for each CoA ester. The changes induced by addition of acetoacetyl-CoA to IVD in the 250 to 375 nm region are different than those induced by this ligand in recombinant human SCAD in which the magnitude of the inversion and increase at 265 nm was almost 5 times the changes at 295 nm [29]. This indicates differences in the binding of the acetoacetyl moiety at the active site of each enzyme. These changes as seen above are related to changes in flavin conformation induced by the CoA esters occupying the substrate binding. This conclusion is based on the observations that: (1) The CD spectra were monitored with the buffer solution without IVD but containing excess ligand being used as baseline, i.e., subtracted from IVD solution containing the ligand. The resulting spectra indicate that the CD changes at 450 nm and the 300 nm regions reflect a change in conformation that is most specifically related to the flavin ring induced by ligand binding. (2) Isobutyryl-CoA, a CoA ester that cannot bind at the IVD active site for steric conflicts, hardly induces any changes to the purified IVD CD or UV visible spectrum indicating lack of significant binding even at a 10:1 ligand to flavin ratio. (3) The changes in CD spectra induced by isovaleryl-CoA, acetoacetyl-CoA and MCPA-CoA are significantly different indicating differences in the mode of interaction with the active site flavin.
Inhibition of IVD activity in the presence of the ETF docking peptide
The inhibition of IVD activity in the presence of the ETF docking peptide using the ETF fluorescence reduction assay indicates that the peptide binds at the IVD ETF docking site. Similar to the CoA esters substrates that have been shown earlier to stabilize IVD and MCAD proteins [26], thermal stability studies are underway to investigate the effect of the ETF docking peptide on protein stability. In addition, ETF docking peptide with point mutations are being tested for improved interaction with IVD. Structure or fragment based ligand design may then be used to search for ligands that bind at the ETF docking site and stabilize naturally occurring IVD mutants that are thermally unstable.
We have presented a detailed description of spectral properties of recombinant human IVD with interacting ligands. Several spectral changes have been observed in the presence of ligands, many of which are consistent with earlier studies on ACADs. Our findings set the stage to better characterize the effects of patient mutations on enzyme function. Understanding the microenvironment of specific residues is crucial for interpreting abnormalities of enzyme function and/or instability induced by mutations that cause isovaleric academia, as well as designing future mutagenesis experiments.
Highlights for Review.
Isovaleryl-CoA dehydrogenase deficiency cause a serious metabolic disorder. Biochemical and biophysical characterization of the enzyme is reported.
Importance of including high concentrations of EDTA to protect residues vulnerable to oxidation induced by O2-free radical generated by sonication.
Novel method for removal of tightly bound inhibitor from the recombinant protein is reported.
A 12-peptide that represents the docking peptide of its redox partner is reported to act as an inhibitor.
IVD W166F mutant is active when measured using ETF as the electron acceptor.
ACKNOWLEGEMENTS
This work has been supported in part by United States Public Health Service National Institutes of Health grant RO1 DK54936 (JV) and the Eunice Kennedy Shriver National Institute of Child Health & Human Development grant R21 HD056004 (A-WM).
ABREVIATIONS
- ACADs
acyl-CoA dehydrogenases
- BCAD
butyryl-CoA dehydrogenase
- DCIP
dichlorophenylindolphenol
- ETF
electron transfer flavoprotein
- CTC
charge transfer complex
- IBD
isobutyryl-CoA dehydrogenase
- IVD
isovaleryl-CoA dehydrogenase
- MCAD
medium chain acyl-CoA dehydrogenase
- LCAD
long chain acyl-CoA dehydrogenase
- PMS
phenazine methosulfate
- SBCAD
short/branched chain acyl-CoA dehydrogenase
- SCAD
short chain acyl-CoA dehydrogenase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Tanaka K, Budd MA, Efron ML, Isselbacher KJ. Isovaleric acidemia: a new genetic defect of leucine metabolism. Proc. Natl. Acad. Sci. USA. 1966;56:236–242. doi: 10.1073/pnas.56.1.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vockley J, Parimoo B, Tanaka K. Identification of the molecular defects responsible for the various genotypes of isovaleric acidemia. Prog. Clin. Biol. Res. 1992;375:533–540. [PubMed] [Google Scholar]
- 3.Zhang J, Zhang W, Zou D, Chen G, Wan T, Zhang M, Cao X. Cloning and functional characterization of ACAD-9, a novel member of human acyl-CoA dehydrogenase family. Biochem. Biophys. Res. Commun. 2002;297:1033–1042. doi: 10.1016/s0006-291x(02)02336-7. [DOI] [PubMed] [Google Scholar]
- 4.Westover JB, Goodman SI, Frerman FE. Binding, hydration, and decarboxylation of the reaction intermediate glutaconyl-Coenzyme A by human glutaryl-CoA dehydrogenase. Biochemistry. 2001;40:14106–14114. doi: 10.1021/bi015637p. [DOI] [PubMed] [Google Scholar]
- 5.Rozen R, Vockley J, Zhou L, Milos R, Willard J, Fu K, Vicanek C, Low-Nang L, Torban E, Fournier B. Isolation and expression of a cDNA encoding the precursor for a novel member (ACADSB) of the acyl-CoA dehydrogenase gene family. Genomics. 1994;24:280–287. doi: 10.1006/geno.1994.1617. [DOI] [PubMed] [Google Scholar]
- 6.Nguyen TV, Andresen BS, Corydon TJ, Ghisla S, Abd-El Razik N, Mohsen A-W, Cederbaum SD, Roe DS, Roe CR, Lench NJ, Vockley J. Identification of isobutyryl-CoA dehydrogenase and its deficiency in humans. Mol Genet Metab. 2002;77:68–79. doi: 10.1016/s1096-7192(02)00152-x. [DOI] [PubMed] [Google Scholar]
- 7.Naito E, Ozasa H, Ikeda Y, Tanaka K. Molecular cloning and nucleotide sequence of complementary DNAs encoding human short chain acyl-coenzyme A dehydrogenase and the study of the molecular basis of human short chain acyl-coenzyme A dehydrogenase deficiency. J Clinical Invest. 1989;83:1605–1613. doi: 10.1172/JCI114058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsubara Y, Kraus JP, Yang-Geng TL, Francke U, Rosenberg LE, Tanaka K. Molecular Cloning of cDNAs Encoding Rat and Human Medium-Chain Acyl-CoA Dehydrogenase and Assignment of the Gene to Human Chromosome 1. Proc. Natl. Acad. Sci. USA. 1986;83:6543–6547. doi: 10.1073/pnas.83.17.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda Y, Okamura-Ikeda K, Tanaka K. Purification and characterization of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases from rat liver mitochondria. Isolation of the holo- and apoenzymes and conversion of the apoenzyme to the holoenzyme. J Biol. Chem. 1985;260:1311–1325. [PubMed] [Google Scholar]
- 10.Andresen BS, Bross P, Vianeysaban C, Divry P, Zabot MT, Roe CR, Nada MA, Byskov A, Kruse TA, Neve S, Kristiansen K, Knudsen I, Corydon MJ, Gregersen N. Cloning and characterization of human very-long-chain acyl-CoA dehydrogenase cDNA, chromosomal assignment of the gene and identification in four patients of nine different mutations within the VLCAD gene. Human Mol. Genet. 1996;5:461–472. doi: 10.1093/hmg/5.4.461. [DOI] [PubMed] [Google Scholar]
- 11.He M, Pei Z, Mohsen A-WA, Watkins P, Murdoch G, Van Veldhoven PP, Ensenauer R, Vockley J. Identification and characterization of new long chain Acyl-CoA dehydrogenases. Mol Genet Met. 2011;102:418–429. doi: 10.1016/j.ymgme.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghisla S, Engst S, Moll M, Bross P, Strauss AW, Kim J-JP. α,β-Dehydrogenation by acyl-CoA dehydrogenases: Role of functional groups at the active center. Prog. Clinical Biol. Res. - New Developments in Fatty Acid Oxidation. 1992;375:127–142. [PubMed] [Google Scholar]
- 13.Nishina Y, Sato K, Hazekawa I, Shiga K. Structural modulation of 2-enoyl-CoA bound to reduced acyl-CoA dehydrogenases: a resonance Raman study of a catalytic intermediate. J Biochemistry. 1995;117:800–808. doi: 10.1093/oxfordjournals.jbchem.a124779. [DOI] [PubMed] [Google Scholar]
- 14.Miura R, Nishina Y, Sato K, Fujii S, Kuroda K, Shiga K. C-13-NMR and N-15-NMR studies on medium-chain acyl-CoA dehydrogenase reconstituted with C-13-Enriched and N-15-Enriched flavin adenine dinucleotide. J. Biochem. Tokyo. 1993;113:106–113. doi: 10.1093/oxfordjournals.jbchem.a123992. [DOI] [PubMed] [Google Scholar]
- 15.Vock P, Engst S, Eder M, Ghisla S. Substrate activation by acyl-CoA dehydrogenases - transition-state stabilization and pKs of involved functional groups. Biochemistry. 1998;37:1848–1860. doi: 10.1021/bi971827h. [DOI] [PubMed] [Google Scholar]
- 16.Engst S, Vock P, Wang M, Kim JJP, Ghisla S. Mechanism of activation of acyl-CoA substrates by medium chain acyl-CoA dehydrogenase: Interaction of the thioester carbonyl with the flavin adenine dinucleotide ribityl side chain. Biochemistry. 1999;38:257–267. doi: 10.1021/bi9815041. [DOI] [PubMed] [Google Scholar]
- 17.Hazekawa I, Nishina Y, Sato K, Shichiri M, Miura R, Shiga K. A Raman study on the C(4)=O stretching mode of flavins in flavoenzymes: hydrogen bonding at the C(4)=O moiety. J. Biochemistry. 1997;121:1147–1154. doi: 10.1093/oxfordjournals.jbchem.a021708. [DOI] [PubMed] [Google Scholar]
- 18.Ikeda Y, Hine DG, Okamura-Ikeda K, Tanaka K. Mechanism of action of short-chain, medium-chain, and long-chain acyl-CoA dehydrogenases: Direct evidence for carbanion formation as an intermediate step using enzyme-catalyzed C-2 proton/deuteron exchange in the absence of C-3 exchange. J. Biol. Chem. 1985;260:1326–1337. [PubMed] [Google Scholar]
- 19.Ghisla S, Thorpe C, Massey V. Mechanistic Studies with General Acyl-CoA Dehydrogenase and Butyryl-CoA Dehydrogenase: Evidence for the Transfer of the b-Hydrogen to the Flavin N(5)-Position as a Hydride. Biochemistry. 1984;23:3154–3161. doi: 10.1021/bi00309a008. [DOI] [PubMed] [Google Scholar]
- 20.Gorelick RJ, Schopfer LM, Ballou DP, Massey V, Thorpe C. Interflavin oxidationreduction reactions between pig kidney general acyl-CoA dehydrogenase and electron-transferring flavoprotein. Biochemistry. 1985;24:6830–6839. doi: 10.1021/bi00345a015. [DOI] [PubMed] [Google Scholar]
- 21.Thorpe C, Kim JJP. Structure and mechanism of action of the acyl-CoA dehydrogenases. FASEB. 1995;9:718–725. doi: 10.1096/fasebj.9.9.7601336. [DOI] [PubMed] [Google Scholar]
- 22.Ghisla S, Thorpe C. Acyl-CoA dehydrogenases. A mechanistic overview. Eur. J. Biochem. 2004;271:494–508. doi: 10.1046/j.1432-1033.2003.03946.x. [DOI] [PubMed] [Google Scholar]
- 23.Toogood HS, Van Thiel A, Basran J, Sutcliffe MJ, Scrutton NS, Leys D. Extensive domain motion and electron transfer in the human electron transferring flavoprotein.medium chain Acyl-CoA dehydrogenase complex. J. Biol. Chem. 2004;279:32904–32912. doi: 10.1074/jbc.M404884200. [DOI] [PubMed] [Google Scholar]
- 24.Toogood HS, Van Thiel A, Scrutton NS, Leys D. Stabilization of non-productive conformations underpins rapid electron transfer to electron-transferring flavoprotein. J. Biol. Chem. 2005;280:30361–30366. doi: 10.1074/jbc.M505562200. [DOI] [PubMed] [Google Scholar]
- 25.Mohsen A-WA, Anderson B, Volchenboum S, Battaile K, Tiffany K, Roberts D, Kim J-J, Vockley J. Characterization of molecular defects in isovaleryl-CoA dehydrogenase in patients with isovaleric acidemia. Biochemistry. 1998;37:10325–10335. doi: 10.1021/bi973096r. [DOI] [PubMed] [Google Scholar]
- 26.Nasser I, Mohsen A-W, Jelesarov I, Vockley J, Macheroux P, Ghisla S. Thermal unfolding of medium-chain acyl-CoA dehydrogenase and iso(3)valeryl-CoA dehydrogenase: study of the effect of genetic defects on enzyme stability. Biochim. Biophys. Acta. 2004;1690:22–32. doi: 10.1016/j.bbadis.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Tiffany KA, Roberts DL, Wang M, Paschke R, Mohsen A-WA, Vockley J, Kim J-JP. Structure of human isovaleryl-CoA dehydrogenase at 2.6 angstrom resolution - basis for substrate specificity. Biochemistry. 1997;36:8455–8464. doi: 10.1021/bi970422u. [DOI] [PubMed] [Google Scholar]
- 28.Mohsen A-WA, Vockley J. High-level expression of an altered cDNA encoding human isovalery-CoA dehydrogenase in Escherichia coli. Gene. 1995;160:263–267. doi: 10.1016/0378-1119(95)00256-6. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen T, Riggs C, Babovic-Vuksanovic D, Kim YS, Carpenter JF, Burghardt TP, Gregersen N, Vockley J. Purification and Characterization of two polymorphic variants of short chain acyl-CoA dehydrogenase reveal reduction of catalytic activity and stability of the Gly185Ser enzyme. Biochemistry. 2002;41:11126–11133. doi: 10.1021/bi026030r. [DOI] [PubMed] [Google Scholar]
- 30.Ibrahim NE, Mohsen A-W. Biochemical studies on recombinant human isobutyryl-CoA dehydrogenase Amer. J. Biochem. Biotech. 2011;7:84–89. [Google Scholar]
- 31.He M, Burghardt TP, Vockley J. A novel approach to the characterization of substrate specificity in short/branched chain Acyl-CoA dehydrogenase. J Biol. Chem. 2003;278:37974–37986. doi: 10.1074/jbc.M306882200. [DOI] [PubMed] [Google Scholar]
- 32.Vockley J, Mohsen A-W, Binzak B, Willard J, Fauq A. Mammalian branched-chain acyl-CoA dehydrogenases: molecular cloning and characterization of recombinant enzymes. Methods Enzymol. 2000;324:241–258. doi: 10.1016/s0076-6879(00)24236-5. [DOI] [PubMed] [Google Scholar]
- 33.Mohsen A-WA, Vockley J. Identification of the active site catalytic residue in human isovaleryl-CoA dehydrogenase. Biochemistry. 1995;34:10146–10152. doi: 10.1021/bi00032a007. [DOI] [PubMed] [Google Scholar]
- 34.McKean MC, Frerman FE, Mielke DM. General acyl-CoA dehydrogenase from pig liver. Kinetic and binding studies. J. Biol. Chem. 1979;254:2730–2735. [PubMed] [Google Scholar]
- 35.Kormanik K, Kang H, Cuebas D, Vockley J, Mohsen A-W. Evidence for involvement of medium chain acyl-CoA dehydrogenase in the metabolism of phenylbutyrate. Mol. Genet. Met. 2013;107:684–689. doi: 10.1016/j.ymgme.2012.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Battaile K, Molin-Case J, Paschke R, Wang M, Bennett D, Vockley J, Kim J-JP. Crystal structure of rat short chain acyl-CoA dehydrogenase complexed with acetoacetyl-CoA; comparison with other acyl-CoA dehydrogenases. J. Biol. Chem. 2002;277:12200–12207. doi: 10.1074/jbc.M111296200. [DOI] [PubMed] [Google Scholar]
- 37.Beckmann JD, Frerman FE. The effects of pH, ionic strength, and chemical modification on the reaction of the electron transfer flavoprotein with an acyl-CoA dehydrogenase. J. Biol. Chem. 1983;258:7563–7569. [PubMed] [Google Scholar]
- 38.Williamson G, Engel PC, Mizzer JP, Thorpe C, Massey V. Evidence that the greening ligand in native butyryl-CoA dehydrogenase is a CoA persulfide. J. Biol. Chem. 1982;257:4314–4320. [PubMed] [Google Scholar]
- 39.Tarnowski GS, Barclay RK, Mountain IM, Nakamura M, Satterwhite HG, Solney EM. Determination of acid-soluble thiols and disulfides in transplanted animal tumors Arch. Biochem. Biophys. 1965;110:210–216. doi: 10.1016/0003-9861(65)90176-1. [DOI] [PubMed] [Google Scholar]
- 40.Williamson G, Engel PC. A convenient and rapid method for the complete removal of CoA from butyryl-CoA dehydrogenase. Biochim. Biophys. Acta. 1982;706:245–248. doi: 10.1016/0167-4838(82)90493-9. [DOI] [PubMed] [Google Scholar]
- 41.Mohsen A-WA, Vockley J. Biochemical characteristics of recombinant human isovaleryl-CoA dehydrogenase pre-treated with ethylenediaminetetraacetate. In: Ghisla S, Kroneck P, Macheroux P, Sund H, editors. Flavins and Flavoproteins 1999. New York: Rudolf Weber; 1999. pp. 515–558. [Google Scholar]
- 42.Battaile KP, Nguyen TV, Vockley J, Kim J-JP. Structures of isobutyryl-CoA dehydrogenase and enzyme-product complex: comparison with isovaleryl- and short-chain acyl-CoA dehydrogenases. J Biol. Chem. 2004;279:16526–16534. doi: 10.1074/jbc.M400034200. [DOI] [PubMed] [Google Scholar]
- 43.Bross P, Engst S, Strauss AW, Kelly DP, Rasched I, Ghisla S. Characterization of wildtype and an active site mutant of human medium-chain acyl-CoA dehydrogenase after expression in E coli . J Biol. Chem. 1990;265:7116–7119. [PubMed] [Google Scholar]
- 44.Djordjevic S, Pace CP, Stankovich MT, Kim J-JP. Three-dimensional structure of butyryl-CoA dehydrogenase from Megasphaera esdenii . Biochemistry. 1995;34:2163–2171. doi: 10.1021/bi00007a009. [DOI] [PubMed] [Google Scholar]
- 45.Ghisla S, Braunwrath A, Vock P. pH and substrate chain length dependence of activity of "short-chain"-, "medium-chain"-and "long-chain"-acyl-CoA dehydrogenase. In: Stevenson Kenneth J, Williams Charles Haddon, Massey Vincent., editors. Flavin and Flavoproteins 1996. University of Calgary Press; 1996. pp. 629–632. [Google Scholar]
- 46.Rupp H, Zarain-Herzberg A, Maisch B. The use of partial fatty acid oxidation inhibitors for metabolic therapy of angina pectoris and heart failure. Herz. 2002;27:621–636. doi: 10.1007/s00059-002-2428-x. [DOI] [PubMed] [Google Scholar]
- 47.Wang WZ, Fu ZJ, Zhou JZ, Kim JJP, Thorpe C. Interaction of 3,4-dienoyl-CoA thioesters with medium chain acyl-CoA dehydrogenase: stereochemistry of inactivation of a flavoenzyme. Biochemistry. 2001;40:12266–12275. doi: 10.1021/bi0109818. [DOI] [PubMed] [Google Scholar]
- 48.Von Holt C. Biochim. Biophys. Acta Methylenecyclopropaneacetic acid, a metabolite of hypoglycin. 1966;125:1–10. doi: 10.1016/0005-2760(66)90138-x. [DOI] [PubMed] [Google Scholar]
- 49.Tanaka K, Kean E, Johnson B. Jamaican vomiting sickness: Biochemical investigation of two cases. N Eng. J. Med. 1976;295:461–467. doi: 10.1056/NEJM197608262950901. [DOI] [PubMed] [Google Scholar]
- 50.Tanaka K. On the mode of action of hypoglycin A. 3. Isolation and identification of cis-4-decene-1,10-dioic, cis, cis-4,7-decadiene-1,10-dioic, cis-4-octene-1,8-dioic, glutaric, and adipic acids N-(methylenecyclopropyl)acetylglycine, and N-isovalerylglycine from urine of hypoglycin A-treated rats. J. Biol. Chemistry. 1972;247:7465–7478. [PubMed] [Google Scholar]
- 51.Tanaka K, Miller EM, Isselbacher KJ. Isovaleric and alpha methylbutyric acidemias induced by hypglycin A: mechanism of Jamaican vomiting sickness. Proc. Natl. Acad. Sci. USA. 1971;68:20–24. doi: 10.1126/science.175.4017.69. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka K, Shih B. Isovaleric and alpha methylbutyric acidemias induced by hypglycin A: mechanism of Jamaican vomiting sickness. Science. 1972;175:69–71. doi: 10.1126/science.175.4017.69. [DOI] [PubMed] [Google Scholar]