Deficits in de novo vitamin B6 biosynthesis impair hormone homeostasis and root development but are a consequence of differential regulation of the genes.
Abstract
Vitamin B6 (pyridoxal 5′-phosphate) is an essential cofactor of many metabolic enzymes. Plants biosynthesize the vitamin de novo employing two enzymes, pyridoxine synthase1 (PDX1) and PDX2. In Arabidopsis (Arabidopsis thaliana), there are two catalytically active paralogs of PDX1 (PDX1.1 and PDX1.3) producing the vitamin at comparable rates. Since single mutants are viable but the pdx1.1 pdx1.3 double mutant is lethal, the corresponding enzymes seem redundant. However, the single mutants exhibit substantial phenotypic differences, particularly at the level of root development, with pdx1.3 being more impaired than pdx1.1. Here, we investigate the differential regulation of PDX1.1 and PDX1.3 by identifying factors involved in their disparate phenotypes. Swapped-promoter experiments clarify the presence of distinct regulatory elements in the upstream regions of both genes. Exogenous sucrose (Suc) triggers impaired ethylene production in both mutants but is more severe in pdx1.3 than in pdx1.1. Interestingly, Suc specifically represses PDX1.1 expression, accounting for the stronger vitamin B6 deficit in pdx1.3 compared with pdx1.1. Surprisingly, Suc enhances auxin levels in pdx1.1, whereas the levels are diminished in pdx1.3. In the case of pdx1.3, the previously reported reduced meristem activity combined with the impaired ethylene and auxin levels manifest the specific root developmental defects. Moreover, it is the deficit in ethylene production and/or signaling that triggers this outcome. On the other hand, we hypothesize that it is the increased auxin content of pdx1.1 that is responsible for the root developmental defects observed therein. We conclude that PDX1.1 and PDX1.3 play partially nonredundant roles and are differentially regulated as manifested in disparate root growth impairment morphologies.
Vitamin B6 is the collective term used to refer to a group of related water-soluble compounds, namely pyridoxine, pyridoxal, pyridoxamine, and their phosphorylated derivatives, also known as vitamers. The importance of vitamin B6 is determined mainly by the involvement of one of its forms, pyridoxal 5′-phosphate (PLP), in amino acid, lipid, and carbohydrate metabolism as an essential cofactor in over 140 enzymatic reactions (Percudani and Peracchi, 2003; Hellmann and Mooney, 2010). Animals and humans cannot biosynthesize vitamin B6 de novo, so they rely on dietary sources for the acquisition of the necessary amounts. Deficiency has been implicated in cardiovascular disease, diabetes, neurological disorders, carpal tunnel syndrome, premenstrual syndrome, and pellagra skin disease (Fitzpatrick et al., 2012). In contrast, plants, fungi, and microorganisms are able to biosynthesize their own vitamin B6 employing one of two mutually exclusive pathways. The deoxyxylulose 5-phosphate (DXP)-dependent pathway, discovered in Escherichia coli, involves seven enzymes (Lam and Winkler, 1992; Zhao and Winkler, 1996; Cane et al., 1998, 1999; Laber et al., 1999) and is only present in a small subset of bacteria (Ehrenshaft et al., 1999; Mittenhuber, 2001). In all plants and fungi, as well as the rest of bacteria, a DXP-independent pathway operates (Tambasco-Studart et al., 2005), employing only two enzymes (pyridoxine synthase1 [PDX1] and PDX2; Ehrenshaft et al., 1999; Ehrenshaft and Daub, 2001; Burns et al., 2005; Raschle et al., 2005).
In Arabidopsis (Arabidopsis thaliana), there are three homologs of PDX1 (PDX1.1, PDX1.2, and PDX1.3) and a single PDX2 homolog. PDX1.1 and PDX1.3 are catalytically active in the biosynthesis of vitamin B6 (Tambasco-Studart et al., 2005; Titiz et al., 2006), whereas PDX1.2 was recently assigned a function of pseudoenzyme involved in enhancing the activity of the catalytic homologs under stress conditions (Moccand et al., 2014). The DXP-independent pathway uses Gln, ribose 5-phosphate, and glyceraldehyde 3-phosphate as precursors to produce PLP (Burns et al., 2005; Raschle et al., 2005; Strohmeier et al., 2006; Zein et al., 2006). Mutants impaired in vitamin B6 biosynthesis de novo have been described in Arabidopsis. Knocking out either the single PDX2 or both PDX1.1 and PDX1.3 homologs was found to be lethal, resulting in an arrest of embryo development at the globular stage (Tambasco-Studart et al., 2005; Titiz et al., 2006). On the other hand, the two single pdx1 mutants are viable (Chen and Xiong, 2005; Titiz et al., 2006; Wagner et al., 2006) but display distinctive phenotypes. Both mutants have been reported to have a short-root phenotype, which is considerably more pronounced in pdx1.3 (Titiz et al., 2006). The pdx1.3 mutant was also shown to be more sensitive to salt, osmotic, and photooxidative stresses compared with pdx1.1 (Titiz et al., 2006). Additionally, pdx1.3 mutants hemizygous for PDX1.1 display more drastic phenotypic differences compared with pdx1.1 mutants hemizygous for PDX1.3, both morphologically and developmentally (Titiz et al., 2006). This despite the fact that both proteins are 87% identical and are able to biosynthesize the vitamin at comparable rates (Tambasco-Studart et al., 2005).
While pdx1.3 was shown to be more deficient in vitamin B6 compared with pdx1.1, a mechanistic explanation for these differences has not been provided. A study of pdx1.3 alone concluded that the stunted root growth of this mutant results from an impairment in local auxin production (Chen and Xiong, 2009a, 2009b). However, direct evidence for this conclusion was lacking. Moreover, the factors behind the root growth impairment of its paralogous knockout mutant, pdx1.1, have not been addressed. Reasons to investigate the homologs more closely derive from recent work that shows that PDX1.1 can be overexpressed while PDX1.3 cannot (Raschke et al., 2011). Indeed, a more strict regulation of the latter has been suggested, as it is a ubiquitination target (Manzano et al., 2008). In addition, data from the Arabidopsis microarray database, Genevestigator, suggest differential expression of PDX1.1 and PDX1.3 under several conditions (Hruz et al., 2008). Notably, all plants for which sequences are available carry at least two copies of PDX1 (Moccand et al., 2014). Taken together, the data suggest that the catalytic PDX1s are not completely redundant in planta and warrant further investigation.
In this study, we provide detailed comparative analyses of pdx1.1 and pdx1.3. The differential sensitivity of both mutants to Suc provides a tool to unravel the consequences of a deficit in vitamin B6. First, an assessment of the promoter regions provides evidence for nonredundant roles of PDX1.1 and PDX1.3. In pdx1.3, impairment in ethylene production plays a critical role in stunted root growth propagated by a deficit in auxin accumulation as well as the SHORT-ROOT (SHR) transcription factor. It has been shown previously that root apical meristem activity is disturbed in pdx1.3 (Chen and Xiong, 2005), and here we also show impaired lateral root formation. PDX1.1 expression, on the other hand, is rapidly down-regulated by Suc. Interestingly, and in stark contrast to pdx1.3, auxin levels are substantially increased in pdx1.1 but confined to the root. Therefore, we conclude that PDX1.1 and PDX1.3 play nonredundant roles and that the phenotypes of the corresponding mutants are a result of differential regulation of the genes.
RESULTS
Differential Impairment of Root Growth and Development in pdx1 Mutants
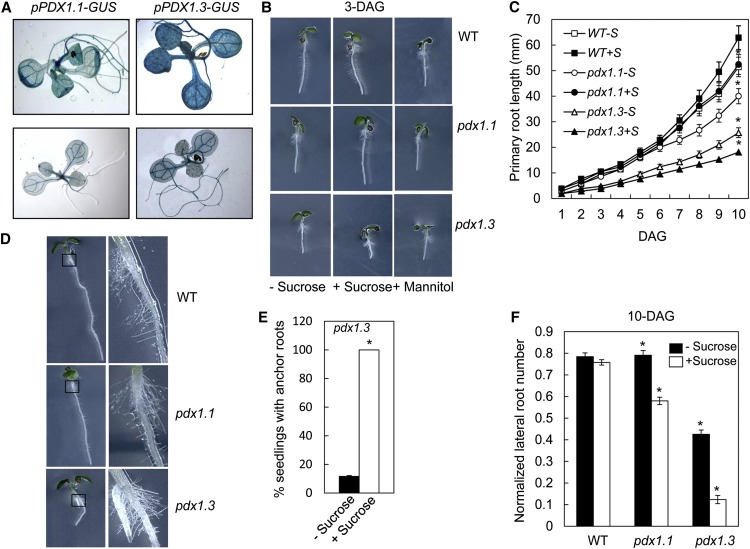
This study was initiated because of the severe root growth defect of the pdx1.3 mutant and the surprising fact that root tissue demonstrates a relatively low level of expression of PDX1.3 in comparison with other tissues, according to two independent quantitative PCR (qPCR) studies (Titiz et al., 2006; Wagner et al., 2006). Furthermore, previous analyses of expression by fusion of the promoter to GUS showed comparatively high activity in leaves with much weaker activity detected in roots (Wagner et al., 2006). In this study, we have reanalyzed the expression of PDX1.3 in more detail in an attempt to explain its disparate phenotype compared with its closest paralog, PDX1.1, assumed to be redundant. Our own analysis with promoter-GUS fusions of the PDX1.3 gene in the wild-type Columbia-0 (Col-0) background corroborates the previous conclusions, although we noted that GUS expression driven by the promoter of PDX1.3 (pPDX1.3:GUS) is much stronger than that of PDX1.1 (pPDX1.1:GUS; Fig. 1A, top row). Upon weaker staining using a lower temperature, we noticed a concentration of GUS activity in the root tip and lateral root emergence areas with pPDX1.3:GUS in particular, which is not observed in pPDX1.1:GUS (Fig. 1A, bottom row). Interestingly, root growth impairment becomes more severe in pdx1.3 in the presence of Suc (Fig. 1B), while it is of benefit to the growth of pdx1.1, similar to that observed in wild-type seedlings. Nonetheless, the growth of both pdx1.1 and pdx1.3 is retarded compared with the wild type in the presence of Suc. However, growth of the seedlings in the presence of mannitol was not significantly different from the control samples, indicating that the phenotype is not a result of osmotic stress. The differential impairments in root growth are clearly discernible in a plot of primary root growth of the mutants and the wild type in the presence and absence of Suc (Fig. 1C). In addition, both pdx1.1 and pdx1.3 have reductions in root hairs compared with the wild type (Fig. 1B). Of note also is that pdx1.3 displays altered root architecture, with anchor roots (adventitious roots emerging from the hypophysis) appearing 5 d after germination (DAG; Fig. 1D), while there are none in pdx1.1 or the wild type under these conditions. In the presence of Suc, all pdx1.3 seedlings developed anchor roots, whereas in its absence, less than 20% formed such structures (Fig. 1E). The pdx1.3 mutant is also significantly impaired in the development of lateral roots, with only 25% to 30% of the seedlings developing such organs at 10 DAG, while all wild-type plants have well-developed lateral roots by this time (Fig. 1E). Notably, the number of lateral roots per plant was normalized against the average length of the differentiation zone of the primary root, according to the recommendations of Dubrovsky and Forde (2012). In the presence of Suc, there is also lateral root growth impairment in pdx1.1, but it is less pronounced than that observed with pdx1.3 (Fig. 1E).
Figure 1.
Gross divergence in the morphology of pdx1.1 and pdx1.3. A, Staining of the promoter-GUS fusion lines pPDX1.1:GUS and pPDX1.3:GUS at 37°C (top row) and 25°C (bottom row) in the Col-0 background. B, Root growth at 3 DAG of pdx1.1 and pdx1.3 plants grown in the presence of 1% (w/v) Suc compared with wild-type (WT) Col-0; pdx1.3 seedlings grown in the presence of the osmoticum mannitol (1%) are also shown. C, Kinetics of root growth of pdx1.1 and pdx1.3 seedlings in the presence or absence of Suc (S) compared with the wild type. The data are from three biological repetitions. Error bars indicate se. Asterisks indicate statistically significant differences (P < 0.001) in comparison with the wild type grown on Suc. D, Morphology of pdx1.1 and pdx1.3 compared with the wild type at 5 DAG. Adventitious roots can be seen in pdx1.3. The black squares outline the areas magnified on the right. E, Number of pdx1.3 seedlings that develop anchor roots in the presence or absence of Suc. The data are derived from three independent experiments. Values display statistically significant differences for P < 0.001. F, Number of lateral roots normalized against the length of the branching zone for each plant. Plants were grown until 10 DAG in the presence or absence of Suc. The data are from three biological repetitions. Error bars indicate se. Asterisks indicate statistically significant differences (P < 0.001) when compared with the wild type.
Evidence for Differential Regulation of PDX1.1 and PDX1.3
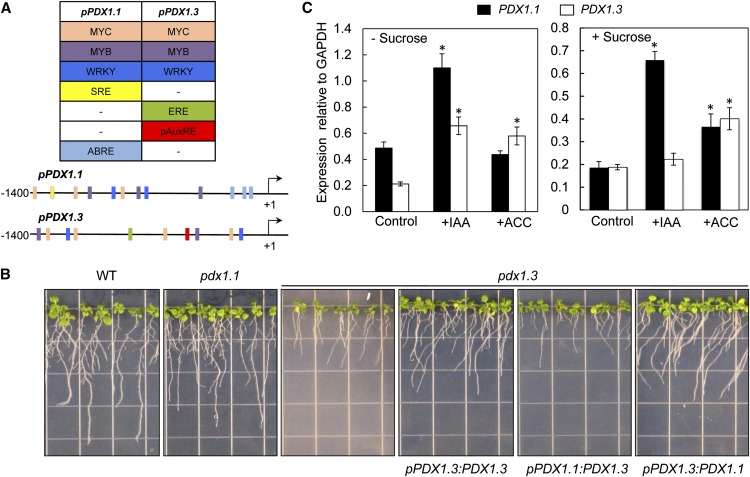
While pdx1.1 and pdx1.3 can be phenotypically distinguished in culture medium by their differential retardation in root growth, they both display a deficiency in total vitamin B6 content, which is especially pronounced in pdx1.3 (Titiz et al., 2006). Root growth is restored for both mutants upon supplementation with the B6 vitamer, pyridoxine (Titiz et al., 2006). In our previous work, combining qPCR and protein expression studies, we demonstrated that PDX1.3 is generally expressed at a higher level than PDX1.1 but is particularly more abundant in roots (Titiz et al., 2006). As judged from the promoter-GUS fusion lines, pPDX1.3:GUS and pPDX1.1:GUS, it becomes apparent that the higher expression level of PDX1.3 compared with PDX1.1 seems to be inherent to the promoter sequence (Fig. 1A). An in silico examination of the upstream regions reveals putative cis-regulatory elements, some of which are disparate between PDX1.1 and PDX1.3 (Fig. 2A). In particular, we found sequences corresponding to the ethylene-responsive element (ERE; AGCCGCC; Shinshi et al., 1995) and a partial auxin response element (AuxRE; TGTCTc; Ulmasov et al., 1997a) in the upstream region of PDX1.3, which are absent in PDX1.1. On the other hand, we noted that the upstream region of PDX1.1 harbors several abscisic acid-responsive elements (CACGT; Iwasaki et al., 1995) as well as a sugar response element (TTATCC; Tatematsu et al., 2005), which are absent from the corresponding region in PDX1.3. Potential binding sites for members of the MYB (WAACCA and CNGTTR), MYC (CANNTG; Abe et al., 2003) and WRKY (TTGAC; Yu et al., 2001) transcription factor families are observed in the upstream regions of both genes. The latter are involved in gene regulation in response to various stresses, including drought, cold, and pathogen attack. With a particular focus on the short-root phenotype and in order to study the functional relevance of the potential differential regulation of expression, we performed promoter-swap experiments. We transformed pdx1.3 with chimeras of the region (approximately 1,400 nucleotides) that lies upstream of the transcriptional start site of either PDX1.1 or PDX1.3 (pPDX1.1 and pPDX1.3, respectively) and fused to either the PDX1.3 or the PDX1.1 coding region. The corresponding lines are named pPDX1.1:PDX1.3 and pPDX1.3:PDX1.1 as well as the control line pPDX1.3:PDX1.3. We observed that while the integration of pPDX1.3:PDX1.1 could complement the pdx1.3 short-root phenotype to the same extent as the control line pPDX1.3:PDX1.3, pPDX1.1:PDX1.3 could not (Fig. 2B). This clearly indicates that the upstream region of PDX1.3 contains specific elements not present in the corresponding region of PDX1.1 and that these elements are responsible for coordinating the proper expression of PDX1.3.
Figure 2.
Evidence for differential regulation of PDX1.1 compared with PDX1.3. A, The scheme depicts elements found in the upstream regions of PDX1.1 (pPDX1.1) and PDX1.3 (pPDX1.3). WRKY, MYB, and MYC correspond to elements recognized by the respective transcription factors. ABRE, Abscisic acid response element; pAuxRE, partial AuxRE; SRE, sugar response element; +1, transcriptional start site. B, Control of the expression of PDX1.3 by the upstream region of PDX1.1 cannot complement the impaired root growth of pdx1.3. Seedlings of wild-type (WT) Col-0, pdx1.1, and pdx1.3 are compared with the pdx1.3 line carrying swapped promoters: the upstream region of PDX1.1 fused to PDX1.3 (pPDX1.1:PDX1.3), the upstream region of PDX1.3 fused to PDX1.1 (pPDX1.3:PDX1.1), and the upstream region of PDX1.3 fused to PDX1.3 (pPDX1.3:PDX1.3) as a control. Seedlings were grown vertically in the presence of 1% (w/v) Suc. Images were captured at 10 DAG. C, Relative expression levels of PDX1.1 and PDX1.3 in Col-0 control plants (whole seedlings) after treatment with IAA and the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC) in the presence or absence of Suc in the growth medium. Values are relative to GAPDH (At1g13440). The data are from at least three biological repetitions. Error bars represent se. Asterisks indicate statistically significant differences (P < 0.05) when treatments are compared with the respective controls.
Since both auxin and ethylene are important determinants of root growth and vitamin B6 is required for their biosynthesis (Mooney and Hellmann, 2010; Fitzpatrick, 2011), we next focused on the involvement of these two hormones in the manifestation of the short-root phenotype. In the first instance, we noted that application of indole acetic acid (IAA) increased the expression in whole seedlings of both PDX1.1 and PDX1.3 in plants grown without Suc but only the levels of PDX1.1 in the presence of Suc (Fig. 2C). Application of the ethylene precursor, ACC, only induced the expression of PDX1.3 in the absence of Suc, albeit to a lesser extent than IAA, while both genes were induced by ACC in the presence of Suc (Fig. 2C). This prompted us to investigate the levels of these hormones in the mutant lines pdx1.1 and pdx1.3 compared with the wild type.
Auxin Levels in Roots of pdx1.1 and pdx1.3
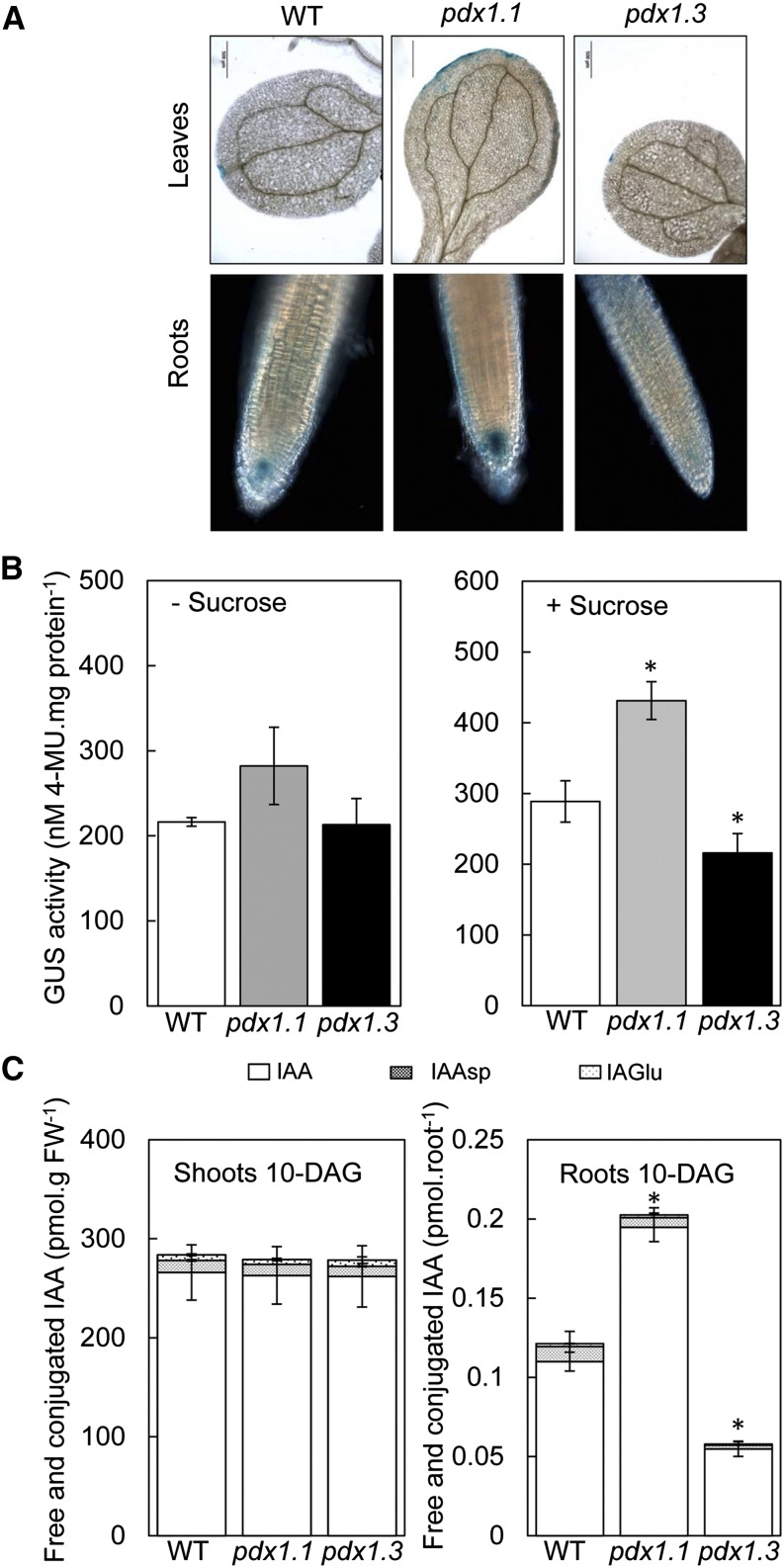
The pdx1.3 mutant has already been investigated with regard to auxin content by Chen and Xiong (2009a, 2009b). In the latter studies, a defect in shoot-to-root transport of auxin was ruled out as an explanation for the short-root phenotype in pdx1.3, because grafting of a wild-type scion on a pdx1.3 rootstock did not rescue the short-root phenotype. Furthermore, in the same studies, the treatment of pdx1.3 harboring DR5-GUS (the artificial AuxRE derived from the natural GH3 element in soybean that reports endogenous IAA sensing/distribution; Ulmasov et al., 1997b) with IAA indicated a functional auxin response (Chen and Xiong, 2009b). Yet, impairment in local auxin biosynthesis in the root and/or transport could not be excluded. The pdx1.1 mutant was not investigated in the latter studies. Therefore, in order to compare the pdx1 mutants and probe their levels and response to auxin, a transgenic line carrying the DR5-GUS fusion gene (Col-0 background) was crossed with both pdx1.1 and pdx1.3, and homozygous mutant lines carrying the transgene were selected. While the GUS staining around the leaf perimeter (as is typically observed with the expression of this construct) was similar for lines carrying the transgene in the wild-type, pdx1.1, and pdx1.3 backgrounds, the staining in the root tip was significantly weaker for pdx1.3 compared with the wild type (Fig. 3A) and was similar to what has been reported previously (Chen and Xiong, 2009b). Surprisingly, on the other hand, the staining was considerably stronger for pdx1.1 in the root tip compared with the wild type (Fig. 3A). Quantitative GUS analysis of the entire seedling substantiated the observations for pdx1.1 in particular (Fig. 3B, right). Interestingly, quantifying the GUS activity driven by DR5 showed that the latter was significantly higher in the pdx1.1 mutant compared with the wild type only in the presence of Suc (Fig. 3B, compare left and right). To corroborate the differential observations in shoot versus root in the presence of Suc, we measured the endogenous auxin levels in the whole root versus shoot of pdx1.1 and pdx1.3 compared with the wild type using gas chromatography-mass spectrometry. Indeed, while the overall levels of IAA and its conjugates were not significantly different from the wild type in the shoot of pdx1.1 or pdx1.3, the levels of free IAA, in particular in the root of pdx1.1 and pdx1.3, were significantly stronger and weaker, respectively, when calculated on a per root basis (Fig. 3C). The latter was used because of the difference in development and size of this tissue.
Figure 3.
Levels of auxin in roots of pdx1.1 and pdx1.3. A, GUS staining of leaves and roots of pdx1.1 and pdx1.3 compared with wild-type (WT) Col-0 carrying the auxin-responsive promoter DR5 fused to GUS. Seedlings were grown in the presence of 1% (w/v) Suc, and images were captured at 10 DAG. B, Quantitative GUS analysis performed with 4-methylumbelliferyl glucuronide (4-MU) in the absence (left) or presence (right) of Suc. Plant lines are at the same age as in A. The data are from at least three biological repetitions. Error bars represent se. Asterisks indicate statistically significant differences (P < 0.05) when compared with the wild type. C, Quantification of free and conjugated auxin in shoots and roots of pdx1.1 and pdx1.3 compared with the wild type. Seedlings were grown on medium in the presence of 1% (w/v) Suc and were analyzed at 10 DAG. The data are from at least three biological repetitions. Error bars represent se. Asterisks indicate statistically significant differences (P < 0.05) when compared with the wild type. FW, Fresh weight.
Addressing pdx1.1 first, several mutants with elevated auxin levels have been studied (e.g. YUC flavin monoxygenase-like enzyme [yucca], superroot1 [sur1], and sur2) and have characteristic phenotypes comprising elongated petioles, narrow epinastic leaves and cotyledons, short roots, but more and longer root hairs (Zhao et al., 2001). The absence of a similar shoot phenotype in pdx1.1 but impaired root growth is consistent with the restriction of auxin accumulation to the root under similar conditions (Fig. 3C) but implies a different mechanism to the reported classical auxin-accumulating mutants. In the case of pdx1.3, our data would appear at the outset to support the conclusion of Chen and Xiong (2009a, 2009b) that local auxin biosynthesis in the root must be impaired. However, expression of the bacterial auxin biosynthetic gene indole acetamide hydrolase under the control of the quiescent center-specific promoter WUSCHEL-related homeobox5 did not alter the root growth of pdx1.3 (Chen and Xiong, 2009a). Therefore, an alternative explanation must be sought.
Ethylene Response Is Impaired in pdx1.1 and pdx1.3
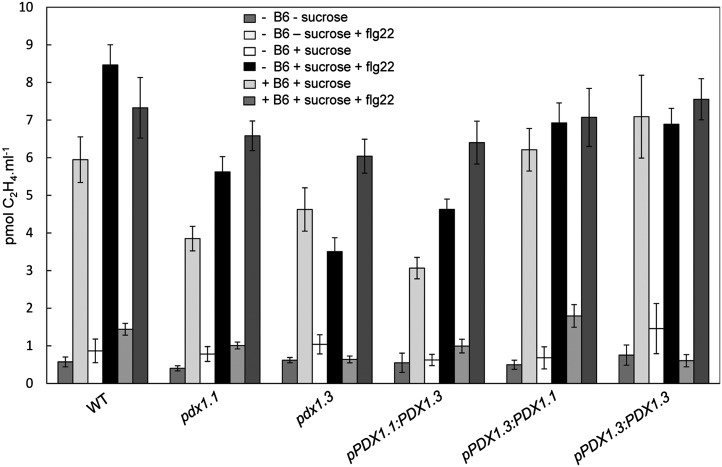
Interactions between ethylene and sugar signaling pathways are well documented. Specifically, several studies have demonstrated that an increased ethylene response or levels can result in increased resistance to Suc and Glc (Zhou et al., 1998; Gibson et al., 2001) and that a decreased ethylene response can lead to Suc and Glc hypersensitivity during early seedling development (Zhou et al., 1998; Gibson et al., 2001). Therefore, the observation of a putative ERE in the promoter region of PDX1.3 as well as its Suc hypersensitivity is interesting. In this context, we sought to test the ethylene biosynthesis and/or responsiveness in relation to the PDX genes. In the first instance, we compared the ability of wild-type plants and pdx1.1 and pdx1.3 mutants to produce ethylene in response to the peptide derived from bacterial flagellin, flg22 (Felix et al., 1999; Zipfel et al., 2004). While both pdx1.3 and pdx1.1 produced lower amounts of ethylene compared with the wild type in the presence of Suc, that of pdx1.3 was more impaired than that of pdx1.1 (Fig. 4). On the other hand, in the absence of Suc, inhibition of ethylene production was less pronounced in pdx1.3 but stronger for pdx1.1 (Fig. 4). A repetition of the experiment with seedlings that had been grown in the presence of pyridoxine supplementation restored flg22-induced ethylene production/responsiveness toward wild-type levels in both pdx1 mutants (Fig. 4). Interestingly, the same experiment with the swapped promoter lines pPDX1.1:PDX1.3 and pPDX1.3:PDX1.1 in the pdx1.3 background demonstrated that the impairment in flg22-induced ethylene production/responsiveness by pdx1.3 can be restored by reintroducing its own promoter sequence to control the expression of either PDX1.1 or PDX1.3 but not the promoter sequence of PDX1.1 (Fig. 4). Therefore, while the PDX1 genes are clearly required for the production and/or response to ethylene under these conditions, the deficits observed must have contributions from the PDX1.3 promoter sequence.
Figure 4.
Ethylene production is impaired in pdx1 mutants. Production of ethylene (C2H4) was induced in the presence of the flg22 peptide elicitor. Seedlings used were grown in the presence or absence of Suc and pyridoxine (B6) as indicated until 10 DAG. The lines used are the wild type (WT), pdx1.1, pdx1.3, the upstream region of PDX1.1 fused to PDX1.3 (pPDX1.1:PDX1.3), the upstream region of PDX1.3 fused to PDX1.1 (pPDX1.3:PDX1.1), and the upstream region of PDX1.3 fused to PDX1.3 (pPDX1.3:PDX1.3). The data are from at least five biological repetitions. Error bars represent se.
Suppression of PDX1.1 Expression by Suc
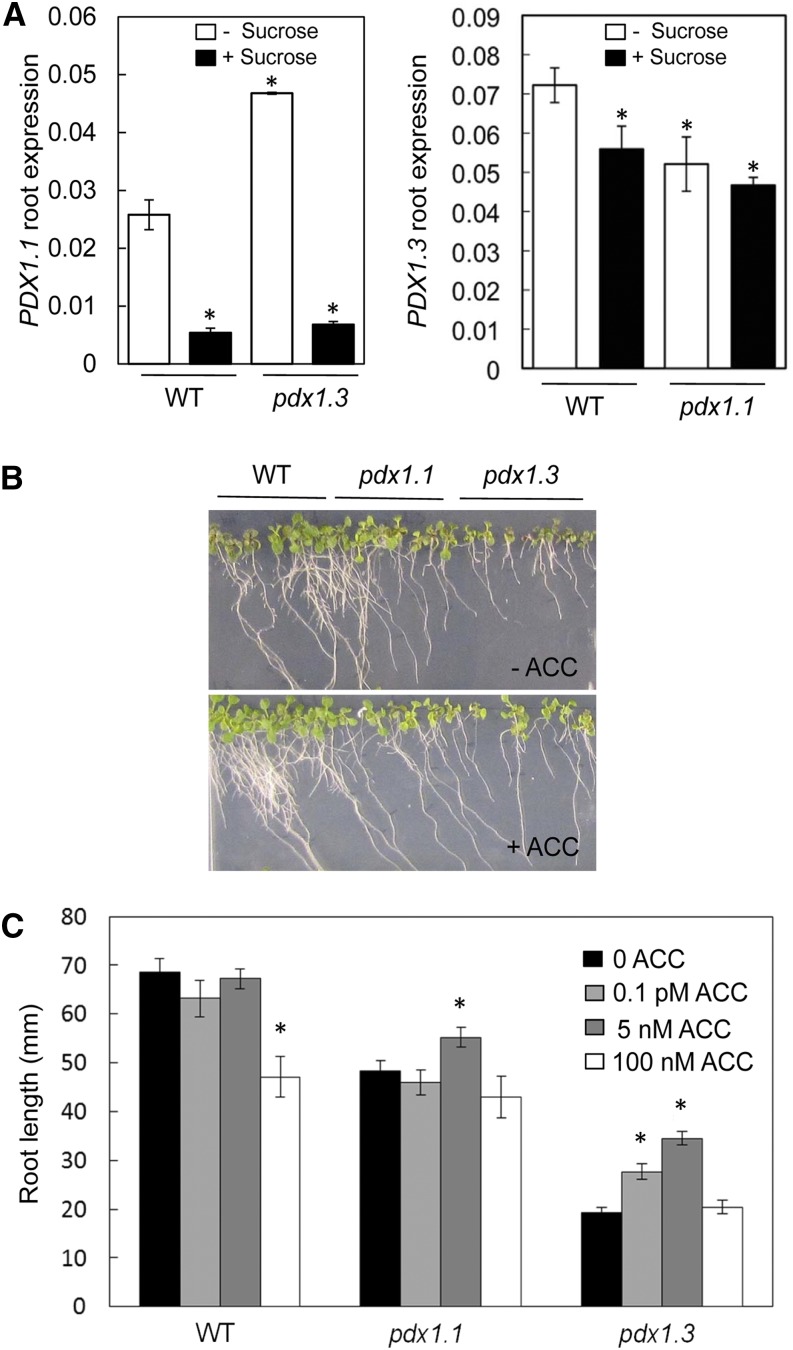
As stated above, vitamin B6 in its form as PLP is required as a cofactor for both auxin and ethylene biosynthesis (Mooney and Hellmann, 2010; Fitzpatrick, 2011). Therefore, the lower abundance and impaired ability to produce these hormones, respectively, in pdx1.3 in particular, may be simply linked to a stronger deficiency in the vitamin related to the fact that PDX1.3 is more abundant than PDX1.1 (Titiz et al., 2006). However, during the course of the studies described here, we observed that the expression of PDX1.1 is considerably down-regulated (approximately 4- to 5-fold) when cultured in medium containing Suc compared with its absence (Fig. 5A). Interestingly, PDX1.1 is up-regulated (approximately 2-fold) in pdx1.3 cultured in medium that does not contain Suc (Fig. 5A). While the expression of PDX1.3 is also slightly down-regulated in the presence of Suc, it is much less pronounced than that observed with PDX1.1 (Fig. 5A). Given that the respective phenotypes can be rescued (albeit not completely; see below) by supplementation with pyridoxine, it would appear that the level of impairment observed in pdx1.3 and pdx1.1 is related to the respective deficiencies in vitamin B6 content (Titiz et al., 2006; Supplemental Fig. S1). In other words, the down-regulation of PDX1.1 expression in the presence of Suc contributes to the strong pdx1.3 phenotype under these conditions. However, the fact that PDX1.1 cannot compensate for the loss of PDX1.3, as it does not rescue the pdx1.3 phenotype even in the absence of Suc (Fig. 1, B and E), still reveals important differences between the two genes and lends support to divergence in their regulation.
Figure 5.
Exogenous Suc represses PDX1.1 expression, but application of ACC can partially rescue the pdx1 root phenotype. A, Expression of PDX1.1 (left) and PDX1.3 (right) in roots of wild-type (WT) Col-0 and pdx1.3 or pdx1.1 in the absence or presence of 1% (w/v) Suc. Seedlings were allowed to grow until 10 DAG. The data are from at least three biological repetitions. Error bars represent se. Asterisks indicate statistically significant differences (P < 0.05) when treatments are compared with the wild type grown in the absence of Suc. B, Growth of the wild type, pdx1.1, and pdx1.3 in the presence of 1% (w/v) Suc as well as in the absence (top) or presence (bottom) of 5 nm ACC. The experiment was done three times, yielding similar results; images were captured at 10 DAG. C, Root length of wild-type, pdx1.1, and pdx1.3 seedlings at 10 DAG grown on 1% (w/v) Suc in the presence or absence of ACC (0–100 nm as indicated). The data are averages of three biological replicates; measurements were performed using ImageJ software (http://imagej.nih.gov/ij/). Error bars represent se. Asterisks indicate statistically significant differences (P < 0.05) when compared with the wild type in the absence of ACC.
In light of the above data and our own studies described so far, we probed the impact of the ethylene precursor, ACC, on the pdx1 mutants. Supplementation with ACC concentrations in the 100 nm range and above inhibits wild-type root growth as reported previously (Rahman et al., 2001; Swarup et al., 2002; Fig. 5C). However, supplementation with ACC at concentrations of 5 nm significantly increased root growth in pdx1.3 and pdx1.1 (Fig. 5B). Indeed, concentrations as low as 0.1 pm enhanced root length in pdx1.3 (Fig. 5C). The promotion of root growth by such low concentrations of ethylene is not without precedence and has been reported previously, where concentrations of 20 nm were observed to promote root elongation (Konings and Jackson, 1979). The increase in root growth was more pronounced for pdx1.3 compared with pdx1.1 or the wild type (Fig. 5C). This suggests that a deficit in ethylene production contributes to the short-root phenotype of pdx1.3. Notably, when ACC was applied in similar concentrations (i.e. 5 nm) to plants grown without Suc, it also appeared to stimulate the root growth of pdx1.3, although the effect was weaker than in the presence of Suc (Supplemental Fig. S2A).
Expression Levels of SHR Are Altered during the Early Developmental Stages of pdx1.3
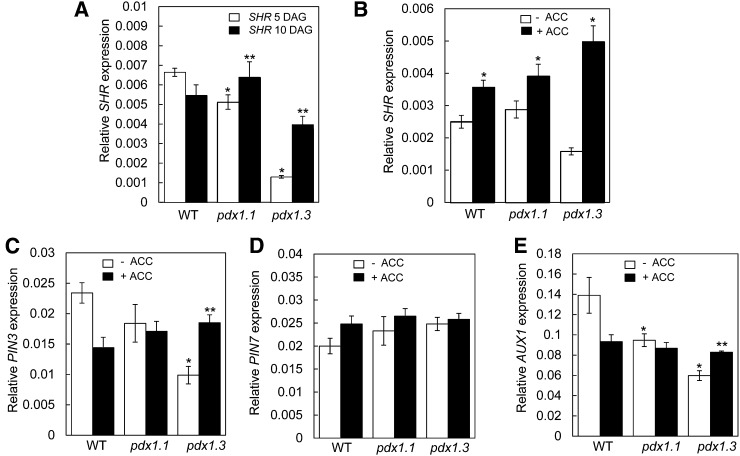
It has been reported previously that pdx1.3 has reduced root apical meristem activity (Chen and Xiong, 2005). The observed differences in root morphology and root apical meristem activity of pdx1.3 in the early stages of development draw some parallels with the shr mutant (Lucas et al., 2011). SHR is a Gibberellic acid insensitive, Repressor of GA1, and SCARECROW family transcription factor involved in root patterning during the early stages of development and is essential for the function of the root apical meristem (Benfey et al., 1993; van den Berg et al., 1995; Blilou et al., 2005). This mutant has a severely shortened main root throughout its life cycle and is impaired in lateral root development, instead forming a large number of anchor roots. Anchor root development was thus suggested to be a compensatory mechanism related to the lack of primary root growth. The latter mechanism was validated in a recent study with wild-type seedlings of Arabidopsis, where removal of the bottom 2 to 3 mm of root apical meristem at 3 DAG dramatically increased anchor root development (Lucas et al., 2011). In this context, we examined the expression level of SHR in the pdx1 mutants. Indeed, in the presence of Suc, we observed that the expression level of SHR is reduced approximately 8-fold in pdx1.3 compared with the wild type at 5 DAG, whereas it is reduced only 1.3-fold in pdx1.1 (Fig. 6A). Expression levels of SHR are also reduced in the mutant lines in the absence of Suc but are less pronounced (Supplemental Fig. S2B). However, we observed that SHR levels are increased in pdx1.1 at a later development stage (i.e. 10 DAG), while expression in pdx1.3 also increases 3-fold toward wild-type levels (Fig. 6A). It is noteworthy that this reflects a developmental time point in pdx1.3 when the rate of root growth accelerates to approach wild-type levels (Fig. 1C). We also examined the effect of 5 nm ACC on the expression of SHR in root tips. While we saw a small induction of SHR in pdx1.1 and the wild type, it was much stronger in pdx1.3 (Fig. 6B). This implies that ethylene is associated with the decrease in SHR expression that is particularly pronounced in pdx1.3 in the presence of Suc. Interestingly, in the absence of Suc, SHR expression levels in pdx1.1 and pdx1.3 are restored to wild-type levels when supplemented with ACC (Supplemental Fig. S2B).
Figure 6.
The altered expression levels of SHR and selected auxin transporters is partially rescued by application of ACC. A, SHR transcript abundance in roots of wild-type (WT) Col-0, pdx1.1, and pdx1.3 seedlings grown in the presence of 1% (w/v) Suc at either 5 or 10 DAG. The data represent means from at least three biological replicates. Error bars represent se. Statistically significant differences (P < 0.05) between plants at different ages compared with the wild type are indicated with asterisks (*, 5 DAG; **, 10 DAG). B, Relative expression of SHR in root tips (10–12 mm) of seedlings grown in the presence of Suc for 10 DAG and in the presence or absence of 5 nm ACC. The data represent means from at least three biological replicates. Error bars represent se. Statistically significant differences (P < 0.05) of treatment groups compared with nontreated controls are indicated by asterisks. C to E, Transcript abundance of PIN3 (C), PIN7 (D), and AUX1 (E) in root tips (10–12 mm) grown on medium containing 1% (w/v) Suc and in the presence or absence of 5 nm ACC. The expression data for each gene were normalized against GAPDH (At1g13440). The data represent means from at least three biological replicates. Error bars represent se. Statistically significant differences (P < 0.05) between wild-type and mutant lines with (**) and without (*) ACC treatment are indicated.
As SHR is associated with specific influx and efflux carriers of auxin (Teale et al., 2006) and as the latter is an important mitotic signal, we monitored the expression of selected auxin transporters in the pdx1 mutants compared with the wild type. In particular, we analyzed the auxin efflux carriers PIN-FORMED3 (PIN3; At1g70940) and PIN7 (At1g23080) and the auxin influx carrier Auxin transporter protein1 (AUX1; At2g38120) in pooled 10- to 12-mm root tips (meristem and elongation zone) at 10 DAG. This part of the root was chosen for expression analysis because it coincided with the area where staining was observed for the DR5-GUS lines at the same age. Both PIN3 and PIN7 are undetectable in shr at this stage of development, while AUX1 is maintained (Lucas et al., 2011). In pdx1.3 (and pdx1.1, albeit to a lesser extent), the transcript level of PIN3 is significantly decreased in the presence of Suc, while that of PIN7 is not significantly changed compared with the wild type under these conditions and at this developmental stage (Fig. 6, C and D). AUX1 expression is also decreased in both pdx1.3 and pdx1.1 (Fig. 6E). Notably, application of ACC down-regulated the expression of AUX1 and PIN3 in the wild type; in contrast, the expression of both increased in pdx1.3 to approach wild-type levels (Fig. 6, C and E). On the other hand, ACC application had no significant effect on the expression of these transporters in pdx1.1 (Fig. 6, C–E). In the absence of Suc, there was no significant induction of PIN3, PIN7, or AUX1 expression by ACC in the three plant lines (Supplemental Fig. S2, C, D, and E, respectively). We compared the above expression data with data on PIN3, PIN7, and AUX1 transcript abundance derived from whole seedlings as well as entire roots of the same plants (Supplemental Fig. S3). A decrease in AUX1 expression can be observed in entire roots in the presence of Suc as well as whole seedlings, but the decrease is negligible in the latter (Supplemental Fig. S3, A and B). The decrease in AUX1 expression in pdx1.1 appears to be independent of Suc. In the case of PIN3, changes can also be observed in these tissues and are affected by Suc but are much less pronounced than those observed in root tips. There are no significant changes in PIN7 expression in all tissues examined. Therefore, we conclude that the expression of these transporters is principally affected in the root tips of pdx1.1 and pdx1.3.
Addressing Anthocyanin Accumulation in pdx1.3
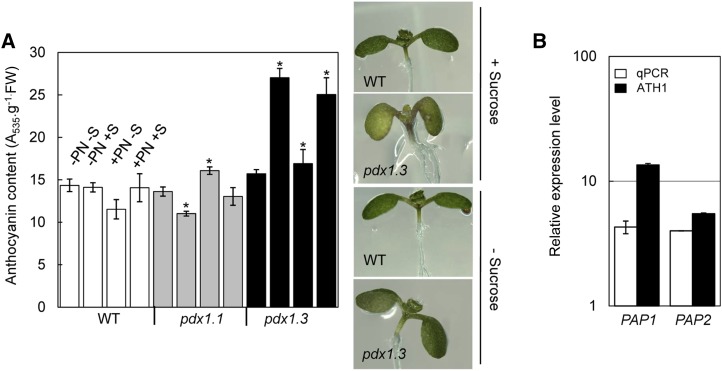
Ethylene inhibits anthocyanin accumulation induced by Suc and light (Jeong et al., 2010). This occurs through the suppression of expression of the transcription factors that positively regulate anthocyanin biosynthesis, such as GLABRA3 (GL3), TRANSPARENT TESTA8 (TT8), and PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP1) and PAP2, while stimulating the expression of the negative R3-MYB regulator MYBL2 (Jeong et al., 2010). We observed hyperaccumulation of anthocyanins in pdx1.3 in the presence of Suc compared with either the wild type or pdx1.1 (Fig. 7A). The impairment in ethylene biosynthesis/signaling observed in pdx1.3 in this study could thereby explain the enhanced accumulation of anthocyanins, as the negative regulation of anthocyanin biosynthesis is likely to be affected in this mutant. Indeed, upon examination of the global transcriptome effect of knocking out pdx1.3, through whole-genome RNA expression analyses performed using the Affymetrix ATH1 DNA microarray (see below), we observed that PAP1 is up-regulated 5.49-fold in shoots of pdx1.3 (Supplemental Table S1). PAP2 is up-regulated 13.51-fold in shoots of pdx1.3 but did not pass our statistical analysis for filtering differential expression. However, differential expression of PAP1 and PAP2 was confirmed by qPCR analysis (Fig. 7B). The gene encoding dihydroflavonol reductase (At5g42800) was also up-regulated in pdx1.3 leaves compared with the wild type (4.52-fold). The latter is considered to be a rate-limiting step (Das et al., 2012) late in anthocyanin biosynthesis and is a target of PAP1 and PAP2 (Yan et al., 2005). On the other hand, no significant changes in the expression of GL3, TT8, or MYBL2 were observed compared with the wild type in the microarray. Ethylene-mediated suppression of anthocyanin accumulation is dependent upon ethylene signaling components responsible for the triple response (Jeong et al., 2010). In this context, we noted that expression of the ethylene receptor ETR2 (At3g23150) is down-regulated 3.3-fold in pdx1.3 shoots compared with the wild type (Supplemental Table S1). Therefore, pdx1.3 is not only impaired in ethylene biosynthesis but most likely also in the propagation of its signaling. The latter statement is corroborated by the fact that pyridoxine supplementation does not completely rescue the short-root phenotype, as would be expected if the phenotype is solely a consequence of a deficit in cofactor requirements (i.e. that compromises ethylene biosynthesis). Moreover, anthocyanin accumulation is observed both in the presence and absence of pyridoxine supplementation, implying a signaling event in addition to a cofactor deficit (Fig. 7A).
Figure 7.
Anthocyanin accumulation in pdx1.3. A, Anthocyanin levels in pdx1.1 and pdx1.3 compared with wild-type (WT) Col-0 in the presence or absence of pyridoxine (PN) and/or Suc (S). Seedlings were grown until 10 DAG. The data represent means from at least three biological replicates. Error bars represent se. Asterisks indicate statistically significant differences (P < 0.001) of mutant lines compared with the corresponding control wild-type line. The morphology of pdx1.3 in the presence or absence of Suc at this stage of growth is shown on the right compared with the wild type. B, Expression levels of PAP1 and PAP2 transcription factors involved in anthocyanin biosynthesis in pdx1.3 compared with wild-type Col-0. Transcript abundance was determined by both an ATH1 microarray (see text) and qPCR. In the case of the qPCR, the data are results of three biological replicates. Error bars represent se.
Global Transcriptome Effect of pdx1.3
As pdx1.3 is more severely affected than pdx1.1 in Arabidopsis, a whole-genome RNA expression analyses was performed on pdx1.3 using the Affymetrix ATH1 DNA microarray in order to ascertain the global transcriptome effect. Total RNAs were extracted from the separated roots and shoots of 10-d-old seedlings of all lines grown in the presence of 1% (w/v) Suc. Significantly differentially regulated genes were clustered according to their transcriptional behavior after statistical analyses (see “Materials and Methods”). A group of 538 and 317 genes were found to be either up-regulated or down-regulated, respectively, in leaves of pdx1.3 compared with the wild type (Supplemental Table S1). The number of genes being significantly altered in the root was substantially lower, with 156 and 162 genes being either up-regulated or down-regulated, respectively (Supplemental Table S1). A comparison of the functional distribution of altered genes according to the Gene Ontology biological process annotation tool at The Arabidopsis Information Resource (http://www.arabidopsis.org/tools/bulk/go/) indicates a significant enrichment of genes involved in metabolism and stress responses in both leaves and roots (Supplemental Fig. S4). In the context of this study, it is notable that several genes involved in ethylene biosynthesis (aminocyclopropane carboxylate synthase [ACS7; At4g26200], ACS8 [At4g37770], ACS11 [At4g08040], and ACC oxidase [At1g62380]) and signaling (ETR2 mentioned above and Ethylene response factor73 [ERF73; At1g72360]) were found to be down-regulated in shoots, while ERF B-2 (At5g61590) was found to be down-regulated in roots. Genes implicated in auxin signaling are also down-regulated, among which are the auxin-regulated gene involved in organ size (At3g59900), Small auxin-upregulated72 [SAUR72; At3g12830), and YUCCA (At5g43890) in leaves as well as SAUR76 (At5g20820) in roots. Many of the differentially expressed genes are of unknown function and may be noteworthy for future studies. In this context, it is interesting that the triterpene synthase, marneral synthase, is the most up-regulated transcript in pdx1.3 roots (118-fold) and that the related thalianol synthase is up-regulated as well (3-fold). Overexpression of these genes causes a dwarf phenotype (Field and Osbourn, 2008; Field et al., 2011). However, the exact role of these molecules in Arabidopsis has not yet been elucidated.
DISCUSSION
In this study, we report on the incomplete redundancy of two genes involved in vitamin B6 biosynthesis de novo in Arabidopsis. The study provides important information on the individual role these genes play and their significance toward maintaining sufficient levels of the phytohormones, ethylene and auxin, and the downstream effects that this has on plant development with a particular focus on the root. The possibility for differential regulation of PDX1.1 and PDX1.3 genes addressed in this work was initially suggested by two earlier studies. The first study, by Titiz et al. (2006), showed that, in spite of the ability of both enzymes to biosynthesize vitamin B6 at comparable rates (at least in vitro), the two respective knockout mutants displayed distinct phenotypes. Moreover, mutants carrying a single functional copy of either PDX1.1 or PDX1.3 were drastically different, with pdx1.3 mutant lines hemizygous for PDX1.1 more severely impaired in development than pdx1.1 mutant plant lines hemizygous for PDX1.3 (Titiz et al., 2006). A second study, describing the overproduction of vitamin B6 in Arabidopsis (Raschke et al., 2011), demonstrated that while both PDX1.1 and PDX1.3 could be overexpressed at the transcript level, only the PDX1.1 paralog could be increased at the protein level. The fact that PDX1.3 had been reported to be a ubiquitination target (Manzano et al., 2008) led to the postulation of a more stringent regulation of this paralog and suggested incomplete redundancy of the PDX1 genes.
Suc can serve as a carbon source, cause osmotic stress (depending on its concentration), or act as a signaling molecule. According to a recent article (Kircher and Schopfer, 2012), sugar produced by newly established photosynthesis during early seedling development is sufficient for the regulation of root elongation in light. Therefore, Suc supplied through the growth medium is beneficial for the root and subsequently for the growth of the entire seedling, since a larger root provides better access to nutrients from the medium. While this proves to be true for wild-type and pdx1.1 seedlings in this study, the growth of pdx1.3 mutant seedlings is severely impaired by the presence of Suc in the culture medium. The stunted growth of the main root was combined with altered morphology, fewer lateral roots, and the appearance of adventitious (anchor) roots, known to serve as a compensatory mechanism when the main root is damaged or missing (Lucas et al., 2011). Features revealed in this study can explain the rather pleiotropic phenotype observed. First, we noted that PDX1.1 expression was down-regulated in the presence of Suc within 30 min (Supplemental Fig. S5). This may be explained by the presence of a sequence (TAACAAA) assigned as a sugar response element (Morita et al., 1998) immediately upstream of PDX1.1 (Fig. 2A). Therefore, the pdx1.3 phenotype in the presence of Suc is exaggerated by the additional depletion of PDX1.1. The consequential deficiency of vitamin B6 leads to reduced amounts of two phytohormones, auxin and ethylene, both of which require the vitamin as a cofactor for their biosynthesis. Several pathways are implicated in auxin biosynthesis (for review, see Korasick et al., 2013), comprising many aminotransferases and decarboxylases, all of which require PLP (the cofactor form of vitamin B6) for catalysis. Likewise, ACC synthase, which is necessary for ethylene biosynthesis, is dependent on PLP for activity (Capitani et al., 1999). The severe depletion in auxin levels in the pdx1.3 mutant could be shown by direct measurements using mass spectrometry as well as indirectly through the use of the synthetic DR5 reporter (Fig. 3).
The role of auxin as a morphogen and the associated generation of local auxin maxima to promote organogenesis have been studied comprehensively (Benková et al., 2003; Heisler et al., 2005; Cederholm et al., 2012). This is exemplified in the regulation of the elaborate network of efflux and influx carriers. As examples, the efflux carriers PIN3 and PIN7 and the influx carrier AUX1 are generally down- and up-regulated, respectively, at the site of lateral root primordia, promoting local auxin accumulation that drives lateral root formation (Lewis et al., 2011). In this study, we observed a down-regulation of AUX1 and PIN3 expression in root tips, which, in the case of pdx1.3, is likely due to deficits in the gradient of auxin accumulation. It is noteworthy that auxin transport is also modulated by anthocyanins (Besseau et al., 2007), which are elevated in pdx1.3 in the presence of Suc and also negatively affect auxin distribution. As a consequence, both primary root and lateral root formation are impaired. The observation that PDX1.3 is highly expressed in the root tip stele region that undergoes rapid elongation and division as well as at the site of lateral root emergence (Fig. 1A) is consistent with this hypothesis. The importance of a gradient of auxin distribution for the establishment and maintenance of the root apical meristem, which is also impaired in pdx1.3, has been discussed previously (Chen and Xiong, 2009a, 2009b). We also noted here that root growth impairment in pdx1.3 draws some parallels with the shr mutant (Lucas et al., 2011). As SHR plays a role in maintaining the root apical meristem (Blilou et al., 2005), we investigated its expression in pdx1.3 and could show that there is a severe deficit in SHR expression during the early stages of root development.
However, it is known that a reduction in ethylene biosynthesis leads to a reduction in auxin biosynthesis, particularly at the tip of the primary root (Stepanova et al., 2005). More explicitly, the effects of ethylene on root tip growth, or its modulation, are thought to lie upstream of the observed effects in auxin biosynthesis and transport (Růzicka et al., 2007). The process depends on the ethylene-signaling pathway, because impaired ethylene perception prevents the accumulation of auxin in the root tip (Růzicka et al., 2007). Flagellin-induced ethylene production is strongly repressed in pdx1.3 (Fig. 4). Significantly for this study, application of the ethylene precursor ACC increases the level and redistribution of auxin, as judged from pdx1.3 harboring the DR5-GUS fusion (Supplemental Fig. S6). Moreover, it increases SHR expression and the deficit in auxin transport expression (Fig. 6, B, C, and E) and, at least partially, rescues root growth (Fig. 5, B and C). In contrast, previous studies on pdx1.3 have shown that the application of auxin or the specific induction of auxin biosynthesis genes in the root meristem did not rescue root growth (Chen and Xiong, 2009a, 2009b). Therefore, we conclude that the principal defect in postembryonic root growth of pdx1.3 as a consequence of vitamin B6 deficiency is impairment in ethylene metabolism. It can be reiterated here that ACC synthase is not only dependent on the B6 vitamer PLP as a cofactor but is a rate-limiting enzyme for ethylene biosynthesis. As a consequence of ethylene misregulation, the downstream effects are manifested in decreased auxin biosynthesis and impairment in its distribution as well as reduced levels of SHR. Notably, SHR expression levels also can be rescued in pdx1.3 by the application of vitamin B6 (Supplemental Fig. S7A). In addition, expression of PDX1.3 is not altered significantly in the shr2 mutant (Levesque et al., 2006) compared with the wild type (Supplemental Fig. S7B), and none of the PDX genes have been identified as direct or indirect targets of SHR in recent studies (Levesque et al., 2006; Cui et al., 2014). Therefore, impairment in SHR expression levels in this case is a consequence of the mutation in pdx1.3. Interestingly, levels of SHR expression are restored in the noncomplemented pdx1.3 line carrying pPDX1.1:PDX1.3 by application of vitamin B6 (Supplemental Fig. S7A). The latter reemphasizes the importance of the promoter region of PDX1.3 to the regulation of these vitamin B6-related parameters. Of note is that we mutated the putative ERE (AGCCGCC to ATCCTCC; Fujimoto et al., 2000) and partial AuxRE (TGTCTc to TGAATc; Ulmasov et al., 1997b) in the promoter region of PDX1.3 fused to the PDX1.3 coding sequence by site-directed mutagenesis and retransformed pdx1.3 to assess for complementation. Lines homozygous for these mutations were analyzed for the rescue of root growth. In almost all cases (six independent lines mutated in the ERE and four in the AuxRE), root growth was still increased compared with the pdx1.3 mutant. However, a comparison of pPDX1.3:PDX1.3 in the pdx1.3 mutant background with the mutated promoter lines indicates that similar root growth is only observed upon enhanced expression of PDX1.3 over that observed in pPDX1.3:PDX1.3 (Supplemental Fig. S8). Therefore, the ERE and AuxRE may contribute (albeit weakly) to the regulation of PDX1.3 expression. The other promoter regions involved in the regulation of PDX1.3 expression remain to be dissected in future experiments.
In pdx1.1, it is intriguing that the elevated auxin content in the presence of Suc is restricted to the root and, similar to other auxin overproduction mutants (Zhao et al., 2001), root growth is impaired. However, in contrast to other auxin overproduction mutants, we do not see a hairy-root phenotype (Fig. 1B). We also do not see epinastic cotyledons, increased apical dominance, or curled leaves, consistent with the restriction of the elevation in auxin content to root tissue. Nonetheless, it is not clear what is triggering elevated auxin production in pdx1.1. Some recent studies have highlighted the influence of Suc on auxin accumulation during seedling development, particularly in the root (Lilley et al., 2012; Sairanen et al., 2012). Specifically, Suc triggers an endogenous carbon-sensing pathway that results in higher free auxin levels and increased rootward auxin transport, dependent, at least in part, on the PHYTOCHOME-INTERACTING FACTOR (PIF) proteins (Lilley et al., 2012). This relay may be misregulated in pdx1.1, such that there is hyperaccumulation of auxin. It is also noteworthy in this context that PDX1.1, but not PDX1.3, expression is induced by IAA (Fig. 2C). Moreover, we have noted that PIF4 expression is up-regulated in pdx1.3 (Supplemental Table S1) in the presence of Suc (conditions where pdx1.1 is also down-regulated), providing support that there may be cross-talk between the PDX1 and PIF genes. We hypothesize that PDX1.1 must be required to prevent the overaccumulation of auxin in the presence of Suc. Whether this is a direct or indirect effect related to the presence of a sugar response element in the promoter region of PDX1.1 remains to be determined and provides a fertile area for future studies.
In summary, the PDX1.1 and PDX1.3 genes are required for postembryonic root development in Arabidopsis. Disruption of either of these genes results in a differential cascade of events as a consequence of vitamin B6 deficiency, which, in the case of pdx1.3, is propagated by a deficit in ethylene production, followed by a dissipation of the auxin gradient in the root tip leading to reduced cell division and expansion. Root growth can be partially rescued in pdx1.3 by the application of the ethylene precursor ACC correcting the cascade of events. PDX1.1 expression is repressed by Suc, and we hypothesize that it plays a role in auxin homeostasis. We conclude that while the biochemical roles of PDX1.1 and PDX1.3 overlap, they are regulated differently and, in turn, influence phytohormone homeostasis. A model summarizing the events as unraveled in this study is depicted in Figure 8. It remains to be explored in future studies what influence the availability of PLP as a cofactor and the vitamin B6 salvage pathway have on the overall homeostasis of this essential nutrient. A short-root phenotype and Suc sensitivity have been reported for mutants of the salvage pathway enzymes PDX3 and salt overly sensitive4 (Shi and Zhu, 2002; González et al., 2007), and the short-root phenotype of the rus (for root ultraviolet B-sensitive) mutants (Tong et al., 2008; Leasure et al., 2009) appears to be suppressed by mutations in the PLP-binding site of Asp aminotransferase as well as by pyridoxine supplementation (Leasure et al., 2011). The rus mutants have defects in polar auxin transport (Ge et al., 2010; Yu et al., 2013) and may provide an interesting link to this study.
Figure 8.
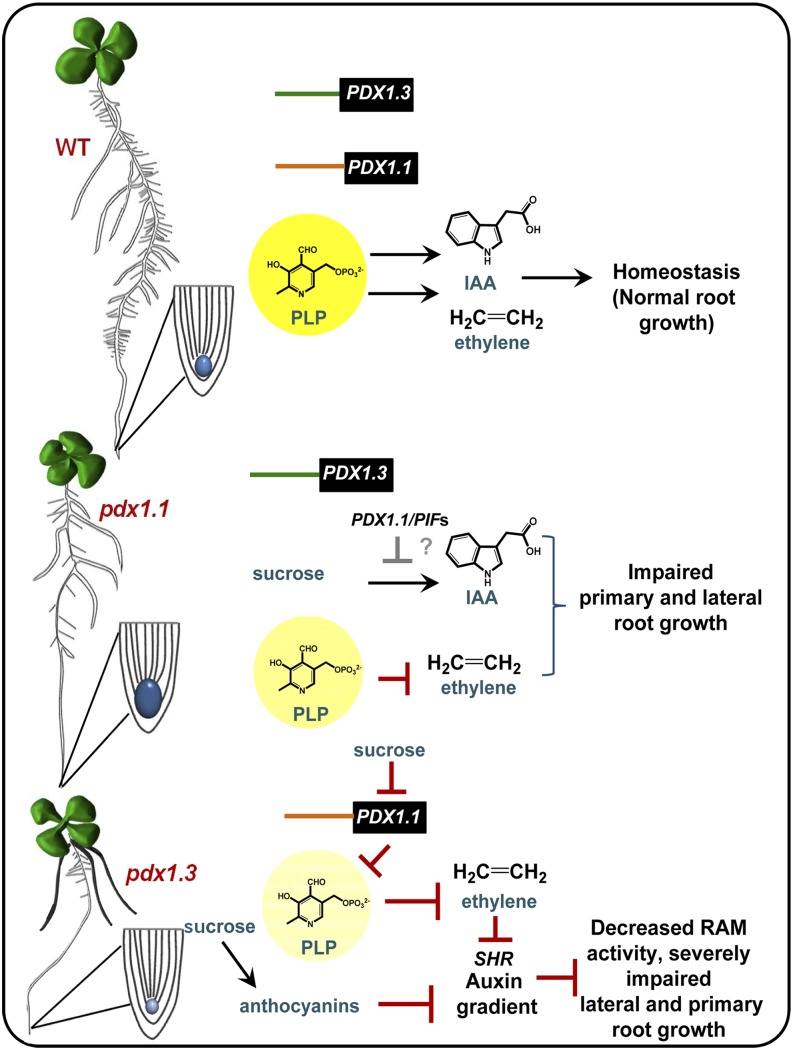
Consequences of a deficit in PLP for hormone homeostasis and root development in Arabidopsis. The top scenario indicates that, in the presence of sufficient PLP levels (yellow), as supplied by the catalytic action of PDX1.1 and PDX1.3, homeostasis of the two phytohormones auxin and ethylene is maintained for normal root development. The region around the root tip is amplified, part of which is shaded in blue to reflect arbitrary auxin levels at this stage of growth. The promoter regions of PDX1.1 and PDX1.3 are depicted by differently colored lines, to reflect their differential expression, while coding regions are shown as black boxes. The middle scenario illustrates observations in the pdx1.1 mutant. In this scenario, there is a mild deficit in the production of PLP (paler yellow; note that PDX1.3 is still expressed), which in turn affects ethylene production. However, auxin levels are much higher (shaded dark blue) than in the wild type, induced by the presence of Suc. The loss of PDX1.1 contributes to the inability to repress the accumulation of auxin as a function of Suc and may be related to the misregulation of PIF proteins. Overall, this results in impaired primary and lateral root growth. The bottom scenario depicts the situation in pdx1.3. In the presence of Suc, the expression of PDX1.1 is down-regulated and anthocyanins accumulate. The decrease in expression of both PDX1.3 and PDX1.1 leads to a major deficit in PLP production (pale yellow), which represses ethylene production. The combined deficit in ethylene production and the accumulation of anthocyanins negatively affects auxin biosynthesis (shaded pale blue) and its distribution. The expression of SHR is also decreased. Root apical meristem (RAM) activity is impaired, as reported (Chen and Xiong, 2005), leading to primary and lateral root growth defects and the appearance of anchor roots (dark gray).
MATERIALS AND METHODS
Plant Material
All Arabidopsis (Arabidopsis thaliana) lines (wild-type Col-0, pdx1.1, and pdx1.3 [Titiz et al., 2006], shr-2 [Benfey et al., 1993; Levesque et al., 2006], as well as the constructed transgenic lines [see below]) used for the described experiments were grown either on soil or in culture (as indicated) under long-day conditions (i.e. a 16-h photoperiod at 100–120 μmol photons m−2 s−1 at 22°C and 8 h in the dark at 18°C). Seedlings grown in culture were cultivated on one-half-strength Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) containing 0.8% (w/v) agar in the presence or absence of 1% (w/v) Suc either with or without pyridoxine supplementation (5 μm) as indicated. Other treatments with mannitol (100 mm), IAA (1 μm), or ACC (5 nm or 1 μm), where specified, were present in the culture medium from the start of the respective experiments.
Root Growth Measurements
Arabidopsis seedlings grown on sterile medium on vertically oriented plates under the conditions described above were used to determine the root length, the root growth curves, and the number of lateral roots in the presence or absence of Suc or after ACC supplementation. Plants were photographed starting at day 1 after germination until 10 DAG, and the root length was subsequently measured using ImageJ software (http://imagej.nih.gov/ij/). The root length and lateral root number data are means from at least two independent experiments with at least 20 technical repetitions, and error bars represent se. Statistical analyses were performed using the ANOVA with Tukey’s test.
Construction of Transgenic Lines
For the construction of the swapped promoter lines, the coding sequences of PDX1.1 and PDX1.3 were amplified from Arabidopsis genomic DNA using the following primer pairs: 5′-CATGCCATGGCAGGAACCGGAGTTGTGGCGGTGTACGGCG-3′ and 3′-GGACTAGTCTCAGAACGACTAGCGAACCTCTC-5′ for the PDX1.1 gene and 5′-CATGCCATGGAAGGAACCGGCGTTGTGGCGGTGTACGGTAACGG-3′ and 3′-GGACTAGTCTCGGAGCGATTAGCGAACCTCTC-5′ for the PDX1.3 gene (the region in underlined italics in each primer pair represents the incorporation of either an NcoI or a SpeI restriction site, respectively). The amplified genes were cloned into the binary vector pCAMBIA 1302 (http://www.cambia.org) using the NcoI and SpeI restriction sites. Subsequently, the 35S cauliflower mosaic virus promoter in the vector was replaced with the PCR-amplified region immediately upstream of the start codon of either PDX1.1 (1,424 bp; primer pair 5′-CATGCCATGGTTTTTTCTAGGGTTTTGAGAGAGTGTG-3′ and 3′-CCGGAATTCGGATTTGAGCATTTGTTAGTTTCTCTG-5′) or PDX1.3 (1,384 bp; primer pair 5′-CATGCCATGGTTATCGGAGATTGAAGAGAAATTTGTG-3′ and 3′-CCGGAATTCTACCATTTTTTTTGTTTGGTCTATATA-5′) using the EcoRI and NcoI restriction sites (underlined and in italics in the primer pairs). In addition to fusing the respective upstream region with the corresponding genes pPDX1.1:PDX1.1 and pPDX1.3:PDX1.3, the PDX1.1 upstream region was fused to the PDX1.3 gene and the PDX1.3 upstream region was fused to the PDX1.1 gene to generate chimeric constructs pPDX1.1:PDX1.3 and pPDX1.3:PDX1.1, respectively. To prevent a cotranslational fusion with GFP in pCAMBIA 1302, either a TAA or a TGA codon was introduced immediately after the coding regions of PDX1.1 and PDX1.3, respectively, using site-directed mutagenesis and the following primer pairs: 5′-GCTAGTCGTTCTGAGTAAACTAGTAAAGGAGAAG-3′ and 3′-CTTCTCCTTTACTAGTTTACTCAGAACGACTAGC-5′ for PDX1.1 and 5′-CGCTAATCGCTCCGAGTGAACTAGTAAAGGAG-3′ and 3′-CTCCTTTACTAGTTCACTCGGAGCGATTAGCG-5′ for PDX1.3. Mutations in the putative ERE and AuxRE of pPDX1.3:PDX1.3 were performed by site-directed mutagenesis with the following primers: 5′-CCTTCCTTTTCAACCACATCCTCCTATTTCTCTATTTAC-3′ and 3′-GTAAATAGAGAAATAGGAGGATGTGGTTGAAAAGGAAGG-5′ for mutating the ERE (AGCCGCC to ATCCTCC; Fujimoto et al., 2000) and 5′-GCTAACATTTTTTCTCCTTGAATTTTATGCAACAATTAATGCGG-3′ and 3′-CCGCATTAATTGTTGCATAAAATTCAAGGAGAAAAAATGTTAGC-5′ for the mutation of the AuxRE (TGTCTc to TGAATc; Ulmasov et al., 1997b), using pPDX1.3:PDX1.3 as a template. For the promoter-GUS fusion lines, the regions immediately upstream of the start codon of either PDX1.1 or PDX1.3, amplified using the primer pairs described above, were cloned into the pCAMBIA 1291Z vector using the EcoRI and NcoI restriction sites (http://www.cambia.org). In all cases, the obtained constructs were transferred into Agrobacterium tumefaciens strain C58, which was used to transform Arabidopsis (Col-0) by the floral dip method (Clough and Bent, 1998). Transformants were selected on the appropriate antibiotic, and only those lines that were homozygous and carrying a single insertion of the respective transgene were used for further analyses. Seeds harboring the DR5-GUS fusion construct originally described by Ulmasov et al. (1997b) were obtained from Jirí Friml, and progeny were crossed with the homozygous knockout mutant lines pdx1.1 and pdx1.3 (Titiz et al., 2006).
Histochemical GUS Expression Analyses
Arabidopsis seedlings grown on one-half-strength MS medium (Murashige and Skoog, 1962) containing 1% (w/v) Suc with or without pyridoxine supplementation for 10 to 12 DAG were harvested into 90% (v/v) acetone on ice and then incubated for 20 min at room temperature. The samples were rinsed three times with 10 mm NaH2PO4/Na2HPO4, pH 7, containing 0.5 mm K3[Fe(CN)6], 0.5 mm K4[Fe(CN)6], 0.1% (w/v) Triton X-100, and 10 mm EDTA-Na2 and then incubated at 37°C in the same solution containing 0.5 mg mL−1 5-bromo-4-chloro-3-indolyl-β-glucuronide. The incubation was done either for 2 or 3 h or overnight depending on the intensity. The excess stain and chlorophyll from the samples were removed by several rinses with 70% (v/v) ethanol. Samples were kept in 70% ethanol until examination.
Quantitative GUS Expression Analyses
Seedlings (approximately 100 mg of tissue) grown as for the histochemical analyses above either in the absence or presence of 1% (w/v) Suc were harvested and immediately frozen in liquid nitrogen. The harvested samples were ground in 50 mm NaH2PO4/Na2HPO4, pH 7, containing 50 mm EDTA-Na2, 0.1% (w/v) Triton X-100, 0.1% (w/v) SDS, 0.025 mg mL−1 phenylmethanesulfonyl fluoride, and 10 mm β-mercaptoethanol (extraction buffer). The extract was centrifuged for 10 min at 13,200g, and the supernatant was collected and used for further analyses. The extraction buffer containing 1 mm 4-methylumbelliferyl-β-d-glucuronide was heated to 37°C (reaction buffer). For each sample to be analyzed, 20 µL of plant extract was added to 1 mL of reaction buffer. After 10 min of incubation, 100 µL of the reaction volume was transferred to a fresh tube containing 900 µL of stop solution (0.2 m Na2CO3), and the fluorescence of the product of the reaction 4-methylumbelliferone was followed at excitation and emission wavelengths of 365 and 455 nm, respectively. The results were derived from a standard curve prepared with serial dilutions of the product. Statistical analyses were performed using the ANOVA with Tukey’s test. The results are mean values of three independent experiments, and error bars represent se.
Free and Conjugated Auxin Analyses
The method used for auxin analysis was described by Pencík et al. (2009). Plant material frozen with liquid nitrogen was ground with a pestle and mortar, and samples of 25 mg average weight were extracted with phosphate buffer (pH 7). The extracts were subjected to a C8-based solid-phase extraction, methylated with ethereal diazomethane, and subsequently purified by immunoaffinity extraction. The final analysis was done by ultra-HPLC coupled to tandem mass detection.
RNA Extraction, Complementary DNA Synthesis, and qPCR Analysis
Arabidopsis seedlings were grown in culture for 10 DAG on one-half-strength MS medium either with or without 1% (w/v) Suc and in the presence or absence of supplementation as indicated. Total RNA was extracted from seedling samples or separated shoots and roots using the Nucleospin RNA kit from Macherey-Nagel and was reverse transcribed with SuperScript II RNase H−, using oligo(dT) primers 12 to 18 according to the manufacturer’s recommendations (Invitrogen), using 2 μg of total RNA and diluted five times. qPCR was performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems) using 3 µL of the diluted complementary DNA samples and SYBR Green as a fluorescence detector (Power SYBR Green PCR master mix; Applied Biosystems) in accordance with the manufacturer’s instructions. Gene-specific primer pairs are listed in Supplemental Table S2. The GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASE (GAPDH) gene was used as a reference to calculate the relative expression levels of the genes under investigation, as its transcript levels did not vary by more than one cycle threshold between treatments. All qPCR values represent three biological replicates with three technical repeats. The ANOVA Tukey or Bonferroni test was used for statistical analysis of the data together with pairwise comparisons of the mean values. The results are means of at least three independent experiments with error bars representing se.
Microarray Analyses
Arabidopsis seedlings were grown in culture on one-half-strength MS medium (Murashige and Skoog, 1962) containing 1% (w/v) Suc without pyridoxine supplementation. Plants were cultivated under a 16-h photoperiod at 100 μmol photons m−2 s−1 at a temperature of 22°C but were maintained at 18°C in the dark. Roots and shoots from the wild type and pdx1.3 (three biological replicates for each line and for each type of tissue) were harvested 10 DAG, and total RNA was extracted using the Macherey-Nagel Nucleic Acid and Protein Purification kit, following the manufacturer’s instructions. The quality of the samples was checked using chip electrophoresis (Bioanalyzer 2100; Agilent). Subsequent biotin-labeled copy RNA synthesis (using 500 ng of total RNA) was done using Ambion MessageAmp II according to the manufacturer’s protocol. Hybridization of the fragmented copy RNA target to GeneChip ATH1 (22,810 probe sets), washing, labeling, and scanning were done according to Affymetrix instructions. The MAS 5.0 algorithm was used for signal expression computation and normalization. To identify differentially expressed transcripts, pairwise comparison analyses were carried out. Each experimental sample was compared with each reference sample, resulting in nine pairwise comparisons. Transcripts were considered differentially expressed if their levels changed in the same direction in seven of nine comparisons. Further data filtering and analyses were performed with the GeneSpring software (Agilent). Only genes displaying at least 2-fold up- or down-regulation in the pdx1.3 mutant versus the wild type were considered further. Additional statistical analyses were performed using the Welch test and Student’s t test.
Ethylene Measurements
The flg22 peptide (corresponding to the sequence of Xanthomonas axonopodis citri, which was fully active to stimulate the pattern-triggered immunity response in Arabidopsis) was obtained from Peptron. Prior to use, the peptide was dissolved in water (stock solutions of 1–10 mm) and diluted to a final concentration of 10 μm in a solution containing 0.1% (w/v) bovine serum albumin and 0.1 m NaCl. For assaying ethylene production, entire seedlings grown for 10 d in sterile culture with and without 5 μm pyridoxine were used. For sampling, five separate seedlings were collected (totaling 20 mg of fresh weight per assay) and transferred to 6-mL glass tubes containing 1 mL of water and the elicitor preparation to be tested. The tubes were closed with rubber septa, and ethylene accumulating in the free air space was measured by gas chromatography after 4 h of incubation. The average of 15 different samples was determined. Samples prepared in an identical fashion were treated with a solution containing only 0.1% (w/v) bovine serum albumin and 0.1 m NaCl (mock treatment) to serve as a control, and ethylene production was assayed in parallel. Statistical analyses were performed using the ANOVA with Tukey’s test. The results are means of three independent experiments, and error bars represent se.
Sequence data from this study can be found in the GenBank/EMBL databases under the following accession numbers: PDX1.1, At2g38230; PDX1.3, At5g01410; AUX1, At2g38120; PIN3, At1g70940; PIN7, At1g23080; GAPDH, At1g13440; PAP1, At1g56650; PAP2, At1g66390; and SHR, At4g37650.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Total B6 vitamer levels in Arabidopsis shoots and roots.
Supplemental Figure S2. Effect of ACC application in the absence of Suc.
Supplemental Figure S3. Relative expression level of polar auxin transporters.
Supplemental Figure S4. Functional categorization of altered expression in pdx1.3.
Supplemental Figure S5. Suc-induced changes in PDX1.1 and PDX1.3 expression.
Supplemental Figure S6. Effect of ACC on DR5 expression.
Supplemental Figure S7. SHR expression in pdx1 lines and PDX1.3 expression in shr-2.
Supplemental Figure S8. Analysis of mutated PDX1.3 promoter lines.
Supplemental Table S1. Microarray analysis of pdx1.3.
Supplemental Table S2. qPCR primers used.
Supplementary Material
Acknowledgments
We thank Jirí Friml (Institute of Science and Technology, Klosterneuburg, Austria) for the kind gift of seeds of the DR5-GUS line; Nadine Müller for technical assistance in the generation and maintenance of Arabidopsis lines at the outset of this study; Olca Titiz, who performed preliminary experiments, and Nikolaus Amrhein (all Eidgenössische Technische Hochschule, Zurich, Switzerland), for support at the beginning of this study; as well as Niko Geldner (University of Lausanne) for provision of the shr2 mutant line.
Glossary
- PLP
pyridoxal-5′-phosphate
- DXP
deoxyxylulose 5-phosphate
- qPCR
quantitative PCR
- Col-0
Columbia-0
- DAG
days after germination
- ERE
ethylene-responsive element
- AuxRE
auxin response element
- IAA
indole acetic acid
- ACC
1-aminocyclopropane-1-carboxylic acid
- MS
Murashige and Skoog
Footnotes
This work was supported by the Swiss National Science Foundation (grant nos. PPOOA_1191186 and 31003A–141117/1 to T.B.F. and the University of Geneva), the Ernest Boninchi Foundation, and the Ernst and Lucie Schmidheiny Foundation.
Articles can be viewed without a subscription.
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15: 63–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Linstead PJ, Roberts K, Schiefelbein JW, Hauser MT, Aeschbacher RA (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119: 57–70 [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J (2003) Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602 [DOI] [PubMed] [Google Scholar]
- Besseau S, Hoffmann L, Geoffroy P, Lapierre C, Pollet B, Legrand M (2007) Flavonoid accumulation in Arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19: 148–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B (2005) The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44 [DOI] [PubMed] [Google Scholar]
- Burns KE, Xiang Y, Kinsland CL, McLafferty FW, Begley TP (2005) Reconstitution and biochemical characterization of a new pyridoxal-5′-phosphate biosynthetic pathway. J Am Chem Soc 127: 3682–3683 [DOI] [PubMed] [Google Scholar]
- Cane DE, Du S, Robinson JK, Hsiung Y, Spenser ID (1999) Biosynthesis of vitamin B6: enzymatic conversion of 1-deoxy-D-xylulose-5-phosphate to pyridoxol phosphate. J Am Chem Soc 121: 7722–7723 [Google Scholar]
- Cane DE, Hsiung Y, Cornish JA, Robinson JK, Spenser ID (1998) Biosynthesis of vitamin B6: the oxidation of 4-(phosphohydroxy)-L-threonine by PdxA. J Am Chem Soc 120: 1936–1937 [Google Scholar]
- Capitani G, Hohenester E, Feng L, Storici P, Kirsch JF, Jansonius JN (1999) Structure of 1-aminocyclopropane-1-carboxylate synthase, a key enzyme in the biosynthesis of the plant hormone ethylene. J Mol Biol 294: 745–756 [DOI] [PubMed] [Google Scholar]
- Cederholm HM, Iyer-Pascuzzi AS, Benfey PN (2012) Patterning the primary root in Arabidopsis. Wiley Interdiscip Rev Dev Biol 1: 675–691 [DOI] [PubMed] [Google Scholar]
- Chen H, Xiong L (2005) Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses. Plant J 44: 396–408 [DOI] [PubMed] [Google Scholar]
- Chen H, Xiong L (2009a) Localized auxin biosynthesis and postembryonic root development in Arabidopsis. Plant Signal Behav 4: 752–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xiong L (2009b) The short-rooted vitamin B6-deficient mutant pdx1 has impaired local auxin biosynthesis. Planta 229: 1303–1310 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui H, Kong D, Liu X, Hao Y (2014) SCARECROW, SCR-LIKE 23 and SHORT-ROOT control bundle sheath cell fate and function in Arabidopsis thaliana. Plant J 78: 319–327 [DOI] [PubMed] [Google Scholar]
- Das PK, Shin DH, Choi SB, Park YI (2012) Sugar-hormone cross-talk in anthocyanin biosynthesis. Mol Cells 34: 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky JG, Forde BG (2012) Quantitative analysis of lateral root development: pitfalls and how to avoid them. Plant Cell 24: 4–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenshaft M, Bilski P, Li MY, Chignell CF, Daub ME (1999) A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc Natl Acad Sci USA 96: 9374–9378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenshaft M, Daub ME (2001) Isolation of PDX2, a second novel gene in the pyridoxine biosynthesis pathway of eukaryotes, archaebacteria, and a subset of eubacteria. J Bacteriol 183: 3383–3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Field B, Fiston-Lavier AS, Kemen A, Geisler K, Quesneville H, Osbourn AE (2011) Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proc Natl Acad Sci USA 108: 16116–16121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B, Osbourn AE (2008) Metabolic diversification: independent assembly of operon-like gene clusters in different plants. Science 320: 543–547 [DOI] [PubMed] [Google Scholar]
- Fitzpatrick TB. (2011) Vitamin B6 in plants: more than meets the eye. InRebeille F, Douce R, eds, Advances in Botanical Research, Vol 59 Elsevier, New York, pp 2–31 [Google Scholar]
- Fitzpatrick TB, Basset GJ, Borel P, Carrari F, DellaPenna D, Fraser PD, Hellmann H, Osorio S, Rothan C, Valpuesta V, et al. (2012) Vitamin deficiencies in humans: can plant science help? Plant Cell 24: 395–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M (2000) Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge L, Peer W, Robert S, Swarup R, Ye S, Prigge M, Cohen JD, Friml J, Murphy A, Tang D, et al. (2010) Arabidopsis ROOT UVB SENSITIVE2/WEAK AUXIN RESPONSE1 is required for polar auxin transport. Plant Cell 22: 1749–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI, Laby RJ, Kim D (2001) The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem Biophys Res Commun 280: 196–203 [DOI] [PubMed] [Google Scholar]
- González E, Danehower D, Daub ME (2007) Vitamer levels, stress response, enzyme activity, and gene regulation of Arabidopsis lines mutant in the pyridoxine/pyridoxamine 5′-phosphate oxidase (PDX3) and the pyridoxal kinase (SOS4) genes involved in the vitamin B6 salvage pathway. Plant Physiol 145: 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler MG, Ohno C, Das P, Sieber P, Reddy GV, Long JA, Meyerowitz EM (2005) Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol 15: 1899–1911 [DOI] [PubMed] [Google Scholar]
- Hellmann H, Mooney S (2010) Vitamin B6: a molecule for human health? Molecules 15: 442–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki T, Yamaguchi-Shinozaki K, Shinozaki K (1995) Identification of a cis-regulatory region of a gene in Arabidopsis thaliana whose induction by dehydration is mediated by abscisic acid and requires protein synthesis. Mol Gen Genet 247: 391–398 [DOI] [PubMed] [Google Scholar]
- Jeong SW, Das PK, Jeoung SC, Song JY, Lee HK, Kim YK, Kim WJ, Park YI, Yoo SD, Choi SB, et al. (2010) Ethylene suppression of sugar-induced anthocyanin pigmentation in Arabidopsis. Plant Physiol 154: 1514–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Schopfer P (2012) Photosynthetic sucrose acts as cotyledon-derived long-distance signal to control root growth during early seedling development in Arabidopsis. Proc Natl Acad Sci USA 109: 11217–11221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings H, Jackson MB (1979) A relationship between rates of ethylene production by roots and the promoting or inhibiting effects of exogenous ethylene and water on root elongation. Z Pflanzenphysiol 92: 385–397 [Google Scholar]
- Korasick DA, Enders TA, Strader LC (2013) Auxin biosynthesis and storage forms. J Exp Bot 64: 2541–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laber B, Maurer W, Scharf S, Stepusin K, Schmidt FS (1999) Vitamin B6 biosynthesis: formation of pyridoxine 5′-phosphate from 4-(phosphohydroxy)-L-threonine and 1-deoxy-D-xylulose-5-phosphate by PdxA and PdxJ protein. FEBS Lett 449: 45–48 [DOI] [PubMed] [Google Scholar]
- Lam HM, Winkler ME (1992) Characterization of the complex pdxH-tyrS operon of Escherichia coli K-12 and pleiotropic phenotypes caused by pdxH insertion mutations. J Bacteriol 174: 6033–6045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leasure CD, Tong H, Yuen G, Hou X, Sun X, He ZH (2009) ROOT UV-B SENSITIVE2 acts with ROOT UV-B SENSITIVE1 in a root ultraviolet B-sensing pathway. Plant Physiol 150: 1902–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leasure CD, Tong HY, Hou XW, Shelton A, Minton M, Esquerra R, Roje S, Hellmann H, He ZH (2011) ROOT UV-B sensitive mutants are suppressed by specific mutations in ASPARTATE AMINOTRANSFERASE2 and by exogenous vitamin B6. Mol Plant 4: 759–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque MP, Vernoux T, Busch W, Cui H, Wang JY, Blilou I, Hassan H, Nakajima K, Matsumoto N, Lohmann JU, et al. (2006) Whole-genome analysis of the SHORT-ROOT developmental pathway in Arabidopsis. PLoS Biol 4: e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Negi S, Sukumar P, Muday GK (2011) Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development 138: 3485–3495 [DOI] [PubMed] [Google Scholar]
- Lilley JL, Gee CW, Sairanen I, Ljung K, Nemhauser JL (2012) An endogenous carbon-sensing pathway triggers increased auxin flux and hypocotyl elongation. Plant Physiol 160: 2261–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Swarup R, Paponov IA, Swarup K, Casimiro I, Lake D, Peret B, Zappala S, Mairhofer S, Whitworth M, et al. (2011) Short-Root regulates primary, lateral, and adventitious root development in Arabidopsis. Plant Physiol 155: 384–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzano C, Abraham Z, López-Torrejón G, Del Pozo JC (2008) Identification of ubiquitinated proteins in Arabidopsis. Plant Mol Biol 68: 145–158 [DOI] [PubMed] [Google Scholar]
- Mittenhuber G. (2001) Phylogenetic analyses and comparative genomics of vitamin B6 (pyridoxine) and pyridoxal phosphate biosynthesis pathways. J Mol Microbiol Biotechnol 3: 1–20 [PubMed] [Google Scholar]
- Moccand C, Boycheva S, Surriabre P, Tambasco-Studart M, Raschke M, Kaufmann M, Fitzpatrick TB (2014) The pseudoenzyme PDX1.2 boosts vitamin B6 biosynthesis under heat and oxidative stress in Arabidopsis. J Biol Chem 289: 8203–8216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney S, Hellmann H (2010) Vitamin B6: killing two birds with one stone? Phytochemistry 71: 495–501 [DOI] [PubMed] [Google Scholar]
- Morita A, Umemura T, Kuroyanagi M, Futsuhara Y, Perata P, Yamaguchi J (1998) Functional dissection of a sugar-repressed alpha-amylase gene (RAmy1 A) promoter in rice embryos. FEBS Lett 423: 81–85 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth of bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Pencík A, Rolcík J, Novák O, Magnus V, Barták P, Buchtík R, Salopek-Sondi B, Strnad M (2009) Isolation of novel indole-3-acetic acid conjugates by immunoaffinity extraction. Talanta 80: 651–655 [DOI] [PubMed] [Google Scholar]
- Percudani R, Peracchi A (2003) A genomic overview of pyridoxal-phosphate-dependent enzymes. EMBO Rep 4: 850–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Amakawa T, Goto N, Tsurumi S (2001) Auxin is a positive regulator for ethylene-mediated response in the growth of Arabidopsis roots. Plant Cell Physiol 42: 301–307 [DOI] [PubMed] [Google Scholar]
- Raschke M, Boycheva S, Crèvecoeur M, Nunes-Nesi A, Witt S, Fernie AR, Amrhein N, Fitzpatrick TB (2011) Enhanced levels of vitamin B6 increase aerial organ size and positively affect stress tolerance in Arabidopsis. Plant J 66: 414–432 [DOI] [PubMed] [Google Scholar]
- Raschle T, Amrhein N, Fitzpatrick TB (2005) On the two components of pyridoxal 5′-phosphate synthase from Bacillus subtilis. J Biol Chem 280: 32291–32300 [DOI] [PubMed] [Google Scholar]
- Růzicka K, Ljung K, Vanneste S, Podhorská R, Beeckman T, Friml J, Benková E (2007) Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sairanen I, Novák O, Pěnčík A, Ikeda Y, Jones B, Sandberg G, Ljung K (2012) Soluble carbohydrates regulate auxin biosynthesis via PIF proteins in Arabidopsis. Plant Cell 24: 4907–4916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Zhu JK (2002) SOS4, a pyridoxal kinase gene, is required for root hair development in Arabidopsis. Plant Physiol 129: 585–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinshi H, Usami S, Ohme-Takagi M (1995) Identification of an ethylene-responsive region in the promoter of a tobacco class I chitinase gene. Plant Mol Biol 27: 923–932 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM (2005) A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17: 2230–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmeier M, Raschle T, Mazurkiewicz J, Rippe K, Sinning I, Fitzpatrick TB, Tews I (2006) Structure of a bacterial pyridoxal 5′-phosphate synthase complex. Proc Natl Acad Sci USA 103: 19284–19289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Parry G, Graham N, Allen T, Bennett M (2002) Auxin cross-talk: integration of signalling pathways to control plant development. Plant Mol Biol 49: 411–426 [DOI] [PubMed] [Google Scholar]
- Tambasco-Studart M, Titiz O, Raschle T, Forster G, Amrhein N, Fitzpatrick TB (2005) Vitamin B6 biosynthesis in higher plants. Proc Natl Acad Sci USA 102: 13687–13692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E (2005) Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiol 138: 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale WD, Paponov IA, Palme K (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7: 847–859 [DOI] [PubMed] [Google Scholar]
- Titiz O, Tambasco-Studart M, Warzych E, Apel K, Amrhein N, Laloi C, Fitzpatrick TB (2006) PDX1 is essential for vitamin B6 biosynthesis, development and stress tolerance in Arabidopsis. Plant J 48: 933–946 [DOI] [PubMed] [Google Scholar]
- Tong H, Leasure CD, Hou X, Yuen G, Briggs W, He ZH (2008) Role of root UV-B sensing in Arabidopsis early seedling development. Proc Natl Acad Sci USA 105: 21039–21044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1997a) ARF1, a transcription factor that binds to auxin response elements. Science 276: 1865–1868 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997b) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B (1995) Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378: 62–65 [DOI] [PubMed] [Google Scholar]
- Wagner S, Bernhardt A, Leuendorf JE, Drewke C, Lytovchenko A, Mujahed N, Gurgui C, Frommer WB, Leistner E, Fernie AR, et al. (2006) Analysis of the Arabidopsis rsr4-1/pdx1-3 mutant reveals the critical function of the PDX1 protein family in metabolism, development, and vitamin B6 biosynthesis. Plant Cell 18: 1722–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Chemler J, Huang L, Martens S, Koffas MAG (2005) Metabolic engineering of anthocyanin biosynthesis in Escherichia coli. Appl Environ Microbiol 71: 3617–3623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Chen C, Chen Z (2001) Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13: 1527–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Karampelias M, Robert S, Peer WA, Swarup R, Ye S, Ge L, Cohen J, Murphy A, Friml J, et al. (2013) ROOT ULTRAVIOLET B-SENSITIVE1/weak auxin response3 is essential for polar auxin transport in Arabidopsis. Plant Physiol 162: 965–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zein F, Zhang Y, Kang YN, Burns K, Begley TP, Ealick SE (2006) Structural insights into the mechanism of the PLP synthase holoenzyme from Thermotoga maritima. Biochemistry 45: 14609–14620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Winkler ME (1996) 4-Phospho-hydroxy-L-threonine is an obligatory intermediate in pyridoxal 5′-phosphate coenzyme biosynthesis in Escherichia coli K-12. FEMS Microbiol Lett 135: 275–280 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J (2001) A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309 [DOI] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA 95: 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.