Abstract
Purpose
The purpose of this study is to describe the Gannet toolkit for the quantitative batch analysis of gamma-aminobutyric acid (GABA) -edited MRS data.
Materials and Methods
Using MEGA-PRESS editing and standard acquisition parameters, four MEGA-PRESS spectra were acquired in three brain regions in 10 healthy volunteers. These 120 datasets were processed without user intervention with Gannet, a Matlab-based tool that takes raw time-domain data input, processes it to generate the frequency-domain edited spectrum, and applies a simple modeling procedure to estimate GABA concentration relative to the creatine or, if provided, the unsuppressed water signal. A comparison of four modeling approaches is also presented.
Results
All data were successfully processed by Gannet. Coefficients of variation across subjects ranged from 11% for the occipital region to 17% for the dorsolateral prefrontal region. There was no clear difference in fitting performance between the simple Gaussian model used by Gannet and the other more complex models presented.
Conclusion
Gannet, the GABA Analysis Toolkit, can be used to process and quantify GABA-edited MRS spectra without user intervention.
Keywords: GABA, editing, magnetic resonance spectroscopy, edited MRS, quantification, MEGA-PRESS
Gamma-aminobutyric acid (GABA), the principal inhibitory neurotransmitter in the human brain, is the focus of widespread interest in the clinical and basic neuroscience community. It is possible to non-invasively detect GABA using in vivo magnetic resonance spectroscopy (MRS) (1,2), and this has been used to investigate the role of GABA in healthy brain function (3–5) and pathological changes in GABA concentration in a range of neurological (6–8), psychiatric (9–15), and developmental disorders (16–18).
GABA signals are present in the MR spectrum at 3.0 ppm, 2.3 ppm, and 1.9 ppm (19), but these are heavily overlapped by signals from more concentrated metabolites such as creatine (Cr), glutamate (Glu), and N-Acetyl Aspartate (NAA). Amongst the numerous methods that have been suggested for separating GABA signals from the larger, overlapping signals in the in vivo spectrum (for a review see Puts and Edden) (2), J-difference editing (20) has emerged as the most widely used, largely due to the ease of incorporating MEGA-editing (21) into the PRESS sequence.
J-difference editing involves the subtraction of two experiments, which differ in their treatment of the GABA spin system. In one, commonly referred to as the ON experiment, frequency-selective editing pulses are applied to GABA spins at 1.9 ppm to refocus evolution of their coupling to the spins of interest at 3 ppm. In the other, commonly referred to as the OFF experiment, no frequency-selective editing pulses are applied (or if applied, at a different frequency) and the coupling evolves for the duration of the echo time. The key result is that the shape of the 3 ppm multiplet is different in those two cases (whereas overlying creatine signals remain the same) and therefore the subtraction removes Cr signals whilst preserving a measurable GABA signal. This difference-editing approach results in a GABA signal that is contaminated by homocarnosine, a GABA derivative, and usually co-edited macromolecular signal (22,23) and that is often referred to as GABA+ for this reason.
Several approaches have been used for the quantitative processing of GABA-edited spectra, including LCModel (24), jMRUI (25), and TARQUIN (26). In this study, we present our processing pipeline ‘Gannet’ (GABA-MRS Analysis Tool), which differs from other approaches in several ways, including: (i) Gannet is specifically targeted for the analysis of GABA-edited MRS data. Tools that were originally designed for the analysis of short-echo-time single-voxel data have a different set of underlying assumptions, for example regarding the nature of the baseline, and the treatment of signals from macromolecules and other metabolites. (ii) Gannet is specifically intended as a tool for the batch analysis of whole datasets with minimum user intervention. (iii) Gannet is coded within Matlab (The Mathworks, Natick, USA), using Optimization and Statistics toolboxes, and is distributed open-source.
MATERIALS AND METHODS
Data Types Supported By Gannet
Gannet is able to process GABA-edited data acquired on each of the three main vendor scanners: GE P-files; Philips .sdat files; Philips .data files; and Siemens .rda files. In so far as is possible, the processing of these datatypes is the same; however there are differences in the degree of preprocessing that is applied before data export, necessitating some vendor-specific steps. Table 1 summarizes the preprocessing steps applied as part of the vendor data export for each data type. The degree of time averaging already applied to exported data varies from none (Philips .data) to full (Siemens .rda), with GE P-files and Philips .sdat files averaging within each phase cycle (Phase-cycle averaging ✓ in Table 1), but storing each phase cycle separately (Time-series averaging × in Table 1).
Table 1.
Summary of Preprocessing Steps Applied During Data Export (Prior to Gannet Processing) for Each Supported Datatype
| GE p-file | Philips .sdat | Philips .data | Siemens .rda | |
|---|---|---|---|---|
| Phased-array channel combination | × | ✓ | ✓ | ✓ |
| Phase-cycle averaging | ✓ | ✓ | × | ✓ |
| Time-series averaging | × | × | × | ✓ |
None of these data types explicitly contain information as to which lines of data correspond to OFF or ON scans with respect to the editing pulse frequency, and therefore assumptions must be made. In general, Gannet assumes that data are stored in the order they are acquired, and that ON-OFF interleaving occurs external to the phase cycling loop used. Thus for GE P-files and Philips .sdat data, the exported time-domain data lines are alternately OFF and ON scans (after phase cycle averaging, if present). The Philips .data file format exports each FID separately, and thus Gannet expects n OFF scans followed by n ON scans followed by n OFF scans etc, where n is the length of the phase cycle used. The Siemens .rda file read is set up to handle the output of Siemens “works-in-progress” (WIP) distribution, which exports fully time-averaged OFF and ON scans, as well as their difference.
Overview of Gannet Modules
Gannet consists of two main modules: GannetLoad, which imports time-domain data from the scanner and processes it into a frequency-domain GABA-edited spectrum; and GannetFit, which uses nonlinear least-squares fitting to integrate the edited GABA peak at 3 ppm and produce GABA concentration estimates. Practically, these two modules are run sequentially from the command-line. First, GannetLoad is called with a list (cell array) of data filenames as the input, creating a single data structure as the output. GannetFit is then called with this structure as the input. There are two levels of output from each module—the data structure itself (described fully in Supp. Table S1, which is available online) and pdf-format summary files.
GannetLoad
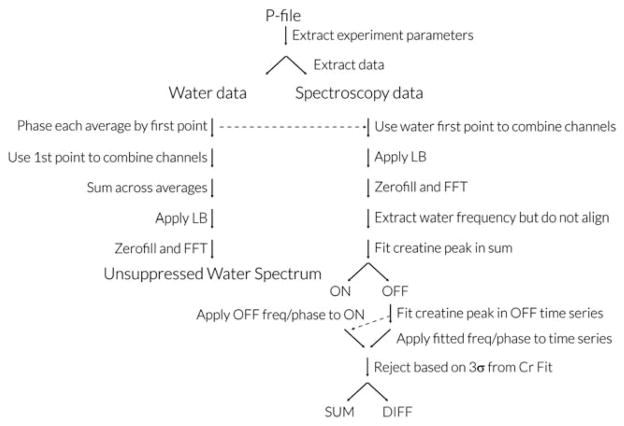
GannetLoad proceeds by parsing certain descriptive variables from the data headers, including the data dimensions, and then reading the time-domain data. The subsequent processing steps are summarized in Figure 1. If unsuppressed water data are available, they are also read in and processed in a similar manner. The main processing steps are as follows:
Figure 1.
Gannet Processing pipeline for GE p-file data. Processing for Philips .sdat and .data files proceeds along similar lines, except in both cases header information is contained in a separate parameter file, .spar and .list, respectively, and phased-array channel combination is performed automatically before data export. Siemens data are exported as time-averaged, time-domain data, so fewer steps are applicable.
1. Phased-Array Channel Combination
The combination of phased-array data is performed using a “signal-weighted” approach (27) based on the first-point of time domain data. If the time domain signal for the kth phased-array channel is expressed as S(t1,t2,k), where t2 represents the acquisition dimension and t1 the time-averaging dimension, channel combination is performed according to:
where K is the number of phased array coils to be combined and the overbar denotes the complex conjugate.
2. Exponential line broadening
The acquisition dimension t2 can be expressed in terms of the jth datapoint divided by the spectral width SW, thus exponential line broadening LB (in Hz) can be applied according to: S(t1,t2) → S(t1,t2) exp(−j π LB/SW), where j is the square root of −1. A default line broadening value of LB = 3 Hz is used.
3. Fourier Transformation
A fast Fourier transformation is applied to the t2 dimension to give an array of time-resolve frequency-domain spectra with zero-filling up to 32k datapoints:
4. Frequency and Phase Correction
Frequency and phase correction of the individual frequency domain spectra is important to maximize the quality of the edited spectrum, particularly for removing subtraction artifacts related to subject movement and scanner drift (28). It is achieved by modeling the creatine signal in the real part of each spectrum with a Lorentzian model (28) (shown below in Eq. (1)), to determine frequency and phase parameters for the correction, with the modification that modeling is only done for the OFF spectra so that the correction does not itself cause a subtraction artifact. Correction parameters from the prior OFF spectrum are applied to each ON spectrum in a pairwise manner (29). Imperfect/incorrect frequency and phase correction results in subtraction artifacts from the choline and creatine signals at 3.2 ppm and 3.0 ppm, respectively. The choline artifact can be used to judge quality of correction, as it occurs in a region of the edited spectrum that should not contain any signals (as highlighted in Figures 2 and 3 of Evans et al (29)), whereas the creatine subtraction overlaps with the edited GABA signal and potentially interferes with quantification.
5. Outlier rejection
Time-resolved spectra are excluded before time averaging on the basis of being greater than three standard deviations from the mean in either the frequency, phase, area or full-width at half maximum (FWHM) of the creatine peak (28). Rejections are applied in a pairwise manner, ie, if an OFF scan meets rejection criteria the subsequent ON scan is also rejected to maintain balance for subtraction. Frequency/phase correction and outlier rejection are not applied to unsuppressed water data (although a phase correction is applied to the sum of the unsuppressed data).
6. Time averaging
Once frequency and phase correction and outlier rejection have been applied, individual dynamics are added to generate, for each dataset, the edited difference spectrum, and the editing OFF spectrum (for Cr peak integration, in which the GABA signal integral is zero).
Variables Stored by GannetLoad
The raw and processed data and several variables descriptive of the experiment are stored in the Gannet-Load output structure. GannetFit subsequently adds several new attributes to the structure. The attributes of this structure are described more fully in Table S1.
GannetFit
Through nonlinear least-squares fitting of the spectra, GannetFit estimates the area under the edited GABA signal at 3 ppm, the Cr signal at 3 ppm and, if provided, the unsuppressed water signal from the same volume. Quantitative results are then presented as the integral ratio between GABA and Cr, and a concentration in “institutional units” relative to water.
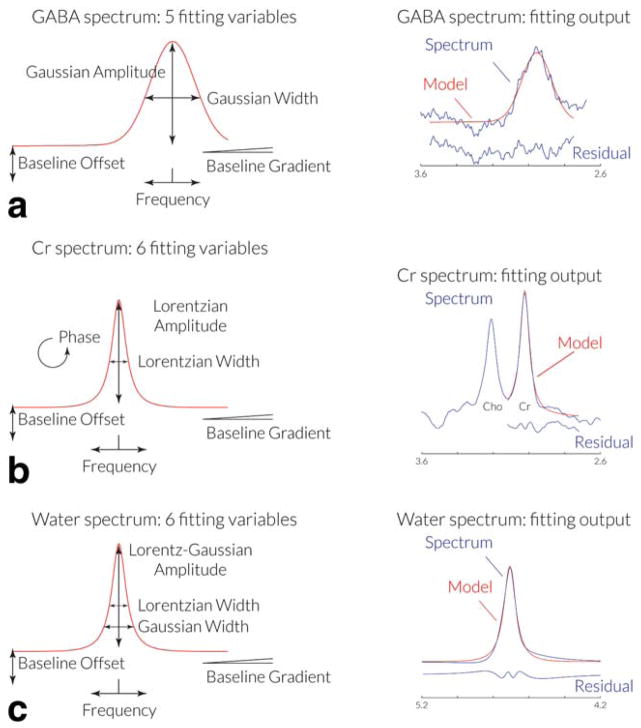
As seen in Figure 2a, fitting of the edited GABA peak in the real spectrum is achieved using a single Gaussian peak with a linear baseline, modeled by the equation:
where f represents the frequency in ppm, A, G, and f0 represent the amplitude, width and center frequency of the Gaussian peak, and C and M represent the baseline offset and slope. Fitting is performed over a range of the spectrum between 2.79 ppm and 3.55 ppm. f0, the GABA peak frequency, is initially estimated as 3.0 ppm, and the baseline parameters C and M as zero.
Figure 2.
GannetFit modeling of spectra. a: A Gaussian model with five variable parameters is used to model the edited GABA signal in the DIFF spectrum. b: A Lorentzian model with six variable parameters is used to model the Cr signal from the OFF spectrum. c: A Gaussian-Lorentzian model with six variable parameters is used to model the water signal from the unsuppressed water spectrum. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
As seen in Figure 2b, fitting of the Cr peak uses the same model that is used for frequency and phase correction above:
| [1] |
where A, L, f0 and ϕ represent the amplitude, width, center frequency, and phase of the Lorentzian peak, and C and M represent the baseline offset and slope. Fitting is performed over a range of the spectrum between 2.72 ppm and 3.12 ppm.
As seen in Figure 2c, fitting of the unsuppressed water peak uses a Gaussian-Lorentzian model:
where A and f0 represent the amplitude and center frequency of the peak, l and G Lorentzian and Gaussian width parameters, and C and M represent the baseline offset and slope. The phase parameter is not required for the water peak fit, as phasing of the unsuppressed reference spectra is applied at the start of GannetLoad processing. Fitting is performed over a range of the spectrum between 3.8 ppm and 5.6 ppm, with f0, the water peak frequency, initially estimated as 4.7 ppm. Fitting in Gannet is performed using the nonlinear least-squares minimization algorithms, lsqcurvefit and nlinfit.
As with all MRS quantified using as internal reference, GABA quantification suffers from the concern that observed effects could be driven by the denominator (ie, the peak that GABA is being quantified relative to) rather than the numerator (ie, GABA itself). Gannet addresses this concern by providing concentration estimates relative to creatine and water, with the expectation that results that hold irrespective of the denominator are more robustly interpretable. Concentration relative to creatine is quoted as a simple integral ratio between the edited GABA signal and the creatine signal in the time-averaged OFF spectrum. Absolute quantification of GABA is a work-in-progress, but Gannet uses the following formula to quantify GABA concentration relative to water:
where κ is the editing efficiency of the sequence (currently set to 0.5 for all data), cw the concentration of MR-visible water, TE and TR the echo time and repetition time of the experiment, T2w T2G T1w and T1G the transverse and longitudinal relaxation times of water (w) and GABA (G), nw and nG the number of scans in the water and GABA-edited experiments (modified to account for averaging during data export and for any scans removed by outlier rejection), and IG and Iw the integrals of the models that best fit the GABA and water spectra.
In addition to generating integrals from the best-fit model peaks, GannetFit also generates the normalized residual for each fit, ie, the standard deviation of the fitting residual (eg, σGABA) divided by the amplitude of the fitted peak (eg, AGABA):
These are then combined to generate an estimate of the overall fit error:
Data Acquisition
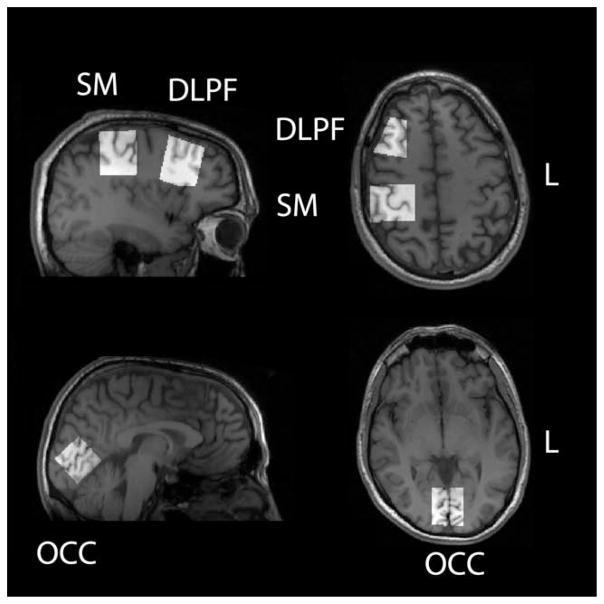
To demonstrate the utility of the Gannet software, GABA-edited spectra were acquired in 10 healthy adult volunteers, who provided written informed consent with the approval of the Cardiff University School of Psychology ethics board. In each subject, spectra (repeated four times) were acquired for each of three brain regions: occipital (OCC; including primary visual cortex); sensorimotor (SM; including primary somatosensory and motor cortex); and dorsolateral prefrontal (DLPF), as shown in Figure 3. Experimental parameters were relatively standard for the field (2), including: GE Signa HDx 3 Tesla (T) scanner using an eight-channel phased-array head coil for receive; repetition time (TR) 1.8 s; echo time (TE) 68 ms; 332 transients of 4k datapoints sampled at 5 kHz; 16 ms Gaussian editing pulse with 95 Hz bandwidth applied at 1.9 ppm in ON scans and 7.46 ppm in OFF scans in an interleaved manner; voxel size 3 × 3 × 3 cm3; acquisition time 10 min. Data were acquired on a GE Signa HDx 3T scanner; a two-step phase cycle, which was time averaged on the scanner so that each FID in the exported data represented two TRs or a period of 3.6 s. OFF/ON interleaving of editing was performed outside the phase cycle loop, so that lines of the exported data were alternately OFF, ON, etc. The unsuppressed water signals from the same volumes were also acquired for quantification.
Figure 3.
Voxel locations. Four repeat GABA-edited MRS spectra were acquired in ten subjects in the occipital (OCC), sensorimotor (SM), and dorsolateral prefrontal (DLPF) voxels shown. The voxel position was determined based upon T1-weighted anatomical images as shown.
As mentioned above these GABA+-edited data contain substantial contributions from macromolecular contaminants, which can be removed by a symmetrical editing scheme (22,23). To demonstrate the ability of Gannet to model MM-suppressed GABA spectra, data were acquired at a TE of 80 ms (22) in a single healthy subject with 20 ms editing pulses at 1.9 ppm (ON scans) and 1.5 ppm (OFF scans). Other experimental parameters include: TR 2 s; 2k datapoints sampled at 2 kHz; 3 × 3 × 3 cm3 voxel in a primary sensorimotor region (4).
Model Choice
As mentioned above, Gannet uses a single-Gaussian model to fit the edited GABA+ signal. Several alternative approaches have been applied in the literature (5,14,17,30–32); here we consider a Gaussian doublet with fixed splitting, a Gaussian doublet with variable splitting and two free Gaussian signals. The single-Gaussian has the five parameters described above (height, width, frequency, and baseline offset and gradient), as does the fixed-Gaussian-doublet model (with a splitting of 14 Hz). The free-Gaussian-doublet model has an additional parameter for the splitting, and the two-free-Gaussians model has two additional parameters representing the height and frequency of the second Gaussian. To investigate the parsimony and robustness of our approach, the fitting section of Gannet has been reproduced incorporating these other fitting options to fit all 120 spectra.
Zero-filling by a factor of eight results in frequency-domain data for which every fourth point is independent (ignoring the impact of line-broadening); therefore, fit quality is assessed using a modified chi-squared, χz2 = χ2/4. The parsimony of model fits can be compared using the Akaike information criterion (AIC), which adds a penalty terms for additional fitted parameters p: AIC = χz2 + 2p.
RESULTS
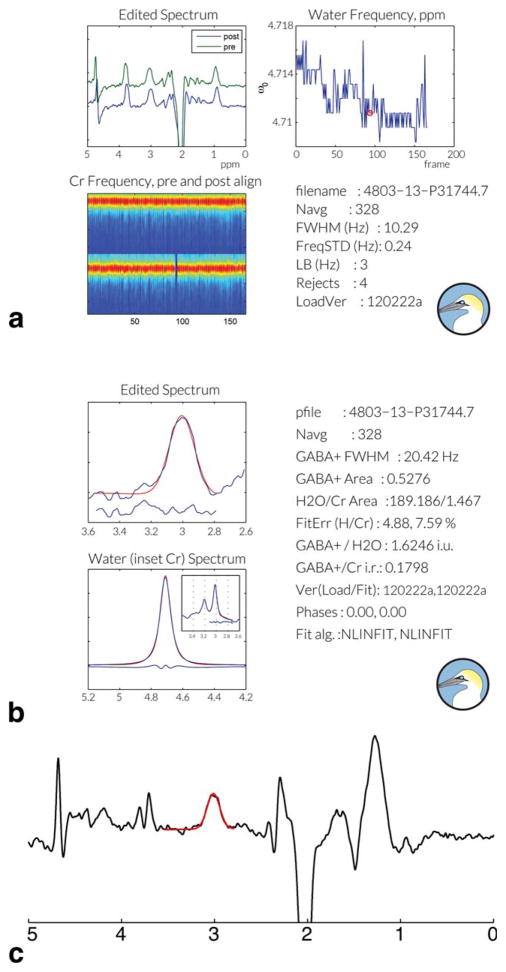
Gannet successfully processed all 120 spectra in this dataset without need for intervention. The output saved from GannetLoad and GannetFit from one dataset are shown in Figure 4. Analysis of the 120 datasets was performed in a total time of 24 min on a 2.2 GHz dual-core processor with 4 Gb of memory. Figure 4c also shows that Gannet successfully processes and fits MM-suppressed GABA-edited spectra.
Figure 4.
Gannet Output. For each dataset analyzed, pdf output is saved by both the (a) GannetLoad and (b) Gannet-Fit modules. Outputs display both graphical and quantitative information to allow users to access data and fit quality. In the GannetLoad output, rejected shots appear as blue lines in the bottom left panel and are marked with red circles in the top right panel. c: Gannet can also be applied to the analysis of MM-suppressed GABA-edited data.
Descriptive Statistics
The GABA concentrations (relative to water, uncorrected for voxel tissue composition) were estimated as 1.42 ± 0.15 i.u., 1.40 ± 0.17 i.u., and 0.95 ± 0.16 i.u. for OCC, SM and DLPF regions respectively. The GABA integral ratios to creatine were 0.15 ± 0.02, 0.17 ± 0.02, and 0.14 ± 0.02 for the same regions. Normalized fitting errors (EGABA,water - combining the residuals from GABA and water data) were 4.0 ± 1.0%, 5.1 ± 1.7%, and 6.6 ± 2.2% for OCC, SM, and DLPF regions respectively. The mean (across all regions) fitting errors (eg, RGABA, etc) for GABA, Cr, and water data were 5.4%, 7.1%, and 0.6%, respectively. Frequency instability during the experiments, whether due to scanner drift or subject motion, was relatively minor in this dataset: the mean standard deviation of the residual water frequency was only 0.57 Hz across all datasets.
Model Comparison
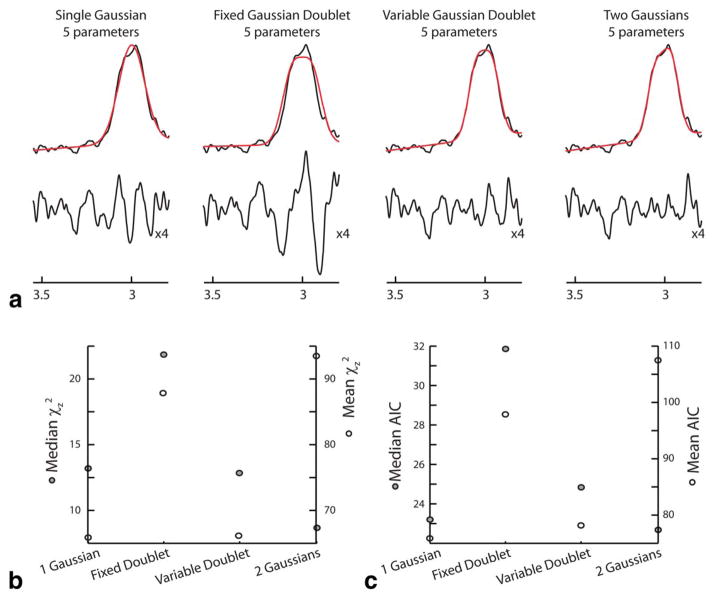
Figure 5a shows the results of fitting one spectrum (of 120) with each of the four models. The models all perform to a similar level, with the addition of more fitting parameters improving fit as expected. In Figure 5b, the mean and median values of χz2 are plotted for each model, with the median (filled points) smallest for the two-free-Gaussians model and the mean (open points) for the single Gaussian. Figure 5c shows the AIC parsimony results, with the two-free-Gaussians model having the best median by a small margin and the single Gaussian the best mean.
Figure 5.
Model Comparison. All data were fit by four models: single Gaussian, fixed Gaussian doublet; variable Gaussian doublet; and two free Gaussians. Data for a representative dataset are shown in (a) with fits in red and residuals (expanded vertically by four) below. b: Mean (white points, right-hand axis) and median (gray points, left-hand axis) χz2 values over all 120 datasets. c: Mean (white points, right-hand axis) and median (gray points, left-hand axis) AIC values over all 120 data-sets. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
DISCUSSION
Edited MRS of GABA has the potential to answer functionally relevant questions in cognitive and clinical neuroscience. Given the level of current interest, it is likely that the main limiting factor in the breadth of application of this methodology will continue to be access to data acquisition sequences and data processing methods that can be applied by the nonexpert spectroscopy user. Being fully automated and implemented as a GUI-less Matlab script without an array of processing parameters to select, Gannet has the potential to fill this processing niche. Indeed, Gannet has already been successfully applied in several cognitive and basic neuroscience studies by local teams with varying amounts of prior local spectroscopy expertise (6,33,34).
Other tools that have been applied for the analysis of GABA-edited MRS data include LCModel, jMRUI, Tarquin. Their performance, in terms of reproducibility when analyzing the same dataset, is approximately equal (1). jMRUI and Tarquin provide a flexible GUI-based tool for the visualization and quantitative analysis of spectra. In contrast, the emphasis of Gannet is on full automation without user intervention (including processing parameter selection), thereby removing rater-dependent variance, preventing the significance-based selection of processing parameters and widening access to the nonexpert user. However, the reports produced by Gannet provide several quality control metrics to allow users to gauge the spectral quality (eg, linewidth, fit error, changes in the water frequency during the scan).
Edited-MRS spectra are different from traditional short-TE single-voxel spectra in several ways that impact the approach and appropriate assumptions for fitting. For example, editing significantly reduces overlap between signals in a spectrum, the main problem addressed by linear combination approaches such as LCModel. Additionally, the individual metabolite components of short-TE spectra have positive multiplets of the same phase, which are superimposed on a positive macromolecular baseline. Using LCModel control parameters typical for short-TE PRESS spectra, when analyzing GABA-edited data, may result in significant edited signal being absorbed into a splined baseline. The linear baseline used by Gannet, appropriate over the narrow frequency range modeled, does not suffer from this same problem.
Comparing different approaches to fitting the data, the clearest outcome is that the fixed-splitting doublet performs consistently badly. This is perhaps surprising, given that the edited peak is sometimes considered as a pseudo-doublet with this splitting (28). However, recent results have suggested that, in some cases, observed splitting in vivo may be the artifactual result of postprocessing (29) and that a more complex lineshape might be expected (35). The median values of chi-squared and the AIC are possibly more representative of the full (chi-squared) distribution than the mean values, and by this metric, the two-Gaussian approach gives the best and (marginally) most parsimonious fit. If the decreased independence of data points that arises from line broadening is taken into account, the chi-squared fit term will be reduced relative to the parameter penalty, and the parsimony of the Two-Gaussian model will be further reduced. In the end, considering the somewhat competing demands of parsimony and robustness, the simple single-Gaussian model adopted by Gannet appears to be sufficient for typical MEGA-PRESS data.
This study has not investigated all possible models and a Gaussian-doublet-plus-Gaussian-singlet (to fit GABA and MM separately) or a model based upon full simulations of the GABA spin system might perform well. It should be also noted that alternative approaches that fit the whole spectrum, such as LCModel, gain SNR from the other GABA signal at 2.3 ppm.
Performing frequency correction based on fitting of individual dynamic scans is a process that relies upon sufficient signal-to-noise ratio (SNR) of individual shots. In the data presented here, this is achieved by fitting two averages of a 3 × 3 × 3 cm3 voxel. There is a lower limit for voxel size below which the SNR of individual shots is insufficient to accurately estimate frequency and phase fluctuations from fitting the creatine resonance, and therefore, this postprocessing step may result in unusable data for small voxels. In this instance, Gannet could still be used by replacing the postalignment corrupted data with the prealignment data before the GannetFit step (see Supp. Table S1 for further details of the output structure).
Although Gannet is focused on the quantification of GABA, it has been applied with minor modifications to glutathione (GSH)-edited spectra, which result in an edited signal with a similar shape and at a similar chemical shift. However, modifications for other edited experiments may require a new model function and/or a new approach to frequency correction, depending on the frequency of the edited signal and the editing target spins. In the case of GSH editing, the NAA peak can be used for frequency correction, with superior performance to Cr. Two additional limitations of Gannet is that it does not currently support analysis of GABA-edited MRSI (eg, Zhu et al) (36) and does not use unsuppressed water data to perform eddy current correction.
Absolute quantification of GABA concentration requires correction for the large differences in GABA concentration between brain tissue (approximately 1 mmol/liter) and cerebrospinal fluid (CSF) (less than 1 μmol/liter) (37), ie, GABA values reported by Gannet should be corrected to account for the tissue composition of the voxel (38). More rigorously, differences in the apparent water concentration between white matter, gray matter, and CSF can be corrected (39). Absolute quantification does not currently form part of Gannet, thus the reported “gabaiu” values should be corrected using structural imaging data, if available. Differences have been reported in the GABA concentration of gray matter and white matter (36,39). Although gray matter concentrations are higher, GABA is also present in white matter, so a ‘correction’ for gray/white matter should be avoided.
In conclusion, we have presented a new tool designed for the analysis of GABA-edited MRS spectra, Gannet. Built in the Matlab programming environment and available open-source, Gannet aims to make the analysis accessible to the nonexpert user and, being fully automated, removes rater-dependent variance. Outputting both pdf-format summaries for each dataset and a data structure containing quantitative results, processed spectra and many processing intermediates, Gannet also provides a useful platform for the development and further optimization of more advanced data processing.
Supplementary Material
Acknowledgments
Contract grant sponsor: NIH; Contract grant numbers: P41 EB015909, R01 EB016089, R21 NS077300, R01 MH096263, R01 MH092443.
The authors thanks Krish Singh for improvements to the fitting within Gannet and scientific discussions. N.A.J.P. holds an Autism Speaks Postdoctoral Fellowship.
Footnotes
Additional Supporting information may be found in the online version of this article.
References
- 1.Mullins PG, McGonigle DJ, O’Gorman RL, et al. Current practice in the use of MEGA-PRESS spectroscopy for the detection of GABA. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2012.12.004. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Puts NA, Edden RA. In vivo magnetic resonance spectroscopy of GABA: a methodological review. Prog Nucl Magn Reson Spectrosc. 2012;60:29–41. doi: 10.1016/j.pnmrs.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muthukumaraswamy SD, Evans CJ, Edden RA, Wise RG, Singh KD. Individual variability in the shape and amplitude of the BOLD-HRF correlates with endogenous GABAergic inhibition. Hum Brain Mapp. 2012;33:455–465. doi: 10.1002/hbm.21223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puts NA, Edden RA, Evans CJ, McGlone F, McGonigle DJ. Regionally specific human GABA concentration correlates with tactile discrimination thresholds. J Neurosci. 2011;31:16556–16560. doi: 10.1523/JNEUROSCI.4489-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stagg CJ, Bachtiar V, Johansen-Berg H. The role of GABA in human motor learning. Curr Biol. 2011;21:480–484. doi: 10.1016/j.cub.2011.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foerster BR, Petrou M, Edden RA, et al. Reduced insular gamma-aminobutyric acid in fibromyalgia. Arthritis Rheum. 2012;64:579–583. doi: 10.1002/art.33339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petroff OA, Rothman DL, Behar KL, Mattson RH. Low brain GABA level is associated with poor seizure control. Ann Neurol. 1996;40:908–911. doi: 10.1002/ana.410400613. [DOI] [PubMed] [Google Scholar]
- 8.Simister RJ, McLean MA, Barker GJ, Duncan JS. Proton MRS reveals frontal lobe metabolite abnormalities in idiopathic generalized epilepsy. Neurology. 2003;61:897–902. doi: 10.1212/01.wnl.0000086903.69738.dc. [DOI] [PubMed] [Google Scholar]
- 9.Sanacora G, Mason GF, Rothman DL, et al. Increased cortical GABA concentrations in depressed patients receiving ECT. Am J Psychiatry. 2003;160:577–579. doi: 10.1176/appi.ajp.160.3.577. [DOI] [PubMed] [Google Scholar]
- 10.Sanacora G, Mason GF, Rothman DL, Krystal JH. Increased occipital cortex GABA concentrations in depressed patients after therapy with selective serotonin reuptake inhibitors. Am J Psychiatry. 2002;159:663–665. doi: 10.1176/appi.ajp.159.4.663. [DOI] [PubMed] [Google Scholar]
- 11.Tayoshi S, Nakataki M, Sumitani S, et al. GABA concentration in schizophrenia patients and the effects of antipsychotic medication: a proton magnetic resonance spectroscopy study. Schizophr Res. 2010;117:83–91. doi: 10.1016/j.schres.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 12.Yoon JH, Maddock RJ, Rokem A, et al. GABA concentration is reduced in visual cortex in schizophrenia and correlates with orientation-specific surround suppression. J Neurosci. 2010;30:3777–3781. doi: 10.1523/JNEUROSCI.6158-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ongur D, Prescot AP, McCarthy J, Cohen BM, Renshaw PF. Elevated gamma-aminobutyric Acid levels in chronic schizophrenia. Biol Psychiatry. 2010;68:667–670. doi: 10.1016/j.biopsych.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kegeles LS, Mao X, Stanford AD, et al. Elevated prefrontal cortex gamma-aminobutyric acid and glutamate-glutamine levels in schizophrenia measured in vivo with proton magnetic resonance spectroscopy. Arch Gen Psychiatry. 2012;69:449–459. doi: 10.1001/archgenpsychiatry.2011.1519. [DOI] [PubMed] [Google Scholar]
- 15.Simpson HB, Shungu DC, Bender J, Jr, et al. Investigation of cortical glutamate-glutamine and gamma-aminobutyric acid in obsessive-compulsive disorder by proton magnetic resonance spectroscopy. Neuropsychopharmacology. 2012;37:2684–2692. doi: 10.1038/npp.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edden RA, Crocetti D, Zhu H, Gilbert DL, Mostofsky SH. Reduced GABA concentration in attention-deficit/hyperactivity disorder. Arch Gen Psychiatry. 2012;69:750–753. doi: 10.1001/archgenpsychiatry.2011.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gaetz W, Bloy L, Wang DJ, et al. GABA estimation in the brains of children on the autism spectrum: measurement precision and regional cortical variation. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.1005.1068. Epub. [DOI] [PMC free article] [PubMed]
- 18.Rojas DC, Singel D, Steinmetz S, Hepburn S, Brown MS. Decreased left perisylvian GABA concentration in children with autism and unaffected siblings. Neuroimage. 2013 doi: 10.1016/j.neuroimage.2013.01.045. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaiser LG, Young K, Meyerhoff DJ, Mueller SG, Matson GB. A detailed analysis of localized J-difference GABA editing: theoretical and experimental study at 4 T. NMR Biomed. 2008;21:22–32. doi: 10.1002/nbm.1150. [DOI] [PubMed] [Google Scholar]
- 20.Rothman DL, Petroff OA, Behar KL, Mattson RH. Localized 1H NMR measurements of gamma-aminobutyric acid in human brain in vivo. Proc Natl Acad Sci U S A. 1993;90:5662–5666. doi: 10.1073/pnas.90.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11:266–272. doi: 10.1002/(sici)1099-1492(199810)11:6<266::aid-nbm530>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 22.Edden RA, Puts NA, Barker PB. Macromolecule-suppressed GABA-edited magnetic resonance spectroscopy at 3T. Magn Reson Med. 2012;68:657–661. doi: 10.1002/mrm.24391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry P-G, Dautry C, Hantraye P, Bloch G. Brain GABA editing without macromolecule contamination. Magn Reson Med. 2001;45:517–520. doi: 10.1002/1522-2594(200103)45:3<517::aid-mrm1068>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 24.Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001;14:260–264. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
- 25.Naressi A, Couturier C, Devos JM, et al. Java-based graphical user interface for the MRUI quantitation package. MAGMA. 2001;12:141–152. doi: 10.1007/BF02668096. [DOI] [PubMed] [Google Scholar]
- 26.Wilson M, Reynolds G, Kauppinen RA, Arvanitis TN, Peet AC. A constrained least-squares approach to the automated quantitation of in vivo (1)H magnetic resonance spectroscopy data. Magn Reson Med. 2011;65:1–12. doi: 10.1002/mrm.22579. [DOI] [PubMed] [Google Scholar]
- 27.Hardy CJ, Bottomley PA, Rohling KW, Roemer PB. An NMR phased array for human cardiac 31P spectroscopy. Magn Reson Med. 1992;28:54–64. doi: 10.1002/mrm.1910280106. [DOI] [PubMed] [Google Scholar]
- 28.Waddell KW, Avison MJ, Joers JM, Gore JC. A practical guide to robust detection of GABA in human brain by J-difference spectroscopy at 3 T using a standard volume coil. Magn Reson Imaging. 2007;25:1032–1038. doi: 10.1016/j.mri.2006.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans CJ, Puts NA, Robson SE, et al. Subtraction artifacts and frequency (Mis-)alignment in J-difference GABA editing. J Magn Reson Imaging. 2013 doi: 10.1002/jmri.23923. Epub. [DOI] [PubMed] [Google Scholar]
- 30.Geramita M, van der Veen JW, Barnett AS, et al. Reproducibility of prefrontal gamma-aminobutyric acid measurements with J-edited spectroscopy. NMR Biomed. 2011;24:1089–1098. doi: 10.1002/nbm.1662. [DOI] [PubMed] [Google Scholar]
- 31.Gabbay V, Mao X, Stanford AD, et al. Anterior cingulate cortex γ-amino butyric acid in depressed adolescents: relationship to anhedonia. Arch Gen Psychiatry. 2012;69:139–149. doi: 10.1001/archgenpsychiatry.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stagg CJ, Wylezinska M, Matthews PM, et al. Neurochemical effects of theta burst stimulation as assessed by magnetic resonance spectroscopy. J Neurophysiol. 2009;101:2872–2877. doi: 10.1152/jn.91060.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petrou M, Pop-Busui R, Foerster BR, et al. Altered excitation-inhibition balance in the brain of patients with diabetic neuropathy. Acad Radiol. 2012;19:607–612. doi: 10.1016/j.acra.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Violante IR, Ribeiro MJ, Edden RA, et al. GABA deficit in the visual cortex of patients with neurofibromatosis type 1: genotype-phenotype correlations and functional impact. Brain. 2013;136(Pt 3):918–925. doi: 10.1093/brain/aws368. [DOI] [PubMed] [Google Scholar]
- 35.Near J, Evans CJ, Puts NA, Barker PB, Edden RA. J-difference editing of gamma-aminobutyric acid (GABA): simulated and experimental multiplet patterns. Magn Reson Med. 2012 doi: 10.1002/mrm.24572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu H, Edden RA, Ouwerkerk R, Barker PB. High resolution spectroscopic imaging of GABA at 3 Tesla. Magn Reson Med. 2011;65:603–609. doi: 10.1002/mrm.22671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wood JH, Glaeser BS, Hare TA, Sode J, Brooks BR, Van Buren JM. Cerebrospinal fluid GABA reductions in seizure patients evoked by cerebellar surface stimulation. J Neurosurg. 1977;47:582–589. doi: 10.3171/jns.1977.47.4.0582. [DOI] [PubMed] [Google Scholar]
- 38.Kreis R, Ernst T, Ross BD. Absolute quantitation of water and metabolites in the human brain. II. Metabolite concentrations. J Magn Reson B. 1993;102:9–19. [Google Scholar]
- 39.Choi C, Bhardwaj PP, Kalra S, et al. Measurement of GABA and contaminants in gray and white matter in human brain in vivo. Magn Reson Med. 2007;58:27–33. doi: 10.1002/mrm.21275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.