Significance
Angiogenin is a stress-activated ribonuclease that cleaves tRNA to produce bioactive small noncoding RNAs [tRNA-derived, stress-induced RNAs (tiRNAs)] that function in a cytoprotective stress response program. Point mutations that reduce its ribonuclease activity are found in a subset of patients with ALS, a fatal neurodegenerative disease. We have found that selected tiRNAs assume G-quadruplex (G4) structures that are necessary for cytoprotective and prosurvival functions. Moreover, stable DNA analogs of these G4-containing tiRNAs spontaneously enter motor neurons and confer cytoprotection against stress. Our results identify tiRNAs as leading compounds for the development of a new class of neuroprotective drugs and give insights into the molecular mechanisms underlying the pathobiology of expanded G4-forming hexanucleotide repeats in the C9ORF72 gene.
Keywords: tRNA, angiogenin, stress, C9ORF72, amyotrophic lateral sclerosis
Abstract
Angiogenin (ANG) is a stress-activated ribonuclease that promotes the survival of motor neurons. Ribonuclease inactivating point mutations are found in a subset of patients with ALS, a fatal neurodegenerative disease with no cure. We recently showed that ANG cleaves tRNA within anticodon loops to produce 5′- and 3′-fragments known as tRNA-derived, stress-induced RNAs (tiRNAs). Selected 5′-tiRNAs (e.g., tiRNAAla, tiRNACys) cooperate with the translational repressor Y-box binding protein 1 (YB-1) to displace the cap-binding complex eIF4F from capped mRNA, inhibit translation initiation, and induce the assembly of stress granules (SGs). Here, we show that translationally active tiRNAs assemble unique G-quadruplex (G4) structures that are required for translation inhibition. We show that tiRNAAla binds the cold shock domain of YB-1 to activate these translational reprogramming events. We discovered that 5′-tiDNAAla (the DNA equivalent of 5′-tiRNAAla) is a stable tiRNA analog that displaces eIF4F from capped mRNA, inhibits translation initiation, and induces the assembly of SGs. The 5′-tiDNAAla also assembles a G4 structure that allows it to enter motor neurons spontaneously and trigger a neuroprotective response in a YB-1–dependent manner. Remarkably, the ability of 5′-tiRNAAla to induce SG assembly is inhibited by G4 structures formed by pathological GGGGCC repeats found in C9ORF72, the most common genetic cause of ALS, suggesting that functional interactions between G4 RNAs may contribute to neurodegenerative disease.
Angiogenin (ANG) is a secreted RNase that was identified as an angiogenic factor produced by tumor cells (1). Point mutations that reduce its RNase activity are found in a small subset of patients with both familial and sporadic ALS (2, 3), a neurodegenerative disease characterized by the progressive loss of motor neurons. Although these results implicate an RNA cleavage event in the pathogenesis of ALS, target RNA(s) whose cleavage promotes motor neuron survival have not been identified.
We discovered that ANG activates a cytoprotective stress response program in eukaryotic cells (4). ANG enters cells via a receptor-mediated process that delivers it to the cytoplasm and the nucleus (5, 6). In the cytoplasm, ANG cleaves the anticodon loops of mature tRNAs to produce 5′- and 3′-fragments that are designated as tRNA-derived, stress-induced RNAs (tiRNAs) (4, 7). We found that a subset of 5′-tiRNAs, but not 3′-tiRNAs, inhibits translation initiation and triggers the assembly of stress granules (SGs) (8, 9), RNA granules implicated in the pathogenesis of ALS (8). These events conspire to enhance the survival of cells subjected to adverse environmental conditions (reviewed in refs. 8, 9).
We have identified two structural features that are required for tiRNAs to inhibit translation initiation (10). The first is a stem–loop structure corresponding to the “D-loop” of tRNA (10). The dimensions of this D-loop structure are similar to the stem–loop structure that allows let-7 microrna precursor to bind the cold shock domain (CSD) of the YB-1 homolog Lin28 (11). Because YB-1 also binds to a stem–loop structure found in the 5′-UTR of Snail1 transcripts (12), we hypothesized that the tiRNA D-loop may bind to the CSD of YB-1. The second required structural feature is a 5′-terminal oligoguanine (TOG) motif that is only found in the translationally repressive 5′-tiRNAs derived from tRNAAla and tRNACys. The 5′-tiRNAAla and 5′-tiRNACys, but not mutants lacking the D-loop or TOG motif, inhibit translation initiation by displacing eIF4F from cap structures (m7G) (10). Mutations in the TOG motif significantly reduce the binding of 5′-tiRNAAla to YB-1 (10), suggesting that this motif is required for the assembly of YB-1/tiRNA complexes that directly target the translation initiation machinery to inhibit protein synthesis.
Guanine-rich (G-rich) oligonucleotides (ODNs) have the potential to assemble intra- or intermolecular G-quadruplex (G4) structures that are resistant to nucleases, and they spontaneously enter cells by a poorly characterized mechanism (13–15). Here, we show that biologically active 5′-tiRNAAla and 5′-tiRNACys form G4 structures similar to G-quadruplexes formed by the hexanucleotide repeats in C9ORF72 (16, 17), the leading genetic cause of ALS and frontotemporal dementia (FTD). The assembly of G4 structures correlates with the ability of DNA analogs of 5′-tiRNAAla and 5′-tiRNACys (5′-tiDNAAla and 5′-tiDNACys) to inhibit translation, promote formation of SGs, and confer neuroprotection to motor neurons exposed to stress. Our results also suggest a new mechanism by which extended G4 structures may be involved in the pathophysiology of repeat-associated neurodegenerative diseases.
Results
Bioactive tiRNAs (and Corresponding tiDNAs) Assume G4 Structures.
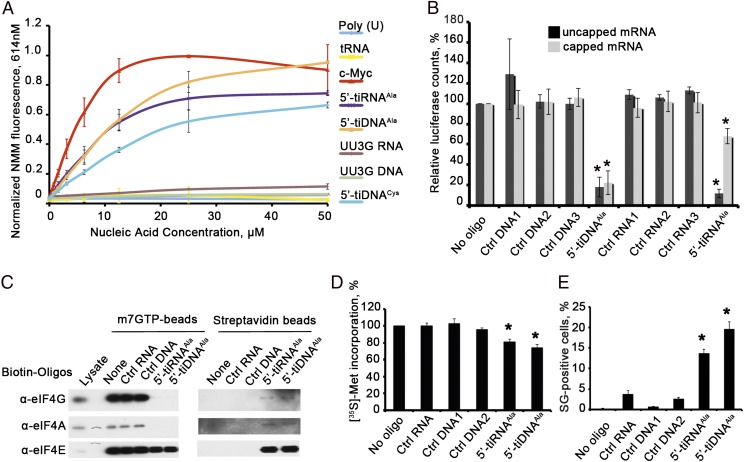
Bioactive tiRNAs and their tiDNA analogs are relatively rich in Guanine residues (Figs. S1A and S2). Because G-rich RNA and DNA ODNs can assemble G4 structures that confer unique biological properties (18), we tested the ability of G-rich tiRNAs and tiDNAs to assemble G4 structures. Binding of N-methyl mesoporphyrin IX (NMM) to G4 structures results in fluorescent emissions at 614 nm (19). A G-rich ODN derived from the c-myc promoter (i.e., c-myc ODN), known to assemble G4 structures (20), binds NMM, resulting in fluorescent emission at 614 nm (Fig. 1A). Similarly, 5′-tiRNAAla, 5′-tiRNACys, and 5′-tiDNAAla also assemble G4 structures, but Poly(U) RNA and tRNA do not. Whereas mutant 5′-tiRNAAla or 5′-tiDNAAla with a single G→U substitution assumes a G4 structure (Fig. S3B), a mutant with two G→U substitutions (UU3G RNA and UU3G DNA) does not (Fig. 1A). The ability of tiRNAAla mutants to inhibit translation initiation (10) correlates with their ability to bind NMM and assume G4 structures (Fig. 1A). When tiRNAAla and tiDNAAla are separated on nondenaturing [20% Tris⋅borate (TB)] or denaturing [15% Tris⋅borate-urea (TBU)] polyacrylamide gels, several molecular species are clearly identified (Fig. S1B). Species migrating faster than expected for 30 nucleotide oligos (Fig. S1B, red arrows) suggest the presence of intramolecular base pairing resulting in compaction of the tertiary structure. Species migrating slower than expected (Fig. S1B, black arrows) are consistent with the presence of intermolecular base pairing indicative of stable multimers.
Fig. 1.
Translationally repressive tiRNAs and tiDNAs assemble G4 structures to inhibit translation. (A) NMM fluorescence analysis. Poly(U) RNA and total yeast tRNA (tRNA) were used as negative controls, and validated G4 oligo derived from the c-myc promoter (c-Myc) was used as a positive control. 5′-tiRNAAla, 5′-tiDNAAla, and 5′-tiRNACys bind NMM in a concentration-dependent manner, in contrast to translationally inactive mutants of 5′-tiR/DNAAla (corresponding to a UU3G mutant with two G→U substitutions at the 5′-end). (B) 5′-tiDNAAla inhibits translation of mRNA reporters in vitro. Uncapped (black) and capped (gray) Firefly luciferase mRNA (Promega) was translated in the RRL in vitro translation system in the presence of the indicated RNA and DNA oligos. Luciferase expression is relative to luciferase expression in the absence of any RNA or DNA (no RNA/DNA oligo = 100%). Means and SDs are from three independent experiments [*P < 0.05, Student t test (n = 3)]. Ctrl, control. (C) 5′-tiDNAAla displaces the eIF4F complex from m7GTP-Sepharose. (Left) Indicated 3′-end biotinylated RNAs or DNAs were added to preformed complexes of m7GTP-Sepharose, and displaced components were quantified by Western blotting. (Right) Streptavidin beads were used to pull down displaced RNA-bound proteins before Western blotting. (D) Quantification of [35S]-Met incorporation in cells transfected with 5′-tiDNAAla. The indicated RNA and DNA oligos were transfected into U2OS cells before pulsing with [35S]-methionine. Total counts per minute per microgram of protein were normalized to cells untreated with any RNA or DNA oligos. Means and SDs are from three independent experiments [*P < 0.05, relative to control without any RNA/DNA, Student t test (n = 3)]. (E) 5′-tiDNAAla induces SG assembly in U2OS cells. U2OS cells were transfected with the indicated RNA or DNA oligos before quantifying SGs by counting 200 cells per experiment. Error bars reflect SDs of the mean [*P < 0.05, relative to control without any RNA/DNA (No Oligo) and all control oligos (Ctrl RNA, Ctrl DNA1, and Ctrl DNA2), Student t test (n = 3)].
Bioactivity of 5′-tiRNAAla and 5′-tiDNAAla.
We previously showed that 5′-tiRNAAla displaces eIF4F from capped mRNA to inhibit translation initiation and induce the assembly of SGs (10). Remarkably, tiDNAs have similar biological activities. Both tiRNAAla and tiDNAAla specifically and significantly inhibit the translation of luciferase transcripts in rabbit reticulocyte lysates (RRLs) (Fig. 1B). To compare the effects of these oligos on eIF4F/cap interactions, we assembled eIF4E-containing complexes from U2OS cell lysates on m7GTP-Sepharose. Sepharose-bound complexes were incubated with 3′-end biotinylated control RNA or DNA, 5′-tiRNAAla, or 5′-tiDNAAla before analyzing retained components of the eIF4E-containing complexes by Western blotting (Fig. 1C). Whereas control oligos do not displace initiation factors from m7GTP-Sepharose, 5′-tiRNAAla and 5′-tiDNAAla completely displace eIF4G and eIF4A and partially displace eIF4E from the beads. The supernatants containing displaced initiation factors were used to capture biotin-RNA/DNA oligos and their associated proteins. Western blotting analysis reveals that biotin–5′-tiRNAAla and 5′-tiDNAAla capture eIF4G/A/E (Fig. 1C). Thus, 5′-tiDNAAla displaces eIF4F from m7G caps like 5′-tiRNAAla. Transfection of 5′-tiRNAAla or 5′-tiDNAAla modestly, but significantly, inhibits incorporation of [35S]-methionine in U2OS cells (Fig. 1D). A subpopulation of these cells also assembles SG, suggesting that these bioactive oligos inhibit translation initiation (Fig. 1E). In all these experiments (Fig. 1 B–E), 5′-tiDNAAla had comparable or even stronger effects on translation inhibition than 5′-tiRNAAla.
tiRNAAla and tiDNAAla Bind the CSD of YB-1.
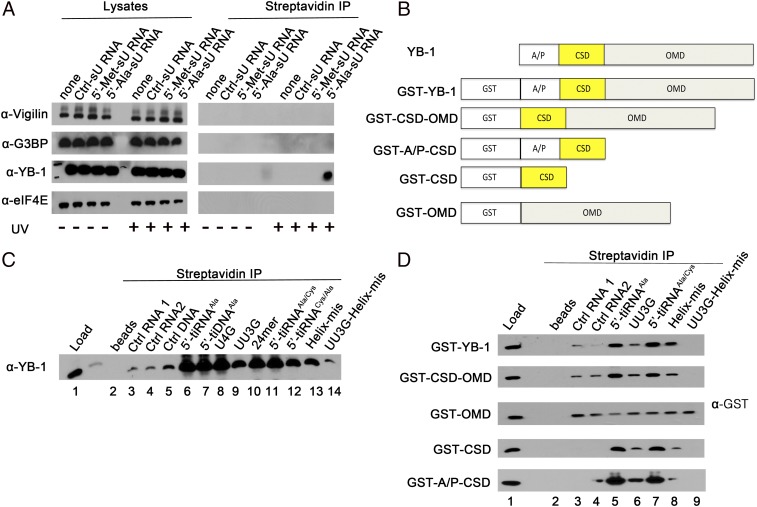
We previously identified YB-1 as a cofactor that is required for tiRNA-mediated translational repression (10). We used 4-thio-uracil (sU)–modified tiRNAs to confirm that bioactive tiRNAs bind directly to endogenous YB-1 in live cells. U2OS cells were transfected with biotinylated oligos [i.e., sU-modified control RNA (Ctrl-sU RNA), inactive 5′-tiRNAMet (5′-Met-sU RNA), or active 5′-tiRNAAla (5′-Ala-sU RNA)] before irradiating cells with UV light to activate sU cross-linking (21). Cells were then solubilized, and streptavidin-Sepharose was used to pull down biotinylated oligos and associated cross-linked proteins for analysis by Western blotting (10). As shown in Fig. 2A, sU modified 5′-tiRNAAla, but not 5′-tiRNAMet, specifically pulled down YB-1, but not other tiRNA-binding proteins, following UV irradiation. Because sU-modified oligos are cross-linked only to proteins in close proximity, this result indicates that 5′-tiRNAAla binds to YB-1 in live cells.
Fig. 2.
YB-1 is a direct interaction partner of 5-tiRNAAla and 5′-tiDNAAla. (A) Modified photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation identifies YB-1 as a direct interaction partner of 5′-tiRNAAla. The indicated RNA oligos were transfected into U2OS cells, irradiated using UV light at 365 nm, solubilized, and immunoprecipitated using streptavidin beads. Precipitated proteins were identified by Western blotting using the indicated antibodies. Cell lysates at a ratio of 1:30 were used as loading controls. (B) Schematic representation of YB-1 protein, its domains, and the N-terminal GST fusion YB-1 variants used in this study. The A/P domain (amino acids 1–59), CSD (amino acids 60–129), and OMD (amino acids 130–324) are shown. (C) Recombinant WT YB-1 directly binds to 5′-tiRNAAla and 5′-tiDNAAla in vitro. Recombinant nontagged YB-1 (full-length) was incubated with the indicated RNA and DNA oligos before affinity precipitation using streptavidin beads and Western blotting analysis using anti–YB-1 antibody. (D) CSD domain of YB-1 is responsible for 5′-tiRNAAla binding specificity. The indicated fusion proteins were incubated with the indicated RNA oligos before affinity precipitation using streptavidin beads and Western blotting using anti-GST antibody.
YB-1 is a multifunctional DNA/RNA binding protein with two distinct nucleic acid binding domains (Fig. 2B). Its oligomerization domain (OMD), containing alternating clusters of positively and negatively charged amino acid residues (Fig. 2B; OMD), binds to single-stranded ssRNA and ssDNA, and its CSD (Fig. 2B) binds to double-stranded oligos or stem–loop structures (reviewed in refs. 22, 23). In addition to the CSD and OMD, YB-1 contains a structurally disordered Ala- and Pro-rich N-terminal domain (the A/P domain) that does not bind nucleic acids (22). We assessed the binding specificity of full-length and truncated YB-1 using biotinylated RNA and DNA oligos (predicted secondary structures are depicted in Fig. S2). Pull downs using streptavidin-Sepharose contain small amounts of YB-1 bound nonspecifically to control RNA and DNA oligos (Fig. 2C, lanes 3–5). Much larger amounts of YB-1 are pulled down with 5′-tiRNAAla (lane 6) and 5′-tiDNAAla (lane 7). Two G→U substitutions in the 5′-TOG motif causing failure in G4 structure formation (Fig. 1A) also reduced binding to YB-1 (UU3G, lane 9), but deletion of the six 3′-terminal nucleotides (following the stem structure; 24mer, lane 10) or single G→U substitutions (U4G, lane 8) did not affect binding. Hybrid oligos composed of the first 17 nucleotides from 5′-tiRNAAla and the last 13 nucleotides from 5′-tiRNACys (Fig. S2C) maintained the 5′-TOG motif, the stem–loop structure, and YB-1 binding (5′-tiRNAAla/Cys, lane 11). In contrast, oligos composed of the first 17 nucleotides from 5′-tiRNACys and the last 13 nucleotides from 5′-tiRNAAla (Fig. S2C) lost the stem–loop structure and had reduced YB-1 binding (5′-tiRNACys/Ala, lane 12). Mutations that disrupted the stem structure of 5′-tiRNAAla (mismatch in the helix structure, Helix-mis) had reduced YB-1 binding (Fig. S2C; Helix-mis, lane 13), and oligos with mutations in both the TOG motif and the stem structure (UU3G-helix-mis, lane 14) bound YB-1 at levels similar to control oligos (lanes 3–5). Thus, the structural features required for translational repression are also required for YB-1 binding.
We used GST-fusion proteins to identify the tiRNA-binding domain in YB-1 (Fig. 2B). GST–YB-1 and a truncation mutant lacking the A/P domain (GST-CSD-OMD) bound strongly to 5′-tiRNAAla and to the active 5′-tiRNAAla/Cys oligo (Fig. 2D, lanes 5 and 7), but weakly to control RNAs (lanes 3 and 4) or biologically inactive mutant oligos (Fig. 2D; UU3G, lane 6), Helix-mis (lane 8), and UU3G-helix-mis (lane 9). GST-OMD bound weakly to all oligos, suggesting sequence nonspecific binding (Fig. 2D, lanes 2–9). In contrast, fusion proteins that include the CSD (GST-CSD or GST-A/P-CSD) bound strongly to bioactive oligos but weakly to inactive mutants and control oligos (Fig. 2D). Similar results were obtained using DNA analogs of 5′-tiRNAAla mutants (5′-tiDNAAla mutants) (Fig. S3A). The binding of the YB-1 CSD to 5′-tiRNAAla mutants correlates with the assembly of G4 structures as assessed by NMM binding (Fig. S3B) and with the ability to inhibit translation of an mRNA reporter in reticulocyte lysates (Fig. S3C). Moreover, recombinant YB-1 binds other well-characterized G4 oligos, including AS1411 (14), c-myc (20), and C9ORF72 (16, 17) (Fig. S3D, Upper). This binding depends on the nature of the monovalent cation used: Whereas sodium ions strongly support the formation of G4 structures (and thus binding to YB-1), lithium ions only weakly support their assembly (reviewed in ref. (24), resulting in less efficient YB-1 binding (Fig. S3D, Lower). Moreover, although recombinant YB-1 strongly binds to the stable G4 derived from the 5′-UTR of MT3-MMP mRNA [M3Q (25)], it does not bind an M3Q mutant (M3Q-Mut) that does not form G4 structures (25) (Fig. S3D, Lower, lanes 11–12). Finally, only RNA/DNA oligos that assemble G4 structures (Fig. 1A and Fig. S3B) exhibit anomalous mobility in gels under denaturing (Fig. S4, Left) or native (Fig. S4, Right) conditions compared with controls or translationally inactive (10), non–G4-containing 5′-tiRNAsVal/Met/Pro (Fig. S3B). In summary, these results suggest that bioactive 5′-tiRNAAla/5′-tiDNAAla oligos directly bind to YB-1. Although the CSD is responsible for the specificity for G4 structures, OMD binds many different nucleic acid structures with little specificity.
Bioactivity of 5′-tiDNAAla.
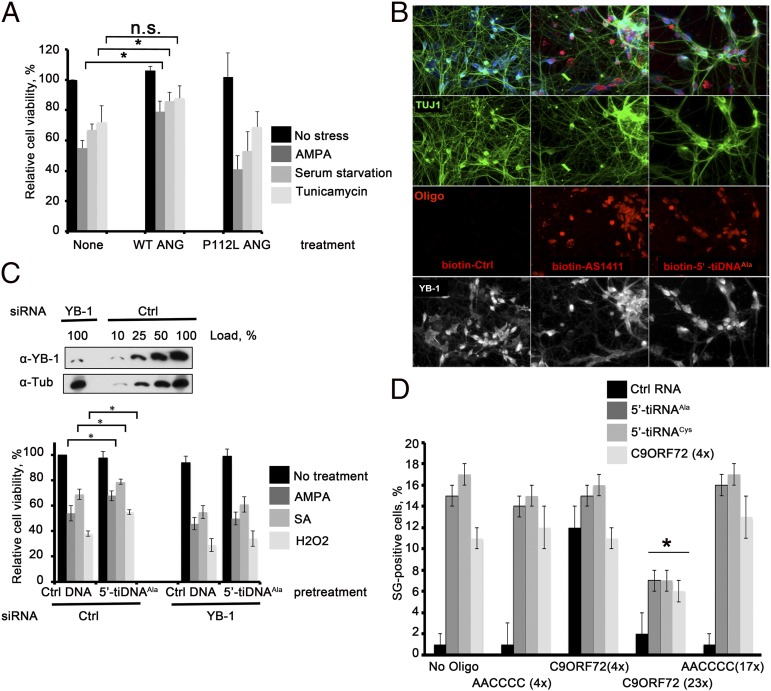
ANG protects mouse P19 embryonal carcinoma cells (26) and cultured mouse motor neurons from stress-induced apoptosis (27, 28). Human motor neurons (hMNs) differentiated from ES cells express the motor neuron markers TUJ1, MAP2, SMI-32, and peripherin (Fig. S5 B and C), as well as YB-1 (Fig. 3B, Lower). Treatment of these cells with WT ANG, but not ALS-associated, RNase-deficient P112L (29) mutant ANG, significantly inhibits excitotoxic (AMPA) treatment (27, 30), and serum withdrawal-induced [serum starvation (27)] death (27) (Fig. 3A). In contrast, knockdown of ANG sensitizes these cells to stress (Fig. S5D) and promotes stress-induced activation of apoptotic caspases (Fig. S5A). Importantly, treatment with recombinant ANG rescues ANG-depleted motor neurons from adverse effects of stress (Fig. S5D).
Fig. 3.
5′-tiDNAAla protects motor neurons from stress-induced death. (A) Recombinant ANG rescues motor neurons from stress-induced death. The hMNs were challenged with the indicated stressors in the absence (None) or presence of recombinant WT or ALS-associated P112L mutant (P112L) ANG (0.5 μg/mL, 24 h) before quantifying cell viability. Means and SDs are from three independent experiments [*P < 0.05, comparison of recombinant ANG WT-pretreated cells under selected stress stimuli with cell, Student t test (n = 3)]. n.s., not significant. (B) 5′-tiDNAAla is spontaneously taken up by hMNs. The indicated biotin oligos were added to hMNs (day 21) for 72 h. Cells were stained with anti-TUJ1 antibody (green), streptavidin-Cy3 (red), and anti–YB-1 (white). (Magnification: 40×.) (C) 5′-tiDNAAla protects motor neurons from stress-induced death in a YB-1–dependent manner. (Upper) hMNs were treated with the indicated siRNAs (YB-1 depletion was verified by Western blotting using a YB-1–specific antibody) and then cultured with the indicated DNA oligos for 72 h before challenge with the indicated stressors. α-Tub, α-tubulin. (Lower) CellTiter-Glo Luminescent Cell Viability Assay (Promega) was used to quantify motor neuron viability. Means and SDs are from four independent experiments [*P < 0.05, comparison of Ctrl- and 5′-tiDNAAla-pretreated cells under selected stress stimuli, Student t test (n = 4)]. (D) Extended C9ORF72 repeats inhibit tiRNA-induced SG assembly. U2OS cells were cotransfected with the indicated RNA oligos before quantifying SG assembly. The percentage of U2OS cells with SGs is shown. Error bars reflect SDs of the mean [*P < 0.05; comparison of SG formation by C9ORF72 (4×), 5′-tiRNAAla, and 5′-tiRNACys cotransfected with C9ORF72 (23×) to cotransfection with AACCCC (17×) or No Oligo].
G-rich oligos that assume G4 structures spontaneously enter cultured cells (18). We confirmed that G4-containing oligos (5′-tiDNAAla and its mutant T4G, 5′-tiDNACys, AS1411, c-myc, and C9ORF72), but not control DNA oligos or translationally inactive/non–G4-containing mutants of 5′-tiDNAAla (TT3G and TT3G-helix-mis), spontaneously enter cultured human U2OS cells (Fig. S6). Upon entering hMNs (Fig. 3B and Fig. S5 B and C), 5′-tiDNAAla is localized to nuclear and cytoplasmic compartments (Fig. S5C). Spontaneous uptake of 5′-tiDNAAla allowed us to test the hypothesis that G4-containing oligos act downstream of ANG to trigger a neuroprotective response. The hMNs were treated with siRNA to reduce the expression of YB-1 protein by ∼90% (Fig. 3C, Upper). Cells were then subjected to excitotoxic (AMPA) (27) or oxidative [sodium arsenite or H2O2 (31)] stress before quantifying viability using the CellTiter-Glo Luminescent Cell Viability Assay (Promega). In cells treated with control siRNA, 5′-tiDNAAla modestly, but significantly, enhances cell survival in response to stress (Fig. 3C, Lower). This effect was eliminated in cells treated with siRNA targeting YB-1, suggesting that 5′-tiDNAAla/YB-1 complexes mediate this cytoprotective response. Interestingly, exposure to 5′-tiDNAAla for 72 h also partially rescues ANG-depleted motor neurons from toxic effects of stress (Fig. S5D).
RNA oligos corresponding to the GGGGCC repeats expanded in the ALS-associated C9ORF72 gene also form G4 structures (32). To determine whether different classes of G4 oligos are capable of functional interactions, we transfected U2OS cells with tiRNAs and GGGGCC repeat oligos alone or in combination and then processed cells for immunofluorescence microscopy to quantify SG assembly. Transfection of 5′-tiRNAAla, 5′-tiRNACys, or C9ORF72 (four repeats) induced SG assembly (Fig. 3D). Cotransfection with control oligos (AACCCC; four or 17 repeats) or with C9ORF72 (4× GGGCC repeats) had no effect on SG assembly. In contrast, cotransfection with C9ORF72 (23× GGGGCC repeats) markedly impaired SG assembly by both tiRNAs and C9ORF72 (4× repeats). The ability of pathological repeats found in C9ORF72 to interfere with tiRNA-induced SG assembly could contribute to reduced viability of motor neurons in patients who have ALS.
Discussion
The demonstration that G4-containing DNA analogs of 5′-tiRNAAla enter cultured motor neurons and trigger a neuroprotective response has important implications for our understanding of both ANG-induced neuroprotection and the pathogenesis of ALS. Although ANG was initially identified as an angiogenic factor found in the conditioned medium of cultured cancer cells (1), it is a component of the acute-phase response that protects the organism from microbial and environmental stress (33). ANG binds to widely expressed, but poorly characterized, receptors to trigger AKT activation (27) and inhibits p53-induced apoptosis (34). Following receptor binding, ANG enters the cell and is translocated to the nucleolus, where it binds to rRNA gene promoters and enhances transcription of rRNA (35). ANG also cleaves tRNA to produce bioactive tiRNAs that reprogram protein synthesis and promote the assembly of SGs (10). The relative contribution of these events to ANG-induced proliferation and survival is not known.
The finding that 5′-tiDNAAla, an analog of a downstream effector of ANG function, spontaneously enters cells and protects motor neurons from adverse effects of stress strongly links tRNA cleavage to the neuroprotective effects of ANG. This conclusion is supported by findings showing that most ALS-associated ANG mutants lack ribonuclease activity (2, 29). Our data suggest that ANG is a neuroprotective factor that functions in an RNase-dependent manner. Because 5′-tiDNAAla is sufficient for neuroprotection, the ANG receptor may not be required for this activity. The remarkable ability of G4-containing oligos to enter cells makes 5′-tiDNAAla a leading compound for the development of a neuroprotective drug.
Recent findings have implicated G4 structures formed by GGGGCC hexanucleotide repeats found in the C9ORF72 gene in the pathogenesis of ALS and FTD (17, 32, 36–38). These structures are postulated to modulate gene expression at transcriptional and posttranscriptional levels, and to promote nucleolar stress in ways that contribute to age-dependent motor neuron dropout (17, 32, 36–38). Together with our results, these studies suggest that various G4 structures may play important roles in ensuring motor neuron viability.
Normal human C9orf72 alleles have two to 20 intronic hexanucleotide repeats, with the majority having fewer than eight repeats (39). The ALS/FTD-associated C9ORF72 alleles contain tens to thousands of hexanucleotide repeats, although the correlation between repeat length and clinical features is not obvious. Several molecular mechanisms have been proposed to contribute to the loss of neurons in C9ORF72 ALS carriers (17, 32, 36–38). We propose that extended C9ORF72 repeats forming G4 structures may interfere with endogenous G4-containing, ANG-induced tiRNAs that are required for motor neuron survival. Molecular details of such interference are not clear but may involve sequestration of RNA-binding proteins [e.g., YB-1, nucleolin (36), hnRNP A3 (16)] that regulate motor neuron survival. Our results add to a growing appreciation for the roles that G4 structures may play in the pathogenesis of motor neuron disease.
Materials and Methods
Tissue Culture, Metabolic Labeling, and Transfection of Cells.
U2OS or NSC34 cells were cultured and labeled with [35S]-methionine as described by Ivanov et al. (10). Cell lines were transfected with RNA or DNA oligos using Lipofectamine 2000 (Invitrogen) as described in SI Materials and Methods. Cryopreserved hMN progenitors derived from human ES cells were purchased from Lonza and differentiated according to the manufacturer’s recommendations. The hMNs were transfected on days 17 and 20 of differentiation by magnetofection using NeuroMag transfection reagent (AMSBio).
Modified Photoactivatable Ribonucleoside-Enhanced Cross-Linking and Immunoprecipitation.
U2OS cells were transfected with 3′-end biotinylated, sU-containing tiRNAs. At 6 h posttransfection [which corresponds to the appearance of SGs (40)], cells were irradiated with UV light (365 nm), lysed, and affinity-precipitated using streptavidin beads. Proteins were eluted, precipitated with trichloroacetic acid, and analyzed by Western blotting (10).
Reagents.
The sources of antibodies and ODNs used in this study are reported in SI Materials and Methods.
Immunofluorescence Microscopy.
Cells were stained with the indicated antibodies before processing for immunofluorescence microscopy as previously described (40).
Preparation of RNA Transcripts.
Firefly luciferase mRNA was purchased from Promega and capped using a ScriptCap m7G Capping System (EPICENTRE Biotechnologies) according to the manufacturer’s recommendations. C9ORF72 (23×) and AAAACC (17×) repeat RNAs were in vitro RNA-transcribed from linearized HindIII plasmids pcDNA3.1-GGGGCC (23×) and pcDNA3.1-AAAACC (17×) (a gift from C. Haass, Ludwig Maximilian University, Munich, Germany) using a RiboProbe In Vitro Transcription System (Promega) as described by the manufacturer.
In Vitro Translation of mRNA Reporters in RRLs.
A Flexi Rabbit Reticulocyte Lysate System (Promega) was used for the in vitro analysis of mRNA translation in RRLs as previously described (10).
m7GTP-Sepharose Chromatography.
Affinity precipitation using m7GTP-Sepharose 4B (m7GTP-Sepharose; GE Healthcare) was performed using previously described methods (10).
Gel Electrophoresis.
ODNs were analyzed by denaturing PAGE wherein 1 μM samples were heated at 85 °C for 5 min and immediately placed on ice before being loaded onto a 15% TBU gel. Nondenaturing PAGE was performed by heating samples (1 μM) at 85 °C for 5 min and slowly cooling them to room temperature (1 °C/min) before being analyzed on a 20% TB polyacrylamide gel. Bands were visualized by SYBR gold staining.
Supplementary Material
Acknowledgments
We thank N. Kedersha, S. Yamasaki, V. Ivanova, and K. Fujimura for technical support and helpful comments. We thank Prof. Guo-fu Hu and Dr. Wenhao Yu (Tufts University School of Medicine) for providing recombinant ANG, Prof. Lev Ovchinnikov and Dr. Sergey Guryanov (Institute of Protein Research, Russian Academy of Sciences) for providing recombinant YB-1 protein, and Prof. Christian Haass and Dr. Kohji Mori (Ludwig Maximilian University) for providing plasmid pcDNA3.1-GGGGCC (×23) and pcDNA3.1-AAAACC (×17) constructs. This work was supported by National Institutes of Health Grant CA168872 (to P.A.), an Investigator Award from the Rheumatology Research Foundation (to P.A.), Research Development Grant ID158521 from the Muscular Dystrophy Association (to P.I.), and ALS Association Grant N7W220 (to P.I.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 18108.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1407361111/-/DCSupplemental.
References
- 1.Fett JW, et al. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry. 1985;24(20):5480–5486. doi: 10.1021/bi00341a030. [DOI] [PubMed] [Google Scholar]
- 2.Greenway MJ, et al. A novel candidate region for ALS on chromosome 14q11.2. Neurology. 2004;63(10):1936–1938. doi: 10.1212/01.wnl.0000144344.39103.f6. [DOI] [PubMed] [Google Scholar]
- 3.Greenway MJ, et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nat Genet. 2006;38(4):411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 4.Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol. 2009;185(1):35–42. doi: 10.1083/jcb.200811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Gf, Xu Cj, Riordan JF. Human angiogenin is rapidly translocated to the nucleus of human umbilical vein endothelial cells and binds to DNA. J Cell Biochem. 2000;76(3):452–462. doi: 10.1002/(sici)1097-4644(20000301)76:3<452::aid-jcb12>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 6.Hu GF, Riordan JF, Vallee BL. A putative angiogenin receptor in angiogenin-responsive human endothelial cells. Proc Natl Acad Sci USA. 1997;94(6):2204–2209. doi: 10.1073/pnas.94.6.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saikia M, et al. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem. 2012;287(51):42708–42725. doi: 10.1074/jbc.M112.371799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolozin B. Regulated protein aggregation: Stress granules and neurodegeneration. Mol Neurodegener. 2012;7(1):56. doi: 10.1186/1750-1326-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolozin B. Physiological protein aggregation run amuck: Stress granules and the genesis of neurodegenerative disease. Discov Med. 2014;17(91):47–52. [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell. 2011;43(4):613–623. doi: 10.1016/j.molcel.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nam Y, Chen C, Gregory RI, Chou JJ, Sliz P. Molecular basis for interaction of let-7 microRNAs with Lin28. Cell. 2011;147(5):1080–1091. doi: 10.1016/j.cell.2011.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evdokimova V, et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15(5):402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 13.Reyes-Reyes EM, Teng Y, Bates PJ. A new paradigm for aptamer therapeutic AS1411 action: Uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010;70(21):8617–8629. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol. 2009;86(3):151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bates PJ, Choi EW, Nayak LV. G-rich oligonucleotides for cancer treatment. Methods Mol Biol. 2009;542:379–392. doi: 10.1007/978-1-59745-561-9_21. [DOI] [PubMed] [Google Scholar]
- 16.Mori K, et al. hnRNP A3 binds to GGGGCC repeats and is a constituent of p62-positive/TDP43-negative inclusions in the hippocampus of patients with C9orf72 mutations. Acta Neuropathol. 2013;125(3):413–423. doi: 10.1007/s00401-013-1088-7. [DOI] [PubMed] [Google Scholar]
- 17.Zamiri B, Reddy K, Macgregor RB, Jr, Pearson CE. TMPyP4 porphyrin distorts RNA G-quadruplex structures of the disease-associated r(GGGGCC)n repeat of the C9orf72 gene and blocks interaction of RNA-binding proteins. J Biol Chem. 2014;289(8):4653–4659. doi: 10.1074/jbc.C113.502336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collie GW, Parkinson GN. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem Soc Rev. 2011;40(12):5867–5892. doi: 10.1039/c1cs15067g. [DOI] [PubMed] [Google Scholar]
- 19.Ren J, Chaires JB. Sequence and structural selectivity of nucleic acid binding ligands. Biochemistry. 1999;38(49):16067–16075. doi: 10.1021/bi992070s. [DOI] [PubMed] [Google Scholar]
- 20.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc Natl Acad Sci USA. 2002;99(18):11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141(1):129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry (Mosc) 2011;76(13):1402–1433. doi: 10.1134/S0006297911130049. [DOI] [PubMed] [Google Scholar]
- 23.Lyabin DN, Nigmatullina LF, Doronin AN, Eliseeva IA, Ovchinnikov LP. Identification of proteins specifically interacting with YB-1 mRNA 3′ UTR and the effect of hnRNP Q on YB-1 mRNA translation. Biochemistry (Mosc) 2013;78(6):651–659. doi: 10.1134/S0006297913060102. [DOI] [PubMed] [Google Scholar]
- 24.Balasubramanian S. Chemical biology on the genome. Bioorg Med Chem. 2014;22(16):4356–4370. doi: 10.1016/j.bmc.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 25.Morris MJ, Wingate KL, Silwal J, Leeper TC, Basu S. The porphyrin TmPyP4 unfolds the extremely stable G-quadruplex in MT3-MMP mRNA and alleviates its repressive effect to enhance translation in eukaryotic cells. Nucleic Acids Res. 2012;40(9):4137–4145. doi: 10.1093/nar/gkr1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li S, Yu W, Kishikawa H, Hu GF. Angiogenin prevents serum withdrawal-induced apoptosis of P19 embryonal carcinoma cells. FEBS J. 2010;277(17):3575–3587. doi: 10.1111/j.1742-4658.2010.07766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kieran D, et al. Control of motoneuron survival by angiogenin. J Neurosci. 2008;28(52):14056–14061. doi: 10.1523/JNEUROSCI.3399-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Subramanian V, Crabtree B, Acharya KR. Human angiogenin is a neuroprotective factor and amyotrophic lateral sclerosis associated angiogenin variants affect neurite extension/pathfinding and survival of motor neurons. Hum Mol Genet. 2008;17(1):130–149. doi: 10.1093/hmg/ddm290. [DOI] [PubMed] [Google Scholar]
- 29.Wu D, et al. Angiogenin loss-of-function mutations in amyotrophic lateral sclerosis. Ann Neurol. 2007;62(6):609–617. doi: 10.1002/ana.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skorupa A, et al. Motoneurons secrete angiogenin to induce RNA cleavage in astroglia. J Neurosci. 2012;32(15):5024–5038. doi: 10.1523/JNEUROSCI.6366-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emara MM, et al. Hydrogen peroxide induces stress granule formation independent of eIF2α phosphorylation. Biochem Biophys Res Commun. 2012;423(4):763–769. doi: 10.1016/j.bbrc.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fratta P, et al. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep. 2012;2:1016. doi: 10.1038/srep01016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson KA, Verselis SJ, Fett JW. Angiogenin is regulated in vivo as an acute phase protein. Biochem Biophys Res Commun. 1998;242(3):480–483. doi: 10.1006/bbrc.1997.7990. [DOI] [PubMed] [Google Scholar]
- 34.Sadagopan S, et al. Angiogenin functionally interacts with p53 and regulates p53-mediated apoptosis and cell survival. Oncogene. 2012;31(46):4835–4847. doi: 10.1038/onc.2011.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu ZP, Tsuji T, Riordan JF, Hu GF. The nuclear function of angiogenin in endothelial cells is related to rRNA production. Biochem Biophys Res Commun. 2002;294(2):287–292. doi: 10.1016/S0006-291X(02)00479-5. [DOI] [PubMed] [Google Scholar]
- 36.Haeusler AR, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507(7491):195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mori K, et al. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339(6125):1335–1338. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 38.Zu T, et al. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110(51):E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rutherford NJ, et al. Length of normal alleles of C9ORF72 GGGGCC repeat do not influence disease phenotype. Neurobiol Aging. 2012;33(12):2950:e5–e7. doi: 10.1016/j.neurobiolaging.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emara MM, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem. 2010;285(14):10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.