Significance
Androgen receptor (AR) signaling is a key driver of prostate cancer (PC), even in the context of resistance to current therapies, creating an unmet need for novel approaches to inhibit AR. We demonstrate that the transcription factor GATA-binding protein 2 (GATA2) is critical for both AR expression and optimal transcriptional activity. GATA2 colocalizes with AR and Forkhead box protein A1 on chromatin to enhance recruitment of steroid receptor coactivators and formation of the transcriptional holocomplex. A GATA2 inhibitor suppressed the expression and transcriptional function of AR (including the constitutively active splice variants) and exerted potent anticancer activity against PC cells. We propose GATA2 inhibition as a previously unexplored approach to extinguish both ligand-dependent and ligand-independent AR transcriptional activity and to improve clinical outcomes for PC patients.
Keywords: prostate cancer, small molecule inhibitor, AR signaling, GATA2, steroid receptor coactivator
Abstract
The androgen receptor (AR) is a key driver of prostate cancer (PC), even in the state of castration-resistant PC (CRPC) and frequently even after treatment with second-line hormonal therapies such as abiraterone and enzalutamide. The persistence of AR activity via both ligand-dependent and ligand-independent mechanisms (including constitutively active AR splice variants) highlights the unmet need for alternative approaches to block AR signaling in CRPC. We investigated the transcription factor GATA-binding protein 2 (GATA2) as a regulator of AR signaling and an actionable therapeutic target in PC. We demonstrate that GATA2 directly promotes expression of both full-length and splice-variant AR, resulting in a strong positive correlation between GATA2 and AR expression in both PC cell lines and patient specimens. Conversely, GATA2 expression is repressed by androgen and AR, suggesting a negative feedback regulatory loop that, upon androgen deprivation, derepresses GATA2 to contribute to AR overexpression in CRPC. Simultaneously, GATA2 is necessary for optimal transcriptional activity of both full-length and splice-variant AR. GATA2 colocalizes with AR and Forkhead box protein A1 on chromatin to enhance recruitment of steroid receptor coactivators and formation of the transcriptional holocomplex. In agreement with these important functions, high GATA2 expression and transcriptional activity predicted worse clinical outcome in PC patients. A GATA2 small molecule inhibitor suppressed the expression and transcriptional function of both full-length and splice-variant AR and exerted potent anticancer activity against PC cell lines. We propose pharmacological inhibition of GATA2 as a first-in-field approach to target AR expression and function and improve outcomes in CRPC.
Despite advances in androgen receptor (AR, NR3C4) targeting in prostate adenocarcinoma (PC) with the androgen synthesis inhibitor abiraterone (1–3) and the second-generation AR antagonist enzalutamide (MDV3100) (2–4), disease progression reoccurs in the form of castration-resistant PCs (CRPCs) that frequently still express prostate-specific antigen (PSA) and other AR-dependent genes, suggesting that the AR axis remains active and highlighting the need for novel approaches to block AR signaling and CRPC growth. Several mechanisms that promote persistent AR axis activation even with castrate levels of peripheral testosterone have been reported (3, 5), including in situ androgen synthesis and metabolism; AR overexpression; missense mutations in the AR ligand-binding domain (LBD); overexpression of AR coactivators; alternatively spliced AR variants (ARVs) lacking the LBD that can signal in a constitutively active, ligand-independent manner (6–8); and ligand-independent, noncanonical AR activation by upstream kinase cascades (such as HER2, IGF1R, IL-6, Src, and Akt) (3). It is of critical significance that these ligand-independent mechanisms of AR activation cannot be inhibited by ligand-depleting or LBD-targeting approaches, such as abiraterone or enzalutamide, respectively (3). As a result, there is an unmet need for novel treatment approaches that can inhibit the expression of both full-length AR (AR-FL) and constitutively active, ligand-independent ARVs and/or the AR coregulators in CRPC.
The p160 steroid receptor coactivators (SRCs): SRC-1 (also known as “NCOA1”), SRC-2 (also known as “TIF2,” “GRIP1,” or “NCOA2”), and SRC-3 (also known as “AIB1,” “ACTR,” or “NCOA3”) are critical components of the AR transcriptional holocomplex (9) and serve as scaffolds for the recruitment of CREB-binding protein (CBP) (10) and the histone acetyltransferase p300 (11). Elevated expression of all three p160 SRCs occurs in PC and is associated with a shorter time to recurrence, resistance to androgen-deprivation therapy, and overall more aggressive disease (12–17). SRC-2 (NCOA2) is frequently subject to gene amplification, somatic point mutations, and widespread overexpression in PC (17). SRC-3 expression is mainly regulated posttranslationally, including via the E3 ubiquitin ligase adaptor speckle-type POZ protein (SPOP), which promotes the cullin 3-dependent ubiquitination and degradation of SRC-3 (18). This tumor-suppressor effect is attenuated by the somatic PC-associated mutations that cluster in the substrate-binding cleft of SPOP (19). These data highlight the p160 SRCs as putative therapeutic targets. In addition to AR, other key transcription factors operate as pioneer factors and are required for AR genomic binding, including Forkhead box protein A1 (FOXA1) (20, 21) and GATA-binding protein 2 (GATA2) (22). ChIP-on-chip analysis revealed that the GATA-binding motif is enriched in the AR-binding chromatin regions in LNCaP cells (23). GATA2 has been reported to facilitate androgen-responsive gene expression (24) and to promote cell migration, tissue invasion, and metastasis (25). It has been reported that GATA2 can directly regulate AR gene expression in a stable LNCaP cell line with doxycycline-inducible GATA2 (26). In the present study, we demonstrate that GATA2 is an important oncogenic driver in PC and is an actionable therapeutic target. Importantly, GATA2 directly promotes the expression of both AR-FL and its ligand-independent ARV and enhances recruitment of p160 SRCs and formation of the active AR transcriptional holocomplex. As a result, GATA2 inhibition can be a surrogate approach for AR inactivation, via both extinguishing AR expression and destabilizing the AR transcriptional holocomplex.
Results
Correlation Between Expression of AR and GATA2 in PC.
Gene-expression datasets from 16 PC cell lines (27) and 150 PC specimens (17) revealed that GATA2 has the highest expression among the six GATA family members and is the only member to exhibit, by linear correlation analysis, a positive and strong correlation with AR mRNA (Fig. S1 A–C). As a result, subsequent studies were focused only on GATA2. Reverse transcription quantitative PCR (RT qPCR) and immunoblot analyses confirmed that full-length or variant AR mRNA and protein are expressed only in GATA2+ lines (Fig. S1 D and E).
Prognostic Significance of GATA2 Expression in Human PC.
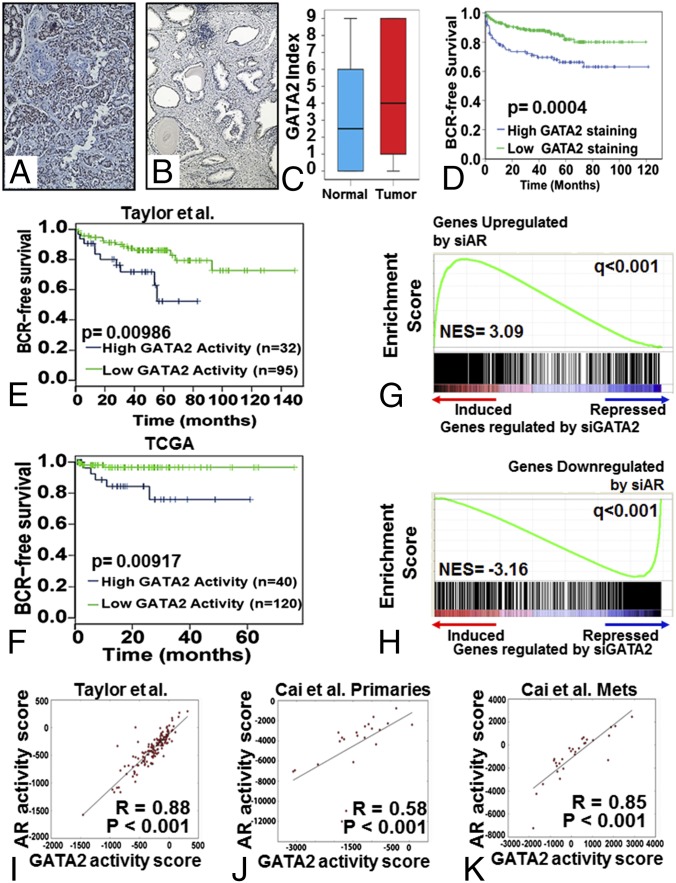
In 383 clinically localized PC samples, GATA2 immunostaining was exclusively nuclear and was present in both malignant and benign luminal epithelial cells (Fig. 1 A and B). Stromal staining was minimal. The median staining index for PCs was 4.0, and 76.5% of cancers showed at least some staining. Levels of GATA2 were significantly higher in tumor than in benign prostate epithelium (P = 0.015; Fig. 1C). GATA2 staining in PC cells was positively correlated with higher Gleason score (R = 0.37, P = 0.008) and higher tumor stage at diagnosis (R = 0.44, P = 0.001). PCs with high GATA2 expression had a significantly shorter time to recurrence (Fig. 1D and Fig. S1 F and G). Patients with a high GATA2 index had higher risk of biochemical recurrence (BCR) than patients with a low GATA2 index, with a hazard ratio (HR) of 1.99 for the upper vs. the lower half of the patients [95% confidence interval (CI): 1.19–3.31, P = 0.0085 by univariate analysis, but P = 0.0767 on multivariate analysis after adjusting for key clinicopathological parameters]. Thus, GATA2 expression is increased significantly in PC cells compared with benign luminal epithelial cells and is a significant predictor of BCR (although it is not an independent predictor). In agreement, we found that GATA2 mRNA expression was increased in metastatic PC compared with primary PC specimens (Fig. S1H), suggesting an important role of GATA2 in PC progression and metastasis.
Fig. 1.
Prognostic significance of GATA2 in PC and its role in AR signaling. Examples of strong (index 9) staining in PC (A) and weak (index 3) staining in benign prostate (B). (Magnification: 200×.) (C) Immunostaining for GATA2 in tumor compared with benign prostate epithelium. (D) PCs with more intense GATA2 immunostaining (upper vs. lower half of the cohort) had shorter BCR-free survival. (E and F) In two publicly available PC gene-expression datasets, Taylor et al. (17) (E) and TCGA (F), patients with high GATA2 transcriptional activity had shorter BCR-free survival (upper 25% vs. lower 75% of each cohort). (G and H) The GATA2 gene signature strongly enriches for AR-regulated genes. GSEA reveals that the transcriptomic footprint of GATA2 significantly enriches for genes regulated by AR. (G) Transcripts induced after treatment with si-GATA2 were enriched for transcripts up-regulated by si-AR. (H) Transcripts repressed after treatment with si-GATA2 were enriched for transcripts down-regulated by si-AR. (I–K) GATA2 activity scores in human PC specimens correlate strongly with AR activity. We computed GATA2 and AR activity scores for all specimens in several publicly available gene-expression datasets from previously reported human PC cohorts: GSE21034 (17) (I), GSE32269 primary tumors (29) (J), and GSE32269 metastatic tumors (29) (K).
We observed a strong correlation of GATA2 expression with expression of AR (Spearman's correlation coefficient: 0.314, P < 0.0001) and a weaker but still significant correlation with the presence of v-ets avian erythroblastosis virus E26 oncogene homolog (ERG) staining (Table S1). We also noted a strong correlation with expression of the AR p160 coactivator SRC-1 and weaker correlations with SRC-2 and SRC-3 expression. There was a relatively weak but statistically significant correlation with proliferation (as determined by Ki67 immunohistochemistry (IHC) (Table S1) but no association with apoptosis (as determined by TUNEL).
GATA2 Activity in PC Is Associated with Enhanced Cell Proliferation, Poor Prognosis, and Increased AR Signaling.
We treated LNCaP cells with GATA2 siRNA for 48 h and analyzed transcript levels on Affymetrix Human Exon 1.0 ST Arrays. Gene Ontology analysis revealed that this GATA2 signature was highly enriched in transcripts involved in key cellular processes such as cellular growth and proliferation (P < 10−108), cell death (P < 10−93), and cell cycle (P < 10−88) (Fig. S1I). Similar results were obtained via Gene Set Enrichment Analysis (GSEA) of the GATA2 transcriptomic response vs. the REACTOME Pathways collection of the Molecular Signatures Database (MSigDB), demonstrating high enrichment for cell cycle and mitotic pathways (Fig. S1 J and K).
We next applied this GATA2 transcriptional activity signature to two commonly used, publicly available PC gene-expression datasets and found that PCs with high expression of GATA2-induced genes and low expression of GATA2-repressed genes had shorter BCR-free survival (log-rank test, P < 0.01) in the Taylor et al. dataset (Fig. 1E) (17) and in The Cancer Genome Atlas (TCGA) (tcga-data.nci.nih.gov/tcga/) (Fig. 1F).
We also used GSEA to compare our si-GATA2–derived gene signature (Fig. S1L, Left) with another gene-expression signature that we obtained by silencing AR in LNCaP cells (Fig. S1L, Right). We discovered that the transcriptomic footprint of GATA2 strongly enriches for genes also regulated by si-AR: Transcripts induced after treatment with si-GATA2 (i.e., repressed by GATA2) were enriched for transcripts up-regulated by si-AR (i.e., repressed by AR) (Fig. 1G), whereas transcripts repressed after treatment with si-GATA2 (i.e., induced by GATA2) were enriched for transcripts down-regulated by si-AR (i.e., induced by AR) (Fig. 1H). We also compared, via GSEA, a previously reported transcriptomic signature of AR3/v7 in LNCaP cells (28) with our GATA2 signature. We discovered that AR3/v7-induced transcripts enrich positively for transcripts induced by GATA2 [i.e., down-regulated by si-GATA2; normalized enrichment score (NES) = 8.02, q < 0.0001] and negatively for genes repressed by GATA2 (i.e., up-regulated by si-GATA2; NES = −3.73, q < 0.0001) in LNCaP cells (Fig. S1M).
Using our GATA2 and AR-FL gene signatures, we calculated the corresponding activity scores in human PC specimens and found, via Pearson correlation analysis, a strong positive correlation in the patient cohort reported by Taylor et al. (17) (r = 0.88, P < 0.001) (Fig. 1I) and in the primary tumors (r = 0.58, P < 0.001) (Fig. 1J) and metastatic tumors (r = 0.85, P < 0.001) (Fig. 1K) reported by Cai et al. (29). Taken together, these results strongly suggest that GATA2 functionally cooperates with both AR-FL and AR3/v7 to determine the transcriptional program of PC cells.
GATA2 Is Critical for AR Expression and Cell Proliferation.
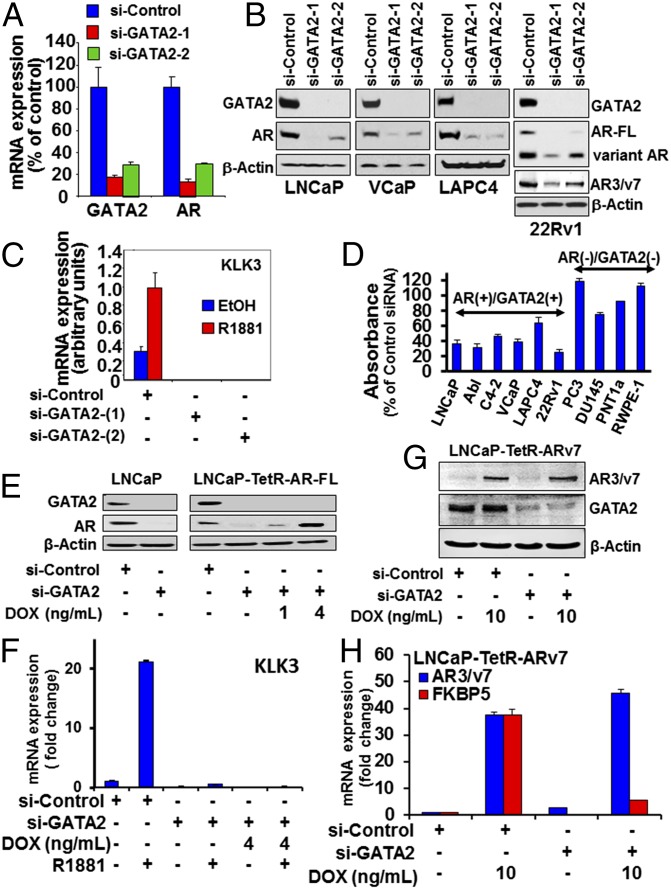
Having demonstrated strong correlation between the GATA2 and AR transcriptional signatures, we next examined the impact of GATA2 on the expression of the AR gene itself. We observed that silencing GATA2 expression markedly depleted AR mRNA (Fig. 2A) and AR protein levels in four PC cell lines (Fig. 2B and Fig. S2A). Interestingly, si-GATA2 also depleted the expression of ligand-independent ARVs (including AR3/AR-v7) in 22Rv1 cells (Fig. 2B) and, expectedly, also depleted the expression of classical AR transcriptional target genes such as KLK3/PSA, TMPRSS2, and FKBP5 (Fig. 2C and Fig. S2 B and C). GATA2 silencing also dramatically suppressed cell proliferation of AR+/GATA2+ PC cell lines (Fig. 2D).
Fig. 2.
GATA2 is critical for the expression and transcriptional function of both AR-FL and ARVs in PC. (A). RT qPCR of GATA2 and AR mRNA expression in LNCaP cells following 72 h of silencing GATA2. (B) Immunoblot analyses of GATA2 and AR or AR3/v7 ARV following 72 h of silencing GATA2. (C) RT qPCR of KLK3 following 72 h of silencing GATA2 in LNCaP cells. Cells were androgen-starved for 48 h in 10% CSS medium and then were treated with 1 nM of R1881 or vehicle for 24 h. (D) Cell viability (MTT absorbance) of PC cell lines 96 h posttransfection with si-nontarget (si-NT) or si-GATA2. Absorbance values are normalized to the corresponding NT siRNA for each cell line. (E and F) Parental LNCaP cells and LNCaP cells stably transfected with tetracycline-inducible AR-FL (LNCaP-TetR-AR-FL) were transfected with si-GATA2 or si-NT for 48 h in the presence or absence of doxycycline (DOX), as indicated. (E) Immunoblot analyses were conducted for the expression levels of GATA2, AR, and β-actin. (F) RT qPCR for KLK3 mRNA following 24 h of treatment with vehicle or 1 nM R1881. (G and H) LNCaP cells stably transfected with tetracycline-inducible AR3/v7 (LNCaP-TetR-ARV7 cells) were transfected with si-NT or si-GATA2 for 48 h in the presence or absence of doxycycline, as indicated. (G) Immunoblot analysis of AR3/v7 and GATA2 expression. (H) RT qPCR was performed for AR3/v7 and FKBP5 (a known AR3/v7 target gene).
Role of GATA2 in a Negative Feedback Loop Regulating AR Expression by Androgen in PC Cells.
Treatment of LNCaP cells with 1 nM R1881 for 24 h suppressed GATA2 and AR mRNA levels but induced the expression of the AR target gene TMPRSS2 (Fig. S2D). In agreement, GATA2 expression was increased in LNCaP cells following treatment with enzalutamide (MDV3100) (Fig. S2E), androgen deprivation [exposure to medium supplemented with 10% charcoal-stripped serum (CSS)] (Fig. S2F), or AR siRNA (Fig. S2G). Collectively, our results indicate that androgens and AR suppress GATA2 gene expression.
Next, we performed ChIP-Seq analyses for GATA2 in LNCaP cells. We used two different anti-GATA2 antibodies, which produced highly similar cistromes (∼97% overlap). Based on our GATA2 ChIP-Seq data, we identified binding of GATA2 at the AR gene locus (Fig. S2H), with a strong ChIP-Seq peak ∼5.5 kb upstream of the gene transcription start site (TSS) and three additional but weaker binding sites within the transcribed gene region. ChIP-qPCR confirmed binding of GATA2 at these binding sites, which decreased upon treatment with androgen (Fig. S2I). We also confirmed binding of AR to a location at ∼102.9 kb downstream of the TSS (Fig. S2J): This site previously was reported to bind AR to repress expression of its own gene through recruitment of lysine-specific demethylase 1 (KDM1A) and demethylation of histone H3 dimethyl Lys4 (H3K4me2) and histone H3 monomethyl Lys4 (H3K4me1) (30).
Our findings suggest that, in addition to the previously reported direct binding of AR protein to the AR gene (30), androgen can suppress AR mRNA expression indirectly by decreasing GATA2 expression and its recruitment to the extended promoter region of the AR gene, constituting another negative feedback loop that regulates AR mRNA expression and can increase it upon androgen deprivation (Fig. S2K).
GATA2 Is Necessary for AR Transcriptional Function.
We next examined whether GATA2 also can regulate the transcriptional activity of the AR protein. Because silencing GATA2 in PC cell lines leads to loss of endogenous AR mRNA and protein expression as well as depletion of the AR target genes KLK3, TMPRSS2, and FKBP5 (Fig. 2 A–C and Fig. S2 A–C), we stably transfected LNCaP cells with tetracycline-inducible vectors for AR-FL or the ARV AR3/ARv7. We determined that restoration of AR-FL expression via treatment with doxycycline (Fig. 2E) failed to restore the expression of KLK3/PSA (Fig. 2F), TMPRSS2, and FKBP5 (Fig. S3) genes when GATA2 is silenced, suggesting a requirement for GATA2. Similarly, tetracycline-inducible exogenous AR3/ARv7 expression (Fig. 2G) failed to induce expression of FKBP5, a known AR3/ARv7-target gene (Fig. 2H). Collectively, these results demonstrate that GATA2 protein is necessary for the transcriptional function of both AR-FL and ARV protein.
Direct Physical Interaction of GATA2 with AR.
Our finding that GATA2 is necessary for the transactivation function of AR proteins led us to determine if there is a direct physical interaction and cooperation between GATA2 and AR-FL or AR3/v7. Sensitized emission FRET (seFRET) showed that, in HeLa cells cotransfected with the corresponding expression vectors, treatment with androgen [2 nM dihydrotestosterone (DHT)] for 2 h significantly promoted the physical interaction between CFP-AR-FL and YFP-GATA2 (Fig. S4 A and C). CFP-ARv7 showed significant FRET with YFP-GATA2 at baseline (without androgen), which was not further increased by androgen treatment, suggesting a constitutive and androgen-insensitive interaction (Fig. S4 B and D).
GATA2 Colocalizes with AR and FOXA1 on Chromatin.
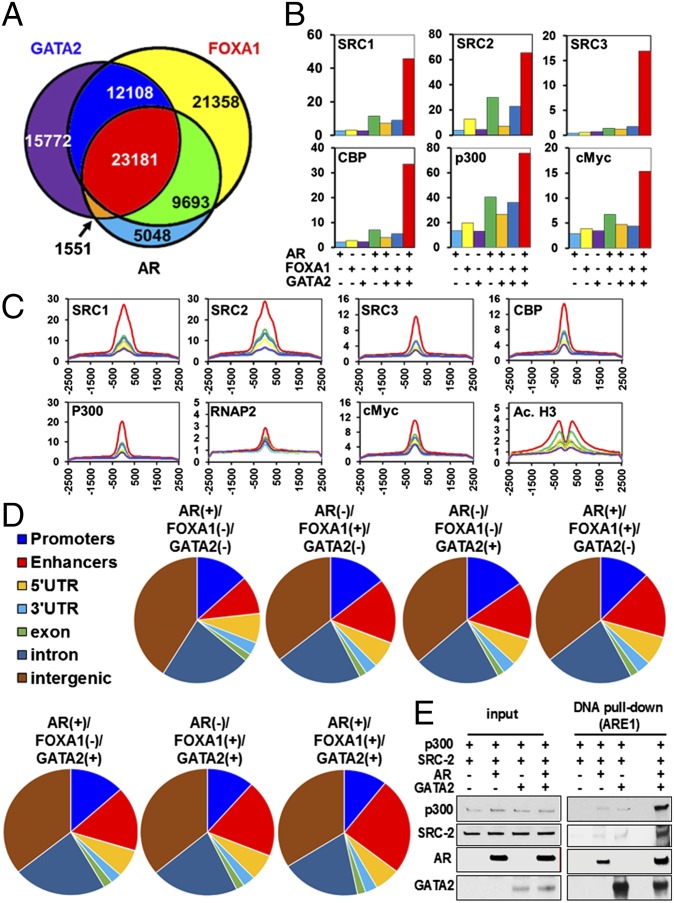
We performed ChIP-Seq in LNCaP cells (cultured at baseline conditions, in medium supplemented with 10% FBS) for AR, GATA2, the steroid receptor coactivators SRC1, SRC2, and SRC3, and p300, CBP, and RNA polymerase 2 (P2). The signals obtained using two different GATA2 antibodies correlated very strongly across the genome (R = 0.94, P < 0.0001); for the remainder of the analysis, we report the ChIP-Seq profile generated using the GATA2-r antibody (sc9008; Santa Cruz). We found that GATA2 binding to chromatin strongly colocalizes with the binding of AR and FOXA1 (Fig. 3A and Fig. S5 A and B). Motif enrichment analysis shows that the overlap of all three peak sets enriches for the FOXA1 (P = 10−4134), GATA (P = 10−502), and AR (P = 10−456) motifs, respectively (Fig. S5 B and C). FOXA1 motifs were detected in all the combinations of FOXA1, GATA2, and AR peaks, even when FOXA1 peaks were not formally detected. Because LNCaP cells express AR-FL almost exclusively, we next used ChIP-Seq data for ARVs and AR-FL generated in 22Rv1 cells by Lu et al. (31). We found that in 22Rv1 cells both ARVs and AR-FL occupy chromatin regions that overlap with regions that recruit GATA2 in our LNCaP dataset (Fig. S5D).
Fig. 3.
GATA2 colocalizes with AR and FOXA1 on the chromatin and cooperates with them to recruit master transcriptional regulators. (A) ChIP-Seq demonstrates that, in LNCaP cells, GATA2 strongly colocalizes on chromatin with AR and FOXA1. (B) Integrative analysis with other ChIP-Seq datasets shows that the steroid receptor coactivators SRC-1, SRC-2, and SRC-3, key transcriptional regulators CBP and p300, and transcription factors such as c-Myc preferentially bind to genomic regions where GATA2, AR, and FOXA1 are colocalized (triple-positive peaks). (C) Distribution of normalized sequence tag density for SRC-1, SRC-2, SRC-3, CBP, p300, cMYC, and RNA pol II confirm the higher recruitment of these key transcriptional regulators at triple-positive peaks. In addition, acetylated histone H3 (Ac. H3) shows higher recruitment at 500 bp upstream and downstream of the triple-positive peaks. Color labeling in B and C follows the pattern shown in A. (D) Peak overlap with gene features (promoter, 5′ UTR, 3′ UTR, exons, introns), distal regulatory elements (enhancers), and intergenic regions shows enrichment of triple-positive peaks at distal enhancers: 25% of triple-positive peaks overlap with enhancers, compared with 10–17% of single-positive peaks (1.82-fold enrichment, P < 0.0001, χ2 test) and 17–20% of double-positive peaks (1.41-fold enrichment, P < 0.0001, χ2 test). (E) Recombinant GATA2 protein increases the recruitment of recombinant AR, SRC-2, and p300 to DNA harboring a classical ARE (from the KLK3 promoter). Immunoblot analyses were conducted following a DNA pull-down assay performed in the presence of R1881.
Colocalization of GATA2, AR, and FOXA1 Is Needed for the Optimal Recruitment of Key Transcriptional Coactivators and Mediators to Enhancer Sites.
Uniform permutation testing of the corresponding ChIP-Seq datasets demonstrated that colocalization of GATA2, AR, and FOXA1 (i.e., a triple-positive peak) is associated with optimal recruitment of key transcriptional mediators SRC1, SRC2, SRC3, CBP, p300, cMyc, mediator complex subunit 12 (MED12), and RNAP2 as well as with increased sensitivity to DNase I and increased recruitment of ERG (Fig. 3B). In addition, we found enrichment for epigenetic marks of active transcription: acetylated histone 3 (H3ac), K27-specific acetylation of histone 3 (H3K27ac), H3K4me1, and H3K4me2 500 bp upstream and downstream of the triple-positive peaks (Fig. 3C and Fig. S6). In particular, H3K4me1 and H3K27ac are known to be associated with enhancer sites (32). Conversely, H3K4me3, a mark enriched at actively transcribed promoters, did not exhibit increased association with the triple-positive peaks. In agreement, at the triple-positive peaks, we did not observe increased presence of WD repeat-containing protein 5 (WDR5) or histone H3 threonine 11 (H3T11) phosphorylation, two epigenetic regulators that recently were shown to facilitate H3K4 trimethylation (H3K4me3) cooperatively at AR target genes (33). As another control, broad transcription inhibition marks (H3K9me3 and H3K27me3), as well as transcription elongation marks (H3K36me3) showed no enrichment at or around the triple-positive GATA2/FOXA1/AR peaks (Fig. S6).
We next quantified the peak distribution with respect to gene features (promoter, 5′ UTR, 3′ UTR, exons, introns), distal regulatory elements (enhancers), and intergenic regions. As shown in Fig. 3D, 25% of triple-positive peaks overlap with enhancers, compared with 10–17% of single-positive peaks (1.82-fold enrichment, P < 0.0001 χ2 test) and 17–20% of double-positive peaks (1.41-fold enrichment, P < 0.0001, χ2 test). Collectively, these data suggest that the triple-positive peaks are more frequently associated with enhancers than with promoters of active genes.
Using a cell-free system, we determined that the presence of GATA2 protein increases the recruitment of AR to DNA harboring a classical AR response element (ARE) from the KLK3 promoter and significantly enhances the recruitment of SRC-2 and p300 proteins (Fig. 3E). Collectively, our data demonstrate that GATA2 is an important component of the AR transcriptional holocomplex.
Potent Anticancer Activity of the GATA2 Small Molecule Inhibitor K7174 Against GATA+/AR+ PC Models in Vitro and in Vivo.
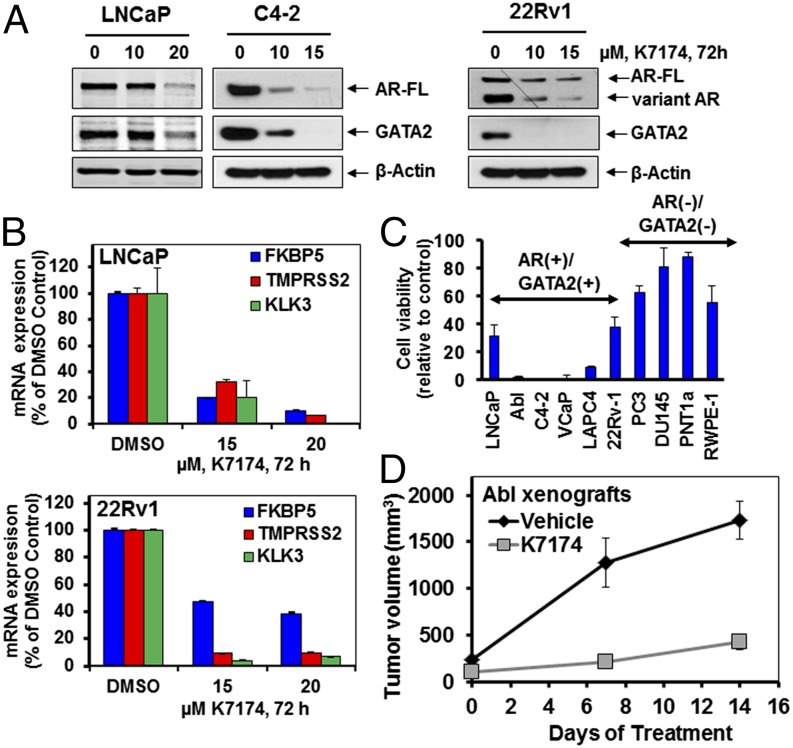
We next examined the effects of the GATA2 small molecule inhibitor (SMI) K7174 in our PC cell line panel. Treatment with K7174 suppressed the expression of AR, including ARVs, in 22Rv1 cells (Fig. 4A). K7174 suppressed the expression of AR target genes in both LNCaP and 22Rv1 cells, indicating inhibition of AR transcriptional activity (Fig. 4B). K7174 treatment exerted potent anticancer activity against GATA2+/AR+ PC cell lines in vitro (Fig. 4C). In our studies, K7174 decreased GATA2 protein levels (Fig. 4A), but we did not detect suppression of GATA2 mRNA, suggesting that the effect on GATA2 protein is caused by a posttranscriptional mechanism. In support, we found that the proteasome inhibitor bortezomib attenuated the marked depletion of GATA2 protein levels that was induced by K7174 (Fig. S7). K7174 also exhibited significant anticancer activity in a PC xenograft model (P < 0.05) (Fig. 4D).
Fig. 4.
The GATA2 SMI K7174 suppresses expression and transcriptional activity of both AR-FL and AR3/ARv7 and suppresses growth of AR+/GATA2+ PC cell lines in vitro and in vivo. (A) Immunoblot analysis for AR and GATA2 expression levels in PC cell lines following treatment with K7174 for 72 h. (B) Relative mRNA expression of AR target genes KLK3, TMPRSS2, and FKBP5 following treatment with K7174, as indicated, for 72 h. Columns show the mean of biologic triplicates ± SD. (C) Cell viability (MTT assay) of PC cell lines following treatment with K7174 (20 μM) for 96 h. Results are reported as percent of corresponding vehicle control. Columns show the mean of biologic quadruplicates ± SD. (D) In vivo activity of K7174 against PC cells. Treatment of mice with vehicle or K7174 (25 mg−1⋅kg−1⋅d, 5 d/wk, i.p. injection, n = 4 per cohort) was initiated when the mean tumor volume was ∼200 mm3. The x axis depicts days after initiation of treatment.
Discussion
There is an urgent need for a new treatment paradigm in CRPC for the suppression of persistent AR signaling. Our present study highlights GATA2 as an actionable therapeutic target in PC. GATA2 inhibition exerts a dual effect on AR signaling by suppressing new AR synthesis and by inactivating the transcriptional activity of already synthesized AR protein. GATA2 enhances the recruitment of SRCs to the AR transcriptional holocomplex, a step necessary for the docking of p300/CBP and for transcriptional activity. Importantly, a GATA2 SMI suppressed the expression and transcriptional function of AR and exerted potent anticancer activity against PC cells in vitro and in vivo. We propose GATA2 inhibition as a previously unexplored approach for PC treatment.
Silencing GATA2 in PC cells suppressed the expression of both AR-FL and ARV mRNA and protein and resulted in a transcriptional footprint highly correlated with that of AR inhibition both in vitro and in human patient samples. This result was validated by a strong positive correlation between the expression of AR and GATA2 in PC, both in vitro and in human patient samples. The direct regulation of AR gene expression by GATA2 is attributable to the recruitment of GATA2 to the extended promoter region of the AR gene (∼5.5 kb upstream of the TSS), which also serves as the basis for a negative autoregulatory feedback loop of AR expression. We observed that androgen suppresses GATA2 mRNA and protein levels, decreasing GATA2 recruitment to the AR gene promoter. Conversely, upon androgen deprivation, GATA2 levels increase, and they, in turn, can up-regulate AR mRNA. AR expression is known to be elevated in many CRPCs. This elevation has been attributed to AR gene amplification in some cases and to derepression of the autoregulatory binding of AR protein to the AR gene (30). We now propose an additional, GATA2-mediated mechanism for this negative feedback loop. That androgen deprivation induces GATA2 and AR expression further supports the need to inhibit GATA2 in CRPC.
In addition to its role in promoting AR expression, GATA2 is required for AR transcriptional function. Both exogenously expressed AR-FL and ARV protein were transcriptionally inactive when GATA2 expression was silenced. Our integrated genomewide cistrome analysis showed that the steroid receptor coactivators SRC1, SRC2, and SRC3 and the key transcriptional regulators CBP and p300 preferentially bind to genomic regions where GATA2, AR, and FOXA1 are colocalized. These AR+/GATA2+/FOXA1+ genomic regions also enrich strongly for cMyc, the mediator complex member MED12, ERG, and RNA polymerase II (pol II), exhibit increased sensitivity to DNase I digestion, and are preferentially located at distal regulatory elements (enhancers). In agreement, we found increased presence of epigenetic marks of active transcription (H3K27ac, H3K4me1, and H3K4me2) that are known to be enriched at enhancer sites (32) at 500 bp upstream and downstream of the triple-positive peaks but no enrichment of marks associated with actively transcribed promoters (H3K4me3, WDR5, or H3T11P), transcription inhibition (H3K9me3 and H3K27me3), or transcriptional elongation (H3K36me3). These data suggest that the role of GATA2 extends beyond the simple facilitation of AR binding to DNA, making a critical contribution to the formation of a higher-order transcription enhancer complex (enhanceosome) that defines the active AR cistrome. These findings were confirmed via FRET, where we documented that AR-FL and GATA2 interact directly upon androgen treatment. The ARV AR3/v7 also interacts with GATA2, albeit in an androgen-independent manner. Furthermore, using a cell-free system, we found that GATA2 increases the recruitment of AR to a classical ARE and significantly promotes the recruitment of SRC-2 and p300. Collectively, our data demonstrate that GATA2 interacts directly with AR and is required for the optimal recruitment of p160 SRCs and other transcriptional cofactors to AR to form a functional enhanceosome. Following the cloning by our laboratory of the first (to our knowledge) bona fide transcriptional steroid receptor coactivator, SRC-1 (34), the designation of transcriptional coregulator generally has been limited to proteins that are recruited to DNA only indirectly (via DNA-bound transcription factors) (35). However, our results now attribute to GATA2 a role in AR complex formation that justifies its classification as an AR coregulator. Because GATA2 interacts with DNA directly, our observations broaden the definition of members of the coregulator category to include DNA-binding proteins that promote the formation and/or stability of the holocomplex.
In summary, we have demonstrated that GATA2 promotes AR signaling in PC by inducing AR gene expression and also by facilitating the recruitment of SRCs to AR, preferably at enhancer sites. A GATA2 SMI suppressed the expression of AR mRNA and protein (including the constitutively active AR3/AR-v7 ARV that is resistant to all current therapies) and AR-dependent target genes and exerted potent anticancer activity against GATA2+/AR+ PC cell lines in vitro and in vivo. Our studies provide an innovative approach to target AR expression and function by pharmacological inhibition of GATA2 and effectively overcome resistance to therapy in CRPC.
Materials and Methods
Animal Studies.
The anticancer activity of the GATA2 SMI K7174 was tested in vivo against xenografts of Abl PC cells. Two million Abl cells (mixed with Matrigel at a volume ratio of 1:1) were s.c. injected in the flank of male SCID-Beige mice (n = 4 per group). Treatment with K7174 (25 mg⋅kg−1⋅d−1, 5 d/wk, i.p.) or vehicle was initiated when the tumors reached ∼200 mm3. Tumor size was compared among cohorts by unpaired t test. All animal studies were conducted in accordance with institutional guidelines. All procedures were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee, which is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care.
Statistical Analysis.
Comparison of GATA2 protein expression among tissue types by IHC was conducted on unrelated observations, i.e., if normal tissue for a patient was included in the analysis, then matching tumor tissue was excluded to preserve independence of samples assumption and maximize the number of samples used. The Mann–Whitney u test was used to evaluate the differences because of the skewness of the data. The association of GATA2 IHC staining with clinical and a preselected panel of pathological variables and other biomarkers was evaluated using the Spearman correlation with no multiple comparison adjustments made. The predictive value of the GATA2 index was analyzed using the Cox proportional hazards regression model univariately and multivariately (after adjusting for key clinicopathological parameters: preoperative PSA, lymph node involvement, seminal vesicle invasion, extraprostatic extension, positive surgical margins, and Gleason score. The HR and 95% CI were computed for each marker. The minimum P value method was used to group patients into “low level” and “high level” categories. Kaplan–Meier survival curves were plotted for each of the level categories. P values of <0.05 were considered significant. All analyses were performed using the SPSS 19.0 software package (IBM).
Additional methods for cell culture, IHC, gene-expression profiling, characterization of the AR transcriptional complex, GSEA, ChIP-Seq and ChIP-PCR, FRET imaging, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, immunoblotting, and RT qPCR are described in SI Materials and Methods. Oligonucleotide sequences used in these studies are shown in Table S2. All prostate cancer specimens from patients were obtained with informed consent as part of an ongoing Baylor College of Medicine IRB approved tissue banking protocol (H-11658).
Supplementary Material
Acknowledgments
We acknowledge the joint participation by the Adrienne Helis Malvin Medical Research Foundation through its direct engagement in the continuous, active conduct of medical research in conjunction with Baylor College of Medicine (BCM), and the assistance of the Shared Resources of the Dan L. Duncan Cancer Center [supported by National Cancer Institute (NCI) Cancer Center Support Grant P30CA125123]. This work was supported by the Prostate Cancer Foundation (B.W.O. and N.M.); Conquer Cancer Foundation of the American Society of Clinical Oncology Young Investigator and Career Development Awards (both to N.M.); Eunice Kennedy Shriver National Institute of Child Health and Human Development Grant 8818 (to B.W.O.); Susan G. Komen for the Cure Foundation Promise Grant 221410 (to B.W.O.); National Institutes of Health Grant 1K01DK096093 (to S.M.H.); pilot grants (to S.M.H. and N.M.) from the Pilot/Feasibility Program of the Diabetes and Endocrinology Research Center at BCM (P30-DK079638); and an Alkek Foundation for Molecular Discovery Pilot grant (to C.C.). N.M. is a Dan L. Duncan Scholar, a Caroline Wiess Law Scholar, and a member of the Dan L. Duncan Cancer Center (supported by NCI Cancer Center Support Grant P30CA125123) and the Center for Drug Discovery at BCM.
Footnotes
The authors declare no conflict of interest.
The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE63539).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421415111/-/DCSupplemental.
References
- 1.de Bono JS, et al. COU-AA-301 Investigators Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364(21):1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan CJ, Tindall DJ. Androgen receptor rediscovered: The new biology and targeting the androgen receptor therapeutically. J Clin Oncol. 2011;29(27):3651–3658. doi: 10.1200/JCO.2011.35.2005. [DOI] [PubMed] [Google Scholar]
- 3.Mitsiades N. A road map to comprehensive androgen receptor axis targeting for castration-resistant prostate cancer. Cancer Res. 2013;73(15):4599–4605. doi: 10.1158/0008-5472.CAN-12-4414. [DOI] [PubMed] [Google Scholar]
- 4.Scher HI, et al. AFFIRM Investigators Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367(13):1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 5.Debes JD, Tindall DJ. Mechanisms of androgen-refractory prostate cancer. N Engl J Med. 2004;351(15):1488–1490. doi: 10.1056/NEJMp048178. [DOI] [PubMed] [Google Scholar]
- 6.Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68(13):5469–5477. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer. 2011;18(5):R183–R196. doi: 10.1530/ERC-11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonarakis ES, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371(11):1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu J, Wu RC, O’Malley BW. Normal and cancer-related functions of the p160 steroid receptor co-activator (SRC) family. Nat Rev Cancer. 2009;9(9):615–630. doi: 10.1038/nrc2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith CL, Oñate SA, Tsai MJ, O’Malley BW. CREB binding protein acts synergistically with steroid receptor coactivator-1 to enhance steroid receptor-dependent transcription. Proc Natl Acad Sci USA. 1996;93(17):8884–8888. doi: 10.1073/pnas.93.17.8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Debes JD, et al. p300 regulates androgen receptor-independent expression of prostate-specific antigen in prostate cancer cells treated chronically with interleukin-6. Cancer Res. 2005;65(13):5965–5973. doi: 10.1158/0008-5472.CAN-04-2837. [DOI] [PubMed] [Google Scholar]
- 12.Zhou HJ, et al. SRC-3 is required for prostate cancer cell proliferation and survival. Cancer Res. 2005;65(17):7976–7983. doi: 10.1158/0008-5472.CAN-04-4076. [DOI] [PubMed] [Google Scholar]
- 13.Yan J, et al. Steroid receptor coactivator-3 and activator protein-1 coordinately regulate the transcription of components of the insulin-like growth factor/AKT signaling pathway. Cancer Res. 2006;66(22):11039–11046. doi: 10.1158/0008-5472.CAN-06-2442. [DOI] [PubMed] [Google Scholar]
- 14.Yan J, et al. Steroid receptor coactivator-3/AIB1 promotes cell migration and invasiveness through focal adhesion turnover and matrix metalloproteinase expression. Cancer Res. 2008;68(13):5460–5468. doi: 10.1158/0008-5472.CAN-08-0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agoulnik IU, et al. Androgens modulate expression of transcription intermediary factor 2, an androgen receptor coactivator whose expression level correlates with early biochemical recurrence in prostate cancer. Cancer Res. 2006;66(21):10594–10602. doi: 10.1158/0008-5472.CAN-06-1023. [DOI] [PubMed] [Google Scholar]
- 16.Agoulnik IU, et al. Role of SRC-1 in the promotion of prostate cancer cell growth and tumor progression. Cancer Res. 2005;65(17):7959–7967. doi: 10.1158/0008-5472.CAN-04-3541. [DOI] [PubMed] [Google Scholar]
- 17.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C, et al. Tumor-suppressor role for the SPOP ubiquitin ligase in signal-dependent proteolysis of the oncogenic co-activator SRC-3/AIB1. Oncogene. 2011;30(42):4350–4364. doi: 10.1038/onc.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geng C, et al. Prostate cancer-associated mutations in speckle-type POZ protein (SPOP) regulate steroid receptor coactivator 3 protein turnover. Proc Natl Acad Sci USA. 2013;110(17):6997–7002. doi: 10.1073/pnas.1304502110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahu B, et al. Dual role of FoxA1 in androgen receptor binding to chromatin, androgen signalling and prostate cancer. EMBO J. 2011;30(19):3962–3976. doi: 10.1038/emboj.2011.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D, et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474(7351):390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Stable CM, Pozas A, Roos BA. A role for GATA transcription factors in the androgen regulation of the prostate-specific antigen gene enhancer. Mol Cell Endocrinol. 2000;167(1-2):43–53. doi: 10.1016/s0303-7207(00)00300-2. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27(3):380–392. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu D, et al. Three-tiered role of the pioneer factor GATA2 in promoting androgen-dependent gene expression in prostate cancer. Nucleic Acids Res. 2014;42(6):3607–3622. doi: 10.1093/nar/gkt1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chiang YT, et al. GATA2 as a potential metastasis-driving gene in prostate cancer. Oncotarget. 2014;5(2):451–461. doi: 10.18632/oncotarget.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Böhm M, Locke WJ, Sutherland RL, Kench JG, Henshall SM. A role for GATA-2 in transition to an aggressive phenotype in prostate cancer through modulation of key androgen-regulated genes. Oncogene. 2009;28(43):3847–3856. doi: 10.1038/onc.2009.243. [DOI] [PubMed] [Google Scholar]
- 27.Wang XD, et al. Identification of candidate predictive and surrogate molecular markers for dasatinib in prostate cancer: Rationale for patient selection and efficacy monitoring. Genome Biol. 2007;8(11):R255.1-255.11. doi: 10.1186/gb-2007-8-11-r255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu R, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72(14):3457–3462. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai C, et al. ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. J Clin Invest. 2013;123(3):1109–1122. doi: 10.1172/JCI66666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cai C, et al. Androgen receptor gene expression in prostate cancer is directly suppressed by the androgen receptor through recruitment of lysine-specific demethylase 1. Cancer Cell. 2011;20(4):457–471. doi: 10.1016/j.ccr.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu J, et al. The cistrome and gene signature of androgen receptor splice variants in castration-resistant prostate cancer cells. J Urol. 2014;S0022-5347(14):04214-1. doi: 10.1016/j.juro.2014.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim JY, et al. A role for WDR5 in integrating threonine 11 phosphorylation to lysine 4 methylation on histone H3 during androgen signaling and in prostate cancer. Mol Cell. 2014;54(4):613–625. doi: 10.1016/j.molcel.2014.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oñate SA, Tsai SY, Tsai MJ, O’Malley BW. Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science. 1995;270(5240):1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 35.Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: A diversity of functions converging on and regulating the AR transcriptional complex. Endocr Rev. 2007;28(7):778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.