Diverse signaling pathways are activated by perturbation of mitochondrial function under different growth conditions.
Abstract
Diverse signaling pathways are activated by perturbation of mitochondrial function under different growth conditions.Mitochondria have emerged as an important organelle for sensing and coping with stress in addition to being the sites of important metabolic pathways. Here, responses to moderate light and drought stress were examined in different Arabidopsis (Arabidopsis thaliana) mutant plants lacking a functional alternative oxidase (alternative oxidase1a [aox1a]), those with reduced cytochrome electron transport chain capacity (T3/T7 bacteriophage-type RNA polymerase, mitochondrial, and plastidial [rpoTmp]), and double mutants impaired in both pathways (aox1a:rpoTmp). Under conditions considered optimal for growth, transcriptomes of aox1a and rpoTmp were distinct. Under adverse growth conditions, however, transcriptome changes in aox1a and rpoTmp displayed a highly significant overlap and were indicative of a common mitochondrial stress response and down-regulation of photosynthesis. This suggests that the role of mitochondria to support photosynthesis is provided through either the alternative pathway or the cytochrome pathway, and when either pathway is inhibited, such as under environmental stress, a common, dramatic, and succinct mitochondrial signal is activated to alter energy metabolism in both organelles. aox1a:rpoTmp double mutants grown under optimal conditions showed dramatic reductions in biomass production compared with aox1a and rpoTmp and a transcriptome that was distinct from aox1a or rpoTmp. Transcript data indicating activation of mitochondrial biogenesis in aox1a:rpoTmp were supported by a proteomic analysis of over 200 proteins. Under optimal conditions, aox1a:rpoTmp plants seemed to switch on many of the typical mitochondrial stress regulators. Under adverse conditions, aox1a:rpoTmp turned off these responses and displayed a biotic stress response. Taken together, these results highlight the diverse signaling pathways activated by the perturbation of mitochondrial function under different growth conditions.
One notable characteristic of plant mitochondria is the possession of a branched electron transport chain. At the input site for electrons deriving from the oxidation of NADH, plant mitochondria contain both types I (NADH-ubiquinone oxidoreductase) and II (alternative) NADH dehydrogenases. At the end of the electron transport chain, oxygen can be reduced to water by either complex IV (cytochrome oxidase) or the alternative oxidase (AOX; Millar et al., 2011). Complexes I and IV each contain several subunits, comprising both mitochondrial- and nuclear-encoded subunits, and electron transport is coupled to the movement of H+ ions across the inner mitochondrial membrane (Millar et al., 2011). In contrast, the alternative NADH dehydrogenases and oxidases can function with a single type of protein (possibly as homodimers), and electron transport does not result in a proton motive force (Vanlerberghe and McIntosh, 1997; Rasmusson et al., 2004). The type II NADH dehydrogenases and AOX are distinguished from complexes I and IV based on the fact that they are insensitive to rotenone and cyanide, respectively (Vanlerberghe and McIntosh, 1997; Rasmusson et al., 2004).
AOX is arguably the most extensively studied component of the plant mitochondrial electron transport chain, and a crystal structure of the AOX from Trypanosoma brucei was published recently (Shiba et al., 2013). Extensive biochemical characterization (Umbach and Siedow, 2000; Millar et al., 2011), investigations into the structure and evolution of genes encoding AOX (McDonald, 2008), and transcriptional and posttranscriptional regulation (Vanlerberghe and McIntosh, 1997; Millar et al., 2011) have been undertaken. In vivo oxygen isotope discrimination measurements revealed that the alternative pathway is active even when the cytochrome pathway is not saturated (Ribas-Carbo et al., 1995). The development of a monoclonal antibody that recognized all AOX proteins studied to date (Elthon et al., 1989; Finnegan et al., 1999) facilitated the cloning of the gene, which allowed genetic approaches to be used to study AOX (Rhoads and McIntosh, 1991). Studies in tobacco (Nicotiana tabacum) with reduced or increased amounts of AOX (Vanlerberghe et al., 1994) showed that AOX suppresses the production of reactive oxygen species (ROS) in mitochondria (Maxwell et al., 1999; Cvetkovska and Vanlerberghe, 2012a, 2012b, 2013; Cvetkovska et al., 2013). Investigations with a variety of abiotic or biotic challenges revealed that AOX acts as a survival protein (Vanlerberghe et al., 2009), in that it allows plants to survive and grow better compared with plants that have reduced amounts of AOX. More detailed studies on the role of AOX in tobacco and Arabidopsis (Arabidopsis thaliana) showed that it not only suppresses the production of ROS (see studies outlined above) but also, by its activity, it mediates or determines signals coming from mitochondria (Arnholdt-Schmitt et al., 2006; Clifton et al., 2006; Vanlerberghe et al., 2009).
Although many studies have examined the role of AOX during stress, the contribution of the cytochrome pathway under stressful conditions has not been as extensively investigated, with the exception of complex I (NADH-ubiquinone oxidoreductase) of the electron transport chain. A variety of mutants that affect the assembly and/or activity of complex I results in the induction of AOX (Gutierres et al., 1997; Karpova et al., 2002; Meyer et al., 2009; Kühn et al., 2011), although the extent differs between studies. Complex I mutants display delayed growth phenotypes and altered stress responses (Dutilleul et al., 2003a, 2003b; Meyer et al., 2009). In addition to the role of complex I in oxidizing NADH and generating a proton motive force for subsequent ATP synthesis, it has been proposed that complex I plays a variety of other roles in plants, including roles in ROS signaling, redox balancing, ascorbate biosynthesis, and recycling of mitochondrial CO2-HCo3− and also, possibly, as a site for metabolite channeling (Braun et al., 2014). It has also been reported to interact with other inner membrane multisubunit complexes, such as the Translocase of the Inner Mitochondrial membrane17:23 (TIM17:23), which may play a role in linking mitochondrial activity with mitochondrial biogenesis (Wang et al., 2012). Thus, as observed with AOX, complex I also seems to have important signaling roles in addition to its enzymatic activity.
Although the various respiratory chain complexes are well studied in terms of activity and composition (Klodmann et al., 2011), the role that these complexes play in signaling is less well understood, primarily because of the fact that mutations leading to the loss of these complexes are often lethal. Lethality of a complete abolition of complex IV activity has been suggested by the characterization of the nonchromosomal stripe mutants in Zea mays (Gu et al., 1994). However, plants with greatly reduced amounts of respiratory chain complexes can be obtained through so-called surrogate mutants. These mutants are disrupted in nuclear genes that are required for the transcription, splicing, editing, translation, or assembly of a mitochondrial-encoded factor (Colas des Francs-Small and Small, 2014). Although complex I deficiencies represent the greatest number of surrogate mutants known to date, surrogate mutants also exist for complex III, cytochrome c, complex IV, and complex V (Colas des Francs-Small and Small, 2014). However, in most cases to date, the description of these mutants has been limited to the characterization of the mitochondrial gene expression defect and its direct phenotypic effects.
A number of studies has carried out meta-analyses on the consequences of mitochondrial dysfunction. A meta-analysis of 11 transcriptome signatures of different mitochondrial functions revealed common responses in genes for the functional categories of plant-pathogen interactions, light reactions of photosynthesis, and protein synthesis (Schwarzländer et al., 2012). However, it was also noticeable that distinct changes did occur depending on the target of the mutation or chemical inhibition (e.g. it was concluded that the loss of AOX1a resulted in changes in transcript abundance that were independent or even inverse compared with the other data sets). Thus, it was suggested that the “particular path of electron flow through the ETC is critical in mounting the underlying signal” (Schwarzländer et al., 2012). Inhibition of the mitochondrial ATP synthase by oligomycin treatment resulted in transcript changes that overlapped with many of the changes usually seen in plants impaired in the electron transport chain (Geisler et al., 2012). Other meta-analyses of transcriptome changes upon perturbation of mitochondrial function have drawn similar conclusions from detecting widespread changes associated with ROS signaling (Van Aken et al., 2009a, 2009b; Umbach et al., 2012; Van Aken and Whelan, 2012). Notably, although perturbation of mitochondrial function results in transcriptome changes affecting many cellular processes, its impact is significantly higher on genes encoding mitochondrial proteins (Van Aken and Whelan, 2012).
A number of studies in Arabidopsis and tobacco have analyzed the response of plants to inactivation of AOX and also, importantly, the response of these plants to stress (Giraud et al., 2008; Cvetkovska and Vanlerberghe, 2012a, 2012b; Cvetkovska et al., 2013). In Arabidopsis, inactivation of the stress-inducible AOX1a gene was shown to increase stress sensitivity and comprehensively alter the transcriptome (Giraud et al., 2008). Comparable studies on plants defective in the cytochrome chain have been lacking because of the fact that plants deficient in the cytochrome chain have not been available. Here, we used T3/T7 bacteriophage-type RNA polymerase, mitochondrial, and plastidial (rpoTmp) mutant lines as surrogate mutants, which lack one of two RNA polymerases in mitochondria and display transcriptional defects leading to a greater than 80% decrease in the activities of mitochondrial complexes I and IV (Kühn et al., 2009), to investigate consequences of defects in the cytochrome pathway. Using Arabidopsis rpoTmp and aox1a mutant lines as well as rpoTmp:aox1a double mutants, we (1) compared the stress responses of plants with limited electron flow through the cytochrome or alternative pathway and (2) analyzed the effects of inhibiting both pathways simultaneously under normal and stress conditions.
RESULTS
Mitochondrial Electron Transport Shows Plasticity in Response to Environmental Challenge
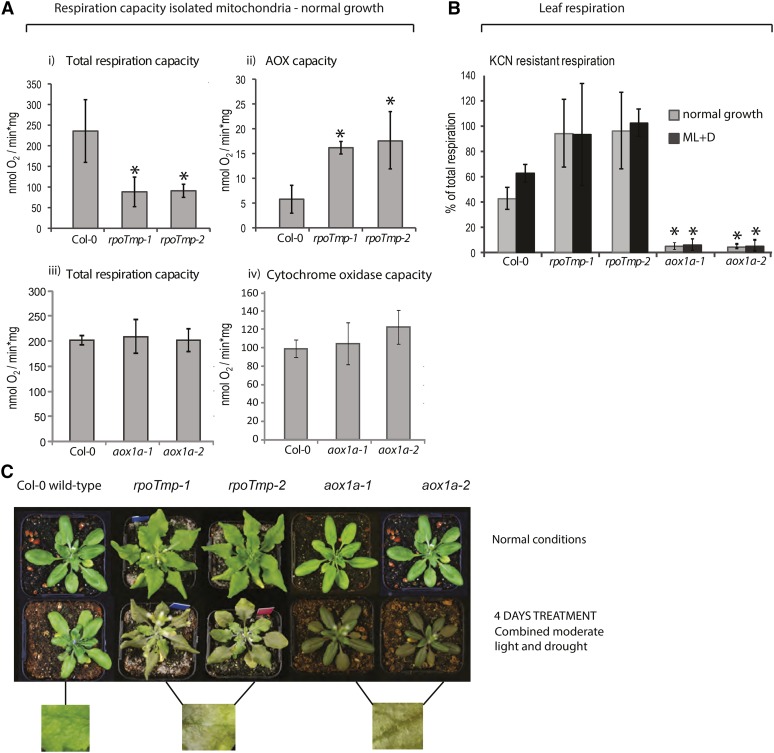
To investigate the effects of limitation of electron flow through the alternative or cytochrome mitochondrial electron transport chain under normal and stress conditions, mutants lacking the AOX (aox1a plants; Giraud et al., 2008) or mutants with dramatically decreased complex I and IV levels (rpoTmp plants; Kühn et al., 2009) were investigated. aox1a mutants lack a functional AOX1a protein (At3g22370), whereas rpoTmp mutants are defective in the dual-targeted mitochondrial and plastidial RNA polymerase (At5g15700). rpoTmp plants display specific defects in transcript abundance of mitochondrial-located genes that cause a reduction in the accumulation of complexes I and IV, and therefore, rpoTmp mitochondria show only approximately 15% of the complex IV capacity measured for the wild type (Kühn et al., 2009). To gain an understanding of the effects that the aox1a and rpoTmp mutations have on respiratory capacity and investigate possible compensatory effects through other pathways of electron transfer, respiratory capacity was analyzed in mitochondria isolated from mutant lines or wild-type (Columbia-0 [Col-0]) plants grown under normal growth conditions (Fig. 1A). Compared with the wild type, mitochondria isolated from rpoTmp mutant plants showed a significant approximately 60% to 70% decrease in the capacity for tricarboxylic acid (TCA) cycle-driven respiration (P < 0.001; Fig. 1A, i). The capacity for oxygen consumption through the alternative respiratory chain was more than double in rpoTmp compared with Col-0 mitochondria (P < 0.001), indicating that substantial compensation is provided by the AOX to maintain electron flow (Fig. 1A, i and ii). In contrast, for mitochondria isolated from aox1a plants, no significant difference in TCA cycle-dependent respiratory capacity was observed compared with the wild type (Fig. 1A, iii). A slight increase in cytochrome oxidase capacity was seen in the two independent aox1a lines compared with wild-type mitochondria; however, this difference was not significant for both lines (Fig. 1A, iv).
Figure 1.
Effects of inhibition of either complexes I and IV or the AOX on respiration and physiology under normal and stress conditions. A, Respiration capacities were compared between mitochondria isolated from Arabidopsis plants lacking either complexes I and IV (rpoTmp mutants) or the AOX (aox1a mutants) and wild-type plants. Rates of TCA cycle-driven respiration were determined for all mutant lines and the wild type (i and iii). The electron transport capacities of the AOX and the cytochrome oxidase in mitochondrial membranes from rpoTmp and aox1a, respectively, were measured in the presence of AOX- or complex IV-specific substrates and compared with wild-type rates (ii and iv). *, Significant differences. B, In vivo KCN-resistant respiratory rates expressed as a percentage of total respiration in leaf discs under normal growth and MLD (ML+D) conditions. Error bars indicate se. *, Significant differences. C, Physiological response of Arabidopsis plants to MLD treatment showing increased anthocyanin pigment accumulation after 4 d of treatment in both rpoTmp and aox1a lines compared with wild-type (Col-0) plants. Plants were grown in normal conditions for approximately 4 (Col-0; aox1a) or 5 weeks (rpoTmp) to reach a comparable developmental stage before being transferred to MLD.
Measurements of respiratory capacities in isolated mitochondria do not necessarily reflect in vivo partitioning or dynamic changes in respiration rates under stress. Therefore, we also sought to investigate partitioning of mitochondrial respiration in leaves in these mutant plant lines under normal growth conditions and after a 4-d treatment of combined moderate light and drought (MLD) stress (Fig. 1B). Before this treatment, plants were grown for 4 weeks under normal conditions, except for rpoTmp mutants, which are delayed in plant development (Kühn et al., 2009) and thus, were grown for 5 weeks to reach the same developmental stage as aox1a and the wild type before treatment. Under this environmental stress, a 50% increase in the potassium cyanide (KCN) -resistant component of respiration was observed in Col-0 leaves, reflecting enhanced AOX contribution to oxygen reduction (Fig. 1B). For rpoTmp leaves, basal rates of respiration under normal conditions showed greater than 90% use of the AOX, and unsurprisingly, this partitioning did not change significantly under stress, presumably because the alternative respiratory pathway was already used at near-maximal capacity in these plants (Fig. 1B). In aox1a plants, most of the electron flow was partitioned through the cytochrome pathway (Fig. 1B). Interestingly, when both rpoTmp and aox1a plants were subjected to conditions of combined MLD, identical physiological responses were observed (Fig. 1C). Both mutant genotypes showed increased sensitivity to stress compared with the wild type and visible purple pigmentation in leaf tissue (Fig. 1C). By approximately 6 d of treatment, both aox1a and rpoTmp plants showed loss of turgor and were not viable upon rewatering and returning to normal light conditions.
Transcriptomic Footprints for aox1a and rpoTmp Plants under Different Growth Conditions
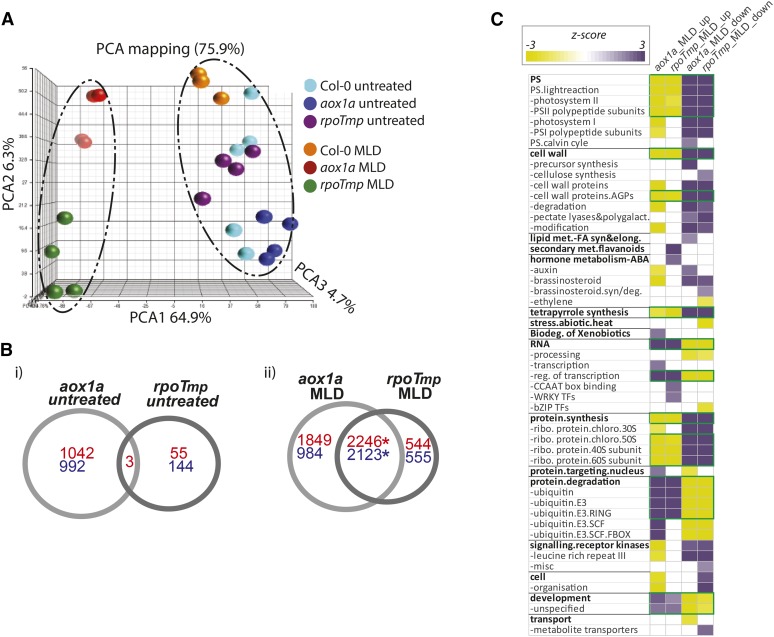
To investigate the extent to which the similar phenotypes in these mutant lines are reflected on a molecular level, the global transcriptome was examined using microarray analyses. Transcriptomic responses were profiled for the wild type (Col-0) as well as independent lines for both aox1a and rpoTmp knockout plants under normal and MLD conditions. A principal component analysis was carried out to reveal any significant differences between the overall transcriptomes of each sample. It was immediately apparent that the untreated aox1a and rpoTmp mutants were significantly distinct from the MLD-treated mutants (Fig. 2A, red and green). In contrast, the untreated and MLD-treated wild-type microarrays still grouped closely together (Fig. 2A, light blue and orange). These trends were further examined by overlapping the number of genes with transcripts abundance that was significantly differentially expressed (P < 0.05; posterior probability of differential expression [PPDE] > 0.95; fold change > 1.5-fold). This revealed that there was little in common between the changes in transcript abundance in aox1a plants compared with rpoTmp plants under normal conditions compared with the wild type (Col-0; Fig. 2B, i). Under normal (untreated) growth conditions, the number of transcripts that changed differed considerably for aox1a (1,045 up and 992 down) compared with rpoTmp (58 up and 144 down; Fig. 2B, i; Supplemental Table S1). Thus, although the growth of the rpoTmp plants was much more restricted compared with aox1a (see below; Giraud et al., 2008; Kühn et al., 2009), the latter displayed far more changes. In fact, under normal growth conditions, there were more antagonistic changes in transcript abundance between aox1a and rpoTmp than similarities (Supplemental Table S2); 48 transcripts of genes were up-regulated in aox1a plants but down-regulated in rpoTmp plants, and 16 transcripts of genes were down-regulated in the aox1a but up-regulated in the rpoTmp mutant. Examination of this list of transcripts of genes did not reveal any distinct pattern, except that many of the genes for transcripts that were up-regulated in abundance in aox1a and down-regulated in rpoTmp encoded ribosomal proteins (Supplemental Table S2A). Thus, the transcriptome responses to limiting the capacity of the alternative pathway or the cytochrome pathway seem quite distinct, which has been previously noted (Schwarzländer et al., 2012).
Figure 2.
Transcriptomic responses to MLD environmental stress in rpoTmp and aox1a plants show common, dramatic, and widespread changes in transcription. A, Three-way principle component analysis (PCA) was carried out for all microarray gene chips, showing a clear separation on PCA1 (64.9%) for aox1a and rpoTmp under MLD conditions. B, Venn diagrams showing the overlap for significantly differentially expressed transcript responses for aox1a and rpoTmp plants compared with wild-type (Col-0) plants (i) and after MLD treatment compared with respective untreated mutant plants (ii). Up-regulated transcripts are shown in red, whereas down-regulated transcripts are shown in blue. *, Significant overlap according to a χ2 test for significance (P < 0.0001). C, Functional categorization analysis (using Pageman) for transcripts changing in abundance in response to stress for both aox1a and rpoTmp plants. Over- and underrepresented categories are shown in purple and yellow, respectively. Boxed in green are the functional categories that are overrepresented in gene sets and show a common response under MLD stress. FA, Fatty acid; syn/deg, synthesis/degradation; bZIP, basic leucine zipper transcription factor; ribo, ribosome; chloro, chloroplast.
To determine if the responses of aox1a and rpoTmp plants to adverse growth conditions also differed, their response to combined light and drought stress was investigated. Previously, it had been reported that aox1a plants display increased sensitivity to combined drought and light stress treatment (Giraud et al., 2008), whereas plants with inactivated complex I have been reported to show higher tolerance to stress (Dutilleul et al., 2003a, 2003b). When aox1a plants were exposed to MLD conditions, 4,095 and 3,107 transcripts were significantly up- and down-regulated, respectively, compared with untreated mutant plants (Fig. 2B, ii; Supplemental Table S1). For rpoTmp plants, 2,790 and 2,678 transcripts were significantly up- and down-regulated, respectively (Fig. 2B; Supplemental Table S1). The overlap in genes with transcript abundance that changed in either mutant compared with the untreated was 2,246 up- and 2,123 down-regulated genes (Fig. 2B, ii). This overlap in responses was dramatically greater than expected by chance (P < 0.00001, χ2 test; Fig. 2B, ii). It is worth noting that, under stress, for transcripts that were significantly altered in the aox1a genetic background but did not seem to be altered in rpoTmp (or vice versa), the transcript changes in the corresponding lines did show similarity in response. However, many of these were filtered out as not significant because of the 1.5-fold change cutoff.
Using Pageman (Usadel et al., 2006), an analysis of the changes in aox1a and rpoTmp plants compared with untreated mutant plants (Fig. 2B, ii) provided insight into the function of the genes responding to MLD stress in these mutants (Fig. 2C). Noticeably, although there were more transcripts changed in abundance in the aox1a mutants compared with the rpoTmp mutants, there was overlap in many functional categories of the differentially expressed genes (Fig. 2C, boxed in green). There was no group of genes that displayed opposite trends of the combined 3,500 distinct changes. Distinct changes observed with aox1a were an underrepresentation of the categories of cell, signaling, and hormone metabolism and an overrepresentation of genes for biodegradation of xenobiotics in up-regulated genes. Lipid metabolism was overrepresented among genes up-regulated in rpoTmp (Fig. 2C).
Only 35 of over 6,000 transcripts changed significantly in response to stress in these two mutant backgrounds that displayed antagonistic responses. Of these, transcripts for 12 genes were down-regulated in aox1a plants but up-regulated in rpoTmp plants, and transcripts of 22 genes were up-regulated in aox1a plants but down-regulated in rpoTmp mutant plants (Supplemental Table S2B). These antagonistic changes did not seem to fall into any distinct class of function, and overall, they were dwarfed in magnitude by the number of common changes.
Thus, although the transcriptome responses of plants with altered respiratory chain capacity through the alternative (aox1a) or cytochrome (rpoTmp) chain are distinct under normal growth conditions, these plants display a highly significant similarity in response upon transfer to adverse growth conditions.
Conserved Transcriptomic Responses to Mitochondrial Respiratory Chain Dysfunction
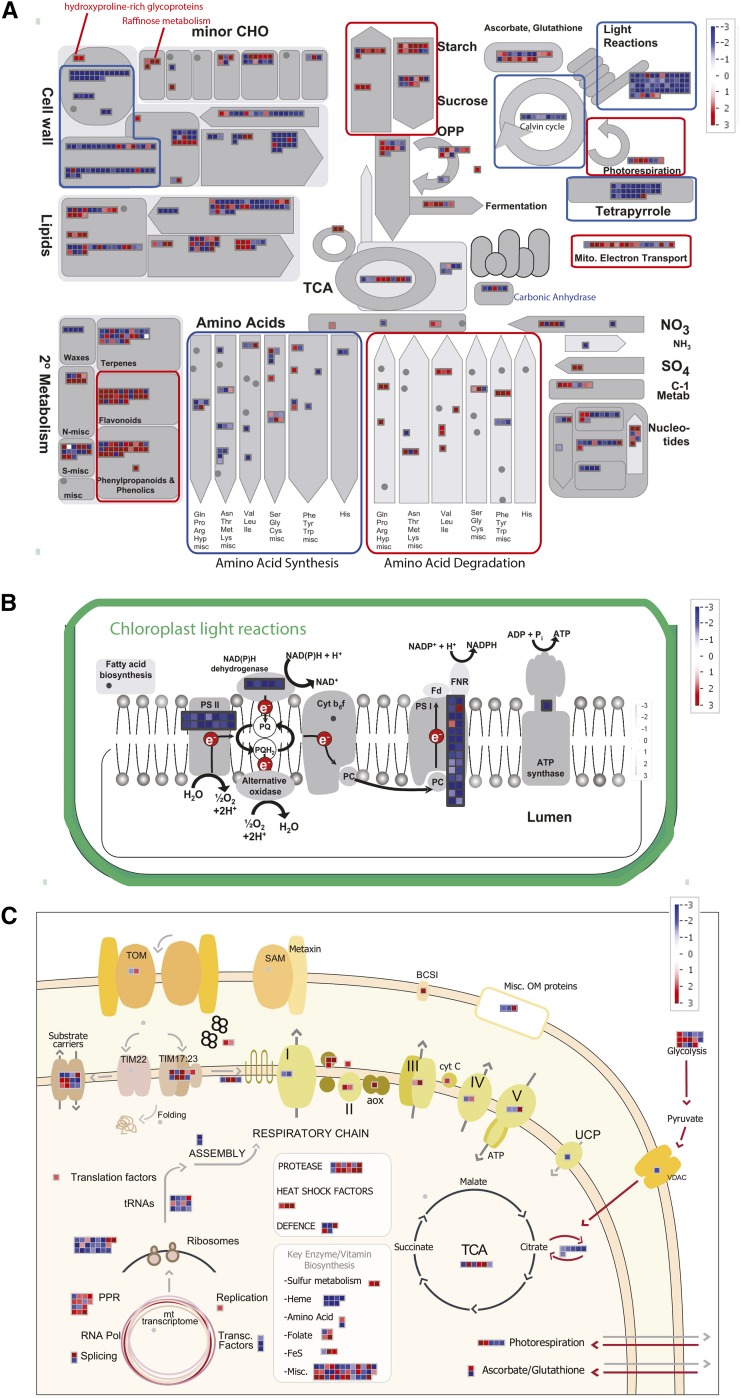
It is worth noting that less than 1% of all significantly differentially expressed transcripts showed antagonistic responses between both mutants, and in fact, most fold changes showed that there was less than 10% difference in the fold change magnitude between the two mutant backgrounds (Supplemental Table S1). Therefore, to further characterize the common transcriptomic response to moderate environmental stress that is observed in both aox1a and rpoTmp mutants, fold changes for transcripts significantly differentially expressed in both genotypes compared with the respective untreated mutant plants with the same direction of change were averaged in magnitude. This common transcriptomic response to restricted mitochondrial electron transfer capacity under stress conditions involved a dramatic up-regulation of transcripts associated with protective secondary metabolism and anthocyanin production (Fig. 3A). Cell wall metabolism was drastically altered as well as processes of degradation and generation of lipids (Fig. 3A). Similar to what had previously been observed in aox1a plants under MLD stress (Giraud et al., 2008), raffinose metabolism seemed to be specifically targeted (Fig. 3A). Even more striking, however, were the effects on plastid function and photosynthesis (Fig. 3, A and B). Transcripts for the chloroplast photosystems PSI and PSII were dramatically, succinctly, and uniformly down-regulated (Fig. 3B) along with transcripts encoding Calvin cycle components and those involved in tetrapyrrole metabolism, whereas photorespiratory enzymes were up-regulated (Fig. 3A). Ascorbate-Glutathione metabolism was also up-regulated (Fig. 3A).
Figure 3.
MapMan overview of transcriptional changes in response to MLD stress in both rpoTmp and aox1a plants compared with respective untreated mutants. Fold changes for transcripts that showed common responses to MLD stress in both aox1a and rpoTmp plants were averaged in abundance and plotted on MapMan images for an overview of metabolism (A), chloroplastic photosynthetic electron transport chain (ETC) function (B), and a custom image generated for a comprehensive overview of mitochondrial function (C; Law et al., 2012). Red and blue boxes indicate up-regulated and down-regulated transcripts, respectively, according to the color scale for log2 fold changes. CHO, Carbohydrate; Fd, ferredoxin; FNR, ferredoxin reductase; PC, plastocyanin; TOM, translocase of the outer membrane; SAM, sorting and assembly machinery of the outer membrane; BCS1, cytochrome bc1 synthesis1; Misc. Om, miscellaneous outer membrane; UCP, uncoupling protein; VDAC, voltage-dependent anion channel; cyt C, cytochrome c; mt, mitochondria; Pol, polymerase.
For the mitochondrion, some changes in transcript abundance of genes encoding electron transport chain components were observed, with a general increase in transcripts encoding alternative respiratory chain components (Fig. 3, A and C). Biogenesis of mitochondrial respiratory complexes requires the coordination of expression in both the nuclear and mitochondrial genomes; this was reflected by up-regulation of some replication factors and pentatricopeptide repeat (PPR) proteins (Fig. 3C), although transcripts of many genes encoding mitochondrial ribosomal proteins were down-regulated along with many tRNA synthetase or ligase genes (Fig. 3C; Supplemental Table S1), suggesting that mitochondrial protein synthesis was down-regulated. The strong reduction in transcripts abundance of genes encoding plastid MITOCHONDRIAL TRANSCRIPTION TERMINATION FACTOR proteins also suggested that plastid gene expression was down-regulated (Supplemental Table S1). Transcripts for small intermembrane chaperones and heat shock proteins were also increased in abundance. Many components involved in mitochondrial heme metabolism were strongly down-regulated, whereas sulfur metabolism and glycolytic processes feeding into mitochondrial metabolism were generally up-regulated (Fig. 3C).
Distinct Transcriptomic Responses to Mitochondrial Respiratory Chain Dysfunction
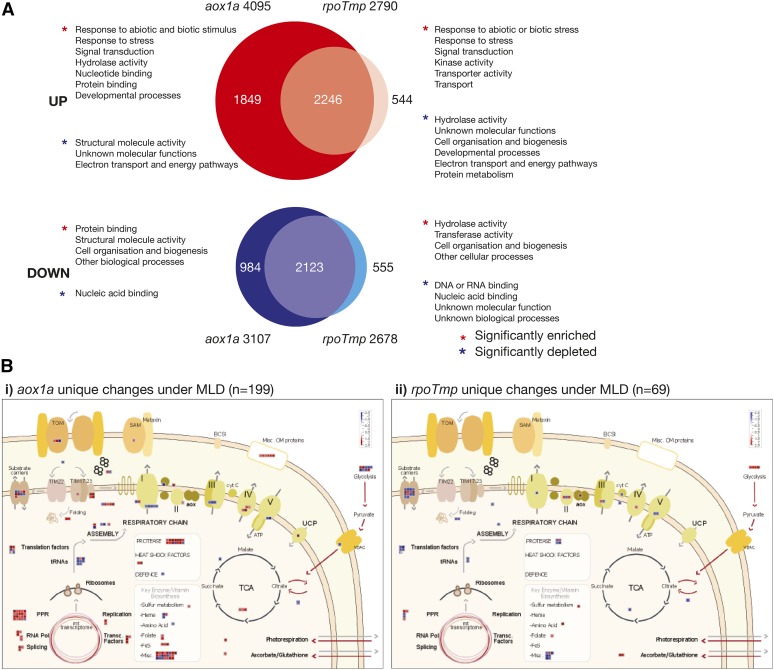
Although there were many common transcript changes in aox1a and rpoTmp upon transfer to adverse growth conditions compared with untreated conditions, distinct changes were also observed in each mutant (MLD; Fig. 2B). These unique changes may be related to the electron transport pathway that was restricted (i.e. cytochrome or alternative pathway) and thus, were examined in more detail. In aox1a, 1,849 and 984 unique transcripts were significantly up-regulated and down-regulated, respectively, in abundance compared with the rpoTmp response after exposure to MLD stress (Fig. 4A; Supplemental Table S3). Gene ontology enrichment analysis of 1,849 genes showed a significant overrepresentation of genes characteristic of a response to stress and abiotic and biotic stimuli, including signal transduction components. Additionally, this aox1a-specific suite of genes was significantly enriched in hydrolase activity, nucleotide binding, and developmental processes (Fig. 4A). In contrast, genes associated with electron transport, energy pathways, and structural molecule activities were significantly underrepresented. This is clearly illustrated by mapping these genes to a custom MapMan pathway (Fig. 4B, i; Supplemental Table S4). This revealed the relatively low transcript abundance of some of the genes encoding components of the electron transport chain (with the exception of complex IV) in the aox1a mutant after MLD treatment (Fig. 4B, i). Thus, the initiation of a stress response in aox1a is accompanied by a suppression of energy production and growth factors, an observation strengthened by the significant enrichment of protein binding, structural molecule activity, and functions associated with cell organization and biogenesis in the list of 984 down-regulated transcripts unique to aox1a (Fig. 4B, i).
Figure 4.
Unique transcript changes in aox1a and rpoTmp under adverse growth conditions. A, Numbers of distinct changes in aox1a and rpoTmp are indicated, and functional categories that are over- and underrepresented among aox1a- or rpoTmp-specific transcript changes are listed. B, The distinct changes in transcripts encoding mitochondrial proteins for aox1a (i) and rpoTmp (ii) under adverse growth conditions. Note that these do not represent the total of changes for genes encoding mitochondrial proteins, because there are many common changes (Fig. 3B). Om, Outer membrane; BCS1, cytochrome bc1 synthesis; SAM, sorting and assembly machinery of the outer membrane; TOM, translocase of the outer membrane; cyt C, cytochrome c; Pol, polymerase; VDAC, voltage-dependent anion channel.
In the rpoTmp lines, 544 and 555 unique transcripts were significantly up-regulated and down-regulated, respectively (Fig. 4A; Supplemental Table S3). Despite the genes for these transcripts not overlapping with the responses in the aox1a plants, the up-regulated set was also significantly overrepresented in genes associated with responses to stress and abiotic/biotic stimuli. In addition to these, this set of genes was also significantly enriched in kinase and transport activity, while showing an underrepresentation of genes encoding functions associated with developmental processes, cell organization and biogenesis, protein metabolism and electron transport, and energy pathways (Fig. 4A). Notably, this underrepresentation of electron transport and energy pathways in the rpoTmp-exclusive gene set further supports the previous observation that the genes encoding these functions are showing overlapping responses to MLD treatment in both aox1a and rpoTmp plants (Figs. 2B and 4A).
A focused analysis of mitochondrial targeted components of this list showed a somewhat similar response to that of the aox1a line, with low transcript abundance of some of the components of the electron transport chain observed with the exception of subunits of complex IV (Fig. 4B, ii; Supplemental Table S4). It differed in that the up-regulation of transcripts for genes encoding protein import components, PPR proteins, and outer membrane proteins was not observed in rpoTmp under adverse growth conditions compared with aox1a (Fig. 4B; Supplemental Table S4). Also, AOX1a was greater than 3-fold up-regulated in rpoTmp, and notably, despite the lack of a functional AOX1a gene in the aox1a background, the up-regulation of AOX1d was barely 2-fold in aox1a compared with a greater than 5-fold increase in rpoTmp (Supplemental Tables S1 and S4).
Overall, however, the functional overlap even among transcripts uniquely changed in aox1a or rpoTmp highlights the highly similar functional response observed between these two mutants (Fig. 4).
aox1a:rpoTmp Double Mutants Display Decreased Biomass Production, Energy Deficits, and Distinct Transcriptome and Proteome Changes Relating to Mitochondrial Biogenesis
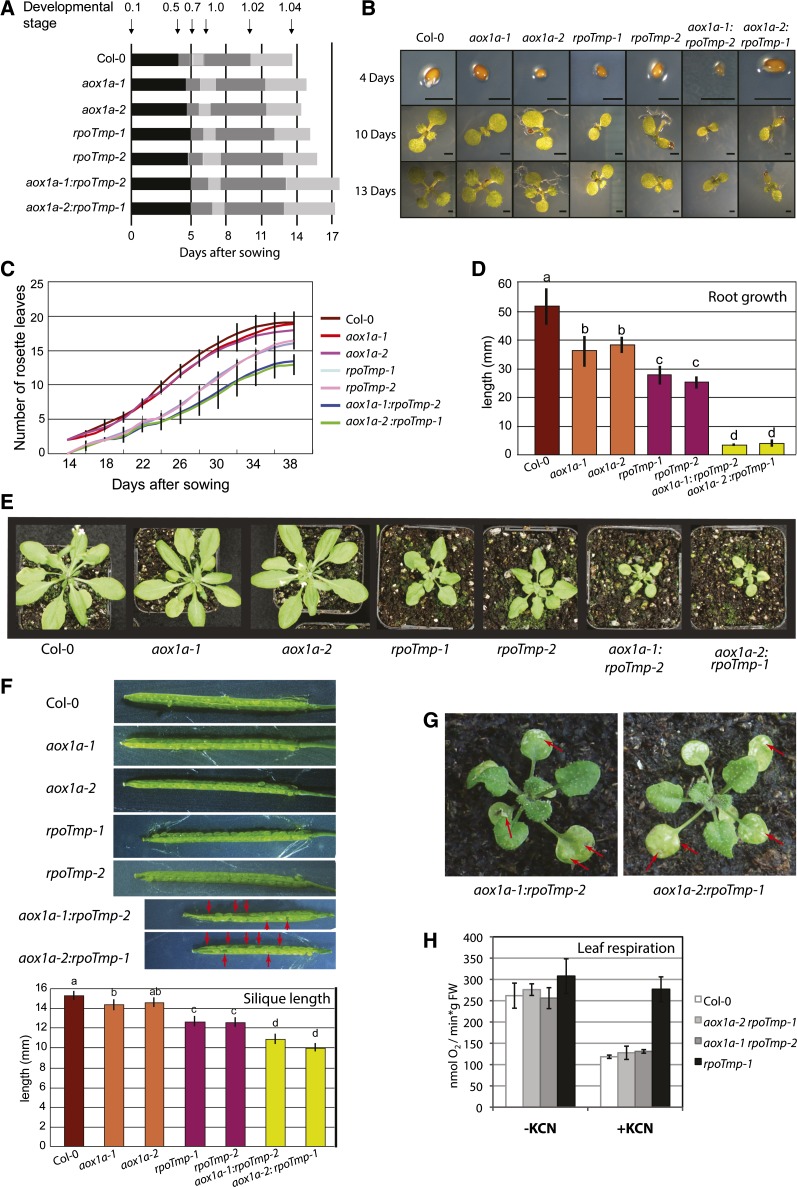
The effect of environmental stress conditions on the rpoTmp and aox1a mutants defective in different components of mitochondrial respiration likely places increased demand on mitochondrial function and respiration. To investigate complementary roles of these components in maintaining respiratory functions, aox1a and rpoTmp lines were crossed to obtain double mutants impaired in both components of the cytochrome and the alternative electron transfer chains. Two independent lines were generated: aox1a-1:rpoTmp-2 and aox1a-2:rpoTmp-1 (Fig. 5). Double-mutant lines showed dramatically reduced rates of germination and growth-stage progression during early seedling development and establishment (Fig. 5, A and B). These growth defects were substantially more pronounced than those observed in either single knockout line. rpoTmp plants showed an approximately 30% reduction in the number of rosette leaves at any particular time point; however, later in development, rosette leaf area reached normal size ranges, albeit with a strong developmental delay (Fig. 5C; Kühn et al., 2009). However, for aox1a:rpoTmp double mutants, this decrease in the number of rosette leaves was greater than 50%, and total rosette leaf area did not recover over development. As has been described for other mitochondrial mutants, root growth was severely compromised to similar levels (approximately 50% of the wild type) for both the aox1a and rpoTmp single mutants and less than 10% of wild-type root length averages for double mutants (Fig. 5D). Even after several weeks postsowing, aox1a:rpoTmp plants were very small, with less than 20% of the total biomass of the corresponding aox1a plants (Fig. 5E).
Figure 5.
Phenotypic growth analyses show defects in biomass production in aox1a rpoTmp double mutants. Phenotypic analysis was carried out according to the work by Boyes et al. (2001) to define growth stages for wild-type plants (Col-0) as well as aox1a-1, aox1a-2, rpoTmp-1, and rpoTmp-2 and double mutants aox1a-1 rpoTmp-2 and aox1a-2 rpoTmp-1. Measurements taken are the average of at least 10 plants grown on plate media (0% Suc). ses are shown. A, Growth stage progression timeline. Highlighted gray horizontal bars show individual growth stages. Time is shown on the x axis. B, Photographic representation at particular time points for the mutant lines. C, Number of rosette leaves over the course of development to 38 d for all plant lines. D, Root growth measures for all lines taken 12 d after sowing. Plants were grown on vertical plates (“Materials and Methods”), and growth analysis was repeated a minimum of three times. In the bar graph, different letters indicate statistically significant differences (P < 0.05). E, Soil-grown aox1a, rpoTmp, and aox1a rpoTmp plants of the same age show that the double mutants do not reach the size of rpoTmp plants, even after several weeks of growth. Aerial parts of aox1a plants were previously shown to match wild-type growth under normal conditions (Giraud et al., 2008). F, Green siliques for all genotypes were dissected to visualize embryo development. Red arrows indicate aborted ovules seen in double mutants. Silique length was measured at the completion of flowering in at least 15 siliques from >10 plants for all genotypes. ses are shown. In the bar graph, different letters indicate statistically significant differences (P < 0.05). G, Five and one-half-week-old aox1a rpoTmp plants grown on soil show yellowing of leaf tissue and lesions. H, In vivo respiratory rates measured in leaf discs from wild-type and aox1a:rpoTmp plants of the same developmental stage grown under normal conditions. rpoTmp-1 grown under the same conditions was included in this experiment for comparison. Oxygen consumption was additionally determined in the presence of the cytochrome oxidase inhibitor KCN. Error bars indicate sds (n = 3). FW, Fresh weight.
These defects in biomass production in the double-mutant background imply that there is a strong energy deficit in these plants. Silique length was significantly shorter in the aox1a:rpoTmp double mutants (by approximately 30% compared with wild-type plants; Fig. 5F). aox1a:rpoTmp plants produced fewer seeds, because on average, 32% of seed embryos showed various stages of embryo abortion. In addition, rpoTmp:aox1a rosette leaves showed leaf yellowing and developed necrotic lesions (Fig. 5G). Leaf respiration rates measured for aox1a:rpoTmp plants were similar to those determined for the wild type and rpoTmp (Fig. 5H). Unlike rpoTmp, the double mutants lacking AOX1a were unable to up-regulate KCN-resistant respiration and displayed the wild type-like leaf respiration rates upon addition of the inhibitor. The recording of the wild type-like oxygen consumption rates for aox1a:rpoTmp both with and without KCN implies that, regardless of the detrimental effects of simultaneous AOX1a and cytochrome oxidase impairment on plant growth, the residual cytochrome oxidase capacity left in double mutants may be sufficient to support normal leaf respiration. Alternatively, other oxygen-consuming activities might be enhanced in aox1a:rpoTmp.
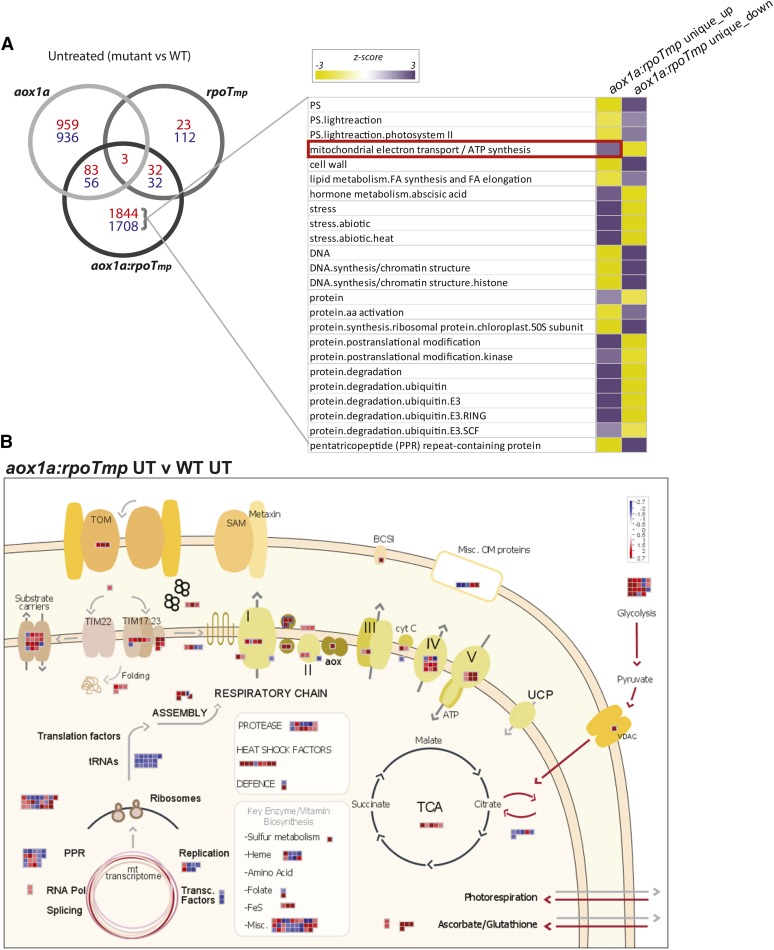
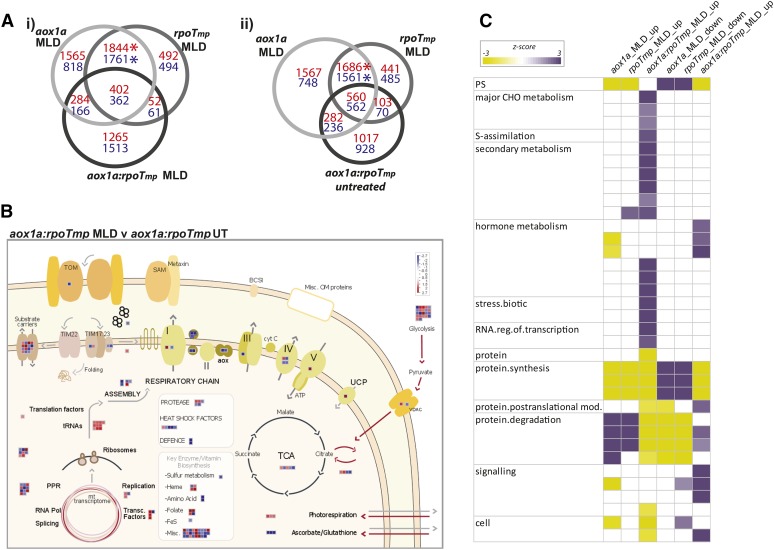
The transcriptome of the aox1a:rpoTmp plants was also analyzed under normal and MLD conditions to compare it with both parental lines (Fig. 6). All comparisons to determine changes in transcript abundances under normal conditions were with the wild type (Col-0), and these sets were then compared between the single and double mutants. Under normal growth conditions, the aox1a:rpoTmp lines displayed a largely unique response (with 1,844 transcripts exclusively up-regulated in abundance and 1,708 transcripts down-regulated in abundance) compared with the aox1a and rpoTmp mutants (Fig. 6A). The overlap in genes changing in the aox1a:rpoTmp mutants compared with aox1a and rpoTmp was very small (Fig. 6A). Thus, the response to restricting both respiratory pathways differed from the response to either alternative or cytochrome pathway disruption. Considering the slow-growth phenotype (Fig. 5), it was not surprising that the transcripts of down-regulated genes were overrepresented in photosynthesis-related genes and genes encoding chloroplast ribosomal proteins and DNA and RNA synthesis, whereas the up-regulated genes were overrepresented in genes encoding mitochondrial electron transport, hormone metabolism related to abscisic acid (ABA), abiotic stress, and protein modification and degradation. The dominance of unique changes in the double mutant means that the transcriptome changes at a functional level were defined by these unique responses. In the aox1a:rpoTmp double mutant, it was also noticeable that abiotic stress and hormone metabolism related to ABA were overrepresented (Fig. 6A), suggesting that, although it was grown under standard growth conditions, it already had triggered a stress response because of the inactivation of the AOX1a and RPOTmp genes.
Figure 6.
Summary of transcript changes in aox1a:rpoTmp under standard growth conditions compared with the wild type (Col-0) and overlap with single mutants. A, The number of transcripts that increased (red) or decreased (blue) in abundance in aox1a:rpoTmp and the overlap with aox1a or rpoTmp. Next to this, a Pageman visualization of the functional groups over- or underrepresented in the genes that were differentially expressed exclusively in the aox1a:rpoTmp mutants, where unique_up indicates the exclusively up-regulated genes and unique_down indicates the exclusively down-regulated genes. B, A customized MapMan overview showing changes in transcripts encoding mitochondrial proteins in aox1a:rpoTmp compared with the wild type (Col-0). The genes encoding proteins involved in glycolysis are also shown outside the mitochondrion. WT, Wild type; TOM, translocase of the outer membrane; SAM, sorting and assembly machinery of the outer membrane; BCS1, cytochrome bc1 synthesis; cyt C, cytochrome c; UCP, uncoupling protein; UT, untreated; VDAC, voltage-dependent anion channel; Pol, polymerase.
Analysis of the aox1a:rpoTmp transcriptome, moreover, revealed that the functional category of mitochondrial electron transport/ATP synthesis was overrepresented in up-regulated transcripts (Fig. 6A, red box), which was not observed with either single mutant under any condition (Fig. 2C; Supplemental Fig. S1). Furthermore, PPR genes, which are intimately linked to organelle gene expression (Barkan and Small, 2014), were overrepresented in down-regulated transcripts (Supplemental Fig. S1). These combined effects indicated that organelle function was likely severely compromised in double mutants. Thus, changes in transcripts encoding mitochondrial proteins were specifically examined (Fig. 6B), which indicated severe mitochondrial dysfunction in aox1a:rpoTmp. Many of the factors with transcript abundance that changed were associated with the mitochondrial stress response (Supplemental Table S5A); they included HEAT SHOCK PROTEIN23 (HSP23; At5g51440; increased 172-fold), AtTIM17-1 (At1g20350; increased 32-fold), BCS1 ATPases associated with diverse cellular activities (At3g50930; increased 24-fold; Zhang et al., 2014), GLUTATHIONE S-TRANSFERASE τ9 (At5g62480; increased 131-fold), and a transcript encoding a CYTOCHROME C OXIDASE19 (COX19)-like CHCH family protein (At5G09570; increased 156-fold). These changes were far in excess compared with what was observed under stress conditions in the wild type, aox1a, or rpoTmp (Supplemental Table S5A).
Other up-regulated transcripts include HSP70 (At3g12580), a protein found in mitochondria (Supplemental Table S5B; Ito et al., 2006) but likely has multiple locations (http://suba.plantenergy.uwa.edu.au), alternative NAD(P)H dehydrogenases, and AOXs (Supplemental Table S5B). Notably, the change in AOX1d was larger in magnitude than usually observed. Changes also encompassed genes for the mitochondrial import apparatus, with 18 transcripts up-regulated. Also, almost 50 transcripts encoding proteins of oxidative phosphorylation (OXPHOS) complexes I to V and a variety of metabolite carriers were up-regulated (Fig. 6B; Supplemental Table S5B) along with 17 transcripts encoding proteins of the glycolytic pathway. Notably, several factors associated with translation in mitochondria were down-regulated in abundance (Fig. 6A; Supplemental Table S5B). Thus, transcripts encoding proteins associated with mitochondrial biogenesis, alternative respiratory chain electron transport, mitochondrial stress response, the cytochrome electron transport chain, and glycolysis were seen to change in aox1a:rpoTmp double mutants. This was a unique response, because it was not a sum of the changes observed in each single mutant (Fig. 6A).
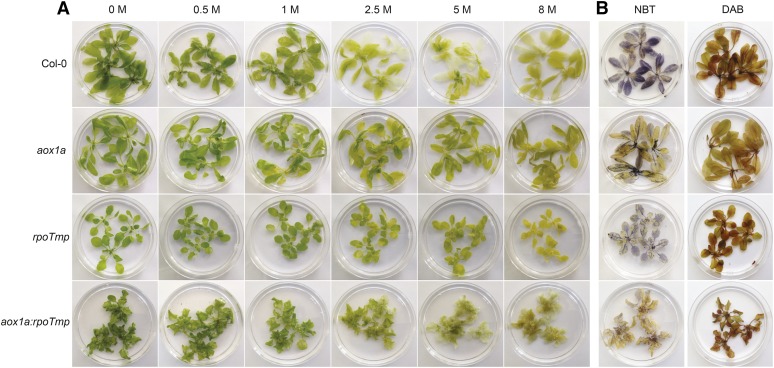
Also, notably, genes encoding stress response functions were overrepresented among the transcripts induced in the untreated aox1a:rpoTmp double mutants (Fig. 6A). Closer examination revealed that transcripts of several genes involved in oxidative stress were induced (Supplemental Table S1). Regardless of the severely delayed growth phenotype shown by the double mutants, they provided an opportunity to study plants in which electron transport through both respiratory pathways was compromised. Therefore, in an effort to determine their ability to withstand oxidative stress, hydrogen peroxide (H2O2) bleaching was tested by incubating whole rosettes overnight in increasing concentrations of H2O2 (Fig. 7). Under these conditions, wild-type plants displayed bleaching at 2.5 m H2O2, whereas the aox1a:rpoTmp double mutants were somewhat resistant even to higher levels of H2O2 (up to 8 m), which was similar to the observations in aox1a mutants (Fig. 7A). Staining for O2− and H2O2 with nitroblue tetrazolium (NBT) and 3,3′diaminobenzidine (DAB), respectively, showed little staining with NBT (O2−) and somewhat reduced staining with DAB (H2O2) in aox1a:rpoTmp plants, again similar to aox1a (Fig. 7B). The difference of aox1a:rpoTmp to the rpoTmp line for both NBT and DAB staining was clear, with the latter showing clear staining, especially with DAB (Fig. 7B). Molecular evidence complementing the observed higher resistance to oxidative stress in the double mutant was also seen when examining the transcripts abundance of genes involved in the oxidative stress response. A previous study had defined five genes as markers of oxidative stress (Gadjev et al., 2006), and when the transcript abundance of these was examined in the single and double mutants, four of five of these marker genes (At1g57630, At1g19020, At1g05340, and At2g21640) were already induced in the untreated aox1a:rpoTmp double mutants, with three of them showing greater than 19-fold induction (Supplemental Table S1). Notably, one of these markers encodes a mitochondrial protein up-regulated in response to oxidative stress (At2g21640). The transcript of this gene was induced 63-fold in the untreated double mutant.
Figure 7.
The ability of aox1a:rpoTmp to withstand oxidative stress. A, Four-week-old Arabidopsis plants were subject to bleaching overnight (16 h) in different concentrations of H2O2 (note that, for rpoTmp and aox1a:rpoTmp, plants were 4 and 5 weeks old, respectively, to match developmental stages as close as possible). B, Four-week-old plants from Col-0, aox1a, rpoTmp, and aox1a:rpoTmp were stained for ROS production using DAB (H2O2) and NBT (O2−).
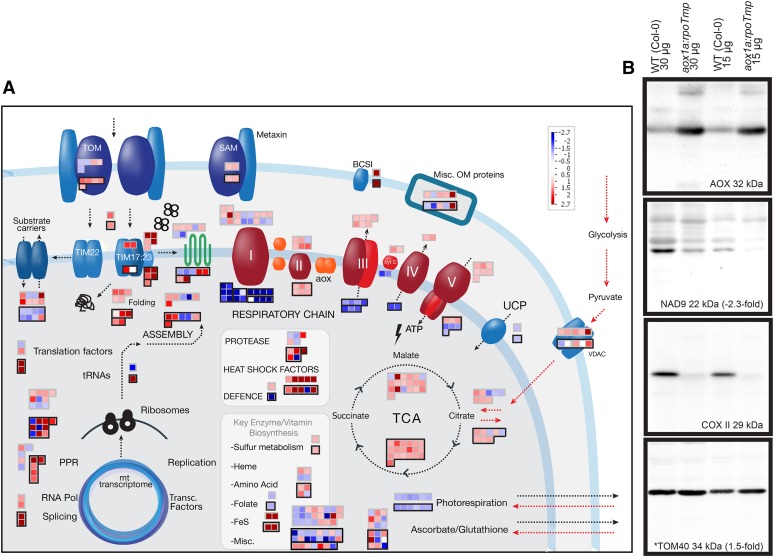
Given the large magnitude of changes in transcript abundance observed in aox1a:rpoTmp plants for genes encoding mitochondrial proteins, quantitative proteomic analysis of purified mitochondria was carried out for the aox1a:rpoTmp double mutant to determine if transcript changes corresponded with changes in the mitochondrial proteome. In total, 210 proteins were detected, and the fold changes in protein abundance were compared with fold changes in transcript abundance (Fig. 8A; Supplemental Table S5D).
Figure 8.
Summary of mitochondrial protein changes in aox1a:rpoTmp under standard growth conditions. A, A customized MapMan image representing various mitochondrial functions is shown. Changes in protein abundance are shown by red (up) or blue (down) squares that are outlined in black, with the corresponding transcripts above. B, Western-blot analysis of selected mitochondrial proteins in aox1a:rpoTmp under standard growth conditions; 15 and 30 μg of purified mitochondrial proteins were isolated from wild-type (Col-0) and aox1a:rpoTmp plants and probed with antibodies as indicated. Representative proteins from various functional groups were examined, and example outputs are shown for Tom40 representing protein import components and AOX from the alternative respiratory pathway, NAD9 (complex I), and COXII (complex IV). *, Proteins were also detected in the quantitative proteomic analysis, and the fold changes in the mutant are indicated. WT, Wild type; TOM, translocase of the outer membrane; SAM, sorting and assembly machinery of the outer membrane; Misc OM, miscellaneous outer membrane proteins; BCS1, cytochrome bc1 synthesis; cyt C, cytochrome c; UCP, uncoupling protein; VDAC, voltage-dependent anion channel; Pol, polymerase.
This revealed that the changes in transcript abundance (Fig. 8A, outlined in gray) and corresponding protein abundances (Fig. 8A, outlined in black) showed the same pattern, with the notable exceptions of complexes I, III, IV, and V of the respiratory chain, where an increase in transcripts but decrease in proteins was observed (Fig. 8A; Supplemental Table S5D). Thus, for components associated with protein import and stress responses, a good correlation was observed between transcript abundance and protein abundances, which were both increased. For five components associated with protein import (AtTim23-2, At1g72750; AtTim44-2, At2g36070; AtTom40, At3g20000; Tim10, At2g29530; and Tom20-4, At5g40930), both transcript and protein abundances were elevated (Fig. 8A; Supplemental Table S5D). Additionally, a small increase in protein abundance for another four import components was seen (Arabidopsis Presequence Degrading Peptidase2 [AtPreP2], At1G49630; AtTom20-2, At1G27390; AtTom20-3, AT3G27080; and AtTom22, AT5G43970), where no change or a decrease in corresponding transcript abundance was observed (Fig. 8A; Supplemental Table S5D). It has been previously documented that changes in abundance in protein import components can occur independently of transcript changes (Law et al., 2012, 2014; Wang et al., 2012), and in this case, both transcriptional and posttranscriptional processes seem to contribute to an increase in the amount of protein import components. For components associated with stress responses, the most induced proteins were chaperones ranging from 14-fold for HSP23.5 (At5g51440) to a 6-fold increase for HSP70 (At4g37910). Additionally, proteases (four proteins), Prohibitins (three proteins), and various components associated with ROS or stress, such as ascorbate peroxidase and aldehyde dehydrogenase, were also induced at the protein level (Supplemental Table S5D), corresponding with the observed differences in oxidative stress tolerance (Figs. 7 and 8A). Also, many components associated with the TCA cycle increased in protein abundance as well as many metabolic enzymes. However, it is worth noting that not all transcript and protein abundances correlated well. For example, the transcript abundance increase of greater than 200-fold observed for an HSP70 gene (At3g12580) was accompanied by a decrease in protein abundance of greater than 2-fold (Supplemental Table S5D). A possible reason for this is that this chaperone protein is dual targeted.
The multisubunit complexes of the OXPHOS system, with the notable exception of complex II (succinate dehydrogenase, a TCA cycle enzyme that is only comprised of nuclear encode subunits in Arabidopsis), were overall decreased in abundance; this included the nuclear-encoded cytochrome c protein. An exception was the ATP synthase g subunit with three isoforms (At4G29480, At2G19680, and At4G26210), which was slightly increased in abundance (Fig. 8A). The g subunit of ATP synthase is not catalytic, and in yeast (Saccharomyces cerevisiae), it is involved in the dimerization of the ATP synthase complex, which is associated with maintaining mitochondrial morphology and also, cytochrome oxidase activity (Boyle et al., 1999). Along with reductions in the ADP/ATP carriers (ADP/ATP carrier 1, At3G08580; ADP/ATP carrier 2, At5G13490; and ADP/ATP carrier 3, At4G28390) that export ATP synthesized in mitochondria, these changes indicated that the cytochrome respiratory chain was severely restricted at a protein level. It should also be noted that detected components of the alternative respiratory pathway showed contrasting trends. Although NAD(P)H DEHYDROGENASE B2 (NDB2; At4G05020) was 5-fold up-regulated in protein abundance, both NDB1 (At4G28220) and NDB3 (At4G21490) were down-regulated by 1.3- and 2.1-fold, respectively (Fig. 8A; Supplemental Table S5D).
The proteomic analysis indicated a large-scale reorganization of the plant mitochondrial proteome. To supplement and verify these observations, western-blot analysis was carried out on selected targets. These included AOX, the alternative NAD(P)H dehydrogenase1 (NDA1), and OXPHOS subunits NAD9 (complex I), Rieske FeS (complex III), COXII (complex IV), and ATP synthase-β (complex V; Fig. 8B; Supplemental Fig. S2). Also, the abundance of a mitochondrial protein import component, TOM40, was determined (Supplemental Fig. S2). In the case of AOX, a large increase in protein abundance was observed compared with the wild type (Fig. 8B). In the aox1a:rpoTmp plants lacking AOX1a, this is attributable to one of four additional AOX proteins present in the Arabidopsis genome, presumably AOX1d, with a transcript that was increased 18-fold. It has been previously reported that AOX1d cannot rescue AOX1a inactivation (Strodtkötter et al., 2009). This is in agreement with the results shown here, where the double mutant aox1a:rpoTmp is significantly smaller and shows considerably different changes in the transcriptome compared with rpoTmp (Fig. 5). Thus, although AOX1d was significantly induced at a transcript and protein level (Fig. 8B), its activity could not fully compensate for the lack of AOX1a. For the respiratory complexes I, III, IV, and V, the decrease in subunits observed in the proteomic analysis was mirrored by that observed with western blotting (Supplemental Fig. S2). For the proteins detected using both proteomics and western-blot analysis, the fold changes observed were in good agreement, Tom40 increased in abundance by 1.45-fold using proteomics and 1.5-fold upon western blotting, and NAD9 was observed to decrease by −2.9-fold using proteomics and −2.3-fold using western blotting (Fig. 8B; Supplemental Fig. S2).
Next, the stress transcriptome of aox1a:rpoTmp was examined and compared with those of aox1a and rpoTmp (Fig. 9A). When the transcriptomes of aox1a:rpoTmp, aox1a, and rpoTmp grown under adverse growth conditions were compared (with their respective untreated mutant plants), the overlap in changes was essentially the same or slightly smaller than the overlap of normally grown aox1a:rpoTmp with stress-exposed aox1a or rpoTmp (compare Fig. 9A, i with Fig. 9A, ii). Thus, the large and highly significant overlap observed between aox1a and rpoTmp grown under adverse conditions (Fig. 2) was not observed with aox1a:rpoTmp (Fig. 9A). Examination of the transcriptomic changes suggested that the mitochondrial stress response observed with aox1a and rpoTmp was not present in aox1a:rpoTmp. In fact, when exposed to MLD stress, the double mutant seemed to turn off or alter the stress response that it showed when grown under standard conditions. This conclusion is based on the observation that many of the NAC (for No Apical Meristem, Arabidopsis Transcription Activation Factor1-2 [ATAF1-2], and Cup-Shaped Cotyledon2) transcription factors that were constitutively expressed in aox1a:rpoTmp under standard conditions decreased in expression under MLD stress (Supplemental Table S1), including NAC013, which is intimately linked with the mitochondrial stress response and mitochondrial oxidative stress (Ng et al., 2014). Furthermore, examination of the transcripts encoding mitochondrial proteins revealed that several components of the alternative respiratory pathway were also decreased in abundance under MLD stress (Fig. 9B; Supplemental Table S1). Consistent with the decrease in the NAC transcription factors mentioned above, specifically, AOX1d and AOX1a were decreased 15- and 1.8-fold, respectively, NDB2, NDB3, and NDB4 were decreased −3.6-, −1.8-, and −7.7-fold, respectively, and NDA1 and NDA2 were decreased −2.4- and −3.1-fold, respectively (Fig. 9B; Supplemental Table S1). Thus, the operation of the alternative respiratory pathways has been down-regulated at the transcript level.
Figure 9.
Overview of the transcriptome response of aox1a:rpoTmp to adverse growth conditions in comparison with untreated mutants, the wild type (Col-0), and single mutants. A, The number of transcripts of genes that increased (red) or decreased (blue) in abundance in aox1a or rpoTmp grown under MLD stress and the overlap with aox1a:rpoTmp grown under MLD stress (i) or untreated conditions (ii). Asterisks indicate a significant overlap as determined by a Pearson's chi-squared test. B, The changes in transcript abundance of genes encoding mitochondrial proteins in aox1a:rpoTmp grown under adverse environmental conditions. C, A Pageman visualization showing the overrepresented functional categories for genes with transcripts that differed in abundance in all three mutants grown under MLD stress. Note that only the functional categories overrepresented in the aox1a:rpoTmp double mutant are shown. The full Pageman output for this analysis is shown in Supplemental Figure S3. CHO, Carbohydrate; TOM, translocase of the outer membrane; SAM, sorting and assembly machinery of the outer membrane; BCS1, cytochrome bc1 synthesis; Misc OM, miscellaneous outer membrane proteins; cyt C, cytochrome c; UCP, uncoupling protein; UT, untreated; VDAC, voltage-dependent anion channel; Pol, polymerase.
To functionally explore the transcript changes in aox1a:rpoTmp grown under adverse growth conditions, Pageman analysis was performed, revealing that the stress response differed dramatically from that of aox1a and rpoTmp (Fig. 9C). Notably, the functional categories of carbohydrate metabolism, secondary metabolism, and response to biotic stress were overrepresented among up-regulated transcripts. This stress response was similar to a response to fungal infection and included the up-regulation of transcripts of many chitin-induced genes. Considering only transcripts that showed a more than 50-fold (Supplemental Table S1) increase in aox1a:rpoTmp revealed a largely unique response, and although many of these genes were up-regulated in aox1a, the magnitude of change in aox1a:rpoTmp was 10- to 100-fold greater. The pathogen response-like changes that have been noted previously in response to AOX1a inactivation (Schwarzländer et al., 2012; Umbach et al., 2012) seemed to be greatly increased in aox1a:rpoTmp under adverse growth conditions. Thus, exposure of plants impaired in both the cytochrome and the alternative respiratory chain to MLD stress seems to turn off the mitochondrial stress response observed in these plants under standard conditions. However, it enhances pathogen response-like (particularly fungal pathogen response-like) transcriptome adaptations.
Specific Regulation of Gene Expression and the Mitochondrial Stress Response
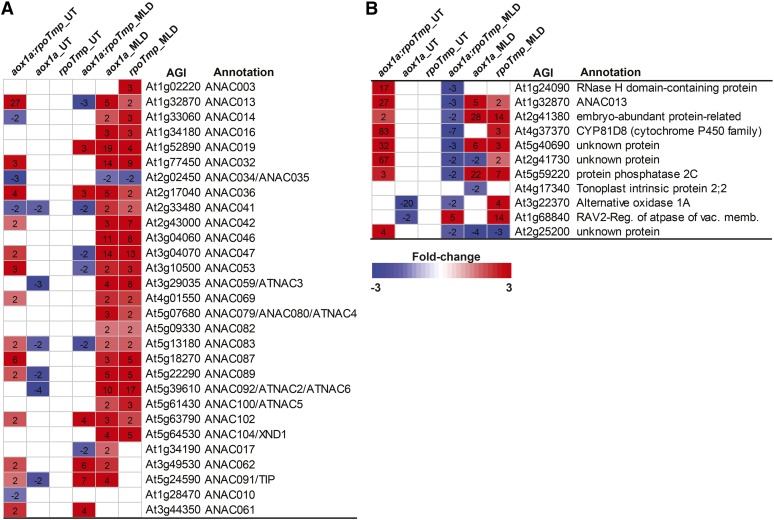
Changes in transcript abundance for transcription factor families that are likely to regulate and underpin these dramatic cell-wide alterations in energy metabolism were also investigated. Broadly, a number of transcription factor families associated with mediating stress responses and integrating these stress responses with energy metabolism were up-regulated in transcript abundance, such as WRKY, bZIP, MYB, AP2-EREBP, and NAC transcription factors (Supplemental Table S1). Although the changes in transcript abundance for all transcription factors will not be detailed individually, the NAC family of transcription factors represents a good example for strongly overlapping transcriptome responses of aox1a and rpoTmp to stress. In Arabidopsis, there are 117 genes annotated as encoding NAC transcription factors (Nuruzzaman et al., 2013); 23 and 25 of these were significantly up-regulated in rpoTmp and aox1a mutants, respectively, with an overlap of 22 in common (Fig. 10A). This overlap included ATAF1 (At1g01720), which is intimately involved in regulation of ABA-related genes and driving expression of ANAC092 (At5g39610). The latter promotes senescence and was commonly up-regulated as well (Fig. 4A). ANAC092 is also antagonistic to Golden-like transcription factor1 (GLK1) and GLK2, which are required for chloroplast development and maintenance (Waters et al., 2009; Jensen et al., 2013). This cascade may underlie the common decrease in transcript abundance for genes encoding photosynthesis observed in both mutants (Fig. 2C) and a common response to mitochondrial perturbation (Schwarzländer et al., 2012). Furthermore, several NAC transcription factors shown to be regulated by ETHYLENE-INSENSITIVE2 and involved in senescence were up-regulated in both mutant lines (Fig. 10A; Kim et al., 2014). Finally, seven membrane-bound NAC transcription factors (ANAC013, At13328700; ANAC014, At1g33060; ANAC016, At1g34180; ANAC042, At2g43000; ANAC053, At3g10500; ANAC069, At4g01550; and ANAC089, At5g22290) that are involved in mediating mitochondrial retrograde signaling (Fig. 10A; De Clercq et al., 2013; Ng et al., 2013a, 2013b) were also commonly up-regulated.
Figure 10.
Changes in aox1a, rpoTmp, and aox1a:rpoTmp transcriptomes under standard and adverse growth conditions compared with wild-type (Col-0) and untreated mutants. A, A list of NAC transcription factors that are changing in all three germplasms under standard and adverse growth conditions. The genotype, growth conditions (untreated [UT] or MLD), Arabidopsis gene identifier (AGI), and gene annotation are indicated. Fold changes highlighted in red are an increase in transcript abundance, and fold changes highlighted in blue are a decrease in transcript abundance. B, Expression of 11 genes defined as being responsive to a variety of mitochondrial perturbations in Schwarzländer et al., 2012 (depicted as in A).
It is important to note that 14 of the NAC transcription factors that were induced in aox1a or rpoTmp under adverse growth conditions were up-regulated in transcript abundance in aox1a:rpoTmp under normal conditions (Fig. 10A). Most notable was NAC013 (At1g32870), which was up-regulated 27-fold in aox1a:rpoTmp under standard conditions compared with just 5- and 2-fold in aox1a and rpoTmp, respectively, under adverse growth conditions (Fig. 10A). It has been shown that NAC013 is responsible for inducing a mitochondrial retrograde response by binding to the promoters of genes that contain a mitochondrial dysfunction motif (De Clercq et al., 2013). NAC013 is a marker for mitochondrial dysfunction (Schwarzländer et al., 2012), and it is induced by H2O2 (Inzé et al., 2012). Evidence that this transcription factor is mediating a response can be seen by analyzing the transcript abundance of genes for mitochondrial proteins that have been previously defined as being responsive to stress (Van Aken et al., 2009a, 2009b; Van Aken and Whelan, 2012). To a great extent, these transcripts were induced in aox1a:rpoTmp in standard growth conditions (Supplemental Table S5A). In many cases, higher fold inductions were observed in normally grown aox1a:rpoTmp than in aox1a and rpoTmp under adverse growth conditions (Supplemental Table S5A). Furthermore, several transcripts defined previously as being markers of mitochondrial dysfunction (Schwarzländer et al., 2012) were already elevated in aox1a:rpoTmp grown under standard conditions (Fig. 10B), whereas they were only induced in aox1a or rpoTmp when grown under adverse conditions (Fig. 10B). Additionally, aox1a:rpoTmp mutants showed some of the changes in auxin-associated genes that are involved in stress responses (Supplemental Fig. S4; Ivanova et al., 2014), particularly for genes involved in auxin transport and conjugation.
Similar observations were made for WRKY transcription factors, with 14 common WRKY genes up-regulated in transcript abundance from 18 and 21 that displayed significant changes in rpoTmp and aox1a mutants, respectively (Supplemental Table S1). Twenty-six Myb transcription factors also showed similar changes (Supplemental Table S1). A number of high-level regulators integrating stress responses were also up-regulated, including the sucrose non-fermenting1 (SNF1)-related protein kinase KIN10 (At3G01090) and OPEN STOMATA1 (SNF-1 related, At4G33950 and SNF7.2, At2G19830). SNF-like kinases were previously shown to interact with NAC transcription factors to mediate stress responses (Jensen et al., 2013). These transcription factors potentially drive the response of both aox1a and rpoTmp plants to MLD stress, which includes the up-regulation of transcripts of many genes previously defined as markers of mitochondrial perturbation (Schwarzländer et al., 2012; Fig. 4B).
Conversely, those transcription factors typically reported in the literature to be involved in growth, cell division, and hormone signaling were down-regulated in both aox1a and rpoTmp under stress. These included TEOSINTE BRANCHED1-CYCLOIDEA-PROLIFERATING CELL FACTOR1 (TCP), basic helix-loop-helix, auxin/indole-3-acetic acid, and ASYMMETRIC LEAVES2 (AS2) transcription factors (Supplemental Table S1). It has been observed by a number of studies that altered auxin signaling (i.e. suppression) seems to accompany mitochondrial perturbation (Tognetti et al., 2010, 2012; Ivanova et al., 2014; Kerchev et al., 2014). An examination of transcript changes for genes involved in auxin metabolism revealed several common changes in auxin efflux and influx proteins, auxin conjugation, and auxin-responsive proteins and transcription factors (Supplemental Fig. S4). We further observed a large-scale reprogramming of histone-related transcripts and global transcriptional regulation machinery (Supplemental Table S1), reflecting the sheer magnitude of transcriptomic responses occurring as a result of mitochondrial respiratory dysfunction under stress.
Together, these results indicate the presence of a common response to mitochondrial dysfunction and induction of a common mitochondrial signal, independent of the specific respiratory complex that is inhibited. The response represents an overall reprogramming of the cellular transcriptome from growth-related processes to stress response and survival. The almost universal decline in transcripts for cell wall and tetrapyrrole synthesis, the general increase in amino acids degradation, and the decrease in amino acid synthesis are consistent with a decrease in proteins synthesis and reveal the extent of the cellular crisis under environmental stress treatment when respiratory capacity is restricted.
DISCUSSION
Although it is recognized that mitochondria (and chloroplasts) can act as sensors and initiate stress responses in plants (Nomura et al., 2012; Suzuki et al., 2012; Vanlerberghe, 2013), roles of the different respiratory pathways in plants (alternative respiratory pathway compared with the cytochrome respiratory pathway) in triggering these responses have remained unclear. This is largely because of the fact that, with the exception of complex I inactivation, total inactivation of the cytochrome pathway is lethal, and plants lacking cytochrome c, which transfers electrons from complex III to IV, or plants deficient in complex IV are not viable (Gu et al., 1994; Welchen et al., 2012). Thus, although a variety of studies have analyzed the effects of the abolition of complex I activity (Dutilleul et al., 2003a, 2003b; Noctor et al., 2004; Meyer et al., 2009; Braun et al., 2014), AOX (Fiorani et al., 2005; Giraud et al., 2008), or alternative NAD(P)H dehydrogenase activities (Wallström et al., 2014a, 2014b), to date, there are a limited number of studies on plants with severely limited electron transport through the cytochrome chain. However, it is possible to decrease the abundance of cytochrome chain components using a surrogate mutant approach (Colas des Francs-Small and Small, 2014), where inactivation of a nuclear gene required for the expression of a mitochondrially encoded subunit can reduce the activity of distinct respiratory chain complexes. Although such mutants can also be lethal (Colas des Francs-Small and Small, 2014), loss of the organellar RNA polymerase RPOTmp has been shown to reduce the activity of complexes I and IV by over 80% but yield viable plants (Kühn et al., 2009). Along with insertional knockout mutants for AOX (Giraud et al., 2008) and aox1a:rpoTmp double mutants generated in this study, these plants were used here to study the effects of stress on plants impaired in the alternative-terminal oxidase, the cytochrome oxidase, or both.
Previous studies have detailed the physiological and molecular aspects of the inactivation of AOX1a in Arabidopsis under both standard and adverse growth conditions (Giraud et al., 2008). Furthermore, a meta-analysis of various mutants affecting mitochondrial functions suggested that inactivation of the alternative or cytochrome chain components lead to a different response at a molecular level (Schwarzländer et al., 2012). Our study confirms this in silico analysis based on the finding that the overlap in transcriptome responses of aox1a and rpoTmp is limited under normal growth conditions (Fig. 2). However, upon growing plants under adverse growth conditions, the response of these mutants is highly similar, with a large overlap in transcriptome changes (Fig. 2), and even transcripts changes that are unique to each mutant fall into the same functional classes (Fig. 5).
There are a number of points that are worth noting about the responses of aox1a and rpoTmp plants to the environmental stress conditions that were applied. The transcriptome changes were remarkably similar, despite the fact that the growth rate and basal transcriptomes differ between the two mutants lines (Figs. 1 and 2). As described previously, under normal growth conditions, aox1a plants show a reprogramming of the transcriptome that involves stress signaling components and transcription factors (Giraud et al., 2008). In contrast, in rpoTmp plants, this basal transcriptional response is much more specific to the mitochondrion (Fig. 2). This suggests that the response elicited by MLD stress in aox1a and rpoTmp represents a defined program that is implemented when respiratory capacity is limited, irrespective of the basal state of the plant. Notably, a defining feature of the response is an accumulation of anthocyanins, which represents a basic and even primitive oxidative stress response in plants that is also induced by accumulation of sugars (Teng et al., 2005). This response is consistent with the idea that, because of their restricted respiratory capacity, aox1a and rpoTmp mitochondria cannot sufficiently oxidize reducing equivalents from the chloroplast, and thus, large-scale transcriptional programming is initiated that, among other pathways, affects photosynthesis. A number of studies have proposed that AOX has a role in maintaining growth and promoting plant survival under adverse conditions. The response to restricting respiratory capacity seems to be a down-regulation of growth, which was evidenced by a down-regulation of Auxin, AS2, and TCP transcription factors (Supplemental Table S1) as well as an up-regulation of many transcription factors associated with stress (Fig. 10A; Supplemental Table S1). Again, because this response is evident irrespective of whether the AOX or the cytochrome pathway is restricted, the overall mitochondrial respiratory capacity seems to play an integral role in signaling growth or defense. The concept of mitochondrial signaling has been proposed by a variety of groups (Arnholdt-Schmitt et al., 2006; Clifton et al., 2006; Van Aken et al., 2009a, 2009b; Vanlerberghe et al., 2009). This study strongly implies that, under environmental stress and with restricted respiratory capacity, signals from mitochondria play an important role to constrain growth and activate stress responses.
In aox1a:rpoTmp mutants, where both respiratory pathways are restricted, the response is not simply the sum of the responses of the two single mutants under either of the growth conditions applied. Under normal growth conditions, aox1a:rpoTmp seems to have already initiated a mitochondrial stress response, which is evidenced by the increase in transcript abundance of several transcription factors from the NAC and WRKY families that have been previously shown to mediate mitochondrial stress responses (Ng et al., 2014). In particular, NAC013, which has been shown to be involved in regulation of the mitochondrial stress response (De Clercq et al., 2013) and also defined as a mitochondrial stress marker (Schwarzländer et al., 2012), was induced 26-fold, far more than what was previously observed after stress treatments. In fact, 8 of 11 transcripts that are markers of mitochondrial stress are all induced in the aox1a:rpoTmp mutant under standard growth conditions (Schwarzländer et al., 2012). It has been previously suggested that this stress response overlaps with ABA signaling responses (Schwarzländer et al., 2012). Given that the response of aox1a:rpoTmp to challenges with H2O2 and superoxide is similar to that of the aox1a mutant (Fig. 7), which has been shown to have altered defense because of modified signaling associated with regulation by ABA INSENSITIVE4 (Giraud et al., 2008, 2009), it is likely that the aox1a:rpoTmp response to the simultaneous restriction of the cytochrome and alternative respiratory pathways overlaps with ABA signaling.
In addition to the activation of a stress response, transcript changes in aox1a:rpoTmp reflected major alterations of mitochondrial function, including an up-regulation of transcripts for mitochondrial protein import components and chaperones, alternative respiratory pathways, the TCA cycle, and glycolysis, which are not normally observed as part of a mitochondrial stress response. These changes in transcript abundance were reflected at a protein level, with the notable exception of the multisubunit inner membrane respiratory complexes that contained both nuclear- and mitochondrial-encoded proteins. For these proteins, we observed relatively large decreases in protein abundance. This is interpreted as a futile attempt to increase the amount of the respiratory complexes, but in the absence of matching amounts of the mitochondrially encoded subunits, these proteins are degraded. The increases in prohibitins, proteases, and chaperones are consistent with this proposal. Overall, the analysis of the aox1a:rpoTmp transcriptome and mitochondrial proteome shows that an additional signal is initiated in the aox1a:rpoTmp background, which signals mitochondrial biogenesis and notably, does not occur in the rpoTmp mutant.
Additionally, in the aox1a:rpoTmp background, the induction of AOX1d at a transcript and protein level is observed. However, although the amount of the AOX1d protein induced seems quite substantial, because it can be readily detected by western blotting compared with AOX1a (Supplemental Fig. S2), the induction of AOX1d cannot compensate for the loss of AOX1a completely in the aox1a:rpoTmp background, which was evidenced by the severe growth retardation of aox1a:rpoTmp compared with rpoTmp (Fig. 6). The reason for this is unclear, because AOX1d would appear to contain all of the residues for activation by α-keto acids and intermolecular disulflide covalent bonding for formation of homodimers (Moore et al., 2013). The observed nonredundant functions of AOX1a and AOX1d merit further investigations into the differential activity of AOX isoforms.
Remarkably, the transcriptomic analysis of aox1a:rpoTmp plants exposed to MLD stress indicates that the mitochondrial stress response is muted, which was evidenced by the down-regulation of transcript abundance for AOX1d, genes for alternative NADP(H) dehydrogenases, and several other genes (Figs. 9 and 10; Supplemental Table S1) that were up-regulated in aox1a:rpoTmp plants grown under normal conditions (Fig. 6), and rather, a reaction akin to a response to pathogen infection is triggered. Note that earlier analyses have shown that responses to pathogen infection overlap with those to mitochondrial dysfunction, such as the response induced by treatment with antimycin A (Schwarzländer et al., 2012; Umbach et al., 2012). The prevalence of pathogen response-like changes could be because of an increase of this response in aox1a:rpoTmp exposed to adverse growth conditions. Alternatively, this response may simply appear more prominent because of the fact that other responses are absent. The physiological significance of this deactivation of the mitochondrial stress response is unclear. It is possible that aox1a:rpoTmp plants are short of resources needed to implement such a response, because these plants are severely slow growing and compromised in development and seed set (Fig. 5), likely because of severe energy limitation.
In conclusion, limitation of the capacity of the alternative, cytochrome, or both termini oxidases results in unique and overlapping responses in Arabidopsis depending on the growth conditions. Most notably, under adverse environmental growth conditions, limitation of either oxidase activates a common mitochondrial signaling pathway. Simultaneous limitation of both oxidases at least partially activates this mitochondrial stress pathway under standard growth conditions, and in addition, a pathway to initiate mitochondrial biogenesis is triggered. However, when plants restricted in both oxidases are exposed to adverse environmental growth conditions, the mitochondrial stress response is diminished, and a unique response is initiated that resembles responses to fungal infection.
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) plants, ecotype Col-0 and transfer DNA insertional lines for aox1a (SALK_084897 and SAIL_030_D08) and rpoTmp (GABI_286E07 and SALK_132842) and double knockout mutants aox1a-1:rpotmp2 (SALK_084897/SALK_132842) and aox1a-2:rpoTmp2 (SAIL_030_D08/ GABI_286E07) were grown at 22°C for 16 h at 100 μE m−2 s−1 light conditions and 8 h of dark (standard conditions). All mutant lines were obtained from the Arabidopsis Biological Resource Center seed stock center or acquired from GABI-Kat (Alonso et al., 2003; Rosso et al., 2003). The isolation and screening of mutant lines for aox1a and rpoTmp have been described previously (Giraud et al., 2008; Kühn et al., 2009). These confirmed transgenic lines were crossed, and double mutants were selected. Combined light and drought treatments (adverse conditions) were applied as follows: 4-week-old plants (5-week-old rpoTmp mutant lines and 6- to 7-week-old aox1a:rpoTmp) grown under normal conditions with watering every 2 d were either transferred to moderate light conditions (approximately 350 μE m−2 s−1) with no water (starting 4 d before transfer to ensure that soil was dry; MLD) or maintained in normal growth conditions. All measurements were taken after 4 d of environmental stress treatment. Phenotypic analyses were carried out according to Boyes et al., 2001. For root growth measurements, seedlings were germinated on vertical Murashige and Skoog agar plates (Gamborg et al., 1968). Appropriate measurements were taken from images using the software ImageJ (http://rsb.info.nih.gov/ij/).
ROS Determination
H2O2 bleaching was performed by excising whole rosettes of Col-0, aox1a, rpoTmp, and aox1a:rpoTmp. Plants grown at 22°C in long-day photoperiods (16 h at 100 μmol photons m−2 s–1 and 8 h of dark) were collected after 4 weeks (or 5 weeks for rpoTmp and 6 to 7 weeks for aox1a:rpoTmp) and submerged in various increasing concentrations of H2O2 for 4 h at 100 μmol photons m–2 s–1 followed by an overnight incubation in the dark. Detection of internal O2− and H2O2 was performed by taking whole excised rosettes and incubating them in 3 mm NBT for O2− or 5 mm DAB for H2O2. After 4 h, the rosettes were placed in 80% (v/v) ethanol and incubated for 24 h to remove the chlorophyll. This step was repeated two times. Three plants per genotype per experiment were assayed for ROS. Each experiment was repeated two times, and representative images are shown.
Mitochondrial Isolation
The aerial parts of plants were ground at 4°C in extraction buffer (0.3 m Suc, 5 mm tetrasodium pyrophosphate, 10 mm KH2PO4 [pH 7.5], 2 mm EDTA, 1% [w/v] polyvinylpyrrolidone 40, 1% [w/v] bovine serum albumin, 5 mm Cys, and 1 mm dithiothreitol) using a Polytron (30 mL of buffer for 10 g of plants, speed 4). The homogenate was centrifuged for 5 min at 3,000g, and the supernatant was centrifuged for 10 min at 20,000g. The pellet was resuspended in wash buffer (0.3 m Suc, 1 mm EGTA, and 10 mm MOPS/KOH, pH 7.2) and subjected to the same low- (3,000g) and high-speed (20,000g) centrifugations. The pellet was resuspended in a small volume of wash buffer and loaded on top of a Percoll step gradient (from bottom to top: 1 volume, 50% [v/v]; 5 volumes, 25% [v/v]; and 1 volume, 18% [v/v] Percoll in wash buffer [equivalent to 20 g of plant loaded per gradient]). The gradient was centrifuged at 40,000g for 45 min. The mitochondrial fraction located at the interface between the 50% and 25% layers was collected and washed three times in wash buffer before being used in respiratory activity measurements.
Respiratory Activity Measurements
All oxygen consumption measurements were performed using a Clark-type oxygen electrode (Hansatech Instrument), and data were collected by Oxygraph Plus, version 1.01 (Hansatech Instruments). The electrode was calibrated according to the manufacturer’s instructions. For measurements on isolated mitochondria, mitochondria equaling 100 μg of total protein were suspended in 1 mL of respiration buffer (0.3 m Suc, 5 mm KH2PO4, 10 mm TES, 10 mm NaCl, 2 mm MgSO4, and 0.1% [w/v] bovine serum albumin, pH 6.8). To determine rates for TCA cycle-dependent total respiration, the following substrates were added: Glu (10 mm) and malate (10 mm) in the presence of the cofactors coenzyme A (12 mm), thiamine pyrophosphate (0.2 mm), NAD+ (2 mm), and ADP (0.3 mm). Oxygen consumption was recorded, and respiration rates were calculated as nanomoles of O2 consumed per minute per milligram of mitochondrial protein. The AOX inhibitor n-Propylgallate (0.5 mm) and the cytochrome oxidase inhibitor KCN (1 mm) were then added to inhibit the terminus oxidases and stop the reaction. Cytochrome oxidase (complex IV) capacity was measured after the addition of the substrates ascorbate (10 mm) and cytochrome c (25 mm) in the presence of Triton X-100 (0.05% [w/v]). The specificity of the reactions was controlled by addition of 1 mm KCN. The maximal capacity of AOX was measured in the respiration buffer detailed above but at pH 7.2 in the presence of succinate (10 mm) and ATP (0.5 mm) and after successive additions of the complex III inhibitor myxothiazol (2.5 mm) and the AOX activators pyruvate (10 mm) and dithiothreitol (5 mm). After recording oxygen consumption, the AOX inhibitor n-Propylgallate (0.5 mm) was added to control the specificity of the reaction. Respiration by Arabidopsis leaves was measured in the dark at 25°C by adding leaf discs (between 15 and 25 mg of fresh weight) in the chamber containing 1 mL of distilled water. The fraction of cyanide-resistant leaf respiration was calculated as the ratio of the rates (per leaf fresh weight) after and before the addition of 1 mm KCN.
Microarray Expression Analysis
Microarrays were carried out using Affymetrix ATH1 microarray gene chips. RNA was isolated from Arabidopsis Col-0, aox1a-1, aox1a-2, rpotmp-1, rpotmp-2, and aox1a-2:rpoTmp2 plants grown under standard/untreated conditions as well as after 4 d of treatment with MLD (adverse conditions) as described above. Three biological replicates were analyzed for each sample/genotype. Tissue was collected 5 h into the light cycle to minimize diurnal effects. Antisense RNA generation, hybridization, washing, and scanning of the gene chips was carried out according to the manufacturers’ instructions. Data quality was assessed using GCOS 1.4 before CEL files were exported into AVADIS Prophetic (version 4.3; Strand Genomics) and Partek Genomics Suite software, version 6.3 for additional analysis. MAS5 normalization was performed only to generate present/absent calls across the arrays. Probe sets that recorded absent calls in two or more of three biological replicates for genotypes and treatment conditions for a particular analysis were removed. Bacterial controls, probe sets for organelle transcripts, and ambiguous probe sets recognizing multiple transcripts were also removed. All CEL files were GC content background Robust Multiarray Average normalized for computing fluorescence intensity values used in differential expression analyses. Correlation plots were examined between all arrays using the scatterplot function in the Partek Genomics Suite, and in all cases, r > 0.98 (data not shown).
Statistical Analysis
Analysis of differential expression was carried out using a regularized t test based on a Bayesian statistical framework using the software program Cyber-T (Baldi and Long, 2001; http://cybert.microarray.ics.uci.edu/). Cyber-T uses a mixture model-based method described in Allison et al., 2006 for the computation of the global false-positive and false-negative levels inherent in a DNA microarray experiment. To compute and show differential expression with false discovery rate and minimize false positives within the differential expression analysis, PPDE (P) values and PPDE (>P) values were calculated. Changes in transcript abundance were considered significant with P < 0.05, PPDE (>P) > 0.95, and a fold change >1.5-fold. These lists of differentially expressed genes were used for the Pageman (Usadel et al., 2006) analysis of gene sets to reveal over-/underrepresentation of genes encoding specific functions compared with the genome. This relies on using Fisher’s test (ORA_Fisher with an ORA cutoff value of 1.0),and significance is indicated by z score, where a score of 1.96 indicates significance of P < 0.05.
Hierarchical clusters were generated using Euclidean distance and average linkage measures in Partek Genomics suite V6.3. Overlaps in the transcript abundance responses for the different genotypes were plotted on Venn diagrams. To determine statistically significant over- or underrepresentation of genes in the Venn diagram overlap compared with that which is expected by random chance, a Pearson’s χ2 test was used. Functional categorization using gene ontology annotations was performed on the Arabidopsis whole-genome set along with gene sets defined as differentially expressed (either positively or negatively) during microarray experiments. Functional categorizations for each gene list were obtained from The Arabidopsis Information Resource using the gene ontology annotations, the functional categorization function (http://www.arabidopsis.org/tools/bulk/go/index.jsp) for cellular component, and the molecular function. The percentage distribution of each category for the various gene lists was compared with that of the whole genome. A Pearson’s χ2 test was performed for each comparison, and percentile distributions were considered to be significantly different at the 98% confidence interval.
Quantitative Proteomic Analysis
Two hundred micrograms of mitochondrial protein isolated from Col-0 and aox1a:rpoTmp was precipitated in 1 mL of acetone at −20°C for 18 h before pelleting at 20,800g for 20 min. Pellets were brought up in 50 µL of 20 mm NH4HCO3 and 30% (v/v) acetonitrile and digested for 18 h with 10 µg of trypsin (Promega). Samples were diluted to 5% (v/v) acetonitrile with HPLC-grade water/0.1% (v/v) formic acid, and the solid phase was extracted on C18 macrospin columns (Nest Group) before drying down in a vacuum centrifuge. Samples were resuspended in 50 µL of 5% (v/v) HPLC acetonitrile/0.1% (v/v) formic acid before passing through 0.22-µm centrifugal filters (Millipore). Samples were analyzed in quadruplicate (8 µg per analysis) on an Agilent 6550 Q-TOF with Chip Cube interface with C18 trapping/analytical Polaris chip using 45 min of 10% to 30% (v/v) acetonitrile gradients in 0.1% (v/v) formic acid. Settings used were positive ion mode, eight mass spectrometry (MS) scans at 250 to 1,400 mass-to-charge ratio per second, maximum of eight precursors per cycle with an absolute threshold of 5,000, scan speed varied according to abundance, and charge state selection set to +2 and +3 and selected by abundance. Resultant spectra were searched against TAIR10 protein sequences using Mascot 2.3.02 (Matrix Science). The resultant *.dat files were imported into Skyline 2.5.0.6079 (MacCoss Lab) with a 0.95 confidence interval and associated with their corresponding *.d folders. Peptides were associated with an in-house curated list of 946 confirmed mitochondrial proteins and filtered to retain the four most intense features per protein and remove empty proteins and repeated peptides. MS1 full-scan features associated with MS/MS identifications were manually checked for appropriate integration, and the area under the curve of the three most intense features with the least interference from coeluting peptides was exported using the MS1 probe input report. In cases where less than three peptides were identified per protein, these were retained. Total peak areas of the mitochondrial proteins were used to create a normalization factor that was applied to each MS1 feature area. Significance was tested using a Student’s t test, a 95% confidence interval was used to filter MS1 features, and a ratio of aox1a:rpoTmp to Col-0 was calculated. A median value of the ratio of MS1 features was then calculated on a per-protein basis and converted to a linear fold change scale by −1/x IF < 1. The MS data are shown in Supplemental Table S6.
Immunodetection
Western-blot analysis and quantification were carried out as previously described (Duncan et al., 2011).
Transfer DNA insertion mutants used were GK-286E07 (rpoTmp-1), SALK_132842 (rpoTtmp-2), SALK_084897 (aox1a-1), and SAIL_030_D08 (aox1a-2).
Microarray data from this article can be found in ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) under accession number E-ATMX-32 (aox1a arrays) and the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE 60960 (rpoTmp and aox1a:rpoTmp arrays).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Pageman representation of the functional categories that are overrepresented or underrepresented among genes whose transcripts changed in abundance in aox1a, rpoTmp, and aox1a:rpoTmp grown under standard conditions.
Supplemental Figure S2. Western-blot analysis of protein abundance in aox1a:rpoTmp.
Supplemental Figure S3. A Pageman visualization showing the overrepresented functional categories for all differentially expressed genes in all three mutants grown under MLD stress.
Supplemental Figure S4. Transcript abundance changes of auxin-related gene ontologies in aox1a, rpoTmp, and aox1a:rpoTmp under standard (untreated) and adverse (MLD) growth conditions.
Supplemental Table S1. Changes in transcript abundance in aox1a, rpoTmp, and aox1a:ropTmp under standard and adverse growth conditions.
Supplemental Table S2. Antagonistic and common changes under standard and adverse growth conditions for aox1a and rpoTmp.
Supplemental Table S3. Unique changes under adverse growth conditions for aox1a and rpoTmp.
Supplemental Table S4. Unique changes in genes encoding mitochondrial proteins under adverse growth conditions for aox1a and rpoTmp.
Supplemental Table S5. Changes in transcripts of genes encoding mitochondrial proteins.
Supplemental Table S6. MS data file for quantitative proteomic data outlined in Figure 8A and Supplemental Table S5D.
Supplementary Material
Glossary
- ABA
abscisic acid
- Col-0
Columbia-0
- DAB
3,3′diaminobenzidine
- H2O2
hydrogen peroxide
- KCN
potassium cyanide
- MLD
moderate light and drought
- MS
mass spectrometry
- NBT
nitroblue tetrazolium
- OXPHOS
oxidative phosphorylation
- PPDE
posterior probability of differential expression
- PPR
pentatricopeptide repeat
- ROS
reactive oxygen species
- TCA
tricarboxylic acid
Footnotes
This work was supported by the Australian Research Council Centre of Excellence (grant nos. CEO561495 and CE140100008).
Articles can be viewed without a subscription.
References
- Allison DB, Cui X, Page GP, Sabripour M (2006) Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet 7: 55–65 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arnholdt-Schmitt B, Costa JH, de Melo DF (2006) AOX—a functional marker for efficient cell reprogramming under stress? Trends Plant Sci 11: 281–287 [DOI] [PubMed] [Google Scholar]
- Baldi P, Long AD (2001) A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17: 509–519 [DOI] [PubMed] [Google Scholar]
- Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415–442 [DOI] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle GM, Roucou X, Nagley P, Devenish RJ, Prescott M (1999) Identification of subunit g of yeast mitochondrial F1F0-ATP synthase, a protein required for maximal activity of cytochrome c oxidase. Eur J Biochem 262: 315–323 [DOI] [PubMed] [Google Scholar]
- Braun HP, Binder S, Brennicke A, Eubel H, Fernie AR, Finkemeier I, Klodmann J, König AC, Kühn K, Meyer E, et al. (February 24, 2014) The life of plant mitochondrial complex I. Mitochondrion 10.1016/j.mito.2014.02.006 [DOI] [PubMed] [Google Scholar]
- Clifton R, Millar AH, Whelan J (2006) Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim Biophys Acta 1757: 730–741 [DOI] [PubMed] [Google Scholar]
- Colas des Francs-Small C, Small I (2014) Surrogate mutants for studying mitochondrially encoded functions. Biochimie 100: 234–242 [DOI] [PubMed] [Google Scholar]
- Cvetkovska M, Alber NA, Vanlerberghe GC (2013) The signaling role of a mitochondrial superoxide burst during stress. Plant Signal Behav 8: e22749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvetkovska M, Vanlerberghe GC (2013) Alternative oxidase impacts the plant response to biotic stress by influencing the mitochondrial generation of reactive oxygen species. Plant Cell Environ 36: 721–732 [DOI] [PubMed] [Google Scholar]
- Cvetkovska M, Vanlerberghe GC (2012a) Alternative oxidase modulates leaf mitochondrial concentrations of superoxide and nitric oxide. New Phytol 195: 32–39 [DOI] [PubMed] [Google Scholar]
- Cvetkovska M, Vanlerberghe GC (2012b) Coordination of a mitochondrial superoxide burst during the hypersensitive response to bacterial pathogen in Nicotiana tabacum. Plant Cell Environ 35: 1121–1136 [DOI] [PubMed] [Google Scholar]
- De Clercq I, Vermeirssen V, Van Aken O, Vandepoele K, Murcha MW, Law SR, Inzé A, Ng S, Ivanova A, Rombaut D, et al. (2013) The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25: 3472–3490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan O, Taylor NL, Carrie C, Eubel H, Kubiszewski-Jakubiak S, Zhang B, Narsai R, Millar AH, Whelan J (2011) Multiple lines of evidence localize signaling, morphology, and lipid biosynthesis machinery to the mitochondrial outer membrane of Arabidopsis. Plant Physiol 157: 1093–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Driscoll S, Cornic G, De Paepe R, Foyer CH, Noctor G (2003a) Functional mitochondrial complex I is required by tobacco leaves for optimal photosynthetic performance in photorespiratory conditions and during transients. Plant Physiol 131: 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Garmier M, Noctor G, Mathieu C, Chétrit P, Foyer CH, de Paepe R (2003b) Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15: 1212–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elthon TE, Nickels RL, McIntosh L (1989) Monoclonal antibodies to the alternative oxidase of higher plant mitochondria. Plant Physiol 89: 1311–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan PM, Wooding AR, Day DA (1999) An alternative oxidase monoclonal antibody recognises a highly conserved sequence among alternative oxidase subunits. FEBS Lett 447: 21–24 [DOI] [PubMed] [Google Scholar]
- Fiorani F, Umbach AL, Siedow JN (2005) The alternative oxidase of plant mitochondria is involved in the acclimation of shoot growth at low temperature: a study of Arabidopsis AOX1a transgenic plants. Plant Physiol 139: 1795–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadjev I, Vanderauwera S, Gechev TS, Laloi C, Minkov IN, Shulaev V, Apel K, Inzé D, Mittler R, Van Breusegem F (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol 141: 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50: 151–158 [DOI] [PubMed] [Google Scholar]
- Geisler DA, Päpke C, Obata T, Nunes-Nesi A, Matthes A, Schneitz K, Maximova E, Araújo WL, Fernie AR, Persson S (2012) Downregulation of the δ-subunit reduces mitochondrial ATP synthase levels, alters respiration, and restricts growth and gametophyte development in Arabidopsis. Plant Cell 24: 2792–2811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Ho LH, Clifton R, Carroll A, Estavillo G, Tan YF, Howell KA, Ivanova A, Pogson BJ, Millar AH, et al. (2008) The absence of ALTERNATIVE OXIDASE1a in Arabidopsis results in acute sensitivity to combined light and drought stress. Plant Physiol 147: 595–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud E, Van Aken O, Ho LH, Whelan J (2009) The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol 150: 1286–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Dempsey S, Newton KJ (1994) Rescue of a maize mitochondrial cytochrome oxidase mutant by tissue culture. Plant J 6: 787–794 [DOI] [PubMed] [Google Scholar]
- Gutierres S, Sabar M, Lelandais C, Chetrit P, Diolez P, Degand H, Boutry M, Vedel F, de Kouchkovsky Y, De Paepe R (1997) Lack of mitochondrial and nuclear-encoded subunits of complex I and alteration of the respiratory chain in Nicotiana sylvestris mitochondrial deletion mutants. Proc Natl Acad Sci USA 94: 3436–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzé A, Vanderauwera S, Hoeberichts FA, Vandorpe M, Van Gaever T, Van Breusegem F (2012) A subcellular localization compendium of hydrogen peroxide-induced proteins. Plant Cell Environ 35: 308–320 [DOI] [PubMed] [Google Scholar]
- Ito J, Heazlewood JL, Millar AH (2006) Analysis of the soluble ATP-binding proteome of plant mitochondria identifies new proteins and nucleotide triphosphate interactions within the matrix. J Proteome Res 5: 3459–3469 [DOI] [PubMed] [Google Scholar]
- Ivanova A, Law SR, Narsai R, Duncan O, Lee JH, Zhang B, Van Aken O, Radomiljac JD, van der Merwe M, Yi K, et al. (2014) A functional antagonistic relationship between auxin and mitochondrial retrograde signaling regulates Alternative Oxidase1a expression in Arabidopsis. Plant Physiol 165: 1233–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen MK, Lindemose S, de Masi F, Reimer JJ, Nielsen M, Perera V, Workman CT, Turck F, Grant MR, Mundy J, et al. (2013) ATAF1 transcription factor directly regulates abscisic acid biosynthetic gene NCED3 in Arabidopsis thaliana. FEBS Open Bio 3: 321–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpova OV, Kuzmin EV, Elthon TE, Newton KJ (2002) Differential expression of alternative oxidase genes in maize mitochondrial mutants. Plant Cell 14: 3271–3284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchev PI, De Clercq I, Denecker J, Mühlenbock P, Kumpf R, Nguyen L, Audenaert D, Dejonghe W, Van Breusegem F (2014) Mitochondrial perturbation negatively affects auxin signaling. Mol Plant 7: 1138–1150 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Hong SH, Kim YW, Lee IH, Jun JH, Phee BK, Rupak T, Jeong H, Lee Y, Hong BS, et al. (2014) Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J Exp Bot 65: 4023–4036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klodmann J, Senkler M, Rode C, Braun HP (2011) Defining the protein complex proteome of plant mitochondria. Plant Physiol 157: 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn K, Carrie C, Giraud E, Wang Y, Meyer EH, Narsai R, des Francs-Small CC, Zhang B, Murcha MW, Whelan J (2011) The RCC1 family protein RUG3 is required for splicing of nad2 and complex I biogenesis in mitochondria of Arabidopsis thaliana. Plant J 67: 1067–1080 [DOI] [PubMed] [Google Scholar]
- Kühn K, Richter U, Meyer EH, Delannoy E, de Longevialle AF, O’Toole N, Börner T, Millar AH, Small ID, Whelan J (2009) Phage-type RNA polymerase RPOTmp performs gene-specific transcription in mitochondria of Arabidopsis thaliana. Plant Cell 21: 2762–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law SR, Narsai R, Taylor NL, Delannoy E, Carrie C, Giraud E, Millar AH, Small I, Whelan J (2012) Nucleotide and RNA metabolism prime translational initiation in the earliest events of mitochondrial biogenesis during Arabidopsis germination. Plant Physiol 158: 1610–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law SR, Narsai R, Whelan J (April 13, 2014) Mitochondrial biogenesis in plants during seed germination. Mitochondrion 10.1016/j.mito.2014.04.002 [DOI] [PubMed]
- Maxwell DP, Wang Y, McIntosh L (1999) The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc Natl Acad Sci USA 96: 8271–8276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald AE. (2008) Alternative oxidase: an inter-kingdom perspective on the function and regulation of this broadly distributed ‘cyanide-resistant’ terminal oxidase. Funct Plant Biol 35: 535–552 [DOI] [PubMed] [Google Scholar]
- Meyer EH, Tomaz T, Carroll AJ, Estavillo G, Delannoy E, Tanz SK, Small ID, Pogson BJ, Millar AH (2009) Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol 151: 603–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Soole KL, Day DA (2011) Organization and regulation of mitochondrial respiration in plants. Annu Rev Plant Biol 62: 79–104 [DOI] [PubMed] [Google Scholar]
- Moore AL, Shiba T, Young L, Harada S, Kita K, Ito K (2013) Unraveling the heater: new insights into the structure of the alternative oxidase. Annu Rev Plant Biol 64: 637–663 [DOI] [PubMed] [Google Scholar]
- Ng S, De Clercq I, Van Aken O, Law SR, Ivanova A, Willems P, Giraud E, Van Breusegem F, Whelan J (2014) Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Mol Plant 7: 1075–1093 [DOI] [PubMed] [Google Scholar]
- Ng S, Giraud E, Duncan O, Law SR, Wang Y, Xu L, Narsai R, Carrie C, Walker H, Day DA, et al. (2013a) Cyclin-dependent kinase E1 (CDKE1) provides a cellular switch in plants between growth and stress responses. J Biol Chem 288: 3449–3459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S, Ivanova A, Duncan O, Law SR, Van Aken O, De Clercq I, Wang Y, Carrie C, Xu L, Kmiec B, et al. (2013b) A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25: 3450–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Dutilleul C, De Paepe R, Foyer CH (2004) Use of mitochondrial electron transport mutants to evaluate the effects of redox state on photosynthesis, stress tolerance and the integration of carbon/nitrogen metabolism. J Exp Bot 55: 49–57 [DOI] [PubMed] [Google Scholar]
- Nomura H, Komori T, Uemura S, Kanda Y, Shimotani K, Nakai K, Furuichi T, Takebayashi K, Sugimoto T, Sano S, et al. (2012) Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat Commun 3: 926. [DOI] [PubMed] [Google Scholar]
- Nuruzzaman M, Sharoni AM, Kikuchi S (2013) Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front Microbiol 4: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmusson AG, Soole KL, Elthon TE (2004) Alternative NAD(P)H dehydrogenases of plant mitochondria. Annu Rev Plant Biol 55: 23–39 [DOI] [PubMed] [Google Scholar]
- Rhoads DM, McIntosh L (1991) Isolation and characterization of a cDNA clone encoding an alternative oxidase protein of Sauromatum guttatum (Schott). Proc Natl Acad Sci USA 88: 2122–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas-Carbo M, Berry JA, Yakir D, Giles L, Robinson SA, Lennon AM, Siedow JN (1995) Electron partitioning between the cytochrome and alternative pathways in plant mitochondria. Plant Physiol 109: 829–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Schwarzländer M, König AC, Sweetlove LJ, Finkemeier I (2012) The impact of impaired mitochondrial function on retrograde signalling: a meta-analysis of transcriptomic responses. J Exp Bot 63: 1735–1750 [DOI] [PubMed] [Google Scholar]
- Shiba T, Kido Y, Sakamoto K, Inaoka DK, Tsuge C, Tatsumi R, Takahashi G, Balogun EO, Nara T, Aoki T, et al. (2013) Structure of the trypanosome cyanide-insensitive alternative oxidase. Proc Natl Acad Sci USA 110: 4580–4585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strodtkötter I, Padmasree K, Dinakar C, Speth B, Niazi PS, Wojtera J, Voss I, Do PT, Nunes-Nesi A, Fernie AR, et al. (2009) Induction of the AOX1D isoform of alternative oxidase in A. thaliana T-DNA insertion lines lacking isoform AOX1A is insufficient to optimize photosynthesis when treated with antimycin A. Mol Plant 2: 284–297 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Koussevitzky S, Mittler R, Miller G (2012) ROS and redox signalling in the response of plants to abiotic stress. Plant Cell Environ 35: 259–270 [DOI] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti VB, Mühlenbock P, Van Breusegem F (2012) Stress homeostasis—the redox and auxin perspective. Plant Cell Environ 35: 321–333 [DOI] [PubMed] [Google Scholar]
- Tognetti VB, Van Aken O, Morreel K, Vandenbroucke K, van de Cotte B, De Clercq I, Chiwocha S, Fenske R, Prinsen E, Boerjan W, et al. (2010) Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell 22: 2660–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbach AL, Siedow JN (2000) The cyanide-resistant alternative oxidases from the fungi Pichia stipitis and Neurospora crassa are monomeric and lack regulatory features of the plant enzyme. Arch Biochem Biophys 378: 234–245 [DOI] [PubMed] [Google Scholar]
- Umbach AL, Zarkovic J, Yu J, Ruckle ME, McIntosh L, Hock JJ, Bingham S, White SJ, George RM, Subbaiah CC, et al. (2012) Comparison of intact Arabidopsis thaliana leaf transcript profiles during treatment with inhibitors of mitochondrial electron transport and TCA cycle. PLoS One 7: e44339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Giraud E, Clifton R, Whelan J (2009a) Alternative oxidase: a target and regulator of stress responses. Physiol Plant 137: 354–361 [DOI] [PubMed] [Google Scholar]
- Van Aken O, Whelan J (2012) Comparison of transcriptional changes to chloroplast and mitochondrial perturbations reveals common and specific responses in Arabidopsis. Front Plant Sci 24: 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aken O, Zhang B, Carrie C, Uggalla V, Paynter E, Giraud E, Whelan J (2009b) Defining the mitochondrial stress response in Arabidopsis thaliana. Mol Plant 2: 1310–1324 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC. (2013) Alternative oxidase: a mitochondrial respiratory pathway to maintain metabolic and signaling homeostasis during abiotic and biotic stress in plants. Int J Mol Sci 14: 6805–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanlerberghe GC, Cvetkovska M, Wang J (2009) Is the maintenance of homeostatic mitochondrial signaling during stress a physiological role for alternative oxidase? Physiol Plant 137: 392–406 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, McIntosh L (1997) ALTERNATIVE OXIDASE: from gene to function. Annu Rev Plant Physiol Plant Mol Biol 48: 703–734 [DOI] [PubMed] [Google Scholar]
- Vanlerberghe GC, Vanlerberghe AE, McIntosh L (1994) Molecular genetic alteration of plant respiration (silencing and overexpression of alternative oxidase in transgenic tobacco). Plant Physiol 106: 1503–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallström SV, Florez-Sarasa I, Araújo WL, Aidemark M, Fernández-Fernández M, Fernie AR, Ribas-Carbó M, Rasmusson AG (2014a) Suppression of the external mitochondrial NADPH dehydrogenase, NDB1, in Arabidopsis thaliana affects central metabolism and vegetative growth. Mol Plant 7: 356–368 [DOI] [PubMed] [Google Scholar]
- Wallström SV, Florez-Sarasa I, Araújo WL, Escobar MA, Geisler DA, Aidemark M, Lager I, Fernie AR, Ribas-Carbó M, Rasmusson AG (2014b) Suppression of NDA-type alternative mitochondrial NAD(P)H dehydrogenases in Arabidopsis thaliana modifies growth and metabolism, but not high light stimulation of mitochondrial electron transport. Plant Cell Physiol 55: 881–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Carrie C, Giraud E, Elhafez D, Narsai R, Duncan O, Whelan J, Murcha MW (2012) Dual location of the mitochondrial preprotein transporters B14.7 and Tim23-2 in complex I and the TIM17:23 complex in Arabidopsis links mitochondrial activity and biogenesis. Plant Cell 24: 2675–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MT, Wang P, Korkaric M, Capper RG, Saunders NJ, Langdale JA (2009) GLK transcription factors coordinate expression of the photosynthetic apparatus in Arabidopsis. Plant Cell 21: 1109–1128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E, Hildebrandt TM, Lewejohann D, Gonzalez DH, Braun HP (2012) Lack of cytochrome c in Arabidopsis decreases stability of Complex IV and modifies redox metabolism without affecting Complexes I and III. Biochim Biophys Acta 1817: 990–1001 [DOI] [PubMed] [Google Scholar]
- Zhang B, Van Aken O, Thatcher L, De Clercq I, Duncan O, Law SR, Murcha MW, van der Merwe M, Seifi HS, Carrie C, et al. (2014) The mitochondrial outer membrane AAA ATPase AtOM66 affects cell death and pathogen resistance in Arabidopsis thaliana. Plant J 80: 709–727 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.