Abstract
Allergic airway inflammation is characterized by marked in situ changes in gene and protein expression, yet the role of microRNAs (miRNAs), a new family of key mRNA regulatory molecules, in this process has not yet been reported. Using a highly sensitive microarray based approach, we identified 21 miRNAs with differential expression between doxycycline-induced lung-specific IL-13 transgenic mice (with allergic airway inflammation) and control mice. In particular, we observed over-expression of miR-21 and under-expression of miR-1 in the induced IL-13 transgenic mice compared to control mice. These findings were validated in two independent models of allergen-induced allergic airway inflammation and in IL-4 lung transgenic mice. While IL-13 induced miR-21 expression was IL-13 receptor alpha 1 dependent, allergen induced miR-21 expression was mediated mainly independent of IL-13 receptor alpha 1 and STAT6. Notably, predictive algorithms identified potential direct miR-21 targets among IL-13-regulated lung transcripts such as IL-12p35 mRNA that was decreased in IL-13 transgenic mice. Introduction of pre-miR-21 dose-dependently inhibited cellular expression of a reporter vector harboring the 3’UTR of IL-12p35. Moreover, mutating miR-21 binding sites in IL-12p35 3’UTR abrogated miR-21 mediated repression. In summary, we have identified a miRNA signature in allergic airway inflammation, which includes miR-21 that modulates IL-12, a molecule germane to T helper cell polarization.
Keywords: Allergy, lung, inflammation, gene regulation
Introduction
Asthma is a chronic inflammatory disease characterized by inflammation of the airways, tissue remodeling, and a decline in respiratory function (1–3). In the United States, 5–10% of the population suffer from asthma, representing a common diagnosis for pediatric hospital admission and a major cause for lost days at work and school (4). Despite intense ongoing research, the incidence of the disease continues to rise, necessitating the need for new scientific inquiry.
MicroRNAs (miRNAs) are single stranded non-coding RNAs of 19–25 nucleotides in length that mediate post-transcriptional silencing of target genes (5, 6). In animals, miRNAs usually bind to complementary sites in the 3' untranslated region (UTR) of target genes and regulate target gene expression by either translational inhibition, mRNA degradation, or both (7). MiRNAs are involved in diverse biological processes, including development, stress response, cancer, and cardiac hypertrophy, implicating them in normal and pathological processes (8). However, to the best of our knowledge, studies relevant to asthma or asthma risk are still lacking, except for a recent report demonstrating that a single nucleotide polymorphism at the 3' untranslated region of HLA-G, an asthma-susceptibility gene, affects the binding of three miRNAs to this gene (9). Allergic airway inflammation may be particularly sensitive to miRNA regulation since it is characterized by marked changes in gene and protein expression in the lung (10–12). For example, lung over-expression of IL-13, a key T helper type 2 (Th2) cell derived effector cytokine in asthma pathogenesis, induces allergic airway inflammation characterized by prominent inflammatory cell accumulation, goblet cell metaplasia (mucus production), smooth muscle hyperplasia, and airway hyperresponsiveness (AHR), processes that are mediated by marked changes in gene and protein expression including cytokines and chemokines (13).
Here, we used both miRNA microarray and qRT-PCR based approaches to assess miRNA expression in murine models of allergic asthma. We define a miRNA signature consisting of 21 differentially regulated miRNAs in IL-13-induced experimental asthma. Focusing on the most highly induced miRNA, we subsequently demonstrate that miR-21 regulates murine IL-12p35, a key cytokine associated with balancing T helper cell polarization.
Material and Methods
Mice
Bitransgenic mice bearing CCSP-rtTA and (tetO)7CMV-IL-13 transgenes were generated in which IL-13 was expressed in a lung-specific manner that allowed for external regulation of transgene expression (14). Transgene expression was induced by feeding bi-transgenic mice doxycycline-impregnated food for 4 weeks. Constitutive IL-4 lung transgenic mice under the control of the CC10 promoter were kindly provided by Dr. Fred Finkelman (15). The IL-13 receptor alpha 1 and STAT6 deficient mice were described previously (16, 17).
Experimental asthma induction
Experimental asthma was induced by injection with 100 µg ovalbumin (OVA) and 1 mg aluminum hydroxide as adjuvant twice, followed by two 50 µg OVA or saline intranasal challenges 3 days apart, starting a least 10 days after the second sensitization. Mice were sacrificed 18–24 hours after the second challenge (10). Aspergillus fumigatus antigen-associated asthma was induced by challenging mice intranasally three times a week for 3 weeks with 100 µg (50 µl) of A. fumigatus extract or 50 µl of saline each time. Mice were sacrificed 48 hours after the last challenge (18). Experimental asthma from IL-13 bitransgenic mice was induced by feeding bi-transgenic mice doxycycline-impregnated food for 4 weeks, as described (19). The control group received no doxycycline. Mice were sacrificed at the end of 4 weeks of IL-13 induction. Intratracheal delivery of IL-13 was performed as previously described (20). All animals were housed under specific pathogen free conditions in accordance with institutional guidelines. The use of animals in these experiments was approved by the Institutional Animal Care and Use Committee of the Cincinnati Children’s Hospital Medical Center.
RNA extraction and microarray experiments
Total RNA was isolated from lung tissue using miRNeasy Mini Kit according to manufacturer’s protocol (Qiagen, Valencia, CA). RNA quality was assessed by using the Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA) and only samples with RNA integrity number >8 were used. RNA samples from these tissues were subsequently fluorochrome labeled by using the miRCURY Hy3/Hy5 labeling kit and hybridized to Exiqon miRCURY Locked Nucleic Acid (LNA) array (Version 10.0), comprising LNA modified probes for all mouse miRNAs in the release 10.0 of the miRBase microRNA Registry (21, 22). The microarray analysis was conducted in the Genomics and Microarray Core Facility at the University of Cincinnati. Data were normalized to a common reference sample using R statistical software (R Foundation for Statistical Computing) (23). MiRNA expression data were analyzed and displayed using Genesis (version 1.7.2) (24). The microarray data have been submitted to ArrayExpress database in compliance with minimum information about microarray experiment (MIAME) standards (ArrayExpress accession number: E-MEXP-1992, www.ebi.ac.uk/arrayexpress).
Quantitative Assessment of miRNA levels
Levels of miRNA expression were measured quantitatively by using TaqMan MicroRNA Assay (Applied Biosystems, Foster City, CA) as described following manufacturer’s protocol and assayed on the Applied Biosystems 7300 Real-Time PCR System (25). Normalization was performed with snoRNA202 (26). Comparative real-time PCR was performed in triplicate, including no-template controls. Relative expression was calculated using the comparative Ct method as previously described (27).
miRNA in situ hybridization
In situ hybridizations were performed in 8 µm cryosections from the lung of saline and Aspergillus fumigatus challenged mice (28). Slides were stained using an automated system, the Discovery XT (Ventana Medical Systems, Tucson, AZ), according to manufacturer’s protocols. Slides were pre-treated with 100 µl of RiboPrep (Ventana Medical Systems, Tucson, AZ) for 20 minutes at 37°C. After rinsing the slides with Reaction Buffer (Ventana Medical Systems, Tucson, AZ), slides were then treated with 100 µl of RiboClear (Ventana Medical Systems, Tucson, AZ) for 12 minutes at 37°C. The slides were rinsed again and then digested with Protease 3 (Ventana Medical Systems, Tucson, AZ) for 12 minutes at 37°C. After protease digestion, the digoxin labeled Locked Nucleic Acid (LNA) scrambled control probe and LNA miR-21 anti-sense probe (Exiqon A/S, Denmark) were hybridized to the slides at 52°C for 6 hours. Following post-hybridization washes with 0.1X SSC buffer at 47°C, 100 µl of rabbit anti-digoxin (Sigma, St. Louis, MO) antibody, diluted 1:2000 in Discovery Antibody Diluent (Ventana Medical Systems, Tucson, AZ), was applied to the slides for 30 minutes at room temperature. The slides were rinsed and then incubated with 100 µl of UltraMap anti-rabbit AP (Ventana Medical Systems, Tucson, AZ) for 16 minutes at room temperature. Color detection was done using a ChromoMap Blue Kit (Ventana Medical Systems, Tucson, AZ). Slides were counterstained with Nuclear Fast Red (Polyscientific, Bayshore, NY), coverslipped and mounted for viewing.
Cell Culture
Raw264.7 (ATCC No.: TIB-71, American Type Culture Collection, Manassas, VA) and NIH 3T3 (ATCC No.: CRL-1658, American Type Culture Collection, Manassas, VA) cells were maintained in DMEM medium supplemented with 10% FBS and 1% penicillin/streptomycin. The murine lung epithelial (MLE) cell line MLE15 and murine fetal lung mesenchyme (MFLM) cell line MFLM4 were kindly provided by Dr. Jeffrey Whitsett (Cincinnati Children’s Hospital) and were maintained in HITES media supplemented with 4% FBS and 1% penicillin/streptomycin (29). Type II alveolar cells were isolated and cultured as previously described (30) and were kindly provided by Dr. Timothy Weaver (Cincinnati Children’s Hospital). Bone marrow derived macrophages were prepared by culturing cells in medium containing 30% L929 conditioned medium (31). Bone marrow derived dendritic cells were prepared by culturing cells in medium supplemented with 40 ng/mL GM-CSF, 20 ng/mL IL-4, 10% FBS and 1% penicillin/streptomycin (32). Neutrophils were generated by using the thioglycollate-induced peritonitis model according to previously published methods (33). After 4 hours, mice were sacrificed, the peritoneal cavity was rinsed with 10 mL PBS and neutrophil purity was 95% as determined by Diff-Quik staining. For LPS activation of Raw264.7 cells, the cells were stimulated with 1 µg/mL LPS (Strain: 055:B5, Sigma, St. Louis, MO) for 24 hours.
Target Predictions
miRNA targets were predicted by using miRanda (34) and TargetScan (35) algorithms. Targets predicted by both algorithms were intersected with IL-13 down-regulated genes (19) and considered for further analysis. NFIB binding site was searched from the 1KB region upstream of 1st exon of all IL-13 induced genes in the lung using the TraFaC program which analyzes non-coding genomic sequences that are evolutionarily conserved between mouse and human (36).
IL-12p35 expression analysis
Total RNA was reverse transcribed using the High Capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA). Samples were analyzed by TaqMan quantitative RT-PCR for IL-12p35 transcripts and normalized to HPRT1 (primer and probe sets from Applied Biosystems, IL-12p35 assay ID: Mm00434169_m1, HPRT1 assay ID: Mm00446968_m1). Relative expression was calculated using the comparative Ct method.
Luciferase Reporter Plasmid Construction
The full length mIL-12p35 3’UTR was amplified with the following primers: mIL-12p35 Forward: ggccactagtGAAAGGCTCAAGGCCCTCT, mIL-12p35 Reverse: ggccaagcttGAACCACAAAATAAGGTATGTTTCAA, and cloned between the Spe I and Hind III sites of the multi cloning region in the 3’UTR of the firefly luciferase expression vector pMIR-Report (Ambion, Austin, TX) and designated pmIL-12p35. The pMIR-21 vector has a perfect miR-21 binding site cloned into the 3’UTR region of the pMIR-Report vector (Ambion, Austin, TX).
Site-directed mutagenesis of miR-21 binding site in pmIL-12p35
The mutations in the miR-21 binding site of pmIL-12p35 were introduced with the QuikChange II XL Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's protocol and designated pmIL-12p35 Mut. The mutagenesis primers are as follows: mIL-12p35 Mut Forward 5’-GGGTGACTGAGTGTTTTCATAAACACTTTGGCACAAAAACAATTCGAATTCAGTTCTTGCTCTTCTGCTAA-3’, mIL12p35 Mut Reverse: 5’-TTAGCAGAAGAGCAAGAACTGAATTCGAATTGTTTTTGTGCCAAAGTGTTTATGAAAACACTCAGTCACCC-3’. The constructs were sequenced to prove sequence integrity.
Transfection of pre-miRNA expression plasmid and reporter plasmids
293T cells were cotransfected with 500 ng of firefly reporter plasmids, 25 ng of reference renilla luciferase reporter plasmid pGL4.73 (Promega, Madison, WI), and 500 ng of pMIRNA1 – Pre-miR-21 or 500 ng of pMIRNA1 control vector (SystemBiosciences, Mountain View, CA) using Lipofectamine reagents as per the recommended conditions (Invitrogen, Carlsbad, CA). Lysates were prepared at 36 h post-transfection.
Dual-Luciferase Reporter Assays
Transfected cells were lysed in 300 µl of Passive Lysis Buffer (Promega, Madison, WI) for 30 min at room temperature. Firefly and renilla luciferase activity were measured using the Promega Dual Luciferase Assay kit following the manufacturer’s instructions and a Veritas Microplate luminometer (Turner Biosystems, Sunnyvale, CA). All measurements were normalized for renilla luciferase activity to correct for variations in transfection efficiencies and non-miR-21-specific effects of miRNA transfection on enzymatic activity.
Results
Expression profiling of miRNA in IL-13 lung transgenic mice
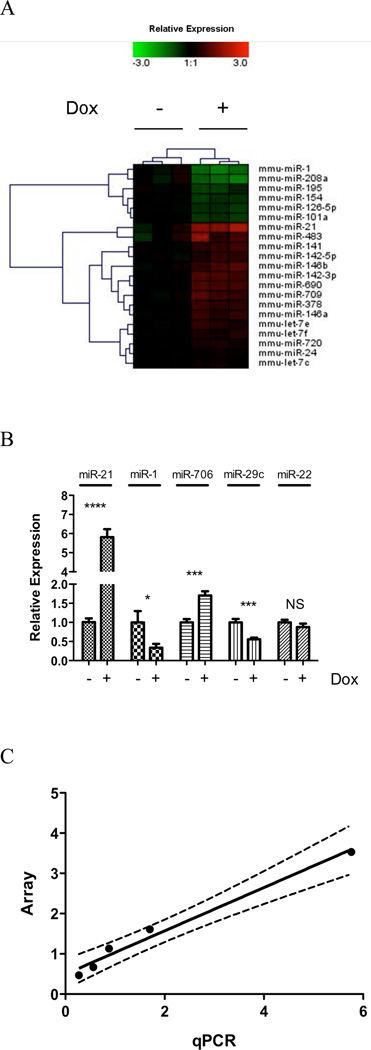
To identify miRNAs differentially expressed after IL-13 induction in IL-13 lung transgenic mice, miRNA expression of total lung tissue was profiled using Exiqon miRCURY LNA array (Version 10.0), comprising LNA modified probes for all mouse miRNAs available in the miRBase microRNA Registry (21, 22). Out of the 579 mouse miRNAs assayed, 131 miRNAs were expressed above background levels. In agreement with previous reports, several miRNAs, including miR-23b, miR-24, miR-30b, miR-451, and members of the let-7 family, were highly enriched in the mouse lung, giving strong hybridization signals on the miRNA arrays (data not shown) (37). Comparing doxycycline-induced IL-13 transgenic mice with control mice that received no doxycycline, 21 miRNAs were found to be differentially expressed at p < 0.01, suggesting that they were regulated directly or indirectly by IL-13 (Fig. 1A). Notably, miR-21 was the most up-regulated miRNA on the array and miR-1 was the most down-regulated miRNA. To validate the results of the microarray platform, we determined the expression of a subset of miRNAs by real time RT-PCR (Fig. 1B). We found strong correlation between our microarray profiling and real-time RT-PCR data (Pearson correlation coefficient: 0.99, p = 0.001, Fig. 1C).
Figure 1. MiRNA expression profile in IL-13 transgenic mice lung.
(A) Heat map of 21 differentially expressed miRNAs following control no doxycycline (−) and doxycycline (+) exposure for 28 days. Relative expression is log2 transformed. (B) A qRT-PCR validation of a selected set of miRNA probes normalized to snoRNA202. NS, not significant; *, p < 0.05; ***, p < 0.001, ****, p < 0.0001. (C) Correlation of miRNA microarray and qRT-PCR validation; dashed line represents 95% confidence interval. Data are represented as mean ± S.E.M; n = 5–7 mice per group; data representative of 3 experiments.
MiR-21 is induced in three separate models of experimental asthma while miR-1 is repressed in the same models
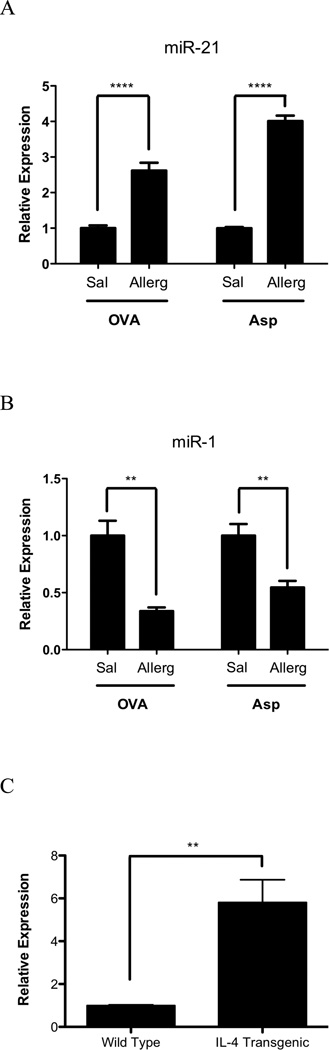
We aimed to determine levels of miR-21 and miR-1 in three independent asthma models (OVA, Aspergillus fumigatus and induced IL-13 bitransgenic mice) using real-time RT-PCR analysis. In the OVA model, mice were sensitized by two i.p. injections of OVA and aluminum hydroxide. The Aspergillus fumigatus model involves a unique mucosal sensitization route (intranasal) compared with the OVA model (10, 18, 20, 38, 39). While the methods of experimental asthma induction are different, all three asthma models have similar phenotypes, including Th2-associated eosinophilic inflammation, mucus production, and AHR (10, 18, 19, 40, 41). First, we examined the inducible IL-13 bitransgenic system and indeed validated that dox-treated IL-13 bitransgenic mice had a 4.62-fold increase in miR-21 (p < 0.01) and a 0.28-fold repression of miR-1 (p < 0.05) compared to control mice that received no doxycycline (Fig. 1B). Second, we examined the OVA-induced asthma model and demonstrated that OVA-challenged mice had a 2.64-fold (p < 0.0001) induction of miR-21 and a 0.34-fold repression of miR-1 (p < 0.001) compared to saline challenged mice (Fig. 2A–B). Third, we examined the Aspergillus fumigatus model of experimental asthma and demonstrated that antigen challenged mice have a 4.01-fold increase in miR-21 level (p < 0.0001) and a 0.55-fold repression of miR-1 (p < 0.01) compared with control mice (Fig. 2A–B). In these models, the BALF eosinophil levels were 1.15 ± 0.34 × 104, 3.39 ± 0.76 × 106 and 4.94 ± 0.88 × 105 cells following IL-13, OVA, and Aspergillus fumigatus challenge, respectively, as reported (39). Finally, to determine if the induction of miR-21 could be mediated by IL-4, we determined miR-21 expression level in IL-4 lung transgenic mice and found that IL-4 transgenic mice had a 5.81 ± 1.06-fold induction of miR-21 (p < 0.01) compared to wild type mice (Fig. 2C).
Figure 2. Expression of miR-21 and miR-1 in experimental asthma models.
MiR-21 (A) and miR-1 (B) expression were assessed in OVA and Aspergillus fumigatus asthma models. (C) MiR-21 expression was determined in IL-4 lung transgenic mice. The relative expression levels were determined by qRT-PCR normalized to snoRNA202; *, p < 0.05; **, p < 0.01, ***, p < 0.001; ****, p < 0.0001. Data are represented as mean ± S.E.M; n = 3–7 mice per group; data representative of 3 experiments.
MiR-21 is induced predominantly by an IL-13 receptor alpha 1 independent pathway
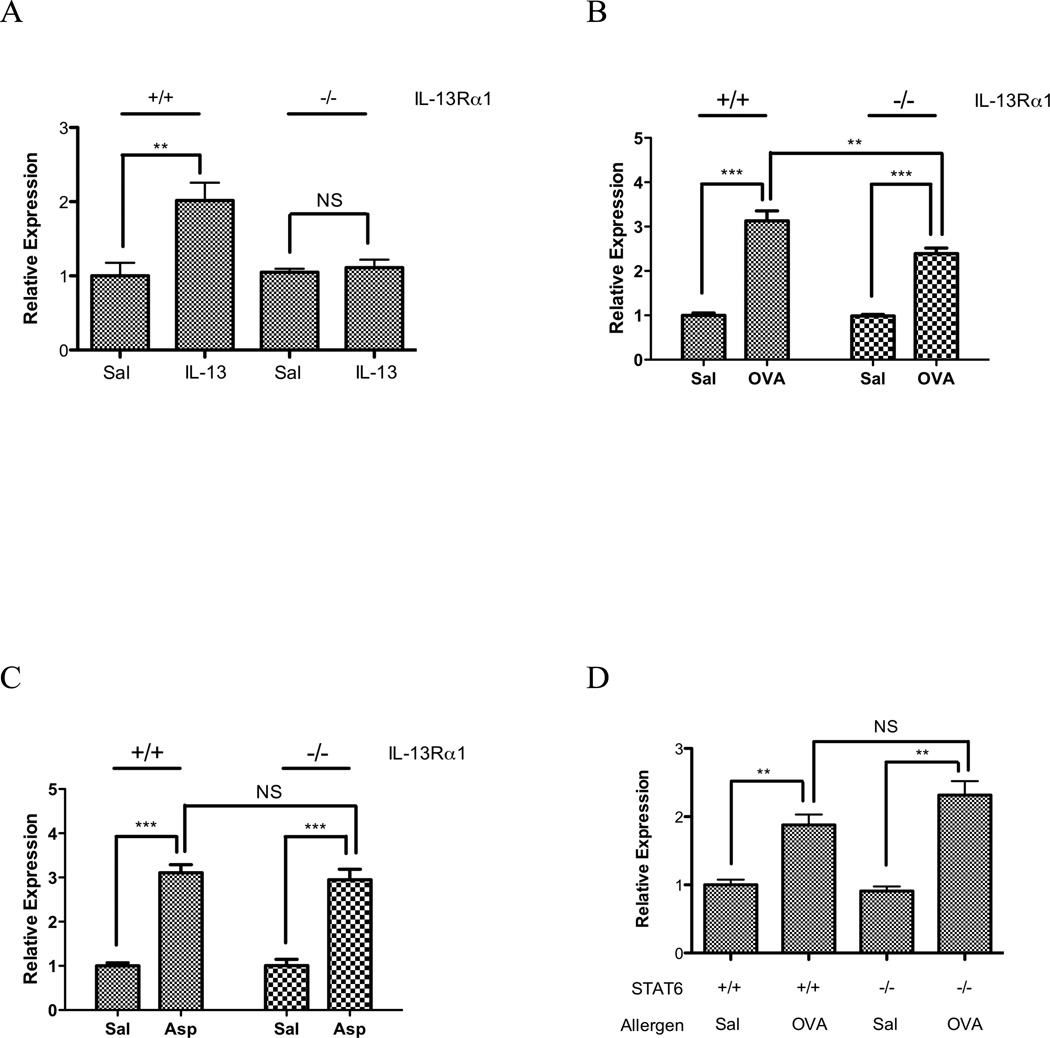
We focused on miR-21 as this was the most markedly changed miRNA and one implicated in processes germane to asthma including cell growth and differentiation, tissue remodeling and myeloid cell function (42–45). Having identified miR-21 as both an IL-13 and allergen induced gene, we were interested in determining if the IL-13 or allergen-induced expression was IL-13Rα1 dependent, as we have recently reported that this receptor mediates some of the key cardinal features of experimental asthma (airway hyperresponsiveness and goblet cell metaplasia), but not leukocyte accumulation (39). First, we delivered IL-13 intratracheally to wild type and IL-13Rα1 gene deficient mice and demonstrated that IL-13 induced miR-21 was IL-13Rα1 dependent (Fig. 3A). We then examined both the OVA-induced model and the Aspergillus fumigatus model of experimental asthma using wild-type and IL-13Rα1 gene deficient mice. Notably, this analysis revealed that both OVA and Aspergillus fumigatus induced miR-21 expression largely through an IL-13Rα1 independent pathway. OVA-challenged wild type mice had a 3.13 ± 0.22-fold induction of miR-21, while OVA challenged gene targeted mice had a 2.39 ± 0.13-fold induction (p < 0.01 for saline vs. OVA in both wild type and gene targeted mice). Aspergillus fumigatus challenged wild type mice had a 3.11 ± 0.10-fold induction of miR-21, while Aspergillus fumigatus challenged gene targeted mice had a 2.95 ± 0.23-fold induction (p < 0.01 for saline vs. Aspergillus fumigatus in both wild type and gene targeted mice) (Fig. 3B–C). We subsequently determined that miR-21 was induced through a STAT6 independent mechanism in OVA challenged mice (Fig. 3D).
Figure 3. Expression of miR-21 in allergen challenged IL-13 receptor alpha 1 deficient mice.
(A) MiR-21 expression was determined in IL-13 challenged wild type and IL-13 receptor alpha 1 deficient mice. (B–C) MiR-21 expression was determined in OVA (B) and Aspergillus fumigatus (Asp) models (C) using IL-13 receptor alpha 1 (−/−) mice and wild type (+/+) controls. (D) MiR-21 expression was determined in OVA challenged wild type and STAT6 deficient mice. The relative expression levels were determined by qRT-PCR normalized to snoRNA202. NS, not significant; **, p < 0.01, ***, p < 0.001. Data are represented as mean ± S.E.M.; n = 3–8 mice per group; data representative of 3 experiments.
MiR-21 is expressed in cells of monocyte/macrophage lineage in the allergic lung
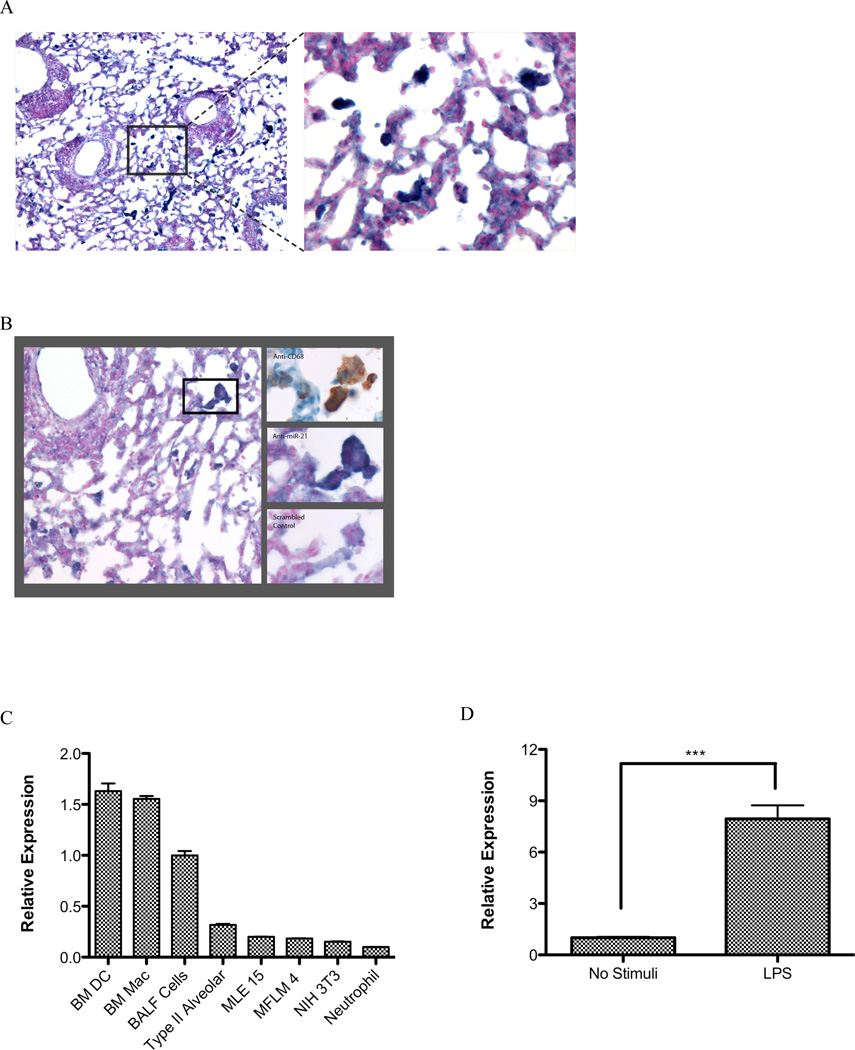
To determine the cell type specific localization of miR-21, in situ hybridization (ISH) was carried out on cryosections of Aspergillus fumigatus challenged lungs using LNA anti-miR-21 and scrambled control probes (28, 46). MiR-21 was primarily detected in the cytoplasm of mononuclear and multi-nucleated myeloid cells with morphology resembling monocytes/macrophages (Fig. 4A). To determine if these cells are indeed in the monocyte/macrophage lineage (both are CD68+), we performed anti-CD68 staining on serial sections. Serial sectioning revealed that anti-CD68 staining was associated with miR-21 expression, at least in part (Fig. 4B). Staining of doxcycline induced IL-13 bitransgenic mouse lungs showed similar results (Supplementary Fig. 1). We quantified the number of miR21 positive cells as a function of total airway macrophages as determined by CD68+ staining in serial sections and found that 64 ± 6.8 % of (mean ± S.D, n=3 mice) CD68+ cells were miR-21 positive. In addition, we determined the expression profile of miR-21 in different cell types and found that bone marrow derived macrophages and dendritic cells expressed relatively high levels of miR-21 compared with lung epithelial cells, fibroblasts and neutrophils (Fig. 4C). The cell composition of BALF cells can be found in Supplementary Fig. 2. We subsequently demonstrated that miR-21 is inducible in a murine macrophage cell line by LPS treatment (Fig. 4D).
Figure 4. In situ hybridization of miR-21 in Aspergillus fumigatus challenged wild type mouse lung.
Expression of miR-21 in (A–B) Aspergillus fumigatus challenged wild type mouse lungs, were determined by LNA-based in situ hybridization. (A) LNA-anti-miR-21, left: 100X field; right: 400× field; (B) Left: 200× field, LNA-anti-miR-21. Right: serial sections at 600×; right top: LNA-anti-miR-21, right middle: anti-CD68, right bottom: LNA-scrambled control probe. (C) Relative expression of miR-21 in different cell types; BM DC: bone marrow derived dendritic cells; BM Mac: bone marrow derived macrophages; BALF: bronchoalveolar lavage fluid cells; MLE15: murine lung epithelial cell line; MFLM4: murine lung mensenchyme cell line; NIH 3T3: murine fibroblasts. (D) Relative expression of miR-21 in LPS stimulated murine macrophage cell line Raw264.7; ***, p < 0.001. Data are representative of three experiments.
MiR-21 targets in the allergic lung
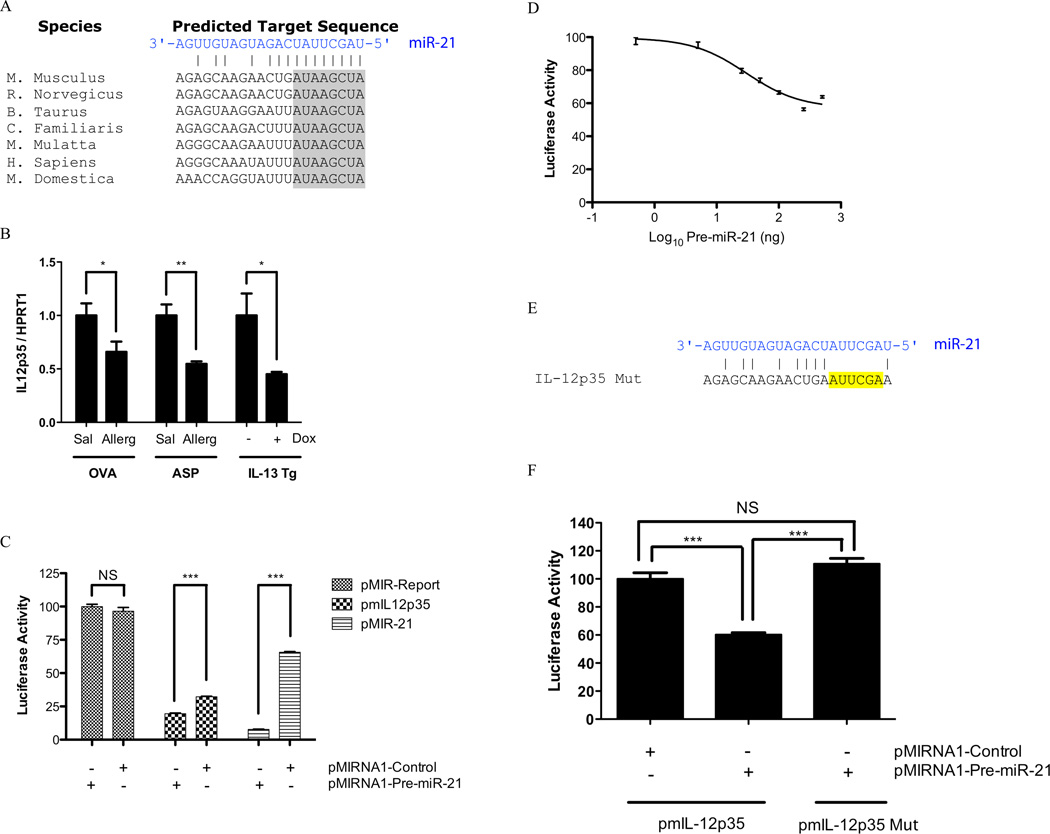
We searched for miR-21 mRNA targets using target-prediction software miRanda and TargetScan. It has been reported that integration of miRNA target predictions from multiple algorithms substantially increases the functional correlations and decreases the false positive rate compared to single algorithms (47, 48). Common predicted targets between miRanda and TargetScan algorithms arrived with a list of 37 predicted targets (Table I). While some of the targets of miRNAs are modulated through translational inhibition only, the majority of targets have mRNA level changes inverse to their respective miRNA regulator (49, 50). We thus intersected the 37 predicted targets with IL-13 down-regulated genes and arrived at a list of IL-13 regulated target genes (Table II). Notably, IL-12p35 was predicted to have a strong miR-21 binding site conserved across species (Fig. 5A). We determined the expression level of IL-12p35 and found that it was indeed down-regulated in all three of the employed asthma models (Fig. 5B).
Table I.
Predicted targets of miR-21 that are common between miRanda and TargetScan algorithms
| Gene Symbol | Description | Transcript ID |
|---|---|---|
| Arhgap24 | Rho GTPase-activating protein 24 | ENSMUST00000094559 |
| Aspn | Asporin | ENSMUST00000021820 |
| Brd1 | Bromodomain containing 1 | ENSMUST00000023022 |
| Cbx4 | Chromobox homolog 4 | ENSMUST00000026665 |
| Ccl1 | Chemokine (C-C motif) ligand 1 | ENSMUST00000021043 |
| Cntfr | Ciliary neurotrophic factor receptor | ENSMUST00000102962 |
| Dazl | Deleted in azoospennia-like | ENSMUST00000010736 |
| Hnrpu | Heterogeneous nuclear ribonucleoprotein U | ENSMUST00000037748 |
| Il12A | IL-I2a | ENSMUST00000029345 |
| Jag1 | Jagged 1 | ENSMUST00000028735 |
| Kcna3 | Potassium voltage-gated channel, shaker-related subfamily, member 3 | ENSMUST00000052718 |
| Krit1 | KRIT1, ankyrin repeat containing | ENSMUST00000080085 |
| Matn2 | Matrilin 2 | ENSMUST00000022947 |
| Mrpl9 | Mitochondrial ribosomal protein L9 | ENSMUST00000029786 |
| Mtap | Methylthioadenosine phosphorylase | ENSMUST00000058030 |
| Nfib | NFIB | ENSMUST00000050872 |
| Ntf3 | Neurotrophin 3 | ENSMUST00000050484 |
| Pcbpl | Poly(rC)-binding protein 1 | ENSMUST00000053015 |
| Pcbp2 | Poly(rC)-binding protein 2 | ENSMUST00000077037 |
| Pdcd4 | Programmed cell death 4 | ENSMUST00000025931 |
| Peli 1 | Pellino 1 | ENSMUST00000093290 |
| Pitx2 | Paired-like homeodomain transcription Factor 2 | ENSMUST00000029657 |
| Plag1 | Pleiomorphic adenoma gene 1 | ENSMUST00000003369 |
| Ppara | Peroxisome proliferator-activated receptor α | ENSMUST00000057979 |
| Psrcl | Proline/serine-rich coiled-coil 1 | ENSMUST00000102630 |
| Reck | Reversion-inducing cysteine-rich protein With kazal motifs | ENSMUST00000030198 |
| Rnf1O3 | Ring finger protein 103 | ENSMUST00000064637 |
| Satbl | Special AT-rich sequence-binding protein 1 | ENSMUST00000024720 |
| Ski | Sloan-Kettering viral oncogene homolog | ENSMUST00000030917 |
| Sox2 | SRY box containing gene 2 | ENSMUST00000099151 |
| Spg20 | Spastic paraplegia 20, spartin (Troyer syndrome) Homolog (human) | ENSMUST00000044116 |
| Stag2 | Stromal Ag 2 | ENSMUST00000069619 |
| Tgfbi | TGF, β induced | ENSMUST00000045173 |
| Tiam1 | T cell lymphoma invasion and metastasis 1 | ENSMUST00000002588 |
| Wwp1 | WW domain containing E3 ubiquitin protein Ligase 1 | ENSMUST00000035982 |
| Xkr6 | X Kell blood group precursor related family Member 6 homolog | ENSMUST00000100485 |
| Zcchc3 | Zinc finger, CCHC domain containing 3 | ENSMUST00000099207 |
Table II.
Predicted targets of miR-21 that are common between miRanda, TargetScan, and IL-13 down-regulated genes
| Name | Transcript ID | TargetScan Prediction | miRanda Prediction |
|---|---|---|---|
| Cntfr | ENSMUST00000102962 |  |
 |
| Il12A | ENSMUST00000029345 |  |
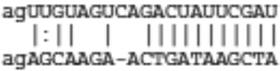 |
| Pitx2 | ENSMUST00000029657 | 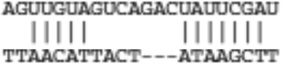 |
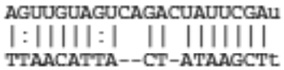 |
| Plag1 | ENSMUST00000003369 |  |
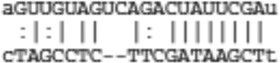 |
| Psrc1 | ENSMUST00000102630 | 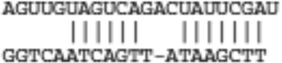 |
 |
| Reck | ENSMUST00000030198 | 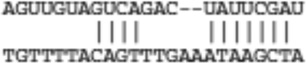 |
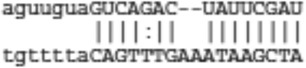 |
| Satb1 | ENSMUST00000024720 | 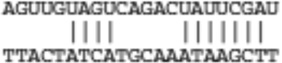 |
 |
| Sox2 | ENSMUST00000099151 |  |
 |
| Spg20 | ENSMUST00000044116 |  |
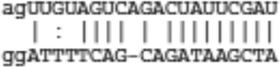 |
| Wwp1 | ENSMUST00000035982 | 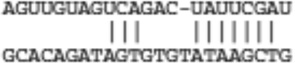 |
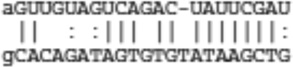 |
Figure 5. MiR-21 targets IL-12p35.
(A) Predicted highly conserved binding site for miR-21 in 3’UTR of IL-12p35. The 8-mer seed sequence is shaded in gray. (B) IL-12p35 expression was determined in OVA, Aspergillus fumigatus (Asp), and doxcycline induced IL-13 bitransgenic models. The relative expression levels were determined by qRT-PCR normalized to HPRT1. *, p < 0.05; **, p < 0.01; n = 5–7 mice per group. (C) Relative luciferase activity in 293T cells co-transfected with control firefly luciferase vector (pMIR-Report), or a firefly luciferase reporter vector containing the 3’UTR of IL-12p35 (pmIL12p35), or a firefly luciferase vector with perfect miR-21 binding site in the 3’UTR (pMIR-21), and either the pre-miR-21 expression vector (pMIRNA1-Pre-miR-21) or control vector (pMIRNA1-Control). Firefly luciferase activity was normalized to the renilla luciferase activity then to the average of the control firefly luciferase reporter; NS, not significant; ***, p < 0.001; n =4 per group, data representative of 3 experiments. (D) A dose response study of pre-miR-21 expression vector on the luciferase activity of the luciferase vector containing the 3’UTR of IL-12p35; n = 4 per group, data representative of 3 experiments. (E) Mutation (yellow-highlighting) in the 3’UTR of mIL-12p35. (F) Relative luciferase activity in 293T cells co-transfected with reporter plasmid containing either the wild type or mutant mIL-12p35 3’UTR. Firefly luciferase activity was normalized to the renilla luciferase activity then to the average of the wild type mIL-12p35 firefly luciferase reporter; NS, not significant; ***, p < 0.001; n = 4 per group, data representative of 3 experiments. All data are represented as mean ± S.E.M.
MiR-21 targets IL-12p35
To determine whether IL-12p35 is a molecular target of miR-21, we constructed a luciferase reporter vector containing the full length mIL-12p35 3’UTR, as well as a positive control vector harboring a perfect miR-21 complementary sequence in the 3’UTR region. Transfecting the miR-21 expression vector inhibited the expression of the luciferase reporter vector containing the mIL-12p35 3’UTR as well as the expression of the positive control vector, while there was no effect in the luciferase vector that did not contain a miR-21 binding site (Fig. 5C). The reduced luciferase activity of mIL-12p35 and pMiR-21 vectors at baseline is likely due to inhibition from both the endogenously expressed miRNAs, as well as other translational inhibition mechanisms such as the 5’-3’ circularization disruption by endogenous translational inhibitor proteins that bind to the 3’UTR region (51). The inhibition of mIL-12p35 reporter was dose dependent with an ED50 of 29.84 ± 1.24 ng (7.19 ± 0.30 nM) and a plateau reached at 250 ng (60 nM) (Fig. 5D). Mutation of the seed sequence of the predicted miRNA-binding site abrogated this effect (Fig. 5E–F).
Discussion
Our study has provided a comprehensive global miRNA expression profile of an IL-13 induced asthma model. Nearly 4% of 579 miRNAs assayed displayed differential expression in IL-13 transgenic mice compared to control mice (Figure 1). This level of miRNA change is consistent with other disease states such as breast cancer, leukemia, and myocardial infarction (52–54). Notably, each miRNA has been predicted to potentially target hundreds of genes (35, 55), indicating the potential significance of small changes in miRNAs. We found that miR-21 and miR-1 were the most induced and repressed miRNAs, respectively. We validated the induction of miR-21 and repression of miR-1 in two additional independent models of allergic airway inflammation as well as the induction of miR-21 by chronic over-expression of the IL-4 transgene in the lung. We analyzed the receptor dependency of miR-21 induction and found that IL-13 induced expression of miR-21 was dependent on IL-13Rα1, but allergen induced miR-21 expression was mediated largely by an IL-13Rα1 independent pathway. Although IL-13 alone completely utilizes IL-13Rα1 to induce experimental features of asthma, this receptor has a key but not complete role in the development of allergen-induced experimental asthma. Notably, leukocyte recruitment to the lung occurs independent of IL-13Rα1 (39). As such, the finding that miR21 induction occurs independent of IL-13Rα1, indicates that miR21 induction likely is derived from (or associated) with the sustained leukocyte recruitment and/or activation in IL-13Rα1 deficient mice. Consistent with this, allergen induced miR-21 was demonstrated to occur independent of STAT6, consistent with sustained leukocyte recruitment in these mice (56). This implies that miR-21 induction may be related to the large portion of asthma signature genes that are STAT6 independent (56). The STAT6 and IL-13Rα1 independent genes that correlate with miR-21 include the C3a receptor and several chemokines (Ccl8, Ccl12, Cxcl10, data not shown) (39, 56). Notably, recent studies demonstrate that certain aspects of asthma occur through a STAT6 independent pathway (57, 58). In situ hybridization of asthmatic lung revealed that miR-21 was expressed by inflammatory leukocytes most consistent with myeloid cells and in agreement with recent studies identifying miR-21 in hematopoietic cells (59–61). Cell type expression profile of miR-21 confirmed that macrophages and dendritic cells indeed had the highest level of expression compared with other cell types analyzed. Although the expression of miR-21 was in cells most consistent with macrophages and/or dendritic cells (both CD68+), this does not rule out expression in other cells that may contain less readily detectable RNA (e.g. eosinophils). Notably, miR-21 has been shown to be inducible in a human promyelocytic cell line HL-60 after phorbol 12-myristate 13-acetate (PMA) treatment, which induces macrophage like differentiation (60). We subsequently demonstrated that miR-21 is inducible in the murine macrophage cell line Raw264.7 by LPS treatment. The relationship between the LPS induced pathway and our in vivo finding has yet to be determined. In addition, miR-21 has been reported to target the transcriptional repressor NFIB (43), providing a mechanism by which miR-21 induction could up-regulate gene expression. We performed in silico analysis of all IL-13 induced genes in the lung, and identified 130 genes with potential NFIB sites in the 1KB region upstream of the 1st exon (Supplementary Table I).
Using bioinformatic approaches, we identified candidate target genes of miR-21. We further intersected this with our previous IL-13-repressed mRNA expression profiling data to identify candidate target genes that were differentially expressed, possibly because of action of these miRNAs. This analysis identified IL-12p35 as a putative target of miR-21. Transfection and reporter assays indeed identified mIL-12p35 as a target gene of miR-21, with miR-21 having the ability to reduce IL-12p35 expression via the 3’UTR. The magnitude of miR-21 mediated repression of the mIL-12p25 reporter vector was about 40%, similar with the reported effects of miRNA-mediated mRNA repression in other systems (44, 49, 50, 62–67). These results imply that the increased levels of miR-21 in experimental asthma contribute to the observed decrease in IL-12. IL-12 is a key cytokine derived from macrophages and dendritic cells (both CD68 positive and consistent with our in situ hybridization results) involved in adaptive immune responses involving Th1 cell polarization (68, 69). The ability of miR-21 to down regulate IL-12 indicates a novel checkpoint for regulating the level of this key immune mediator. These results suggest that strategies designed to deliver miR-21 may prime for Th2 and IL-13 associated responses; conversely, miR-21 inhibition has potential to drive Th1 polarization, promoting cellular responses (and classic adjuvant activity) (70, 71). Down-regulation of IL-12p35 could also affect IL-35 production and Treg cell function, leading to a pro-inflammatory phenotype (72). In addition to regulating IL-12 levels, miR-21 may regulate other processes germane to allergic airway inflammation. Notably, MiR-21 has been shown to regulate the level of the matrix metalloprotease (MMP) inhibitor RECK (42, 73, 74), and indeed we have identified this gene to be down regulated by IL-13 in the lung (Table II); it is interestingly to speculate that this could account for some of increased MMP activity seen in asthma (75, 76).
In our study, we identified miR-1 as the most down-regulated gene in the IL-13 lung transgenic mice. MiR-1 is considered to be a muscle specific miRNA (77–79), implicated in the determination of the differentiated state of muscle cells, in myogenesis and in cardiogenesis (78, 80). Down-regulation of miR-1 has been associated with cardiac and skeletal muscle hypertrophy (81, 82). Notably, disruption of just one of the two miR-1 family members, miR-1-2, has profound consequences for development and maintenance of the heart, as miR-1-1 did not compensate for loss of miR-1-2 (80). This indicates that a stable level of miR-1 is very important for normal muscle physiology. MiR-1 is down-regulated in all three of our asthma models ranging from 1.82- to 3.57-fold. It is interesting to speculate that down-regulation of miR-1 contributes to the smooth muscle hypertrophy and re-modeling seen in asthma (83).
In conclusion, we report a miRNA signature of experimental asthma. We found miR-21 is the most up-regulated miRNA and the up-regulation is largely through an IL-13Rα1 independent pathway. Using computational and tissue culture-based assays, we identify mIL-12p35 as a molecular target of miR-21. As such, miR-21 induction in experimental asthma likely leads to a concomitant decrease in mIL-12p35, which likely contributes to polarization of T helper cells toward a Th2 response. This increase in the expression of miR-21 likely contributes to the action of IL-13 in the lung (and possibly asthma pathogenesis). Taken together, these results suggest a key role for miRNA in allergic lung inflammation.
Supplementary Material
Acknowledgements
We thank Maureen A. Sartor for help with microarray data analysis. We are grateful to Drs. Pablo Abonia, Bruce Aronow, Charles DeBrosse, Fred Finkelman, Patricia Fulkerson, Leigh Grimes, Keith Stringer, Timothy Weaver, Jeffrey Whitsett and Nives Zimmermann, for helpful discussions, technical expertise and/or review of this manuscript.
Footnotes
This work was supported by NIH P01 HL076383 (M.E.R.), R01 AI057803 (M.E.R.). and the Organogenesis Training Grant (NIH T32 HD046387 supporting T.X.L.). This work was also supported by Medical Scientist Training Program training grant (T32 GM063483 supporting T.X.L) from the National Institute of General Medical Sciences.
References
- 1.Elias JA, Zhu Z, Chupp G, Homer RJ. Airway remodeling in asthma. J Clin Invest. 1999;104:1001–1006. doi: 10.1172/JCI8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosse Y, Pare PD, Seow CY. Airway wall remodeling in asthma: from the epithelial layer to the adventitia. Curr Allergy Asthma Rep. 2008;8:357–366. doi: 10.1007/s11882-008-0056-0. [DOI] [PubMed] [Google Scholar]
- 3.Vignola AM, Mirabella F, Costanzo G, Di Giorgi R, Gjomarkaj M, Bellia V, Bonsignore G. Airway remodeling in asthma. Chest. 2003;123:417S–422S. doi: 10.1378/chest.123.3_suppl.417s. [DOI] [PubMed] [Google Scholar]
- 4.Platts-Mills TA, Carter MC, Heymann PW. Specific and nonspecific obstructive lung disease in childhood: causes of changes in the prevalence of asthma. Environ Health Perspect. 2000;108(Suppl 4):725–731. doi: 10.1289/ehp.00108s4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 6.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 7.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 8.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 9.Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, Ober C. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmermann N, King NE, Laporte J, Yang M, Mishra A, Pope SM, Muntel EE, Witte DP, Pegg AA, Foster PS, Hamid Q, Rothenberg ME. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest. 2003;111:1863–1874. doi: 10.1172/JCI17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuperman DA, Lewis CC, Woodruff PG, Rodriguez MW, Yang YH, Dolganov GM, Fahy JV, Erle DJ. Dissecting asthma using focused transgenic modeling and functional genomics. Journal of Allergy and Clinical Immunology. 2005;116:305–311. doi: 10.1016/j.jaci.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 12.Lewis CC, Yang JY, Huang X, Banerjee SK, Blackburn MR, Baluk P, McDonald DM, Blackwell TS, Nagabhushanam V, Peters W, Voehringer D, Erle DJ. Disease-specific gene expression profiling in multiple models of lung disease. Am J Respir Crit Care Med. 2008;177:376–387. doi: 10.1164/rccm.200702-333OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, Zhang Y, Elias JA. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wan H, Kaestner KH, Ang SL, Ikegami M, Finkelman FD, Stahlman MT, Fulkerson PC, Rothenberg ME, Whitsett JA. Foxa2 regulates alveolarization and goblet cell hyperplasia. Development. 2004;131:953–964. doi: 10.1242/dev.00966. [DOI] [PubMed] [Google Scholar]
- 15.Finkelman FD, Yang M, Perkins C, Schleifer K, Sproles A, Santeliz J, Bernstein JA, Rothenberg ME, Morris SC, Wills-Karp M. Suppressive effect of IL-4 on IL-13-induced genes in mouse lung. J Immunol. 2005;174:4630–4638. doi: 10.4049/jimmunol.174.8.4630. [DOI] [PubMed] [Google Scholar]
- 16.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 17.Ramalingam TR, Pesce JT, Sheikh F, Cheever AW, Mentink-Kane MM, Wilson MS, Stevens S, Valenzuela DM, Murphy AJ, Yancopoulos GD, Urban JF, Jr, Donnelly RP, Wynn TA. Unique functions of the type II interleukin 4 receptor identified in mice lacking the interleukin 13 receptor alpha1 chain. Nat Immunol. 2008;9:25–33. doi: 10.1038/ni1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fulkerson PC, Fischetti CA, McBride ML, Hassman LM, Hogan SP, Rothenberg ME. A central regulatory role for eosinophils and the eotaxin/CCR3 axis in chronic experimental allergic airway inflammation. Proc Natl Acad Sci U S A. 2006;103:16418–16423. doi: 10.1073/pnas.0607863103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fulkerson PC, Fischetti CA, Hassman LM, Nikolaidis NM, Rothenberg ME. Persistent effects induced by IL-13 in the lung. Am J Respir Cell Mol Biol. 2006;35:337–346. doi: 10.1165/rcmb.2005-0474OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishra A, Wang M, Schlotman J, Nikolaidis NM, DeBrosse CW, Karow ML, Rothenberg ME. Resistin-like molecule-beta is an allergen-induced cytokine with inflammatory and remodeling activity in the murine lung. Am J Physiol Lung Cell Mol Physiol. 2007;293:L305–L313. doi: 10.1152/ajplung.00147.2007. [DOI] [PubMed] [Google Scholar]
- 21.Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. A sensitive array for microRNA expression profiling (miChip) based on locked nucleic acids (LNA) RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dessau RB, Pipper CB. "R"--project for statistical computing. Ugeskr Laeger. 2008;170:328–330. [PubMed] [Google Scholar]
- 24.Sturn A, Quackenbush J, Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18:207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 25.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, Tommerup N, Kauppinen S. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14:432–444. doi: 10.1261/rna.783108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Obernosterer G, Martinez J, Alenius M. Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat Protoc. 2007;2:1508–1514. doi: 10.1038/nprot.2007.153. [DOI] [PubMed] [Google Scholar]
- 29.Besnard V, Wert SE, Kaestner KH, Whitsett JA. Stage-specific regulation of respiratory epithelial cell differentiation by Foxa1. Am J Physiol Lung Cell Mol Physiol. 2005;289:L750–L759. doi: 10.1152/ajplung.00151.2005. [DOI] [PubMed] [Google Scholar]
- 30.Rice WR, Conkright JJ, Na CL, Ikegami M, Shannon JM, Weaver TE. Maintenance of the mouse type II cell phenotype in vitro. Am J Physiol Lung Cell Mol Physiol. 2002;283:L256–L264. doi: 10.1152/ajplung.00302.2001. [DOI] [PubMed] [Google Scholar]
- 31.Pan Q, Kravchenko V, Katz A, Huang S, Ii M, Mathison JC, Kobayashi K, Flavell RA, Schreiber RD, Goeddel D, Ulevitch RJ. NF-kappa B-inducing kinase regulates selected gene expression in the Nod2 signaling pathway. Infect Immun. 2006;74:2121–2127. doi: 10.1128/IAI.74.4.2121-2127.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mayordomo JI, Zorina T, Storkus WJ, Zitvogel L, Garcia-Prats MD, DeLeo AB, Lotze MT. Bone marrow-derived dendritic cells serve as potent adjuvants for peptide-based antitumor vaccines. Stem Cells. 1997;15:94–103. doi: 10.1002/stem.150094. [DOI] [PubMed] [Google Scholar]
- 33.Smith ML, Olson TS, Ley K. CXCR2- and E-selectin-induced neutrophil arrest during inflammation in vivo. J Exp Med. 2004;200:935–939. doi: 10.1084/jem.20040424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Jegga AG, Sherwood SP, Carman JW, Pinski AT, Phillips JL, Pestian JP, Aronow BJ. Detection and visualization of compositionally similar cis-regulatory element clusters in orthologous and coordinately controlled genes. Genome Res. 2002;12:1408–1417. doi: 10.1101/gr.255002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams AE, Perry MM, Moschos SA, Lindsay MA. microRNA expression in the aging mouse lung. BMC Genomics. 2007;8:172. doi: 10.1186/1471-2164-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurup VP, Seymour BW, Choi H, Coffman RL. Particulate Aspergillus fumigatus antigens elicit a TH2 response in BALB/c mice. J Allergy Clin Immunol. 1994;93:1013–1020. doi: 10.1016/s0091-6749(94)70050-8. [DOI] [PubMed] [Google Scholar]
- 39.Munitz A, Brandt EB, Mingler M, Finkelman FD, Rothenberg ME. Distinct roles for IL-13 and IL-4 via IL-13 receptor alpha1 and the type II IL-4 receptor in asthma pathogenesis. Proc Natl Acad Sci U S A. 2008;105:7240–7245. doi: 10.1073/pnas.0802465105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Z, Homer R, Wang Z, Chen Q, Geba G, Wang J, Zhang Y, Elias J. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–788. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kurup V, Choi H, Murali S, Coffman RL. IgE and eosinophil regulation in a murine model of allergic aspergillosis. J. Leukocyte Biology. 1994;56:593–598. doi: 10.1002/jlb.56.5.593. [DOI] [PubMed] [Google Scholar]
- 42.Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, Iba H. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- 44.Sayed D, Rane S, Lypowy J, He M, Chen IY, Vashistha H, Yan L, Malhotra A, Vatner D, Abdellatif M. MicroRNA-21 Targets Sprouty2 and Promotes Cellular Outgrowths. Mol Biol Cell. 2008;19:3272–3282. doi: 10.1091/mbc.E08-02-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, Cvijic H, Ullmann AK, Stadler PF, Horn F. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 46.Kloosterman WP, Wienholds E, de Bruijn E, Kauppinen S, Plasterk RH. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- 47.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 48.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: A comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 50.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazumder B, Seshadri V, Fox PL. Translational control by the 3'-UTR: the ends specify the means. Trends Biochem Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 52.Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garzon R, Volinia S, Liu CG, Fernandez-Cymering C, Palumbo T, Pichiorri F, Fabbri M, Coombes K, Alder H, Nakamura T, Flomenberg N, Marcucci G, Calin GA, Kornblau SM, Kantarjian H, Bloomfield CD, Andreeff M, Croce CM. MicroRNA signatures associated with cytogenetics and prognosis in acute myeloid leukemia. Blood. 2008;111:3183–3189. doi: 10.1182/blood-2007-07-098749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 56.Zimmermann N, Mishra A, King NE, Fulkerson PC, Doepker MP, Nikolaidis NM, Kindinger LE, Moulton EA, Aronow BJ, Rothenberg ME. Transcript signatures in experimental asthma: identification of STAT6-dependent and -independent pathways. J Immunol. 2004;172:1815–1824. doi: 10.4049/jimmunol.172.3.1815. [DOI] [PubMed] [Google Scholar]
- 57.Foster PS, Webb DC, Yang M, Herbert C, Kumar RK. Dissociation of T helper type 2 cytokine-dependent airway lesions from signal transducer and activator of transcription 6 signalling in experimental chronic asthma. Clin Exp Allergy. 2003;33:688–695. doi: 10.1046/j.1365-2222.2003.01647.x. [DOI] [PubMed] [Google Scholar]
- 58.Blease K, Schuh JM, Jakubzick C, Lukacs NW, Kunkel SL, Joshi BH, Puri RK, Kaplan MH, Hogaboam CM. Stat6-deficient mice develop airway hyperresponsiveness and peribronchial fibrosis during chronic fungal asthma. Am J Pathol. 2002;160:481–490. doi: 10.1016/S0002-9440(10)64867-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jin P, Wang E, Ren J, Childs R, Shin JW, Khuu H, Marincola FM, Stroncek DF. Differentiation of two types of mobilized peripheral blood stem cells by microRNA and cDNA expression analysis. J Transl Med. 2008;6:39. doi: 10.1186/1479-5876-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kasashima K, Nakamura Y, Kozu T. Altered expression profiles of microRNAs during TPA-induced differentiation of HL-60 cells. Biochem Biophys Res Commun. 2004;322:403–410. doi: 10.1016/j.bbrc.2004.07.130. [DOI] [PubMed] [Google Scholar]
- 61.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 63.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 64.Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, Li Y. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 66.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 67.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gately MK, Renzetti LM, Magram J, Stern AS, Adorini L, Gubler U, Presky DH. The interleukin-12/interleukin-12-receptor system: role in normal and pathologic immune responses. Annu Rev Immunol. 1998;16:495–521. doi: 10.1146/annurev.immunol.16.1.495. [DOI] [PubMed] [Google Scholar]
- 69.Trinchieri G. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat Rev Immunol. 2003;3:133–146. doi: 10.1038/nri1001. [DOI] [PubMed] [Google Scholar]
- 70.Love TM, Moffett HF, Novina CD. Not miR-ly small RNAs: big potential for microRNAs in therapy. J Allergy Clin Immunol. 2008;121:309–319. doi: 10.1016/j.jaci.2007.12.1167. [DOI] [PubMed] [Google Scholar]
- 71.Gumireddy K, Young DD, Xiong X, Hogenesch JB, Huang Q, Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew Chem Int Ed Engl. 2008;47:7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Collison LW, Workman CJ, Kuo TT, Boyd K, Wang Y, Vignali KM, Cross R, Sehy D, Blumberg RS, Vignali DA. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450:566–569. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 73.Hu SJ, Ren G, Liu JL, Zhao ZA, Yu YS, Su RW, Ma XH, Ni H, Lei W, Yang ZM. MicroRNA expression and regulation in mouse uterus during embryo implantation. J Biol Chem. 2008;283:23473–23484. doi: 10.1074/jbc.M800406200. [DOI] [PubMed] [Google Scholar]
- 74.Zhang Z, Li Z, Gao C, Chen P, Chen J, Liu W, Xiao S, Lu H. miR-21 plays a pivotal role in gastric cancer pathogenesis and progression. Lab Invest. 2008;88:1358–1366. doi: 10.1038/labinvest.2008.94. [DOI] [PubMed] [Google Scholar]
- 75.Mattos W, Lim S, Russell R, Jatakanon A, Chung KF, Barnes PJ. Matrix metalloproteinase-9 expression in asthma: effect of asthma severity, allergen challenge, and inhaled corticosteroids. Chest. 2002;122:1543–1552. doi: 10.1378/chest.122.5.1543. [DOI] [PubMed] [Google Scholar]
- 76.Atkinson JJ, Senior RM. Matrix metalloproteinase-9 in lung remodeling. Am J Respir Cell Mol Biol. 2003;28:12–24. doi: 10.1165/rcmb.2002-0166TR. [DOI] [PubMed] [Google Scholar]
- 77.Rao PK, Kumar RM, Farkhondeh M, Baskerville S, Lodish HF. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc Natl Acad Sci U S A. 2006;103:8721–8726. doi: 10.1073/pnas.0602831103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A. 2007;104:20844–20849. doi: 10.1073/pnas.0710558105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 81.McCarthy JJ, Esser KA. MicroRNA-1 and microRNA-133a expression are decreased during skeletal muscle hypertrophy. J Appl Physiol. 2007;102:306–313. doi: 10.1152/japplphysiol.00932.2006. [DOI] [PubMed] [Google Scholar]
- 82.Luo X, Lin H, Pan Z, Xiao J, Zhang Y, Lu Y, Yang B, Wang Z. Down-regulation of miR-1/miR-133 contributes to re-expression of pacemaker channel genes HCN2 and HCN4 in hypertrophic heart. J Biol Chem. 2008;283:20045–20052. doi: 10.1074/jbc.M801035200. [DOI] [PubMed] [Google Scholar]
- 83.Barrios RJ, Kheradmand F, Batts L, Corry DB. Asthma: pathology and pathophysiology. Arch Pathol Lab Med. 2006;130:447–451. doi: 10.5858/2006-130-447-APAP. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.