Significance
Essential tremor is one of the most frequent movement disorders of humans, but its causes remain largely unknown. In a six-generation family with both essential tremor and Parkinson disease, we identified a rare missense mutation of HTRA2 as the causative allele. Family members homozygous for this allele were more severely affected than those heterozygous for this allele. The same mutation had been associated with Parkinson characteristics in mouse mutants and with Parkinson disease in some, but not all, epidemiologic studies. Our results suggest that HTRA2 may be responsible for essential tremor in some families and that homozygosity for damaging alleles of HTRA2 may be responsible for Parkinson disease.
Keywords: gene identification, neurodegenerative disease, mitochondrial dysfunction, DNA sequencing, mutation
Abstract
Essential tremor is one of the most frequent movement disorders of humans and can be associated with substantial disability. Some but not all persons with essential tremor develop signs of Parkinson disease, and the relationship between the conditions has not been clear. In a six-generation consanguineous Turkish kindred with both essential tremor and Parkinson disease, we carried out whole exome sequencing and pedigree analysis, identifying HTRA2 p.G399S as the allele likely responsible for both conditions. Essential tremor was present in persons either heterozygous or homozygous for this allele. Homozygosity was associated with earlier age at onset of tremor (P < 0.0001), more severe postural tremor (P < 0.0001), and more severe kinetic tremor (P = 0.0019). Homozygotes, but not heterozygotes, developed Parkinson signs in the middle age. Among population controls from the same Anatolian region as the family, frequency of HTRA2 p.G399S was 0.0027, slightly lower than other populations. HTRA2 encodes a mitochondrial serine protease. Loss of function of HtrA2 was previously shown to lead to parkinsonian features in motor neuron degeneration (mnd2) mice. HTRA2 p.G399S was previously shown to lead to mitochondrial dysfunction, altered mitochondrial morphology, and decreased protease activity, but epidemiologic studies of an association between HTRA2 and Parkinson disease yielded conflicting results. Our results suggest that in some families, HTRA2 p.G399S is responsible for hereditary essential tremor and that homozygotes for this allele develop Parkinson disease. This hypothesis has implications for understanding the pathogenesis of essential tremor and its relationship to Parkinson disease.
Essential tremor is one of the most frequent movement disorders in humans (1). It is characterized primarily by postural or kinetic tremor of the arms and hands, but head, legs, voice, and other regions of the body may also be affected (2). The worldwide prevalence is 0.9%, increasing to more than 4% in elderly populations (1). Familial essential tremor is genetically heterogeneous. Genetic linkage studies of multiply affected families revealed three genomic regions segregating with the condition, on chromosomes 3q13 [ETM1; Online Mendelian Inheritance in Man (OMIM) 190300], 2p22-24 (ETM2; OMIM 602134), and 6p23 (ETM3; OMIM 611456) (3–5). No clearly causal mutations have been identified in these regions, although the common variant DRD3 p.S9G in the ETM1 region has been proposed as a risk factor and HS1BP3 p.A265G in the ETM2 region appeared in two multiply affected families (6, 7). Genomewide association studies of essential tremor reported associations with common variants in an intron of LINGO1 and in an intron of SLC1A2 (8–10). Recently, DNAJC13 p.N855S, which had been identified in Parkinson disease patients, was also found in two unrelated patients with essential tremor (11). Nonsense mutation p.Q290X in the RNA-binding protein FUS was identified by whole exome sequencing in a large family with essential tremor (ETM4; OMIM 614782) (12). Screening other subjects with essential tremor for FUS revealed two rare missense variants, suggesting that mutations in FUS explain a subset of cases with the condition (13, 14).
In this study, we examined a six-generation family segregating essential tremor, and in multiple relatives, essential tremor as a feature of Parkinson disease. We carried out whole exome sequencing of genomic DNA from three severely affected family members and subsequent pedigree analysis to identify the genetic basis of essential tremor and Parkinson disease in the family.
Results
Clinical Features.
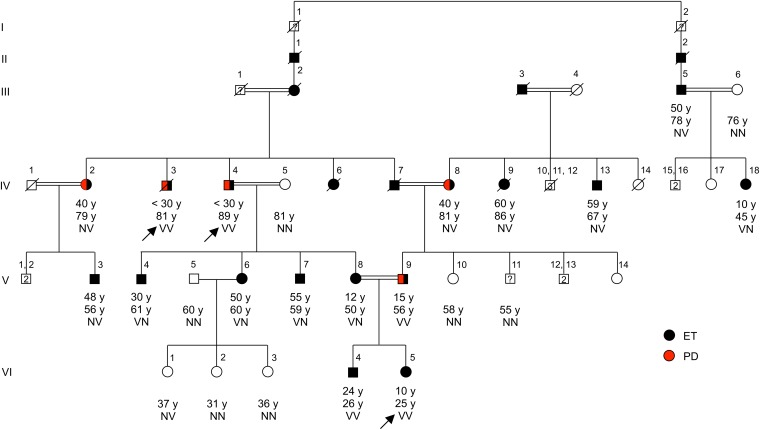
The ET-1 family is from central Anatolia, where consanguineous marriages are common practice. Ancestors of the extended family have lived in the same area for more than 400 y. Essential tremor is known to have segregated in the family for generations. For this study, 24 individuals from the family were clinically assessed (Fig. 1). Diagnosis of essential tremor was based on criteria of the consensus statement on tremor of the movement disorder society (15). Diagnosis of Parkinson disease required presence of bradykinesia plus at least one of muscular rigidity, resting tremor, or postural instability (16). Based on these criteria, 11 relatives were diagnosed with essential tremor and five relatives were diagnosed with essential tremor coexisting with Parkinson disease (Table 1). All five relatives with both essential tremor and Parkinson disease reported having tremors for multiple years before appearance of clinically apparent Parkinson signs (Table S1). In the family as a whole, ages of onset of tremor ranged from approximately 10 to 60 y, although some family members were not able to determine the exact age of onset as their tremor developed over many years.
Fig. 1.
Pedigree of family ET-1 segregating essential tremor, with genotypes at HTRA2 p.G399S. Individuals with essential tremor (ET) are shown with black symbols, and those with Parkinson disease (PD) with red symbols. Age at onset of tremor for affected individuals, current ages, and genotypes at HTRA2 p.G399S are indicated in this order under the symbols. N indicates the wild-type allele, glycine; V indicates the variant allele, serine, at HTRA2 p.G399S. Individuals who underwent exome sequencing (IV:3, IV:4, VI:5) are indicated with arrows. Subject VI:1, who is unaffected and heterozygous for HTRA2 p.G399S, is presently 37 y old, younger than the mean age at onset of essential tremor among heterozygotes in the family. Phenotypes of four relatives are unknown: I:1, I:2, and III:1 are deceased, and V:11 refused clinical examination.
Table 1.
Clinical diagnosis of individuals of family ET-1
| Individual | Age at onset of tremor | Age at examination | Essential tremor | Parkinson disease |
| III:5 | 50 | 78 | Severe | No |
| IV:2 | 40 | 79 | Severe | Yes |
| IV:3 | <30 | 81 | Severe | Yes |
| IV:4 | <30 | 89 | Severe | Yes |
| IV:8 | 40 | 81 | Severe | Yes |
| IV:13 | 59 | 67 | Severe | No |
| IV:18 | 10 | 45 | Severe | No |
| V:3 | 48 | 56 | Mild | No |
| V:4 | 30 | 61 | Mild | No |
| V:6 | 50 | 60 | Mild | No |
| V:7 | <55 | 59 | Mild | No |
| V:8 | 12 | 50 | Severe | No |
| V:9 | 15 | 56 | Severe | Yes |
| VI:5 | 10 | 25 | Severe | No |
See Table S1 for detailed clinical characteristics.
Gene Discovery.
To identify the gene responsible for essential tremor in the family, we carried out whole exome sequencing of three severely affected relatives, IV:3, IV:4, and VI:5 (Table S2 and Fig. S1). Given that the kindred include multiple consanguineous marriages, we first considered the possibility of recessive inheritance of essential tremor as the result of homozygosity for a critical mutation that was identical by descent from a common ancestor. To evaluate this possibility, we identified homozygous genomic regions greater than 1 MB shared by the three affected relatives. There were three such regions, on chromosomes 2p13.1-p12, 14q32.13, and 22q11 (Table S3). We then identified, in each of these regions, all variants predicted to be damaging (Methods). The only potentially damaging variant was HTRA2 (high temperature requirement protein A2) p.G399S (c.1195G > A, NM_013247) at chr2:74,759,825 G > A (rs72470545). HTRA2 p.G399S was predicted to be damaging by bioinformatics prediction tools Polyphen2 (score 0.986), SIFT (P = 0.02), and MutationAssessor (score 2.39).
We next genotyped HTRA2 p.G399S in all family members. Of the 16 individuals with essential tremor in the family, five were homozygous and 11 were heterozygous for the variant (Fig. 1). The occurrence of both heterozygotes and homozygotes for the mutation among affected relatives precluded recessive inheritance via identity by descent. However, genotypes of the family were consistent with dominant inheritance of essential tremor due to this allele, possibly with a dosage effect.
To evaluate the kindred for the possibility of dominant inheritance of any mutation in the genome, we next identified all potentially damaging variants, whether heterozygous or homozygous, shared by the three affected relatives with exome sequence. Including HTRA2 p.G399S, there were 13 such variants, all missenses (Table S4). We genotyped all 13 variants in all family members. The only variant segregating with essential tremor in the family was the HTRA2 allele (Fig. 1 and Table S4). The logarithm of odds (LOD) score for linkage of HTRA2 p.G399S under an autosomal dominant model of inheritance was 5.27.
We next considered the possibility that affected family members heterozygous for HTRA2 p.G399S might carry a second damaging allele of HTRA2 on their other copy of chromosome 2. To evaluate this possibility, two tests were carried out. First, HTRA2 was fully sequenced in all family members. No rare variants other than p.G399S were identified. Second, to test the possibility of a critical noncoding regulatory mutation of HTRA2 shared by affected family members, extended (3.7 mb) haplotypes flanking HTRA2 were determined for all relatives by using informative polymorphic markers (Fig. S2). Subjects heterozygous for HTRA2 p.G399S did not share a second haplotype. We concluded that the possibility of a second pathogenic HTRA2 allele segregating in this family is extremely unlikely. We also evaluated and excluded the possibility of linkage of essential tremor to a mutation in any of the genomic regions previously reported to be associated with the disorder (Fig. S3).
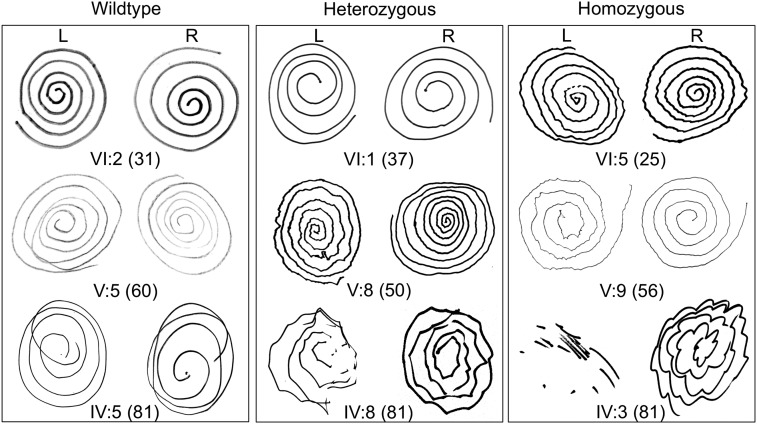
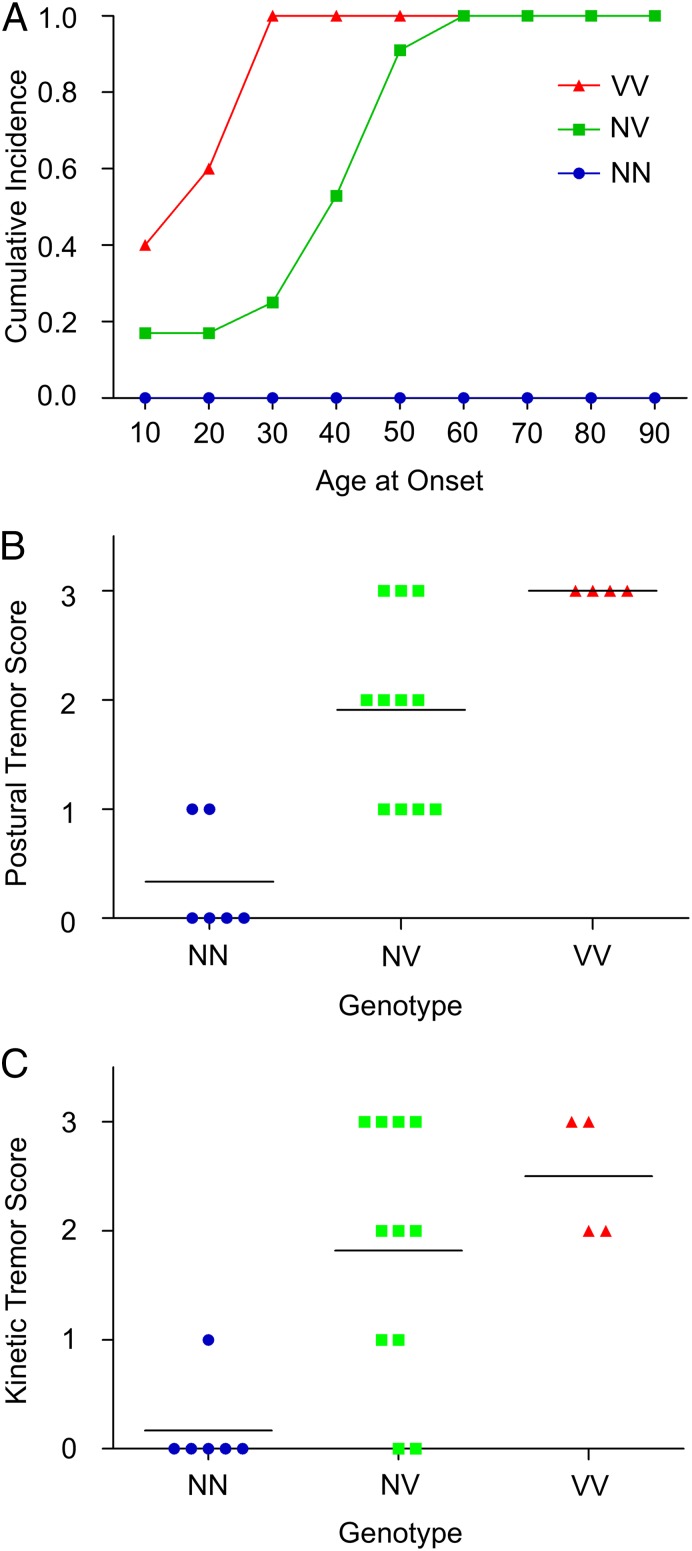
Several lines of evidence suggested that the number of copies of HTRA2 p.G399S influenced severity of the phenotype. First, results of the Archimedes spiral test (17) suggested that severity of action tremor was influenced by both genotype and age (Fig. 2). Second, homozygosity versus heterozygosity at HTRA2 p.G399S was significantly associated with age at onset of tremor [F = 28.99 (2, 24 df), P < 0.0001; Fig. 3A]. Mean ages at onset of tremor were 21.4 y and 41.3 y for subjects homozygous and heterozygous for the mutation, respectively. Third, homozygosity versus heterozygosity at HTRA2 p.G399S was associated with severity of both postural tremor [F = 18.68 (2, 17 df)], P < 0.0001; Fig. 3B) and kinetic tremor [F = 9.24 (2, 17 df)], P = 0.0019; Fig. 3C).
Fig. 2.
Archimedes spiral tests of individuals of various ages and genotypes at HTRA2 p.G399S. For all individuals, R was the dominant right hand and L was the nondominant left hand.
Fig. 3.
Relationship between HTRA2 genotype and age at onset of essential tremor and severity of tremors. V indicates the variant allele serine, and N indicates the wild-type allele glycine at HTRA2 p.G399S. Subjects heterozygous for the variant allele are indicated NV, and those homozygous for the variant allele are indicated VV. (A) Essential tremor age at onset varies significantly by genotype, P < 0.0001. (B) Severity of postural tremor of homozygous and heterozygous subjects differs significantly; analysis of covariance by genotype with age at examination as covariate yields F = 18.68, (2, 17 df), P < 0.0001. (C) Severity of kinetic tremor of homozygous and heterozygous subjects differs significantly; analysis of covariance by genotype with age at examination as covariate yields F = 9.24, (2, 17 df), P = 0.0019. Individuals with NN genotype and +1 postural or kinetic tremor scores did not fulfill the criteria for essential tremor diagnosis.
Of 59 other Turkish individuals with Parkinson Disease, none carries HTRA2 p.G399S. In 25 other Turkish families, each including multiple relatives with essential tremor, complete sequencing of HTRA2 did not reveal any damaging mutations. Of 364 unrelated Turkish controls, two were heterozygous and none were homozygous for HTRA2 p.G399S, yielding an allele frequency of 0.0027 for this population. The reported allele frequency among persons of various ancestries is ∼0.0034 (Table S4). Of the two Turkish controls heterozygous for the allele, one was anonymous and the other had given permission to be recontacted. The recontacted control is presently 27 y old. Upon neurological examination, she had no signs of essential tremor.
Discussion
Whole exome sequencing of three severely affected relatives of family ET-1, with no prior hypothesis about a causal gene, revealed HTRA2 p.G399S as the only potentially damaging allele cosegregating with essential tremor in the extended kindred. Homozygosity versus heterozygosity for this allele was associated with earlier age at onset and increased severity of essential tremor. Homozygotes for the mutation expressed a more severe phenotype, including signs of Parkinson disease at middle age, suggesting a dosage effect for this allele. Even among heterozygotes, age at onset of tremor was variable, due to genetic or environmental modifiers or stochastic effects (18).
HTRA2 encodes a serine protease of 458 aa that localizes to the intermembrane space of mitochondria (19). Upon an apoptotic stimulus, the HTRA2 protein is released from the mitochondria into the cytosol and binds to inhibitor-of-apoptosis proteins to initiate apoptosis (19). HTRA2 proteolytic activity also triggers caspase-independent cell death (20).
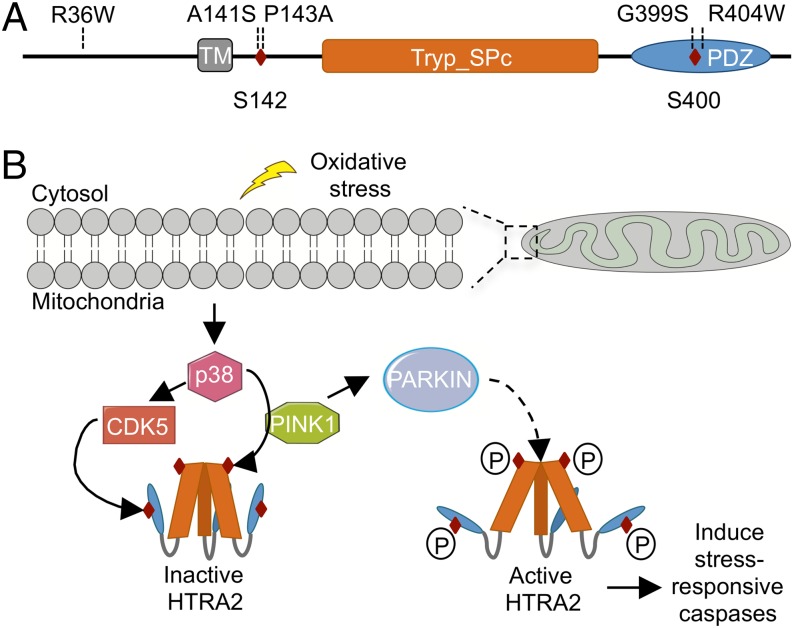
Several lines of evidence suggest involvement of HTRA2 in Parkinson disease. In the mnd2 mouse model, HtrA2 p.S275C leads to loss of protease activity and to a motor neuron degeneration phenotype with ataxia, repetitive movements, and akinesis (21). In addition, HtrA2 knockout mice show parkinsonian features due to the loss of neurons in striatum (22). Based on these observations, Strauss et al. sequenced HTRA2 in a series of Parkinson disease patients and controls from Germany, and identified HTRA2 p.G399S (the mutation of family ET-1) and HTRA2 p.A141S as associated with the disorder (Fig. 4A) (23). They also identified HTRA2 in Lewy bodies of Parkinson disease patients (23). Their study suggested that HTRA2 p.G399S leads to loss of function, because both missense mutations (HTRA2 p.G399S and HTRA2 p.A141S) led to mitochondrial dysfunction, altered mitochondrial morphology, and decreased protease activity, and HTRA2 p.G399S resulted in increased sensitivity to toxicity (23). A recent study of transgenic mice also suggested that HtrA2 p.G399S is a loss-of-function allele, because mice overexpressing a wild-type allele of HtrA2 showed significant motor impairments, whereas mice overexpressing HtrA2 p.G399S had normal motor function (24).
Fig. 4.
Schematic representation of the HTRA2 protein and its activation against mitochondrial stress. (A) Locations of all reported mutations in HTRA2 in persons with essential tremor or Parkinson disease. The full-length HTRA2 protein consists of a transmembrane domain (TM; residues 105–124), a conserved catalytic trypsin-like serine protease domain (Tryp_SPc; residues 178–342), and a C-terminal PDZ domain (residues 363–445). Phosphorylation sites are at Ser142 and Ser400. (B) Oxidative stress results in the activation of p38 stress kinase pathway. In HTRA2, p38 phosphorylates serine at residue 142 in a PINK1-dependent manner; CDK5 phosphorylates serine at residue 400, increasing the proteolytic activity of HTRA2. Active HTRA2 induces stress-responsive caspases. PINK1 also recruits Parkin, an E3 ubiquitin protein ligase, from cytosol to mitochondria to induce mitophagy. Red diamonds represent phosphorylation sites at residues 142 and 400. P, phosphorylation. Adapted by permission from Macmillan Publishers Ltd: Nature Cell Biology (20), copyright (2007).
To experimentally evaluate a dosage effect for this allele, it would be ideal to compare animal models heterozygous and homozygous for HtrA2 p.G399S. These animals have not been developed, but a clue to a possible dosage effect appears from the comparison of wild-type, HtrA2+/−, and HtrA2−/− mice (22). Compared with wild-type mice, HtrA2+/− mice appear to show a subtle, although not statistically significant, decrease in performance on sensory-motor tests (22). Increasing the number of HtrA2+/− mice in these experiments and extending their evaluation over a wider age range would provide valuable information on the possibility of a dosage effect for HTRA2 loss-of-function mutations.
Three other rare missense alleles, HTRA2 p.R36W, p.P143A, and p.R404W, have been reported in patients with Parkinson disease (Fig. 4A) (25–27). HTRA2 p.A141S and p.P143A lie in close proximity to serine at residue 142, which is phosphorylated upon the activity of p38, dependent on PTEN-induced-putative-kinase (PINK1) (Fig. 4B) (28, 29). HTRA2 p.G399S and p.R404W lie in close proximity to serine at residue 400, which is phosphorylated upon activation of cyclin-dependent kinase-5 (CDK5) (28, 29). In transgenic mice expressing HtrA2 p.G399S in cortex, phosphorylation at residue 400 was significantly reduced (29). Phosphorylation of HTRA2 at both sites is important for cellular stress response (29). Furthermore, PINK1 and CDK5 kinases are both known to be associated with Parkinson disease (30, 31). However, despite the biological plausibility of a role for HTRA2 in Parkinson disease, subsequent epidemiologic studies did not detect higher frequencies of any of the rare missense alleles of HTRA2 among Parkinson disease patients than among controls (25–27, 32).
The role of HTRA2 in essential tremor may resolve this paradox. The individuals in family ET-1 who developed features of Parkinson disease exhibited these symptoms decades after onset of essential tremor. These subjects included all three individuals homozygous for HTRA2 p.G399S and older than age 55, and two individuals heterozygous for HTRA2 p.G399S and older than age 78. These observations suggest that HTRA2 has a causal role in essential tremor and in the subset of Parkinson disease preceded by essential tremor. Among essential tremor patients generally, the incidence of Parkinson disease is increased four- to fivefold, and essential tremor and Parkinson disease have been observed in the same families (33–35). We speculate that case-control studies of that subset of Parkinson disease preceded by essential tremor would reveal associations with functional missense alleles of HTRA2.
Although essential tremor is one of the most common inherited neurologic disorders, identifying the responsible underlying genes has been challenging. Complexities of essential tremor include genetic heterogeneity, age-dependent penetrance, and variable expressivity, leading to difficulties both in differential diagnosis and in genetic analysis (18). Our results suggest that mutation of HTRA2 can be responsible for essential tremor in some families and that parkinsonian features may develop in these patients, after age 70 in heterozygotes and in middle age in homozygotes. These observations reveal one cause of essential tremor and may illuminate some of the shared features of essential tremor and Parkinson disease phenotypes.
Methods
Subjects.
Family ET-1 is of Turkish origin. The proband was first evaluated at Ankara University Medical School. He and his informative relatives were followed at Ankara University Medical School and Hacettepe University Medical School. This project was approved by the ethics committees of all participating universities, and informed consent was obtained from all individuals. Each participant was examined for essential tremor by using the criteria of both the Washington Heights-Inwood Genetic Study of Essential Tremor and the Consensus Statement of the Movement Disorder Society on Tremor (Table 1 and Table S1) (15, 17). Each participant was rated for rest and postural tremors and was asked to perform four different tasks (pouring water, drinking water from a cup, finger-to-nose movement, and drawing spirals) to elicit kinetic tremor. During the examination, severity of tremor was rated during each task (17). Participants were evaluated for features of Parkinson disease by using the diagnostic criteria of the UK Parkinson Disease Society Brain Bank (16). Diagnosis of Parkinson disease required presence of bradykinesia plus at least one of muscular rigidity, resting tremor, or postural instability (16). We collected histories about distribution and severity of tremor and change of these parameters over time. Participants were interviewed about concurrent use of drugs that might cause action or resting tremor and for symptoms of hyperthyroidism, which was ruled out by thyroid-stimulating hormone tests as needed. Clinical assessments were carried out by at least two neurologists without knowledge of participants’ genotypes. An additional 25 families with hereditary essential tremor, 59 patients with Parkinson disease, and 364 healthy controls, ages 20–30 and from the same central Anatolian region as family ET-1, were recruited for genetic analysis from Ankara University Medical School, Hacettepe University Medical School, and Bilkent University. Unaffected individuals showed no signs of disease at the time of examination. Because controls were young adults, they serve as population controls. DNA was extracted from blood by using Nucleospin Blood Kit (Macherey-Nagel) according to manufacturer’s protocol.
Genomics.
Three severely affected individuals were selected for whole exome sequencing. Library construction and sequencing was carried out as described (36). Isolated genomic DNA was randomly sheared into 200–300 bp followed by end repair, A-tailing, and indexed paired-end adapter ligation. Exomes were captured by SeqCap EZ Exome v2 (Roche) and hybridized to biotinylated capture probes. Libraries were sequenced on an Illumina HiSeq2500.
Bioinformatics.
Paired-end sequence reads were aligned to the human reference genome (hg19) by using Burrows-Wheeler Aligner (v0.6.1-r104) (37). Removal of PCR duplicates, sorting, and indexing were done by using SAMtools v0.1.18 (38). Indel realignments and base quality score recalibration were done with Genome Analysis Tool Kit (GATK; v3.0–0-g6bad1c6; broadinstitute.org/gatk) by using recommended parameters (39). Genotypes were called and filtered by using GATK Unified Genotyper and Variant Filtration tools. Variants were annotated by using our in-house pipeline. Common SNPs and artifacts were excluded by using dbSNP v138, the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (evs.gs.washington.edu/EVS), the 1000 Genomes Project (1000genomes.org), and 700 exomes previously sequenced in our laboratory. Variants were defined as potentially damaging if they led to a premature stop codon or were missense mutations with scores on in silico prediction tools of SIFT P ≤ 0.05, PolyPhen2 ≥ 0.8, and MutationAssessor ≥ 1.95 (Table S4) (sift.jcvi.org; genetics.bwh.harvard.edu/pph2; mutationassessor.org) (40–42).
Sanger Sequencing.
Genotypes for candidate variants for 24 informative relatives of family ET-1 were determined by capillary sequencing (ABI 3130xl Genetic Analyzer). All coding regions, potential regulatory regions, and miRNA binding sites of HTRA2 were sequenced in probands from 25 unrelated families with multiple relatives with essential tremor. Primers were designed by using Primer3 (Table S5) (43). Products were analyzed via gel electrophoresis and Sanger sequenced. Sanger traces were analyzed with CLCBio Main Workbench software package (CLCBio). TaqMan genotyping assays (Life Technologies) were used for screening HTRA2 p.G399S in Parkinson disease patients and controls (ABI 7900HT Fast Real-Time PCR System). Family ET-1 was genotyped with FAM- and HEX-labeled primers for polymorphic markers on chromosome 2 flanking HTRA2 (ABI 3130xl Genetic Analyzer). Data were analyzed with GeneMapper v4.0 software package (Applied Biosystems).
Linkage Analysis.
A LOD score for linkage of HTRA2 to essential tremor in the ET-1 family was calculated by using LINKAGE v6.0 (44) under an autosomal dominant mode of inheritance with penetrance for homozygous or heterozygous genotypes of 1.0 at age 40 and older and 0.6 before age 40, no phenocopies, and a mutant allele frequency of 0.01 in the general population.
Statistical Analysis.
Statistical significance was evaluated by one-way analysis of variance, or analysis of covariance, as appropriate.
Supplementary Material
Acknowledgments
We thank the families for participating in the study. The study is supported by The Scientific and Technological Research Council of Turkey (TUBITAK) Research Project 113S959 (to A.B.T.), Turkish Academy of Sciences (TUBA) support (to T.O.), TUBA-Young Scientists Award Programme (GEBIP) support (to A.B.T.), TUBITAK-Department of Science Fellowships and Grant Programmes (BIDEB) 2214/A doctoral fellowship (to H.U.G.), and by unrestricted gifts to the M.-C.K. laboratory.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419581111/-/DCSupplemental.
References
- 1.Louis ED, Ferreira JJ. How common is the most common adult movement disorder? Update on the worldwide prevalence of essential tremor. Mov Disord. 2010;25(5):534–541. doi: 10.1002/mds.22838. [DOI] [PubMed] [Google Scholar]
- 2.Benito-León J, Louis ED. Essential tremor: Emerging views of a common disorder. Nat Clin Pract Neurol. 2006;2(12):666–678, quiz 2p following 691. doi: 10.1038/ncpneuro0347. [DOI] [PubMed] [Google Scholar]
- 3.Gulcher JR, et al. Mapping of a familial essential tremor gene, FET1, to chromosome 3q13. Nat Genet. 1997;17(1):84–87. doi: 10.1038/ng0997-84. [DOI] [PubMed] [Google Scholar]
- 4.Higgins JJ, Pho LT, Nee LE. A gene (ETM) for essential tremor maps to chromosome 2p22-p25. Mov Disord. 1997;12(6):859–864. doi: 10.1002/mds.870120605. [DOI] [PubMed] [Google Scholar]
- 5.Shatunov A, et al. Genomewide scans in North American families reveal genetic linkage of essential tremor to a region on chromosome 6p23. Brain. 2006;129(Pt 9):2318–2331. doi: 10.1093/brain/awl120. [DOI] [PubMed] [Google Scholar]
- 6.Jeanneteau F, et al. A functional variant of the dopamine D3 receptor is associated with risk and age-at-onset of essential tremor. Proc Natl Acad Sci USA. 2006;103(28):10753–10758. doi: 10.1073/pnas.0508189103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higgins JJ, et al. A variant in the HS1-BP3 gene is associated with familial essential tremor. Neurology. 2005;64(3):417–421. doi: 10.1212/01.WNL.0000153481.30222.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stefansson H, et al. Variant in the sequence of the LINGO1 gene confers risk of essential tremor. Nat Genet. 2009;41(3):277–279. doi: 10.1038/ng.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thier S, et al. Polymorphisms in the glial glutamate transporter SLC1A2 are associated with essential tremor. Neurology. 2012;79(3):243–248. doi: 10.1212/WNL.0b013e31825fdeed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhlenbäumer G, Hopfner F, Deuschl G. Genetics of essential tremor: Meta-analysis and review. Neurology. 2014;82(11):1000–1007. doi: 10.1212/WNL.0000000000000211. [DOI] [PubMed] [Google Scholar]
- 11.Rajput A, et al. VPS35 and DNAJC13 disease-causing variants in essential tremor. Eur J Hum Genet. 2014 doi: 10.1038/ejhg.2014.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merner ND, et al. Exome sequencing identifies FUS mutations as a cause of essential tremor. Am J Hum Genet. 2012;91(2):313–319. doi: 10.1016/j.ajhg.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y-R, et al. Identification of a novel risk variant in the FUS gene in essential tremor. Neurology. 2013;81(6):541–544. doi: 10.1212/WNL.0b013e31829e700c. [DOI] [PubMed] [Google Scholar]
- 14.Rajput A, et al. Identification of FUS p.R377W in essential tremor. Eur J Neurol. 2014;21(2):361–363. doi: 10.1111/ene.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deuschl G, Bain P, Brin M. Ad Hoc Scientific Committee Consensus statement of the movement disorder society on tremor. Mov Disord. 1998;13(Suppl 3):2–23. doi: 10.1002/mds.870131303. [DOI] [PubMed] [Google Scholar]
- 16.Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: A clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 1992;55(3):181–184. doi: 10.1136/jnnp.55.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Louis ED, Ford B, Lee H, Andrews H, Cameron G. Diagnostic criteria for essential tremor: A population perspective. Arch Neurol. 1998;55(6):823–828. doi: 10.1001/archneur.55.6.823. [DOI] [PubMed] [Google Scholar]
- 18.Louis ED. ‘Essential tremor’ or ‘the essential tremors’: Is this one disease or a family of diseases? Neuroepidemiology. 2014;42(2):81–89. doi: 10.1159/000356351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hegde R, et al. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J Biol Chem. 2002;277(1):432–438. doi: 10.1074/jbc.M109721200. [DOI] [PubMed] [Google Scholar]
- 20.Alnemri ES. HtrA2 and Parkinson’s disease: Think PINK? Nat Cell Biol. 2007;9(11):1227–1229. doi: 10.1038/ncb1107-1227. [DOI] [PubMed] [Google Scholar]
- 21.Jones JM, et al. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature. 2003;425(6959):721–727. doi: 10.1038/nature02052. [DOI] [PubMed] [Google Scholar]
- 22.Martins LM, et al. Neuroprotective role of the Reaper-related serine protease HtrA2/Omi revealed by targeted deletion in mice. Mol Cell Biol. 2004;24(22):9848–9862. doi: 10.1128/MCB.24.22.9848-9862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strauss KM, et al. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson’s disease. Hum Mol Genet. 2005;14(15):2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- 24.Sood P, et al. Characterization of transgenic mice overexpressing wild type and G399S mutant HtrA2/Omi – Implications for PD. Basal Ganglia. 2013;3(1):41. [Google Scholar]
- 25.Bogaerts V, et al. Genetic variability in the mitochondrial serine protease HTRA2 contributes to risk for Parkinson disease. Hum Mutat. 2008;29(6):832–840. doi: 10.1002/humu.20713. [DOI] [PubMed] [Google Scholar]
- 26.Lin C-H, Chen M-L, Chen GS, Tai C-H, Wu R-M. Novel variant Pro143Ala in HTRA2 contributes to Parkinson’s disease by inducing hyperphosphorylation of HTRA2 protein in mitochondria. Hum Genet. 2011;130(6):817–827. doi: 10.1007/s00439-011-1041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C-M, et al. HTRA2 variations in Taiwanese Parkinson’s disease. J Neural Transm. 2014;121(5):491–498. doi: 10.1007/s00702-013-1131-9. [DOI] [PubMed] [Google Scholar]
- 28.Plun-Favreau H, et al. The mitochondrial protease HtrA2 is regulated by Parkinson’s disease-associated kinase PINK1. Nat Cell Biol. 2007;9(11):1243–1252. doi: 10.1038/ncb1644. [DOI] [PubMed] [Google Scholar]
- 29.Fitzgerald JC, et al. Phosphorylation of HtrA2 by cyclin-dependent kinase-5 is important for mitochondrial function. Cell Death Differ. 2012;19(2):257–266. doi: 10.1038/cdd.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fitzgerald JC, Plun-Favreau H. Emerging pathways in genetic Parkinson’s disease: Autosomal-recessive genes in Parkinson’s disease—a common pathway? FEBS J. 2008;275(23):5758–5766. doi: 10.1111/j.1742-4658.2008.06708.x. [DOI] [PubMed] [Google Scholar]
- 31.Nakamura S, Kawamoto Y, Nakano S, Akiguchi I, Kimura J. p35nck5a and cyclin-dependent kinase 5 colocalize in Lewy bodies of brains with Parkinson’s disease. Acta Neuropathol. 1997;94(2):153–157. doi: 10.1007/s004010050687. [DOI] [PubMed] [Google Scholar]
- 32.Simón-Sánchez J, Singleton AB. Sequencing analysis of OMI/HTRA2 shows previously reported pathogenic mutations in neurologically normal controls. Hum Mol Genet. 2008;17(13):1988–1993. doi: 10.1093/hmg/ddn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benito-León J, Louis ED, Bermejo-Pareja F. Neurological Disorders in Central Spain Study Group Risk of incident Parkinson’s disease and parkinsonism in essential tremor: A population based study. J Neurol Neurosurg Psychiatry. 2009;80(4):423–425. doi: 10.1136/jnnp.2008.147223. [DOI] [PubMed] [Google Scholar]
- 34.Yahr MD, Orosz D, Purohit DP. Co-occurrence of essential tremor and Parkinson’s disease: Clinical study of a large kindred with autopsy findings. Parkinsonism Relat Disord. 2003;9(4):225–231. doi: 10.1016/s1353-8020(02)00057-3. [DOI] [PubMed] [Google Scholar]
- 35.Farrer M, et al. A chromosome 4p haplotype segregating with Parkinson’s disease and postural tremor. Hum Mol Genet. 1999;8(1):81–85. doi: 10.1093/hmg/8.1.81. [DOI] [PubMed] [Google Scholar]
- 36.Walsh T, et al. Whole exome sequencing and homozygosity mapping identify mutation in the cell polarity protein GPSM2 as the cause of nonsyndromic hearing loss DFNB82. Am J Hum Genet. 2010;87(1):90–94. doi: 10.1016/j.ajhg.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 41.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Untergasser A, et al. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ott J. Analysis of Human Genetic Linkage. Johns Hopkins Univ Press; Baltimore: 1999. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.