Abstract
Background
The 2-ΔΔCT method has been extensively used as a relative quantification strategy for quantitative real-time polymerase chain reaction (qPCR) data analysis. This method is a convenient way to calculate relative gene expression levels between different samples in that it directly uses the threshold cycles (CTs) generated by the qPCR system for calculation. However, this approach relies heavily on an invalid assumption of 100% PCR amplification efficiency across all samples. In addition, the 2-ΔΔCT method is applied to data with automatic removal of background fluorescence by the qPCR software. Since the background fluorescence is unknown, subtracting an inaccurate background can lead to distortion of the results. To address these problems, we present an improved method, the individual efficiency corrected calculation.
Results
Our method takes into account the PCR efficiency of each individual sample. In addition, it eliminates the need for background fluorescence estimation or subtraction because the background can be cancelled out using the differencing strategy. The DNA amount for a certain gene and the relative DNA amount among different samples estimated using our method were closer to the true values compared to the results of the 2-ΔΔCT method.
Conclusions
The improved method, the individual efficiency corrected calculation, produces more accurate estimates in relative gene expression than the 2-ΔΔCT method and is thus a better way to calculate relative gene expression.
Background
Quantitative real-time polymerase chain reaction (qPCR) has been extensively used to quantify gene expression levels. The two strategies for analyzing qPCR data are absolute and relative quantification (1–3). Absolute quantification identifies the input gene amount based on a standard curve. In contrast, relative quantification determines changes in gene expression relative to a reference sample. Relative quantification is easier to perform than absolute quantification, and it requires fewer reagents, since there is no need to generate a standard curve (4). Errors caused by standard dilutions when creating a standard curve can also be avoided. In addition, sometimes the relative gene amount between two treatment groups is of more interest than exact DNA/RNA molecular numbers. Therefore, relative quantification is widely performed.
The 2-ΔΔCT method is the method of relative quantification that is most frequently found in popular software packages for qPCR experiments (1,5–6). The threshold cycle (CT) is the cycle at which the fluorescence level reaches a certain amount (the threshold). This method directly uses the CT information generated from a qPCR system to calculate relative gene expression in target and reference samples, using a reference gene as the normalizer. Table 1 illustrates a typical study design related to samples and genes. A target sample may be, for instance, a treated sample, while a reference sample is an untreated control. Target and reference samples can also be samples from different tissues, samples treated at different time points, or samples from other distinct groups. To correct for differences in the amount of DNA/RNA added for each sample and to reduce variation caused by PCR set-up and the cycling process, reference genes or internal control genes have been used to normalize the PCRs (2). Housekeeping genes, such as glyceraldehyde 3-phosphate dehydrogenase (GAPDH), (β-actin, and 18S rRNA, are commonly used reference genes because their expression levels remain relatively stable in response to any treatment (1,7–8). With respect to the ΔΔCT of the 2- ΔΔCT method, the first ΔCT is the difference in threshold cycle between the target and reference genes:
Table 1. An Illustration of the Experimental Design.
| Reference sample | Target sample | |
|---|---|---|
| Reference gene | A | B |
| Target gene | C | D |
Note: In the experiment described in the Methods section, there are four combinations of samples and genes in the experimental design, referred as A, B, C, and D. In each of the four combinations, there are six replicates. The reference samples in this study are those without dilution. The target samples are those with dilutions of 1:10, 1:100, and 1:1000, respectively. The reference gene is human GAPDH, and the target gene is human FAM73B.
Using the notation in Table 1, the ΔCT for the target sample is CTD −CTB, and the ΔCT for the reference sample is CTC −CTA. The ΔΔCT is the difference in ΔCT as described in the above formula between the target and reference samples, which is
The final result of this method is presented as the fold change of target gene expression in a target sample relative to a reference sample, normalized to a reference gene. The relative gene expression is usually set to 1 for reference samples because ΔΔCT is equal to 0 and therefore 20 is equal to 1.
The 2-ΔΔCT method assumes a uniform PCR amplification efficiency of 100% across all samples (1,9). The value 2 is 1 plus a PCR amplification efficiency of 1 (100%). This assumption makes the method easy to perform, and it can be valid under optimal conditions. However, PCR efficiency cannot be 100% because of factors such as the presence of PCR inhibitors or enhancers, RNA extraction, and different uses of probes, primers, and enzymes. These factors also contribute to variations in efficiency (2,10–11). Therefore, the assumption becomes problematic when PCR efficiencies vary among samples, which is always the case in reality. Previous studies have suggested that PCR efficiencies vary from 60% to 110% (2), from 80% to 100% (12–13), and from 65% to 90% (14). Variations in PCR efficiency then lead to distortion of 2-ΔΔCT results. For example, when efficiencies vary over a range as small as 0.04, from 1.78 to 1.82, it results in a 4-fold error in fold difference (13). A difference in the PCR efficiency of 5% between a target gene and a reference gene can lead to a miscalculated difference in expression ratio by 432% (15). Hence, it is necessary to assess sample-specific PCR efficiencies before doing relative quantification.
Another problem of PCR data analysis is determining the background fluorescence level. Background fluorescence may originate from unbound SYBR Green dye or from fluorochrome bound to the template cDNA or primers with nontarget DNA binding (16). Thus, failing to remove background fluorescence or incorrectly subtracting it will lead to invalid results (17–20). First, subtracting an incorrect background can cause miscalculation of both the gene amount and PCR efficiency (16,21). Second, different model fittings result in very different background fluorescence estimates (Table 1; 22). Third, it is unreasonable to apply a constant background to all samples in a PCR experiment or to different PCRs. Therefore, comparisons among many samples can be difficult (2). Finally, Rebrikov and Trofimov suggested that background fluoresence can be divided into constant and variable components, the latter of which is a function of the cycle number (18). If this is the case, it greatly increases the complexity of estimating the background fluorescence.
In the present study, we propose a new algorithm, the individual efficiency corrected calculation method that takes into account the PCR efficiency of an individual sample and uses sample-specific efficiencies to calculate the fold changes in gene expression between samples. Additionally, the differencing strategy of this method cancels out the background fluorescence and therefore avoids the errors caused by either not or incorrectly removing it. The results of this study suggest that the individual efficiency corrected calculation method provides more accurate estimates of relative gene expression than the commonly used 2-ΔΔCT method.
Results
The current study compares the accuracy and precision of DNA amounts estimated using the 2-ΔΔCT method and the individual efficiency corrected calculation method. The accuracy of these methods was assessed by two criteria. First, the estimates of the initial DNA amount for the four dilutions should be close to the ratios of 1, 1:10 (0.1), 1:100 (0.01), and 1:1000 (0.001) for either FAM73B or GAPDH. The results of the two methods are shown in Table 3. For FAM73B, the 2-ΔΔCT method gave the estimated ratios of 1, 0.44 (far from 0.1), 0.038 (far from 0.01), and 0.0013 (close to 0.001). CVs for the 6 replicates in each dilution ranged from 11% to 21%. For the control gene, GAPDH, the estimates were 1, 0.06, 0.005, and 0.0005, which were close to the true ratios. CVs were smaller, with a range of 5%–12%. By contrast, the individual efficiency corrected calculation method gave estimated ratios of 1, 0.36, 0.025, and 0.0017 for FAM73B and 1, 0.09, 0.005, and 0.0036 for GAPDH. These values were overall closer to 1, 0.1, and 0.01, respectively, but further from 0.001 than the 2-ΔΔCT method. CVs were 12%–20% for FAM73B and 4%–9% for GAPDH, which were comparable to those of the 2-ΔΔCT method. These results suggest that the individual efficiency corrected calculation method results in a more accurate estimation of the DNA amount for a certain gene with similar variation than does the 2-ΔΔCT method.
Table 3. Comparisons between the Method and the Individual Efficiency Corrected Calculation Method.
| FAM73B | GAPDH | FAM73B/GAPDH | ||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| True ratio | Estimated ratio | CV | Estimated ratio | CV | True relative gene expression | Estimated relative gene expression | CV | |
| 2−ΔΔCT method | 1 | 1 | 11.2% | 1 | 4.7% | 1 | 1 | 11.2% |
| 0.1 | 0.44 | 18.9% | 0.06 | 7.0% | 1 | 7.52 | 18.9% | |
| 0.01 | 0.038 | 12.6% | 0.005 | 8.3% | 1 | 7.42 | 12.6% | |
| 0.001 | 0.0013 | 20.9% | 0.0005 | 11.6% | 1 | 2.70 | 20.9% | |
|
| ||||||||
| Individual efficiency corrected calculation method | 1 | 1 | 12.0% | 1 | 3.9% | 1 | 1 | 12.0% |
| 0.1 | 0.36 | 15.9% | 0.09 | 5.5% | 1 | 3.82 | 15.9% | |
| 0.01 | 0.025 | 11.9% | 0.005 | 6.3% | 1 | 4.83 | 11.9% | |
| 0.001 | 0.0017 | 19.5% | 0.0036 | 8.7% | 1 | 0.47 | 19.5% | |
The second criterion for assessing the accuracy of the two methods relies on the idea that the relative gene amounts (FAM73B/GAPDH) among the four dilution conditions should be almost the same. If the value of the original sample without dilution was set to 1, then the true ratio would be 1: 1: 1: 1 for all dilution conditions. The ratio for the 2-ΔΔCT method was 1: 7.52: 7.42: 2.7, with CVs ranging from 11% to 21%. By contrast, the individual efficiency corrected calculation method gave a better estimated ratio of 1: 3.82: 4.83: 0.47, with comparable CVs of 12%–20%. These values are much closer to 1 than those of the 2-ΔΔCT method. Collectively, our method produced more accurate estimates in relative gene expression level with similar variation than did the 2-ΔΔCT method.
Discussion
The results of this study suggest that the individual efficiency corrected calculation method is an improved algorithm over the 2-ΔΔCT method in terms of estimates of relative gene amounts that are closer to true values. Our method is advantageous in that it involves calculation of PCR amplification efficiency in an individual sample and it avoids potential errors related to background fluorescence subtraction.
The PCR efficiencies estimated using the individual efficiency corrected calculation method ranged from 1.73 to 1.88 for the FAM73B gene and from 1.55 to 1.74 for the GAPDH gene. These numbers obviously conflict with the uniform efficiency among all samples assumed by the 2-ΔΔCT method. Therefore, it is not surprising to see the big variation between the ratio of relative gene amount, 1: 7.52: 7.42: 2.7, estimated using the 2-ΔΔCT method and the true ratio of 1: 1: 1: 1. Yet we cannot rule out the possibility of experimental errors or sample variation.
The assumption of the same sample-to-sample efficiency for the 2-ΔΔCT method can only be relaxed if very specific primers are designed and the experimental conditions are approximately the same for every sample. However, this is hard to achieve. Different approaches have been used to calculate the PCR amplification efficiency. Pfaffl presented efficiency-corrected calculation models based on multiple samples and/or multiple reference genes, taking into account the differences in amplification efficiency between target and reference genes (2). But these methods assumed equal efficiencies between control and treated samples. This assumption is not true according to our data. In fact, every sample has its own amplification efficiency. Another method is the dilution method (23), which calculated the amplification rate E based on the slope of a linear curve of a series of dilutions (E =10-1/slope). This approach has been found to be highly reproducible, but it tends to overestimate efficiency (by sometimes more than 100%). Besides this, the dilution method can be laborious and time- and reagent-consuming if it is conducted for different samples (15). The linear regression model has also been used to fit data points within the exponential phase of the PCR amplification curves, and the estimated parameter has been used to calculate the individual efficiency in the formula of E =10slope (13). This efficiency estimation is similar to ours, but our linear regression model incorporates the differencing strategy that removes any potential influence of background fluorescence. Nonlinear regression models, such as sigmoidal curve fittings, have also been used to estimate PCR efficiency. In these models, all data points in an individual PCR run were used in the curve fittings, and individual efficiencies could be calculated using estimated parameters and information about the exponential amplification phase (14,24). However, background fluorescence was included in the models. Hence, incorrect values for unknown background fluorescence may affect the whole sigmoidal curve fitting and identification of the exponential phase and thereafter distort the result (14,25–26). Additionally, Schefe et al. developed the gene expression CT difference (GED) method to compute individual efficiencies for all samples separately (27). They found that the 2-ΔΔCT method tended to overestimate differences in gene expression and that it was crucial to detect efficiency outliers. But there was no consideration of background fluorescence during the derivation of the GED formula. Considering the constant and variable components of the background fluorescence (18), our method is superior in that it eliminates the need to estimate the background, whereas all the above methods either do not consider the background or estimate a problematic background.
Another assumption of the 2-ΔΔCT method is that internal control genes are constantly expressed without being affected by treatment. However, variations in the expression of these genes may depend on species, tissue types, disease conditions, and other potential factors (28). It has been shown that the hydroxymethylbilane synthase, beta-2-microglobulin, and β-actin genes are appropriate normalizers in osteoclasts (29). Another study suggested that the combination of GAPDH and mitogen-activated protein kinase 1 genes had the highest stability for most experimental sets (30). In addition, 18S rRNA was found to be the best invariant internal control gene in grass carp (31). Thus, it may be necessary to determine whether candidate genes have a stable expression level among different samples before conducting a qPCR experiment.
Conclusions
Collectively, the individual efficiency corrected calculation method is an improvement of the commonly used 2-ΔΔCT method in that it calculates an individual efficiency for each sample and prevents potential problems related to background fluorescence estimation. Thus, this new approach is more accurate for such relative quantification.
Methods
The expression levels of human family with sequence similarity 73, member B (FAM73B) and GAPDH genes in limited dilution samples (1:10, 1:100, and 1:1000) were analyzed using qPCR. GAPDH was used as an internal control. Referring to the study design in Table 1, FAM73B is the target gene, and GAPDH is the reference gene. The original sample without any dilution is the reference, and a 1:10, 1:100, and 1:1000 dilution of the original sample are the target samples. There are 6 replicates in each combination of gene and dilution. The gene-specific primers were as follows: hgapdh-5′-ATGGAAATCCCATCACCATCTT-3′ and hgapdh-5′-CGCCCCACTTGATTTTGG-3′; hfam73b-5′-CTCCTGCAGGTGGTAGGC-3′ and hfam73b-5′-CAGAGACTGCATCAGAGCCA-3′. mRNA was extracted from human hepatoma (Huh7) cells and used as a template for reverse transcription by superscript III reverse transcriptase purchased from Invitrogen (Carlsbad, CA). All qPCR experiments were performed using the Applied Biosystems Stepone and StepOnePlus Real-Time system (Perkin-Elmer Applied Biosystems). All the amplifications were done using SYBR Green PCR Master Mix (Applied Biosystems). The thermal cycling conditions included an initial denaturation step at 95°C for 10 min, followed by 40 cycles at 95°C for 30s, 60°C for 30s, and 72°C for 30s. Melting curve analysis of every qPCR was conducted after each cycle.
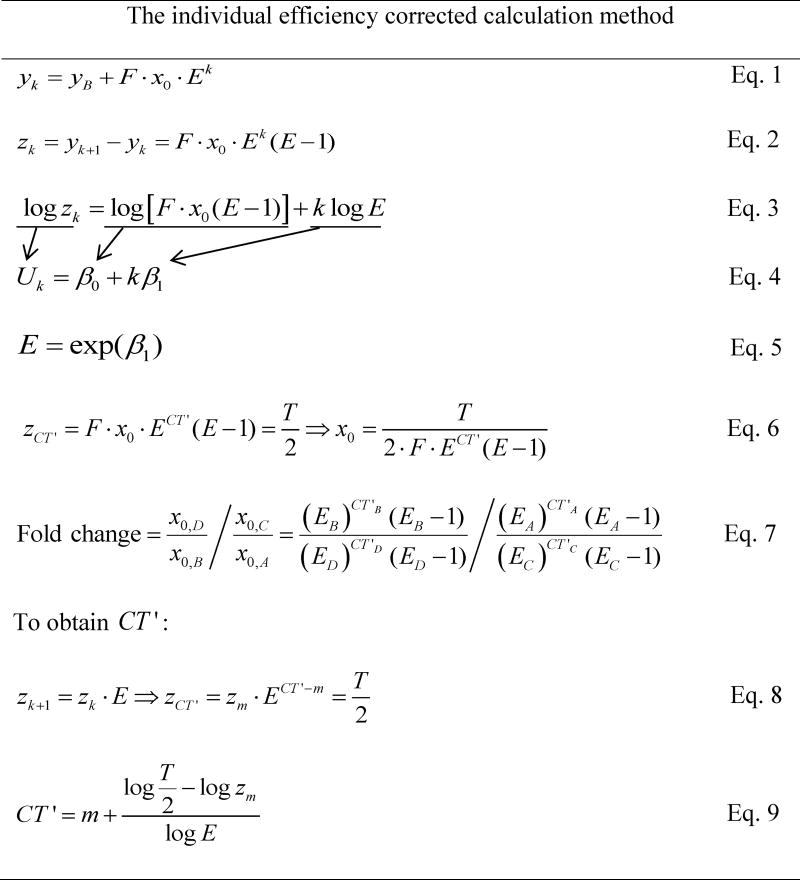
In this study, we try to improve the 2-ΔΔCT method. Our method, called the individual efficiency–corrected calculation method, is shown in Table 2. Unlike the 2-ΔΔCT method, our method accounts for individual efficiencies of samples. We computed the amplification rate E (1 + efficiency) for each sample (Table 2, Eqs. 1-5). Specifically, the method was based on an exponential function, and background fluorescence was included in this function (Eq. 1). Then we took the difference between two consecutive PCR cycles by subtracting the fluorescence of the former cycle from that of the later cycle (Eq. 2). Therefore, the data with n cycles were transformed to data with n−1 cycles. Importantly, background fluorescence was removed. After that, a simple linear regression model (Eq. 4) was applied to the log-transformed equation (Eq. 3). The parameter (β1) estimated using linear regression can be used to calculate E (Eq. 5). To calculate the starting DNA amount (x0), we need to find out the new threshold cycle, CT', and we set the new threshold to T/2 (Eqs. 2 and 6). The fold change of gene expression level was calculated as the relative DNA amount of a target gene in a target sample and a reference sample, normalized to a reference gene (Eq. 7). The DNA amounts of a reference gene in reference and target samples are denoted as x0,A and x0,B, and the amounts of a target gene in the two groups are denoted as x0,C and x0,D, respectively. The derivation of CT' was based on the equal-ratio property of the difference value zk and the cycle m, which is an integer cycle right before CT' (Eqs. 8-9). The fluorescence value zm should be less than the new threshold T/2 because the selected data points had monotone increasing values of zk.
Table 2. Equations Used in the Individual Efficiency Corrected Calculation Method.
|
Note: Eq. 1 states that the observed fluorescence level after k cycles (yk) is the background fluorescence (yB) plus the initial DNA amount (x0) times the conversion factor F and Ek. F is the conversion factor between the number of target molecules and observed fluorescence, and E is 1 + amplification efficiency. Eq. 2 indicates that we take the difference in fluorescence for each of two consecutive cycles. After taking the natural logarithm on both sides of Eq. 2, we get Eq. 3. Then we apply linear regression in the form of Eq. 4 and calculate E using the estimated slope according to Eq. 5. Basically, equations 1–5 show the derivation of E. Thereafter, we plug the new threshold cycle (CT') into Eq. 2 and derive gene expression or starting DNA (x0) as indicated in Eq. 6. Fold change is computed by comparing the relative gene expression between target and reference samples (Eq. 7). The A, B, C, and D that are used to denote x0 refer to different experimental combinations described in Table 1. Eqs. 8–9 are employed to obtain CT'. Eq. 8 is derived from Eq. 2, and the new threshold is set to be half of the threshold (T). Here, m is the integer cycle right before CT', therefore, its corresponding fluorescence value (zm) is smaller than the new threshold (T/2). Thus, CT' can be obtained (Eq. 9).
For the 2-ΔΔCT method, we directly used the threshold cycle values automatically generated by the qPCR system. For the individual efficiency corrected calculation method, we selected four successive cycles for every PCR run, the first three of which have the fluorescence values below the threshold and the last of which has a fluorescence value larger than the threshold. Therefore, the target cycles are the rounded threshold cycle and the former three cycles, or the rounded threshold cycle and the former two cycles plus the latter one.
In the individual efficiency corrected calculation method, we calculated PCR amplification efficiency for every sample. To reduce potential variation, we then took the mean of the efficiencies for the 6 replicates under each condition, which is a combination of gene and dilution. Hence, the 6 replicates had the same efficiency for further calculation, but each combination (for example, the combination of FAM73B and a 1:10 dilution) had a different efficiency.
Because the 2-ΔΔCT method and our method are relative quantification strategies, it is difficult to assess their accuracy. This is why we planned to use a series of dilutions of the original sample to evaluate the accuracy of these two methods based on the pattern of the estimates. According to the experimental design, there were two trends in the estimates. First, for each gene, the ratios of the initial DNA amount were 1, 1:10 (0.1), 1:100 (0.01), and 1:1000 (0.001), corresponding to the four dilution conditions. The second trend was that the relative gene amounts (FAM73B/GAPDH) with respect to the four dilution conditions were the same, with a ratio of 1: 1: 1: 1 if the original sample without dilution was set to 1. The precision of the methods was then analyzed by computing coefficients of variation (CVs). The equation is:
where s and x̄ are the standard deviation and the mean of the 6 replicates in each combination of gene and dilution.
Acknowledgments
This research was supported by the U.S.A. National Institutes of Health grants 5P50 CA100632 and 5PO1 CA055164. Funding for open access charge: National Institutes of Health.
Footnotes
Authors' contributions: XR participated in the design of the study and performed the statistical analysis, and drafted the manuscript. XH conceived of the study, and supervised the study design and development of the statistical method, and revised the manuscript. ZZ designed and carried out the qPCR experiment. XL supervised the design and conduction of the qPCR experiment.
Contributor Information
Xiayu Rao, Email: rxy712@gmail.com.
Xuelin Huang, Email: xlhuang@mdanderson.org.
Zhicheng Zhou, Email: zzhou2@mdanderson.org.
Xin Lin, Email: xllin@mdanderson.org.
References
- 1.Livak KJ, Schmittgen TD. Methods. Vol. 25. San Diego, CA: 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method; pp. 402–408. [DOI] [PubMed] [Google Scholar]
- 2.Pfaffl MW. Quantification strategies in real-time PCR. In: Bustin SA, editor. The Real-Time PCR Encyclopedia A–Z of Quantitative PCR. International University Line; La Jolla, CA: 2004. pp. 87–112. Published by. [Google Scholar]
- 3.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 4.Bustin SA. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 5.Schmittgen TD, Zakrajsek BA, Mills AG, Gorn V, Singer MJ, Reed MW. Quantitative reverse transcription-polymerase chain reaction to study mRNA decay: comparison of endpoint and real-time methods. Analytical biochemistry. 2000;285:194–204. doi: 10.1006/abio.2000.4753. [DOI] [PubMed] [Google Scholar]
- 6.Winer J, Jung CK, Shackel I, Williams PM. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Analytical biochemistry. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 7.Bas A, Forsberg G, Hammarstrom S, Hammarstrom ML. Utility of the housekeeping genes 18S rRNA, beta-actin and glyceraldehyde-3-phosphate-dehydrogenase for normalization in real-time quantitative reverse transcriptase-polymerase chain reaction analysis of gene expression in human T lymphocytes. Scand J Immunol. 2004;59:566–573. doi: 10.1111/j.0300-9475.2004.01440.x. [DOI] [PubMed] [Google Scholar]
- 8.Morse DL, Carroll D, Weberg L, Borgstrom MC, Ranger-Moore J, Gillies RJ. Determining suitable internal standards for mRNA quantification of increasing cancer progression in human breast cells by real-time reverse transcriptase polymerase chain reaction. Analytical biochemistry. 2005;342:69–77. doi: 10.1016/j.ab.2005.03.034. [DOI] [PubMed] [Google Scholar]
- 9.Arocho A, Chen B, Ladanyi M, Pan Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol. 2006;15:56–61. doi: 10.1097/00019606-200603000-00009. [DOI] [PubMed] [Google Scholar]
- 10.Liu W, Saint DA. A new quantitative method of real time reverse transcription polymerase chain reaction assay based on simulation of polymerase chain reaction kinetics. Analytical biochemistry. 2002;302:52–59. doi: 10.1006/abio.2001.5530. [DOI] [PubMed] [Google Scholar]
- 11.Mygind T, Birkelund S, Birkebaek NH, Ostergaard L, Jensen JS, Christiansen G. Determination of PCR efficiency in chelex-100 purified clinical samples and comparison of real-time quantitative PCR and conventional PCR for detection of Chlamydia pneumoniae. BMC Microbiol. 2002;2:17. doi: 10.1186/1471-2180-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamphuis W, Schneemann A, van Beek LM, Smit AB, Hoyng PF, Koya E. Prostanoid receptor gene expression profile in human trabecular meshwork: a quantitative real-time PCR approach. Invest Ophthalmol Vis Sci. 2001;42:3209–3215. [PubMed] [Google Scholar]
- 13.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neuroscience letters. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 14.Tichopad A, Dilger M, Schwarz G, Pfaffl MW. Standardized determination of real-time PCR efficiency from a single reaction set-up. Nucleic acids research. 2003;31:e122. doi: 10.1093/nar/gng122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaffl MW. Relative quantification. In: Dorak T, editor. Real Time PCR BIOS Advanced Methods. Taylor & Francis; 2006. pp. 63–82. Vol. [Google Scholar]
- 16.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, van den Hoff MJ, Moorman AF. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic acids research. 2009;37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bar T, Stahlberg A, Muszta A, Kubista M. Kinetic Outlier Detection (KOD) in real-time PCR. Nucleic acids research. 2003;31:e105. doi: 10.1093/nar/gng106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rebrikov DV, Trofimov D. Real-time PCR: approaches to data analysis (a review) Prikl Biokhim Mikrobiol. 2006;42:520–528. [PubMed] [Google Scholar]
- 19.Rutledge RG. Sigmoidal curve-fitting redefines quantitative real-time PCR with the prospective of developing automated high-throughput applications. Nucleic acids research. 2004;32:e178. doi: 10.1093/nar/gnh177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rutledge RG, Stewart D. A kinetic-based sigmoidal model for the polymerase chain reaction and its application to high-capacity absolute quantitative real-time PCR. BMC Biotechnol. 2008;8:47. doi: 10.1186/1472-6750-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shain EB, Clemens JM. A new method for robust quantitative and qualitative analysis of real-time PCR. Nucleic acids research. 2008;36:e91. doi: 10.1093/nar/gkn408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guescini M, Sisti D, Rocchi MB, Stocchi L, Stocchi V. A new real-time PCR method to overcome significant quantitative inaccuracy due to slight amplification inhibition. BMC bioinformatics. 2008;9:326. doi: 10.1186/1471-2105-9-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rasmussen R. Quantification on the LightCycler. In: Meuer S, Wittwer C, Nakagawara K, editors. Rapid cycle real-time PCR, methods and applications. Springer Press; Heidelberg: 2001. [Google Scholar]
- 24.Liu W, Saint DA. Validation of a quantitative method for real time PCR kinetics. Biochem Biophys Res Commun. 2002;294:347–353. doi: 10.1016/S0006-291X(02)00478-3. [DOI] [PubMed] [Google Scholar]
- 25.Swillens S, Dessars B, Housni HE. Revisiting the sigmoidal curve fitting applied to quantitative real-time PCR data. Analytical biochemistry. 2008;373:370–376. doi: 10.1016/j.ab.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 26.Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H. Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression's CT difference” formula. J Mol Med (Berl) 2006;84:901–910. doi: 10.1007/s00109-006-0097-6. [DOI] [PubMed] [Google Scholar]
- 28.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 29.Stephens AS, Stephens SR, Morrison NA. Internal control genes for quantitative RT-PCR expression analysis in mouse osteoblasts, osteoclasts and macrophages. BMC Res Notes. 2011;4:410. doi: 10.1186/1756-0500-4-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rocha-Martins M, Njaine B, Silveira MS. Avoiding pitfalls of internal controls: validation of reference genes for analysis by qRT-PCR and Western blot throughout rat retinal development. PLoS One. 2012;7:e43028. doi: 10.1371/journal.pone.0043028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su J, Zhang R, Dong J, Yang C. Evaluation of internal control genes for qRT-PCR normalization in tissues and cell culture for antiviral studies of grass carp (Ctenopharyngodon idella) Fish Shell fish Immunol. 2011;30:830–835. doi: 10.1016/j.fsi.2011.01.006. [DOI] [PubMed] [Google Scholar]


