Wheat transcription factors located on chromosome group 2 drive the yield-related production of supernumerary spikelets.
Abstract
Bread wheat (Triticum aestivum) inflorescences, or spikes, are characteristically unbranched and normally bear one spikelet per rachis node. Wheat mutants on which supernumerary spikelets (SSs) develop are particularly useful resources for work towards understanding the genetic mechanisms underlying wheat inflorescence architecture and, ultimately, yield components. Here, we report the characterization of genetically unrelated mutants leading to the identification of the wheat FRIZZY PANICLE (FZP) gene, encoding a member of the APETALA2/Ethylene Response Factor transcription factor family, which drives the SS trait in bread wheat. Structural and functional characterization of the three wheat FZP homoeologous genes (WFZP) revealed that coding mutations of WFZP-D cause the SS phenotype, with the most severe effect when WFZP-D lesions are combined with a frameshift mutation in WFZP-A. We provide WFZP-based resources that may be useful for genetic manipulations with the aim of improving bread wheat yield by increasing grain number.
Bread wheat (Triticum aestivum; genome nomenclature, BBAADD) is one of the most important food crops in the world. Its yield in grain production per plants largely depends on the architecture of the inflorescences. Studies on genetic determinism of inflorescence development may allow new spike architectures to be designed, with the aim of enhancing grain production. The wheat inflorescence classically consists in a spike with a main axis (the spike rachis) carrying lateral sessile spikelets that are directly attached to the rachis and also a terminal spikelet (Fig. 1A). The spikelet constitutes the basal unit of the spike inflorescence and is a characteristic of all modern grasses, except one lineage that diverged a long time ago (Malcomber et al., 2006). The number of spikelets per rachis node is a key taxonomic trait of the Triticeae tribe (Muramatsu, 2009; Sakuma et al., 2011).
Figure 1.
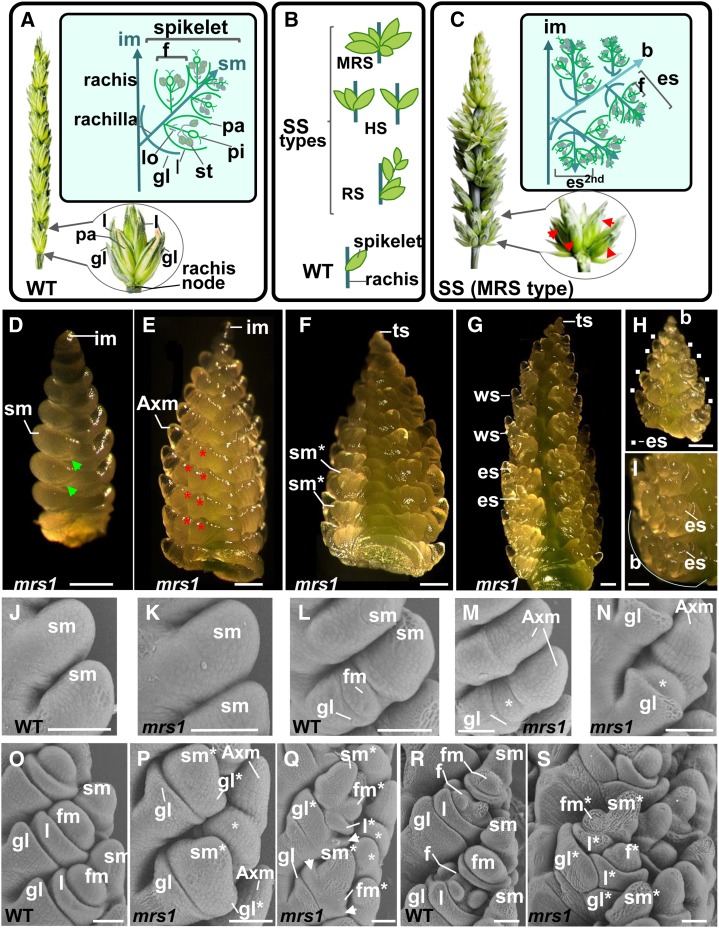
SS phenotypes in bread wheat. A, Schematic representations of a spike (left) and a spikelet (right) from bread wheat N67 and of a theoretical wild-type (WT) spikelet (boxed). B, Schematic illustration of various SS structures: a cluster of spikelets at a rachis node referred to as a MRS, three spikelets (triple spikelet), and two spikelets in horizontal positions at a rachis node referred to as HSs; additional sessile spikelets with lateral branch bearing spikelets at a node referred to as a RS; and a single spikelet at a node for the wild-type spike. C, Illustration of the spike structure in the line NIL-mrs1 harboring numerous SSs (left), a rachis node with additional spikelets (right) where SSs are indicated with red arrows, and schematic representation of a branch-like structure (b) bearing ectopic spikelets at a rachis node in the lower part of the NIL-mrs1 spike (boxed). D to G, Light microscopy images of the NIL-mrs1 inflorescence at several developmental stages. D, Illustration of the spikelet differentiation stage; green arrows indicate glume primordia. E and F, Early floret differentiation stage. E, Secondary AxMs that later produce ectopic spikelets are indicated with asterisks. F, Spikelet meristems of ectopic spikelets form glume primordia. G, Late floret differentiation stage when all floral organs of wild-type spikelets (ws) are differentiated in the upper part of the inflorescence. H, A branch-like structure dissected from a rachis node of the NIL-mrs1 inflorescence. White squares indicate the ectopic spikelet (es). I, Location of ectopic spikelets at rachis nodes of the NIL-mrs1 inflorescence. Bars = 0.25 mm. J to S, Scanning electron microscopy images of N67 and NIL-mrs1 inflorescences at various developmental stages. J and K, Spikelet differentiation stage in the wild type (J) and mrs1 (K). L, Early floret differentiation stages when the spikelet meristem produces the FM in the wild type. M and N, Differentiation of secondary AxMs (indicated by asterisks) in the mrs1 mutant. O, Early floret differentiation stage showing lemmas. P and Q, The development of glumes (gl* and indicated by white arrows), lemma, and FMs by secondary AxMs (indicated by asterisks) in the mrs1 mutant. R, Floret differentiation stage showing differentiated floral organs in a basal floret of the wild type. S, The development of ectopic spikelets in mrs1. Bars =100 μm. es2nd, Ectopic spikelet of the second order, developing at the place of a floret of the first order ectopic spikelet; f , floret with floret organ primordia; f*, floret of an ectopic spikelet; Fm, floret meristem; Fm*, floret meristem of an ectopic spikelet; gl, glume; im, inflorescence meristem; l, lemma; l*, lemma of an ectopic spikelet; lo, lodicule; pa, palea; pi, pistil; ts, terminal spikelet; sm, spikelet meristem; sm*, spikelet meristem of an ectopic spikelet; st, stamen.
A wheat spike normally bears one spikelet per rachis node, which arises directly on the main inflorescence axis; the formation of supernumerary spikelets (SS) is rare. The term SS includes any additional sessile spikelets developed at a rachis node, such as multirow spike (MRS), horizontal spikelets (HSs), and the ramified spike (RS) or branched spike (Fig. 1B; Supplemental Fig. S1). In tetraploid and hexaploid wheat species, the development of SS is a recessive trait (Pennell and Halloran, 1983; Klindworth et al., 1990b; Dobrovolskaya et al., 2009) and is modulated by a number of environmental factors (Sharman, 1944; Pennell and Halloran, 1983). In tetraploid wheat (genome nomenclature, BBAA), the SS trait is quantitatively inherited, controlled by a single major gene acting in concert with an unknown number of minor genes (Klindworth et al., 1990b; Haque et al., 2012). It has been suggested that the SS trait in hexaploid wheat (BBAADD) is determined by either two (Pennell and Halloran, 1983) or three (Koric, 1980) loci. The SS trait in hexaploid wheat has been reported to be a consequence of the absence of a complete chromosome (Swaminathan et al., 1966). The involvement of the homoeologous chromosome group 2 in the control of SS has been suggested in both tetraploid and hexaploid wheats (Sears, 1954; Klindworth et al., 1990a; Peng et al., 1998; Laikova et al., 2005; Yang et al., 2005). Genetic loci linked to the SS trait have been located on the chromosome 2AS in hexaploid (qTS2A-1 locus), tetraploid (branched head [bh] gene), and diploid wheats (bhm gene) and on the chromosome 2D in hexaploid bread wheat (Dobrovolskaya et al., 2009; Li et al., 2011; Haque et al., 2012; Amagai et al., 2014).
The MRS phenotype is one of the SS traits of the bread wheat spike (Fig. 1C; Supplemental Fig. S1). The primary feature of MRS lines is that they have a large number of spikelets emerging from each rachis node, in most cases in the lower third of the spike (Fig. 1, B and C). The central and upper thirds of the spike generally have only three spikelets per node, similar to the spike architecture of six-rowed barley (Hordeum vulgare) lines (Tanno and Takeda, 2004). The mrs1 gene, on chromosome 2DS, may be the major determinant of the MRS trait in two genetically related bread wheat lines derived from genotypes obtained by chemical mutation (Dobrovolskaya et al., 2008, 2009). This gene is also known as bh-D1 (Jia et al., 2013), as it is considered to be a mutant allele of the Bh-D1 locus (http://ccg.murdoch.edu.au/cmap/ccg-live).
Here, we report the characterization of genetically unrelated MRS mutants that led to the identification of the wheat FRIZZY PANICLE (FZP) homoeologous genes; we provide evidence that these genes are responsible for SS traits in bread wheat. Structural and functional characterization of the three wheat FZP homoeologous genes (WFZP) implicates WFZP as a key player in spikelet development during the floret meristem (FM) transition phase.
RESULTS
The mrs1 Mutant Allele Affects Spikelet Development
The 13 MRS lines used in this work all have numerous SSs emerging from a rachis node (Fig. 1B). We developed a near-isogenic line (NIL), NIL-mrs1, which carries a 17-centimorgan (cM) segment that includes the mrs1 locus in bread wheat ‘Novosibirskaya 67’ (N67) background (Supplemental Figs. S1 and S2). NIL-mrs1 and N67 only differ in inflorescence structure, with NIL-mrs1 showing severe SS phenotypes with numerous SSs emerging at rachis nodes in the lower half of the spike (Supplemental Fig. S1); the additional spikelets were not sessile as they developed at very short branches (Fig. 1C). At the earliest stages of spike development, NIL-mrs1 inflorescence appeared phenotypically normal (Fig. 1, D, E, K, M, and N). At the spikelet differentiation stage, the inflorescence meristem produced primary axillary meristems (AxMs), which first initiated glume primordia (Fig. 1D); at the early floret differentiation stage (Fig. 1, E, M, and N), secondary AxMs were produced that could not be distinguished from normal FM in the wild-type inflorescence (Fig. 1L). In the wild type, the FM first initiated lemma and then formed the other floral organs (Fig. 1, O and R). By contrast, in the NIL-mrs1 inflorescence, basal secondary AxMs developed as ectopic spikelet meristems and first produced spikelet organs, the glumes (Fig. 1, F and P), and then generated the other spikelet organs (Fig. 1, G, Q, and S).
Thus, SSs in NIL-mrs1 developed exactly at the location of florets of primary spikelets. The development of the basal florets was replaced by ectopic spikelets in the middle part of the spike rachis, whereas the development of most or all florets of primary spikelets were replaced by ectopic spikelets in the lower part of the spike rachis, and thus, branch-like structures were formed (Fig. 1, H and I). These branch-like structures were not encompassed by normal glumes, and rudimentary glumes could be distinguished only at early stages of the spike development (Fig. 1, P and Q). Most ectopic spikelets developed as wild-type spikelets and produced florets. Within the ectopic spikelets, located in the basal part of the branch-like structure, the development of basal florets could be replaced by secondary ectopic spikelets (Fig. 1C). The meristems of ectopic spikelets were formed at 90° (Fig. 1, P and S) to normal spikelets or to the previous order ectopic spikelet. In line MC1611, with a weak SS phenotype, ectopic spikelets developed at the location of basal florets of the lower part of the spike (Supplemental Fig. S3). The other genetically unrelated SS lines used in this work, Skle128, Ruc204, and So164, showed phenotypes similar to NIL-mrs1 and the line MC1611 (next section; Supplemental Fig. S1).
The 2DS Locus Is the Main Contributor to the SS Trait in Bread Wheat
In addition to the MRS lines (mrs1) described above, we also genetically characterized four unrelated SS bread wheat lines: MC1611, Skle128, Ruc204, and So164 (Supplemental Fig. S1). Line MC1611 is a mutant produced by NitrosoMethylUrea (NMU) mutagenesis, line Skle128 was derived from spontaneous mutation(s), and line So164 was derived from a cross with a branched tetraploid wheat (see Supplemental Text S1 for detailed descriptions of these lines). C-banding and Simple Sequence Repeat genotyping characterization revealed structural rearrangements of chromosome 2D in three of these four lines (Supplemental Figs. S4 and S5). The C-banding analysis revealed structural rearrangements on the short arm of the chromosome 2D in both Skle128 and Ruc204 (Supplemental Fig. S4) and that chromosome 2D was completely absent from So164, making it nulli2D-tetra2A (Supplemental Fig. S5). The short arm of chromosome 2D in the line Ruc204 was clearly short (approximately 10% shorter than controls), associated with a diagnostic C-band at the 2DS termini, proof of an interstitial deletion. The other chromosomes in Ruc204 were morphologically indistinguishable from those in normal reference lines (Supplemental Fig. S4). The deletion in the short arm of chromosome 2D in line Skle128 was much larger than that in line Ruc204, such that the arm was one-half the size of that in controls (Supplemental Fig. S4). The presence of these deletions was confirmed, and the location of deletion breakpoints was determined precisely by Simple Sequence Repeat genotyping of the 2DS chromosomes in these lines (Supplemental Fig. S4).
We performed genetic mapping with F2 populations derived from Ruc204 × bread wheat ‘Saratovskaya 29’ (S29) and Skle128 × S29 crosses to investigate the contributions of these deletions to the SS trait (Supplemental Text S1). The two deletions identified were found to be related to the SS phenotype and explained approximately 50% of the phenotypic variation (Supplemental Fig. S6). The deletions observed on the chromosome 2D genetic map colocate with the mrs1 locus (Supplemental Fig. S7). In addition to the 2D quantitative trait loci (QTL), the 2A QTL, which explained approximately 16% of the phenotypic variation, was detected in Ruc204. The 2A QTL, also responsible for the SS trait, has also been described in the Tibetan Triple-Spikelet Wheat line (with Skle128 originating from Tibetan Triple-Spikelet Wheat; Li et al., 2011). NIL-mrs1 was crossed to each of the two SS deletion lines to conduct genetic complementation tests. All the F2s produced expressed the SS phenotype, demonstrating that the SS phenotypes of the three lines share the same genetic determinant (Supplemental Table S1). The segregation ratios in F2 and back cross1 (BC1) progeny from crosses between S29 and the MC1611 mutant line, which was obtained in a S29 background by NMU mutagenesis, clearly indicate that the SS trait in MC1611 is due to the mutation of a single gene (Supplemental Table S1). Similarly, MC1611 was crossed with standard spiked bread wheat ‘Skala,’ and the observed F2 segregation was close to monogenic (Supplemental Table S1). Genetic mapping data indicated that the gene responsible for SS was on 2DS and colocated with the mrs1 locus (Supplemental Fig. S7). Genetic complementation tests between MC1611 and NIL-mrs1 revealed that, in addition to the 2DS gene, other secondary genetic factors may also contribute to the SS trait (Supplemental Table S1). Nevertheless, our findings for genetically unrelated SS genetic materials (NIL, bread wheat lines, biparental populations, and independent mutants) implicate the 2DS gene/locus as the main determinant of the SS trait in bread wheat, with the driving gene (mrs1 in this study) still to be rigorously identified.
Dissection of the FZP Locus and Identification of the Gene in Bread Wheat
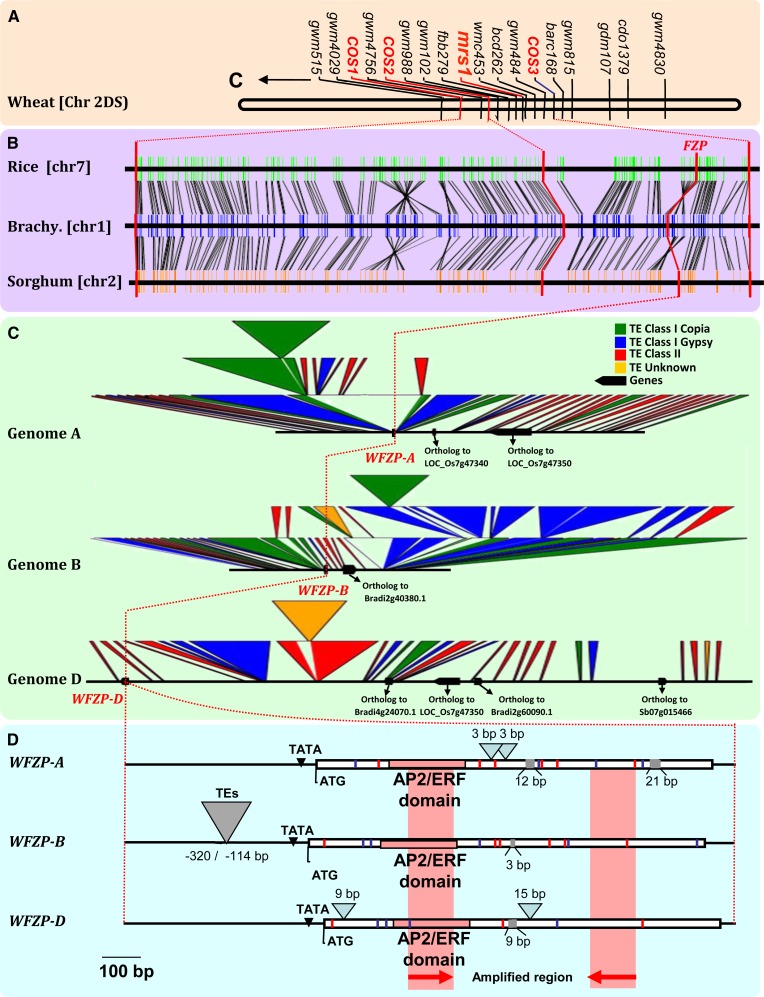
Accurate genetic maps that included the mrs1 gene (Dobrovolskaya et al., 2009) were used for further synteny-based identification of SS candidate markers using the COS genes available in bread wheat (Quraishi et al., 2009; Pont et al., 2013). The mrs1 locus is flanked by the COS1 (LOCOs07g38530-Bradi1g23970-Sb02g037070), COS2 (LOCOs07g45064), and COS3 (LOCOs07g48200-Bradi1g17680-Sb02g043000) markers (Fig. 2A). The COS2-COS3 region shares synteny with the rice (Oryza sativa) chromosome 7, B. distachyon chromosome 1, and sorghum (Sorghum bicolor) chromosome 2 regions (Fig. 2B). The identification of unique orthologous regions in rice, sorghum, and B. distachyon provided a list of candidate genes (Supplemental Table S2). The rice region hosts a FZP homolog that encodes a transcription factor of the APETALA2 (AP2)/Ethylene Response Factor (ERF; Tanaka et al., 2013) family; these factors contribute to determining inflorescence development and spikelet meristem identity in rice. In rice fzp mutants, the formation of florets is replaced by sequential rounds of branching (Komatsu et al., 2003). The fzp mutant phenotype suggests that FZP is required to prevent the formation of AxMs within the spikelet meristem and to allow the subsequent establishment of floral meristem identity (Komatsu et al., 2003). The maize (Zea mays) gene branched silkless1 (bd1) is an FZP ortholog, and the BD1 protein is associated with a conserved function in spike development (Chuck et al., 2002; Komatsu et al., 2003). Based on shared synteny between grasses at the mrs1 locus and the observed similarity in mutant phenotypes between fzp/bd1 and mrs1, the bread wheat FZP gene (namely, WFZP) appeared to be the strongest candidate for the MRS trait.
Figure 2.
WFZP gene characterization in bread wheat. A, Composite genetic map of the wheat chromosome 2D, including the mrs1 locus and flanking conserved orthologous set (COS) markers. B, Orthologous chromosomes in rice (Chr7), sorghum (Chr2), and Brachypodium distachyon (Brachy.; Chr1) are shown, with conserved genes linked with black lines. C, Annotated BAC clones are illustrated with conserved genes (black boxes) linked with black connecting lines. Copia (green), Gypsy (blue), uncharacterized (yellow), and class II (red) TEs are indicated. D, Comparison of WFZP homoeologous gene structures with red boxes highlighting the conserved AP2/ERF domain. Gray boxes indicate small deletions, light blue triangles indicate small insertions, and gray triangles indicate insertion of TEs. Red and blue lines indicate nonsynonymous and synonymous single nucleotide substitutions, respectively. Red arrows indicate the positions of primers designed from the B. distachyon (Bradi1g18580) gene sequence and that were used for BAC library screening.
Homoeologous WFZP genes were isolated from a bread wheat ‘Chinese Spring’ bacterial artificial chromosome (BAC) library (Supplemental Text S1) using B. distachyon as a model genome for the Triticeae to develop a new set of COS markers for further gene cloning and sequencing. The BLAST search did not find significant orthologous wheat sequences in public databases, and therefore primers were designed from the conserved regions of the orthologous FZP genes in grasses (Supplemental Table S2). These primers were used to screen the bread wheat BAC library from the ‘Chinese Spring’ genotype (http://cnrgv.toulouse.inra.fr/fr/library/genomic_resource/Tae-B-Chinese%20spring). BAC clones harboring WFZP genes were identified, and three carrying the WFZP homoeologs from the A, B, and D subgenomes were sequenced. All the WFZP genes consist of single exons with an AP2/ERF domain that is conserved in grass relatives (Fig. 2D; Supplemental Fig. S8). The WFZP genes showed organ-specific and stage-specific expression patterns. WFZP transcripts were detected in young spikes, with the highest level of expression observed at the early floret developmental stage (Fig. 3D). Homoeologous WFZP genes were found in the 2AS, 2BS, and 2DS chromosome bins with genome-specific primer pairs (Supplemental Fig. S9) and DNA from bread wheat ‘Chinese Spring’ deletion lines. Genetic mapping indicated that WFZP-D was at the expected position on 2DS, colocating with the mrs1/quantitative trait loci for the SS trait on all genetic maps constructed (Supplemental Figs. S7–S9).
Figure 3.
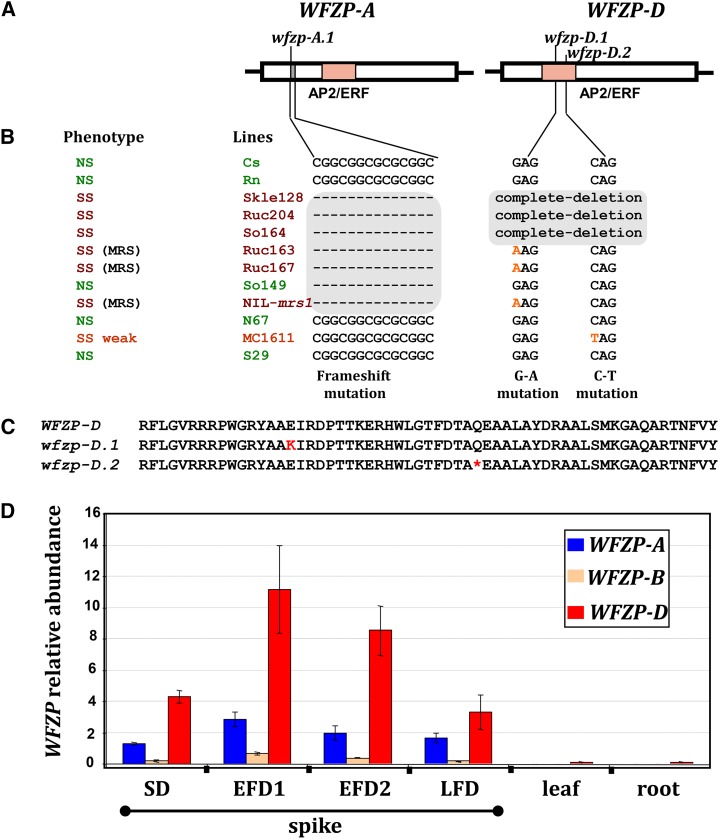
Structural and functional characterization of WFZP genes. A, Schematic representation of the WFZP-A and WFZP-D gene structures and the mutations identified. The light red box indicates the AP2/ERF domain, and gray boxes indicate deletions. B, WFZP haplotypes of SS (red) and standard spiked (normal spiked [NS], green) lines. C, Alignment of the amino acid sequences of the AP2/ERF domains of wild type, WFZP-D, and wfzp-D.1, wfzp-D.2 mutants. Black asterisks indicate amino acids that confer specificity to the GCC-box binding site. D, Relative abundance of WFZP homoeolog mRNAs as determined by quantitative reverse transcription (qRT)-PCR with samples from spikes of bread wheat ‘Chinese Spring’ at various developmental stages of the spike (see below) and from roots and leaves. The values shown correspond to the mean values for three biological and three technical replicates, normalized to the value for the Ta.304.3 gene used as a reference. Error bars represent ses between replicates. Spike developmental stages were assigned as follows: spikelet differentiation stage (SD; when spikelets are differentiated to form two opposite rows at the rachis and glume primordia are initiated), early floret differentiation stage 1 (EFD1; spikelet meristems initiate floral meristems and lemma primordia are apparent), early floret differentiation stage 2 (EFD2; floral organ primordia are apparent in basal florets), and late floret differentiation stage (LFD; floral organs are differentiated in each floret).
Coding and Null Mutations in WFZP-D Are Associated with the SS Phenotype
We investigated the relationship between the WFZP sequences in the genetically characterized SS lines and the SS trait. Analysis of the WFZP BAC clone sequences allowed us to develop a set of sequence-specific primers for further resequencing of WFZP homoeologous genes in bread wheat cultivars (Supplemental Table S3; Supplemental Fig. S10). Direct Sanger sequencing was used to determine the sequences of the PCR products of WFZP-A, WFZP-B, and WFZP-D genes in 11 bread wheat lines, including seven mutants (Ruc163, Ruc167, NIL-mrs1, MC1611, Ruc204, Skle128, and So164) and four normal spiked lines (Renan, S29, N67, and So149). Thus, we obtained the sequences of the FZP genes in SS and normal spiked parents of the populations used for mapping purposes, NIL-mrs1 and its parent (N67), the NMU-induced mutant (MC1611), and the wild-type line (S29).
WFZP-D
The WFZP-D gene structure differed between mutant and normal spiked lines (Fig. 3, A–C). The WFZP-D gene was completely deleted from three of the seven SS lines (Fig. 3B), due either to a deletion from 2DS or a substitution of the entire 2D chromosome (Supplemental Figs. S4 and S5). A mutation in the GCC-box binding site of the AP2/ERF functional domain of WFZP-D was found in three genetically related mutant MRS lines, including the NIL-mrs1 (Fig. 3, A–C; Supplemental Fig. S10). Genotyping, using sequence-specific primers (Supplemental Table S3), revealed this nonsynonymous substitution in the other 10 MRS lines (listed in Supplemental Text S1) and in all of the MRS F2 plants resulting from Ruc167 × So149 and Ruc163 × So149 crosses. This allele was named wfzp-D.1. The NMU-induced mutant MC1611 was found to carry a premature stop codon in the AP2/ERF functional domain of WFZP-D (Fig. 3, B and C; Supplemental Fig. S10). The WFZP-D sequences in 96 BC1s derived from the cross between MC1611/S29//MC1611 were determined: all the SS plants harbored the mutant allele, now named wfzp-D.2, whereas the standard spiked plants were heterozygous for the WFZP-D wfzp-D.2 alleles. Thus, the mutant allele wfzp-D.2 is associated with the SS phenotype in the MC1611 line (obtained by NMU mutagenesis of S29 considered here as the wild type). Overall, SS lines were either null mutants for WFZP-D or harbored G-A or C-T mutations in the functional AP2/ERF domain of the FZP gene (Fig. 3B).
WFZP-B
The sequencing of WFZP-B in bread wheat lines revealed that, independent of their spike morphology, all contained miniature inverted-repeat TEs inserted (114 and 320 bp upstream from the TATA-box) into the proximal promoter region of FZP; this was associated with major reorganizations in the promoter structure. Expression tests revealed that the abundance of WFZP-B transcripts was very low in the developing spikes of bread wheat ‘Chinese Spring’ (as little as 5% of the amount of WFZP-D transcript; Fig. 3D). Thus, in bread wheat, the WFZP-B gene is largely inactive. These observations are consistent with the possibility that the insertion of the miniature inverted-repeat TE into the promoter region is responsible for the loss of WFZP-B homoeolog expression and even its silencing.
WFZP-A
In WFZP-A, there was a 14-bp deletion 40 bp upstream from the functional AP2/ERF domain, which resulted in a frameshift, in six of the seven SS lines (the exception was MC1611) investigated and in one normal spiked line, So149 (Fig. 3, A and B). Primers were designed (Supplemental Table S3) to test for this deletion and were used to screen 48 bread wheat cultivars with standard spike morphology (listed in Supplemental Table S4): 6.3% of these cultivars carried the deletion. Thus, the deletion is part of the natural variation in bread wheat and does not by itself cause phenotypic changes; this allele was named wfzp-A.1. However, we observed that the combination of wfzp-A.1 with wfzp-D.1 or with wfzp-D.null alleles resulted in a severe SS phenotype in MRS lines, SS deletion lines, and SS substitution line; the combination of the wild allele WFZP-A and the mutant allele wfzp-D.2 in line MC1611 led to only a weak SS phenotype (Figs. 1 and 3, A–C). The segregation patterns observed in the crosses between standard spiked lines and SS/MRS lines confirmed the WFZP-A and WFZP-D allele composition of the lines (Supplemental Table S1). Thus, F2s derived from the crosses between the standard spiked parental line So149 (wfzp-A.1 WFZP-D) and the MRS lines Ruc163 and Ruc167 (wfzp-A.1 wfzp-D.1) showed segregation of 3 (standard spike) to 1 (MRS). Unlike the parental line N67 (WFZP-A WFZP-D), NIL-mrs1 harbors mutant alleles at both the wfzp-A.1 and wfzp-D.1 loci. The various mapping and sequencing results provide evidence that WFZP-D is synonymous with MRS1 and that the wfzp-D.1 and wfzp-D.2 alleles are associated with the MRS/SS phenotype in bread wheat. The WFZP-D transcript was four times more abundant than the WFZP-A/WFZP-B transcripts (Fig. 3D), consistent with the WFZP-D homeoallele making the largest contribution to the MRS/SS trait.
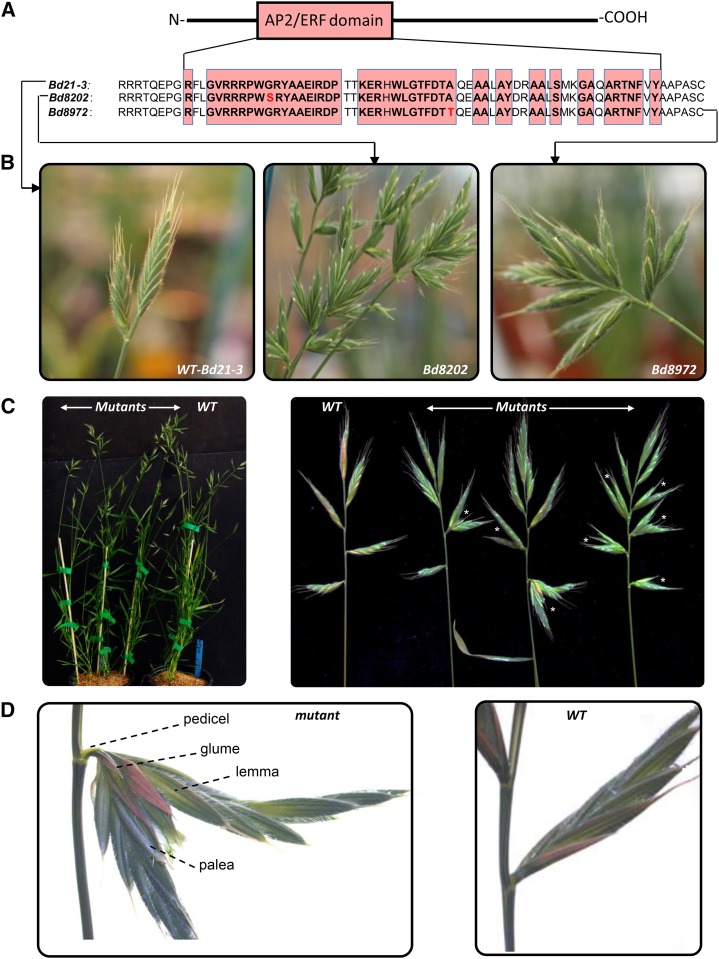
Finally, we screened the collection of mutagenized B. distachyon lines from Institut National de la Recherche Agronomique-Versailles (http://www-ijpb.versailles.inra.fr/en/plateformes/crb/index.html) for aberrant spikelets as described in Dalmais et al. (2013). Two lines, Bd8202 and Bd8972, showed an SS phenotype but produced viable seeds (Fig. 4). The full sequence of Bradi1g18580 gene was amplified by PCR from the genome of Bd8202, Bd8972, and wild-type (Bd21-3) lines and determined. Two transitions were detected: G-232-A in Bd8972 and G-163-A in Bd8202. These mutations lead to amino acid substitutions in the conserved region of the AP2/ERF domain (Fig. 4). These results are consistent with the report that deletion in the Bradi1g18580 promoter causes inflorescence branching (Derbyshire and Byrne, 2013). Our findings with the closely related B. distachyon as a validation system confirm the role of FZP and more precisely AP2/ERF domain function in the determination of spike architecture in Triticeae.
Figure 4.
Functional validation of FZP in B. distachyon. FZP mutant screening identified two alleles with independent mutations (Bd8202 and Bd8972) located in the AP2/ERF domain (A), both exhibiting SSs not present in the wild type (WT, Bd 21-3; B). B. distachyon fzp mutants and wild-type plants show similar growth and architecture (C, left). Spikes with different penetrance of the SS phenotype can be observed in mutants (C, right; stars indicate SSs). D, Spikes in fzp mutants show spikelets with a single pedicel as observed in the wild type but with dichotomous or trichotomous rachilla.
DISCUSSION
The inflorescence of cereals includes a unique structure called the spikelet. In bread wheat, spikelets are arranged as two opposite rows on the main axis, the rachis. Each spikelet is composed of florets joined at the indeterminate spikelet axis (rachilla), alternately on opposite sides, and flanked by two bracts, the glumes. Normally, the spikelet meristem gives rise to glume primordia and then to the FM. The FM, in turn, differentiates into floret organs such as a lemma, a palea, a pair of lodicules, three stamens, and a pistil. Spikes in the SS mutant lines display SSs developed on very short branches. We compared the anatomy of young spikes of the line NIL-mrs1 presenting a severe SS phenotype of the MRS type and its normal spiked parent, N67: differences in their morphological development were investigated by light microscopy and scanning electron microscopy. Ectopic spikelets developed at the location of florets in NIL-mrs1, in all cases at 90° to the normal spikelets, or to ectopic spikelets of the previous order, suggesting that the defect in the mrs1 mutant occurs soon after the acquisition of the spikelet meristem identity. Our observations suggest that the mrs1 mutant allele affects spikelet development and disturbs the establishment of the floral meristem identity.
In all the SS wheat lines studied in this work, the SSs develop in the same way, at the location of florets, at 90° to normal spikelets. The number of additional spikelets and thus the severity of the SS phenotype of these lines, depend on the number of florets that are replaced by ectopic spikelets, the number of mature ectopic spikelets, and the number of spikelets with hypoplasia. Similar SS phenotypes have been reported in tetraploid wheats. Coffmann (1924) described spontaneous mutants of the Mindum durum (Triticum turgidum ssp. durum) wheat variety characterized by the presence of one or two additional spikelets at several rachis nodes in the lower part of the spike. The spike phenotype of these spontaneous mutants is similar to SS in bread wheat. The Four-Rowed Spike (FRS) trait has been described in T. turgidum (Zhang et al., 2013), where in FRS plants, two spikelets develop at a rachis node in the tetrastichon manner. It seems likely that the same developmental mechanisms are involved in the appearance of SSs in both hexaploid and tetraploid wheats.
The MRS trait has been shown to be determined by a single recessive mrs1 gene on chromosome 2DS (Dobrovolskaya et al., 2009). Here, we genetically characterized four bread wheat SS lines, in addition to the mrs1 mutant, that included both induced and spontaneous mutants. We show that different SS phenotypes in fact reflect the same series of related traits with a common genetic determinism (Supplemental Table S5) and that the 2DS locus is the main contributor to the SS trait in bread wheat. Sears (1954) described the reduplication of spikelets in hexaploid wheat plants nullisomic for chromosomes 2A or 2D. The involvement of the homoeologous chromosome group 2 in the control of SS in both tetraploid and hexaploid wheat was subsequently suggested (Klindworth et al., 1990a, 1997; Peng et al., 1998; Laikova et al., 2005; Muramatsu, 2009). The frs1 gene for the FRS trait and the bh gene have now been mapped on chromosome 2AS of tetraploid wheats T. turgidum (Zhang et al., 2013) and Triticum durum Desf. var. ramosoobscurum Jakubz. (Haque et al., 2012). Consequently, the general view is that the 2AS and 2DS homoeologous loci are responsible for the SS trait in bread wheat.
We used a combination of molecular genetic mapping analyses based on the conservation of synteny, and phenotypic characterization of independent SS mutants to study the gene underlying SS-related traits. We mapped the mrs1 locus on the chromosome 2DS using COS markers and showed that the mrs1 region shared synteny with the rice chromosome 7 carrying the FZP gene. In rice fzp mutants, supernumerary AxMs are formed in axils of bracts in young spikelet meristems, and the formation of florets is replaced by sequential rounds of branching (Komatsu et al., 2003). Rao et al. (2008) confirmed that FZP represses AxM formation. The gene bd1 is the maize ortholog of FZP (Chuck et al., 2002), and bd1 mutants show a similar phenotype involving the initiation of extra spikelets in the tassel, and in the ear, spikelet meristems are replaced by branch meristems (Chuck et al., 2002). Derbyshire and Byrne (2013) isolated a B. distachyon mutant more spikelets1 (mos1) with similar defects in spikelet architecture; although no mutation in the coding region of FZP was found in this mutant, the authors proposed that the larger number of AxMs is due to a slightly but significantly lower expression of this AP2/ERF transcription factor than in the wild type (20% less transcripts in the mos1 mutant). Derbyshire and Byrne (2013) suggested that this abnormal expression may be the consequence of a chromosome rearrangement affecting the promoter region of FZP. Here, we reported two new mutants sharing mutations in the highly conserved AP2/ERF domain of FZP and associated with an increased number of lateral floral meristems. These findings are consistent with the FZP gene acting in the same way as bd1 and the FZP gene in B. distachyon. The identification of two independent mutant alleles (Bd8202 and Bd8972) within the AP2/ERF domain associated with a SS phenotype provides clues about the role of FZP and, more precisely, implicates AP2/ERF domain functionality in the maintenance of spike architecture. Homologs of the BD1 gene have been isolated from sorghum BAC libraries and from foxtail millet (Setaria italica), common millet (Panicum miliaceum), oats (Avena sativa), and finger millet (Eleusine coracana) by PCR with degenerate primers (Chuck et al., 2002). FZP (Triticeae), bd1 (maize), and mos1 (B. distachyon) genes encode orthologous transcription factors of the AP2/ERF family (Chuck et al., 2002; Komatsu et al., 2003), showing complete amino acid identity within the AP2/ERF domain and with up to 75% identity (Chuck et al., 2002) with Bradi1g18580, 59% with BD1, and 54% with FZP (Derbyshire and Byrne, 2013).
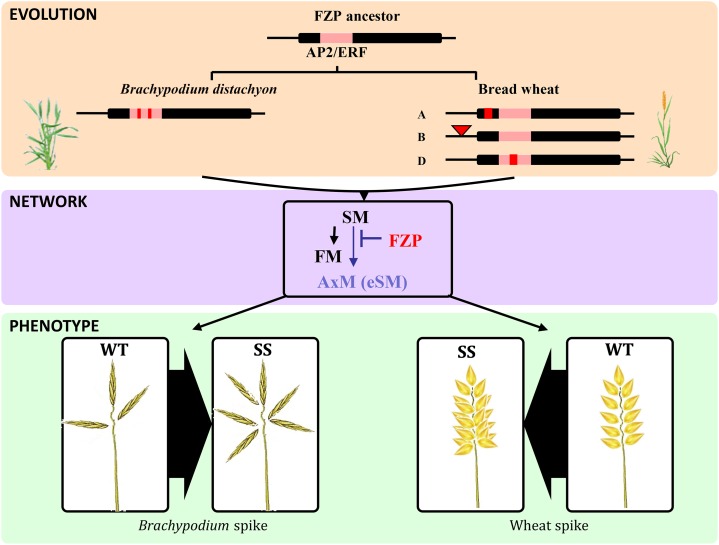
Bread wheat (AABBDD) is a natural allohexaploid species originating from three diploid ancestral species: the A genome is derived from Triticum urartu, the D genome is from Aegilops tauschii, and the B genome is from an unknown progenitor closely related to Aegilops speltoides (Feldman, 2001; Feldman and Levy, 2005). We isolated and sequenced WFZP-A/WFZP-B/WFZP-D gene copies and thereby established that they are associated with a single exon structure as in rice, maize, and B. distachyon. Interestingly, the loci that were genetically associated with the SS trait in tetraploid and hexaploid wheats map on chromosomes 2A and 2D, consistent with our finding that WFZP-B is not functional. qRT-PCR showed consistently stronger expression (more abundant mRNA) of WFZP-D than WFZP-A, also in agreement with the 2DS locus being the main contributor to the SS/MRS phenotypes in hexaploid wheat. Thus, our analyses revealed that the three homoeologous gene copies in wheat do not all have the same roles in spike morphology. In rice, the alteration of one of six conserved amino acids of the GCC-box binding site (referenced as fzp-2 and fzp-7 mutants) in the AP2/ERF domain or the formation of premature stop codons (referenced as fzp-1 and fzp-4 mutants) resulted in severe fzp phenotypes (Komatsu et al., 2003). By contrast, we report that, in bread wheat, a G-A mutation in the GCC-box binding site of the AP2/ERF functional domain (wfzp-D.1) of WFZP-D, a C-T mutation in AP2/ERF (wfzp-D.2) of WFZP-D, and complete deletions of the WFZP-D locus (null mutations) are associated with SS phenotypes. In bread wheat, the wfzp-D.1 and wfzp-D null mutations in combination with a frameshift mutation in the WFZP-A gene copy resulted in severe SS phenotypes, whereas the wfzp-D.2 mutation in combination with the wild-type WFZP-A allele resulted in only a weak SS phenotype. These results agree with the 2DS locus being the main contributor to the SS phenotype. The WFZP mutations described here control the development of SSs from ectopic spikelet meristems in all the SS-related bread wheat mutants investigated in this study. However, the number of SSs and length of branch-like structures, which distinguish the different SS phenotypes (HS, RS, and MRS), may be determined by other, as yet unknown, genes. Note also that the function of the FZP orthologs appears to be conserved among cereal species, including bread wheat, rice, maize, and B. distachyon (Fig. 5).
Figure 5.
Evolutionary and functional model of FZP in grasses. FZP genes are illustrated as black horizontal bars showing substitutions in the AP2/ERF domain (red boxes), frameshift mutations (red bars), and TE insertions in the promoter region (red triangle) leading to FZP nonfunctionality and driving SS phenotypes in B. distachyon and wheat. In wild-type (WT) bread wheat, FZP represses the formation of AxMs from spikelet meristems (SMs) and controls the establishing FM identity. By contrast, in SS mutants, AxM is not suppressed and leads to ectopic spikelet meristem (eSM) formation.
Finally, we report structural and functional characterization of the WFZP gene in wheat and demonstrate that it is one of the key regulators of inflorescence development and a determinant of SS formation. Therefore, it may serve as a basis for the development of new resources for the genetic manipulation of the grain number, one of the major components of yield. Standard wheat morphotypes respond to environmental changes, such as the availability of resources, using three levels of branching: tillers, spikelets in the spike, and grains in the spikelet. These metameric organs allow plants to respond to environmental constraints during growth and development (Acevedo et al., 2002). The hierarchical organization of plants (Nicolis et al., 1977) ensures their reproduction even in unfavorable conditions where increases in apical dominance, such as inter- and intraplant competition, decreases crop yield (Miralles and Slafer, 2007). The clusters of spikelets in MRS lines constitute another metameric structure: the increased numbers of spikelets may contribute to yield stability, especially under the influence of environmental constraints. The yields of MRS lines remain more stable than those of standard varieties if the doses of fertilizers and pesticides applied during wheat development are altered (Martinek and Váňová, 2012). Repetitive marker-assisted backcrossing of the MRS donors with selected elite varieties with standard spike structure, using the WFZP markers described here, will rapidly lead to the development of new breeding lines with MRSs and improved genetic backgrounds and yield potential.
MATERIALS AND METHODS
The experimental procedures may be found in Supplemental Information S1 which details plant materials, light microscopy and SEM analysis, C-banding procedure, primers, mapping procedures, COS-Single-Strand Conformation Polymorphism analysis, BAC clone screening, sequencing, and annotation, resequencing, qRT-PCR, Brachypodium distachyon mutant identification and sequencing, qSS mapping, and WFZP expression pattern analysis.
Sequence data from this article can be found in the GenBank/EMBL datalibraries under accession numbers BAC-clone-WFZP-A (KP276772), BAC-clone-WFZP-B (KP276773), and BAC-clone-WFZP-D (KP276774).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Spike morphology of bread wheat lines with different SS phenotypes.
Supplemental Figure S2. Genetic mapping of the NIL-mrs1 line on the chromosome 2D.
Supplemental Figure S3. Light microscopy image of the MC1611 inflorescence.
Supplemental Figure S4. Deletion breakpoint mapping and ideograms for the Skle128 and Ruc204 lines on chromosome 2D.
Supplemental Figure S5. Ideogram of the So164 line.
Supplemental Figure S6. Frequency distribution of SSs in Skle128 × S29 and Ruc204 × S29 F2 mapping populations.
Supplemental Figure S7. mrs1 gene and QTL mapping in five F2 mapping populations.
Supplemental Figure S8. Alignment of wheat WFZP gene copies with rice and maize orthologs.
Supplemental Figure S9. Deletion bins and genetic positions of WFZP homoeologous genes.
Supplemental Figure S10. Alignment of deduced WFZP-A, WFZP-B, and WFZP-D amino acid sequences.
Supplemental Table S1. Segregation data for the numbers of normal spiked and SS plants in the F2 and BC1 progenies.
Supplemental Table S2. List of orthologous genes (rice, sorghum, maize, and B. distachyon) at the MRS1 locus.
Supplemental Table S3. Primer sequences derived from WFZP-A/WFZP-B/WFZP-D gene copies.
Supplemental Table S4. WFZP-A alleles characterized in bread wheat cultivars.
Supplemental Table S5. SS phenotypes and associated mutations in WFZP-D.
Supplemental Text S1. Additional and background information.
Supplementary Material
Acknowledgments
We thank Dr. Vyacheslav M. Melnick (Altai Research Institute of Agriculture, Barnaul, Russia) for providing the MC1611 mutant line, Olga M. Popova (Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences) and Pierre Desray (Institut National de la Recherche Agronomique-UBP Unité Mixte de Recherche-1095) for technical assistance, Dr. Pierre Sourdille (Institut National de la Recherche Agronomique-UBP Unité Mixte de Recherche-1095) for providing DNA of F2s from the cv Chinese Spring × Renan cross, Dr. Audrey Didier (Institut National de la Recherche Agronomique-UBP Unité Mixte de Recherche-1095) for providing information about normal spiked bread wheat cultivars used in this research, Tatiana I. Aksenovich (Institute of Cytology and Genetics, Siberian Branch of the Russian Academy of Sciences) for helpful suggestions on the article, and Sébastien Antelme, Philippe Le Bris, and Aurélie Lemaire (Institut Jean-Pierre Bourgin) for taking care of plant cultures and genotyping B. distachyon mutants referenced in the article.
Glossary
- SS
supernumerary spikelet
- HS
horizontal spikelet
- RS
ramified spike
- MRS
multirow spike
- NIL
near-isogenic line
- N67
cv Novosibirskaya 67
- AxM
axillary meristem
- FM
floret meristem
- NMU
NitrosoMethylUrea
- S29
cv Saratovskaya 29
- QTL
quantitative trait loci
- BAC
bacterial artificial chromosome
- TE
transposable element
- FRS
Four-Rowed Spike
- RT
reverse transcription
Footnotes
This work was supported by grants from the Russian Foundation for Basic Research (project nos. 10–04–01458–a and 12–04–00897–a), the Agence Nationale de la Recherche (programs ANRjc-PaleoCereal [reference no. ANR–09–JCJC–0058–01] and ANR Blanc-PAGE [reference no. ANR–2011–BSV6-00801]), the program of the Presidium of Russian Academy of Sciences Molecular and Cell Biology (project no. 6.14), the French Embassy in Russia, Department for Science, Technology, and Space (Mobility grant in 2012 to O.D.), and the Ministry of Agriculture of the Czech Republic (project no. RO0211 to P.M.).
References
- Acevedo E, Silva P, Silva H (2002) Wheat growth and physiology. InCurtis B, Rajaram S, Gomez Macpherson H, eds, Bread Wheat Improvement and Production. Food and Agriculture Organization of the United Nations, Rome [Google Scholar]
- Amagai Y, Martinek P, Watanabe N, Kuboyama T (2014) Microsatellite mapping of genes for branched spike and soft glumes in Triticum monococcum L. Genet Resour Crop Evol 61: 465–471 [Google Scholar]
- Chuck G, Muszynski M, Kellogg E, Hake S, Schmidt RJ (2002) The control of spikelet meristem identity by the branched silkless1 gene in maize. Science 298: 1238–1241 [DOI] [PubMed] [Google Scholar]
- Coffman FA. (1924) Supernumerary spikelets in Mindum wheat. J Hered 5: 187–192 [Google Scholar]
- Dalmais M, Antelme S, Ho-Yue-Kuang S, Wang Y, Darracq O, d’Yvoire MB, Cézard L, Légée F, Blondet E, Oria N, et al. (2013) A TILLING platform for functional genomics in Brachypodium distachyon. PLoS ONE 8: e65503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire P, Byrne ME (2013) MORE SPIKELETS1 is required for spikelet fate in the inflorescence of Brachypodium. Plant Physiol 161: 1291–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrovolskaya O, Martinek P, Röder MS, Börner A (2008) Microsatellite mapping of a mutant gene (mrs) for multirow spike in wheat (T. aestivum). Conf Proc Breeding 2008: 133–136 [Google Scholar]
- Dobrovolskaya O, Martinek P, Voylokov AV, Korzun V, Röder MS, Börner A (2009) Microsatellite mapping of genes that determine supernumerary spikelets in wheat (T. aestivum) and rye (S. cereale). Theor Appl Genet 119: 867–874 [DOI] [PubMed] [Google Scholar]
- Feldman M. (2001) The origin of cultivated wheat. InBonjean A, Angus W, eds, The Wheat Book. Lavoisier, Paris, pp 1–56 [Google Scholar]
- Feldman M, Levy AA (2005) Allopolyploidy—a shaping force in the evolution of wheat genomes. Cytogenet Genome Res 109: 250–258 [DOI] [PubMed] [Google Scholar]
- Haque MA, Martinek P, Kobayashi S, Kita I, Ohwaku K, Watanabe N, Kuboyama T (2012) Microsatellite mapping of genes for semi-dwarfism and branched spike in Triticum durum Desf. var. ramosoobscurum Jakubz. “Vetvistokoloskaya.” Genet Resour Crop Evol 59: 831–837 [Google Scholar]
- Jia J, Zhao S, Kong X, Li Y, Zhao G, He W, Appels R, Pfeifer M, Tao Y, Zhang X, et al. (2013) Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature 496: 91–95 [DOI] [PubMed] [Google Scholar]
- Klindworth DL, Klindworth MM, Williams ND (1997) Telosomic mapping of four genetic markers in durum wheat. J Hered 88: 229–232 [Google Scholar]
- Klindworth DL, Williams ND, Joppa LR (1990a) Chromosomal location of genes for supernumerary spikelets in tetraploid wheat. Genome 33: 515–520 [Google Scholar]
- Klindworth DL, Williams ND, Joppa LR (1990b) Inheritance of supernumerary spikelets in a tetraploid wheat cross. Genome 33: 509–514 [Google Scholar]
- Komatsu M, Chujo A, Nagato Y, Shimamoto K, Kyozuka J (2003) FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130: 3841–3850 [DOI] [PubMed] [Google Scholar]
- Koric S. (1980) Study of branched gene complex of T. aestivum ssp. vulgare and its significance for wheat breeding. J Sci Agric Res 142: 271–282 [Google Scholar]
- Laikova LI, Arbuzova VS, Popova OM, Efremova TT, Melnick VM (2005) Study on spike branching in the T. aestivum mutant lines (cv Saratovskaya29). In PL Goncharov, RA Zilke, TN Gordeeva, eds, Proceeding of the IX Workshop on Genetics and Breeding. SB RAAS, Novosibirsk, Russia, pp 388–393 [Google Scholar]
- Li J, Wang Q, Wei H, Hu X, Yang W (2011) SSR mapping for locus conferring on the triple-spikelet trait of the Tibetan triple-spikelet wheat (Triticum aestivum L. concv. tripletum). Triticeae Genomics Genet 2: 1–6 [Google Scholar]
- Malcomber ST, Preston JC, Reinheimer R (2006) Developmental gene evolution and the origin of grass inflorescence diversity. Adv Bot Res 44: 423–479 [Google Scholar]
- Martinek P, M Váňová (2012) The effect of crop management practice and year on yield traits and grain quality in selected varieties and lines of winter wheat differing in spike morphology. Obilnářské Listy 4: 90–97 [Google Scholar]
- Miralles DJ, Slafer GA (2007) Sink limitations to yield in wheat: how could it be reduced? J Agric Sci 145: 139–149 [Google Scholar]
- Muramatsu M. (2009) A presumed genetic system determining the number of spikelets per rachis node in the tribe Triticeae. Breed Sci 59: 617–620 [Google Scholar]
- Nicolis JS, Protonotarios EN, Voulodemou I (1977) Control Markov chain models for biological hierarchies. J Theor Biol 68: 563–581 [DOI] [PubMed] [Google Scholar]
- Peng ZS, Yen C, Yang JL (1998) Chromosomal location of genes for supernumerary spikelet in bread wheat. Euphytica 103: 109–114 [Google Scholar]
- Pennell AL, Halloran GM (1983) Inheritance of supernumerary spikelets in wheat. Euphytica 32: 767–776 [Google Scholar]
- Pont C, Murat F, Guizard S, Flores R, Foucrier S, Bidet Y, Quraishi UM, Alaux M, Doležel J, Fahima T, et al. (2013) Wheat syntenome unveils new evidences of contrasted evolutionary plasticity between paleo- and neoduplicated subgenomes. Plant J 76: 1030–1044 [DOI] [PubMed] [Google Scholar]
- Quraishi UM, Abrouk M, Bolot S, Pont C, Throude M, Guilhot N, Confolent C, Bortolini F, Praud S, Murigneux A, et al. (2009) Genomics in cereals: from genome-wide conserved orthologous set (COS) sequences to candidate genes for trait dissection. Funct Integr Genomics 9: 473–484 [DOI] [PubMed] [Google Scholar]
- Rao NN, Prasad K, Kumar PR, Vijayraghavan U (2008) Distinct regulatory role for RFL, the rice LFY homolog, in determining flowering time and plant architecture. Proc Natl Acad Sci USA 105: 3646–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma S, Salomon B, Komatsuda T (2011) The domestication syndrome genes responsible for the major changes in plant form in the Triticeae crops. Plant Cell Physiol 52: 738–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears ER. (1954) The Aneuploids of Common Wheat. University of Missouri, Columbia, MO, pp 3–58 [Google Scholar]
- Sharman BC. (1944) Branched head in wheat and wheat hybrids. Nature 153: 497–498 [Google Scholar]
- Swaminathan MS, Chopra VL, Sastry GRK (1966) Expression and stability of an induced mutation for ear branching in bread wheat. Curr Sci 35: 91–92 [Google Scholar]
- Tanaka W, Pautler M, Jackson D, Hirano HY (2013) Grass meristems II: inflorescence architecture, flower development and meristem fate. Plant Cell Physiol 54: 313–324 [DOI] [PubMed] [Google Scholar]
- Tanno K, Takeda K (2004) On the origin of six-rowed barley with brittle rachis, agriocrithon (Hordeum vulgare ssp. vulgare f. agriocrithon [Aberg] Bowd.), based on a DNA marker closely linked to the vrs1 (six-row gene) locus. Theor Appl Genet 110: 145–150 [DOI] [PubMed] [Google Scholar]
- Yang WY, Lu BR, Hu XR, Yu Y, Zhang Y (2005) Inheritance of the triple-spikelet character in a Tibetan landrace of common wheat. Genet Resour Crop Evol 52: 847–851 [Google Scholar]
- Zhang RQ, Wang XE, Chen PD (2013) Inheritance and mapping of gene controlling four-rowed spike in tetraploid wheat (Triticum turgidum L.). Acta Agron Sin 39: 29–33 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.