Significance
We have solved a long-standing mystery of the biosynthetic origin of 1-undecene, a ubiquitous hydrocarbon semivolatile metabolite of Pseudomonas. Our study revealed an unprecedented family of nonheme oxidases that specifically convert medium-chain fatty acids into the corresponding terminal olefins using an oxygen-activating, nonheme iron-dependent mechanism. Our findings unveil previously unidentified chemistry in the nonheme Fe(II) enzyme family, aid the functional study of this ubiquitous metabolite in Pseudomonas, expand the scarce enzyme inventory for the transformation of fatty acid precursors to hydrocarbons, and serve as the basis for engineering efforts to establish bioprocesses to produce medium-chain terminal olefins, useful as fuels and chemical building blocks, from renewable resources.
Keywords: hydrocarbon, terminal olefin, iron-dependent desaturase/decarboxylase, biofuel, biosynthesis
Abstract
Aliphatic medium-chain 1-alkenes (MCAEs, ∼10 carbons) are “drop-in” compatible next-generation fuels and precursors to commodity chemicals. Mass production of MCAEs from renewable resources holds promise for mitigating dependence on fossil hydrocarbons. An MCAE, such as 1-undecene, is naturally produced by Pseudomonas as a semivolatile metabolite through an unknown biosynthetic pathway. We describe here the discovery of a single gene conserved in Pseudomonas responsible for 1-undecene biosynthesis. The encoded enzyme is able to convert medium-chain fatty acids (C10–C14) into their corresponding terminal olefins using an oxygen-activating, nonheme iron-dependent mechanism. Both biochemical and X-ray crystal structural analyses suggest an unusual mechanism of β-hydrogen abstraction during fatty acid substrate activation. Our discovery unveils previously unidentified chemistry in the nonheme Fe(II) enzyme family, provides an opportunity to explore the biology of 1-undecene in Pseudomonas, and paves the way for tailored bioconversion of renewable raw materials to MCAE-based biofuels and chemical commodities.
Surging energy consumption and environmental concerns have stimulated interest in the production of chemicals and fuels through sustainable and renewable approaches. Medium-chain 1-alkenes (MCAEs) are of particular interest because they are “drop-in”–ready next-generation fuels with superior properties such as low freezing point compared with long-chain diesels, high energy content compared with short-chain fuels, easy product recovery due to insolubility in water, and compatibility with the existing engine systems and transportation infrastructure (1, 2). Because of a readily derivatized terminal functionality, MCAEs are also valuable precursors to commodity chemicals such as lubricants, pesticides, polymers, and detergents (3, 4). Biological production of MCAEs from renewable resources holds promise for mitigating dependence on fossil hydrocarbons. Although MCAEs are naturally produced by diverse species as semivolatile metabolites (5, 6), little is known about the genetic and molecular basis for MCAE biosynthesis. Elucidation of MCAE biosynthetic pathway will serve as the basis for engineering efforts to establish bioprocesses for producing MCAE-based biofuels and chemical commodities from renewable resources.
1-Undecene, an MCAE with 11 carbons, was identified as a biomarker of Pseudomonas aeruginosa, one of the most significant human pathogens (7–10). However, the biology of this characteristic semivolatile metabolite in P. aeruginosa remains enigmatic, and the biosynthetic pathway of 1-undecene has not previously been explored. It was also reported that some species of Pseudomonas produce 1-undecene, whereas some species do not (8), inspiring us to use a comparative genomics approach to reveal the genetic basis for 1-undecene biosynthesis (11). It is notable that one of the major challenges for MCAE biosynthetic study is the detection and quantification of MCAE production. MCAEs, such as 1-undecene, are only produced in trace amounts by their native producers (5, 6). In addition, MCAEs are insoluble in water, less dense than water, and readily escape the cell culture as semivolatile metabolites (8). Therefore, a reliable and sensitive method for MCAE detection is a prerequsite for their biosynthetic study. In this work, we used a headspace solid phase microextraction (SPME)–gas chromatography mass spectrometry (GCMS) analysis to detect the semivolatile metabolite 1-undecene, and we revealed that the production of 1-undecene is ubiquitous in the genus of Pseudomonas. We further discovered that a novel enzyme conserved in Pseudomonas is responsible for 1-undecene biosynthesis through an unusual catalytic mechanism.
Results
Analysis of 1-Undecene Production in Microbes.
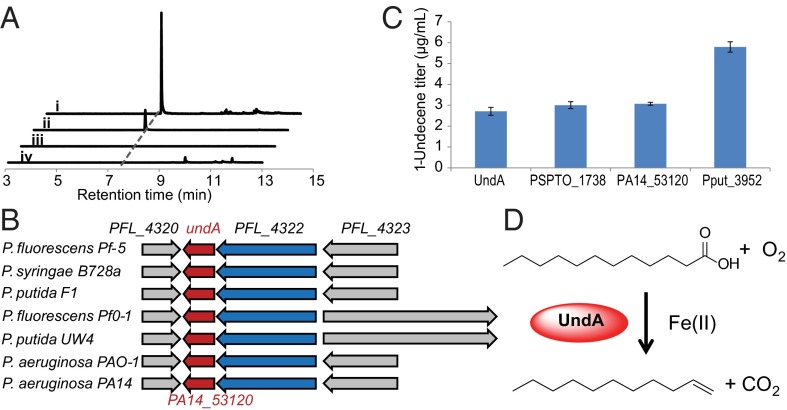
Hydrocarbon metabolites produced in cell cultures are typically extracted by organic solvents (11, 12), but the low titer and volatility of 1-undecene resulted in ambiguous signals during our initial GCMS analysis using this extraction method. Alternatively, SPME allows the efficient extraction of volatile metabolites directly from the headspace. Based on headspace SPME–GCMS analysis, we developed a sensitive detection method for volatile metabolites with a detection limit of ∼1 ng of total 1-undecene in cell cultures (including both liquid and gas phases). All of the tested Pseudomonas strains produced 1–100 ng/mL of 1-undecene (SI Appendix, Materials and Methods), demonstrating the robustness of this method for semivolatile MCAE analysis. Although 1-undecene has also been reported to be produced by Shewanella (6), we did not detect the production of 1-undecene by seven selected Shewanella strains. In addition, none of Escherichia coli strains tested produced a detectable amount of 1-undecene (Fig. 1A).
Fig. 1.
Identification of the enzyme responsible for 1-undecene biosynthesis in Pseudomonas. (A) Demonstration of 1-undecene production from headspace SPME–GCMS analysis. (i) 1-undecene production was observed during the library screening in the E. coli EPI300 expressing the gene undA. (ii) P. aeruginosa PA14 naturally produces 1-undecene. (iii) Disruption of the gene PA14_53120 (an undA homolog) completely abolished 1-undecene production in the P. aeruginosa ∆PA14_53120 mutant. (iv) E. coli EPI300 does not naturally produce 1-undecene. (B) UndA-containing two-gene operons conserved in Pseudomonas. (C) Production titer of extracellular 1-undecene by overexpressing undA (P. fluorescens Pf-5), PSPTO_1738 (P. syringae pv. tomato DC3000), PA14_53120 (P. aeruginosa PA14), and Pput_3952 (P. putida F1) in E. coli BL21 Star. Error bars represent SDs from at least three independently performed experiments. (D) The biosynthesis of 1-undecene from LA catalyzed by UndA, an oxygen-activating, nonheme, iron-dependent desaturase/decarboxylase.
1-Undecene Biosynthetic Gene Discovery.
We hypothesized that 1-undecene originates from the fatty acid metabolism. Feeding [12-13C]lauric acid (LA) and [1-13C]LA to Pseudomonas cultures resulted in the production of [11-13C]undecene and [U-12C11]undecene, respectively (SI Appendix, Fig. S1), implying that LA is the 1-undecene biosynthetic precursor and the terminal carboxylic acid moiety is removed during 1-undecene formation. At least two mechanisms were recently characterized to yield long-chain 1-alkenes from fatty acyl precursors with a loss of CO2. These reported mechanisms include the cytochrome P450 fatty acid peroxygenase-catalyzed synthesis of 8-methyl-1-nonadecene and 17-methyl-1-nonadecene in the Jeotgalicoccus bacteria species (12, 13), and the multidomain polyketide synthase-promoted production of very long-chain 1-alkenes in the Synechococcus cyanobacteria species (14). Bioinformatics analysis revealed that no homologs of these enzymes are encoded in the genomes of Pseudomonas, suggesting different biochemistry underlying 1-undecene synthesis.
The comparative genomics analysis yielded thousands of gene candidates for 1-undecene biosynthesis due to the ubiquitous production of 1-undecene in Pseudomonas and a few related bacteria species, demonstrating the limitation of this approach. Alternatively, we attempted to locate the biosynthetic genes by heterologously expressing a genomic library of Pseudomonas fluorescens Pf-5 in E. coli followed by phenotype screening of the 1-undecene producing clones (SI Appendix, Fig. S2). After screening ∼6,000 fosmids, we determined that one single gene, designated as undA, is necessary and sufficient to confer 1-undecene production in E. coli (Fig. 1 A and B). To further probe the function of undA homologs in the biosynthesis of 1-undecene, we heterologously expressed three additional undA homologs from various Pseudomonas species. The overexpression of all gene candidates in E. coli conferred 1-undecene production, confirming the proposed function of these genes (Fig. 1C). Additionally, we examined the ∆PA14_53120 mutant of P. aeruginosa PA14, which bears a transposon insertion in the gene PA14_53120 homologous to undA (15). This gene disruption completely abolished 1-undecene production in P. aeruginosa PA14 (Fig. 1 A and B), confirming the necessity of this gene in 1-undecene biosynthesis by P. aeruginosa.
Biochemical Analysis of UndA.
To establish the precise role of UndA in biosynthesis of 1-undecene, we performed in vitro enzymatic assays using the purified recombinant enzyme from E. coli. UndA is a small protein (261 amino acids) with low sequence homology (identity/similarity = 13%/23%) to TenA, a thiaminase II from Bacillus subtilis (16), but the essential catalytic cysteine of TenA is missing in UndA. In addition, the structure of an UndA homolog from Pseudomonas syringae pv. tomato DC3000 (identity/similarity = 79%/86%) [Protein Data Bank (PDB) 3OQL] was resolved by the Joint Center for Structural Genomics, but no enzymatic activity was reported and no cofactor was found in the crystal structure. Because in silico analysis failed to predict any possible activity of UndA, we first tested a panel of substrates in the UndA biochemical assays, including LA, α-hydroxydodecanoic acid (AHDA), β-hydroxydodecanoic acid (BHDA), 2,3-dodecenoic acid (DEA), methyl laurate, ethyl laurate, and lauroyl-CoA. All attempts failed to produce 1-undecene. Although alternative substrates could have been possible, we reasoned that a critical factor was likely missing in the in vitro assays. We then screened common enabling factors in combination with the substrate panel for 1-undecene production. These tested factors include Fe2+, Fe3+, Mg2+, Ca2+, Mn2+, Co2+, Ni2+, Cu2+, Zn2+, nicotinamides, pyridoxal phosphate, flavins, S-adenosyl methionine, pyrroloquinoline quinone (PQQ), and ATP. After an extensive number of trials, we discovered that only one combination, LA and Fe2+, elicited 1-undecene production in the UndA assay. Notably, all other tested metal ions including Fe3+ failed to enable 1-undecene production. These results demonstrated that UndA is a nonheme iron (II)-dependent enzyme that converts LA to 1-undecene (Fig. 1D).
After reconstituting the activity of UndA in vitro, we performed detailed biochemical analyses of this enzyme. The iron content of UndA after anaerobic reconstitution of purified apo-UndA with Fe2+ was determined to be 82% by inductively coupled plasma mass spectrometry. Using [1-13C]LA as a substrate, formation of [13C]CO2 was detected (Fig. 2 and SI Appendix, Fig. S3), confirming that UndA catalyzes production of 1-undecene from LA through oxidative decarboxylation. Because the in vitro aerobic reaction mixture contained only desalted UndA–Fe(II) and LA with no additional oxidant/reductant, we reasoned that molecular oxygen served as the oxidant of the reaction, which was supported by the observations that 1-undecene production was nearly abolished in anaerobic enzymatic assays, and that O2 was consumed in aerobic assays (SI Appendix, Fig. S4). The measured stoichiometry of the reaction showed that one molecule of UndA–Fe(II) consumed approximately one equivalent of O2 to produce close to one equivalent of 1-undecene (SI Appendix, Fig. S4). O2 was presumably reduced to hydrogen peroxide or water by UndA. We proposed that H2O was a likely end product, as no production of H2O2 was detected in the UndA assays by two independent sensitive analytical methods, and the addition of catalase to the reaction system had no detectable effect.
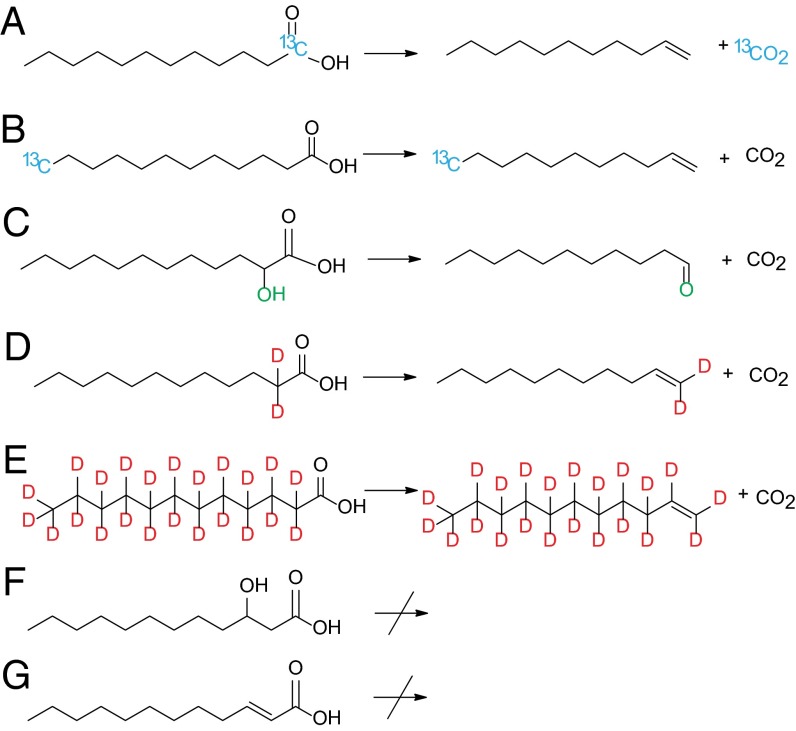
Fig. 2.
UndA activity toward selected substrates: A, [1-13C]LA; B, [12-13C]LA; C, AHDA; D, [α,α-D2]LA; E, [D23]LA; F, BHDA; G, DEA. Both deuterium atoms of [α,α-D2]LA are proposed to retain at the α-carbon position forming [1,1-D2]undecene in D.
Only single turnover reactions were obtained in the UndA in vitro assays, probably due to an electron imbalance, where two electrons are donated by one molecule of LA while four electrons are required to reduce one molecule of O2 to water. This electron imbalance stalls the enzyme, presumably with an inactive oxidized iron species. This hypothesis is supported by an UndA recycling experiment in which apo-UndA recycled after the single turnover reaction maintained over 80% activity upon reconstitution with fresh Fe2+. We next surveyed a group of possible reductive cosubstrates based on those used by well-studied oxygen-activating iron-dependent oxygenases (17–20). These reductive cosubstrates included α-ketoglutarate (α-KG), ascorbic acid, glutathione, cysteine, DTT, Tris(2-carboxyethyl)phosphine, nicotinamides, flavins, ferredoxin, tetrahydropterin, phenazine methosulfate (PMS)/NADH, and PQQ/cysteine. Initially, none of the tested reducing systems promoted 1-undecene production over the stoichiometric amount of one turnover. We suspected that the limiting factor was likely the consumption of dissolved O2 by both the enzymatic reaction and some of the tested reducing agents in the sealed vial without replenishment. Indeed, through coupling with the chlorite dismutase reaction to generate O2 in situ (21), we found that ascorbate enabled multiple turnovers for UndA in vitro, but the rate (four turnovers per hour) was much slower than the initial rate of 1-undecene production under the single turnover condition [0.06 ± 0.002 mol 1-undecene/(mol UndA-Fe(II) second)] (SI Appendix, Figs. S5 and S6). Other electron donors such as PMS/NADH and PQQ/cysteine also enabled multiple turnovers, although the resulting rates and yields were lower than when ascorbate was the reductant (SI Appendix, Fig. S5).
Because HppE, a long recognized nonheme iron (II) oxidase, was recently characterized to be a peroxidase (22), we then extensively tested the possibility that H2O2, rather than O2, is the real substrate for UndA. The reduction of H2O2 to water requires only two electrons, matching the two-electron requirement for the overall conversion of LA to 1-undecene. However, in the presence of H2O2, no 1-undecene was formed in anaerobic reactions with either UndA–Fe(III) or UndA–Fe(II). In the presence of both H2O2 and O2, no 1-undecene was formed with UndA–Fe(III), further confirming the requirement of Fe(II) for the activity of UndA. In addition, H2O2 did not promote the 1-undecene formation in the reaction containing O2 and UndA–Fe(II), indicating that H2O2 is not the substrate for UndA. These results are consistent with the observations that addition of dithionite to reduce O2 to H2O2 in situ inhibited 1-undecene production by UndA, but the addition of dithionite increased the rate of HppE up to 1,000 times (22).
In addition to LA (C12:0), UndA converted myristic acid (C14:0) and capric acid (C10:0) to their corresponding “[M-1]-carbon” 1-alkenes (SI Appendix, Figs. S7 and S8), and the initial rates of 1-alkene production at varying substrate concentrations were further measured (SI Appendix, Fig. S6). UndA failed to act on palmitic acid (C16:0) or caprylic acid (C8:0), indicating that the substrate binding pocket of the enzyme accepts a range of medium chain-length fatty acid substrates. Interestingly, UndA transformed AHDA to 1-undecanal, but exhibited no activity toward DEA and BHDA (Fig. 2 and SI Appendix, Fig. S9), suggesting that the β-carbon of LA, rather than the α-carbon, is the site of activation during enzymatic catalysis. This is further confirmed by the UndA assay with [α,α-D2]LA, which led to the production of 1-undecene retaining both deuterium atoms, and the UndA assay with [D23]LA, which led to the production of [D22]1-undecene (Fig. 2 and SI Appendix, Figs. S10 and S11).
Structural Analysis of UndA.
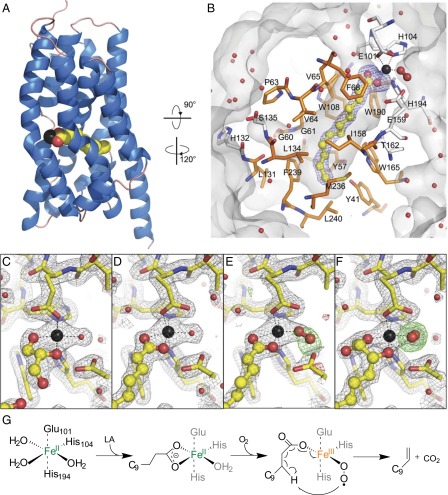
To understand the molecular basis of catalysis and the substrate recognition mechanism, we determined crystal structures of UndA in its holo form (PDB ID code 4WWJ) and in complex with two substrate analogs, either DEA (PDB ID code 4WWZ) or BHDA (PDB ID code 4WX0), with resolutions ranging from 1.7 to 1.9 Å (Fig. 3 and SI Appendix, Fig. S12 and Table S2). The active site of UndA contains a single iron center in an octahedral configuration with three sites of coordination from the enzyme side chains of Glu101, His104, and His194, and three variable sites of coordination, two facing the pocket and one exposed to exterior. The three variable sites are coordinated by oxygen atoms from the substrate analogs, solvent molecules such as water or glycerol, or O2. Different from typical nonheme mononuclear Fe(II) enzymes, the His194–His104–Glu101 residues of UndA are not arranged as a “facial” triad (20). The critical roles of these three residues were confirmed by site-directed mutagenesis, which resulted in an over 1,000-fold drop in 1-undecene production (SI Appendix, Fig. S13). The substrate analogs DEA or BHDA reside in a deep hydrophobic pocket that extends from the surface to the center of the enzyme (Fig. 3 A and B). The depth of the pocket limits the length of the fatty acid substrate to an ∼14-carbon chain, consistent with the biochemical results. Whereas the holo-structure represents the resting state of the active site with three iron coordination positions occupied by solvent molecules (one from water and two from glycerol) (Fig. 3C), cocrystallization with the substrate analogs DEA and BHDA seemed to trap the enzyme in different oxygen activation steps of the catalytic cycle. In crystals with DEA-bound enzymes, two different proposed intermediates are present in the crystallographic asymmetric unit. One subunit contains a water molecule coordinated to the iron, demonstrating an intermediate stage with a bound fatty acid substrate before O2 binding (Fig. 3D). In the other subunit, a diatomic molecule, assigned as molecular oxygen, replaces the water and occupies the iron coordination site opposite to the carboxyl oxygen of the fatty acid substrate in an end-on fashion (Fig. 3E). Dioxygen also appears in the BHDA-bound structure, which differs from the DEA-bound structures by the coordination of the β-hydroxyl group opposite to His104 (Fig. 3F). The refined distance between the oxygen atoms in the BHDA-bound structure converges to 1.45 Å, suggesting the presence of a peroxide species, although this assignment is tentative due to the limit of resolution of 1.7 Å. The β-hydroxyl oxygen of BHDA is positioned only 2.45 Å from the distal oxygen atom (SI Appendix, Fig. S12), implying a possible β-hydrogen abstraction from the native substrate LA during the catalysis.
Fig. 3.
Structures of UndA and proposed 1-undecene synthesis mechanism. (A) Overall structure of UndA with helices in blue and loops in salmon. Iron is shown in black, and DEA is shown in yellow for carbons and red for oxygens. (B) Substrate binding pocket of UndA. DEA, presented as a ball-and-stick model, is surrounded by a simulated annealing omit map in blue, contoured at 3.0 σ. Pocket-forming residues are displayed as sticks and the hydrophobic residues are colored in orange. (C–F) Weighted electron density maps surrounding the active site of the holo-, DEA-, and BHDA-bound structures with 2mFo-DFc (in gray, at 1.8 σ) and mFo-DFc (green and red, at ±3.0 σ). The distal oxygen atoms of the dioxygen species in E and F are surrounded by the simulated annealing omit maps in green, contoured at 3.0 σ. (G) Proposed mechanism for 1-undecene biosynthesis by UndA. The molecular oxygen is proposed to be reduced to H2O likely by some reductant as shown in SI Appendix, Fig. S14A.
Discussion
Based on the biochemical and structural analyses of UndA, we postulate the following O2-activating, nonheme Fe(II)-dependent mechanism for the oxidative decarboxylation of LA (Fig. 3G and SI Appendix, Fig. S14A). First, LA binds to the ferrous iron of the enzyme through its carboxylate moiety and organizes the iron center to coordinate molecular oxygen. This sequential binding is similar to that in many other nonheme iron enzymes, such as naphthalene dioxygenase and superoxide reductase (18, 20, 23–26). Next, O2 binds, generating a putative Fe(III)–superoxide complex that abstracts the unactivated β-hydrogen of LA. Single electron transfer at the stage of a highly reactive radical intermediate leads to the formation of 1-undecene, CO2, H2O, and a possible unstable Fe(IV)=O species that can be reduced back to the ferrous state by a reducing cosubstrate to regenerate the active ferrous form of the enzyme. This proposed mechanism is consistent with the observed stoichiometry under single turnover conditions where one molecule of UndA–Fe(II) consumed approximately one equivalent of O2 to produce close to one equivalent of 1-undecene. Although the α-hydrogen of LA is generally considered to be more reactive, we propose β-hydrogen abstraction of LA based on several observations: (i) The UndA assay with [α,α-D2]LA led to the production of 1-undecene retaining both deuterium atoms (Fig. 2); (ii) UndA accepts AHDA, but not BHDA, as a substrate (Fig. 2); and (iii) the β-hydroxyl oxygen of BHDA is only 2.45 Å away from the distal oxygen atom in the BHDA-bound structure, suggesting that the activated O2 can reach the β-hydrogen of LA for hydrogen abstraction (SI Appendix, Fig. S12). The analogous β-hydrogen abstraction in substrate activation has also been proposed during porphyrin biosynthesis, in which the propionate side chains of coproporphyrinogen III are converted into the corresponding vinyl groups through oxidative decarboxylation. Interestingly, two structurally unrelated enzymes, one oxygen-dependent (HemF) and one oxygen-independent (HemN), are able to catalyze this reaction using a possible Mn(III)–superoxo species and a 5′-deoxyadenosyl radical to initiate β-hydrogen abstraction, respectively (27, 28).
It is notable that the high-valent Fe(IV)=O species is canonically considered to be sufficiently reactive for hydrogen abstraction from an unactivated carbon center, such as the β-hydrogen of fatty acids. The observed multiple turnover reaction of UndA in the presence of ascorbate raises the possibility of an alternative mechanism, in which ascorbate serves as the two-electron donating cosubstrate to form the Fe(IV)=O species for the subsequent hydrogen abstraction of LA, similar to the catalytic mechanism used by 1-aminocyclopropane-1-carboxylic acid oxidase (ACCO) (SI Appendix, Fig. S14B) (29, 30). However, unlike the ACCO-catalyzed reaction, the rate of 1-undecene formation under single turnover conditions in the absence of ascorbate was not significantly lower than the rate under multiple turnover conditions in the presence of ascorbate, and nearly a stoichiometric amount of 1-undecene was formed in the absence of ascorbate, suggesting that ascorbate is not needed for O2 activation by UndA–Fe(II) to catalyze 1-undecene formation (29, 30). In addition, ACCO is found in plants, which naturally produce ascorbate, but most Pseudomonas species lack the ability to synthesize ascorbate (31, 32), suggesting that ascorbate is probably not the cognate-reducing cosubstrate for UndA. We propose that in the in vitro assays, ascorbate acts mainly as a reductant after 1-undecene formation by restoring the ferrous species, and this role of ascorbate has previously been reported (SI Appendix, Fig. S14A) (33–36). We thus favor the mechanism in which the initial two-electron donating cosubstrate is LA in the case of UndA (SI Appendix, Fig. S14A), analogous to ascorbate in the case of ACCO and α-KG in many other known examples (35). This mechanism is further supported by the observation that H2O2/Fe3+ failed to elicit UndA activity, in contrast to the P450 fatty acid peroxygenase (13). We postulate that the proposed reaction of Fe(III)–superoxide-promoted β-hydrogen abstraction might be thermodynamically possible in concert with a single electron transfer yielding CO2, an excellent leaving group. Experiments to further characterize the catalytic mechanism are in progress.
Phylogenetic analysis revealed that undA is ubiquitous and well conserved in all of the sequenced Pseudomonas species and several other closely related species, including species from Acinetobacter, Burkholderia, and Myxococcus genera, with a total of more than 1,500 homologs identified from published genomes (SI Appendix, Fig. S15). No homolog of undA could be found in the sequenced genomes of Shewanella, consistent with the observations that all tested Pseudomonas spp. produced 1-undecene, but Shewanella spp. did not. Examination of the genomic contexts in Pseudomonas further revealed that undA belongs to a conserved two-gene operon (Fig. 1B), and the other conserved ORF (such as PFL_4322 in P. fluorescens Pf-5 and PA0861 in P. aeruginosa PAO1) encodes a protein with three domains: a sensory PAS domain, a GGDEF-class diguanylate cyclase, and an EAL-class phosphodiesterase. These domain activities are related to signal recognition, and synthesis and degradation of cyclic di-GMP, an important and ubiquitous second messenger in bacteria (37). Recent studies revealed that homologs of PFL_4322 are related to biofilm formation and dispersal in P. aeruginosa PAO1 and P. fluorescens Pf0–1 (38, 39), and it is yet to be determined if 1-undecene, synthesized through the function of undA homolog in the same operon, is also a semivolatile signaling molecule related to biofilm formation and dispersal. 1-Undecene may also play a role in bacterial–plant interactions, as many of the producing organisms are plant-associated bacteria (40). Our discovery of these conserved operons provides an intriguing opportunity to study the physiological role of this ubiquitous hydrocarbon metabolite in Pseudomonas, the abundant and widespread bacteria.
In summary, we revealed the genetic basis and molecular mechanism for biosynthesis of 1-undecene, a ubiquitous hydrocarbon metabolite of Pseudomonas. Our study discovered a previously unidentified family of nonheme oxidases that specifically convert medium-chain fatty acids (C10–C14) into the corresponding terminal olefins using an oxygen-activating, nonheme, iron-dependent mechanism. Both biochemical and structural analyses suggest an unusual mechanism of β-hydrogen abstraction by a “less reactive” iron center during fatty acid activation. It is notable that without strain optimization, overexpression of undA homologs in E. coli produced 1-undecene at a titer over 25-fold higher than the best titer in Pseudomonas (Fig. 1C). This result lays an advanced foundation for pathway engineering, such as improving the availability of medium-chain fatty acids or cognate-reducing cosubstrate for the enzyme, to increase the production titer and yield of MCAEs. Our findings expand the scarce enzyme inventory for the transformation of fatty acid precursors to hydrocarbons (2) and open the road for producing MCAEs, useful as fuels and chemical building blocks, from renewable resources.
Materials and Methods
Bacterial Growth and Hydrocarbon Analysis.
For 1-undecene production, 50 μL of seed culture was used to inoculate 5 mL of LB medium. The culture was shaken in a sealed 20-mL headspace vial containing a stir bar at 30 °C for 36 h. After incubation, an SPME fiber (30 μm polydimethylsiloxane, Supelco, Sigma-Aldrich Group) was manually inserted into the headspace vial and incubated at 25 °C for 12.5 min for GCMS analysis. For details, see SI Appendix, Materials and Methods.
Identification of the 1-Undecene Biosynthetic Gene.
The fosmid genomic library of P. fluorescens Pf-5 was constructed using a pCC2Fos CopyControl library kit (Epicentre Biotechnologies) (SI Appendix, Fig. S2). Pools of 10 clones were first analyzed for 1-undecene production, followed with another round of screening to identify single 1-undecene–producing clones. Three identified fosmids—6F8, 6E2, and 4F3—were sequenced to reveal a 15-kb overlapping region that was further analyzed to reveal gene(s) related to 1-undecene production. For details, see SI Appendix, Materials and Methods.
Biochemical Analysis of UndA.
UndA purification, screening of possible substrates and products, in vitro single-turnover and multiple-turnover reactions, quantification of substrates and products, rate analysis, and so forth are detailed in SI Appendix, Materials and Methods.
Crystallography.
UndA crystals were grown at room temperature using the hanging-drop vapor diffusion method in 0.1 M Mes, 1.8 M (NH4)2SO4, 0.2 mM (NH4)2Fe(SO4)2, pH 6.5–7.0. To obtain ligand-bound structures, UndA was incubated with 2.5 mM DEA or BHDA on ice for 15 min before crystallization. X-ray diffraction data were collected at beamline 8.3.1 at the Advanced Light Source at Lawrence Berkeley National Laboratory. SI Appendix, Table S2 summarizes the statistics of data collection and refinement. For details, see SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank J. Klinman (University of California) for discussions regarding reaction mechanisms, S. Bauer (University of California) for assisting with GCMS analysis, N. Gilbert (Vanderbilt University) for early structural modeling, E. Kemper and W. Bao for library screening, A. Arkin (University of California) for providing Shewanella strains, C. Walsh (Harvard Medical School) and S. Lindow (University of California) for providing Pseudomonas strains, and D. Hung (Massachusetts General Hospital) for providing P. aeruginosa PA14 nonredundant transposon insertion mutants. This work was funded by grants from the Energy Biosciences Institute (to W.Z. and J.H.D.C.).
Footnotes
Conflict of interest statement: Z.R. and W.Z. have filed a provisional patent application on aspects of this research.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4WWJ, 4WWZ, and 4WX0).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419701112/-/DCSupplemental.
References
- 1.Peralta-Yahya PP, Zhang F, del Cardayre SB, Keasling JD. Microbial engineering for the production of advanced biofuels. Nature. 2012;488(7411):320–328. doi: 10.1038/nature11478. [DOI] [PubMed] [Google Scholar]
- 2.Lennen RM, Pfleger BF. Microbial production of fatty acid-derived fuels and chemicals. Curr Opin Biotechnol. 2013;24(6):1044–1053. doi: 10.1016/j.copbio.2013.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ray S, Rao PVC, Choudary NV. Poly-alpha-olefin-based synthetic lubricants: A short review on various synthetic routes. Lubr Sci. 2012;24(1):23–44. [Google Scholar]
- 4.Moiseenkov AM, Schaub B, Margot C, Schlosser M. A new stereoselective synthesis of (Z)-9-tricosene, the sex attractant of the common housefly. Tetrahedron Lett. 1985;26(3):305–306. [Google Scholar]
- 5.Kai M, et al. Bacterial volatiles and their action potential. Appl Microbiol Biotechnol. 2009;81(6):1001–1012. doi: 10.1007/s00253-008-1760-3. [DOI] [PubMed] [Google Scholar]
- 6.Schulz S, Dickschat JS. Bacterial volatiles: The smell of small organisms. Nat Prod Rep. 2007;24(4):814–842. doi: 10.1039/b507392h. [DOI] [PubMed] [Google Scholar]
- 7.Labows JN, McGinley KJ, Webster GF, Leyden JJ. Headspace analysis of volatile metabolites of Pseudomonas aeruginosa and related species by gas chromatography-mass spectrometry. J Clin Microbiol. 1980;12(4):521–526. doi: 10.1128/jcm.12.4.521-526.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zechman JM, Labows JN., Jr Volatiles of Pseudomonas aeruginosa and related species by automated headspace concentration—Gas chromatography. Can J Microbiol. 1985;31(3):232–237. doi: 10.1139/m85-045. [DOI] [PubMed] [Google Scholar]
- 9.Graham JE. Bacterial volatiles and diagnosis of respiratory infections. Adv Appl Microbiol. 2013;82:29–52. doi: 10.1016/B978-0-12-407679-2.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bos LD, Sterk PJ, Schultz MJ. Volatile metabolites of pathogens: A systematic review. PLoS Pathog. 2013;9(5):e1003311. doi: 10.1371/journal.ppat.1003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB. Microbial biosynthesis of alkanes. Science. 2010;329(5991):559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- 12.Rude MA, et al. Terminal olefin (1-alkene) biosynthesis by a novel p450 fatty acid decarboxylase from Jeotgalicoccus species. Appl Environ Microbiol. 2011;77(5):1718–1727. doi: 10.1128/AEM.02580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belcher J, et al. Structure and biochemical properties of the alkene producing cytochrome P450 OleTJE (CYP152L1) from the Jeotgalicoccus sp. 8456 bacterium. J Biol Chem. 2014;289(10):6535–6550. doi: 10.1074/jbc.M113.527325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mendez-Perez D, Begemann MB, Pfleger BF. Modular synthase-encoding gene involved in α-olefin biosynthesis in Synechococcus sp. strain PCC 7002. Appl Environ Microbiol. 2011;77(12):4264–4267. doi: 10.1128/AEM.00467-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liberati NT, et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci USA. 2006;103(8):2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkins AL, Zhang Y, Ealick SE, Begley TP. Mutagenesis studies on TenA: A thiamin salvage enzyme from Bacillus subtilis. Bioorg Chem. 2008;36(1):29–32. doi: 10.1016/j.bioorg.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaillancourt FH, Yeh E, Vosburg DA, O’Connor SE, Walsh CT. Cryptic chlorination by a non-haem iron enzyme during cyclopropyl amino acid biosynthesis. Nature. 2005;436(7054):1191–1194. doi: 10.1038/nature03797. [DOI] [PubMed] [Google Scholar]
- 18.Chang WC, et al. Mechanistic studies of an unprecedented enzyme-catalysed 1,2-phosphono-migration reaction. Nature. 2013;496(7443):114–118. doi: 10.1038/nature11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirica LM, McCusker KP, Munos JW, Liu HW, Klinman JP. 18O kinetic isotope effects in non-heme iron enzymes: Probing the nature of Fe/O2 intermediates. J Am Chem Soc. 2008;130(26):8122–8123. doi: 10.1021/ja800265s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruijnincx PC, van Koten G, Klein Gebbink RJ. Mononuclear non-heme iron enzymes with the 2-His-1-carboxylate facial triad: Recent developments in enzymology and modeling studies. Chem Soc Rev. 2008;37(12):2716–2744. doi: 10.1039/b707179p. [DOI] [PubMed] [Google Scholar]
- 21.Dassama LMK, et al. O(2)-evolving chlorite dismutase as a tool for studying O(2)-utilizing enzymes. Biochemistry. 2012;51(8):1607–1616. doi: 10.1021/bi201906x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang C, et al. Evidence that the fosfomycin-producing epoxidase, HppE, is a non-heme-iron peroxidase. Science. 2013;342(6161):991–995. doi: 10.1126/science.1240373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karlsson A, et al. Crystal structure of naphthalene dioxygenase: Side-on binding of dioxygen to iron. Science. 2003;299(5609):1039–1042. doi: 10.1126/science.1078020. [DOI] [PubMed] [Google Scholar]
- 24.Katona G, et al. Raman-assisted crystallography reveals end-on peroxide intermediates in a nonheme iron enzyme. Science. 2007;316(5823):449–453. doi: 10.1126/science.1138885. [DOI] [PubMed] [Google Scholar]
- 25.Jeoung JH, Bommer M, Lin TY, Dobbek H. Visualizing the substrate-, superoxo-, alkylperoxo-, and product-bound states at the nonheme Fe(II) site of homogentisate dioxygenase. Proc Natl Acad Sci USA. 2013;110(31):12625–12630. doi: 10.1073/pnas.1302144110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Donk WA, Krebs C, Bollinger JM., Jr Substrate activation by iron superoxo intermediates. Curr Opin Struct Biol. 2010;20(6):673–683. doi: 10.1016/j.sbi.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breckau D, Mahlitz E, Sauerwald A, Layer G, Jahn D. Oxygen-dependent coproporphyrinogen III oxidase (HemF) from Escherichia coli is stimulated by manganese. J Biol Chem. 2003;278(47):46625–46631. doi: 10.1074/jbc.M308553200. [DOI] [PubMed] [Google Scholar]
- 28.Layer G, et al. The substrate radical of Escherichia coli oxygen-independent coproporphyrinogen III oxidase HemN. J Biol Chem. 2006;281(23):15727–15734. doi: 10.1074/jbc.M512628200. [DOI] [PubMed] [Google Scholar]
- 29.Rocklin AM, Kato K, Liu HW, Que L, Jr, Lipscomb JD. Mechanistic studies of 1-aminocyclopropane-1-carboxylic acid oxidase: Single turnover reaction. J Biol Inorg Chem. 2004;9(2):171–182. doi: 10.1007/s00775-003-0510-3. [DOI] [PubMed] [Google Scholar]
- 30.Mirica LM, Klinman JP. The nature of O2 activation by the ethylene-forming enzyme 1-aminocyclopropane-1-carboxylic acid oxidase. Proc Natl Acad Sci USA. 2008;105(6):1814–1819. doi: 10.1073/pnas.0711626105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smirnoff N. Vitamin C: The metabolism and functions of ascorbic acid in plants. Adv Bot Res. 2011;59:107–177. [Google Scholar]
- 32.Bremus C, Herrmann U, Bringer-Meyer S, Sahm H. The use of microorganisms in L-ascorbic acid production. J Biotechnol. 2006;124(1):196–205. doi: 10.1016/j.jbiotec.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 33.Myllylä R, Kuutti-Savolainen ER, Kivirikko KI. The role of ascorbate in the prolyl hydroxylase reaction. Biochem Biophys Res Commun. 1978;83(2):441–448. doi: 10.1016/0006-291x(78)91010-0. [DOI] [PubMed] [Google Scholar]
- 34.Clifton IJ, et al. Structural studies on 2-oxoglutarate oxygenases and related double-stranded beta-helix fold proteins. J Inorg Biochem. 2006;100(4):644–669. doi: 10.1016/j.jinorgbio.2006.01.024. [DOI] [PubMed] [Google Scholar]
- 35.Costas M, Mehn MP, Jensen MP, Que L., Jr Dioxygen activation at mononuclear nonheme iron active sites: Enzymes, models, and intermediates. Chem Rev. 2004;104(2):939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 36.Puistola U, Turpeenniemi-Hujanen TM, Myllylä R, Kivirikko KI. Studies on the lysyl hydroxylase reaction. I. Initial velocity kinetics and related aspects. Biochim Biophys Acta. 1980;611(1):40–50. doi: 10.1016/0005-2744(80)90040-6. [DOI] [PubMed] [Google Scholar]
- 37.Hengge R. Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol. 2009;7(4):263–273. doi: 10.1038/nrmicro2109. [DOI] [PubMed] [Google Scholar]
- 38.An S, Wu J, Zhang LH. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-Di-GMP phosphodiesterase with a putative hypoxia-sensing domain. Appl Environ Microbiol. 2010;76(24):8160–8173. doi: 10.1128/AEM.01233-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O’Toole GA. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. J Bacteriol. 2011;193(18):4685–4698. doi: 10.1128/JB.05483-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blom D, et al. Production of plant growth modulating volatiles is widespread among rhizosphere bacteria and strongly depends on culture conditions. Environ Microbiol. 2011;13(11):3047–3058. doi: 10.1111/j.1462-2920.2011.02582.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.