Summary
Background
A selective susceptibility of certain individuals to form multiple alloantibodies in response to red cell transfusion is well-recognized in clinical practice, and is a particular problem in persons with sickle cell disease (SCD). The reason for this differential susceptibility is unclear, but inter-individual genetic differences are likely to contribute.
Methods
We conducted a pilot case-control genome-wide association study using 1,000,000 SNPs in 94 alloimmune responders (cases) and non-responders (controls) with SCD in order to identify loci of large effect size associated with alloimmunization.
Results
No loci showed evidence of association at a genome-wide significance cut-off (p < 0.5 × 10–8). SNPs in the ARAP1/STARD10 region showed suggestive association (p < 1 × 10–6), but no association was observed at previously implicated loci TRIM21 or HLA. In analyses of the number of accumulated antibodies, a modest association was found with SNPs in the Toll-like receptor gene TLR10 (p < 1 × 10–4).
Conclusions
Alloimmunization in persons with SCD is unlikely to be mediated by loci of very large effect size; however, larger and more comprehensive studies are required to fully evaluate loci with more moderate effects. This study provides a working approach to such future studies in SCD.
Keywords: Genome-wide association studies, GWAS, African American, Responders, Genomics
Introduction
Transfusion of allogenic blood products is an essential part of modern medicine; however, this process is also an immunologic challenge to the recipient, who is exposed to multiple foreign antigens as a consequence. In the setting of red cell transfusions, this exposure sets the stage for the development of alloantibodies, the presence of which creates the considerable challenge of providing compatible blood products while avoiding the attendant problem of hemolytic transfusion reactions [1]. Given that some 5 million patients receive blood products every year in the USA, including those who receive multiple transfusions as a consequence of inherited or acquired hematological disorders, post-transfusion alloimmunization is a problem of major public health significance [2].
In clinical practice, it is well established that some individuals will form multiple antibodies with almost each transfusion; such ‘responders’ appear immunologically distinct from ‘nonresponders’ for whom the development of antibodies is a rare and unusual occurrence. This phenotypic dichotomy of ‘responders’ and ‘non-responders’ is commonly observed in persons with sickle cell disease (SCD), who often require red cell transfusions both in the acute setting of sickle crises and for their long-term disease management. The rate of red cell alloimmunization in SCD patients is empirically quoted in the range of 30–50% [3, 4] – several orders of magnitude higher than the ˜5% figure observed in the general population [2, 5]. Alloimmune responders with SCD are also at increased risk of hyperhemolysis [6], an uncommon but potentially devastating type of delayed transfusion reaction.
The reason some individuals become responders while others do not is not well-understood, but is likely to be multifactorial, involving donor, recipient, and external factors [5, 6, 7]. For instance, there is support for genetic ancestry contributing to alloimmunization in SCD patients – the genetic ancestry of individuals with SCD is predominantly African American, whereas blood donors of similar ethnicity are underrepresented in the national donor pool [7, 8, 9, 10]. This potentiates the mismatches between donor and recipient red cell antigens that are necessary to stimulate an alloimmune response. Accordingly, expanded red cell antigen phenotyping and closer matching of common antigens between donor and recipient has decreased the incidence of alloimmunization in susceptible groups [11, 12], but a significant rate of alloimmunization remains, strongly implicating other mechanisms in the pathogenesis [13].
Among the potential additional factors is the well-known genetic variation that exists between individuals; for instance, possession of human leukocyte antigen (HLA) types HLA-B35 and HLA-Cw4 alleles have been linked to ‘responder’ status in persons with SCD [14, 15], and a single nucleotide polymorphism (SNP) in the erythrocyte antigen Ro52 (TRIM21) has been associated with the induction and efficiency of alloimmunization in both a transfusion-dependent and age-dependent fashion in patients with SCD [16]. These and other similar reports [17] validate a genetic underpinning to the alloimmune response, but they do so in a disparate, ad hoc fashion, and the primary results have not been replicated in other cohorts.
Advances in microarray technologies designed to interrogate SNPs across the genome have made it possible to perform genome-wide association studies (GWAS). The GWAS approach has emerged as a powerful way of agnostically identifying disease susceptibility loci and has facilitated the identification of biologically important loci in multiple diseases across a range of disciplines [18, 19] including disease severity in SCD [20, 21]. Many of the associations with the greatest magnitudes of effect have occurred in diseases with an immune or autoimmune basis [22, 23, 24, 25] or in disorders with a strong gene-environment component [26, 27, 28]. These associations have fundamentally changed our understanding of the underlying disease processes [29] and in some cases have facilitated clinically useful genetic testing [30, 31]. Therefore, GWAS are an attractive and robust way of identifying biologically relevant genes and gene pathways contributing to the alloimmune responder phenotype in individuals with SCD; this in turn holds the ultimate promise of new therapeutic targets for those at higher risk. Further, because the GWAS approach includes the identification of both susceptibility and protection alleles, relevant loci could directly impact the ability of blood centers to preemptively identify both susceptible recipients for whom extended phenotyping is especially indicated as well as likely non-responders to whom random units can be provided, thereby increasing the number of donors who are available to supply phenotype-matched blood to SCD patients.
As a first foray into alloimmunization genomics, we undertook a pilot case-control GWAS of alloimmune responder status in a cohort of multiply transfused participants with SCD, with the express goal of identifying large-effect susceptibility loci. The genome-wide coverage achieved also allowed us to evaluate previously associated loci and to perform subgroup analyses of allo-antibody accumulation.
Material and Methods
Subjects
The study protocol was approved by the Institutional Review Board of St Luke's Episcopal Hospital and of Baylor College of Medicine. DNA samples used were those taken from patients with a diagnosis of SCD referred to LifeShare for blood group genotyping prior to receiving a red cell transfusion. DNA remaining after completion of the clinical test was retained and de-identified. As these clinical refuse samples were fully de-identified, only limited clinical data was available and the total (or lifetime) number of transfusion units for each individual is unknown; however, all patients had to have received a minimum of two transfusions in the past to be included. Each patient's hemoglobinopathy status was confirmed by genotyping. Samples were obtained from within the LifeShare service area which includes Southeast Texas, the northern half of Louisiana, and Southern Arkansas. Patients were also screened for antibodies using routine serologic techniques. This included incubations at room temperature and 37 ° C using Lo-Ion (Immucor, Norcross, GA, USA) and an AHG phase using polyspecific antiglobulin reagent. Patients were characterized as ‘responders’ (N = 198) if they developed clinically significant alloantibodies after two or more packed red cell transfusions, where clinically significant was defined as ‘an antibody that is frequently associated with HDFN (Hemolytic Disease of the Fetus or Newborn), hemolytic transfusion reactions, or a notable decrease in the survival of transfused RBCs’ [32]. ‘Non-responders’ (N = 186) were those with no alloantibodies after a similar exposure (table 1). We obtained funding to analyze 109 of these DNA samples in the first phase pilot GWAS – 55 ‘responders’ and 54 ‘non-responders’ homozygous for the sickle cell (HbS) mutation. The responder group also included 17 persons with a history of hyperhemolysis syndrome, defined as a post-transfusion drop in hemoglobin below pre-transfusion levels without evidence of bleeding.
Table 1.
Cohort (n = 384) and pilot study demographics
| Cohort | 384 |
| Alloimmune category | |
| Non-responder | 186 |
| Responder | 198 |
| Alloantibodies, median (min, max, IQR) Gender | 2 (1, 11, 1–4) |
| Male | 165 |
| Female | 211 |
| Unknown | 8 |
| Sickle genotype | |
| HbSS | 373 |
| HbSC | 11 |
| Age group | |
| Adult (>18 years) | 346 |
| Child (<18 years) | 36 |
| Unknown | 2 |
| Genotyped individuals | 104 |
| Alloimmune category | |
| Non-responder (males) | 54 (23) |
| Responder (males) | 50 (20) |
| Alloantibodies, median (min, max, IQR) Sickle genotype | 4 (1, 11, 2–5) |
| HbSS | 104 |
IQR = Interquartile range.
DNA Extraction and Genotyping
DNA was extracted from leukocyte buffy coats or whole blood using either a manual method (PureGene®; Qiagen, Venlo, the Netherlands) or the QIACube® (Qiagen). The quantity of double-stranded DNA was assessed in each sample using the PicoGreen (Invitrogen, Carlsbad, CA, USA) assay. All assays were performed according to manufacturer's specifications. Five samples had a DNA concentration below the 50 vg/μl concentration recommended for optimal microarray genotyping and were omitted from further study.
Genotyping was performed according to manufacturer's instructions using the Bead-Station system and HumanOmni1-Quad Bead Chips (Illumina, San Diego, CA, USA), which was specifically designed to provide improved SNP coverage in people of West African heritage, as might be expected for participants with SCO. The GenomeStudio software (Illumina) was used to interpret normalized fluorescent intensities as genotypes. HumanOmni1-Quad BeadChips contain approximately 1,140,419 SNPs. SNPs with genotyping efficiency < 90% or out of Hardy-Weinberg equilibrium in controls (at the p < 10–5 level) were removed. The final analysis incorporated genotype data from 1,008,655 SNPs.
Sample Quality Control
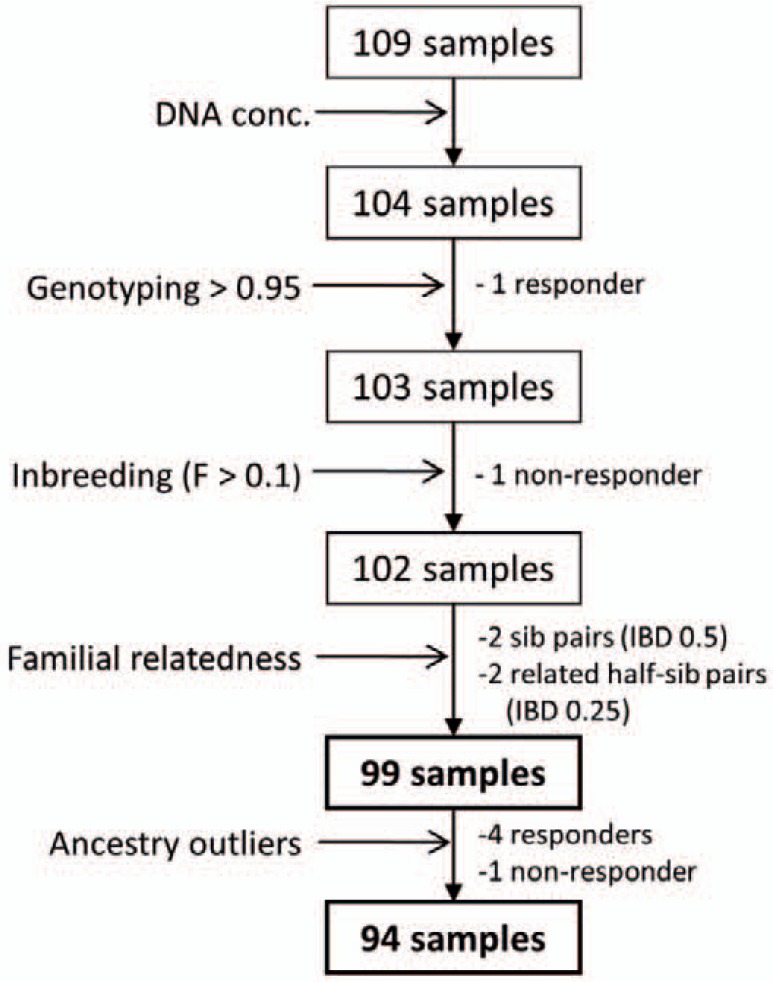
Known siblings or other related samples were excluded from the initial sample selection. In addition, samples with more than 5% missing data (genotyping efficiency < 0.95) or with evidence of excessive inbreeding (inbreeding coefficient F > 0.1) were removed. Pairs of samples with excess allele sharing suggestive of close familial relationships (parent-offspring, siblings, or 1st degree relatives; PI_HAT > 0.1) were identified by identity-by-descent (IBD); in such cases one member of the pair was removed from analysis (fig. 1).
Fig. 1.
Sample quality control (QC); Samples were removed from analysis if they i) failed to meet the minimum DNA concentration for the assay according to manufacturer's recommendations (DNA conc); ii) were successfully genotyped at less than 95% of included SNPs (Genotyping); iii) had an inbreeding coefficient (F) suggestive of an excess of homozygotes; iv) had evidence of close-familial pairing (identify by-decent (IBD) or were ancestry population outliers on principal components analysis (see fig. 2).
Identification of Population Stratification
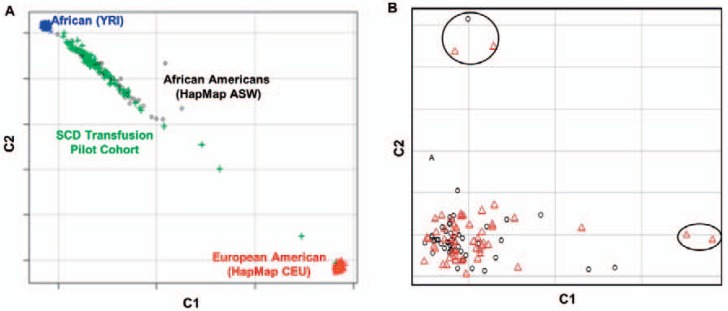
Prior to analysis, the remaining individuals were evaluated for shared ancestry using multidimensional scaling (MDS). The principle of MDS is to cluster individuals of similar ancestry by repeatedly adjusting individual genotypes by the amounts attributable to ancestry. The resulting dimensions can be plotted to visualize individuals whose inferred ancestry is outside that of the sample. Clustering was performed with a subset of SNPs pruned using a sliding window to exclude SNPs with strong nonrandom association (linkage disequilibrium; LD) with surrounding SNPs (r2 > 0.2). Five outliers were identified using this approach (fig. 2). Exclusion of these outliers resulted in a final cohort of 48 ‘responder’ cases and 46 ‘non-responder’ controls.
Fig. 2.
Multidimensional scaling (MDS) cluster plot of study participants for principal components C1 and C2. A Clustering of SCD pilot cohort with HapMap African Americans from Oklahoma and in relation to major continental HapMap groups (YRI = Yoruba from Nigeria; CEU = Ceph from Utah). B MDS clustering of cases (black circles) and controls (red triangles) in the pilot cohort. PCA study outliers are circled.
Statistical Analyses
MDS clustering, IBD analysis, and F statistics (inbreeding) were carried out using PLINK [33], as were basic statistical analyses (χ2 and permutation). After primary analysis, label swapping adaptive permutation was performed for each SNP. In label swapping, the case/control label is swapped for each individual, and the association statistic is derived multiple times to create a new null distribution with which to compare with the observed data. The ‘adaptive’ mode essentially curtails the run length by removing SNPs that are highly unlikely to attain statistically significance at each permutation, until a user-defined maximum number of permutations (in this case 1,000,000) is reached for SNPs that continue to be statistically significant. Genome-wide inflation was estimated using the genABEL package [34] in R. Manhattan and MDS plots were generated using a webbased R source code (http://GettingGeneticsDone.com) and LocusZoom [35]. Shapeit2 [36] and Impute-2 [37] were used to impute genotype data from the HapMap [38] and the 1000 Genomes Project Consortium [39]. Imputation uses derived patterns of population-specific LD between SNPs to infer genotypes at sites that have not directly been genotyped. This can be a particularly useful approach in populations of African ancestry, where genotyped SNPs may not provide true ‘genome-wide’ coverage.
In subgroup analyses of the cumulative number of antibodies developed among responders, the distribution of alloantibodies was slightly skewed to the right. A power transformation (x1/4) was performed to better fit a normal distribution. Unadjusted linear regression models were fit to the data to derive beta statistics and p values.
Quanto 1.2.4 [40] was used to estimate study power. The study sample size employed had 80% power to determine genetic risk ratios (GRRs) greater than 10 at genome-wide significance (p < 0.5 × 10–8) using SNPs with minor allele frequency (MAF) of 0.15 or greater under an additive disease model.
Results
No Large-Effect Responder Loci, but Suggestive Association in ARAP1/STARD10
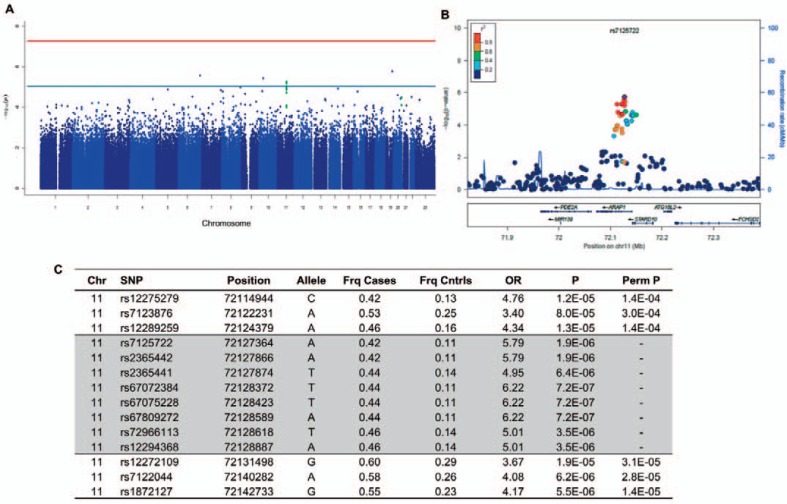
For our primary analysis, each SNP with MAF > 0.10 was tested for association (χ2) under an allelic trend model using 48 responders as cases and 46 non-responders as controls. The genome-wide inflation factor in this analysis was 0.985, suggesting little residual inflation of the test statistic from genotyping error, cryptic relatedness of individuals, or population stratification. We did not observe any SNPs or clusters of SNPs at or below the accepted threshold for statistical significance in genome-wide association studies (p < 0.5 × 10–8) (fig. 3a).
Fig. 3.
Case control GWAS of responder status. A Manhattan plot showing no SNPs below genome-wide significance (red line, p = 0.5 × 10–8), but suggestive SNPs (blue line, p = 1 × 10–4) in the ARAP1/STARD10 gene region (green dots). B Local Manhattan plot of 250 kb region around ARAP1/ STARD10 including genotyped and imputed SNPs. Pairwise LD (r2) is derived from the HapMap YRI population. C GWAS results for genotyped and imputed (shaded) SNPs in the ARAP1/STARD10 region. OR = Odds ratio; Perm P = permuted p value.
Next, we sought to evaluate SNPs with p values that were below the ‘background’ association of most SNPs, but did not meet genome-wide significance. We focused on regions of the genome that had more than one SNP within 100 kb with p < 1 × 10–4; this threshold was deemed as ‘suggestive’ association. One region met this criteria (fig. 3a) – a cluster of three SNPs (lowest p = 1.2 × 10–5) on the long arm of chromosome 11 at the 3 end of ARAP1, extending 5 of STARD10 (fig. 3c). This modest association persisted after permutation testing (lowest p = 1.4 × 10–4). Manual inspection of the genotyping clusters and the relative local LD pattern in the HapMap YRI from Yoruba, Nigeria [38], suggested that technical (genotyping) error was unlikely to be a significant contributing factor.
Having observed moderately suggestive association with SNPs across this region, we then performed statistical imputation of ˜17,983 variants within in a 5 megabase (MB) region centered upon the SNP with the best association in the original genotyping. We then filtered for common variants (MAF > 0.05) with imputation certainty greater than 95%. This resulted in a final test set of 3,795 SNPs. Using a χ2 test for allelic trend under the same case-control model, five additional SNPs showed similar and, in some cases, stronger association than the genotyped SNPs at this candidate locus (fig. 3b, c), although none reached genome-wide significance.
No Association with Responder Status at Previously Reported Loci
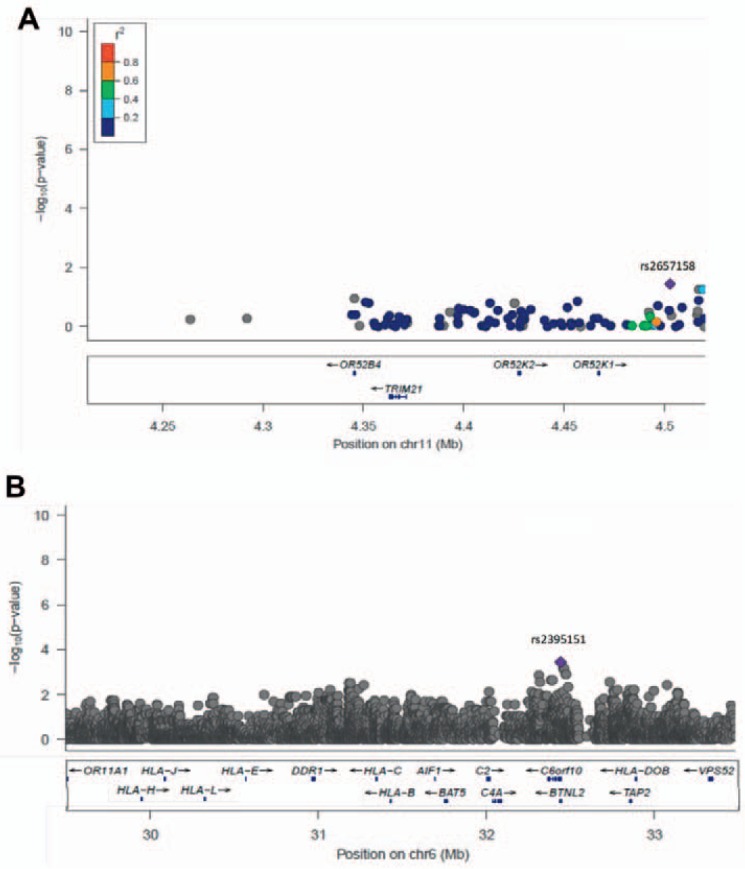
Next, we focused on two regions that have been previously associated with the development of alloantibodies in other studies – TRIM21 on chromosome 11 and the major histocompatibility complex (MHC) on chromosome 6. At TRIM21, none of the genotyped SNPs had a p value less than 10–3 (fig. 4a). A similar pattern was observed across the4 MB of the MHC; most SNPs had p values greater than 1 × 10–3 (fig. 4b), with the strongest SNP association (p < 0.0004) being found in c6orf10, upstream of the MHC class II region some distance from the major HLA types previously associated with the phenotype.
Fig. 4.
Local Manhattan plots for previously associated loci TRIM21 (A) and the MHC (B). In each figure the highlighted SNP (purple diamond) is the strongest association in the region. In A pairwise LD (r2) is derived from the Hapmap YRI population. In B the gene list is restricted for space considerations.
Subgroup Analysis of Alloantibody Accumulation Reveals an Additional Suggestive Locus
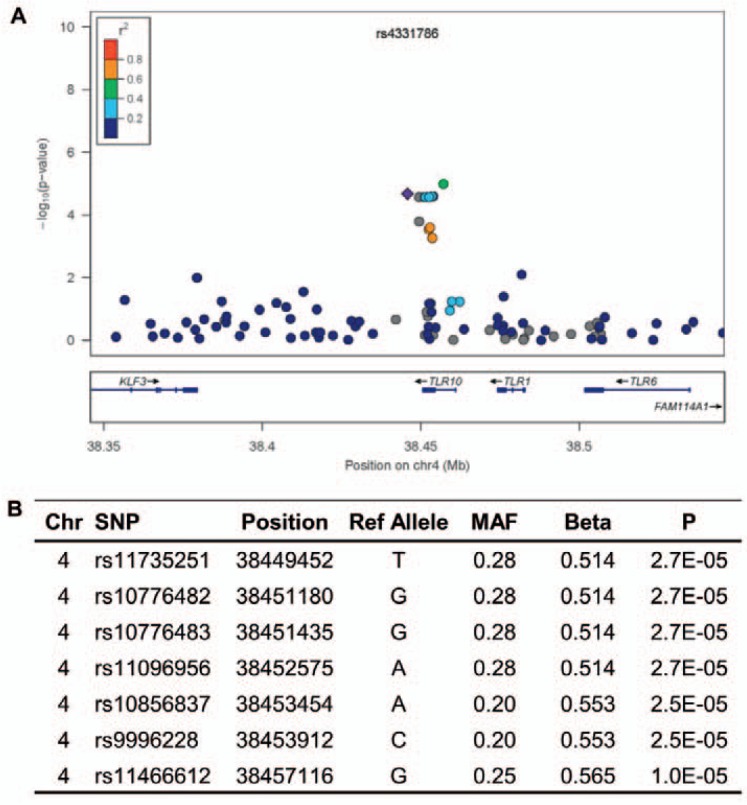
As an exploratory subgroup analysis, we incorporated a quantitative approach that focused on SNPs influencing the number of antibodies formed by each responder (N = 48) using unadjusted linear regression models. In this analysis there were no SNPs with evidence of association at or below genome-wide significance; however, using the same criteria for suggestive loci employed in our primary analysis, three regions had more than two SNPs with p < 1 × 10–4 within 100 kb. The most convincing of these included a cluster of eight SNPs in an 11 kb region on chromosome 4p involving the Toll-receptor 10 (TLR10) gene (fig. 5).
Fig. 5.
Suggestive association between SNPs across TLR10 and number of alloantibodies formed. A Local Manhattan plot of SNPs within 250 kb of TLR10. B SNPs showing suggestive association.
Discussion
We did not observe any SNP associations with responder status at or below genome-wide significance in our cohort. In the small sample size of this pilot study, genome-wide significance represents a conservative cut-off p value below which false-positive results are highly unlikely. Thus, we can conclude that in this SCD cohort, becoming an alloimmune responder is not influenced by a locus with very large effect size (GRR > 10). Like many other complex traits, the contribution of common genetic variation to responder status is more likely to be the result of loci with more moderate effects. This class of effects is difficult to confidently discern in a pilot GWAS such as this one, and is further compounded by the African ancestry of the cohort. African populations have greater genetic diversity and less LD between SNPs. Because genome-wide genotyping will rarely, if ever, genotype the true causal variant, GWAS studies are dependent upon LD for genotyped SNPs to act as ‘proxies’ for ungenotyped SNPs. The relative lack of LD in African and African-derived populations thus makes finding more moderate association signals using conventional microarray genotyping platforms more difficult [41], despite the availability of statistical imputation. Recent interest in the development of Afro-centric SNP genotyping microarray chips should make future GWAS in the African-American SCD population more robust.
The use of a less conservative cut-off allowed us to evaluate evidence for loci with more moderate effects on the phenotype, although such SNPs have a higher possibility of being false-positives. A cluster of SNPs spanning ARAP1 and the adjacent STARD10 genes showed suggestive association with responder status in our cohort. ARAP1 (MIM# 606646) encodes a Rho/Arf GTPase-activating protein (GAP) postulated to play a role in regulating endocytic trafficking of epidermal growth factor receptors (EGFRs) to the cell membrane. STARD10 (Gene ID:10809) encodes one of a group of proteins characterized by incorporation of a steroidogenic acute regulatory protein(StAR)-related lipid transfer (START) domain that binds specific lipid motifs and implicated in the control of lipid biology and cell trafficking. The related STARD11 protein interacts with the Goodpasture antigen and is known to play a role in the autoimmune response seen in Goodpasture syndrome [42]; there is thus a potential, albeit speculative, biological role for STARD10 in the processing and presentation of red cell membrane antigens. Larger, well-powered replication studies, utilizing more comprehensive genotyping platforms that are better suited to the African American population, are necessary to validate these findings.
In keeping with recent murine studies [43], our findings suggest that there is little evidence to support a major role for variants in TRIM21 in responder status. Our sample size was larger than that employed in previous studies; however, our study was not designed to look for more subtle determinants or known covariates of alloimmunization (e.g. age or transfusion dependency) and thus we could not confirm the postulated role of the rs660 SNP in this cohort. It should be noted that rs660 is not represented on the HumanOmni1-quad chip, but also maps to several sites in the current version of the human genome reference sequence (www.ensembl.org). The fact that rs660-flanking sequences do not map uniquely in the current genome assemblies raises the possibility that technical factors may underlie the previously reported association. Similarly, we did not find strong evidence for association with SNPs across the MHC in our analyses of this cohort. This would be in line with earlier studies suggesting that restricted HLA allelotypes may not play a significant role in susceptibility to alloimmune transfusion tolerance [44]. However, because our pilot study did not include extended haplotype analyses of the MHC and current genotyping microarrays do not necessarily fully capture African diversity at the MHC, this result does not completely preclude a role for the MHC in the responder phenotype.
In subgroup analysis we employed a quantitative approach to look at potential genetic factors influencing the development of alloantibodies. A quantitative approach for continuous and ordinal traits has increased power to detect association in genetic studies and consequently generally requires a smaller sample size to find similar effect sizes. This approach has been successfully employed in other GWAS [45, 46], including the association of SNPs in BCL11A with fetal hemoglobin levels in SCD patients, which was accomplished with a few hundred cases and controls [20]. We did not find evidence for large-effect loci controlling the development of multiple antibodies; however, several SNPs around TLR10 showed modest association with increasing alloantibodies. Toll-like receptors (TLRs) have a well-established role in the early immune response to invading pathogens, although a specific role for the TLR10 protein has not yet been identified. Recently, murine models of cardiac and skin transplantation have implicated TLRs in the alloimmunity of graft rejection [47], and polymorphisms in TLR4 have been associated with lung transplant rejection in humans [48]. There is thus growing interest in the role of innate immunity and TLRs in alloimmunization. This again makes it imperative that larger and more comprehensive genomics studies are undertaken to replicate the preliminary results presented here.
This pilot GWAS of alloimmune responder status and alloantibody number did not reveal evidence of a large-effect locus in this SCD cohort, including previously identified loci TRIM21 and the MHC. However, suggestive association at the putative candidate loci ARAP1/STARD10 and TLR10 advocate for future genomics studies that will not only evaluate these preliminary results but also engender novel approaches to understanding differential alloantibody susceptibility in SCD. A genomics approach to the problem has recently been endorsed as a viable framework in which to study various aspects of the SCD responder phenotype [49]. The challenge demonstrated by this study is that success in such a venture will require larger cohorts built around a broad expertise and strong collaboration.
Disclosure Statement
JMM serves is currently an employee of Grifols Inc. and previously served as the principal investigator for grants and research agreements between LifeShare and Novartis Diagnostics or BioArray/Immucor. No other authors have conflicts to declare.
Acknowledgements
This work was funded in part by a grant from Gulf Coast Regional Blood Center to AC. NH is supported by a Clinical Scientist Development Award from the Doris Duke Charitable Foundation. We would also like to thank Kristine Bucasas for help with implementing the statistical imputation.
References
- 1.Schonewille H, Haak HL, van Zijl AM. Alloimmunization after blood transfusion in patients with hematologic and oncologic diseases. Transfusion. 1999;39:763–771. doi: 10.1046/j.1537-2995.1999.39070763.x. [DOI] [PubMed] [Google Scholar]
- 2.Heddle NM, Soutar RL, O'Hoski PL, Singer J, McBride JA, Ali MA, Kelton JG. A prospective study to determine the frequency and clinical significance of alloimmunization post-transfusion. Br J Haematol. 1995;91:1000–1005. doi: 10.1111/j.1365-2141.1995.tb05425.x. [DOI] [PubMed] [Google Scholar]
- 3.Rosse WF, Gallagher D, Kinney TR, Castro O, Dosik H, Moohr J, Wang W, Levy PS. Transfusion and alloimmunization in sickle cell disease. The Cooperative Study of Sickle Cell Disease. Blood. 1990;76:1431–1437. [PubMed] [Google Scholar]
- 4.Aygun B, Padmanabhan S, Paley C, Chandrasekaran V. Clinical significance of RBC alloantibodies and autoantibodies in sickle cell patients who received transfusions. Transfusion. 2002;42:37–43. doi: 10.1046/j.1537-2995.2002.00007.x. [DOI] [PubMed] [Google Scholar]
- 5.Bauer MP, Wiersum-Osselton J, Schipperus M, Vandenbroucke JP, Briet E. Clinical predictors of alloimmunization after red blood cell transfusion. Transfusion. 2007;47:2066–2071. doi: 10.1111/j.1537-2995.2007.01433.x. [DOI] [PubMed] [Google Scholar]
- 6.Talano JA, Hillery CA, Gottschall JL, Baylerian DM, Scott JP. Delayed hemolytic transfusion reaction/hyperhemolysis syndrome in children with sickle cell disease. Pediatrics. 2003;111:e661–665. doi: 10.1542/peds.111.6.e661. [DOI] [PubMed] [Google Scholar]
- 7.Luban NL. Variability in rates of alloimmunization in different groups of children with sickle cell disease: effect of ethnic background. Am J Pediatr Hematol Oncol. 1989;11:314–319. [PubMed] [Google Scholar]
- 8.Vichinsky EP, Earles A, Johnson RA, Hoag MS, Williams A, Lubin B. Alloimmunization in sickle cell anemia and transfusion of racially unmatched blood. N Engl J Med. 1990;322:1617–1621. doi: 10.1056/NEJM199006073222301. [DOI] [PubMed] [Google Scholar]
- 9.Olujohungbe A, Hambleton I, Stephens L, Serjeant B, Serjeant G. Red cell antibodies in patients with homozygous sickle cell disease: a comparison of patients in Jamaica and the United Kingdom. Br J Haematol. 2001;113:661–665. doi: 10.1046/j.1365-2141.2001.02819.x. [DOI] [PubMed] [Google Scholar]
- 10.Murphy EL, Shaz B, Hillyer CD, Carey P, Custer BS, Hirschler N, Fang J, Schreiber GB. Minority and foreign-born representation among us blood donors: Demographics and donation frequency for 2006. Transfusion. 2009;49:2221–2228. doi: 10.1111/j.1537-2995.2009.02271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ness PM. To match or not to match: the question for chronically transfused patients with sickle cell anemia. Transfusion. 1994;34:558–560. doi: 10.1046/j.1537-2995.1994.34794330007.x. [DOI] [PubMed] [Google Scholar]
- 12.Vichinsky EP, Luban NL, Wright E, Olivieri N, Driscoll C, Pegelow CH, Adams RJ. Prospective RBC phenotype matching in a stroke-prevention trial in sickle cell anemia: a multicenter transfusion trial. Transfusion. 2001;41:1086–1092. doi: 10.1046/j.1537-2995.2001.41091086.x. [DOI] [PubMed] [Google Scholar]
- 13.Chou ST, Jackson T, Vege S, Smith-Whitley K, Friedman DF, Westhoff CM. High prevalence of red blood cell alloimmunization in sickle cell disease despite transfusion from Rh-matched minority donors. Blood. 2013;122:1062–1071. doi: 10.1182/blood-2013-03-490623. [DOI] [PubMed] [Google Scholar]
- 14.Alarif L, Castro O, Ofosu M, Dunston G, Scott RB. HLA-B35 is associated with red cell alloimmunization in sickle cell disease. Clin Immunol Immunopathol. 1986;38:178–183. doi: 10.1016/0090-1229(86)90136-4. [DOI] [PubMed] [Google Scholar]
- 15.Noizat-Pirenne F, Tournamille C, Bierling P, Roudot-Thoraval F, Le Pennec PY, Rouger P, Ansart-Pirenne H. Relative immunogenicity of Fya and K antigens in a Caucasian population, based on HLA class II restriction analysis. Transfusion. 2006;46:1328–1333. doi: 10.1111/j.1537-2995.2006.00900.x. [DOI] [PubMed] [Google Scholar]
- 16.Tatari-Calderone Z, Minniti CP, Kratovil T, Stojakovic M, Vollmer A, Barjaktarevic I, Zhang E, Hoang A, Luban NL, Vukmanovic S. Rs660 polymorphism in Ro52 (SSA1 TRIM21) is a marker for age-dependent tolerance induction and efficiency of alloimmunization in sickle cell disease. Mol Immunol. 2009;47:64–70. doi: 10.1016/j.molimm.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Tatari-Calderone Z, Tamouza R, Le Bouder GP, Dewan R, Luban NL, Lasserre J, Maury J, Lionnet F, Krishnamoorthy R, Girot R, Vukmanovic S. The association of CD81 polymorphisms with alloimmunization in sickle cell disease. Clin Dev Immunol. 2013;2013:937846. doi: 10.1155/2013/937846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hindorff LA, MacArthur J, Morales J, Junkins HA, Hall PN, Klemm AK, Manolio TA. A Catalog of Published Genome-Wide Association Studies. 2011. www.genome.gov/gwastudies/ Accessed May 27, 2011.
- 19.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhatnagar P, Purvis S, Barron-Casella E, Debaun MR, Casella JF, Arking DE, Keefer JR. Genome-wide association study identifies genetic variants influencing F-cell levels in sickle-cell patients. J Hum Genet. 2010;56:316–323. doi: 10.1038/jhg.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, Usala G, Busonero F, Maschio A, Albai G, Piras MG, Sestu N, Lai S, Dei M, Mulas A, Crisponi L, Naitza S, Asunis I, Deiana M, Nagaraja R, Perseu L, Satta S, Cipollina MD, Sollaino C, Moi P, Hirschhorn JN, Orkin SH, Abecasis GR, Schlessinger D, Cao A. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci U S A. 2008;105:1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt KA, Zhernakova A, Turner G, Heap GA, Franke L, Bruinenberg M, Romanos J, Dinesen LC, Ryan AW, Panesar D, Gwilliam R, Takeuchi F, McLaren WM, Holmes GK, Howdle PD, Walters JR, Sanders DS, Playford RJ, Trynka G, Mulder CJ, Mearin ML, Verbeek WH, Trimble V, Stevens FM, O'Morain C, Kennedy NP, Kelleher D, Pennington DJ, Strachan DP, McArdle WL, Mein CA, Wapenaar MC, Deloukas P, McGinnis R, McManus R, Wijmenga C, van Heel DA. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fellay J, Shianna KV, Ge D, Colombo S, Ledergerber B, Weale M, Zhang K, Gumbs C, Castagna A, Cossarizza A, Cozzi-Lepri A, De Luca A, Easterbrook P, Francioli P, Mallal S, Martinez-Picado J, Miro JM, Obel N, Smith JP, Wyniger J, Descombes P, Antonarakis SE, Letvin NL, McMichael AJ, Haynes BF, Telenti A, Goldstein DB. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Behrens EM, Finkel TH, Bradfield JP, Kim CE, Linton L, Casalunovo T, Frackelton EC, Santa E, Otieno FG, Glessner JT, Chiavacci RM, Grant SF, Hakonarson H. Association of the TRAF1-C5 locus on chromosome 9 with juvenile idiopathic arthritis. Arthritis Rheum. 2008;58:2206–2207. doi: 10.1002/art.23603. [DOI] [PubMed] [Google Scholar]
- 25.Hakonarson H, Grant SF, Bradfield JP, Marchand L, Kim CE, Glessner JT, Grabs R, Casalunovo T, Taback SP, Frackelton EC, Lawson ML, Robinson LJ, Skraban R, Lu Y, Chiavacci RM, Stanley CA, Kirsch SE, Rappaport EF, Orange JS, Monos DS, Devoto M, Qu HQ, Polychronakos C. A genome-wide association study identifies KIAA0350 as a type 1 diabetes gene. Nature. 2007;448:591–594. doi: 10.1038/nature06010. [DOI] [PubMed] [Google Scholar]
- 26.Tekola Ayele F, Adeyemo A, Finan C, Hailu E, Sinnott P, Burlinson ND, Aseffa A, Rotimi CN, Newport MJ, Davey G. HLA class II locus and susceptibility to podoconiosis. N Engl J Med. 2012;366:1200–1208. doi: 10.1056/NEJMoa1108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daly AK, Donaldson PT, Bhatnagar P, Shen Y, Pe'er I, Floratos A, Daly MJ, Goldstein DB, John S, Nelson MR, Graham J, Park BK, Dillon JF, Bernal W, Cordell HJ, Pirmohamed M, Aithal GP, Day CP, Study D International SAEC. HLA-B*5701 genotype is a major determinant of druginduced liver injury due to flucloxacillin. Nat Genet. 2009;41:816–819. doi: 10.1038/ng.379. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K, Sakamoto N, Nakagawa M, Korenaga M, Hino K, Hige S, Ito Y, Mita E, Tanaka E, Mochida S, Murawaki Y, Honda M, Sakai A, Hiasa Y, Nishiguchi S, Koike A, Sakaida I, Imamura M, Ito K, Yano K, Masaki N, Sugauchi F, Izumi N, Tokunaga K, Mizokami M. Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet. 2009;41:1105–1109. doi: 10.1038/ng.449. [DOI] [PubMed] [Google Scholar]
- 29.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hetherington S, Hughes AR, Mosteller M, Shortino D, Baker KL, Spreen W, Lai E, Davies K, Handley A, Dow DJ, Fling ME, Stocum M, Bowman C, Thurmond LM, Roses AD. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet. 2002;359:1121–1122. doi: 10.1016/S0140-6736(02)08158-8. [DOI] [PubMed] [Google Scholar]
- 31.Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomazic J, Jagel-Guedes E, Rugina S, Kozyrev O, Cid JF, Hay P, Nolan D, Hughes S, Hughes A, Ryan S, Fitch N, Thorborn D, Benbow A, Team P-S. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358:568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 32.American Association of Blood Banks. AABB Technical Manual. 17th ed. Bethesda: American Association of Blood Banks (AABB); 2011. [Google Scholar]
- 33.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. Plink: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. Genabel: An R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–1296. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- 35.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. Locuszoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 37.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Genomes Project C, Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gauderman W, Morrison J. Quanto 1.1: A Computer Program for Power and Sample Size Calculations for Genetic-Epidemiology Studies. 2006. http://hydra.Usc.Edu/gxe.
- 41.Jallow M, Teo YY, Small KS, Rockett KA, Deloukas P, Clark TG, Kivinen K, Bojang KA, Conway DJ, Pinder M, Sirugo G, Sisay-Joof F, Usen S, Auburn S, Bumpstead SJ, Campino S, Coffey A, Dunham A, Fry AE, Green A, Gwilliam R, Hunt SE, Inouye M, Jeffreys AE, Mendy A, Palotie A, Potter S, Ragoussis J, Rogers J, Rowlands K, Somaskantharajah E, Whittaker P, Widden C, Donnelly P, Howie B, Marchini J, Morris A, SanJoaquin M, Achidi EA, Agbenyega T, Allen A, Amodu O, Corran P, Djimde A, Dolo A, Doumbo OK, Drakeley C, Dunstan S, Evans J, Farrar J, Fernando D, Hien TT, Horstmann RD, Ibrahim M, Karunaweera N, Kokwaro G, Koram KA, Lemnge M, Makani J, Marsh K, Michon P, Modiano D, Molyneux ME, Mueller I, Parker M, Peshu N, Plowe CV, Puijalon O, Reeder J, Reyburn H, Riley EM, Sakuntabhai A, Singhasivanon P, Sirima S, Tall A, Taylor TE, Thera M, Troye-Blomberg M, Williams TN, Wilson M, Kwiatkowski DP. Genome-wide and fine-resolution association analysis of malaria in West Africa. Nat Genet. 2009;41:657–665. doi: 10.1038/ng.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alpy F, Tomasetto C. Give lipids a start: The star-related lipid transfer (start) domain in mammals. J Cell Sci. 2005;118:2791–2801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- 43.Patel SR, Hendrickson JE, Smith NH, Cadwell CM, Ozato K, Morse HC, 3rd, Yoshimi R, Zimring JC. Alloimmunization against RBC or PLT antigens is independent of TRIM21 expression in a murine model. Mol Immunol. 2011;48:909–913. doi: 10.1016/j.molimm.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brantley SG, Ramsey G. Red cell alloimmunization in multitransfused HLA-typed patients. Transfusion. 1988;28:463–466. doi: 10.1046/j.1537-2995.1988.28588337338.x. [DOI] [PubMed] [Google Scholar]
- 45.Sanna S, Busonero F, Maschio A, McArdle PF, Usala G, Dei M, Lai S, Mulas A, Piras MG, Perseu L, Masala M, Marongiu M, Crisponi L, Naitza S, Galanello R, Abecasis GR, Shuldiner AR, Schlessinger D, Cao A, Uda M. Common variants in the SLCO1B3 locus are associated with bilirubin levels and unconjugated hyperbilirubinemia. Hum Mol Genet. 2009;18:2711–2718. doi: 10.1093/hmg/ddp203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hazra A, Kraft P, Lazarus R, Chen C, Chanock SJ, Jacques P, Selhub J, Hunter DJ. Genome-wide significant predictors of metabolites in the one-carbon metabolism pathway. Hum Mol Genet. 2009;18:4677–4687. doi: 10.1093/hmg/ddp428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goldstein DR. Toll-like receptors and other links between innate and acquired alloimmunity. Curr Opin Immunol. 2004;16:538–544. doi: 10.1016/j.coi.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Palmer SM, Burch LH, Davis RD, Herczyk WF, Howell DN, Reinsmoen NL, Schwartz DA. The role of innate immunity in acute allograft rejection after lung transplantation. Am J Respir Crit Care Med. 2003;168:628–632. doi: 10.1164/rccm.200303-447OC. [DOI] [PubMed] [Google Scholar]
- 49.Yazdanbakhsh K, Ware RE, Noizat-Pirenne F. Red blood cell alloimmunization in sickle cell disease: Pathophysiology, risk factors, and transfusion management. Blood. 2012;120:528–537. doi: 10.1182/blood-2011-11-327361. [DOI] [PMC free article] [PubMed] [Google Scholar]