Significance
We demonstrate, to our knowledge for the first time, that bacterial biofilms are associated with colorectal cancers, one of the leading malignancies in the United States and abroad. Colon biofilms, dense communities of bacteria encased in a likely complex matrix that contact the colon epithelial cells, are nearly universal on right colon tumors. Most remarkably, biofilm presence correlates with bacterial tissue invasion and changes in tissue biology with enhanced cellular proliferation, a basic feature of oncogenic transformation occurring even in colons without evidence of cancer. Microbiome profiling revealed that biofilm communities on paired normal mucosa cluster with tumor microbiomes but lack distinct taxa differences. This work introduces a previously unidentified concept whereby microbial community structural organization exhibits the potential to contribute to disease progression.
Keywords: colorectal cancer, microbiome, biofilm, bacterial communities, adenoma
Abstract
Environmental factors clearly affect colorectal cancer (CRC) incidence, but the mechanisms through which these factors function are unknown. One prime candidate is an altered colonic microbiota. Here we show that the mucosal microbiota organization is a critical factor associated with a subset of CRC. We identified invasive polymicrobial bacterial biofilms (bacterial aggregates), structures previously associated with nonmalignant intestinal pathology, nearly universally (89%) on right-sided tumors (13 of 15 CRCs, 4 of 4 adenomas) but on only 12% of left-sided tumors (2 of 15 CRCs, 0 of 2 adenomas). Surprisingly, patients with biofilm-positive tumors, whether cancers or adenomas, all had biofilms on their tumor-free mucosa far distant from their tumors. Bacterial biofilms were associated with diminished colonic epithelial cell E-cadherin and enhanced epithelial cell IL-6 and Stat3 activation, as well as increased crypt epithelial cell proliferation in normal colon mucosa. High-throughput sequencing revealed no consistent bacterial genus associated with tumors, regardless of biofilm status. However, principal coordinates analysis revealed that biofilm communities on paired normal mucosa, distant from the tumor itself, cluster with tumor microbiomes as opposed to biofilm-negative normal mucosa bacterial communities also from the tumor host. Colon mucosal biofilm detection may predict increased risk for development of sporadic CRC.
When healthy, the colon is covered by a mucus layer that segregates the microbiota from direct contact with the host colonic epithelium (1). Breaches of this protective mucus layer with resulting increased contact between mucosal microbiota and the colonic epithelial cells have been proposed as a critical first step in inciting changes in tissue biology and/or inflammation that yield inflammatory bowel disease (2–4). Concomitant with increased access to the mucosal epithelium, microbial community communication (such as quorum sensing) is predicted to change, thereby modifying microbial structure and function and often resulting in biofilm formation (5). Biofilms are defined as aggregations of microbial communities encased in a polymeric matrix that adhere to either biological or nonbiological surfaces. Biofilms that invade the colonic mucus layer and come into direct contact with mucosal epithelial cells indicate a pathologic state (6, 7). Biofilms characterize numerous chronic mucosal disease states in and outside of the colon (including inflammatory bowel diseases, pharyngo-tonsillitis, otitis media, rhinosinusitis, urethritis, and vaginitis), where direct bacterial contact with epithelial cells results in perturbed epithelial function and chronic inflammation (8). However, no association of biofilms with colorectal cancer (CRC) pathologic states has been reported.
Results
Herein, we systematically studied the microbial communities associated with surgically resected colorectal tumors (CRC and adenomas) compared with paired pathologically tumor-free mucosa (herein referred to as “normal”) [SI Appendix, Tables S1 and S2, Johns Hopkins Hospital (JHH) and University of Malaya Medical Centre (UMMC), respectively]. The normal colon tissues were obtained from the margins of the resected specimens furthest from the site of the tumor (SI Appendix, Fig. S1). In addition, we studied colon biopsies obtained from individuals without colorectal tumors and without a diagnosis of inflammatory colonic disease undergoing routine screening colonoscopy at JHH (SI Appendix, Table S3).
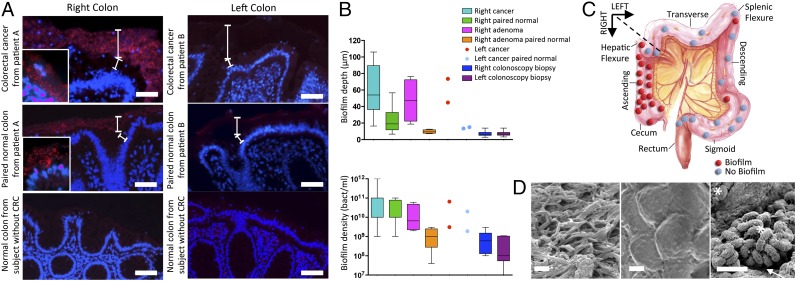
Using samples from JHH, we first compared the spatial relationship of the microbiota with the host mucus layer and colonic epithelium using FISH. Carnoy’s solution-fixed, paraffin-embedded tissues, known to preserve the mucus layer, were uniquely suited for this purpose and were used whenever possible. To detect all bacterial populations, we hybridized the tissues with a probe (Eub338) targeting the conserved 16S ribosomal RNA bacterial domain (9). Bacterial biofilms were defined as massive bacterial invasions (>109 bacteria/mL) of the mucus layer spanning at least a linear distance of 200 µm of the epithelial surface. Bacterial biofilms were identified by FISH analysis on 50% (15 of 30) and 67% (4 of 6) of all evaluated CRCs and adenomas, respectively (Fig. 1A and SI Appendix, Fig. S2). Bacterial biofilm presence on tumors was ordered by geographical location along the colonic axis. Unexpectedly, tumors in the ascending colon and hepatic flexure were biofilm-positive in 87% (13 of 15) and 100% (4 of 4) of CRCs and adenomas, respectively, whereas tumors located in the transverse and descending colon displayed biofilms in 13% (2 of 15) and 0% (0 of 2) of CRCs and adenomas, respectively (P = 0.0001 for carcinomas and P = 0.067 for adenomas, Fisher’s exact test; Fig. 1C). Surgical resection samples from the UMMC confirmed this geographical ordering of bacterial biofilm presence on tumors. Namely, in this Malaysian population, all four tumors harvested from the ascending colon and hepatic flexure were biofilm positive, whereas 22% of tumors (4 of 17 CRCs and 0 of 1 adenoma) from the transverse and descending colon were biofilm positive (SI Appendix, Table S2). Biofilm presence was not associated with age, sex, race, CRC stage, tumor size, bowel preparation, or histopathologic classification.
Fig. 1.
Detection and quantification of bacterial biofilms on colon tumors. (A) FISH of all bacteria (red) on cancers (Top), paired normal tissue from patients with CRC (Middle), and colonoscopy biopsies from healthy individuals without CRC (Bottom). All were counterstained with the nuclear stain, DAPI (blue). The top white brackets demarcate the mucus layer and the bottom white brackets denote the cytoplasm separating the nucleus (blue) of the colorectal epithelium from the mucus layer. PAS stains (SI Appendix, Fig. S2) further delineate the mucus layer on these samples. (Insets) Closeup (100×) showing close contact between bacteria and epithelial cells in patient A. The pale, nonpunctate red staining of the mucus layer in patients without biofilms (patient B) represents nonspecific binding to the mucus layer, which is easily demarcated from the bright red punctate staining of the bacteria infiltrating the mucus layer in patients with biofilms. (Scale bars, 50 µm.) (B) Biofilm depth and density measurements from right CRCs/surgical normal pairs (n = 15), right adenomas/surgical normal pairs (n = 4), left CRCs/surgical normal pairs (n = 15), left adenomas/surgical normal pairs (n = 2), and right/left paired normal colonoscopy biopsies from healthy individuals without CRC (n = 60). Data displayed as bar and whisker graphs, where line designates the median, boxes the 25/75th percentile, and whiskers the 95th percentile. (C) Geographical distribution of tumors (CRC, n = 30, and adenomas, n = 6) with biofilm designation. (D) SEM images. (Left) Biofilm on a right colon cancer dominated by filamentous bacteria. (Center) Biofilm-negative left colon cancer where no bacteria are visualized. (Right) Image of bacterial contact with host epithelium (white arrow) on biofilm-covered right colon adenoma. Mixed bacterial morphology (*rods and cocci) is seen. (Scale bars, 2 µm.)
All biofilm-covered CRCs and adenomas exhibited the remarkable feature of bacterial invasion into the tumor mass (Fig. 2A and SI Appendix, Fig. S3 A and B, white arrows) not detected in biofilm-negative tumors. SEM of a tumor sample subset was consistent with the FISH results, revealing both direct bacterial–epithelial surface contact and a dense biofilm comprising mixed bacterial morphologies on all ascending colon tumors, with few mucosal bacteria detected on tumors distal to the hepatic flexure (Fig. 1D). These data further confirm that a breach of the colonic protective mucus layer is strikingly dictated by colon geographic location. Embryologic development is often used to define the right colon as comprising the cecum through most of the transverse colon and the left colon as the distal transverse colon (near the splenic flexure) through the rectum (10). However, on the basis of the discrete anatomical distribution of biofilms identified in our study, we defined right colon cancer as proximal to the hepatic flexure and left colon cancer as distal to the hepatic flexure (Fig. 1C).
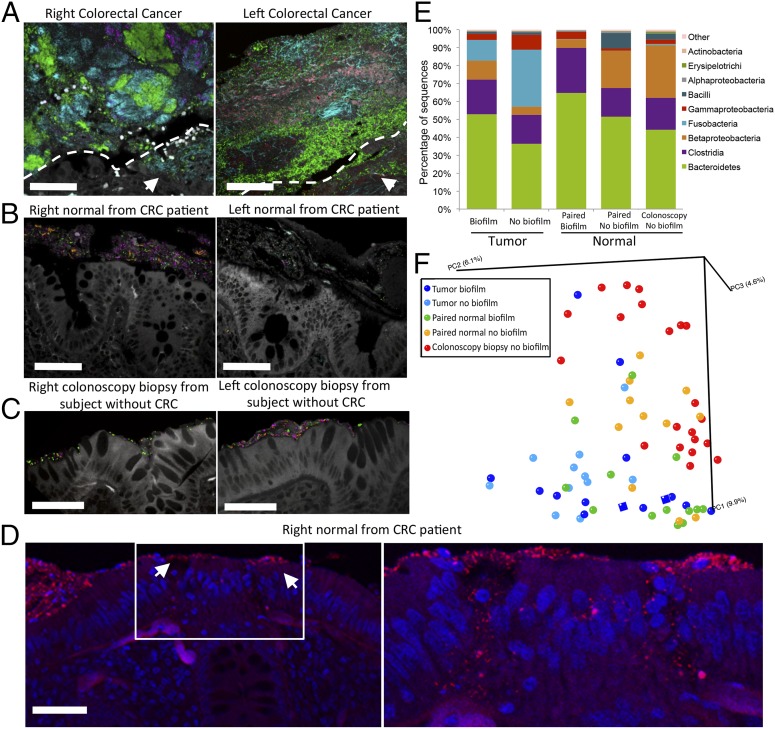
Fig. 2.
FISH and sequencing analysis of tissue reveal invasive polymicrobial biofilms and transitioning microbial populations. (A–C) Multiprobe spectral images of FISH-targeted bacterial groups (40×). Bacteroidetes (green), Lachnospiraceae (magenta), Fusobacteria (cyan), Enterobacteriaceae (orange), Bacteroides fragilis (red) are represented within the biofilms, and tissue autofluorescence is white. (A) Multigroup bacterial biofilm with invasion of cancer tissue (white arrows). Dotted white line depicts margin between bacterial biofilm and cancer tissue in lower right portion of image. Right cancer with a Fusobacteria dominant polymicrobial biofilm (by sequencing analysis; see text) also containing Bacteroidetes, Lachnospiraceae, and Enterobacteriaceae. Dominant group in left cancer is Bacteroidetes. B. fragilis, Lachnospiraceae, and Fusobacteria are also present. Cancer-invading bacteria represent a subset of biofilm community members. (B) Bacterial biofilms on paired surgical normal tissue from CRC patients, comprising Lachnospiraceae, Bacteroidetes, and Enterobacteriaceae. (C) Thin bacterial biofilms detected on right (Bacteroidetes, Lachnospiraceae, and Enterobacteriaceae) and left (Bacteroidetes and Lachnospiraceae) normal colonoscopy biopsies from two different individuals without CRC. (D, Left) All bacteria FISH (red) with DAPI nuclear counterstain (blue) of surgically resected normal tissue covered by a biofilm (20×) from a patient with CRC. White arrows mark two sites of biofilm infiltration of the epithelial tissue (20×). (D, Right) Confocal z-stack of tissue bacterial (red) invasion (40×) denoted by white box in D, Left. Disordered epithelial cells and leukocytes are visible at the infiltrated sites, whereas surrounding epithelial cells are intact and ordered as seen in D, Left. (Scale bars, 50 µm in A–D.) (E) Histogram of bacterial classes represented on biofilm-positive and -negative samples as defined by sequence analysis. Tumor denotes 23 CRCs and two adenomas. (F) PCoA plot (based on unweighted UniFrac distances) displaying mucosa community structure of all samples (each point reflects an individual sample). Normal colonoscopy biopsies from healthy individuals without CRC (n = 21, red) and surgically resected normal tissues without a biofilm from patients with CRC (n = 12, orange) transition to normal tissues with a biofilm from patients with CRC (n = 13, green) that cluster more closely to biofilm-positive adenomas (n = 2, dark blue squares) and CRCs with (n = 12, dark blue) and without (n = 11, light blue) biofilms.
To determine whether biofilm formation was specific for the tumor microenvironment, we next used FISH to examine the paired normal colon tissues obtained from the surgical resection margin furthest from the tumor mass (SI Appendix, Fig. S1). No biofilms were detected on the paired normal surgically resected colon tissues from patients with biofilm-negative tumors (adenomas or CRCs). In striking contrast, all but one normal colon tissue sample from patients with biofilm-covered tumors were biofilm positive. This was true for patients with both adenomas and carcinomas, regardless of their location within the colon. Of note, the single surgically resected normal tissue on which we failed to detect a biofilm was fixed in formalin rather than Carnoy’s and thus not optimized for mucus preservation (1). Although biofilm bacterial density did not differ between tumors (CRCs or adenomas) and their paired normal colon tissues, biofilm depth was significantly increased on tumor samples compared with their paired normal colon tissues (P = 0.001 for CRCs, P = 0.028 for adenomas; Fig. 1B). These findings demonstrate that biofilm formation represents a broad regional alteration in host epithelial–microbiota association not restricted to tumor tissue.
Screening colonoscopy biopsies from healthy individuals were typically covered with a mucus layer devoid of bacteria (Fig. 1A and SI Appendix, Fig. S2). A subset of colonoscopy biopsies (15 of 120, 13%) revealed thin bacterial biofilms with an average density of 108 bacteria/mL (Fig. 1B). Biofilm formation on colonoscopy biopsy tissues did not differ by colon location (8 of 60, right colon vs. 7 of 60, left colon) (SI Appendix, Fig. S1B). Thus, the right colon does not have a greater likelihood of bacterial biofilm development in a cancer-free host. These findings in the healthy host are consistent with past reports detecting biofilms on ∼15% of biopsies from asymptomatic individuals (11).
We next evaluated the composition and spatial organization of specific bacteria within the biofilms using fluorescence spectral imaging using 11 group- and species-specific FISH probes to target the majority (85%) of the major groups of bacteria identified by sequencing (SI Appendix, Table S4 and Datasets S1–S3). Combinations of nine probes were selected for simultaneous hybridization to the tissues. Sixteen biofilm-covered tumors (three adenomas from two patients, 13 CRCs) and their paired normal mucosa were available for analysis along with normal mucosa from five right and four left colonoscopy biopsies (in three cases, biopsies were available from both the right and left colon). All biofilms, whether associated with normal colon mucosa or tumor tissue, were polymicrobial (Fig. 2 A–C). Predominant bacterial phyla associated with adenomas and CRCs were Bacteroidetes and Firmicutes (family Lachnospiraceae including Clostridium, Ruminococcus, and Butyrivibrio). A subset of tumors harbored predominant populations of Fusobacteria (4 of 16) or Gammaproteobacteria (1 of 16) (determined to be the Enterobacteriaceae family) (SI Appendix, Fig. S3). Using this multiprobe method, the tumor-invading bacterial groups present in all CRC and adenoma tissues were also identified in the biofilm bacterial composition on the tumor surface consistent with tissue invasion by a subset of these bacterial groups (Fig. 2A and SI Appendix, Fig. S3 A and B). Biofilms identified on surgically resected, normal tissues were also consistently diverse, composed of Bacteroidetes, Lachnospiraceae, and Gammaproteobacteria (Fig. 2B). Biofilms detected on normal mucosa obtained at colonoscopy from patients without CRC were similarly composed of Bacteroidetes and Lachnospiraceae (Fig. 2C). A subset of biofilm-positive surgically resected normal mucosa from patients with tumors (4 of 16, three tissues from CRC patients and one from an adenoma patient) also revealed bacterial invasion into the colonic epithelial cells or submucosa (Fig. 2D and SI Appendix, Fig. S4). No mucosal biopsies with or without biofilms from healthy individuals undergoing colonoscopy revealed invasive bacteria.
To further evaluate the colonic microbiota associated with these samples, high-throughput 454 pyrosequencing targeting the hypervariable V3–V5 region of the 16S ribosomal RNA gene was performed on DNA extracted from the mucosa of 23 CRCs, two adenomas and their paired surgically resected normal tissues, and 22 biopsies obtained on colonoscopy of healthy control patients (11 right and left-matched pairs, none biofilm positive) (SI Appendix, Datasets S1–S3).
Sequence analysis revealed substantial overlap between tumors (adenomas or CRCs) and paired normal tissue bacterial membership at the genus level; tumor bacterial membership was a complete subset of their normal pair in 52% of tumor–normal sets (SI Appendix, Dataset S3). Among the 25 tumor–normal paired tissue samples, eight CRCs (32%, four right and four left), but not their paired surgically resected normal tissues, were Fusobacteria dominant (≥25% of total sequences) (Fig. 2E and SI Appendix, Fig. S5), a finding compatible with recent reports (12–15). No biopsies from healthy controls displayed dominant membership of Fusobacteria (Fig. 2E and SI Appendix, Fig. S5). Collectively, our data are consistent with the concept of “on tumor:off tumor” communities, as previously reported (16).
We detected nine differentially abundant genus-level groups in colonoscopy biopsies compared with paired normal biofilm-positive tissue samples from tumor patients (false discovery rate <3%), including significant enrichment of Lactococcus, Leuconostoc, and Comamonas and other Burkholderiales members in colonoscopy biopsies, and conversely, a 10-fold relative increase of a candidate Ruminococcaceae member in surgically resected paired normal biofilm-positive tissue samples. In contrast, we detected significant depletion of Bacilli and some Bacteroidetes members in surgically resected normal biofilm-positive tissue samples, with on average 28- and sevenfold lower relative abundance than surgically resected normal biofilm-negative samples, respectively (false discovery rate <5%).
The differences between tissues with and without a biofilm from the tumor host were highlighted by unweighted Unifrac distance analysis and principal coordinate analysis (PCoA), which revealed a striking progression of bacterial dysbiosis in biofilm-positive relative to biofilm-negative mucosa, despite the minimal differentially abundant taxa between the two groups (Fig. 2 E and F and SI Appendix, Fig. S6 and Dataset S3). These analyses revealed discrete clustering of normal colonoscopy biopsies relative to tumor-associated communities (Fig. 2F and SI Appendix, Fig. S6); microbial populations from these two sample types were the most structurally divergent (P < 8e-7). Communities from CRC-associated normal mucosa without biofilm were on average significantly closer in overall structure to healthy colonoscopy biopsy populations than to CRC-associated communities (P = 0.001). In striking contrast, biofilm-positive normal tissue communities were significantly closer in structure to CRC-associated populations than to those from healthy biopsies (P = 1e-8). This distinction supports the notion that biofilm presence correlates with the dysbiosis detected within the tumor-associated microbiota. Our findings suggest that stepwise colon mucosal microbial community dysbiosis, largely with depletion of common microbiota community members, parallels the transition from normal colon mucosa to CRC.
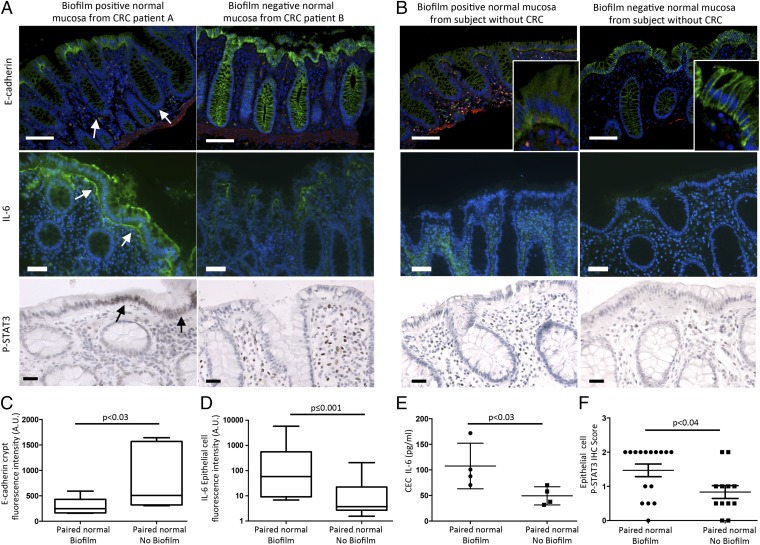
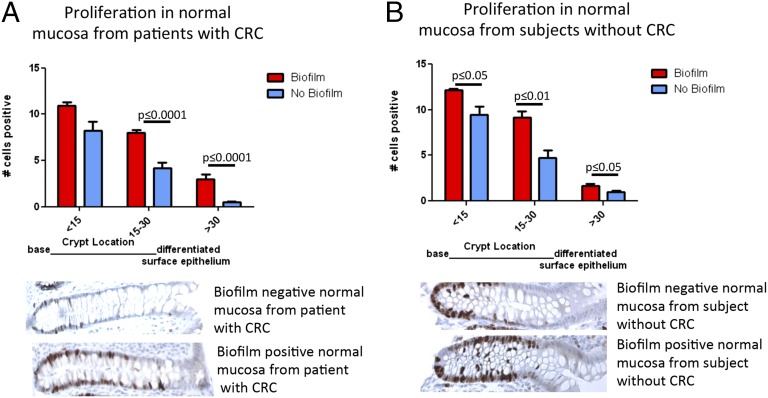
Can biofilms modify epithelial biology before initiation of transformation? To evaluate this conjecture, we conducted analyses of colonic epithelial cell biologic changes relevant to carcinogenesis, including barrier permeability (using the cell–cell adhesion molecule epithelial cadherin (E-cadherin) detection as a marker) (17, 18), interleukin 6 (IL-6), signal transducer and activator of transcription 3 (STAT3) activation (19, 20), proliferation, and apoptosis in normal tissues from CRC patients as well as from healthy individuals. Loss of E-cadherin activates Wnt signaling in colon cancer and IL-6–driven Stat3 activation in colonic epithelial cells is critical to colon carcinogenesis in multiple murine models. These analyses showed marked differences between biofilm-positive and biofilm-negative normal colon tissues from the CRC host. Namely, biofilm-positive normal tissues in the CRC host displayed significantly reduced crypt cell E-cadherin (Fig. 3 A and C), with significantly increased epithelial cell IL-6 (Fig. 3 A, D, and E and SI Appendix, Fig. S7). Consistent with the IL-6 colonic epithelial cell localization, Stat3 activation (measured by pStat3 immunohistochemistry) was significantly increased in the epithelial cells (Fig. 3 A and F). Consistent with the action of Stat3 to promote epithelial cell proliferation and survival (19), we further detected a significant increase in crypt epithelial cell proliferation, as measured by Ki67 staining, in normal tissues covered with biofilms from CRC patients compared with normal tissues without a biofilm, also from CRC patients (P ≤ 0.0001; Fig. 4A). In contrast, biofilm-negative normal colon tissues from the CRC host displayed intact E-cadherin (Fig. 3 A and C and SI Appendix, Fig. S9). Lamina propria IL-6 and Stat3 activation did not differ between biofilm-positive and -negative normal colon tissues, both from the CRC host (Fig. 3A and SI Appendix, Figs. S8 and S10A). Notably, although epithelial cell E-cadherin was quantitatively unchanged (Fig. 3B and SI Appendix, Fig. S11 A and B) in biofilm-positive vs. biofilm-negative colonoscopy biopsies from healthy subjects without CRC, E-cadherin localized to the basal pole of the epithelial cells in biofilm-positive colonoscopy biopsies from healthy subjects without CRC (Fig. 3B, Insets). IL-6 detection was also significantly increased in biofilm-positive colonoscopy biopsies from healthy subjects without CRC (Fig. 3B and SI Appendix, Fig. S11C), whereas Stat3 activation was similar in biofilm-positive and -negative colonoscopy biopsies from healthy subjects (Fig. 3B and SI Appendix, Fig. S10B). Nonetheless, epithelial cell proliferation was significantly increased in biofilm-positive colonoscopy biopsies from healthy individuals (P ≤ 0.01; Fig. 4B). In parallel, TUNEL staining was performed to determine whether increased proliferation was simply a byproduct of increased cell turnover (SI Appendix, Fig. S12). Importantly, epithelial cell apoptosis was not increased in biofilm-positive tissues, suggesting that the increased epithelial proliferation measured is a prooncogenic state. This contrasts with the biofilm-negative normal tissues from the CRC host, where lower proliferation (Fig. 4A) was associated with significantly increased apoptosis (SI Appendix, Fig. S12). Spearman correlations between Ki67 staining and genus-level relative abundances were sought in samples stratified by biofilm positive and negative status. No genera significantly correlated with the Ki67 counts in any subgroup.
Fig. 3.
Biofilms are associated with changes in E-cadherin, IL-6, and Stat3 activation. (A and B) Evaluation of E-cadherin and IL-6 by immunofluorescence (green) and activated Stat3 (pStat3, brown nuclei) by immunohistochemistry. Blue, nuclear DAPI counterstain; red, smooth muscle antigen. [Scale bars, 100 μm (E-cadherin) and 50 μm (IL-6, pStat3).] (A) Normal colonic tissues associated with a biofilm from patients with CRC (Left), obtained during surgery, display diminished crypt colonic epithelial cell E-cadherin (white arrows, n = 7 biofilm-positive or -negative tissues) and increased epithelial cell IL-6 (white arrows, n = 13 biofilm-positive or -negative tissues), as well as epithelial cell pStat3 (black arrows, n = 16 biofilm-positive and n = 12 biofilm-negative tissues). Normal colonic tissues without a biofilm from patients with CRC (Right), likewise obtained during surgery, display intact E-cadherin. IL-6 and pStat3 are detected in the lamina propria. (B) Biofilm-positive colonoscopy biopsies from subjects without CRC (Left) display epithelial cell E-cadherin redistribution (Inset) and increased tissue IL-6, whereas biofilm-negative colonoscopy biopsies (Right) display intact E-cadherin and modest lamina propria IL-6 expression. pStat3 is observed in the lamina propria immune cells in both biofilm-positive and biofilm-negative colonoscopy biopsies. (C–F) Quantification of crypt cell E-cadherin (fluorescence intensity), epithelial cell IL-6 [fluorescence intensity and isolated colonic epithelial cells (CEC) by ELISA], and epithelial cell pStat3 (immunohistochemistry) from A are shown in C–F, respectively. Data displayed as bar and whisker graphs, where line designates the median, boxes the 25/75th percentile, and whiskers the 95th percentile (C and D) or mean ± SD (E and F). Details in SI Appendix, Materials and Methods. SI Appendix, Figs. S10 and S11 show additional E-cadherin, IL-6, and pStat3 quantification from A and B.
Fig. 4.
(A and B) Scoring of Ki67-positive cells from the base of the crypt to the luminal surface. Normal tissues from patients with CRC obtained at surgery (A) with (n = 17) and without (n = 18) a biofilm, as well as normal mucosa from healthy subjects obtained via colonoscopy (B) with (n = 7) and without (n = 10) a biofilm, displayed increased proliferation in a biofilm setting. Data displayed as mean ± SEM in groups based on distance from crypt base (<15 cells, 15–30 cells, >30 cells).
Discussion
Although it has been long suspected that bacteria contribute to chronic inflammation leading to CRC, to our knowledge this is the first time that bacterial biofilms, a known driver of tissue inflammation (3), have been identified in CRC. Further, our data show that biofilm formation in both the colon cancer host and healthy subjects is associated with reduced or redistributed colonic epithelial cell E-cadherin, consistent with increased epithelial permeability. Our detection of enhanced IL-6 associated with biofilm formation even in healthy subjects without CRC suggests that early biofilm formation can initiate procarcinogenic tissue inflammation; in the cancer host with biofilm formation, IL-6 is notably localized in colonic epithelial cells with Stat3 activation. The IL-6 family of proinflammatory cytokines and their downstream effector Stat3 have been shown to promote CRC through increased epithelial proliferation, diminished apoptosis, and/or angiogenesis (19, 20). Thus, our data support a model whereby biofilm formation enhances epithelial permeability that increases direct access of bacterial antigens/mutagens to an unshielded epithelial surface and promotes procarcinogenic tissue inflammation. Collectively these events are predicted to induce epithelial cell mutations with consequent increased proliferation of colonic epithelial cells. In this regard, a key observation linking biofilm formation to tumor biology is our identification of the tight association between mucosal biofilm formation and the procancerous state of increased pithelial proliferation. Individual genetic polymorphisms likely govern the composition of the mucosal immune response to the mucosal biofilms, with Th17-dominant mucosal immune responses increasingly associated with oncogenesis and poor outcomes in CRC (21, 22).
It has been hypothesized that differences in diet have significant effects on the gut microbiome and, thus, whether diet relates to biofilm status is an interesting question. Definitive diet–microbiome correlations require large epidemiologic studies that are difficult to control. We do note that the primary associations between biofilms on tumor and associated normal mucosa predominantly in right-sided colon cancers are faithfully reproduced in a population of CRC patients from Malaysia, which, in general, has a very different diet and environmental exposures than the United States. That biofilm patterns in colon cancer are so similar in such different populations, despite likely differences in microbiota, emphasizes the importance of microbiota structure.
The primary findings of this study are that the vast majority of right-sided CRCs are associated with a dense bacterial biofilm and that the normal colonic mucosa from patients whose tumors are covered with biofilms (whether right- or left-sided) are biofilm positive. None of the normal mucosa from patients with biofilm-negative CRCs possessed a biofilm. These findings introduce the concept that the organization, as opposed to the composition per se, of the mucosa-associated microbial community is an important factor in CRC pathogenesis, particularly in the proximal colon. Microsatellite instability, hypermethylation, hypermutation (not all correlating with the presence of microsatellite instability), and the BRAF(V600E) mutation have also been associated with right colon cancer (23). On the basis of the numbers of patients and normal individuals with colonic biofilms, we speculate that colorectal cancers develop in two different settings: individuals with biofilms and individuals without them. Based on the data described here, the risk of developing CRC is more than fivefold higher in the patients with biofilms compared with those without biofilms. This risk is considerably higher than that reported for other environmental associations with CRCs (24, 25). One can envision minimally invasive assays to evaluate the presence of these biofilms, as well as probiotic treatments that could eliminate them. On the basis of these findings, prospective epidemiologic studies to directly test these hypotheses are currently being designed.
Materials and Methods
This study was approved by the Johns Hopkins Institutional Review Board and the Medical Institutional Review Board and the UMMC Medical Ethics Committee at the University of Malaya. All samples were obtained in accordance with the Health Insurance Portability and Accountability Act. Excess colon tumor (adenomas and cancers) and paired normal tissues were collected for analysis from patients undergoing surgery at JHH or UMMC in Kuala Lumpur, Malaysia. Healthy control patients undergoing screening colonoscopy or colonoscopy for diagnostic workup at JHH were also enrolled. Colon tissues were analyzed with up to 11 FISH probes for bacterial composition and biofilm bacterial density and depth; by SEM, fluorescence microscopy (IL-6, E-cadherin), ELISA (IL-6), and immunohistochemistry (pStat3, Ki67). Extracted sample DNA was amplified and sequenced to determine the V3–V5 region of 16S rDNA using the Roche 454 method and procedures described by the Human Microbiome Project standard protocols. Raw sequence reads were processed to yield a final high-quality contaminant-free dataset analyzed by the CloVR-16S pipeline (v1.1) (26). Unweighted UniFrac distances (27) and principal coordinate analysis plots were computed in QIIME. The nonparametric Mann-Whitney U test [performed in R (v2.15.1)] was used to analyze all data except as noted in the text. Pairwise β-diversity comparisons also used the nonparametric Mann-Whitney U test. Detailed methods are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health through Grants R01 CA151393 (to C.L.S. and D.M.P.), R21 CA170492 (to C.L.S.), K08 DK087856 (to E.C.W.), 5T32CA126607-05 (to E.M.H.), P30 DK089502 (GI Core), P30 CA006973 (Sidney Kimmel Comprehensive Cancer Center core), U54 CA091409 (Howard-Hopkins Partnership), T32AI007417 (to the Department of Molecular Microbiology and Immunology, Bloomberg School of Public Health, for C.M.D.), and R01 DE022586 (to G.G.B.); 300-2344 (Alexander and Margaret Stewart Trust, Johns Hopkins); GSRRIG-015 (American Society of Colon and Rectal Surgeons, to E.M.H.); and a grant from the Mérieux Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.W.W. is a guest editor invited by the Editorial Board.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession no. PRJNA258534).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406199111/-/DCSupplemental.
References
- 1.Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8(6):458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 3.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347(6):417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 4.Johansson ME, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63(2):281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh PK, et al. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature. 2000;407(6805):762–764. doi: 10.1038/35037627. [DOI] [PubMed] [Google Scholar]
- 6.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43(7):3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 8.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2(2):95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 9.Ahmed S, et al. Mucosa-associated bacterial diversity in relation to human terminal ileum and colonic biopsy samples. Appl Environ Microbiol. 2007;73(22):7435–7442. doi: 10.1128/AEM.01143-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bufill JA. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med. 1990;113(10):779–788. doi: 10.7326/0003-4819-113-10-779. [DOI] [PubMed] [Google Scholar]
- 11.Swidsinski A, et al. Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut. 2007;56(3):343–350. doi: 10.1136/gut.2006.098160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostic AD, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22(2):292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kostic AD, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Castellarin M, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCoy AN, et al. Fusobacterium is associated with colorectal adenomas. PLoS ONE. 2013;8(1):e53653. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dejea C, Wick E, Sears CL. Bacterial oncogenesis in the colon. Future Microbiol. 2013;8(4):445–460. doi: 10.2217/fmb.13.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grivennikov SI, et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature. 2012;491(7423):254–258. doi: 10.1038/nature11465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soler AP, et al. Increased tight junctional permeability is associated with the development of colon cancer. Carcinogenesis. 1999;20(8):1425–1431. doi: 10.1093/carcin/20.8.1425. [DOI] [PubMed] [Google Scholar]
- 19.Bromberg J, Wang TC. Inflammation and cancer: IL-6 and STAT3 complete the link. Cancer Cell. 2009;15(2):79–80. doi: 10.1016/j.ccr.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, et al. IL-17 can promote tumor growth through an IL-6-Stat3 signaling pathway. J Exp Med. 2009;206(7):1457–1464. doi: 10.1084/jem.20090207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morikawa T, et al. STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin Cancer Res. 2011;17(6):1452–1462. doi: 10.1158/1078-0432.CCR-10-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tosolini M, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71(4):1263–1271. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colditz GA, Wolin KY, Gehlert S. Applying what we know to accelerate cancer prevention. Sci Transl Med. 2012;4(127):127rv4. doi: 10.1126/scitranslmed.3003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Key TJ, et al. Diet, nutrition and the prevention of cancer. Public Health Nutr. 2004;7(1A):187–200. doi: 10.1079/phn2003588. [DOI] [PubMed] [Google Scholar]
- 26.Angiuoli SV, et al. CloVR: A virtual machine for automated and portable sequence analysis from the desktop using cloud computing. BMC Bioinformatics. 2011;12:356. doi: 10.1186/1471-2105-12-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011;5(2):169–172. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.