Significance
Expansion of a poly-glutamine (polyQ) repeat is the causal mutation of several inherited neurological disorders, including Huntington’s disease. In a yeast genetic screen, we identified several proteins with Q-rich, prion-like domains that reduce the toxicity of mutant polyQ proteins when overexpressed. One of these, glycine threonine serine repeat protein (Gts1p), was characterized in more detail. Association with Gts1p did not prevent aggregation but altered the physical properties and the interactome of the aggregates. Specifically, Gts1p expression reduced the sequestration of other prion-like proteins into the polyQ aggregates. These findings link polyQ toxicity in yeast with the coaggregation of prion proteins and show that short Q-rich peptides are able to shield toxic forms of polyQ proteins, directing them into nontoxic aggregates.
Keywords: protein aggregation, protein misfolding, neurodegeneration, prion, polyglutamine proteins
Abstract
Expansion of a poly-glutamine (polyQ) repeat in a group of functionally unrelated proteins is the cause of several inherited neurodegenerative disorders, including Huntington’s disease. The polyQ length-dependent aggregation and toxicity of these disease proteins can be reproduced in Saccharomyces cerevisiae. This system allowed us to screen for genes that when overexpressed reduce the toxic effects of an N-terminal fragment of mutant huntingtin with 103 Q. Surprisingly, among the identified suppressors were three proteins with Q-rich, prion-like domains (PrDs): glycine threonine serine repeat protein (Gts1p), nuclear polyadenylated RNA-binding protein 3, and minichromosome maintenance protein 1. Overexpression of the PrD of Gts1p, containing an imperfect 28 residue glutamine-alanine repeat, was sufficient for suppression of toxicity. Association with this discontinuous polyQ domain did not prevent 103Q aggregation, but altered the physical properties of the aggregates, most likely early in the assembly pathway, as reflected in their increased SDS solubility. Molecular simulations suggested that Gts1p arrests the aggregation of polyQ molecules at the level of nonfibrillar species, acting as a cap that destabilizes intermediates on path to form large fibrils. Quantitative proteomic analysis of polyQ interactors showed that expression of Gts1p reduced the interaction between polyQ and other prion-like proteins, and enhanced the association of molecular chaperones with the aggregates. These findings demonstrate that short, Q-rich peptides are able to shield the interactive surfaces of toxic forms of polyQ proteins and direct them into nontoxic aggregates.
Expansion of a poly-glutamine (polyQ) repeat in otherwise unrelated proteins is the cause of several inherited neurological disorders, including Huntington’s disease (HD), spinobulbar muscular atrophy, dentatorubral-pallidoluysian atrophy, and spinocerebellar ataxias 1, 2, 3, 6, 7, and 17 (1). In all these cases, increasing the length of the polyQ repeat over a critical threshold (above 37 Q in HD) (2) results in disease manifestation, with the length of the repeat correlating inversely with age of disease onset (3).
According to the gain-of-toxic function theory, the polyQ expansions increase aggregation propensity and confer to the disease proteins the ability to populate one or more toxic conformations, most likely including various oligomeric and higher-order aggregate states. These aggregate species vary greatly in number of monomeric units, detergent solubility, binding of dyes, and identity and mobility of interacting proteins (4–6). A prominent hypothesis suggests that the pathologic protein aggregates expose novel, highly interactive surfaces that mediate aberrant interactions with other proteins, resulting in their functional impairment and sequestration (7–9). Moreover, the aggregation process is thought to interfere with general protein quality control pathways, including protein folding and the clearance of misfolded proteins (10–13). The yeast S. cerevisiae has been used extensively as a model to explore the basic mechanisms of toxicity mediated by polyQ expansions. The polyQ length dependence of aggregation has been reproduced in yeast upon expression of N-terminal fragments of huntingtin containing the polyQ stretch (N-Htt) (14, 15). Interestingly, toxicity, as measured by growth impairment, was found to depend critically on the properties of the sequences flanking the polyQ region (16). Like mammalian prions, yeast prions are able to cause detectable phenotypes without transmitting any genetic material (17), and both N-Htt aggregation and toxicity were shown to require the [RNQ+] prion (18, 19). Although there are numerous proteins in the yeast proteome that contain Q-rich regions (20), there is no Htt homolog in yeast. Therefore, any toxic effects observed in this model system on N-Htt expression can be attributed to a gain-of-toxic function.
Although polyQ aggregation is strongly associated with cell toxicity, the exact nature and the conformational properties of the toxic polyQ species remain elusive. To gain insight into the basic mechanisms of polyQ toxicity, we performed an unbiased yeast genetic screen for suppressors of the growth defect caused by the expression of polyQ expanded N-Htt. We identified six genes that reproducibly restored cell growth when overexpressed. Surprisingly, these suppressors include several proteins with Q-rich sequences, nuclear polyadenylated RNA-binding protein 3 (Nab3p), minichromosome maintenance protein 1 (Mcm1p), and glycine threonine serine repeat protein (Gts1p), which have been suggested to contain candidate prion domains (PrDs) (21). Consistent with results from molecular modeling, quantitative proteomic analysis revealed that these proteins act by shielding the interactive surfaces of polyQ aggregates, thereby reducing their aberrant interactions with endogenous proteins, including multiple bona fide yeast prions, while promoting their association with molecular chaperones.
Materials and Methods
Suppressor Screen.
A yeast genomic DNA multicopy library cloned in YEp13 (22, 23) was transformed into YPH499 + pYES2L-103Q-GFP using a high efficiency transformation protocol (24). The cells were plated on synthetic complete (SC) selection medium with galactose to induce 103Q-GFP expression. Of 150,000 transformants, about 200 colonies grew. Each of these colonies was then grown on SC selection medium without 103Q-GFP induction with glucose as the sole carbon source. False positives were eliminated by three successive rounds of screening and positive clones were sequenced (SI Materials and Methods).
Interactome Analysis.
Yeast cells expressing 103Q-GFP and the suppressor proteins were harvested and lysed. 103Q-GFP was isolated with anti-GFP magnetic beads (Miltenyi). Eluates were desalted and subsequently analyzed by LC coupled to a Q Exactive mass spectrometer (Thermo Fisher) via a nano electrospray source. Data were analyzed using the MaxQuant software (v 1.4.3.19) (25) and label-free algorithms (26). Raw data were searched against the UniProtKB Yeast FASTA database (06/2012). For more details, see SI Materials and Methods.
Results
A Yeast Screen for Suppressors of polyQ Toxicity.
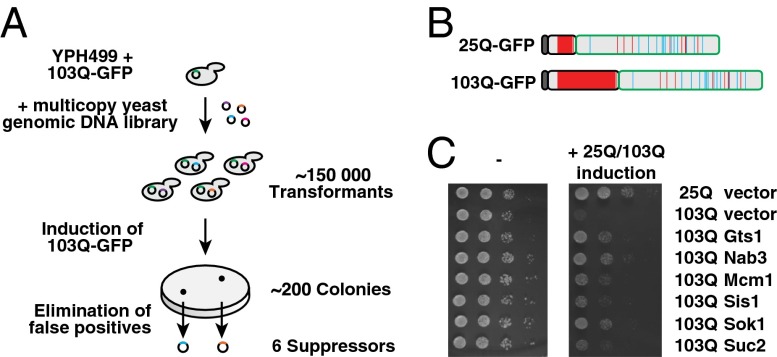
Previous genetic screens in yeast identified 52 nonessential gene deletions that enhanced the toxicity of a polyQ expanded Htt fragment (27), 28 genes that suppressed toxicity when deleted (28), and 317 genes that suppressed toxicity when overexpressed under control of a GAL1 promoter (29). However, overexpression of potential suppressors under the strong GAL1 promoter may lead to variations in copy number of the toxic protein (30) or result in toxicity so that some suppressors may be missed. To overcome these problems, we used a library of genes under the control of their endogenous promoter on a multicopy plasmid, thus resulting in an intermediate expression level (Fig. 1A). To this end, we expressed a toxic N-Htt-GFP fusion protein with 103 Q, lacking the poly-proline region and containing a N-terminal Flag-tag (103Q-GFP) (Fig. 1B) (16), under control of a galactose inducible promoter in yeast positive for the [RNQ+] prion. Expression of 103Q-GFP caused a strong growth defect, whereas an otherwise identical protein containing 25 Q (25Q-GFP) caused no growth impairment (Fig. 1C) (16, 19).
Fig. 1.
Identification of suppressors of polyQ toxicity. (A) Workflow of overexpression suppressor screen. A yeast genomic DNA library was transformed into cells harboring the inducible 103Q-GFP construct. About 200 colonies were able to grow on induction of 103Q-GFP. False positives were eliminated by three successive rounds of screening. Sequencing of DNA inserts showed that inserts contained 10 different genomic regions. After elimination of inserts that encoded genes involved in the metabolism of galactose, six suppressors of polyQ-mediated toxicity remained. (B) Schematic representation of the 25Q/103Q-GFP constructs used in this study. Dark gray indicates the N-terminal Flag-tag, light gray indicates the N terminus of Htt, and green represents the GFP-tag. Glutamines (Q) are shown in red; asparagines (N) in cyan. (C) Expression of Gts1p, Nab3p, Mcm1p, Sis1p, Sok1p, and Suc2p suppresses polyQ toxicity. The potential suppressors were preinduced for 48 h before serial dilutions of cells were spotted on galactose medium without doxycycline to coexpress 25Q/103Q-GFP and the suppressors (Right) or on glucose plates to control for equal spotting (Left).
A yeast genomic DNA multicopy library (22) was transformed into cells harboring the inducible 103Q-GFP construct (Fig. 1A). Of 150,000 transformants, ∼200 colonies were able to grow on induction of 103Q-GFP. After elimination of colonies that had lost either the expression of 103Q-GFP or the [RNQ+] prion, 40 positive clones remained and were analyzed in more detail. Sequencing of the DNA inserts showed that most inserts were selected several times and mapped to 10 different genomic regions, suggesting that the screen was saturated. Four of the isolated inserts encoded genes involved in the metabolism of galactose. These genes were not further analyzed as their expression may have interfered with the expression of 103Q-GFP.
Overview of Screen Results.
We identified six different genes that were able to suppress polyQ mediated toxicity (Fig. 1C), including the Hsp40 chaperone SIS1, the invertase SUC2, a gene with unknown function, SOK1, and three genes that encode proteins with Q-rich domains: NAB3, MCM1, and GTS1.
Sis1p is an essential member of the Hsp40 chaperone family. It functionally cooperates with Hsp70 proteins of the Ssa family and was shown to modulate the aggregation and toxicity of Htt polyQ proteins (14, 19, 31). We recently identified Sis1p as a strong interactor of mutant Htt and as a critical factor in the degradation of misfolded proteins (32). The nonessential protein Sok1p has been shown to suppress mutations in a cAMP-dependent protein kinase, but its exact function is not known (33). SUC2 has previously been identified as a suppressor of polyQ-mediated toxicity (29) and encodes a nonessential invertase, which hydrolyzes sucrose into glucose and fructose (34). Its mechanism of suppression remains unclear.
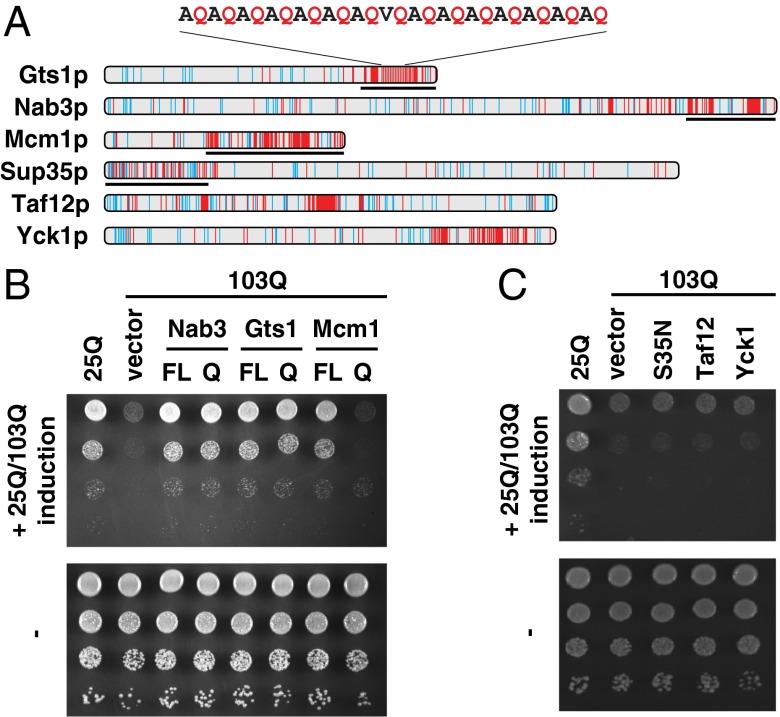
The identification of Q-rich, but otherwise unrelated, proteins as suppressors of polyQ-mediated toxicity was intriguing. Notably, a recent bioinformatics screen identified Nab3p, Mcm1p, and Gts1p as potentially prion like (21). Nab3p is a known interactor of expanded polyQ-containing proteins (32, 35) and has an essential role in the termination of RNA polymerase II transcripts (36). Mcm1p is an essential transcription factor involved in pheromone response and regulation of mating type-specific genes (37). Strong overexpression of Mcm1p causes growth arrest (38), but when expressed under control of its endogenous promoter in the multicopy plasmid pRS426, cells grew normally and the growth defect conferred by 103Q-GFP was suppressed (Fig. 1C). Gts1p is a nonessential transcription factor that has been implicated in several processes including the stress response, sporulation, and the cell cycle (39, 40). Gts1p was erroneously thought to contain glycine/threonine-serine repeats, but instead contains a Q-rich region including an imperfect glutamine-alanine repeat (QAR) in its C-terminal domain (Fig. 2A). Gts1p also contains a Zn-finger motif and an ubiquitin-associated (UBA) domain (40). The Q-rich sequences of the three suppressors differ from those of bona fide prion domains in proteins such as the eukaryotic release factor Sup35p in that they are not enriched in asparagine (N) (Fig. 2A).
Fig. 2.
Effect of Q-rich regions on 103Q-GFP–mediated toxicity. (A) Distribution of glutamine residues in Q-rich proteins. Schematic representation of Q and N distribution in the sequences of Q-rich constructs used in this study. N residues, cyan; Q residues, red. Sequences of truncation constructs used in B and C are indicated by black lines. (B) The Q-rich regions of Gts1p and Nab3p alone are sufficient to suppress 103Q-mediated toxicity. Full-length (FL) and truncated Q-rich regions (Q) of GTS1 and NAB3 were cloned in pCM190; MCM1 FL and MCM1 Q were cloned under the control of the MCM1 promoter in pRS426. Growth tests were performed as described in Fig. 1C. (C) Compositional bias toward glutamines is not sufficient to suppress 103Q-GFP toxicity. The Q-rich proteins Taf12p and Yck1p, as well as the Q-rich N terminus of Sup35p (S35N) were cloned under the control of a tetracycline repressible promoter in pCM190. Growth tests were performed as described in Fig. 1C.
Q-Rich Domains of Gts1p and Nab3p Are Sufficient to Suppress polyQ Toxicity.
The finding that three of the identified suppressors contain uninterrupted polyQ repeats between 9 and 16 residues in length, as well as discontinuous polyQ segments, led us to investigate whether the Q-rich domains of Gts1p, Nab3p, and Mcm1p are sufficient for suppression (Fig. 2A). Expression of the Q-rich C-terminal regions of Gts1p or Nab3p was sufficient to rescue cell growth to a similar extent as expression of the full-length proteins. In contrast, the corresponding construct of Mcm1p failed to suppress (Fig. 2B). Similarly, the Q-rich prion-like proteins Yck1p and Taf12p (21), as well as the prion domain of Sup35p (S35N) (Fig. 2A), also failed to suppress 103Q-GFP toxicity, at least at the expression levels tested (Fig. 2C). Thus, specific sequence features beyond a compositional bias toward glutamine or the presence of a candidate prion domain must be required for suppression of polyQ toxicity.
The Q-rich domains of Nab3p and Gts1p comprise less than 25% of the sequence of the full-length proteins and contain neither the RNA recognition motif of Nab3p nor the Zn-finger and UBA domain of Gts1p. Thus, it seemed likely that these domains suppress the toxicity of 103Q-GFP by a mechanism unrelated to the biological functions of the full-length proteins. In the remainder of this study, we used Gts1p and its C-terminal region (Gts1Q) as a model to investigate this mechanism.
Gts1p Expression Does Not Change the Prion Status of the Prion Protein Rnq1p.
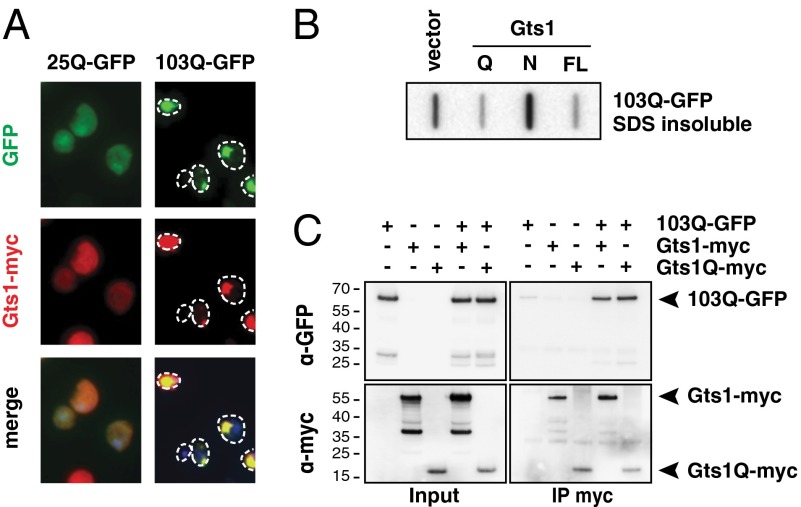
103Q-GFP toxicity can only be observed in yeast cells when Rnq1p is present in the aggregated prion form [RNQ+] (19). Curing the [RNQ+] status will suppress 103Q-GFP–mediated toxicity, and indeed two previously identified loss-of-function suppressors, rnq1Δ and hsp104Δ, cause the loss of [RNQ+] (28). Heat shock protein 104 levels have been reported to be altered by Gts1p expression (41), which might influence the Rnq1p prion status and subsequently reduce 103Q-GFP toxicity. To address this possibility, we investigated whether overexpression of Gts1p alters the prion status of Rnq1p, using Rnq1-GFP as a reporter protein (42). We found that expression of Gts1p neither cured the [RNQ+] prion nor did it induce [RNQ+] in cells that had previously been cured by treatment with guanidinium chloride (GdmCl) (Fig. S1A). This conclusion is supported further by the observation that 103Q-GFP remains localized in fluorescent foci when Gts1p is overexpressed (Fig. 3A and Fig. S1B). In contrast, cells cured of the [RNQ+] prion by GdmCl treatment showed nearly no visible 103Q-GFP foci (Fig. S1B) (19).
Fig. 3.
Gts1p interacts with 103Q-GFP and modulates its aggregation status. (A) 103Q-GFP and Gts1-myc colocalize in vivo. YPH499 was transformed with plasmids encoding 25Q/103Q-GFP and Gts1-myc. 25Q/103Q-GFP localization is observed via the fluorescent GFP tag (green). Gts1-myc is observed via indirect fluorescence (red). (B) Gts1p expression reduces the amount of SDS insoluble 103Q-GFP. Full-length Gts1p (FL), an N-terminal fragment (N), or a fragment containing the Q-rich C terminus (Q) were preinduced before induction of 103Q-GFP. Cells were lysed and analyzed for SDS insoluble 103Q-GFP by filter retardation assay. (C) Gts1-myc and 103Q-GFP physically interact. YPH499 was transformed with plasmids encoding 103Q-GFP and Gts1-myc or Gts1Q-myc. After lysis, anti-myc antibodies were used for immunoprecipitation (IP), followed by SDS/PAGE and Western blotting using anti-GFP or anti-myc antibodies.
Gts1p Expression Changes the Aggregation Status of 103Q-GFP.
The finding that 103Q-GFP is still present in visible foci on Gts1p overexpression does not exclude the possibility that the presence of Gts1p modifies biophysical properties of the aggregates. The polyQ inclusions that are virtually indistinguishable by light microscopy may nevertheless differ in their physical properties: they may be detergent (SDS) insoluble, containing ordered polyQ fibrils, or detergent soluble, containing structurally amorphous aggregates (15, 43). Using an established filter trap assay for the detection of SDS insoluble aggregates larger than 0.2 μm (44), we found that expression of Gts1p or Gts1Q strongly reduced the amount of SDS insoluble 103Q-GFP present in cells, whereas expression of the N-terminal Gts1p fragment was without effect (Fig. 3B). Thus, Gts1p shifts the aggregation properties of the protein from SDS insoluble to SDS soluble aggregate species that still form visible inclusions (Fig. 3A), without reducing the amount of 103Q-GFP that can be detected by gel electrophoresis (Fig. 3C, input panel).
To demonstrate that Gts1p interacts directly with 103Q-GFP, we added a myc-tag to the N terminus of Gts1p and Gts1Q (Gts1-myc and Gts1Q-myc) to enable detection. Addition of this tag did not change the Gts1p-mediated rescue of 103Q-GFP toxicity (Fig. S2). Although Gts1-myc was diffusely distributed in cells expressing 25Q-GFP (Fig. 3A), in 103Q-GFP expressing cells, Gts1-myc was present in bright fluorescent foci colocalizing with 103Q-GFP. Coimmunoprecipitation experiments confirmed the association of Gts1-myc and Gts1Q-myc with 103Q-GFP (Fig. 3C), but showed only little association with 25Q-GFP (Fig. S3).
Molecular Modeling of Gts1p–polyQ Interactions.
Further evidence for direct interactions between the QA-repeat region of Gts1p and the polyQ tract of Htt was obtained by performing atomistic Monte Carlo simulations using the CAMPARI simulation engine (campari.sourceforge.net) and ABSINTH implicit solvation model and forcefield paradigm (45). This approach has recently been used to generate accurate insights regarding the early, nonspecific steps that lead to polyQ aggregation (46, 47) and their modulation by sequences flanking the polyQ tract within Htt (48).
We performed simulations with a polyQ sequence of 35 Q and the imperfect 28 residue QA-region denoted as QAR of Gts1p, present either at a ratio of two molecules of QAR to one molecule of 35Q (a continuous polyQ repeat of 35 Q) or the reverse ratio. The choice of a 35 residue polyQ molecule was made because this allows for simulations that afford reliable and reproducible statistics, and as shown in previous studies, the simulation results are unlikely to vary by increasing or decreasing the polyQ length by five residues vis-à-vis 35Q (46, 47).
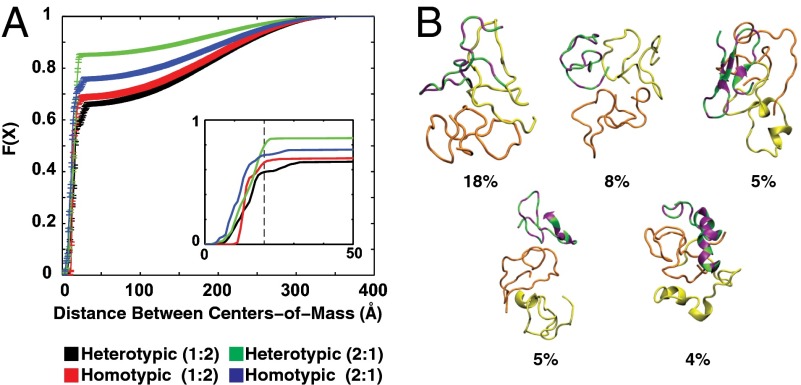
Quantifying the statistics for the occurrence of specific distances of separation between the centers of mass of pairs of molecules provided a measure of interaction strength that can be converted into estimates of dissociation constants. We used cumulative distribution functions (CDFs) to quantify the probability that a pair of molecules will assume a specific intermolecular separation at a temperature of 315 K. According to these CDFs, in simulations with a QAR:35Q stoichiometry of 1:2, we find that the probability of heterotypic (QAR-35Q) interactions is equivalent to the probability of homotypic interactions between 35Q molecules (Fig. 4A). This result suggests that QAR competes just as effectively for interactions with polyQ as do other polyQ molecules: a finding that is borne out further in simulations with a QAR:35Q stoichiometry of 2:1. Here, we find that QAR molecules prefer interactions with the surfaces of polyQ molecules even in the presence of an additional QAR molecule. Representative conformations drawn from simulations with a QAR:35Q stoichiometry of 1:2 demonstrate the range of interaction modes that are available for QAR to cover the surface of polyQ molecules (Fig. 4B). It is worth noting that the ensemble is characterized by considerable structural heterogeneity implying that 35Q and QAR molecules can associate while assuming a range of different conformations. This finding provides a rationale for the observation that Gts1p arrests the aggregation of polyQ molecules at the level of SDS soluble, nonfibrillar species. Taken together, the simulations suggest that QAR is a trans-acting cap of polyQ associations that destabilizes intermediates en route to fibrillar species. Interactions of polyQ with QAR presumably generate SDS soluble oligomers with less reactive surfaces. The ability of QA repeats to compete for the surface of polyQ globules within these simulations, in contrast to homotypic interactions between QAR molecules, is supported by the finding that trimers are the dominant species in simulations with a QAR:35Q ratio of 2:1, whereas trimers make up only 50% of the population when the QAR:35Q ratio is 1:2. Importantly, homodimers are prominent when the QAR:35Q ratio is 1:2, whereas heterodimers are preferred over homodimers when the QAR:35Q ratio is 2:1 (Fig. S4). A more extensive analysis of details that emerge from the simulations coupled with in-depth biophysical investigations along the lines of recent efforts (49) is beyond the scope of the current study and will be published elsewhere.
Fig. 4.
Molecular modeling of Gts1p-polyQ interactions. (A) Cumulative distribution functions quantifying the probabilities for homotypic vs. heterotypic intermolecular interactions. The abscissa denotes the distance (in Angstroms) between the centers of mass of a pair of molecules. The ordinate F(X) is the cumulative distribution function (CDF). For a given value of X, F(X) quantifies the probability that a specific intermolecular separation will be less than or equal to X. The black and red curves were generated using statistics from simulations with one molecule containing the imperfect glutamine-alanine repeat of Gts1p (QAR) and two 35Q molecules, respectively. The green and blue curves were generated using statistics from simulations with two QAR molecules and one 35Q molecule. (Inset) Zoomed in version of the CDFs to illustrate the quantitative differences and similarities among different curves. It also shows that the distances between the centers of mass are equivalent when each of the F(X) curves reaches their respective plateau value. This result is true irrespective of the stoichiometry in the simulations and the type of interactions, heterotypic vs. homotypic, that are being analyzed. (B) Representative conformations of trimers formed in simulations with a QAR:35Q ratio of 1:2. The molecules are drawn using backbone traces. The 35Q molecules are shown in orange and yellow, respectively, whereas the QAR molecule is colored using green for the alanine (A)/valine (V) residues and purple for the Q residues. The conformations represent the centers of distinct conformational clusters. Below each representative conformation we show the percent probability of sampling the conformational type within ensembles generated at 315 K. Details of the move sets utilized for the simulations analyzed in A and B can be found in Table S1.
Gts1p Alters the Interaction Properties of 103Q-GFP.
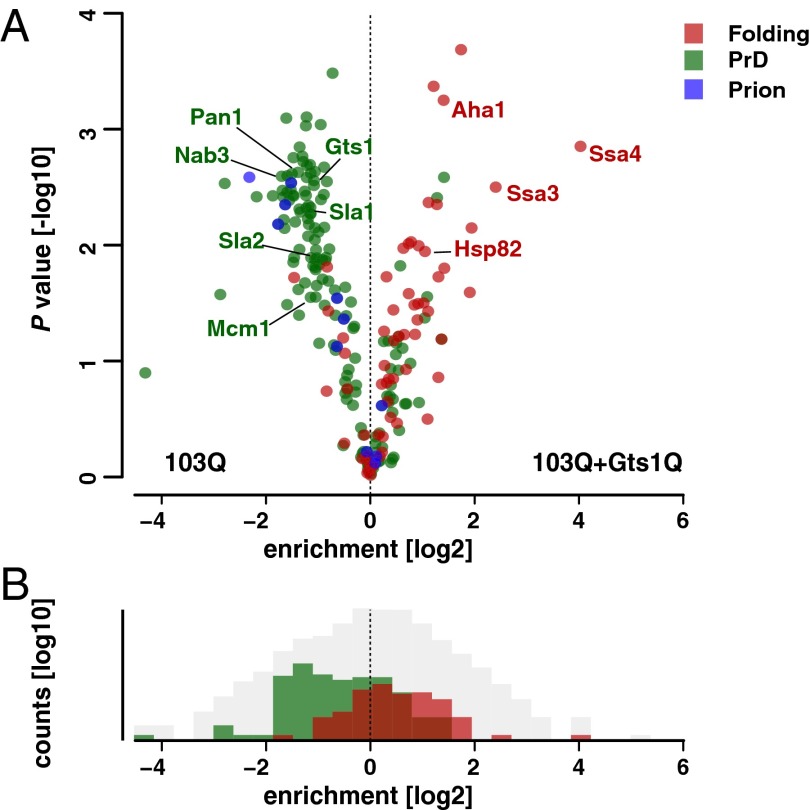
PolyQ proteins are widely thought to exert toxicity by mediating aberrant interactions with other proteins, resulting in their functional impairment and sequestration (7, 8, 32, 50). Based on the finding that Gts1p coaggregates with 103Q and consistent with the results of the atomistic simulations, it seemed plausible that Gts1p and Gts1Q suppress polyQ toxicity by interfering with such aberrant interactions. To investigate this possibility, we applied MS-based proteomics. We established a quantitative, label-free workflow that allows an unbiased comparison of multiple samples (25, 26). Using these high accuracy proteomics tools, we quantified the interactome of 103Q-GFP in the absence and presence of Gts1Q expression. Proteins associated with 103Q-GFP were isolated using an anti-GFP antibody coupled to magnetic beads, followed by LC-MS analysis. Interestingly, the group of 103Q-GFP interactors that was most predominantly reduced by expression of Gts1Q (P = 1.28 × 10−30) are known yeast prions or proteins containing PrDs (Fig. 5 and Dataset S1) (21). Sequestration of the PrD containing proteins Sla1p, Sla2p, and Pan1p into aggregates has been suggested to elicit polyQ toxicity by seeding the formation of toxic aggregates (51, 52). Thus, the reduced interaction of these proteins with 103Q-GFP in the presence of Gts1Q can contribute to the suppression of 103Q-GFP toxicity. It is interesting to note that expression of Gts1Q also reduces the interaction of 103Q-GFP with the three prion-like suppressors identified in our screen (Nab3p, Mcm1p, and endogenous Gts1p), thereby also reversing effects possibly due to functional depletion of these proteins, as has been suggested for Nab3p (35).
Fig. 5.
Gts1Q changes the 103Q-GFP interactome. (A) Gts1Q induced alteration of 103Q-GFP interactors. The comparison of P value and difference of means from t test statistics shows proteins that are enriched, unchanged, or depleted in the comparison of the two interactomes. Interactors involved in protein folding are labeled red, interactors with a predicted prion domain (PrD) are shown in green, and proteins annotated as prions in blue. (B) Gts1Q expression increases the number of 103Q-GFP interactors involved in protein folding and decreases the number of interactors with a PrD. The binned distribution of proteins in the pairwise comparison of interactomes shows converse shifts for proteins with a predicted PrD and those involved in protein folding. Interactors involved in protein folding are labeled red, interactors with a predicted prion domain (PrD) are shown in green, and all other identified proteins are shown in gray.
More than half of the proteins reduced in abundance on 103Q-GFP are nuclear proteins. Many are involved in transcription and RNA metabolic processes, suggesting that interference with these functions contributes to polyQ toxicity. Among the proteins that show an increased interaction with 103Q-GFP in the presence of Gts1Q are several chaperone proteins such as the Hsp70s Ssa3p and Ssa4p, the Hsp90 Hsp82p, and its cochaperone Aha1p. However, there was no change in the interaction of 103Q-GFP with Sis1p, which is the only chaperone identified in our suppressor screen. Comparable results were achieved by overexpression of full-length Gts1p (Fig. S5 and Dataset S1). In this case, the function of Gts1p as transcription factor may alter the composition of the proteome in a manner unrelated to the mechanism described here. The use of Gts1p instead of Gts1Q, for which no tryptic peptides can be detected, allows us to calculate the relative abundances of 103Q-GFP and Gts1p in the interactome. As expected, Gts1p and 103Q-GFP are among the most abundant proteins quantified. The similarity of the normalized MaxLFQ values for Gts1p and 103Q-GFP is consistent with the formation of a 1:1 stoichiometric complex (Fig. S5C).
The displacement of functionally critical nuclear components from the polyQ aggregates by Gts1p in combination with the more effective shielding of the aggregates by molecular chaperones can explain how Gts1p suppresses polyQ toxicity. It is also possible that the reduced interaction of the polyQ aggregates with known prions and PrD containing proteins contributes to the suppression of toxicity.
Discussion
Neutralizing the toxic effects of protein aggregates is a rational strategy in the development of treatments for various neurodegenerative disorders, including Huntington’s, Parkinson’s, and Alzheimer’s diseases. In this study, we performed a yeast screen for suppressors of the toxicity of polyQ protein aggregation without making assumptions about the nature of the toxic aggregate species. Among the six suppressors identified were one Hsp40 chaperone, Sis1p, and three proteins that contain C-terminal domains with interrupted polyQ repeats and a compositional bias toward glutamine (Fig. 2A). Interestingly, the C-terminal regions of these proteins were previously predicted to contain PrDs (21). For two of the proteins, Gts1p and Nab3p, we demonstrated that expression of the Q-rich C terminus alone is sufficient to prevent polyQ mediated toxicity (Fig. 2B). Thus, these proteins use some shared property of their Q-rich domains to modulate polyQ toxicity, apparently by interacting directly with the polyQ sequence of Htt. Association with the suppressor did not prevent 103Q-GFP aggregation but altered the physical properties of the aggregates. As a result, the association of molecular chaperones of the Hsp70 and Hsp90 systems with the polyQ aggregates was enhanced, and their ability to sequester nuclear proteins was strongly reduced. Protein sequestration into aggregates may lead to an impairment of multiple key cellular pathways and is considered an important mechanism of aggregate toxicity (7, 9, 10, 32, 50, 53–58).
The observed ability of Gts1p and Nab3p to suppress polyQ toxicity was not shared by all proteins containing a PrD (Fig. 2C) (30), suggesting that it is based on specific sequence properties of the Q-rich domain and its context with flanking sequences. Our observation that the three PrDs of the suppressors identified in our screen are not enriched for N is in agreement with a recent study that demonstrates that the relative Q and N content of overexpressed PrDs is critical for their ability to suppress polyQ-mediated toxicity (30). Our molecular simulations indicated that the QA stretch might interact with early polyQ intermediates, thereby modulating their surface properties and preventing the formation of large fibrillar aggregates.
Expression of Gts1p markedly reduced the association of yeast prion proteins with the polyQ aggregates. The prion status has been shown to be required for the efficient aggregation of polyQ expansion proteins in yeast, and it has been suggested that the prion aggregates function by seeding toxic polyQ aggregation (18, 19). It seems plausible that Gts1p competes with the yeast prions for interaction with polyQ sequences, thereby shifting the aggregation pathway toward formation of more benign aggregates. Two recent screens suggest that expression of a subset of PrD containing proteins additionally modulates the spatial organization of 103Q-GFP foci (30, 35). At the same time, Gts1p enhances the association of polyQ aggregates with chaperones, which may then cooperate with Gts1p in shielding potentially dangerous surfaces of the polyQ aggregates. Indeed, Gts1Q also reduced the interaction of 103Q-GFP with PrD-containing proteins. A functional depletion of several of these proteins by sequestration into the aggregates has been invoked as a cause of polyQ toxicity (35, 51, 52).
The striking effect of Gts1p in reducing polyQ toxicity may suggest a biological role for proteins with similar QA-repeat sequence motifs in mitigating potentially toxic polyQ interactions, comparable to what has been described for dominant prion mutants (59). Although there is no direct mammalian homolog of Gts1p, the human transcription elongation regulator TCERG1/CA150 also contains a long QA-repeat. Interestingly, TCERG1/CA150 was reported to bind to Htt (60), and overexpression of this transcription factor was shown to protect striatal cells from mutant Htt toxicity in a QA-repeat–dependent manner (61). Expression of peptides with a specific Q-rich region, like the QA-repeat of Gts1p or TCERG1/CA150, might present a strategy to interfere with Htt toxicity. However, regions with QA-repeats may themselves tend to aggregate, as reflected by the aggregation propensity of Gts1p (21, 62) and Cyc8p, another yeast prion with a QA-repeat (63). Indeed, expansion of the QA-repeat of TCERG1/CA150 can lead to an earlier onset of HD (60), presumably by placing a burden on the cellular proteostasis system (10). Nevertheless, controlled expression of peptides containing specific Q-rich domains may be a way to influence Htt toxicity. Future experiments will have to show whether specific interactions between short peptides containing regions similar to the ones discovered in this study can prevent the formation of toxic aggregates and whether this approach can be developed to benefit patients.
Supplementary Material
Acknowledgments
We thank R. Wedlich-Söldner for help with light microscopy and Kristina Weber for assistance during the initial phases of this project. The research leading to these results has received funding from the European Commission under Grant FP7 GA ERC-2012-SyG_318987–ToPAG (to R.K., D.H., M.M., F.U.H., and M.S.H.), the Munich Cluster for Systems Neurology (F.U.H. and M.S.H.), the European Molecular Biology Organization (L.R.), and National Institutes of Health Grant 5R01NS056114 (to R.V.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421313111/-/DCSupplemental.
References
- 1.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 2.Duyao M, et al. Trinucleotide repeat length instability and age of onset in Huntington’s disease. Nat Genet. 1993;4(4):387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- 3.Gusella JF, MacDonald ME. Molecular genetics: Unmasking polyglutamine triggers in neurodegenerative disease. Nat Rev Neurosci. 2000;1(2):109–115. doi: 10.1038/35039051. [DOI] [PubMed] [Google Scholar]
- 4.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: New insights into protein folding, misfolding diseases and biological evolution. J Mol Med (Berl) 2003;81(11):678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 5.Winklhofer KF, Tatzelt J, Haass C. The two faces of protein misfolding: Gain- and loss-of-function in neurodegenerative diseases. EMBO J. 2008;27(2):336–349. doi: 10.1038/sj.emboj.7601930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross CA, Poirier MA. Opinion: What is the role of protein aggregation in neurodegeneration? Nat Rev Mol Cell Biol. 2005;6(11):891–898. doi: 10.1038/nrm1742. [DOI] [PubMed] [Google Scholar]
- 7.Perez MK, et al. Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J Cell Biol. 1998;143(6):1457–1470. doi: 10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 9.Olzscha H, et al. Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell. 2011;144(1):67–78. doi: 10.1016/j.cell.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 10.Hipp MS, Park SH, Hartl FU. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 2014;24(9):506–514. doi: 10.1016/j.tcb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Gidalevitz T, Kikis EA, Morimoto RI. A cellular perspective on conformational disease: The role of genetic background and proteostasis networks. Curr Opin Struct Biol. 2010;20(1):23–32. doi: 10.1016/j.sbi.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Biological and chemical approaches to diseases of proteostasis deficiency. Annu Rev Biochem. 2009;78:959–991. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- 13.Holmes WM, Klaips CL, Serio TR. Defining the limits: Protein aggregation and toxicity in vivo. Crit Rev Biochem Mol Biol. 2014;49(4):294–303. doi: 10.3109/10409238.2014.914151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krobitsch S, Lindquist S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci USA. 2000;97(4):1589–1594. doi: 10.1073/pnas.97.4.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muchowski PJ, et al. Hsp70 and hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA. 2000;97(14):7841–7846. doi: 10.1073/pnas.140202897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duennwald ML, Jagadish S, Muchowski PJ, Lindquist S. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci USA. 2006;103(29):11045–11050. doi: 10.1073/pnas.0604547103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wickner RB. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264(5158):566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 18.Duennwald ML, Jagadish S, Giorgini F, Muchowski PJ, Lindquist S. A network of protein interactions determines polyglutamine toxicity. Proc Natl Acad Sci USA. 2006;103(29):11051–11056. doi: 10.1073/pnas.0604548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meriin AB, et al. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol. 2002;157(6):997–1004. doi: 10.1083/jcb.200112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc Natl Acad Sci USA. 2000;97(22):11910–11915. doi: 10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137(1):146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nasmyth KA, Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- 23.Nasmyth KA, Reed SI. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci USA. 1980;77(4):2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gietz RD, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 25.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 26.Cox J, et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willingham S, Outeiro TF, DeVit MJ, Lindquist SL, Muchowski PJ. Yeast genes that enhance the toxicity of a mutant huntingtin fragment or alpha-synuclein. Science. 2003;302(5651):1769–1772. doi: 10.1126/science.1090389. [DOI] [PubMed] [Google Scholar]
- 28.Giorgini F, Guidetti P, Nguyen Q, Bennett SC, Muchowski PJ. A genomic screen in yeast implicates kynurenine 3-monooxygenase as a therapeutic target for Huntington disease. Nat Genet. 2005;37(5):526–531. doi: 10.1038/ng1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mason RP, et al. Glutathione peroxidase activity is neuroprotective in models of Huntington’s disease. Nat Genet. 2013;45(10):1249–1254. doi: 10.1038/ng.2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kayatekin C, et al. Prion-like proteins sequester and suppress the toxicity of huntingtin exon 1. Proc Natl Acad Sci USA. 2014;111(33):12085–12090. doi: 10.1073/pnas.1412504111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Douglas PM, Summers DW, Ren HY, Cyr DM. Reciprocal efficiency of RNQ1 and polyglutamine detoxification in the cytosol and nucleus. Mol Biol Cell. 2009;20(19):4162–4173. doi: 10.1091/mbc.E09-02-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park SH, et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154(1):134–145. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Ward MP, Garrett S. Suppression of a yeast cyclic AMP-dependent protein kinase defect by overexpression of SOK1, a yeast gene exhibiting sequence similarity to a developmentally regulated mouse gene. Mol Cell Biol. 1994;14(9):5619–5627. doi: 10.1128/mcb.14.9.5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trumbly RJ. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992;6(1):15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- 35.Wolfe KJ, Ren HY, Trepte P, Cyr DM. Polyglutamine-rich suppressors of huntingtin toxicity act upstream of Hsp70 and Sti1 in spatial quality control of amyloid-like proteins. PLoS ONE. 2014;9(5):e95914. doi: 10.1371/journal.pone.0095914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conrad NK, et al. A yeast heterogeneous nuclear ribonucleoprotein complex associated with RNA polymerase II. Genetics. 2000;154(2):557–571. doi: 10.1093/genetics/154.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elble R, Tye BK. Both activation and repression of a-mating-type-specific genes in yeast require transcription factor Mcm1. Proc Natl Acad Sci USA. 1991;88(23):10966–10970. doi: 10.1073/pnas.88.23.10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Espinet C, de la Torre MA, Aldea M, Herrero E. An efficient method to isolate yeast genes causing overexpression-mediated growth arrest. Yeast. 1995;11(1):25–32. doi: 10.1002/yea.320110104. [DOI] [PubMed] [Google Scholar]
- 39.Yaguchi S, Mitsui K, Kawabata K, Xu Z, Tsurugi K. The pleiotropic effect of the GTS1 gene product on heat tolerance, sporulation and the life span of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1996;218(1):234–237. doi: 10.1006/bbrc.1996.0041. [DOI] [PubMed] [Google Scholar]
- 40.Bossier P, Goethals P, Rodrigues-Pousada C. Constitutive flocculation in Saccharomyces cerevisiae through overexpression of the GTS1 gene, coding for a ‘Glo’-type Zn-finger-containing protein. Yeast. 1997;13(8):717–725. doi: 10.1002/(SICI)1097-0061(19970630)13:8<717::AID-YEA132>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 41.Yaguchi S, Tsurugi K. Gts1p activates SNF1-dependent derepression of HSP104 and TPS1 in the stationary phase of yeast growth. J Biol Chem. 2003;278(32):29760–29768. doi: 10.1074/jbc.M301441200. [DOI] [PubMed] [Google Scholar]
- 42.Sondheimer N, Lindquist S. Rnq1: An epigenetic modifier of protein function in yeast. Mol Cell. 2000;5(1):163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 43.Warrick JM, et al. Suppression of polyglutamine-mediated neurodegeneration in Drosophila by the molecular chaperone HSP70. Nat Genet. 1999;23(4):425–428. doi: 10.1038/70532. [DOI] [PubMed] [Google Scholar]
- 44.Wanker EE, et al. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol. 1999;309:375–386. doi: 10.1016/s0076-6879(99)09026-6. [DOI] [PubMed] [Google Scholar]
- 45.Vitalis A, Pappu RV. ABSINTH: A new continuum solvation model for simulations of polypeptides in aqueous solutions. J Comput Chem. 2009;30(5):673–699. doi: 10.1002/jcc.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vitalis A, Wang X, Pappu RV. Atomistic simulations of the effects of polyglutamine chain length and solvent quality on conformational equilibria and spontaneous homodimerization. J Mol Biol. 2008;384(1):279–297. doi: 10.1016/j.jmb.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vitalis A, Lyle N, Pappu RV. Thermodynamics of beta-sheet formation in polyglutamine. Biophys J. 2009;97(1):303–311. doi: 10.1016/j.bpj.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Williamson TE, Vitalis A, Crick SL, Pappu RV. Modulation of polyglutamine conformations and dimer formation by the N-terminus of huntingtin. J Mol Biol. 2010;396(5):1295–1309. doi: 10.1016/j.jmb.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crick SL, Ruff KM, Garai K, Frieden C, Pappu RV. Unmasking the roles of N- and C-terminal flanking sequences from exon 1 of huntingtin as modulators of polyglutamine aggregation. Proc Natl Acad Sci USA. 2013;110(50):20075–20080. doi: 10.1073/pnas.1320626110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schaffar G, et al. Cellular toxicity of polyglutamine expansion proteins: Mechanism of transcription factor deactivation. Mol Cell. 2004;15(1):95–105. doi: 10.1016/j.molcel.2004.06.029. [DOI] [PubMed] [Google Scholar]
- 51.Meriin AB, et al. Aggregation of expanded polyglutamine domain in yeast leads to defects in endocytosis. Mol Cell Biol. 2003;23(21):7554–7565. doi: 10.1128/MCB.23.21.7554-7565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meriin AB, et al. Endocytosis machinery is involved in aggregation of proteins with expanded polyglutamine domains. FASEB J. 2007;21(8):1915–1925. doi: 10.1096/fj.06-6878com. [DOI] [PubMed] [Google Scholar]
- 53.Jana NR, Zemskov EA, Wang Gh, Nukina N. Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum Mol Genet. 2001;10(10):1049–1059. doi: 10.1093/hmg/10.10.1049. [DOI] [PubMed] [Google Scholar]
- 54.Yamanaka T, et al. Mutant Huntingtin reduces HSP70 expression through the sequestration of NF-Y transcription factor. EMBO J. 2008;27(6):827–839. doi: 10.1038/emboj.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao X, et al. Sequestration of Sup35 by aggregates of huntingtin fragments causes toxicity of [PSI+] yeast. J Biol Chem. 2012;287(28):23346–23355. doi: 10.1074/jbc.M111.287748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steffan JS, et al. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci USA. 2000;97(12):6763–6768. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gong H, et al. Polyglutamine toxicity is controlled by prion composition and gene dosage in yeast. PLoS Genet. 2012;8(4):e1002634. doi: 10.1371/journal.pgen.1002634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kochneva-Pervukhova NV, Alexandrov AI, Ter-Avanesyan MD. Amyloid-mediated sequestration of essential proteins contributes to mutant huntingtin toxicity in yeast. PLoS ONE. 2012;7(1):e29832. doi: 10.1371/journal.pone.0029832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DiSalvo S, Derdowski A, Pezza JA, Serio TR. Dominant prion mutants induce curing through pathways that promote chaperone-mediated disaggregation. Nat Struct Mol Biol. 2011;18(4):486–492. doi: 10.1038/nsmb.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holbert S, et al. The Gln-Ala repeat transcriptional activator CA150 interacts with huntingtin: Neuropathologic and genetic evidence for a role in Huntington’s disease pathogenesis. Proc Natl Acad Sci USA. 2001;98(4):1811–1816. doi: 10.1073/pnas.041566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Arango M, et al. CA150 expression delays striatal cell death in overexpression and knock-in conditions for mutant huntingtin neurotoxicity. J Neurosci. 2006;26(17):4649–4659. doi: 10.1523/JNEUROSCI.5409-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanada M, Kuroda K, Ueda M. GTS1 induction causes derepression of Tup1-Cyc8-repressing genes and chromatin remodeling through the interaction of Gts1p with Cyc8p. Biosci Biotechnol Biochem. 2011;75(4):740–747. doi: 10.1271/bbb.100860. [DOI] [PubMed] [Google Scholar]
- 63.Patel BK, Gavin-Smyth J, Liebman SW. The yeast global transcriptional co-repressor protein Cyc8 can propagate as a prion. Nat Cell Biol. 2009;11(3):344–349. doi: 10.1038/ncb1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.