Abstract
Age-related variations in DNA methylation have been reported; however, the functional relevance of these differentially methylated sites (age-dMS) are unclear. Here we report potentially functional age-dMS, defined as age- and cis-gene expression-associated methylation sites (age-eMS), identified by integrating genome-wide CpG methylation and gene expression profiles collected ex vivo from circulating T cells (227 CD4+ samples) and monocytes (1,264 CD14+ samples, age range: 55–94 years). None of the age-eMS detected in 227 T cell samples are detectable in 1,264 monocyte samples, in contrast to the majority of age-dMS detected in T cells that replicated in monocytes. Age-eMS tend to be hypomethylated with older age, located in predicted enhancers, and preferentially linked to expression of antigen processing and presentation genes. These results identify and characterize potentially functional age-related methylation in human T cells and monocytes, and provide novel insights into the role age-dMS may play in the aging process.
Advancing age is associated with extensive changes in human physiology, and is the most important risk factor for many diseases. Age-related changes in gene expression are thought to underlie many of these physiologic and pathologic consequences of aging 1. To better understand age-related changes in gene expression, it is important to consider changes in mechanisms that regulate gene expression, such as epigenetic modifications including DNA methylation of cytosines in CpG dinucleotides and histone modifications 2, 3. Previously, we investigated differentially methylated CpG sites (dMS) in 1,264 CD14+ monocyte samples for potential functional relationships with cis-gene (± 1Mb) expression 4, and uncovered many significant (false discovery rate (FDR) <0.001) gene expression-associated methylation sites (eMS). Some of these eMS were very strongly correlated with cis-gene expression, such as CpG site cg17005068. Located in the GSTT1 (glutathione S-transferase theta 1) promoter, methylation of cg17005068 was highly correlated with GSTT1 expression (prho = -0.86, p<2.2×10−308), and in combination with 15 other GSTT1-eMS, methylation accounted for 77% of the variance of GSTT1 expression in monocytes 4.
Other recent studies have identified dMS associated with age (age-dMS), including regions with decreased (hypo age-dMS) and increased (hyper age-dMS) methylation with older age 5-10. However, the results from previous studies investigating the relationships between agedMS and gene expression are inconclusive 11-15. One of the most comprehensive studies measured methylation and gene expression in the whole blood of 168 individuals, and reported significant negative correlations between age-dMS and gene expression 12, while another study measuring methylation and gene expression in different samples reported negligible relationships between age-dMS and gene expression 15. Small sample sizes, mixed cell samples, and gene expression and methylation data measured in different samples makes findings from previous studies difficult to interpret. Overall, there is still a lack of clear understanding of the effects of age-dMS on the transcriptome 16.
To better understand the functional implications of age-dMS, and to identify age-dMS that potentially mediate the relationship between age and gene expression for future in vitro functional evaluation, here we utilized methylomic and transcriptomic data from 1,264 CD14+ purified monocyte samples, collected from a large population of community-dwelling participants in the Multi-Ethnic Study of Atherosclerosis (MESA), ranging in age from 55 to 94 years (Supplementary Table 1 and Methods), as well as methylomic and transcriptomic data from 227 CD4+ T cell samples, from a subset of the population. We identified, cross-sectionally, potentially functional age-associated methylation signals that were correlated with cis-gene expression and clinical measures of vascular aging (pulse pressure-eMS), and provide detailed functional annotation from publicly available datasets (e.g. ENCODE) characterizing the genomic landscape surrounding these potentially functional methylation sites.
Results
Identification of Age-dMS
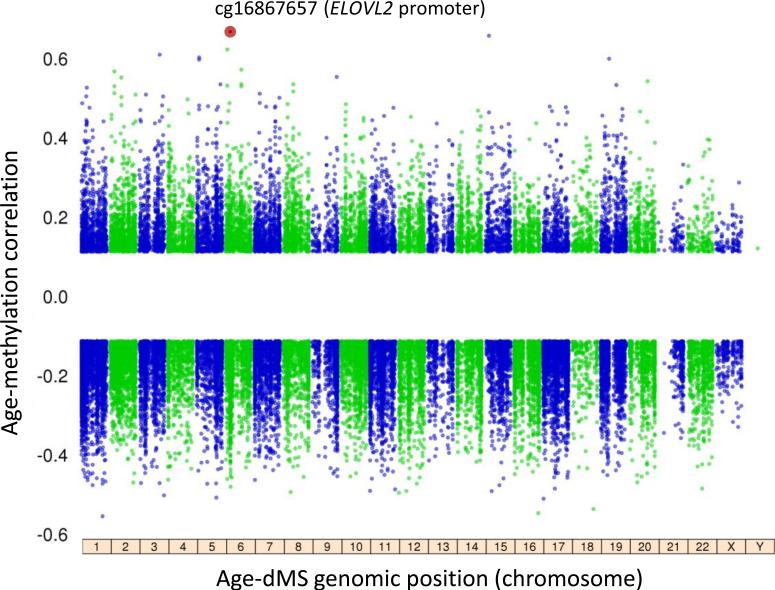
We first characterized DNA methylation at ~450,000 CpG sites across the genome in CD14+ purified cells (predominately monocytes) and CD4+ purified cells (T cells) collected from 227 MESA individuals. Using association analysis with a false discovery rate (FDR) threshold of 0.001, and adjusting for biological and technical covariates (Methods), we identified 2,285 monocyte specific age-dMS, 2,023 T cell specific age-dMS, and 572 overlapping age-dMS across the two cell types. We then expanded our monocyte sample size to 1,264 MESA individuals, and identified 37,911 CpG sites with age-associated methylation (~8% of all CpG sites, FDR<0.001; Fig. 1). The majority of age-dMS we detected in 227 T cell samples shared a similar effect direction between methylation and age in the 1,264 monocyte samples (Supplementary Fig. 1a and Supplementary Data 1). Many of the most significant age-dMS detected in both monocytes and T cells were previously reported to have age-associated methylation measured in whole blood 17, including CpG sites in ELOVL2 (ELOVL fatty acid elongase 2; cg16867657, prho = 0.66, FDR = 3.65×10−140), FHL2 (four and a half LIM domains 2; cg06639320, partial correlation (prho) = 0.55, FDR = 4.45×10−88), and PENK (proenkephalin; cg16419235, prho = 0.52, FDR = 2.85×10−75).
Figure 1. The aging methylome in 1,264 monocyte samples.
The analysis of age and methylation in 1,264 CD14+ monocyte samples included 448,523 CpG sites, of which 37,911 had methylation associated with age (age-dMS; 26,159 negatively associated, 11,752 positively associated, FDR<0.001). The most significant age-dMS (red circle, cg16867657, prho = 0.66, FDR = 3.65×10−140) was detected on chromosome 6 in the ELOVL2 (ELOVL fatty acid elongase 2) promoter. The partial correlation between CpG methylation and age is shown on y-axis, compared to CpG genomic location (by chromosome, x-axis). Linear regression analysis also included the following covariates: race, sex, site of data collection, microarray chip, and residual sample contamination with non-targeted cells (see Methods).
Characterization of Hyper and Hypo Age-dMS
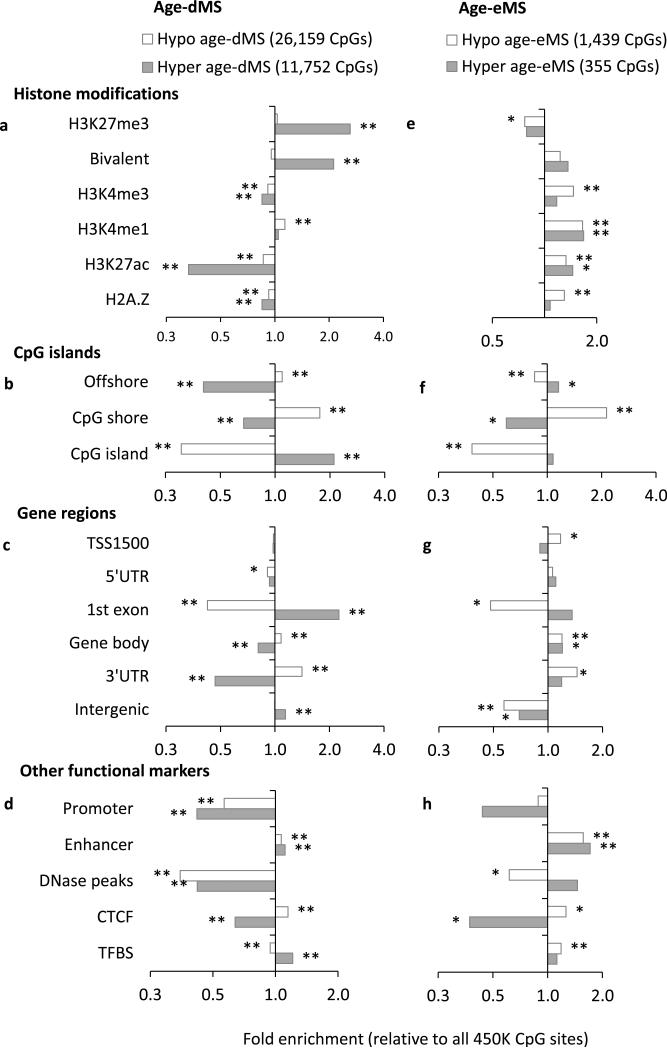
We next examined the enrichment of 37,911 monocyte age-dMS from our 1,264 monocyte samples, within genomic regions with predicted roles in regulating gene expression (e.g. enhancers, insulators, etc.) based on histone modifications, CTCF binding, and DNase hypersensitivity reported in a monocyte sample by ENCODE 18, 19. Age-dMS exhibiting increased methylation with age (hyper age-dMS) were located in distinctly different functional domains than age-dMS exhibiting decreased methylation with age (hypo age-dMS), consistent with previous reports 6, 10, 20. Compared to all CpG sites tested, hyper age-dMS were significantly enriched for inactive/repressive histone modifications 18 (H3K27me3, bivalent H3K27me3/H3K4me3), while being depleted for active chromatin marks 3, 18, 21 (H3K4me3, H3K27ac (Fig. 2a). However, there was no clear preference for hypo age-dMS among inactive vs. active histone modifications (fold enrichments ranging 0.9 – 1.1). We also replicated previous findings 10, 14, 22 that hyper age-dMS are enriched among CpG islands (Fig. 2b) and 1st exons (Fig. 2c), while hypo age-dMS are enriched among CpG island “shores”, and the 3’ untranslated regions (3’ UTR) of genes.
Figure 2. Enrichment of Age-dMS and Age-eMS monocytes for regulatory features in 1,264 monocyte samples.
Fold enrichment of age-associated CpG sites (37,911 age-dMS, FDR<0.001, left), and fold enrichment of age and cis-gene expression-associated methylation sites (FDR<0.001, 1,794 age-eMS, right), stratified by methylation positively associated with age (hyper age-dMS, shaded) and negatively associated with age (hypo age-dMS, white) for a,e) histone modifications reported by ENCODE in a monocyte sample, b,f) CpG islands and ‘shores’, c,g) gene regions, including: within 1.5kb upstream of the transcription start site (TSS1500), the 5’ untranslated region (5’ UTR), the 1st gene exon, the gene body, the 3’ UTR, or intergenic, and d,h) predicted gene expression regulatory regions based on histone modifications (enhancer and promoter based on H3K4me1/3, H3K27ac), CTCF binding, and DNase peaks reported in a monocyte sample (ENCODE/UCSC browser), and transcription factor binding sites (TFBS) reported in any cell type available from the UCSC Genome Browser. Fold enrichments presented are from 1,264 monocyte samples, and are relative to all 448,523 CpG sites tested (y-axis); * 1×10−6 ≤ p < 0.01, ** p < 1×10−6, chi-squared test
Histone modifications (H3K4me1/3, H3K27ac), CTCF binding, and DNase peaks previously reported in a monocyte (CD14+) sample 19 were also used to predict monocyte specific functional regions, including promoters, enhancers, and insulators. Both hyper and hypo age-dMS were depleted among predicted promoter regions and DNase peaks, while being enriched among enhancer regions (Fig. 2d). Hypo age-dMS were also enriched for CTCF binding sites; however, hyper age-dMS were not. We also report the enrichment of age-dMS among transcription factor binding sites (TFBS) reported in any cell type available from the UCSC Genome Browser 23 (due to the lack of monocyte specific TFBS). Hyper age-dMS showed some enrichment among these sites, while hypo age-dMS did not.
Identification of Age-eMS
Potentially functional age-dMS were defined as CpG sites whose % methylation was associated with age (FDR<0.001) and with mRNA expression of any gene within one megabase of the CpG site in question (FDR<0.001). Among 227 T cell samples, 44 age-, and expression-associated methylation sites (age-eMS) were detected (2% of the 2,595 T cell age-dMS), with methylation correlated with age (prho ranging -0.54 – 0.70) and with cis-gene expression (prho ranging -0.62 – 0.56). Half of these T cell age-eMS (22 CpG sites) had methylation profiles associated with age in 1,264 monocyte samples; however, there was no replication of the association between methylation and gene expression for these 22 CpG sites in monocyte samples (Supplementary Fig. 1b). Due to the lack of age-eMS detected in both cell types, combined with our larger monocyte sample size and the availability of monocyte specific histone modification data from ENCODE 19, we focus the rest of our age-eMS investigations on findings from 1,264 monocyte samples.
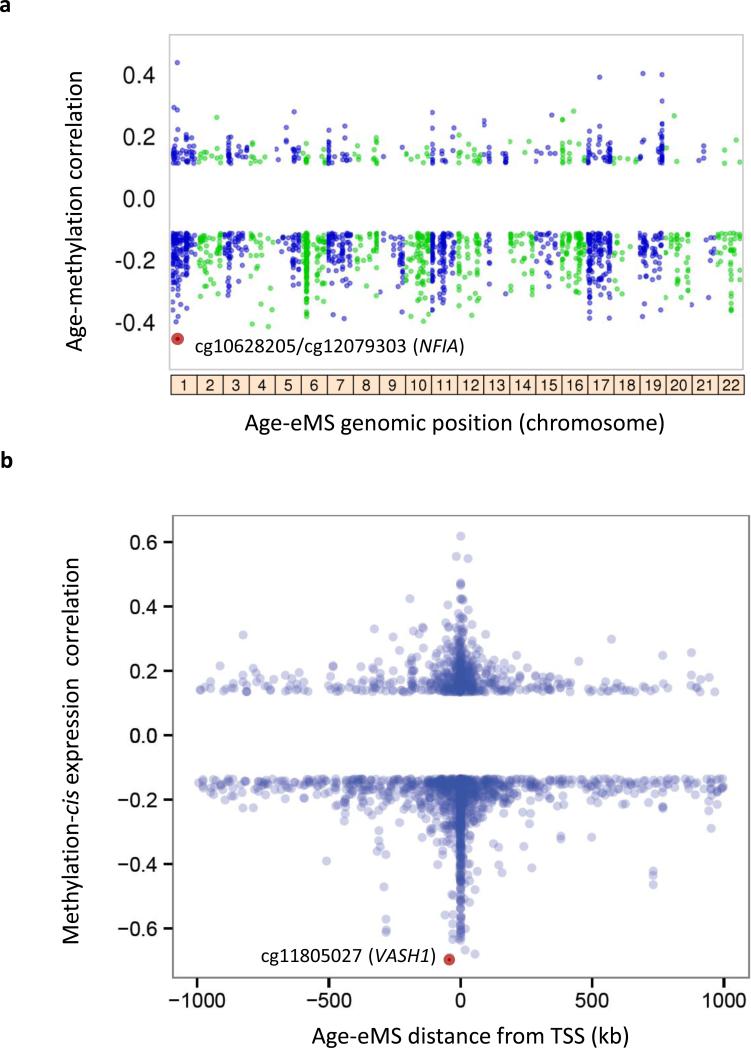
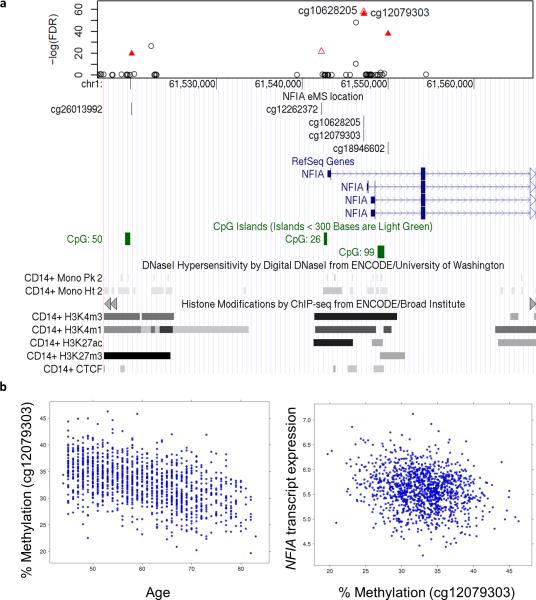
We detected 1,794 age-eMS among the 1,264 monocyte samples (4.7% of 37,911 monocyte age-dMS; reported in Supplementary Data 2), with methylation correlated with age (prho ranging: -0.46 – 0.44; Fig. 3a), and cis-gene expression (prho ranging -0.69 – 0.62; Fig. 3b). The most significant age-dMS identified as an age-eMS (Fig. 3a) in monocytes was detected within a predicted enhancer region of the NFIA (nuclear factor I-A) gene body (Fig. 4). NFIA is a transcription factor whose expression drives erythropoiesis, while its silencing drives granulopoiesis 24. Hypomethylation with age of cg10628205 (partial correlation (prho) = -0.46; FDR = 1.06×10−58) correlated with increased NFIA expression (prho = -0.16; FDR = 1.97×10−5). Five nearby CpG sites were also identified as age-eMS linked with NFIA expression. Using multiple regression analysis including all six NFIA age-eMS, four were found to be independently associated with NFIA expression. In total, methylation of these four CpG sites explained 7.5% of the variance of NFIA gene expression. However, some age-dMS identified as age-eMS had stronger correlations between methylation and gene expression (Fig. 3b and Supplementary Data 2). For instance, the strongest correlation detected was between age-dMS cg10628205 (methylation and age phro = -0.15) and expression of VASH1 (vasohibin 1, phro = -0.69). This age-eMS is 42 kb upstream of the VASH1 transcription start site. VASH1 is an angiogenesis inhibitor that has previously been reported to suppress monocyte and macrophage infiltration of the kidney 25.
Figure 3. Correlations between age-methylation and methylation-gene expression for AgeeMS in 1,264 monocyte samples.
CpG sites with methylation associated with age (FDR<0.001) and cis-gene expression (1,794 age-eMS; FDR<0.001; Supplementary Data 2). a) The partial correlation of age-eMS methylation with age (y-axis), compared to age-eMS genomic location (by chromosome, x-axis); the strongest correlations (prho = -0.46) were between age and methylation of cg10628205 and cg12079303 (red circle), which were also correlated with expression of NFIA. b) The partial correlation between age-eMS methylation and cis-gene expression (y-axis), compared to age-eMS distance to the associated gene transcription start site (TSS, x-axis); the strongest correlation (prho = -0.69) was between methylation of cg11805027 and expression of VASH1. Linear regression analysis also included the following covariates: race, sex, site of data collection, microarray chip, and residual sample contamination with non-targeted cells (see Methods).
Figure 4. Methylation of NFIA is associated with age and with NFIA expression in 1,264 monocyte samples.
(a) Regional association plot showing the significance (−log10 FDR, top panel y-axis) of single CpG methylation associations with age surrounding NFIA (nuclear factor I/A) by genomic position on chromosome 1 (x-axis). Six CpG sites near NFIA (red triangles) have methylation associated with age (FDR ranging 1.1×10−58 – 1.2×10−16), and with increased expression of NFIA in monocytes (age-eMS, FDR ranging 2.0×10−11 to 3.2×10−4). Methylation of four age-eMS have independent effects on NFIA expression from multiple regression analysis (filled-in triangles, cg00165994 downstream, not shown) with cg12079303 remaining the most significant. NFIA age-eMS overlap histone modification marks indicative of active regulatory regions (i.e. H3K4me1/3, H3K27ac by ChIP-seq from ENCODE, bottom panel). (b) cg12079303 methylation is negatively correlated with age (left; prho = -0.46, rho = -0.48) and negatively correlated with NFIA mRNA expression (right; prho = -0.16, rho = -0.12).
Characterization of Hyper and Hypo Age-eMS
The majority of age-eMS were located in close proximity to the associated gene transcription start site (TSS; Fig. 3b), with 71% of age-eMS in monocytes located within 100 kb of the TSS. Additionally, the correlations between methylation and gene expression of age-eMS located within 100 kb of the TSS tended to be stronger (absolute prho: average = 0.22, median 0.18) than age-eMS located further than 100 kb from the TSS (phro: average = 0.17, median = 0.16).
Age-eMS enriched in open chromatin and enhancer regions
In an effort to further explore the potential functionality of the age-eMS identified, we examined the enrichment of age-eMS within genomic regions with predicted roles in regulating gene expression based on histone modifications, CTCF binding, and DNase hotspots reported in a monocyte sample by ENCODE 18, 19. The most prominent features of age-eMS were their enrichment for histone modifications indicative of open/active chromatin (H3K4me1 and H3K27ac, Fig. 2e) and predicted enhancer regions (Fig. 2h), while being depleted among repressed genomic regions (H3K27me3), for both hypo and hyper age-eMS.
Age-eMS linked to antigen processing and presentation genes
The presence of predicted functional regions overlapping the identified age-eMS provides additional support for these genomic regions being important regulatory regions. Therefore, we consider age-eMS overlapping enhancers, insulators, or promoters (669 age-eMS) as top candidates for potentially functional age-dMS (Supplementary Data 2). These 669 CpG sites overlapping potentially functional regulatory regions are associated with the expression of 403 different genes, which are significantly enriched 26 with antigen processing and presentation genes (GO:0019882, FDR = 9.60×10−4). Table 1 shows associations between age, methylation, and expression of 13 antigen processing and presentation genes including major histocompatibility complex (MHC) class I and II genes. Supporting previous findings 27, we observe an up-regulation of all MHC class I and II genes with expression associated with age (FDR < 0.01) (Supplementary Table 2).
Table 1.
Antigen processing and presentation genes enriched among genes with expression linked to potentially functional age-eMS
| Age-CpG methylation | CpG methylation-gene expression | |||||||
|---|---|---|---|---|---|---|---|---|
| CpG ID: | CpG location | prho | rho | FDR | Gene | prho | rho | FDR |
| cg26350754 | 1stExon (HLA-DPB1) | −0.38 | −0.39 | 1.2E-38 | HLA-DPB1 | −0.17 | −0.13 | 3.6E-06 |
| cg04856022 | Body (PPT2) | −0.37 | −0.39 | 1.3E-36 | HLA-DRB1 | −0.15 | −0.13 | 1.2E-04 |
| cg08998192 | Body (TAP2) | −0.33 | −0.34 | 1.9E-27 | TAP2 | −0.14 | −0.15 | 3.3E-04 |
| cg08998192 | Body (TAP2) | −0.33 | −0.34 | 1.9E-27 | PSMB9 | 0.21 | 0.16 | 2.4E-10 |
| cg13007871 | Body (HLA-E) | −0.30 | −0.30 | 5.0E-23 | HLA-E | −0.22 | −0.19 | 3.3E-11 |
| cg13007871 | Body (HLA-E) | −0.30 | −0.30 | 5.0E-23 | HLA-F | −0.18 | −0.16 | 6.5E-07 |
| cg08818207 | Body (TAP1) | −0.28 | −0.27 | 1.3E-20 | PSMB8 | −0.17 | −0.13 | 2.3E-06 |
| cg08009669 | TSS1500 (MICB) | −0.23 | −0.24 | 1.1E-13 | MICB | −0.19 | −0.15 | 5.4E-08 |
| cg25843003 | 3'UTR (HCP5) | −0.23 | −0.21 | 2.3E-13 | HLA-B | −0.17 | −0.13 | 3.9E-06 |
| cg25925210 | 5'UTR (TTLL4) | −0.20 | −0.21 | 1.7E-10 | SLC11A1 | −0.19 | −0.12 | 9.4E-08 |
| cg01309328 | Body (PSMB8) | −0.19 | −0.19 | 1.9E-09 | HLA-DPA1 | −0.14 | −0.15 | 7.8E-04 |
| cg26083458 | Body (TAPBP) | −0.17 | −0.18 | 8.4E-08 | TAPBP | 0.14 | 0.09 | 6.2E-04 |
| cg21470947 | TSS1500 (HLA-DMB) | 0.12 | 0.15 | 2.7E-04 | HLA-DMB | −0.14 | −0.11 | 2.3E-04 |
Thirteen antigen processing and presentation genes (from Gene Ontology gene set GO:0019882) harbored age- and cis-gene expression-associated methylation sites (age-eMS) which overlapped predicted functional regions (enhancer, promoter, or insulator, based on histone modifications and CTCF binding reported by ENCODE in a monocyte sample). Shown above are the unique CpG IDs, CpG locations relative to associated gene [within gene body (gene containing CpG in parentheses), untranslated regions (UTR) of genes, or within 1,500 base pairs of a transcription start site (TSS1500)], and the correlations (prho and rho) and significance (FDR<.001) between CpG methylation and age/gene expression (sorted by association between age and methylation). Total effects of age on gene expression were significant (FDR≤0.05) for 6 genes (HLA-DPB1, TAP2, HLA-E, HLA-B, HLA-DPA1, and TAPBP) which all increased with age (see Supplementary Table 2). Covariates included: race, sex, site of data collection, microarray chip, and residual sample contamination with non-targeted cells (Methods).
Age-eMS linked with vascular aging
To further explore the biological relevance of age-eMS, we identified a subset of 186 age-eMS that were associated with vascular age (FDR<0.001), measured by pulse pressure (Supplementary Data 2). After adjusting for chronological age, 42 age-eMS remained nominally associated with pulse pressure (pulse pressure-eMS), some of which were associated with expression of genes that have biologically plausible roles in vascular aging, such as ARID5B (AT rich interactive domain 5B (MRF1-like)), and GSN (gelsolin). ARID5B is a transcription factor that has been implicated in the pathogenesis of coronary artery disease 28. Gelsolin is an actin binding protein which has been linked to vascular permeability 29, cell motility, and the development of many pathological processes, including cardiovascular diseases 30.
Discussion
Here we report, for the first time, the relationship between aging, DNA methylation, and cis-gene expression at the global level, using two purified cell types from the same individuals in a large cohort study. We find that only a small fraction of dMS (~2%) associated with aging were also associated with differential expression of nearby genes in 227 T cell samples. In parallel, we saw a slightly larger proportion of age-dMS (~5%) associated with cis-gene expression in a larger sample of monocytes (1,264). However, in contrast to age-dMS, no ageeMS were detected in both cell types. The lack of replication of any T cell age-eMS within monocytes could reflect a cell-specific nature of age-eMS, demonstrating the importance of using purified cells for methylomic and transcriptomic studies.
In addition to their association with cis-gene expression, we also found that age-eMS were enriched in enhancers and active chromatin, and are more likely to be hypo- than hypermethylated with older age. Notably, age-associated hypomethylation of an enhancer region of NFIA was the most significant age-dMS with cis-gene expression-associated methylation. Supporting the potential functional nature of these sites, decreased NFIA expression has been linked with increasing methylation of these CpG sites in response to all-trans retinoic acid treatment in a myeloid leukemia cell line 24.
Our age-dMS results are in agreement with many previous findings. Hyper age-dMS are more likely to occur in CpG islands 5, 5, 10, 12-14 and inactive chromatin 6, 10, 12, 13, 15, 31. Conversely, we found hypo age-dMS are more likely to occur in CpG shores and insulator regions 10, 14, 15, 31.
Why are so few age-dMS associated with gene expression? The occurrence of dMS has been proposed to be a highly polymorphic and stochastic process occurring during cell division 32, yet one that still results in consistent dMS across the genome due to chromatin state and sequence specific factors (e.g. CpG density, transcription factor binding sites) which may influence the likelihood of differential methylation 33. Thus, it is plausible that age-dMS may form stochastically as function of cell division (mitotic age), and are allowed to accumulate at specific, permissive locations. However, age-dMS have been reported in post-mitotic tissues 5, 10, 15, and across cells showing a wide range of proliferative capabilities 15, suggesting mitotic age alone does not explain the occurrence of age-dMS. Indeed, we found substantial overlap between age-dMS identified in two cell types with highly distinct replicative profiles (monocytes and T cells). Whether they occur as a function of mitotic age, or some other age-associated factors, the robust occurrence of age-dMS could be linked to their preference for inactive chromatin states and subsequent unimportance for nearby gene expression, thus allowing them to accumulate without harming cellular function. Further, cumulative effects of multiple agedMS on cis-gene expression 11 may have also been missed by investigating the relationship between individual CpGs with nearby gene expression. Age-eMS, on the other hand, may have more heterogeneous origins, representing both stochastic events that are allowed to occur and persist after loss of cellular homeostasis with age (e.g. cell-specific changes in histone modification patterns and transcription factor expression and activity), and also compensatory mechanisms by the cell in response to other age-related changes.
Supporting the link between age-eMS and biological aging, we found that genes with expression linked to potentially functional age-related methylation sites are enriched with antigen processing and presentation genes (MHC class I & II). Up-regulation of MHC class II signaling has been implicated in “para-inflammation” and the development of age-related chronic inflammatory diseases and autoimmune diseases 27, 34. Our present work supports previous findings that the MHC class II antigen presentation pathway is up-regulated with age 27, and suggests that methylation based regulatory mechanisms may contribute to the up-regulation of this pathway.
In summary, we identified and characterized an important subset of age-related methylation patterns that were associated with cis-gene expression in purified cells. These potentially functional methylation sites, particularly those associated with vascular aging or linked with expression of previously implicated “aging genes”, may facilitate the prioritization of biologically relevant age-associated methylation loci for future interrogation.
Methods
Study Population
The Multi-Ethnic Study of Atherosclerosis (MESA) was designed to investigate the prevalence, correlates, and progression of subclinical cardiovascular disease in a population cohort of 6,814 participants. Since its inception in 2000, five clinic visits collected extensive clinical, socio-demographic, lifestyle and behavior, laboratory, nutrition, and medication data 35. The present analysis is primarily based on analyses of purified monocyte samples from the April 2010-February 2012 examination of 1,264 randomly selected MESA participants (55-94 years old, Caucasian (47%), African American (21%) and Hispanic (32%), female 51%) from four MESA field centers (Baltimore, MD; Forsyth County, NC; New York, NY; and St. Paul, MN). T cells were also purified from 225 randomly selected samples which already had purified monocytes. The study protocol was approved by the Institutional Review Boards at Johns Hopkins Medical Institutions, University of Minnesota, Columbia University Medical Center, and Wake Forest University Health Sciences. All participants signed informed consent.
Power calculation
We estimate >80% power to detect age explaining as little as 3.0% of the variance of CpG methylation in monocytes, based on a simple linear regression (alpha = 0.05, two-sided test) with a Bonferroni correction to adjust for 448,523 CpG sites tested with age, estimated using QUANTO v1.2 (N=1,264, average age ± SD = 60.2 ± 9.5) 36. In T cells we estimate > 80% power to detect age explaining as little as 16% of the variance of CpG methylation in age (N=227).
Purification of Monocytes and T cells
Centralized training of technicians, standardized protocols, and extensive quality control (QC) measures were implemented for collection, on-site processing, and shipment of MESA specimens and routine calibration of equipment. Blood was initially collected in sodium heparin-containing Vacutainer CPT™ cell separation tubes (Becton Dickinson, Rutherford, NJ) to separate peripheral blood mononuclear cells from other elements within 2 hours from blood draw. Subsequently, monocytes and T cells were isolated with anti-CD14 and anti-CD4 monoclonal antibody coated magnetic beads, respectively, using AutoMACs automated magnetic separation unit (Miltenyi Biotec, Bergisch Gladbach, Germany). Based on flow cytometry analysis of 18 specimens, monocyte samples were consistently > 90% pure. T cells were isolated from a subset of our population (N=227) with anti-CD4 monoclonal antibody coated magnetic beads, respectively, using AutoMACs automated magnetic separation unit (Miltenyi Biotec, Bergisch Gladbach, Germany). Based on flow cytometry analysis of 18 specimens, T cell samples were consistently > 90% pure.
DNA and RNA Extraction
DNA and RNA were isolated from samples simultaneously using the AllPrep DNA/RNA Mini Kit (Qiagen, Inc., Hilden, Germany). DNA and RNA QC metrics included optical density (OD) measurements, using a NanoDrop spectrophotometer and evaluation of the integrity of 18s and 28s ribosomal RNA using the Agilent 2100 Bioanalyzer with RNA 6000 Nano chips (Agilent Technology, Inc., Santa Clara, CA) following manufacturer's instructions. RNA with RIN (RNA Integrity) scores > 9.0 was used for global expression microarrays. The median of RIN for all the analyzed samples was 9.9.
Epigenome-Wide Methylation Quantification
The Illumina HumanMethylation450 BeadChip and HiScan reader were used to perform the epigenome-wide methylation analysis. The EZ-96 DNA Methylation™ Kit (Zymo Research, Orange, CA) was used for bisulfate conversation with 1ug of input DNA (at 45 ul). 4 μl of bisulfite-converted DNA were used for DNA methylation assays, following the Illumina Infinium HD Methylation protocol. This consisted of a whole genome amplification step followed by enzymatic end-point fragmentation, precipitation, and resuspension. The resuspended samples were hybridized on HumanMethylation 450 BeadChips at 48°C for 16 h. The individual samples were assigned to the BeadChips and to chip position using the same sampling scheme as that for the expression BeadChips.
Global Expression Quantification
The Illumina HumanHT-12 v4 Expression BeadChip and Illumina Bead Array Reader were used to perform the genome-wide expression analysis, following the Illumina expression protocol. The Illumina TotalPrep-96 RNA Amplification Kit (Ambion/Applied Biosystems, Darmstadt, Germany) was used for reverse transcription, and amplification with 500 ng of input total RNA (at 11ul). 700 ng of biotinylated cRNA was hybridized to a BeadChip at 58°C for 16 – 17 hours. To avoid potential biases due to batch, chip, and position effects, a stratified random sampling technique was used to assign individual samples (including five common control sample for the first 480 samples) to specific BeadChips (12 samples/chip) and chip position.
Quality Control and Pre-Processing of Microarray Data
Data pre-processing and quality control (QC) analyses were performed in R (http://www.r-project.org/) using Bioconductor (http://www.bioconductor.org/) packages. For expression data, data corrected for local background were obtained from Illumina's proprietary software GenomeStudio. QC analyses and bead type summarization (average bead signal for each type after outlier removal) were performed using the beadarray package 37. Detection P-values were computed using the negative controls on the array. The neqc function of the limma 38 package was used to perform a normal-exponential convolution model analysis to estimate non-negative signal, quantile normalization using all probes (gene and control, detected and not detected) and samples, addition of a recommended (small) offset, log2 transformation, and elimination of control probe data from the normalized expression matrix. Multidimensional scaling plots showed the five common control samples were highly clustered together and identified three outlier samples, which were excluded subsequently.
Bead-level methylation data were summarized in GenomeStudio. Because the Illumina HumanMethylation450 BeadChip technology employs a two-channel system and uses both Infinium I and II assays, normalization was performed in several steps using the lumi package 39. We first adjusted for color bias using “smooth quantile normalization”. Next, the data were background adjusted by subtracting the median intensity value of the negative control probes. Lastly, data were normalized across all samples by standard quantile normalization applied to the bead-type intensities and combined across Infinium I and II assays and both colors. QC measures included checks for sex and race/ethnicity mismatches, and outlier identification by multidimensional scaling plots. The final methylation value for each methylation probe was computed as the M-value, essentially the log ratio of the methylated to the unmethylated intensity 40. The M-value is well suited for high-level analyses and can be transformed into the beta-value, an estimate of the percent methylation of an individual CpG site that ranges from 0 to 1 (thus M is logit(beta-value)).
The Illumina HumanMethylation450 BeadChip included probes for 485K CpG sites. Of these 485K CpG sites, 448,523 passed the following filters: “detected” methylation levels <90% of MESA samples using a detection p-value cut-off of 0.05, 65 control probes which assay highly-polymorphic single nucleotide polymorphisms (SNPs) rather than DNA methylation 41, or overlap with a repetitive element or region.
Pre-processing with global normalization removed large position and chip effects across all probes; however, probe-specific chip effects were found for some CpG sites and gene transcripts, while probe-specific position effects existed for some CpG sites but were ignorable for all gene transcripts. These probe-specific effects were included as covariates in all subsequent analyses.
The Illumina HumanHT-12 v4 Expression BeadChip included 48K transcripts. Statistical analyses were limited to probes retained after applying the following filters: non-detectable expression in ≥90% of MESA samples using a detection p-value cut-off of 0.0001, overlap with a repetitive element or region, low variance across the samples (<10th percentile), or putative and/or not well-characterized genes, i.e. gene names starting with KIAA, FLJ, HS, Cxorf, MGC, or LOC.
Association Analyses
The overall goal of the association analysis was to identify associations, at the genome-wide level, between age and CpG site methylation, and transcript expression and CpG site methylation. Association analyses were performed using the linear model (lm) function of the Stats package and the stepAIC function of the MASS package in R. To identify gene transcripts or methylation sites associated with age, we fit separate linear regression models with age as a predictor of transcript expression or the M-value for each gene transcript or CpG site, respectively. Covariates were sex, race/ethnicity, study site, expression/methylation chip, methylation position (for age-CpG methylation analyses only), and residual sample contamination with non-targeted cells (e.g. non-monocytes, see below). To identify methylation sites associated with gene expression in cis, we fit separate linear regression models with the M-value for each CpG site (adjusted for methylation chip and position effects) as a predictor of transcript expression for any autosomal gene within 1 Mb of the CpG site in question. Covariates were age, sex, and race/ethnicity, study site, expression chip, and residual sample contamination with non-targeted cells. Sex- and ethnicity-stratified analyses were performed as an internal validation and check of generalizability. To look for potential population stratification, we used EIGENSTRAT 42 to compute principal components (PCs) for each race, based on Affymetrix 6.0 array genotype data 43, and examined the association between the first five PCs and gene expression, as well as CpG methylation, in race stratified analyses. Less than 1% of expression transcripts and CpG methylation sites were associated with PCs in the Caucasian and African American populations (FDR<0.05). However, 14.7% of gene expression transcripts and 3.1% of methylation sites in the Hispanic population were associated with the first two PCs (FDR<0.05); therefore, analyses in the Hispanic population were adjusted for the first two PCs. Normality-based P-values were obtained for all tests (they were highly similar to permutation-based P-values and produced essentially the same ranking in a subset of 360 samples). P-values were adjusted for multiple testing using the q-value FDR method 44, separately for each of the three association analyses. To minimize false-positive results, we used the stringent FDR threshold of 0.001. All the reported FDR was calculated at the epigenome-wide level for the entire gene transcripts or methylation probes that were tested.
Although other phenotypic traits are available in MESA, only pulse pressure was tested as an age-associated outcome. Association analyses for individual gene transcripts and pulse pressure were performed using the linear model (lm) function in R. We fit separate linear regression models with of transcript expression as a predictor of pulse pressure. Covariates included sex, race/ethnicity, study site, expression/methylation chip, methylation position (for age-CpG methylation analyses only), and residual sample contamination. One set of analyses was performed with this set of covariates, and another set of analyses also included age as a covariate.
To estimate residual sample contamination for monocyte data analysis, we generated separate enrichment scores for neutrophils, B cells, T cells, and natural killer cells. We implemented a Gene Set Enrichment Analysis 45 to calculate the enrichment scores using the gene signature of each blood cell type in the ranked list of expression values for each MESA sample. The cell type-specific signature genes were selected from previously defined lists 46 and passed the following additional filters: at least four-fold more highly expressed in the targeted cell type than in other cell populations and low expression levels in the targeted cells (here, monocytes).
To minimize spurious associations due to a few highly influential data points, we calculated Cook's distance 47 for each data point and repeated the analysis after removing the top four samples with the highest Cook's distance. We then removed associations whose p-values no longer fell below the FDR-based significance threshold.
Functional Annotation Analysis
Histone modifications and CTCF binding reported in a CD14+ sample by ENCODE 48, and transcription factor binding sites in all available cell types were downloaded from the UCSC Genome Browser 19. Fold enrichment of CD14+ histone markers (H3K27me3, bivalent (H3K27me3/H3K4me3), H3K4me3, H3K4me1, H3K27ac, and H2A.Z) among monocyte agedMS and age-eMS were reported relative to all 448,523 CpG sites tested. Other predicted functional loci, such as promoters, enhancers, and insulators were predicted based on proximity to a transcription start sites, as well as the presence of overlapping histone modifications 18 (H3K4me1/3 and H3K27ac), or CTCF binding data available from ENCODE in a CD14+ sample. DAVID Bioinformatics Resources 6.7 49, 50 was used to examine the enrichment (FDR < 0.05) of GO (Gene Ontology) pathways for genes with expression associated (FDR<0.001) with methylation of monocyte age-eMS detected among predicted CD14+ promoter, enhancer, and insulator regions, against a background of all monocyte expressed genes (14,619 mRNA transcripts).
Supplementary Material
Acknowledgements
This research was supported by contracts N01-HC-from the National Heart, Lung and Blood Institute, by grants UL1-RR-024156 and UL1-RR-025005 from the National Center for Research Resources, and T32AG033534 from the National Institute of Aging. The MESA Epigenomics & Transcriptomics Study was funded by NHLBI grant R01HL101250 to Wake Forest University Health Sciences. The authors thank the other investigators, the staff, and the participants of the MESA study for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org.
Footnotes
Authors’ contributions
Y.L. oversaw the study, carried out experiments, researched the data, and wrote the manuscript. L.M.R. and J.R.T. carried out experiments, researched the data, and wrote the manuscript. J.D., D.S., I.H., C.E.M, D.H., R.P.T., carried out experiments, researched the data, and helped write the manuscript. K.L., C.J., G.B., W.P., S.S., D.R.J.J., H.S., and S.B.K. carried out experiments and researched the data.
Conflict of interest statement
The authors declare that they have no conflicts of interests.
Accession codes:
Microarray data presented in this manuscript have been deposited in the Gene Expression Omnibus (GEO) repository under the accession code GSE56047.
References
- 1.de Magalhães JP, Curado J, Church GM. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics. 2009;25:875–881. doi: 10.1093/bioinformatics/btp073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gravina S, Vijg J. Epigenetic factors in aging and longevity. Pflugers Arch. 2010;459:247–258. doi: 10.1007/s00424-009-0730-7. [DOI] [PubMed] [Google Scholar]
- 3.Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Human Molecular Genetics. 2009;18:R195–R201. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, et al. Methylomics of gene expression in human monocytes. Human Molecular Genetics. 2013;22:5065–5074. doi: 10.1093/hmg/ddt356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hernandez DG, et al. Distinct DNA methylation changes highly correlated with chronological age in the human brain. Hum Mol Genet. 2011;20:1164–1172. doi: 10.1093/hmg/ddq561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rakyan VK, et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20:434–439. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alisch RS, et al. Age-associated DNA methylation in pediatric populations. Genome Res. 2012;22:623–632. doi: 10.1101/gr.125187.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hannum G, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ong ML, Holbrook JD. Novel region discovery method for Infinium 450K DNA methylation data reveals changes associated with aging in muscle and neuronal pathways. Aging Cell. 2013 doi: 10.1111/acel.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day K, et al. Differential DNA methylation with age displays both common and dynamic features across human tissues that are influenced by CpG landscape. Genome Biology. 2013;14:R102. doi: 10.1186/gb-2013-14-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zykovich A, et al. Genome-wide DNA methylation changes with age in disease free human skeletal muscle. Aging Cell. 2013 doi: 10.1111/acel.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bell JT, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS. Genet. 2012;8:e1002629. doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyn H, et al. Distinct DNA methylomes of newborns and centenarians. Proc. Natl. Acad. Sci. U. S. A. 2012;109:10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson A, Enroth S, Gyllensten U. Continuous Aging of the Human DNA Methylome Throughout the Human Lifespan. PLoS One. 2013;8:e67378. doi: 10.1371/journal.pone.0067378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horvath S. DNA methylation age of human tissues and cell types. Genome Biology. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Winnefeld M, Lyko F. The aging epigenome: DNA methylation from the cradle to the grave. Genome Biology. 2012;13:165. doi: 10.1186/gb-2012-13-7-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garagnani P, et al. Methylation of ELOVL2 gene as a new epigenetic marker of age. Aging Cell. 2012;11:1132–1134. doi: 10.1111/acel.12005. [DOI] [PubMed] [Google Scholar]
- 18.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenbloom KR, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic Acids Res. 2013;41:D56–D63. doi: 10.1093/nar/gks1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schellenberg A, et al. Replicative senescence of mesenchymal stem cells causes DNA-methylation changes which correlate with repressive histone marks. Aging (Albany NY) 2011;3:873–888. doi: 10.18632/aging.100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christensen BC, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS. Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karolchik D, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zardo G, et al. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood. 2012;119:4034–4046. doi: 10.1182/blood-2011-08-371344. [DOI] [PubMed] [Google Scholar]
- 25.Nasu T, et al. Vasohibin-1, a negative feedback regulator of angiogenesis, ameliorates renal alterations in a mouse model of diabetic nephropathy. Diabetes. 2009;58:2365–2375. doi: 10.2337/db08-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dennis G, Jr., et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 27.VanGuilder HD, et al. Concurrent hippocampal induction of MHC II pathway components and glial activation with advanced aging is not correlated with cognitive impairment. J. Neuroinflammation. 2011;8:138. doi: 10.1186/1742-2094-8-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang G, et al. Genetic variations of Mrf-2/ARID5B confer risk of coronary atherosclerosis in the Japanese population. Int. Heart J. 2008;49:313–327. doi: 10.1536/ihj.49.313. [DOI] [PubMed] [Google Scholar]
- 29.Becker PM, et al. Pulmonary vascular permeability and ischemic injury in gelsolin-deficient mice. Am J Respir. Cell Mol Biol. 2003;28:478–484. doi: 10.1165/rcmb.2002-0024OC. [DOI] [PubMed] [Google Scholar]
- 30.Li GH, Arora PD, Chen Y, McCulloch CA, Liu P. Multifunctional roles of gelsolin in health and diseases. Med Res Rev. 2012;32:999–1025. doi: 10.1002/med.20231. [DOI] [PubMed] [Google Scholar]
- 31.Raddatz G, et al. Aging is associated with highly defined epigenetic changes in the human epidermis. Epigenetics & Chromatin. 2013;6:36. doi: 10.1186/1756-8935-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landan G, et al. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat Genet. 2012;44:1207–1214. doi: 10.1038/ng.2442. [DOI] [PubMed] [Google Scholar]
- 33.Lienert F, et al. Identification of genetic elements that autonomously determine DNA methylation states. Nat Genet. 2011;43:1091–1097. doi: 10.1038/ng.946. [DOI] [PubMed] [Google Scholar]
- 34.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 35.Bild DE, et al. Multi-ethnic study of atherosclerosis: objectives and design. Am. J. Epidemiol. 2002;156:871–881. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 36.Gauderman W, Morrison J. A computer program for power and sample size calculations for genetic-epidemiology studies. 2006 http://hydra.usc.edu/gxe.
- 37.Dunning MJ, Smith ML, Ritchie ME, Tavare S. beadarray: R classes and methods for Illumina bead-based data. Bioinformatics. 2007;23:2183–2184. doi: 10.1093/bioinformatics/btm311. [DOI] [PubMed] [Google Scholar]
- 38.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 39.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 40.Du P, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC. Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pidsley R, et al. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 43.Fox CS, et al. Genome-wide association of pericardial fat identifies a unique locus for ectopic fat. PLoS. Genet. 2012;8:e1002705. doi: 10.1371/journal.pgen.1002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U. S. A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Abbas AR, et al. Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun. 2005;6:319–331. doi: 10.1038/sj.gene.6364173. [DOI] [PubMed] [Google Scholar]
- 47.Henderson AR. Information for authors: Is the advice regarding the reporting of residuals in regression analysis incomplete? Should Cook's distance be included? Clin. Chem. 2006;52:1848–1850. doi: 10.1373/clinchem.2006.068296. [DOI] [PubMed] [Google Scholar]
- 48.ENCODE Project Consortium A user's guide to the encyclopedia of DNA elements (ENCODE). PLoS. Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang d.W., Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 50.Huang d.W., Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.