Preface
A role for somatic mutations in carcinogenesis is well accepted, but the degree to which mutation rates influence cancer initiation and development is under continuous debate. Recently accumulated genomic data has revealed that thousands of tumour samples are riddled by hypermutation, broadening support that cancers acquire a mutator phenotype. This major expansion of cancer mutation data sets has provided unprecedented statistical power for the analysis of mutation spectra, which has confirmed several classical sources of mutation in cancer, highlighted new prominent mutation sources and empowered the search for cancer drivers. The confluence of cancer mutation genomics and mechanistic insight provides great promise for understanding the basic development of cancer through mutations.
Mutations are among the usual suspects for causing cancer being found in oncogenes and tumour suppressors in malignant tumours. Moreover, there are several classical cases in which increased spontaneous or environmentally enhanced mutagenesis correlates with increased mutation load and cancer risk. Such instances of high mutation load, which we shall refer to as hypermutation, have served as a fundamental support for the hypothesis that cancer involves the establishment of a mutator phenotype1, where mutations occur at elevated rates. Despite the general observation that tumours often contain a large number of mutations, neither how these mutations accumulate (i.e. through higher mutation rates or increased number of replications in highly proliferative cancer cells)1-3 nor whether they accelerate cancer or are merely a by-product of immortalization has yet to be established.
Resequencing of cancer genomes have revealed that mutation loads can differ by several orders of magnitude 4, 5, with a wide variety of tumour types, such as melanoma, lung, stomach, colorectal, endometrial, and cervical cancers, displaying high mutation loads consistent with hypermutation, which may generate drivers of malignancy. Evaluating this contribution by cataloguing cancer genes frequently affected by hypermutation and determining the mechanisms of hypermutation may further our understanding of cancer biology, through which new therapeutic targets may be identified. This review will access the current understanding of hypermutation in cancer and speculate on future advances in this field facilitated by the rapidly evolving area of cancer genomics, where the analysis of vast whole genome and exome mutation datasets merges with detailed knowledge about DNA transactions to identify new mutagenic mechanisms and find new cancer drivers.
Hypermutation in cancer
Scientists have long understood that the root causes of cancer lie in the dysregulation of cell survival and proliferation often as the result of multiple genetic alterations that accumulate within a cell despite a normally low mutation rate. However, 40 years after the initial suggestion of the cancer mutator phenotype, this hypothesis remains supported primarily by the increased cancer predisposition of individuals deficient in a variety of DNA replication and repair processes as well as limited experimental observation of usually large numbers of mutations in a variety of tumour samples. The number of cancer genomes and exomes (currently exceeding ten thousand and growing fast) sequenced by the collective efforts of individual groups as well as The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) has provided the ability to have a much broader assessment of the sources and consequences of hypermutation in cancer development, largely thorough statistical analysis of patterns within the mutation data. In these studies, the sequence of tumour DNA is compared to the DNA sequence of either the patient's matched normal tissue or blood to identify tumour-specific mutations that occur at an allele fraction >5%. The requirement for a mutation to be seen in >5% of available reads limits the contribution of mutations in neighbouring stromal cells but allows the detection of mutations occurring within a small sub-clone of a heterogeneous tumour. As a consequence, these mutation lists represent a composite image of the mutagenesis occurring in all sub-clones of the tumour.
The philosophy and statistical approaches for extracting useful information from catalogues of mutations in cancer genomes are overall analogous to the analysis of mutation spectra obtained in experiments with mutation reporters – the classical approach in molecular genetics6, 7. Apparent “irregularities” in distribution of mutation types and position as compared to the null hypothesis of random mutation spectrum are matched against mechanistic knowledge about the chemistry of a mutagenic factor and genetic systems expected to repair the resulting DNA lesions. For example, mutation spectra of ultraviolet radiation (UV) are in good agreement with its capability to cause bulky lesions (cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts (6-4PPs)) in adjacent pyrimidine nucleotides8, 9. However, where the analysis of mutation spectra from reporters in model systems is greatly aided by defined experimental conditions and genotypes, the background information for cancer genome mutation catalogues is much less defined. In part this is compensated for by the large number of mutations within individual cancer genomes, sometimes greater than 105 allowing statistical analysis of cancer mutation spectra unrestricted by mechanistic hypotheses.
De novo genome- or exome-wide patterns
The first mutation patterns within whole genome sequenced cancers were detected in relation to the distribution of mutations in genomic space (TABLE 1). Universally, mutation frequency was observed to be increased near breakpoints of structural rearrangements10, 11, which are present in large numbers in many cancer genomes and are unique in each sample. Studies in model microbial systems established that this relationship likely results from either error-prone DNA synthesis associated with double-strand-break (DSB) repair 12-15 or an increased number of unrepaired lesions in long single-strand (ss) DNA created around the break site16-20 (also see below). Collectively, the experimental evidence showing that regions of DNA associated with DSB repair are prone to mutation and the bioinformatics analysis describing similar effects within clinical tumour samples suggested that intrinsic chromosomal features could themselves alter the rate of mutation and serve as one source of hypermutation in cancer.
Table 1.
Genomic features affecting mutation rate in cancer genomes.
| Genomic feature | Type of feature | Association with increased mutation density | Potential mechanism | Examples of affected cancer types | References* |
|---|---|---|---|---|---|
| Rearrangement breakpoints | Sample-specific | Vicinity to a breakpoint | Hypermutation in ssDNA, error-prone DNA synthesis | Breast, head and neck, colorectal, prostate, melanoma, multiple myeloma, chronic lymphocytic leukemia | REFS. 10, 11, 18, 65 |
| Replication timing | Universal | Late replication | Unknown | Melanoma, pan-cancer studies of up to 27 cancer types | REFS. 5, 24, 25 |
| Transcription | Static | Low transcription and/or non-transcribed strand | TCR removal of mutagenic lesions | Urothelial upper tract, lung, melanoma, pan-cancer study of 30 cancer types | REFS. 4, 28-30, 147 |
| Heterodiromatin marks | Universal or cancer type-specific | H3K9me3, H3K9me2, H4K20me3 | Absence of TCR, poor access of GGR | Leukemia, melanoma, lung, prostate | REF. 32 |
| Nucleosome density | Universal or cancer type-specific | DNAse I resistant regions with higher nucleosome content | Poor access of GGR complexes | Melanoma | REF. 25 |
| Local sequence | Cancer type-specific | Trinucleotide context | DNA repair, replication, endogenous lesions | Multiple types of cancer | see text |
Other genomic features, such as replication timing, transcription levels, and chromatin organisation also affect mutagenesis and are considered to occur universally among normal somatic cells and various cancer types. They have non-uniform profiles across the genome, however each profile is relatively constant between cells of the same type and even, in part, between different cancer cell lines. The stratification of these features across genomic space, initially developed by individual groups and subsequently expanded by the ENCODE consortium21-23, has generated a large database of profiles that can be correlated with densities of somatic mutations in cancer genomes. The most robust and reproducible correlation was documented for late replicating regions accumulating higher mutation densities than in the rest of the cancer genomes5, 24, 25. Interestingly, late replicating regions appear to be also more mutable over an evolutionary time scale26, 27. A greater chance of replication fork uncoupling leading to formation of hypermutable ssDNA in late replicating regions was suggested as a mechanism underlying this hypermutability27. A second, relatively invariant, feature associated with increased mutation density is non-transcribed or low transcribed regions as compared to highly transcribed sections of the genome5. High transcription may prevent mutations by enabling transcription-coupled repair (TCR), a form of nucleotide excision repair initiated by lesion-stalled RNA polII, which together with global genome repair (GGR), would remove more mutation-generating DNA lesions than the GGR alone would do in non-transcribed or low transcribed regions. In support of a TCR role, mutation densities were often lower on the transcribed strand, versus non-transcribed strand, where TCR does not operate4, 28-31. Transcription levels may also in part explain the increase in mutation density associated with heterochromatin marks32. Regions with condensed chromatin, as determined by resistance to S1 nuclease indicating a higher density of nucleosomes, had increased mutation density in melanoma genomes25. This could be also due to more active TCR in more highly transcribed open chromatin. However, even non-transcribed regulatory, regions with low nucleosome density had low mutation density. Supporting an alternative explanation, melanoma samples with mutations in nucleotide excision repair (NER) genes display higher mutation densities in S1-sensitive nucleosome deprived regions than in samples with wild type NER suggesting that the increase in mutation density in nucleosome-rich regions can be, in part, due to lower accessibility to NER complexes.
In addition to regional mutation patterns in cancer, patterns of sequence specificity can also be observed. Initial model studies sequencing mutation reporters established that mutation rates can vary substantially for different “mutation signatures” i.e., the choices of mutated nucleotide, kind of nucleotide resulting from mutation and immediate sequence context. These choices may be defined by one or several factors, such as a mutagen's DNA lesion spectrum, replication fidelity, and the relative contribution of DNA repair pathways in an individual or cell type7 (see also next section). Therefore, mutation signatures in a cancer sample may carry useful information about the history of a tumour's development which can be likened to an archaeological record33.
A powerful analytical tool, non-negative matrix factorisation (NMF)34, 35, which proved to be useful for a variety of bioinformatics analyses of large datasets, has likewise been productive in decrypting mutation signatures in cancer genomes. A general approach has been to apply NMF to catalogues of somatic mutations accumulated in genomes or exomes of multiple cancer samples belonging to the same cancer type to find motifs that are usually defined by a mutation and its two flanking nucleotides. Lawrence, Getz and colleagues5 highlighted six prevailing mononucleotide or dinucleotide mutation signatures in over 3000 samples of 27 different cancer types. Some signatures were highly concordant with exposures to specific mutagens, such as tobacco or UV radiation. Stratton and colleagues employed the NMF algorithm to all possible combinations of trinucleotides and base substitutions of the central base. Their analysis identified 21 mutation signatures, each consisting of several simple trinucleotide mutation motifs4, 36, 37. A somewhat different version of NMF led Zhao and colleagues to identify 20 mutation signatures, most of which overlapped with signatures found by other groups. They also explored the heterogeneity of mutation signatures within cancer types38. They concluded that the mutation spectrum in cancers is generated by a complex mix of mechanisms resulting in a signature profile unique for each individual cancer. The advantage of the NMF-based approach is that it is unbiased and hypothesis-free, so that, in principle, it can extract any, even previously unsuspected, mutational signature. However, as indicated in36 the number of extracted signatures depends on the number of samples being analysed and the diversity and frequency of mutation signatures. Hence low frequency signatures will be difficult to identify. Still, with a large number of cancer samples, many signatures can be detected and then compared to experimental and mechanistic knowledge supporting attempts to decipher multiple mutagenic mechanisms that occur during cancer development. NMF-generated signatures are usually complex, and may result from a mix of different mechanisms. Further analysis can be greatly facilitated by identifying a single-mechanism component(s) within a signature.
De novo patterns in mutation clusters
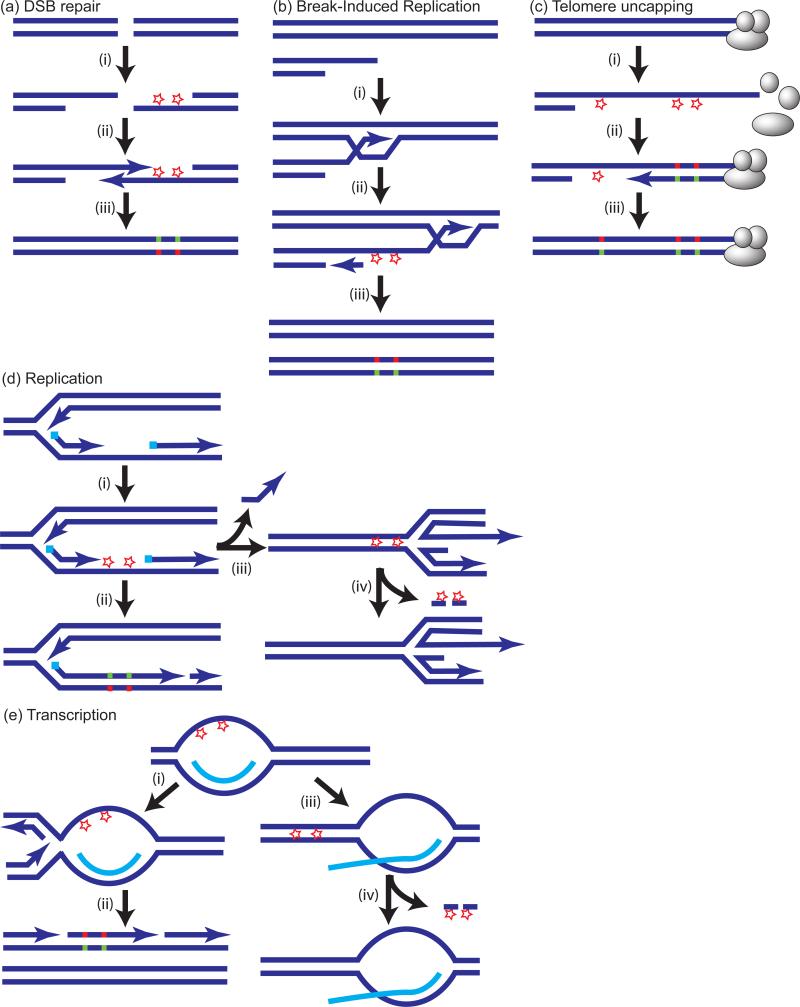
Single-mechanism components of a mutation signature can be pinpointed by statistical analysis of spectra in groups of mutations located too close to each other to be attributed to random independent events, i.e. representing mutation clusters. The phenomenon of unusually close positioning of several mutations was initially detected by Sommer and colleagues39 using the Big Blue mouse mutation reporter system. These unusually spaced mutations were called mutation showers by analogy with a meteor shower on the dark sky of the night39. Later, indications of mutation showers were also found within the EFGR gene in lung cancers40. We found that in yeast model systems clusters of up to 30 mutations spanning 200 kb could arise from unrepaired lesions in transient long regions of ssDNA formed at DSBs41, at uncapped telomeres42, during break-induced replication43 and at dysfunctional or uncoupled replication forks16, 18, 20, 44-48 (FIG. 1): all common events occurring in evolving cancer cells49-53. The specificity of mutation clusters for ssDNA results in some cases from either the robust removal of lesions in dsDNA by various excision repair systems, leaving only ssDNA lesions remaining or from lesions caused by agents that specifically affect ssDNA17. An important condition for clustered hypermutation is that the lesions in transient regions of ssDNA are not repaired until this strand is copied during synthesis of a nascent complementary strand with the help of error-prone trans-lesion synthesis (TLS) polymerases. Such repair may happen if a ssDNA region is annealed with an undamaged complementary strand for example by reannealing of R-loops54 or replication fork regression47 (FIG. 1).
Figure 1.
Lesions in single-strand (ss) DNA can result in clusters of strand coordinated mutations. Lesions are shown as stars. In the case of base specific damage, e.g. cytidine deamination by APOBEC enzyme(s), lesions would be in the same type of DNA base (i.e., C) of the same strand. Trans-lesion DNA synthesis (TLS) will introduce mutations in the complementary strand, which can be then fixed in DNA by a subsequent round of synthesis (step shown in a(iii), b(iii), c(iii), d(ii), e(ii)). This will result in strand-coordination of mutations changing only Cs (red) of the initially damaged strand and only Gs (green) mutated in the complementary strand. (a). (i) ssDNA formed by 5’→3’ resection at double strand DNA breaks (DSBs); (ii) one of several DSB repair mechanisms150 restores double-strand (ds) DNA at the position of break. (b). ssDNA formed by migrating loop and uncoupled strand copying during break-induced replication43, 48. (c). Telomere uncapping (REFS17, 69 and therein). (d). ssDNA formed during replication (shown is ssDNA in lagging strand; a similar chain of events may be associated with the leading strand). Lesions may result in mutations (ii) or be repaired via mechanism of fork regression which displaces a short stretch of complementary strand and pairs damaged ssDNA of the gap with the intact region of the complementary nascent strand (iii)47. The latter provides a template for excision repair of lesions generated in the ssDNA gap (iv). (e). (i) and (ii) Lesions in transient ssDNA formed by mRNA-DNA pairing (R-loops54) can lead to mutation clusters. (iii) and (iv) Mutations will be prevented, if re-annealing of DNA strands followed by excision repair occurs before replication.
The probability of mutation in transient ssDNA can be as high as several thousand fold greater than in the rest of the genome, leading to the incidence of mutation clusters containing multiple closely spaced mutations. Since all mutations of a cluster occurred simultaneously, presumably from lesions in the single DNA strand, they exhibit “strand-coordination”16, 18, 20 (FIG. 1). Moreover, DNA damaging agents often exhibit preference for certain nucleotide(s) or even short nucleotide motif(s). Therefore, simultaneous mutations in a cluster occur primarily at the same kind of nucleotide or motif. The combined result of the stand-coordination and homogeneous motif specificity of clustered mutations is a pure mutation spectrum stemming from a single mutagenic process. The original proof-of-principle for this concept was for lesions in ssDNA caused by UVC-radiation16,20. Strand-coordination was also demonstrated for other lesions (induced by methyl methanesulfonate (MMS), sulphites, and the cytidine deaminase apolipoprotein B mRNA editing enzyme catalytic polypeptide-like 3G (APOBEC3G)) in artificially created ssDNA17, 19. However, it is important to note that although strand-coordinated clusters have so far been associated with lesions in ssDNA, they can also stem from lesions in dsDNA as long as the lesions occur simultaneously (see also Figure 2 in55)).
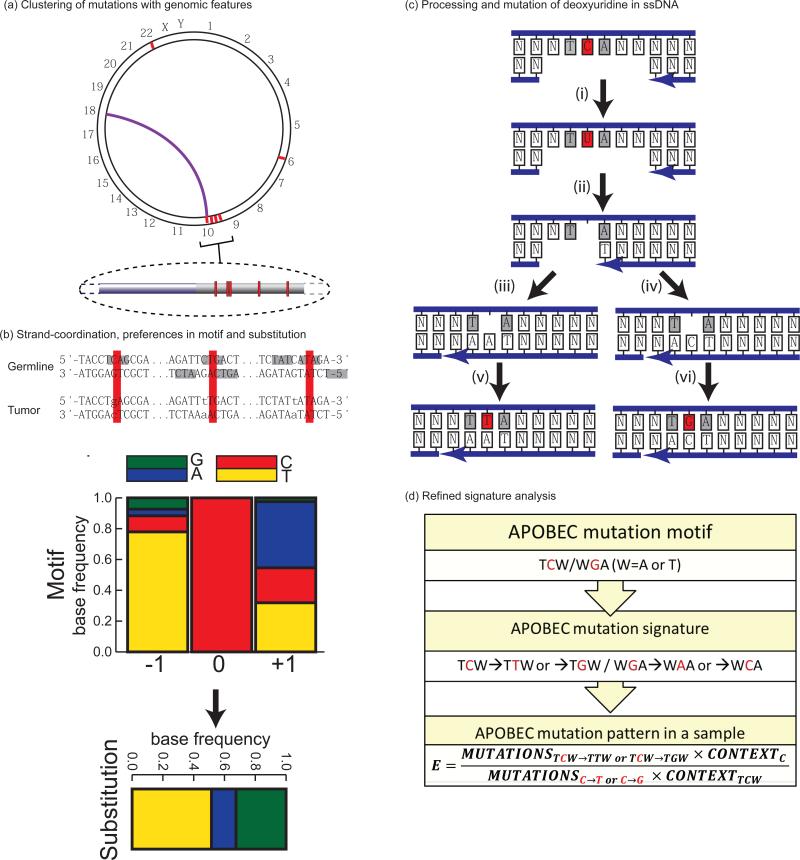
Figure 2.
Mutation patterns and mechanistic knowledge used to define an APOBEC mutation signature and produce sample-specific statistics evaluating mutagenesis. Robust mutation signatures can be developed by identifying groups of spatially clustered mutations (red lines and highlights on (b) and (c)) likely to have been induced by a single mutagenic mechanism. Additional features used to implicate the causative factor: (a). Proximity to sites of chromosomal rearrangement (purple connector line). (b). Strand-coordination (example with sequence context of a C-coordinated cluster from multiple myeloma18,see also Figure 1 and text), motif preference (grey fill), and substitution specificity. For example, the co-localization of strand-coordinated clustered cytosine substitutions with rearrangement breakpoints implicates the involvement of double strand DNA break (DSB) repair in formation of the mutations. The frequent involvement of single-strand (ss) DNA intermediates during such DSB repair events combined with an over-representation of TCA and TCT sequences among the mutations corresponds to the biochemical characteristics of a subset of APOBEC cytidine deaminases within ssDNA. Both cytosine to thymine and cytosine to guanine substitutions are also over-represented. (c). Mechanism of downstream processing of deoxyuridine (deamination product of deoxycytidine) explaining mutations specificity in clusters 69: (i). Glycolytic conversion of deoxyuridine to an abasic site (ii) creates a block to DNA synthesis during gap filling. The concerted action of DNA Pol delta and DNA Pol zeta (iii) or DNA Pol delta, DNA Pol zeta, and REV1 (iv) makes mutagenic insertion of either adenine or cytosine opposite of the abasic site, respectively, resulting in C to T and C to G mutations (v,vi). (d) Combining APOBEC enzymes’ favoured sequence motifs and base substitution preferences creates a refined mutation signature and allows calculation of sample-specific statistics evaluating mutagenesis – enrichment (E) by APOBEC signature mutations over the presence of APOBEC mutation motif in surrounding nucleotide context. TCW/WGA indicates the APOBEC targeted sequences of TCT, TCA, and their complements as in56. Nucleotides involved in mutation event are shown in red.
Based on these and other findings, Roberts et al.18 reasoned that if strand-coordinated clusters could be found in cancer genomes, the mutations in each cluster would likely have occurred simultaneously and resulted from one mechanism. Consequently, analysis of mutations in these clusters would enable evaluation of a single mutagenic mechanism singled out from a complex mix of mutation causes in cancer. Indeed, many mutation clusters (containing up to 34 mutations and spanning several kilobases) were found in whole genome sequenced (WGS) multiple myelomas, head and neck, prostate, and colorectal cancers, and around 30% of clusters were completely strand-coordinated18, 56. Completely strand-coordinated clusters of cytosines or guanines (C- or G-coordinated) prevailed over A- or T-coordinated clusters. A- or T-coordinated clusters were found mostly in multiple myelomas and were clearly associated with the mutation motif [T(A|T)] (mutated base underlined; ambiguous nucleotides shown in parentheses separated by “|”). This mutation signature has been reported for the error-prone DNA Pol eta participating in gap-filling DNA synthesis associated with non-canonical excision repair of U:G mismatches during somatic hypermutation (SHM) of immunoglobulin genes57, 58.
Abnormal targeting of activation induced cytidine deaminase (AID) (which induces the U:G mismatches during somatic hypermutation) to random chromosomal locations could be the source of A- or T-coordinated clusters59. However, if AID-induced U:G mismatches are not repaired they would result in a C to T or C to G substitutions at AID's preferred DNA motif for cytidine deamination: [(A|T)(A|G)C→(T|G)]60, 61 (short), or [(A|T)(A|G)C(A|T|C)→(T|G)]62 (extended). Consequently, AID-generated clusters normally carry a mix of mutations in C and A60. Indeed, such mixed clusters were found in immunoglobulin loci and a small number of secondary targets (including potential cancer genes59, 63, 64) known to be targeted by AID mutagenesis with lower efficiency59 in multiple myelomas18, 65 and chronic leukemic leukaemias66. Further supporting a SHM origin of these clusters, both cancer types originate from immune cells that have experienced SHM and the mutations were enriched with the AID mutation signature18, 65. AID mutagenesis, however, appears to occur primarily within the context of SHM as no enrichment for the AID signature was found in randomly located clusters or in WGS datasets within the same myeloma genomes18 and the AID deamination signature was absent from pan-cancer signature decomposition by NMF4. Similarly, C- or G-coordinated clusters as well as genome-wide mutagenesis across all cancer types analysed were depleted for AID signature18.
C- or G-coordinated clusters were more frequent than A- or T-coordinated clusters in WGS as well as in whole-exome sequence (WES) datasets18, 56 were frequently co-localised with breakpoints of rearrangements found within the same cancer sample while no colocalisation of A- or T-coordinated clusters detected18. Inspection of the nucleotides immediate flanking strand coordinated C or G mutations revealed high enrichment with a mutation signature [TC(A|T)→(T|G)], characteristic of a subclass of APOBEC cytidine deaminases (FIG. 2). Similar clusters enriched with mutations in C and G were also found using NMF by Nik-Zainal et al. in WGS of breast cancer samples37. Sizes and mutation densities of individual clusters (called micro-clusters in that study) were comparable to those found by Roberts et al.18 by mutated motif and co-localization with rearrangements. It was also noted that some larger areas of the cancer genome (up to 1 Mb) have a high density of mutations. Nik-Zainal et al. named this phenomenon kataegis (Greek word for thunderstorm by analogy with previously suggested term - mutation shower). In a later work4 they defined kataegis (or kataegic foci) as 6 or more consecutive mutations with an average inter-mutation distance of less than or equal to 1 kb. This study found several kataegic foci not only in breast cancer but also in cancers of pancreas, lung, liver, meduloblastomas, lymphomas and leukaemias4. The high prevalence of the [TC(A|T)→(T|G)] mutation signature in C- or G-kataegic foci suggested that APOBEC mutagenesis could be the major component of some signatures revealed by NMF, where each signature listed several trinucleotide motifs. Indeed [TC(A|T)→(T|G)] mutations prevailed in signatures 2 and 13 observed in several cancer types4.
Pairing mutation signatures with their source: APOBEC example
Several key experimentally determined characteristics of APOBEC cytidine deaminases enabled attribution of the mutation signature [TC(A|T)→(T|G)] to these enzymes (FIG. 2). The human genome encodes 8 APOBEC polypeptides (7 are located in the APOBEC3 cluster of highly homologous genes), which normally serve to restrict viral infection and retrotransposon mobility by deaminating cytosines during the ssDNA stage of their replication cycle67, 68. These enzymes have exquisite preference for ssDNA where they deaminate C that resides in TCA and TCT trinucleotides to U. This U can be directly copied to produce C to T substitutions. However uracil bases in ssDNA are often excised by uracil DNA glycosylase leaving abasic sites which after copying by TLS polymerases results in both C to T and C to G mutations17, 69-72. Moreover, some APOBECs are highly processive, allowing direct formation of mutation clusters73, 74. These biochemical properties correlated well with the sequence motif and base substitution spectrum of mutations in C-and G-coordinated clusters, as well as the colocalisation with rearrangement breakpoints where ssDNA is expected to have formed, either because ssDNA is more fragile or because regions of ssDNA would be created around DSBs by end resection41.
The primary benefit of associating a mutagenic process with a stringent signature, such as [TC(A|T)→(T|G)], is that this signature can be used to develop a measure of mutagenesis in the entire genome of individual samples – enrichment over expected presence of the signature if caused by random mutagenesis18, 56. This allowed computing sample specific p-values as well as q-values corrected for multiple testing errors. In the case of the [TC(A|T)→(T|G)] signature, analysis of about a million TCGA mutations in 14 types of cancer revealed that bladder, cervical, breast, head and neck and lung cancers have a high prevalence of APOBEC mutated samples. The same cancer types showed the presence of signatures 2 and/or 13 in the NMF-based analysis4. However, only the capacity to compute sample-specific q-values and enrichments enabled the finding that tumours in the HER2-enriched subtype of breast cancer are much more likely to be mutated by APOBEC as compared with three other breast cancer subtypes56.
APOBEC1 as well as six of the APOBEC3 proteins have the ability to generate the [TC(A|T)→(T|G)] mutation signature in vitro and in model in vivo studies55, 67. A high level of APOBEC3B expression in several cancer types and weak but statistically significant correlation of its’ mRNA with the load of APOBEC signature mutations prompted Burns et al. to suggest that this enzyme is the likely cause of mutagenesis75, 76. However, another study implicated APOBEC3A as well as APOBEC3B as possible mutagenic enzymes in breast cancer77. Interestingly, APOBEC3B homozygous deletion is polymorphic within the human population78 and individuals homozygous-null and even heterozygous have a detectable increase in frequencies of breast or liver cancers79-81 and can show a high frequency of mutations fitting the APOBEC signature82. Identification of the real culprit through analysis of cancer genomics datasets is complicated by the level of mutagenesis likely depending on additional factors besides mRNA levels of APOBECs, such as the active protein level, accessibility of chromosomal DNA, availability of ssDNA substrate (discussed in 56). It is also worth noting that many mutations detected in cancers may have occurred long before tumour mRNA was extracted for RNAseq.
While several questions about APOBEC mutagenesis in cancers remain unresolved, it is a useful example of a strategy leading to the construction of a rigid mutation signature. This in turn allows statistical power sufficient for computing sample-specific p-values, which are not available, if complex signatures derived from NMF are used for sample-by-sample analysis directly. In addition to APOBEC mutagenesis, cancer genome re-sequencing identified other mutation signatures occurring both in mutation clusters and scattered across the genome. While many of these signatures cannot yet be linked to a specific source, correlations with mechanism-based hypotheses have enabled the likely causative agents for the following signature which can be broadly separated according to the source – exposure to a mutagen, or resulting from defective DNA repair. (TAB. 2 and below).
Table 2.
Signatures associated with different causes of hypermutation in human cancer genomes.
| Source of mutations | Mutated motif | Resulting base(s) | Common base change in a group | Hypermutation load; percent (reference)** | Additional features |
|---|---|---|---|---|---|
| UV-radiation-1 | (T|C) C (A|T|C|G) | T | C-->T | 30,000-50,000; 40-50% (REF. 91) | Exposure history; somatic mutations, transcription |
| UV-radiation-2 | (T|C) C (G) | T | C-->T | 3,000-5,000; 5-10% (REF. 91) | Exposure history; somatic mutations, transcription |
| UV-radiation-3 | CC | TT | C-->T | 500-1,000; 1% (REF. 91) | Exposure history; somatic mutations, transcription |
| 5-meCpG deamination | (A|T|C|G) C (G) | T | C-->T | 1,000-10,000; 5-10% (REF. 4) | Age dependence |
| APOBEC | (T) C (A|T) | T|G | C-->T | 50,000-100,000; 30-70%; (REF. 56) | C- or G-strand-coordinated clusters |
| AID* | (A| G) C | t|g | C-->T | Region-specific | Clusters in AID genomic targets |
| DNA Pol epsilon-1 | (T) C (G) | T | C-->T | 10,000-20,000; 40-60% (REF. 128) | Somatic mutations, MSS |
| DNA Pol epsilon-2 | (T) C (T) | A | C-->A | Total for Pol epsilon is in the above cell | Somatic mutations, MSS |
| ROS | (A|T|C|G) C (A|T|C|G) | A | C-->A | TBD | TBD |
| Tobacco | (A|T|C|G) C (A|T|C|G) | A | C-->A | 30,000-100,000; 20-90% (REF. 4) | Exposure history |
| Temozolimide (TMZ) | (A|T|C|G) C (T|C) | A | C-->A | 30,000-200,000; 30-90% (REF. 114) | Treatment record |
| Aristolochic acid | (C) T (A| G) | A | T-->A | 50,000-200,000; 30-70% (REF. 28, 31) | Exposure history |
| DNA Pol eta | (A|T|C|G) T (A|T) | A|T|C|G | T-->A | TBD | TBD |
| MMR defects | Microsatellite instability | NA | NA | - | Somatic mutations, MLH1 hypermethylation |
Shown for each motif is the mutated nucleotide (underlined) as well as 5′ and 3′ flanking nucleotides. Ambiguous nucleotides for a position are shown in parentheses divided by “|”. Same types of nucleotides in a position are aligned to aid visual comparison between motifs.
Motifs produced by the same mutagenic mechanism or source are numbered.
*** An additional 5′ nucleotide in AID [(A|T)(A|G)C] motif is not shown to allow alignment with the table space
**** Signatures are grouped by common type of base change, since this feature has been reported in the vast majority of mutagenesis publications
***** Provided are representative examples; WES data are prorated to whole genome scale in assumption of 100× more mutations in WGS
# values indicate the combined contribution of both signatures
Hypermutation by increase in lesions or by decrease in error-free repair
This group of mutagenic mechanisms rely upon exogenous or endogenous (such as the AID and APOBEC enzymes discussed above) DNA lesions which are turned into mutations by downstream DNA repair and replication (FIG. 3 and TAB. 2).
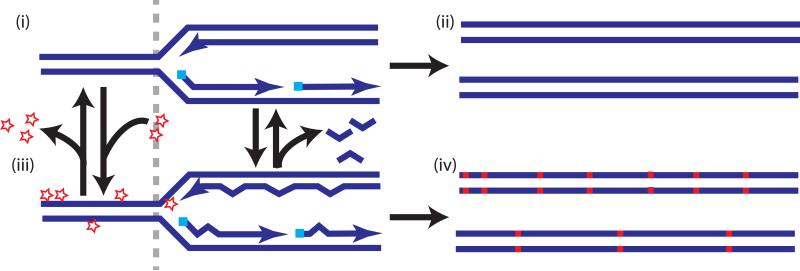
Figure 3.
Sources of hypermutation in cancer. (i and iii) Hypermutation can occur through alterations in the equilibrium between the formation of lesions (stars) and error-free lesion repair (left of grey dashed line) or changes in the equilibrium between the rate of replication errors and DNA polymerase proofreading (star at the 3’end of the leading strand) or mismatch repair (MMR) (shown as removal of bulged mis-pairing behind the fork) (right of grey dashed line). Low levels of lesions and replication errors and/or high efficiency of error free lesion repair, proofreading and MMR results in low mutation frequency (ii), however reduction in repair and/or increase in lesions or in replication error levels may lead to hypermutation (iv).
Mutations in CpG motifs
Cytosines in CpG are often methylated at the C5 carbon producing 5-methylcytosines (5-meC). Deamination of 5-meC produces T and often C→T mutations. Such mutations can be prevented if a T:G mispair is recognised by the T:G-specific thymine DNA glycosylase (TDG) and the resulting abasic site is repaired by base excision repair (BER)83, 84. Unlike mutations caused by C-deamination in the APOBEC motif, CpG mutations are depleted within C- or G-coordinated mutation clusters, expected from simultaneous mutation events 18, 56. Frequent conversion of 5-meC to T leads to depletion of CpG dinucleotides from genomes, except within regulatory CpG islands or rare coding positions, where methylation is limited and they are maintained by positive selection85, 86. When mutation rates in CpG are normalised for their presence in the genome or in exome, this mutagenesis becomes detectable in many WES and WGS cancer mutation datasets4, 5. Unlike other mutation signatures, CpG mutagenesis correlates with patient's age at cancer detection4.
Ultaviolet (UV) radiation
UVB (280-320 nm) and UVA (320-400 nm) components of sunlight are established risk factors for melanomas and head and neck squamous cell cancers87. The spectrum and signature of UV-mutagenesis is defined by the spectrum of lesions that arise and the interplay of repair pathways and TLS across unrepaired lesions8, 9. CPDs and 6-4PPs in dsDNA are normally repaired by NER using the undamaged DNA strand as a template. Unrepaired photoproducts are converted into mutations by error-prone TLS during replication88, 89. The specific UV mutagenesis signature relies on a highly increased rate of deamination of cytosines in CPDs8, 90. The resulting Py-U CPD is copied by error-free TLS polymerase DNA Pol eta, resulting in C→T mutation in [(T|C) C(A|T|G|C)] context. When CC dinucleotides form a CPD, deamination of both cytosines results in CC→TT dinucleotide substitution. Indeed, both of these mutation signatures, are prevalent in melanomas and to a lesser extent in squamous cell carcinomas of the head and neck4, 5, 29, 91.
Oxidative DNA damage
Free radical (hydroxyl, superoxide) and non-radical (hydrogen peroxide) reactive oxygen species (ROS) that are constantly produced by cell metabolism and enter the cell from the environment, can cause various kinds of oxidative damage to all DNA bases, which can turn into mutations92-94. Studies with mutation reporters have indicated that oxidative damage can lead to a higher prevalence of mutations in G:C than in A:T base pairs95-98, which could contribute to the bias towards G:C mutations observed in many cancers4, 5. Highly mutagenic 8-oxo-7,8 dihydroguanine (8-oxoG) could be the reason for such a bias, however other products of reactions of guanine and cytosine could also contribute94. Vasquez and colleagues 99, 100 found the mutation motif [(A|T|G|C)(T|C)C(A|T|C)]) is enriched in several cancers. Experimentally, they found that guanines in the complementary sequence context have a higher potential to trap electrons and thus have an increased chance of chemical modification. Partial overlap of this motif with the APOBEC mutation motif [(A|T)GA] in the G-containing strand) suggests that additional steps should be taken at statistical analysis of either motif.
Tobacco
Tobacco contains multiple ingredients that are capable of forming mutagenic DNA adducts101, 102. Among those, benzo[a]pyrene (BaP), is the most studied experimentally. Its adduct with guanine predominantly induces G:C→T:A mutations. However, the same class of mutations is expected from 8-oxoG, which can result from ROS that are generated by smoking. Regardless, G:C→T:A mutations are the major class of base substitutions observed in TP53 from lung cancers and in WGS and WES of lung cancers, and the occurrence of this class of mutation correlates with smoking history30, 103-105. Interestingly in bladder cancer, for which smoking is known to be a strong risk factor, the WES of 130 samples did not contain an increased number of G:C→T:A mutations95. Instead, bladder cancers showed enrichment for the APOBEC mutation signature.
Temozolomide
The major mutagenic product of the SN1 type alkylating agents including the anticancer drug temozolimide (TMZ), is O6-methylguanine (O6-meG). This DNA lesion can be copied by DNA polymerase, inserting either the correct C-nucleotide or the incorrect T. The latter would cause a C:G→T:A mutation. O6-meG:C as well as O6-meG:T are recognised by the mismatch repair (MMR) pathway, which functions in conjunction with DNA replication. Recognition triggers removal of a nascent DNA stretch across from the O6-meG lesion and reinitiation of DNA synthesis. Subsequent synthesis past the same O6-meG frequently regenerates the mismatch, which is in turn recognized by MMR and establishes a repetitive repair cycle106-108. This ‘futile repair cycle’ is mostly toxic to proliferating cells, which underlies the effect of TMZ on tumours. However, resistance to TMZ as well as to other SN1 alkylating agents can occur as long as cancer cells acquire additional defects in MMR or overexpresses O6-methylguanine-DNA methyltransferase (MGMT)109-112. This results in a high frequency of G:C→A:T mutations which occurs in the genomes of TMZ-treated gliomas and melanomas4, 113, 114. Importantly these G:C→A:T mutations were predominantly in CC or (to a lesser extent) in CT motifs, which allows distinguishing them from C→T mutations associated with 5meCG.
Aristolochic Acid (AA)
. This carcinogenic substance is contained in the East Asian traditional medicinal plant Aristolochicia sp.115, 116. Its metabolized derivative forms mutagenic aristolactam (AL)-DNA adducts. AL-dA adducts are refractory to the global genome branch of NER (GGR) and can be repaired only by TCR in the transcribed strand117, which can explain a transcriptional strand bias and high frequency of A:T→TA mutations in cancer genes such as TP53118, 119. The mutation load in upper tract urothelial cancers induced by AA is one of the highest known so far. A:T→TA mutations in these cancers were enriched with [(C/T)AG]28 (Note: Table 2 shows this motif as in the T strand) or even with a more stringent [A(C/T)AGG]31 motif.
Hypermutation by increased DNA synthesis errors
The chances for mutations to arise from DNA synthesis copying a normal template during genome replication are limited by replicase accuracy in base selection, nearly instant proofreading of replication errors and post-replicative MMR120, 121. Inactivation of any of these safeguards can lead to hypermutation. Similarly, mutations also can be introduced during replication when error prone TLS polymerases get to copy small stretches of DNA that do not contain lesions122.
MMR defects
Hereditary predisposition to non-polyposis colorectal cancer (HNPCC) as well as to several other types of cancers is the consequence of germline genetic defects in one of the MMR genes: MSH2, MSH6, PMS2 and MLH1121. The most easily detected result of MMR defects, even in WGS and WES datasets, is an increase in microsatellite instability (MSI) – frequent deletions and insertions in arrays of very short direct repeat sequences (microsatellites)123. In sporadic colorectal, gastric and endometrial cancers124-126, genome-wide MSI often coincides with MLH1 hyper-methylation or with somatic mutations in MMR genes. Samples with MSI also have an increased frequency of various base substitutions. They form a category of hypermutated cancers, which is distinct from the hypermutation that occurs in cancer cells with somatic mutations in replicative DNA polymerases.
DNA pol epsilon and delta mutations
Somatic mutations in the respective catalytic subunits of replicative DNA polymerases epsilon (POLE1) and delta (POLD1) have been recently associated with familial predisposition to colorectal, endometrial and ovarian endometroid cancers127-130. Based on the budding yeast model, DNA pol delta synthesises the lagging strand and DNA pol epsilon is responsible for the synthesis of the leading strand in a bi-directional replication fork131. In WES analysis, samples with somatic mutations in POLE1 are ‘ultramutated’ and carry even higher mutation loads than MSI MMR-deficient tumours. It was noted that sporadic mutations in POLD1 were less frequent among ultramutated tumours, which mostly carried POLE1 mutations in the proofreading exonuclease domain (EDM) 127, 132-134. Samples with EDM mutations are particularly enriched with two separate signatures [TCT→A] and [TCG→T]4.
TLS polymerases
TLS DNA pol eta can perform error free copying of CPDs, however it has a high error rate when copying an undamaged template in vitro135. The POL eta [T(A|T)] mutation motif was significantly enriched in A- or T-coordinated clusters found in multiple myelomas18 (see also above section “De novo patterns in mutation clusters”). Enrichment with this motif was also documented for chronic leukaemias and in B-cell lymphomas4, 66. Another mutation signature, [AT→C], detected in HV-C positive hepatocellular carcinomas was suggested to originate from error prone synthesis by one of the TLS polymerases based on overexpression of POL zeta and POL iota in this type of cancer136, 137.
In search of carcinogenic potential of hypermutation
A high level of cancer genome and epigenome instability is a well-accepted fact138, 139, but the relative roles and specific contribution of each type of instability is still a question of today's cancer research. Not surprisingly, the relative carcinogenic role of hypermutation is still under debate1-3. However, it is clear that hypermutated cancer genomes can contain prevailing mutation signatures in genes that are important for cancer initiation and development 5, 28, 31, 56, 76, 140. Recent bioinformatics and experimental analyses indicate that in at least some of these cases, hypermutation is inducing mutations that actively drive carcinogenesis. For example, mutations occurring in the gene PIK3CA are more common in APOBEC hypermutated tumours, regardless of tumour type. Moreover, the PIK3CA mutations in APOBEC hypermutated tumours are skewed toward 2 hotspot locations in the regulatory helical domain over the third canonical mutation hotspot located directly in the kinase domain. The over-representation of the tumour-selected helical domain mutations, which exist in APOBEC target motifs, over an equally selectable, non-APOBEC mutation in the APOBEC hypermutated cancers suggests strongly that APOBEC enzymes were likely causative in these cancers141. In addition, Marais and colleagues have shown that UV induced mutations accelerate carcinogenesis in BRAF deficient mice by inducing selected mutations in TP53142. Identifying similar examples of hypermutation induced cancer will likely become easier as our knowledge of the key genes responsible for tumourigenesis becomes more complete. Lawrence et al.5 found that accounting for mutational heterogeneity associated with various functional and structural features across the genome (TAB. 1) increased the statistical power for detecting significantly mutated genes (SMGs) that are potentially important for cancer and eliminated apparent false positives, such as olfactory receptor genes. These genes reside in late replicating, low transcribed sections of the genome that display an elevated mutation rate. After correcting for regional differences in mutation rate, these genes fell out from the SMG category. We anticipate that the accumulation of mechanistic knowledge as well as WES and WGS cancer mutation data will increase the accuracy of correction factors by accounting for specific features of mutagenic mechanisms that are operating in individual hypermutated cancer samples. For example, calculating enrichments for a simple mutation signature allows selecting for analysis those samples, where a significant part of mutations carrying the signature really have risen from a mutagen. Groups of samples with statistically evaluated high presence of specific mutation signatures can be used as a discovery set for defining genomic profile of hypermutation caused by a signature-generating mutagen leading to development of mutagen-specific correction factors in searching for SMGs.
Perspectives – actions and outcomes
Despite the current successes of re-sequencing genomes in understanding the impact of mutation on cancer and genome dynamics in general, a number of the mutation signatures highlighted by NMF36 cannot so far be associated with any known mutagenic pathway or agent. Likewise, the mechanism underlying other forms of hypermutation, such as the mutation signature [(A|T|C|G)TT→G] in oesophageal cancer143, hypermutation in female inactivated X chromosome144 and hypermutated prostate cancer145, 146 remain unknown. Combining statistical signature analysis with mechanistic research could bring clarity to these issues (Box 1). Moreover, statistical evaluation of specific mutagenesis signatures among an expanded sequencing effort that includes more preneoplastic lesions and relapsed tumour samples may eventually enable analyses correlating these mutations to clinical phenotypes and provide a better understanding on the role of hypermutation in cancer development as well as potentially lead to new cancer prevention, early diagnostics and therapy strategies. These perspectives can become closer with concerted effort of the bioinformatics and mechanistic fields.
Box1: Merging statistical pattern analysis with mechanistic information.
Although de novo pattern recognition approaches are useful for highlighting mutation signatures prevailing in large groups of cancers, they can only identify samples enriched with a given signature after the principal component of a signature is extracted and used for repeated analysis. Such an extraction can be done on the basis of the prevalence of a signature derived by NMF, however an independent statistical analysis, such as the presence of a principal component of a signature in clusters and/or correlation with mechanistic information on a suggested source of mutagenesis may result in more rigid definition of a signature, thereby increasing the power of the statistical analysis.
Key benefits of utilising experimentally derived mechanistic knowledge to generate rigid mutation signatures:
➢ The information about mutagenic specificity of environmental and endogenous causes of mutations can be used for reducing each multicomponent signature derived from NMF to a less complex one, which is better suited for sample-to-sample analysis.
➢ Mechanistic information can help avoid over-simplification of mutation signatures which would result in excessive overlap.
➢ As long as two simple signatures are known to come from the same mechanism they can be used within a single more focused and powerful statistical hypothesis. Examples listed in TAB. 2 and below (see also 99, 147) can be used to illustrate these lines of analysis.
Example 1. The UV signature, [(T|C)C(A|T|C|G)]→T, overlaps with the [TC(A|T)→T] part of the APOBEC signature. However, the other part of the APOBEC signature [TC(A|T)→G] is absent in UV mutagenesis. Further supporting the lack of [TC(A|T)→G] mutations stemming from UV radiation, in melanoma genomes, the density of [TC(A|T)→T] mutations showed reverse dependence on nucleosome density, whereas [TC(A|T)→G] did not25. Another feature distinguishing UV from APOBEC mutagenesis is the lack of preference in the choice of the 3’-nucleotide setting the mutation motif as [(T|C)C(A|T|C|G) →T]. Moreover, the third nucleotide position for a UV induced signature should be enriched with G, because methylation of cytosine increases the chance of CPD formation8, 90. Indeed, high prevalence of [(T|C)CG→T] mutations are a common feature of many UV-associated cancers4, 5, 29, 91. Together with CC→TT enrichment in a sample, these attributes provide a good tool for highlighting cancer samples with a strong component of UV-mutagenesis. In addition the partial overlap between the part of UV mutation signature [(T|C)C→T] and the APOBEC signature can also be resolved by UV-mutagenesis occurring more frequently in the non-transcribed strand (TAB. 1), while APOBEC signature mutations do not show any transcriptional bias4.
Example 2. The partial overlap between the APOBEC mutation signature [(T)C(A|T)→T|G] and the DNA POL epsilon mutation signature [TCT→A] can be resolved by accounting for the difference in the base resulting from a mutation (T or G vs. A) which is in agreement with mechanistic information about TLS across abasic sites - the primary lesion generated in place of uracils created by APOBEC cytidine deamination in ssDNA69.
Example 3. The overlap between the tobacco signature, [(A|T|C|G)C(A|T|C|G)→A], and the TMZ signature, [(A|T|C|G)C(T|C)→A], can be resolved using the clinical history of exposure.
Since mechanism-based mutation signatures are confined to simple nucleotide motifs, it is possible to calculate enrichment for the motif in mutations over its presence in a sequenced part of the genome (see FIG. 2 for APOBEC example). These calculations are useful for discerning between mutagenic processes that generate overlapping signatures (see e.g. Fig. 1 in56) as the more likely process would generate a higher enrichment.
Both, the bioinformatics and mechanistic sides of the field will benefit from a growing number of analysed samples, especially if the output data would be expanded and brought to the uniform format between different project and data depositories. A common organisation would greatly facilitate analyses conducted on different cancer datasets (Box 2). Unification and supplementing information in datasets could be matched by mechanistic research commonly addressing questions of mutation signatures and developing clear mutation motifs as long as such can be derived from a study. In other words, an effort should be made, where possible, to organise conclusions of mechanistic research in a form as convenient as possible for immediate statistical exploration in cancer mutation datasets. These simple steps may result in drafting many researchers with all kinds of expertise, whom would otherwise be deterred by the perspective of an indefinite time needed for mining and organising the data before performing pilot analyses, which only infrequently lead to a finding. Altogether, merging mechanistic and bioinformatics approaches suggest realistic modifications to study formats and data organisation with the payoff in more discoveries that could be important for the fight against cancer.
Box 2: Establishing a uniform organisation among mutation databases.
Unifying the organization of the large tables containing mutation lists (called Mutation Annotation Files (MAF) in TCGA data depository) and supplementing them with additional information could greatly facilitate the use of genomics data by a large number of mechanistic experts whom do not have sufficient bioinformatics expertise available. Below are several suggestions in this regard:
➢ Unifying the column titles and data values designations is a simple first step to facilitate data analysis by multiple groups outside the data generation consortia.
➢ Unifying sample IDs so they are the same for a given sample between all platforms (clinical records, expression, methylation, mutation, rearrangements, copy number variation etc.) and data sets within a study.
➢ Providing the values for allele fractions and coverage for each mutation call, which is a natural requirement for accurate representation of mutation data in a cancer sample usually consisting of more than one clone. Analysis of allelic fractions provides an understanding of cancer development history in the sense of mutation incidence, selection and fixation148, 149.
➢ So far, statistical analyses of hypermutation have been based on tumour-specific mutation calls. Adding the data about germline genotype for each patient with a sporadic tumour analysed by genomics platforms would allow genetic analysis of mutation spectra and mutation signatures as quantitative genetic traits. Although these data are available from the same sequencing effort that produces tumour-specific mutation calls, they are not provided in sample depositories or by individual studies.
Glossary
• DNA
- Genome
DNA of all chromosomes; note that whole-genome re-sequencing skips a significant part of highly repeated genomic DNA
- Exome
part of the genome coding for known mRNAs; sequence is determined after pull-down with specially designed hybridization kits
- Genomic coordinate
chromosome number and a nucleotide number in a database.
- pol DNA damage
changes in chemical structure of DNA.
- DNA lesion
specific change in DNA structure resulting from DNA damage, e.g. cytidine deaminiation, base alkylation, base oxidation, strand crosslinks, UV-dimers, DNA breakage.
- SN1 type alkylating agents
Chemicals that form a reactive intermediate in the body which can attack DNA bases to covalently link an organic group (e.g. a methyl group).
• Genome Features
- Late replicating regions
Regions of the genome where DNA is synthesized near the end of S-phase during replication.
- R-loops
Stable hybrids of RNA and DNA formed during transcription.
• DNA Repair Pathways
- Nucleotide excision repair (NER)
A DNA repair pathway that removes bulky DNA lesions by removing a stretch of the DNA strand containing the lesion and then subsequently using the remaining undamaged DNA strand as a template to synthesize a new DNA stretch to replace the excised one.
- Transcription-coupled repair (TCR)
A form of nucleotide excision repair that utilizes a stalled RNA polymerase as a means to recognize a DNA lesion in the transcribed DNA strand.
- Trans-lesion synthesis (TLS)
A form of lesion tolerance that involves the insertion of a new nucleotide across from a DNA lesion, usually by a specialized DNA polymerase.
- Non-canonical excision repair
An excision of a DNA lesion or modification that does not completely follow the pathway mechanisms of base excision repair, nucleotide excision repair, or mismatch repair.
- Mismatch repair (MMR)
A DNA repair pathway that removes mismatched nucleotides (e.g. C:T base pairs) in DNA by removing a stretch of the DNA strand containing the lesion and then subsequently using the remaining undamaged DNA strand as a template to synthesize a new DNA stretch to replace the excised one.
- Break-induced replication (BIR)
A DNA double strand break repair mechanism involving the invasion of one DNA end into a homologous locus on a sister chromatid or homologous chromosome. Once invaded, the broken DNA is used to prime replication to the end of the unbroken sister chromatid or homologous chromosome to replace DNA sequence lost due to the DNA double strand break.
• Structural variation
- Structural rearrangement
change in location of genomic blocks relative to each other
- Rearrangement breakpoints
pair of distant genomic coordinates brought to immediate vicinity by a rearrangement
• Mutations
- Mutation reporters
DNA sequences that when mutated in a cell gives the cell a phenotype that can be selected. In most cases the mutation either restores the function of an inactive protein or provides resistance to a drug.
- Mutation frequencies and rates
measured by counting the number of mutant cells or individuals within a group or population
- Mutation load
number of mutations in the genome or part of the genome of a cell, group of cells (tumour, tissue, etc.) or an organism.
- Mutation spectrum
list of mutation types and coordinates within a mutation load
- Mutation signature
characteristics of a mutation, such as mutated base, resulting base(s), nucleotides in the immediate vicinity that occur more frequently than expected with random mutation of genomic DNA
- Cancer driver mutation
a mutation increasing the probability of tumour incidence or
- Mutation cluster
a group of mutations spaced more closely than expected by random distribution of mutations in a genome
- Strand coordination
a phenomenon where clustered mutations involve changes of only one kind of base within the same DNA strand (e.g., C-coordination - only cytosines mutated in the top DNA strand)
- Kataegis or Mutation shower
a group of clustered mutations carrying additional similarity features unlikely to be random (e.g., changes of the same nucleotide in the given strand – strand-coordination)
REFERENCES
- 1.Fox EJ, Prindle MJ, Loeb LA. Do mutator mutations fuel tumorigenesis? Cancer metastasis reviews. 2013 doi: 10.1007/s10555-013-9426-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox EJ, Beckman RA, Loeb LA. Reply: Is There Any Genetic Instability in Human Cancer? DNA Repair (Amst) 2010;9:859–860. doi: 10.1016/j.dnarep.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shibata D, Lieber MR. Is there any genetic instability in human cancer? DNA Repair (Amst) 2010;9:858. doi: 10.1016/j.dnarep.2010.04.011. discussion 859-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawrence MS, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benzer S, Freese E. Induction of Specific Mutations with 5-Bromouracil. Proc Natl Acad Sci U S A. 1958;44:112–9. doi: 10.1073/pnas.44.2.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogozin IB, Pavlov YI. Theoretical analysis of mutation hotspots and their DNA sequence context specificity. Mutat Res. 2003;544:65–85. doi: 10.1016/s1383-5742(03)00032-2. [DOI] [PubMed] [Google Scholar]
- 8.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 9.Sage E. Distribution and repair of photolesions in DNA: genetic consequences and the role of sequence context. Photochem Photobiol. 1993;57:163–74. doi: 10.1111/j.1751-1097.1993.tb02273.x. [DOI] [PubMed] [Google Scholar]
- 10.De S, Babu MM. A time-invariant principle of genome evolution. Proc Natl Acad Sci U S A. 2010;107:13004–9. doi: 10.1073/pnas.0914454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drier Y, et al. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res. 2013;23:228–35. doi: 10.1101/gr.141382.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–5. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malkova A, Haber JE. Mutations arising during repair of chromosome breaks. Annu Rev Genet. 2012;46:455–73. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- 14.Deem A, et al. Break-induced replication is highly inaccurate. PLoS Biol. 2011;9:e1000594. doi: 10.1371/journal.pbio.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shee C, Gibson JL, Rosenberg SM. Two mechanisms produce mutation hotspots at DNA breaks in Escherichia coli. Cell Rep. 2012;2:714–21. doi: 10.1016/j.celrep.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burch LH, et al. Damage-induced localized hypermutability. Cell Cycle. 2011;10:1073–85. doi: 10.4161/cc.10.7.15319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan K, et al. Base damage within single-strand DNA underlies in vivo hypermutability induced by a ubiquitous environmental agent. PLoS Genet. 2012;8:e1003149. doi: 10.1371/journal.pgen.1003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts SA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–35. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Gordenin DA, Resnick MA. A single-strand specific lesion drives MMS-induced hyper-mutability at a double-strand break in yeast. DNA Repair (Amst) 2010;9:914–21. doi: 10.1016/j.dnarep.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y, Sterling J, Storici F, Resnick MA, Gordenin DA. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rhind N, Gilbert DM. DNA replication timing. Cold Spring Harb Perspect Biol. 2013;5:a010132. doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stamatoyannopoulos JA. What does our genome encode? Genome Res. 2012;22:1602–11. doi: 10.1101/gr.146506.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thurman RE, Day N, Noble WS, Stamatoyannopoulos JA. Identification of higher-order functional domains in the human ENCODE regions. Genome Res. 2007;17:917–27. doi: 10.1101/gr.6081407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu L, De S, Michor F. DNA replication timing and higher-order nuclear organization determine single-nucleotide substitution patterns in cancer genomes. Nat Commun. 2013;4:1502. doi: 10.1038/ncomms2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Polak P, et al. Reduced local mutation density in regulatory DNA of cancer genomes is linked to DNA repair. Nat Biotechnol. 2014;32:71–5. doi: 10.1038/nbt.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koren A, et al. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am J Hum Genet. 2012;91:1033–40. doi: 10.1016/j.ajhg.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stamatoyannopoulos JA, et al. Human mutation rate associated with DNA replication timing. Nat Genet. 2009;41:393–5. doi: 10.1038/ng.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoang ML, et al. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci Transl Med. 2013;5:197ra102. doi: 10.1126/scitranslmed.3006200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–90. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poon SL, et al. Genome-wide mutational signatures of aristolochic acid and its application as a screening tool. Sci Transl Med. 2013;5:197ra101. doi: 10.1126/scitranslmed.3006086. [DOI] [PubMed] [Google Scholar]
- 32.Schuster-Bockler B, Lehner B. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature. 2012;488:504–7. doi: 10.1038/nature11273. [DOI] [PubMed] [Google Scholar]
- 33.Stratton MR. Exploring the genomes of cancer cells: progress and promise. Science. 2011;331:1553–8. doi: 10.1126/science.1204040. [DOI] [PubMed] [Google Scholar]
- 34.Devarajan K. Nonnegative matrix factorization: an analytical and interpretive tool in computational biology. PLoS Comput Biol. 2008;4:e1000029. doi: 10.1371/journal.pcbi.1000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee DD, Seung HS. Learning the parts of objects by non-negative matrix factorization. Nature. 1999;401:788–91. doi: 10.1038/44565. [DOI] [PubMed] [Google Scholar]
- 36.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–59. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nik-Zainal S, et al. Mutational Processes Molding the Genomes of 21 Breast Cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia P, Pao W, Zhao Z. Patterns and processes of somatic mutations in nine major cancers. BMC Med Genomics. 2014;7:11. doi: 10.1186/1755-8794-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, et al. Evidence for mutation showers. Proc Natl Acad Sci U S A. 2007;104:8403–8. doi: 10.1073/pnas.0610902104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Z, Feng J, Buzin CH, Sommer SS. Epidemiology of doublet/multiplet mutations in lung cancers: evidence that a subset arises by chronocoordinate events. PLoS One. 2008;3:e3714. doi: 10.1371/journal.pone.0003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mimitou EP, Symington LS. DNA end resection--unraveling the tail. DNA Repair (Amst) 2011;10:344–8. doi: 10.1016/j.dnarep.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dewar JM, Lydall D. Similarities and differences between “uncapped” telomeres and DNA double-strand breaks. Chromosoma. 2012;121:117–30. doi: 10.1007/s00412-011-0357-2. [DOI] [PubMed] [Google Scholar]
- 43.Saini N, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–92. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 45.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 46.McInerney P, O'Donnell M. Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J Biol Chem. 2004;279:21543–51. doi: 10.1074/jbc.M401649200. [DOI] [PubMed] [Google Scholar]
- 47.Yeeles JT, Poli J, Marians KJ, Pasero P. Rescuing stalled or damaged replication forks. Cold Spring Harb Perspect Biol. 2013;5:a012815. doi: 10.1101/cshperspect.a012815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sakofsky C.J.e.a. Break-induced replication is a source of mutation clusters underlying kataegis. Cell Rep. 2014;7:1640–1648. doi: 10.1016/j.celrep.2014.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bartek J, Lukas J, Bartkova J. DNA damage response as an anti-cancer barrier: damage threshold and the concept of ‘conditional haploinsufficiency’. Cell Cycle. 2007;6:2344–7. doi: 10.4161/cc.6.19.4754. [DOI] [PubMed] [Google Scholar]
- 50.Costantino L, et al. Break-induced replication repair of damaged forks induces genomic duplications in human cells. Science. 2014;343:88–91. doi: 10.1126/science.1243211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chin K, et al. In situ analyses of genome instability in breast cancer. Nat Genet. 2004;36:984–8. doi: 10.1038/ng1409. [DOI] [PubMed] [Google Scholar]
- 52.Vega F, et al. Splenic marginal zone lymphomas are characterized by loss of interstitial regions of chromosome 7q, 7q31.32 and 7q36.2 that include the protection of telomere 1 (POT1) and sonic hedgehog (SHH) genes. Br J Haematol. 2008;142:216–26. doi: 10.1111/j.1365-2141.2008.07176.x. [DOI] [PubMed] [Google Scholar]
- 53.Poncet D, et al. Changes in the expression of telomere maintenance genes suggest global telomere dysfunction in B-chronic lymphocytic leukemia. Blood. 2008;111:2388–91. doi: 10.1182/blood-2007-09-111245. [DOI] [PubMed] [Google Scholar]
- 54.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–24. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 55.Roberts SA, Gordenin DA. Clustered and genome-wide transient mutagenesis in human cancers: Hypermutation without permanent mutators or loss of fitness. Bioessays. 2014 doi: 10.1002/bies.201300140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roberts SA, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–6. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nature immunology. 2001;2:530–6. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 58.Neuberger MS, Rada C. Somatic hypermutation: activation-induced deaminase for C/G followed by polymerase eta for A/T. J Exp Med. 2007;204:7–10. doi: 10.1084/jem.20062409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu M, Schatz DG. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 2009;30:173–81. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 60.Maul RW, Gearhart PJ. AID and somatic hypermutation. Adv Immunol. 2010;105:159–91. doi: 10.1016/S0065-2776(10)05006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peled JU, et al. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- 62.Rogozin IB, Diaz M. Cutting edge: DGYW/WRCH is a better predictor of mutability at G:C bases in Ig hypermutation than the widely accepted RGYW/WRCY motif and probably reflects a two-step activation-induced cytidine deaminase-triggered process. J Immunol. 2004;172:3382–4. doi: 10.4049/jimmunol.172.6.3382. [DOI] [PubMed] [Google Scholar]
- 63.Migliazza A, et al. Frequent somatic hypermutation of the 5' noncoding region of the BCL6 gene in B-cell lymphoma. Proc Natl Acad Sci U S A. 1995;92:12520–4. doi: 10.1073/pnas.92.26.12520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pasqualucci L, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412:341–6. doi: 10.1038/35085588. [DOI] [PubMed] [Google Scholar]
- 65.Bolli N, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Puente XS, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–5. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Refsland EW, Harris RS. The APOBEC3 Family of Retroelement Restriction Factors. Curr Top Microbiol Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM. Functions and regulation of the APOBEC family of proteins. Semin Cell Dev Biol. 2012;23:258–68. doi: 10.1016/j.semcdb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chan K, Resnick MA, Gordenin DA. The choice of nucleotide inserted opposite abasic sites formed within chromosomal DNA reveals the polymerase activities participating in translesion DNA synthesis. DNA Repair (Amst) 2013 doi: 10.1016/j.dnarep.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gibbs PE, Lawrence CW. Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. J Mol Biol. 1995;251:229–36. doi: 10.1006/jmbi.1995.0430. [DOI] [PubMed] [Google Scholar]
- 71.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–82. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krokan HE, et al. Error-free versus mutagenic processing of genomic uracil-Relevance to cancer. DNA Repair (Amst) 2014 doi: 10.1016/j.dnarep.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 73.Pham P, Chelico L, Goodman MF. DNA deaminases AID and APOBEC3G act processively on single-stranded DNA. DNA Repair (Amst) 2007;6:689–92. doi: 10.1016/j.dnarep.2007.01.001. author reply 693-4. [DOI] [PubMed] [Google Scholar]
- 74.Chelico L, Pham P, Calabrese P, Goodman MF. APOBEC3G DNA deaminase acts processively 3' -- > 5' on single-stranded DNA. Nat Struct Mol Biol. 2006;13:392–9. doi: 10.1038/nsmb1086. [DOI] [PubMed] [Google Scholar]
- 75.Burns MB, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–70. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013 doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taylor BJ, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kidd JM, Newman TL, Tuzun E, Kaul R, Eichler EE. Population stratification of a common APOBEC gene deletion polymorphism. PLoS Genet. 2007;3:e63. doi: 10.1371/journal.pgen.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Long J, et al. A common deletion in the APOBEC3 genes and breast cancer risk. J Natl Cancer Inst. 2013;105:573–9. doi: 10.1093/jnci/djt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xuan D, et al. APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry. Carcinogenesis. 2013 doi: 10.1093/carcin/bgt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang T, et al. Evidence of associations of APOBEC3B gene deletion with susceptibility to persistent HBV infection and hepatocellular carcinoma. Hum Mol Genet. 2013;22:1262–9. doi: 10.1093/hmg/dds513. [DOI] [PubMed] [Google Scholar]
- 82.Nik-Zainal S, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet. 2014 doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pfeifer GP. Mutagenesis at methylated CpG sequences. Curr Top Microbiol Immunol. 2006;301:259–81. doi: 10.1007/3-540-31390-7_10. [DOI] [PubMed] [Google Scholar]
- 84.Visnes T, et al. Uracil in DNA and its processing by different DNA glycosylases. Philos Trans R Soc Lond B Biol Sci. 2009;364:563–8. doi: 10.1098/rstb.2008.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Antequera F. Structure, function and evolution of CpG island promoters. Cell Mol Life Sci. 2003;60:1647–58. doi: 10.1007/s00018-003-3088-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schmidt S, et al. Hypermutable non-synonymous sites are under stronger negative selection. PLoS Genet. 2008;4:e1000281. doi: 10.1371/journal.pgen.1000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Humans, I.W.G.o.t.E.o.C.R.t Radiation. IARC Monogr Eval Carcinog Risks Hum. 2012;100:7–303. [PMC free article] [PubMed] [Google Scholar]
- 88.Plosky BS, Woodgate R. Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr Opin Genet Dev. 2004;14:113–9. doi: 10.1016/j.gde.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 89.Sale JE. Translesion DNA synthesis and mutagenesis in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012708. doi: 10.1101/cshperspect.a012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ikehata H, Ono T. The mechanisms of UV mutagenesis. J Radiat Res. 2011;52:115–25. doi: 10.1269/jrr.10175. [DOI] [PubMed] [Google Scholar]
- 91.Berger MF, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–6. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free Radic Biol Med. 1995;18:1033–77. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 93.Cadet J, Wagner JR. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: induction, repair and significance. Mutat Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 95.Cancer Genome Atlas Research, N Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Degtyareva NP, et al. Oxidative stress-induced mutagenesis in single-strand DNA occurs primarily at cytosines and is DNA polymerase zeta-dependent only for adenines and guanines. Nucleic acids research. 2013;41:8995–9005. doi: 10.1093/nar/gkt671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Moraes EC, Keyse SM, Tyrrell RM. Mutagenesis by hydrogen peroxide treatment of mammalian cells: a molecular analysis. Carcinogenesis. 1990;11:283–93. doi: 10.1093/carcin/11.2.283. [DOI] [PubMed] [Google Scholar]
- 98.Tkeshelashvili LK, McBride T, Spence K, Loeb LA. Mutation spectrum of copper-induced DNA damage. J Biol Chem. 1991;266:6401–6. [PubMed] [Google Scholar]
- 99.Bacolla A, Cooper DN, Vasquez KM. Mechanisms of base substitution mutagenesis in cancer genomes. Genes (Basel) 2014;5:108–46. doi: 10.3390/genes5010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bacolla A, et al. Guanine holes are prominent targets for mutation in cancer and inherited disease. PLoS Genet. 2013;9:e1003816. doi: 10.1371/journal.pgen.1003816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.DeMarini DM. Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat Res. 2004;567:447–74. doi: 10.1016/j.mrrev.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 102.Pfeifer GP, et al. Tobacco smoke carcinogens, DNA damage and p53 mutations in smoking-associated cancers. Oncogene. 2002;21:7435–51. doi: 10.1038/sj.onc.1205803. [DOI] [PubMed] [Google Scholar]
- 103.Govindan R, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–34. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee W, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–7. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 106.Bodell WJ, Gaikwad NW, Miller D, Berger MS. Formation of DNA adducts and induction of lacI mutations in Big Blue Rat-2 cells treated with temozolomide: implications for the treatment of low-grade adult and pediatric brain tumors. Cancer Epidemiol Biomarkers Prev. 2003;12:545–51. [PubMed] [Google Scholar]
- 107.Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair (Amst) 2004;3:1091–101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 108.Tomita-Mitchell A, et al. Mismatch repair deficient human cells: spontaneous and MNNG-induced mutational spectra in the HPRT gene. Mutat Res. 2000;450:125–38. doi: 10.1016/s0027-5107(00)00020-8. [DOI] [PubMed] [Google Scholar]
- 109.Cahill DP, et al. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13:2038–45. doi: 10.1158/1078-0432.CCR-06-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hunter C, et al. A hypermutation phenotype and somatic MSH6 mutations in recurrent human malignant gliomas after alkylator chemotherapy. Cancer Res. 2006;66:3987–91. doi: 10.1158/0008-5472.CAN-06-0127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Moen EL, Stark AL, Zhang W, Dolan ME, Godley LA. The role of gene body cytosine modifications in MGMT expression and sensitivity to temozolomide. Mol Cancer Ther. 2014 doi: 10.1158/1535-7163.MCT-13-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang J, et al. Certain imidazotetrazines escape O6-methylguanine-DNA methyltransferase and mismatch repair. Oncology. 2011;80:195–207. doi: 10.1159/000327837. [DOI] [PubMed] [Google Scholar]
- 113.Cancer Genome Atlas Research, N Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Johnson BE, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–93. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Arlt VM, Stiborova M, Schmeiser HH. Aristolochic acid as a probable human cancer hazard in herbal remedies: a review. Mutagenesis. 2002;17:265–77. doi: 10.1093/mutage/17.4.265. [DOI] [PubMed] [Google Scholar]
- 116.National Toxicology, P Aristolochic acids. Rep Carcinog. 2011;12:45–9. [PubMed] [Google Scholar]
- 117.Sidorenko VS, et al. Lack of recognition by global-genome nucleotide excision repair accounts for the high mutagenicity and persistence of aristolactam-DNA adducts. Nucleic acids research. 2012;40:2494–505. doi: 10.1093/nar/gkr1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Chen CH, et al. Aristolochic acid-associated urothelial cancer in Taiwan. Proc Natl Acad Sci U S A. 2012;109:8241–6. doi: 10.1073/pnas.1119920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hollstein M, Moriya M, Grollman AP, Olivier M. Analysis of TP53 mutation spectra reveals the fingerprint of the potent environmental carcinogen, aristolochic acid. Mutat Res. 2013;753:41–9. doi: 10.1016/j.mrrev.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 121.Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 122.Shcherbakova PV, Fijalkowska IJ. Translesion synthesis DNA polymerases and control of genome stability. Front Biosci. 2006;11:2496–517. doi: 10.2741/1985. [DOI] [PubMed] [Google Scholar]
- 123.Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol. 2010;7:153–62. doi: 10.1038/nrclinonc.2009.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nagarajan N, et al. Whole-genome reconstruction and mutational signatures in gastric cancer. Genome Biol. 2012;13:R115. doi: 10.1186/gb-2012-13-12-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.TCGA Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kim TM, Laird PW, Park PJ. The landscape of microsatellite instability in colorectal and endometrial cancer genomes. Cell. 2013;155:858–68. doi: 10.1016/j.cell.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Briggs S, Tomlinson I. Germline and somatic polymerase epsilon and delta mutations define a new class of hypermutated colorectal and endometrial cancers. J Pathol. 2013;230:148–53. doi: 10.1002/path.4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Palles C, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat Genet. 2013;45:136–44. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Valle L, et al. New insights into POLE and POLD1 germline mutations in familial colorectal cancer and polyposis. Hum Mol Genet. 2014 doi: 10.1093/hmg/ddu058. [DOI] [PubMed] [Google Scholar]
- 130.Zou Y, et al. Frequent POLE1 p.S297F mutation in Chinese patients with ovarian endometrioid carcinoma. Mutat Res. 2014;761:49–52. doi: 10.1016/j.mrfmmm.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 131.Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–144. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cancer Genome Atlas, N Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cancer Genome Atlas Research, N et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Donehower LA, et al. MLH1-silenced and non-silenced subgroups of hypermutated colorectal carcinomas have distinct mutational landscapes. J Pathol. 2013;229:99–110. doi: 10.1002/path.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Matsuda T, Bebenek K, Masutani C, Hanaoka F, Kunkel TA. Low fidelity DNA synthesis by human DNA polymerase-eta. Nature. 2000;404:1011–3. doi: 10.1038/35010014. [DOI] [PubMed] [Google Scholar]
- 136.Machida K, et al. Hepatitis C virus induces a mutator phenotype: enhanced mutations of immunoglobulin and protooncogenes. Proc Natl Acad Sci U S A. 2004;101:4262–7. doi: 10.1073/pnas.0303971101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Totoki Y, et al. High-resolution characterization of a hepatocellular carcinoma genome. Nat Genet. 2011;43:464–9. doi: 10.1038/ng.804. [DOI] [PubMed] [Google Scholar]
- 138.Burrell RA, Swanton C. The evolution of the unstable cancer genome. Curr Opin Genet Dev. 2013;24C:61–67. doi: 10.1016/j.gde.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 139.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. APOBEC-Mediated Cytosine Deamination Links PIK3CA Helical Domain Mutations to Human Papillomavirus-Driven Tumor Development. Cell Rep. 2014 doi: 10.1016/j.celrep.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 142.Viros A, et al. Ultraviolet radiation accelerates BRAF-driven melanomagenesis by targeting TP53. Nature. 2014 doi: 10.1038/nature13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dulak AM, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013;45:478–86. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jager N, et al. Hypermutation of the inactive X chromosome is a frequent event in cancer. Cell. 2013;155:567–81. doi: 10.1016/j.cell.2013.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kumar A, et al. Exome sequencing identifies a spectrum of mutation frequencies in advanced and lethal prostate cancers. Proc Natl Acad Sci U S A. 2011;108:17087–92. doi: 10.1073/pnas.1108745108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–77. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Poon S, McPherson J, Tan P, Teh B, Rozen S. Mutation signatures of carcinogen exposure: genome-wide detection and new opportunities for cancer prevention. Genome Medicine. 2014;6:24. doi: 10.1186/gm541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012 doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nik-Zainal S, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]