An iron-binding protein causes degradation of proteins involved in the iron deficiency response.
Abstract
Iron uptake and metabolism are tightly regulated in both plants and animals. In Arabidopsis (Arabidopsis thaliana), BRUTUS (BTS), which contains three hemerythrin (HHE) domains and a Really Interesting New Gene (RING) domain, interacts with basic helix-loop-helix transcription factors that are capable of forming heterodimers with POPEYE (PYE), a positive regulator of the iron deficiency response. BTS has been shown to have E3 ligase capacity and to play a role in root growth, rhizosphere acidification, and iron reductase activity in response to iron deprivation. To further characterize the function of this protein, we examined the expression pattern of recombinant ProBTS::β-GLUCURONIDASE and found that it is expressed in developing embryos and other reproductive tissues, corresponding with its apparent role in reproductive growth and development. Our findings also indicate that the interactions between BTS and PYE-like (PYEL) basic helix-loop-helix transcription factors occur within the nucleus and are dependent on the presence of the RING domain. We provide evidence that BTS facilitates 26S proteasome-mediated degradation of PYEL proteins in the absence of iron. We also determined that, upon binding iron at the HHE domains, BTS is destabilized and that this destabilization relies on specific residues within the HHE domains. This study reveals an important and unique mechanism for plant iron homeostasis whereby an E3 ubiquitin ligase may posttranslationally control components of the transcriptional regulatory network involved in the iron deficiency response.
Iron deficiency is a critical issue for most living organisms because iron is an essential component of many metabolic processes. Excess iron can be equally problematic due to its potential to react with oxygen and form damaging reactive oxygen species. Consequently, plants and animals tightly regulate their responses to iron bioavailability. Plants have evolved two primary ways to uptake iron from soil. Under low-iron conditions, grasses such as rice (Oryza sativa) and maize (Zea mays) primarily utilize the Strategy II response, whereby they release phytosiderophores into the rhizosphere that bind to ferric iron with high affinity. Phytosiderophore-ferric iron complexes are transported into the root via membrane-localized yellow stripe and yellow stripe-like (YSL) transporters (Curie et al., 2001; Inoue et al., 2009), although several studies suggest that rice is also able to directly uptake ferrous iron (Ishimaru et al., 2006; Cheng et al., 2007).
Dicots and nongraminaceous monocots utilize the Strategy I response under low-iron conditions. In Arabidopsis (Arabidopsis thaliana), the Strategy I response involves an increase in the expression and activity of proton-translocating adenosine triphosphatases (H+-ATPases), most notably AHA2, which acidifies the rhizosphere and increases the solubility of ferric iron oxides near the root epidermis (Römheld et al., 1984; Santi and Schmidt, 2009). There is also an increase in the expression and activity of the membrane-bound iron reductase FERRIC REDUCTASE OXIDASE2 (FRO2), which reduces ferric iron to ferrous iron (Robinson et al., 1999). Ferrous iron is then transported into epidermal cells via IRON-REGULATED TRANSPORTER1 (IRT1), a membrane-localized metal ion transporter that transports iron and other metal ions, including zinc, manganese, and cadmium (Eide et al., 1996; Vert et al., 2002; Colangelo and Guerinot, 2004). After uptake, iron is bound to chelators such as nicotianamine and citrate and subsequently translocated into and throughout the vasculature (Durrett et al., 2007; Curie et al., 2009). This coordinated iron deficiency response is controlled by the basic helix-loop-helix (bHLH) transcription factor FER-LIKE FE DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT; Colangelo and Guerinot, 2004). FIT acts in concert with two other bHLH proteins (Yuan et al., 2008), ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 (Lingam et al., 2011), and is posttranslationally regulated by the presence of nitric oxide via 26S proteasome-dependent degradation (Meiser et al., 2011; Sivitz et al., 2011). A FIT-independent iron homeostasis pathway has also been identified involving bHLH100 and bHLH101 (Sivitz et al., 2012). Recently, it has been shown that root exudates including phenolics and riboflavin derivatives function as iron-binding compounds in Arabidopsis and Medicago truncatula, similar to those in the chelation-based Strategy II response. The induction of phenylpropanoid and flavin pathway genes is tightly linked to core genes of the iron acquisition machinery, such as FIT, FRO2, IRT1, and AHA2, and thus appears to constitute an integral component of the Strategy I response in Arabidopsis and M. truncatula (Rodríguez-Celma et al., 2013).
We reported that the bHLH protein POPEYE (PYE) plays a critical role in iron homeostasis by positively controlling the iron deficiency response in Arabidopsis. PYE interacts with several close homologs, namely IAA-LEUCINE RESISTANT3 (ILR3), bHLH104, and bHLH115, hereafter PYE-like (PYEL) proteins. These PYELs are up-regulated during iron deficiency (Long et al., 2010), and one of them, ILR3, plays a known role in modulating auxin-conjugate hydrolysis in an iron-dependent manner, perhaps by transcriptionally controlling the expression of iron transporters (Rampey et al., 2006). Unlike PYE, all three of the PYEL proteins interact with a second protein, BRUTUS (BTS).
BTS is induced in response to low iron, and loss of BTS induction leads to increased tolerance to iron deficiency, as evident by increased root growth and rhizosphere acidification compared with wild-type plants (Long et al., 2010). BTS encodes a unique plant protein consisting of several conserved domains, including three hemerythrin (HHE) cation-binding domains located near the N terminus, a CHY zinc-finger domain, and a Really Interesting New Gene (RING) domain near the C terminus (Long et al., 2010; Kobayashi et al., 2013). The conserved HHE domains are left-twisted 4-α-helical bundles that provide a hydrophobic pocket in which O2 binds diiron. Iron is typically coordinated within the HHE domain via the carboxylate side chains of a Glu, an Asp, and five His residues, but other means of iron coordination have been described (Holmes et al., 1991; Holmes and Stenkamp, 1991; Shu et al., 2012). The presence of a RING domain near the BTS C terminus suggests that it may have E3 ligase capability. BTS and the BTS orthologs in rice, Oryza sativa Hemerythrin motif-containing RING and Zinc-Finger Protein1 (OsHRZ1) and OsHRZ2, were shown to bind iron at the HHE domains and have ubiquitination capacity (Kobayashi et al., 2013). However, no study has examined whether the iron-responsive PYEL proteins that BTS interacts with are targeted by this E3 ligase activity. We hypothesize that BTS negatively regulates iron homeostasis by targeting PYEL transcription factors for 26S proteasome-mediated degradation via its RING (E3 ligase) domain. This activity is dependent on alterations in BTS stability, which is mediated by the HHE domains and therefore responsive to fluctuations in iron availability.
We explored BTS expression in different tissues throughout plant development and analyzed the implications of altered BTS expression on the iron deficiency response and on reproductive growth. We also examined the capacity of BTS to bind iron through the HHE domains. We demonstrate that iron affects the stability of BTS in a concentration-dependent manner and that specific residues within the HHE domains are critical for protein stability. We also found that the interaction between BTS and PYEL proteins relies on the presence of the RING domain and that BTS facilitates 26S proteasome-mediated degradation of at least two PYEL proteins. Taken together, our results show that BTS plays an important role in maintaining iron homeostasis in plants, potentially by regulating bHLH transcription factors.
RESULTS
BTS Is Expressed throughout Plant Development
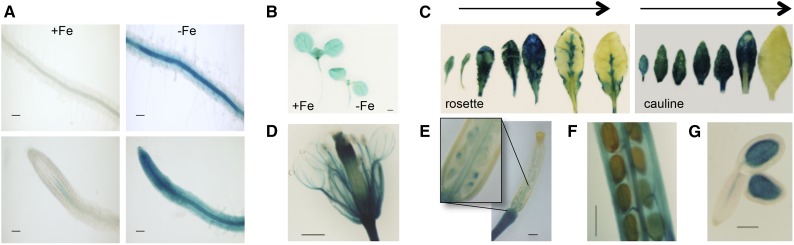
BTS is induced in response to iron deficiency in the root vasculature according to microarray analysis and confocal microscopy of lines expressing ProBTS::GFP (Long et al., 2010). To verify BTS expression throughout growth and development, we generated wild-type plants expressing ProBTS::GUS. Similar to ProBTS::GFP lines (Long et al., 2010), a strong GUS signal was detected in the root vasculature in response to iron deficiency (Fig. 1A), further validating the up-regulation of BTS under iron deficiency. Microarray analysis has revealed that apart from roots, BTS is also expressed in shoots in Arabidopsis (Sivitz et al., 2012). To characterize BTS expression in the shoot and during reproductive growth, we performed GUS staining on young seedlings and mature plants. GUS activity was observed in cotyledons of seedlings regardless of iron availability when grown on medium (Fig. 1B). When ProBTS::GUS plants were grown on soil, GUS expression was observed in both rosette and cauline leaves, with highest expressions in young leaves (Fig. 1C; Supplemental Fig. S1A). We further examined BTS expression in reproductive tissues of ProBTS::GUS-expressing wild-type plants and found that it is expressed in floral vasculature, developing and mature embryos, funiculus, septum, and gynoecium valves (Fig. 1, D–G; Supplemental Fig. S1, B–D).
Figure 1.
BTS expression throughout plant development. A and B, GUS activity in roots (A) and shoots (B) of 7-d-old wild-type ecotype Columbia (Col-0) seedlings grown on +Fe medium for 4 d and then transferred to ±Fe medium for 3 d expressing ProBTS::GUS. The results shown are representative of three independent lines. C, GUS activity in leaves from 4-week-old soil-grown plants. Arrows indicate youngest to oldest. D to G, GUS activity in flower (D), young silique (E), mature green silique (F), and mature embryo (G). Bars = 100 μm (A), 5 mm (B–F), and 2 mm (G).
BTS Controls Root Iron Responsiveness and Embryo Development
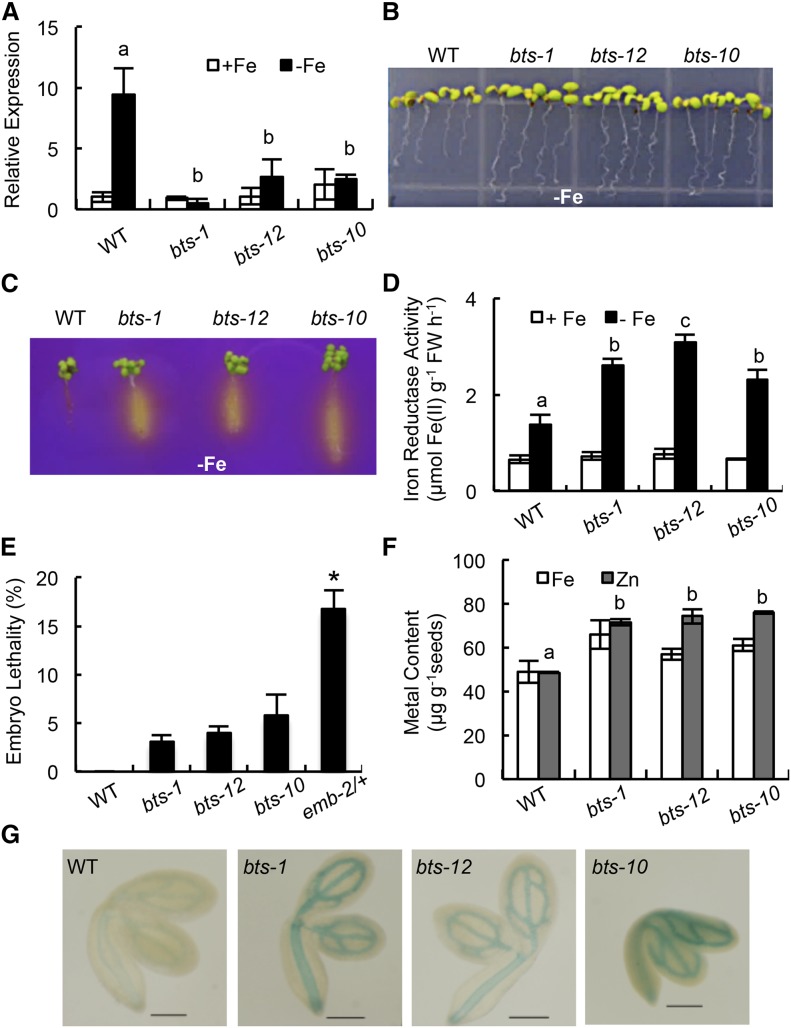
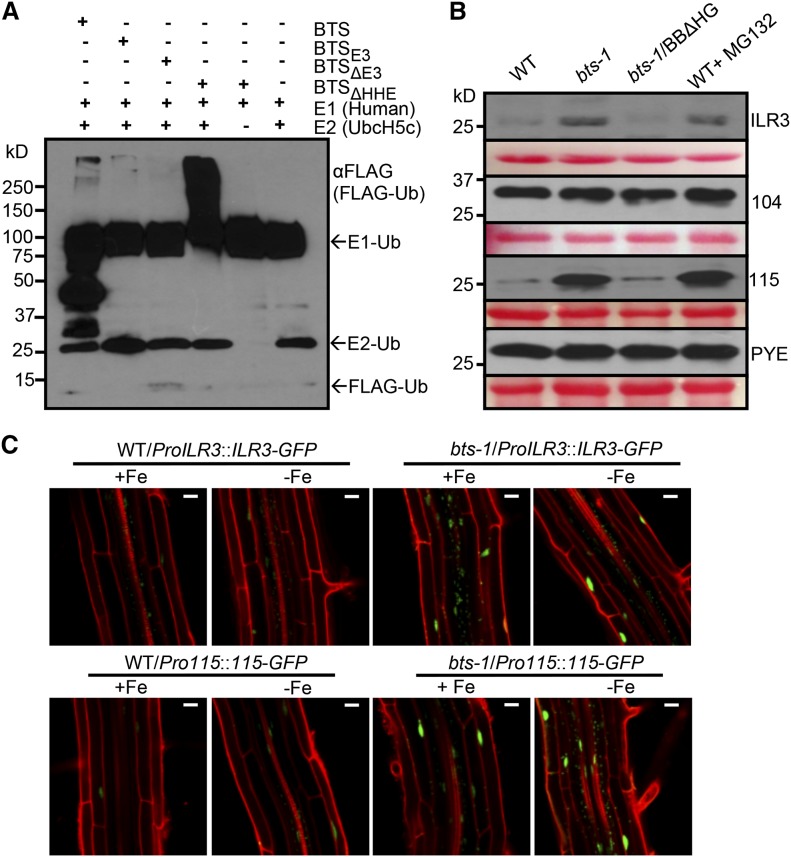
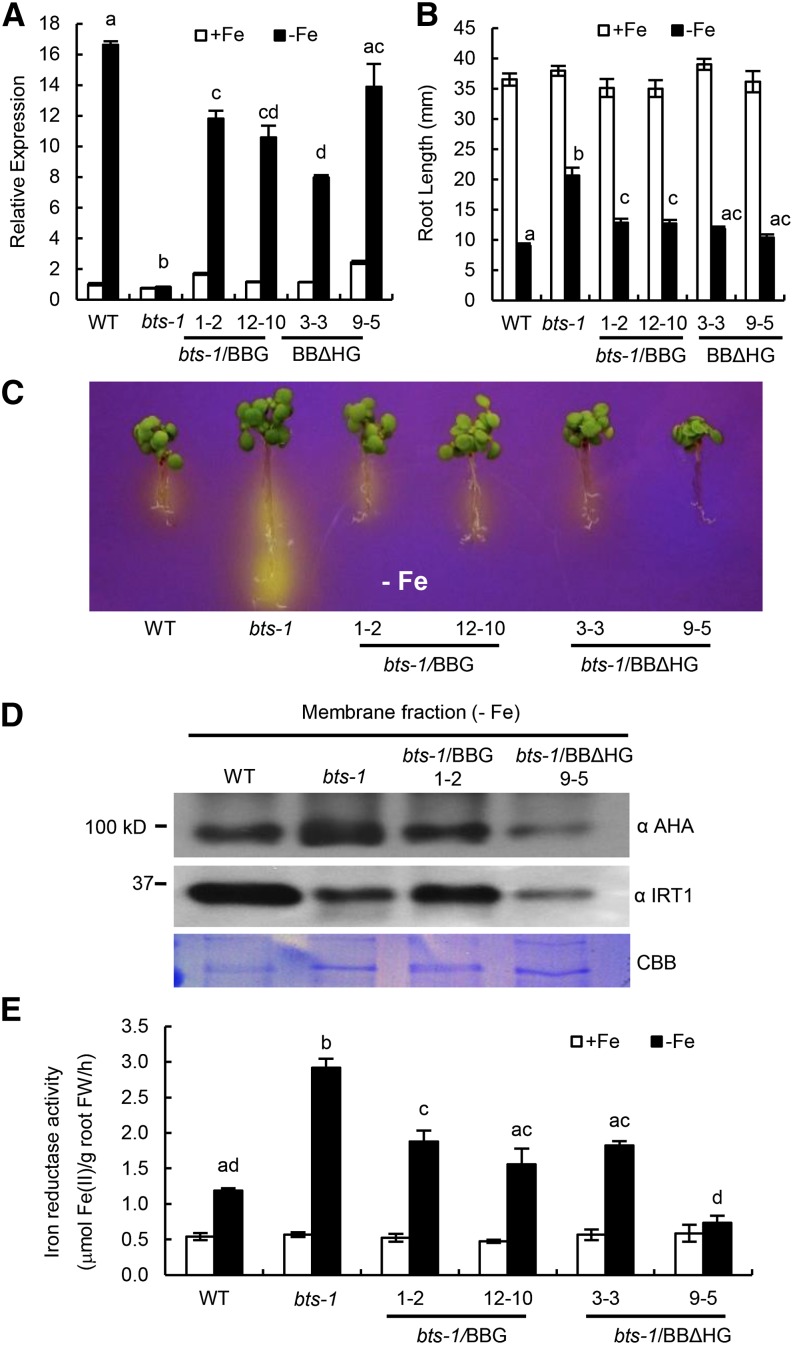
Putative BTS orthologs play diverse roles in other plant species, including inhibition of Tobacco mosaic virus infection in Nicotiana tabacum (Yamaji et al., 2010) and root nodulation in Lotus japonicus (Shimomura et al., 2006). Furthermore, BTS is critical for embryogenesis, as evidenced from the embryonic lethal bts alleles emb2454-1 and emb2454-2 (emb-2/+; McElver et al., 2001; Tzafrir et al., 2004). Because emb (–/–) lines are embryonic lethal, we analyzed the iron deficiency response of several other bts alleles resulting from transfer DNA (T-DNA) insertions in the BTS promoter and 5′ untranslated region sequence (Supplemental Fig. S2). These alleles exhibit loss of transcript induction under iron-deficient conditions but maintain expression levels similar to wild-type under iron sufficiency (Fig. 2A). The loss of BTS induction in homozygous bts-1, bts-12, and bts-10 alleles is correlated with increased root elongation, rhizosphere acidification, and iron reductase activity under iron deficiency compared with wild-type plants (Fig. 2, B–D; Supplemental Fig. S3, A and B). These data support the hypothesis that BTS negatively regulates key responses to iron deprivation, as well as overall growth (Long et al., 2010).
Figure 2.
BTS affects root iron response and embryogenesis. A, Relative BTS expression in root tissue from 7-d-old seedlings grown on +Fe medium for 4 d and then transferred to ±Fe medium for 3 d. Error bars indicate ± se of the mean (n = 4), and columns with different letters are significantly different from each other (P < 0.05). B, Root growth of 11-d-old seedlings grown on –Fe medium (4 d +Fe and 7 d –Fe). C, Rhizosphere acidification of 8-d-old seedlings grown on –Fe medium (4 d +Fe, 3 d –Fe, and 1 d bromocresol purple). Eight plants per genotype were grouped on bromocresol purple agar medium. Results shown represent four independent assays. D, Iron reductase activity of 10-d-old seedlings grown on ±Fe medium (7 d +Fe and 3 d ±Fe). Error bars indicate ± se of the mean (n = 4), and columns with different letters are significantly different from each other (P < 0.05). E, Embryo lethality in developing siliques. Error bars indicate ± se of the mean (n = 5). The asterisk indicates significant difference from the wild type (WT; P < 0.05). F, Iron and zinc content in seeds (150 mg per replicate) analyzed by ICP-OES. Error bars indicate ± se of the mean (n = 3), and columns with different letters are significantly different from each other (P < 0.05) G, Iron (Fe3+) localization in germinating seeds visualized by Perls staining. Results shown represent 10 independent assays. Bars = 2 mm. FW, Fresh weight.
Because the emb2/+ allele of BTS exhibits elevated embryonic lethality (Fig. 2E), we quantified embryo lethality in the homozygous bts alleles. Approximately 3% to 6% of embryos from bts-1, bts-12, and bts-10 alleles were defective compared with approximately 20% of embryos from emb-2/+ (Fig. 2E). This finding indicates that BTS is expressed in the shoot and plays a critical role during embryo development, perhaps by affecting iron mobilization into developing siliques and embryos. Inductively coupled plasma (ICP)-optical emission spectrometry (OES) analysis of iron content from bts-1 seedling shoots and roots revealed slight increases in iron content under both iron-replete and -deplete conditions (Supplemental Fig. S3C). Seeds showed overall increased accumulation of iron (16%–34%) and zinc (20%–60%) compared with wild-type seeds (Fig. 2F). The relatively higher content of iron in bts mutant seeds, although not statistically significant, was further validated by histochemical analysis of iron localization in germinating seeds by Perls staining, which revealed enhanced iron accumulation in the bts mutants compared with the wild type (Fig. 2G).
PYELs Interact with BTS in the Nucleus
There are 133 bHLH genes within the Arabidopsis genome grouped into 12 subfamilies (Heim et al., 2003). According to yeast (Saccharomyces cerevisiae) two-hybrid (Y2H) analysis, BTS does not interact with the group IVb iron homeostasis bHLH protein PYE. However, it does physically interact with group IVc PYEL proteins ILR3, bHLH104, and bHLH115 (Long et al., 2010). ILR3, bHLH104, and bHLH115 are transcriptionally induced under iron-deficient conditions in wild-type roots (Dinneny et al., 2008). Moreover, microarray analysis indicates that ILR3 directly or indirectly targets genes involved in metal ion homeostasis (Rampey et al., 2006). Loss of ILR3 function leads to increased sensitivity to indole-3-acetic acid (IAA)-Leu in an iron-dependent manner, leading to the theory that ILR3 controls metal-dependent IAA-conjugate hydrolysis by controlling the expression of iron transporters (Rampey et al., 2006).
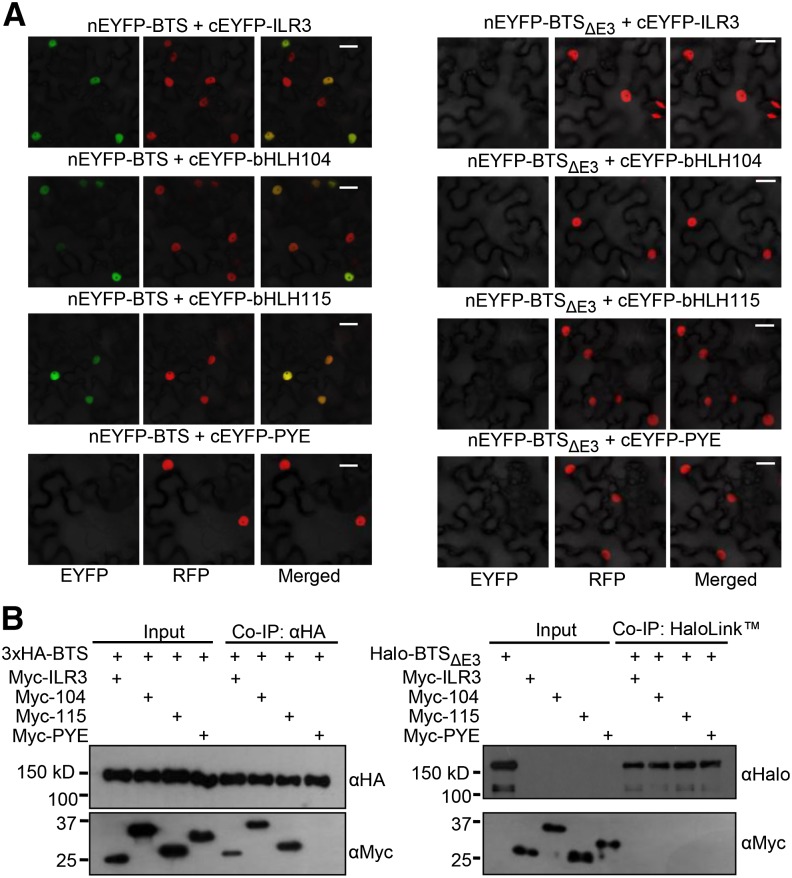
We hypothesize that PYEL proteins ILR3, bHLH104, and bHLH115 may be downstream targets of BTS. Transient expression of GFP fusions of BTS, PYE, and PYEL proteins using the constitutive 35S promoter in Nicotiana benthamiana leaves suggests that these proteins are colocalized in the nucleus (Supplemental Fig. S4A). We used bimolecular fluorescence complementation (BiFC) assays to further assess the interaction between BTS and the PYEL proteins. BiFC assays were conducted by transiently coexpressing proteins using the constitutive 35S promoter in N. benthamiana leaf epidermal cells. In planta, BTS interacted with ILR3, bHLH104, and bHLH115 but not PYE (Fig. 3A). These interactions occur in the nucleus and are dependent upon the presence of the RING domain (E3 ligase), as is evident from the loss of these interactions with BTSΔE3 deletion protein (Fig. 3A; Supplemental Fig. S5). This finding suggests that the BTS RING domain may act to facilitate its interaction with PYEL proteins but does not exclude the possibility that loss of the RING domain affects the folding of BTS, disrupting its capacity to interact with PYEL proteins. BiFC assay results were validated through in vitro coimmunoprecipitation (Co-IP) studies. Epitope-tagged fusions of BTS, BTSΔE3, PYE, and PYEL proteins were in vitro translated using a wheat (Triticum aestivum) germ extract system, and Co-IP assays were performed between interacting proteins. ILR3, bHLH104, and bHLH115 coimmunoprecipitated with full-length BTS but not BTSΔE3 (Fig. 3B), further supporting the BiFC results.
Figure 3.
Interaction of BTS with PYEL depends on the presence of the RING (E3 ligase) domain. A, BiFC assay showing in planta interactions of ILR3, bHLH104, bHLH115, and PYE with BTS and BTSΔE3 deletion protein in leaf epidermal cells. Indicated proteins are transiently coexpressed in red fluorescent protein (RFP)-Histone2B (nuclear marker) transgenic N. benthamiana plants. Enhanced YFP (EYFP) fluorescence indicates interaction between proteins in the nucleus. All interactions were tested using both combinations of N-/C-EYFP-fused reciprocal proteins. Results shown represent three independent assays. B, Immunoblot analysis showing Co-IP of in vitro-translated Myc-tagged ILR3, bHLH104, and bHLH115 by 3xHA-tagged BTS (full length) but not by Halo-tagged BTSΔE deletion protein. Results shown represent two independent assays. Bars = 20 μm.
PYE and PYELs Form Heterodimer Complexes in Planta
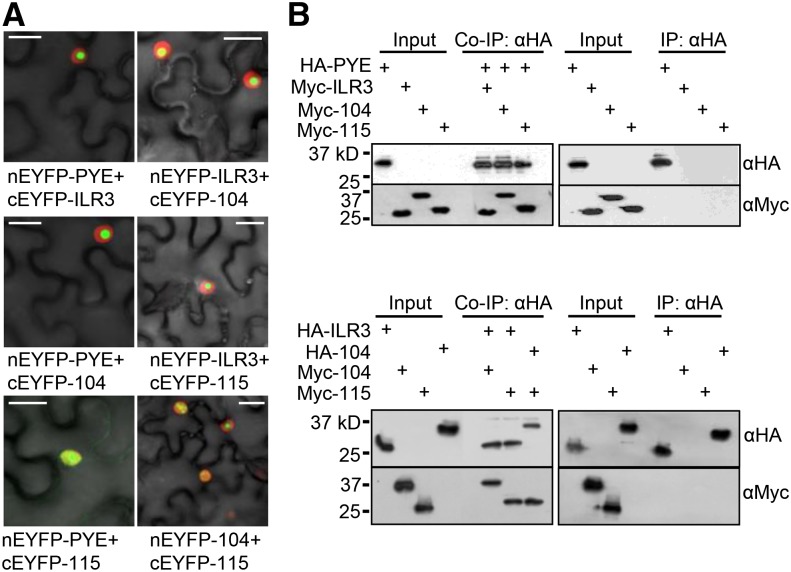
bHLH transcription factors within a subgroup are thought to share very similar or related functions by making homo- or heterodimer complexes with their family members (Heim et al., 2003). Previously, we identified interactions between PYE and two of its family members, ILR3 and bHLH115, using Y2H analysis (Long et al., 2010). We validated Y2H interactions in planta using BiFC assays in N. benthamiana leaves, confirming that PYE forms heterodimer complexes with ILR3, bHLH115, and bHLH104 in the nucleus (Fig. 4A). We also tested for interactions among PYEL proteins and observed that ILR3 interacts with both bHLH104 and bHLH115 and that bHLH104 interacts with bHLH115 in the nucleus (Fig. 4A). None of these bHLH proteins form homodimer complexes (Supplemental Fig. S6). BiFC results were confirmed by Co-IP assays using in vitro-translated epitope-tagged bHLH transcription factors PYE and PYEL (Fig. 4B).
Figure 4.
PYE and PYEL proteins form heterodimers in the nucleus. A, BiFC assay showing in planta interactions between PYE, ILR3, bHLH104, and bHLH115 proteins in the epidermal cells. Indicated proteins are transiently coexpressed in RFP-Histone2B (nuclear marker) transgenic N. benthamiana plants. EYFP fluorescence indicates interaction between indicated proteins. All interactions were tested using both combinations of N-/C-EYFP-fused reciprocal proteins. Results shown represent three independent assays. B, Immunoblot analysis showing Co-IP of in vitro-translated HA-tagged PYE, ILR3, or bHLH104 and Myc-tagged ILR3, bHLH104, and bHLH115 proteins. Results shown represent two independent assays. Bars = 20 μm.
BTS Targets PYELs for 26S Proteasome-Mediated Degradation
The interactions of BTS with PYEL proteins led us to speculate that BTS regulates its interacting proteins via RING (E3 ligase) domain-mediated ubiquitination and subsequent 26S proteasome-mediated degradation. Previously, BTS and the BTS rice orthologs OsHRZ1 and OsHRZ2 were shown to exhibit some level of ubiquitination capacity (Kobayashi et al., 2013). The E3 ligase activity of BTS protein and its derivatives (Supplemental Fig. S5) were tested through an in vitro ubiquitination assay using purified in vitro-translated proteins obtained from wheat germ extract. Our results indicate that BTS has intrinsic ubiquitin ligase activity. This ligase activity relies on the presence of the E3 domain, as evident from the loss of ubiquitination capacity of truncated BTSΔE3 protein (Fig. 5A). Along with full-length BTS, the truncated BTSΔHHE protein is also capable of E3 ligase-mediated ubiquitination in an E1- and E2-dependent manner (Fig. 5A). The E3 ligase activity of BTS, BTSE3, and BTSΔHHE, as shown by immunoblots under nonreducing conditions, generated a high Mr (>100 kD) smear of ubiquitinated proteins. The presence of several prominent bands below 100 kD in the lane containing BTS is consistent with the observation that full-length BTS is less stable than BTSΔHHE protein (Fig. 5A; Supplemental Fig. S8E).
Figure 5.
BTS targets ILR3 and bHLH115 for 26S proteasome-mediated degradation. A, E3 ligase activity of BTS and its derivatives. In vitro-translated and purified 3xHA-tagged BTS, BTSE3 domain, and BTSΔE3 and BTSΔHHE deletion proteins were subjected to in vitro ubiquitination assay containing ATP, FLAG-tagged ubiquitin (Ub), and human E1 and E2 (UbcH5c). Proteins were immunodetected using anti-FLAG antibody. Results shown represent two independent assays. B, Cell-free 26S proteasome-mediated degradation of PYEL proteins by BTS and BTSΔHHE. In vitro-translated and purified Myc-tagged ILR3, bHLH104, bHLH115, and PYE proteins were incubated with cell-free protein extracts prepared from 7-d-old seedlings of the wild type (WT), bts-1, and bts-1/ProBTS::BTSΔHHE-GFP (BBΔHG) grown on –Fe medium (4 d +Fe and 3 d –Fe). Reactions were performed with and without 160 µm MG132 (proteasome inhibitor). Proteins were immunodetected using anti-Myc antibody. Ponceau S staining indicates equal amount of cell-free protein extract loaded. C, Stability of ILR3-GFP and bHLH115-GFP in wild-type and bts-1 seedlings expressing either ProILR3::ILR3-GFP (top) or Pro-115::115-GFP (bottom) in roots. Plants were grown for 6 d on ±Fe medium (4 d +Fe and 2 d ±Fe). Results shown represent two independent assays. Bars = 20 μm.
Next, we investigated the physiological relevance of BTS-PYEL interactions within the context of BTS E3 ligase activity. We were unable to detect either native or recombinant GFP fusion of PYEL and PYE proteins in Arabidopsis seedlings by immunoblot analysis and therefore could not test whether BTS facilitates ubiquitination of PYEL proteins in planta. Using a cell-free degradation assay (Wang et al., 2009), we tested whether BTS plays a role in the 26S proteasome-mediated degradation of PYEL and PYE proteins in vitro. For the assay, Myc-tagged PYEL and PYE proteins were purified from in vitro translation reactions. The purified proteins were quantified, equally distributed, and incubated with cell-free extracts prepared from wild-type and bts-1 seedlings grown under iron deficiency. After treatment, the effect of endogenous BTS on PYEL and PYE protein content was assessed by immunoblot analysis. ILR3 and bHLH115 protein levels are stable when incubated with bts-1 extract, but both proteins appear to be degraded when incubated with wild-type extract (Fig. 5B). Accordingly, protein stability of ILR3 and bHLH115 in bts-1 extract are relatively similar to those in wild-type extract supplemented with MG132 (N-(benzyloxycarbonyl)leucinylleucinylleucinalZ-Leu-Leu-Leu-al), a specific 26S proteasome inhibitor. Differences in protein stability were not observed with bHLH104 and PYE proteins. Overall, these data provide evidence that BTS is involved in the 26S proteasome-mediated degradation of ILR3 and bHLH115. These results were confirmed in planta by expressing translational GFP fusions to ILR3 (ProILR3::ILR3-GFP) and bHLH115 (Pro-115::115-GFP) in both wild-type and bts-1 plants. Confocal microscopy analysis of roots after 2 d of exposure to iron deficiency revealed enhanced fluorescence associated with ILR3-GFP and bHLH115-GFP in the bts-1 mutant under iron-sufficient as well as iron-deficient conditions (Fig. 5C). The dramatic differences in protein accumulation between wild-type and bts-1 backgrounds could be due to higher stability of the PYEL proteins in presence of low levels of BTS in roots of bts-1 rather than modest transcriptional changes (Supplemental Fig. S7). Our in vitro data shows that BTS has ubiquitination capacity through the E3 domain (Fig. 5A), which is also critical for the physical interaction of BTS with two PYEL proteins (Fig. 3).
HHE Domains Bind Iron and Affect BTS Protein Stability
BTS is structurally and functionally similar to the essential mammalian protein F-box and leucine-rich repeat protein5 (FBXL5). FBXL5 contains a single HHE domain near the N terminus of the protein and assembles into the SKP1-CUL1-F-box-protein [(SCF)FBXL5] SCFFBXL5 E3 ligase complex that interacts with iron homeostasis proteins, facilitating their ubiquitin-mediated degradation (Salahudeen et al., 2009; Vashisht et al., 2009). In the absence of iron, conformational changes in FBXL5’s HHE domain lead to the degradation of FBXL5 and a subsequent increase in overall iron uptake and metabolism (Salahudeen et al., 2009; Ruiz et al., 2013). Deletion of the FBXL5 HHE domain leads to constitutive accumulation of the truncated protein, while iron and oxygen binding to the HHE domain cause increased FBXL5 protein accumulation (Salahudeen et al., 2009).
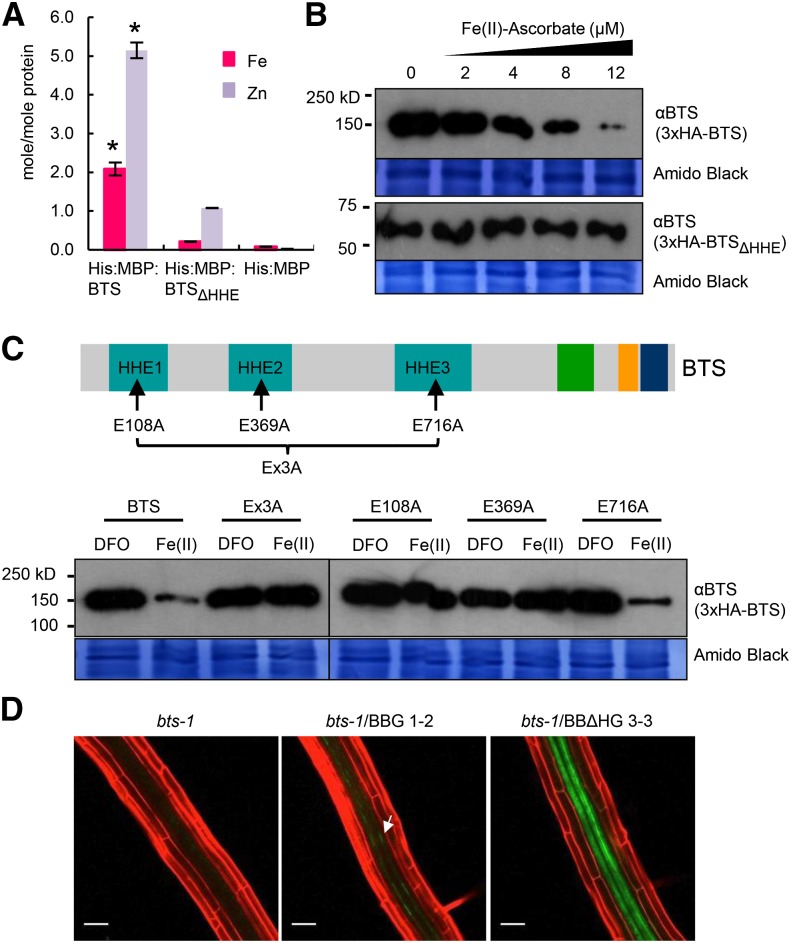
Notably, both BTS and the BTS rice orthologs OsHRZ1 and OsHRZ2 bind iron and zinc (Kobayashi et al., 2013), and deletion of the entire first two-thirds of the OsHRZ1 protein region containing the HHE domains decreases iron as well as zinc binding of the truncated OsHRZ1 and OsHRZ2 protein (Kobayashi et al., 2013). We hypothesized that iron binding at the HHE domains could cause a conformational change in BTS structure and activity similar to that exhibited by FBXL5. We measured iron and zinc content of BTS and BTSΔHHE, which lacks all three HHE domains but maintains the N-terminal end of the protein (Supplemental Fig. S5). Recombinant His:Maltose-Binding Protein (His:MBP) fusions to BTS and BTSΔHHE as well as His:MBP alone were expressed in Escherichia coli, purified, and subjected to metal ion analysis. ICP-mass spectrometry (MS) analysis of the recombinant protein revealed that each molecule of protein binds two molecules of iron, which is consistent with iron binding through one HHE domain (Fig. 6A). Deletion of the three HHE domains resulted in an approximately 10-fold reduction in the amount of iron associated with each molecule of protein (Fig. 6A). These results are consistent with the amount of iron associated with BTS rice ortholog OsHRZ1 and the OsHRZ1ΔH constructs described by Kobayashi et al. (2013). Residual iron present in BTSΔHHE protein samples might be due to the presence of other cation-binding sites such as the CHY zinc-finger domain, which is capable of binding other metals, including iron (Besold et al., 2010). The full-length BTS protein also contained significant amounts of zinc metal ion, and deletion of HHE domains resulted in a 5-fold reduction in zinc content associated with BTSΔHHE protein (Fig. 6A). This indicates that BTS, similar to its rice orthologs OsHRZ1 and OsHRZ2 (Kobayashi et al., 2013), may bind both iron and zinc.
Figure 6.
BTS binds iron through HHE domains, negatively affecting BTS protein stability. A, Iron and zinc content of His:MBP fusions of BTS and HHE deleted (BTSΔHHE) proteins or His:MBP expressed in E. coli and purified. Error bars indicate ± se of the mean (n = 3 for His:MBP and His:MBP:BTS and n = 2 for His:MBP:BTSΔHHE). Asterisks indicate significant difference from BTSΔHHE and His:MBP (P < 0.05). B, In vitro translation of 3xHemagglutinin (3xHA)-tagged BTS and BTSΔHHE proteins performed in the presence of increasing concentration of ferrous iron (Fe(II)-ascorbate). Proteins were immunodetected using anti-BTS antibody. Amido Black staining indicates equal amount of wheat germ extract were loaded from in vitro protein translations. Results shown represent two independent assays. C, In vitro translation of 3xHA-tagged wild-type BTS, single amino acid substitutions (E108A, E369A, and E716A), and triple amino acid substitution (Ex3A) BTS proteins were performed in presence of ferrous iron chelator (100 μm DFO) and ferrous iron [12 μm Fe(II)-ascorbate]. Proteins were immunodetected using anti-BTS antibody. Amido Black staining indicates that equal amounts of wheat germ extract were loaded from in vitro protein translations. Results shown represent two independent assays. D, In planta stability of BTSΔHHE protein. Confocal microscopy images of roots of 7-d-old (4 d +Fe and 3 d –Fe) bts-1 seedlings expressing ProBTS::BTS-GFP (BBG, arrows) and ProBTS::BTSΔHHE-GFP (BBΔHG) stained with propidium iodide (red). Results shown represent four independent assays. Scale bars = 50 μm.
We investigated the effect of iron on BTS stability in planta by Agrobacterium tumefaciens-mediated transient expression of BTS-Enhanced GFP (EGFP) protein in N. benthamiana leaves. Protein expression was driven under the constitutive 35S promoter, and leaves were infiltrated with either an iron source (400 µm ferric ammonium citrate [FAC]) or a ferrous iron chelator (400 µm deferoxamine-B [DFO]). The presence of excess iron caused a visible decrease in BTS-EGFP signals, while the chelation of ferrous iron led to significantly higher visual BTS-EGFP signals (Supplemental Fig. S8A). We were unable to detect BTS protein under any condition in the native system by immunoblot analysis, and there are no previous reports of immunodetection of the protein in the native system in the literature. Therefore, to determine whether the presence of iron affects BTS stability, we translated 3xHA-tagged BTS and BTSΔHHE proteins in vitro in the presence of increasing concentrations of ferrous iron [0–12 µm Fe(II)-ascorbate] using a wheat germ extract system. Using an antibody that recognizes both BTS- and BTSΔHHE-deleted proteins, we determined that the levels of full-length BTS protein accumulation were dramatically decreased in a dose-dependent manner with the inclusion of Fe(II)-ascorbate compared with the control (Fig. 6B). Similar results were obtained with ferric ammonium citrate (FAC), but significantly more FAC was needed to achieve the same effect (Supplemental Fig. S8B). BTS protein accumulation was not affected in the presence of ferrous iron chelators DFO and ferrozine (Supplemental Fig. S8B). It has been previously reported that wheat germ extract has negligible 26S proteasome activity (Takahashi et al., 2009). However, in vitro translation reactions performed in the presence and absence of MG132 produced no changes with respect to BTS accumulation (Supplemental Fig. S8B), indicating that MG132 alone also has no effect on BTS stability. While the nucleus is known to contain high concentrations of iron (Roschzttardtz et al., 2011a), we note that the physiological effects of high concentrations of iron on BTS accumulation should be viewed with caution, as the exact concentration of iron that accumulates within cellular compartments under varying iron conditions is unknown. Importantly, the levels of truncated BTSΔHHE protein accumulation were unaffected by the presence of an iron source and appear identical after control and Fe(II)-ascorbate, FAC, and iron chelator treatments (Fig. 6B; Supplemental Fig. S8B). The iron status also had no effect on in vitro-translated bHLH transcription factors PYE and PYEL (Supplemental Fig. S8B), suggesting that the observed effect of iron on BTS is specific and addition of excess iron or iron chelators does not alter the Salmonella typhimurium phage6 (SP6) promoter-mediated in vitro transcription or translation of proteins using wheat germ extract.
Because the HHE domains are required for iron binding and deletion of the HHE domains corresponds to increased accumulation of truncated BTS (Fig. 6D; Supplemental Fig. S8E), we hypothesized that binding of iron through these domains decreases the stability of BTS and conversely, in the absence of iron, BTS is stabilized and able to accumulate. A point mutation in the mammalian FBXL5 HHE domain resulting in substitution of an iron-coordinating Glu to Ala (E61A) eliminates FBXL5 iron-binding capacity and prevents accumulation (Salahudeen et al., 2009). Amino acid sequence alignment of the FBXL5 and BTS HHE domains revealed conserved Glu residues in the same position as the FBXL5 E61 in all three BTS HHE domains (Supplemental Fig. S8C). To test whether these three conserved residues were critical for the stability of BTS, we in vitro translated recombinant 3xHA-BTS proteins containing single amino acid substitutions (E108A, E369A, and E716A) and BTS protein containing all three amino acid substitutions (Ex3A) using a wheat germ system in the presence of Fe(II)-ascorbate, FAC, and DFO (Fig. 6C; Supplemental Fig. S8D). The E108A and E369A substitutions in BTS HHE domains 1 and 2, respectively, as well as the combination of the three-amino acid substitution (Ex3A) resulted in proteins that were unaffected by the presence of the iron source [Fe(II)-ascorbate and FAC] or iron chelator (DFO; Fig. 6C; Supplemental Fig. S8D). Substitution of E716A in HHE domain 3, however, resulted in protein content similar to that of wild-type BTS, suggesting instability in the presence of the iron source (Fig. 6C; Supplemental Fig. S8D). These results indicate that E108 and E369 are critical amino acid residues for iron-dependent BTS instability and may further explain the inability to detect full-length BTS protein in plant extracts by immunoblot analysis. The Ex3A, E108A, and E369A point mutations are unlikely to cause the observed increased protein accumulation of BTS (compared with the wild type) due to increased in vitro protein translation in the presence of physiologically relevant levels of iron. Moreover, because transcript synthesis in the in vitro translation system is driven by a constitutive promoter, it is unlikely the alterations observed in protein accumulation result from changes in levels of transcription but more likely reflect actual changes in protein stability in response to iron binding.
The HHE Domain Mediates BTS Accumulation and Function in Planta
To further verify the role of the HHE domains in planta, we generated bts-1 lines expressing ProBTS::BTS-GFP (bts-1/BBG) and bts-1 lines expressing ProBTS::BTSΔHHE-GFP (bts-1/BBΔHG), a truncated version of BTS lacking the HHE domains (Supplemental Fig. S5). We exposed these plants to both iron-replete and -deplete conditions. Relative transcript levels of BTS were determined for the wild type, bts-1, and the complemented lines. Both of the constructs used to complement bts-1 are capable of being induced under iron deficiency, but only BBΔHG line 9-5 expresses at a level that does not differ significantly from the wild type (Fig. 7A).
Figure 7.
Complementation of bts-1 mutant by ProBTS::BTS-GFP (BBG) and ProBTS::BTSΔHHE-GFP (BBΔHG). A, Relative BTS expression in root tissue of 7-d-old seedlings grown on +Fe medium for 4 d and then transferred to ±Fe medium for 3 d. Error bars indicate ± se of the mean (n = 4), and columns with different letters are significantly different from each other (P < 0.05). B, Root length of 11-d-old seedlings grown on ±Fe medium (4 d +Fe and 7 d ±Fe). Error bars indicate ± se of the mean (n = 32), and columns with different letters are significantly different from each other (P < 0.05). C, Rhizosphere acidification of 8-d-old seedlings grown on –Fe medium (4 d +Fe, 3 d –Fe, and 1 d bromocresol purple). Eight plants per genotype were grouped on bromocresol purple agar medium. Results shown represent four independent assays. D, Levels of AHA and IRT1 proteins in membrane fractions isolated from the roots of wild-type (WT), bts-1 mutant, and BBG and BBΔHG complemented lines. Coomassie Brilliant Blue (CBB) R-250 staining shows equal loading of protein. Results shown represent two independent assays. E, Iron reductase activity of 10-d-old seedlings grown on ±Fe medium (7 d +Fe and 3 d ±Fe). Error bars indicate ± se of the mean (n = 4), and columns with different letters are significantly different from each other (P < 0.05). FW, Fresh weight.
BTS-GFP protein is barely detected by confocal microscopy and undetectable by immunoblot analysis irrespective of iron status (Fig. 6D; Supplemental Fig. S8, E and F). However, lines expressing BBΔHG accumulate the truncated protein under both conditions, with substantially more accumulation under iron deficiency in both the vascular nucleus and cytosol (Fig. 6D; Supplemental Fig. S8, E and F). Similar localization is also seen in N. benthamiana leaf epidermal cells transiently expressing GFP fusions of BTS lacking the HHE domains (BTSΔHHE; Supplemental Figs. S4B and S5). Note that the increased protein accumulation of BBΔHG compared with BBG in Arabidopsis roots is not due to differences in transcript accumulation between these two lines (Fig. 7A), suggesting that accumulation of BBΔHG is posttranscriptionally regulated.
To verify that both BBG and BBΔHG constructs are biologically relevant, we examined bts-1 lines expressing BBG and BBΔHG for complementation of the mutant phenotypes observed in bts-1 (Fig. 2, A–D). Relative to the bts-1 mutant, all of the BBG and BBΔHG lines mostly complement root length, rhizosphere acidification, and iron reductase activity to wild-type levels (Fig. 7, B, C, and E; Supplemental Fig. S9), with BBΔHG line 9-5 (Fig. 7A) complementing most fully to or beyond wild-type levels. Rhizosphere acidification levels correspond to the accumulation of AHA protein in the root membrane fractions derived from plants after transfer to iron-depleted medium (Fig. 7D). Immunoblot analysis also revealed lower IRT1 protein level in bts-1, with bts-1/BBG line 1-2 exhibiting recovery of IRT1 protein levels similar to wild-type levels (Fig. 7D). bts-1/BBΔHG 9-5 plants exhibited decreased levels of IRT1 protein compared with both the wild type and bts-1. Such a response could be due to the complex transcriptional and posttranslational regulation of IRT1 (Barberon et al., 2011), which may be indirectly affected by other alterations in iron homeostasis caused by increased stability of BTS. Overall, these results suggest that the artificial stability of BBΔHG is able to complement many aspects of the bts-1 phenotype, likely as a result of increased BTS protein accumulation and functional E3 ligase activity. This is further supported by the result that cell-free degradation assays using a cell-free extract from bts-1/BBΔHG 9-5 complemented lines resulted in degradation of ILR3 and bHLH115 proteins similar to the wild type (Fig. 5B).
DISCUSSION
In this study, we further characterize the role of BTS in iron homeostasis in Arabidopsis. We provide evidence that BTS functions as an iron-binding protein that regulates the accumulation of PYEL bHLH transcription factors in response to iron deficiency. Because these proteins form heterodimers with PYE, we propose that the interaction between BTS and the PYEL proteins could mediate PYE activity and, thus, response to iron deprivation, including rhizosphere acidification, iron reductase activity, and iron content in plants. The finding that accumulation of the PYEL protein ILR3 controls iron availability for auxin homeostasis (Rampey et al., 2006) also provides evidence that BTS could mediate iron homeostasis through interaction with other PYEL proteins, independent of PYE. Further analysis of ILR3, bHLH115, and bHLH104 within the context of iron homeostasis is needed to determine how their interplay with PYE, BTS, and each other is involved in metal ion homeostasis.
Disruption of BTS leads to increased root elongation compared with wild-type plants (Fig. 2B; Supplemental Fig. S3A). Mutations in BTS also result in increased levels of embryo lethality (Fig. 2E; McElver et al., 2001; Tzafrir et al., 2004). This suggests that BTS plays an important role in certain aspects of plant growth and development, which corresponds with BTS expression in roots and reproductive tissues (Figs. 1 and 2). The effect of loss of BTS function on embryonic development (Fig. 2E) may be due to perturbations in the regulation of iron translocation into developing siliques and embryos, as is evident from relatively high iron content in bts seeds (Fig. 2, F–G). The existence of ferritin-mediated checkpoints for iron transport from hull to the seed and proper iron storage in the seeds is critical for healthy embryo development (Ravet et al., 2009). Several reports have indicated the importance of proper loading of metal ions into seeds for successful plant reproduction. The role of YSL (AtYSL1 and AtYSL3) and the oligopeptide transporter (AtOPT3) protein family in iron translocation to developing embryo and into the seed coat is critical for reproductive growth (Le Jean et al., 2005; Stacey et al., 2008; Chu et al., 2010; Mendoza-Cózatl et al., 2014; Zhai et al., 2014). Additionally, the role of Arabidopsis thaliana FERRIC REDICTASE DEFECTIVE3 (AtFRD3) in iron acquisition in reproductive tissues such as pollen grain and embryos suggests a complex mechanism of iron nutrition during embryogenesis (Roschzttardtz et al., 2011b). Examination of ysl, opt3, and frd3 mutations in combination with the bts mutant background could shed new light on the role of BTS during seed development. The observed increased iron accumulation during embryogenesis could contribute to lethality (Fig. 2, E and F), most likely due to the iron-mediated production of reactive oxygen species leading to oxidative stress during the crucial period of embryo development (Ravet et al., 2009). Exposing the BTS embryonic lethal knockdown allele emb-2/+ to iron deficiency during embryogenesis could potentially rescue the bts effect on embryogenesis.
Rhizosphere acidification coupled with higher FRO2 and IRT1 activity leads to increased solubility and transport of iron into root epidermal cells (Marschner, 1995; Eide et al., 1996; Robinson et al., 1999; Vert et al., 2002). Similar to seed iron content (Fig. 2F), we observed slight, although statistically insignificant, increases in shoot and root iron content in bts-1 seedlings (Supplemental Fig. S3C), which could be attributed to enhanced rhizosphere acidification (Fig. 2C; Supplemental Fig. S3B) facilitated by increased AHA protein levels (Fig. 7D) and higher FRO2 activity (Fig. 2D). Although reduced IRT1 protein levels (Fig. 7D) could be a rate-limiting step for iron uptake in bts mutants, it is likely that the elevated rhizosphere acidification, iron reductase activity, and iron uptake are a result of decreased BTS activity, which leads to the phenotype of increased growth of bts mutants under low iron, as evident by increased root length (Fig. 2B; Supplemental Fig. S3A).
Because full-length BTS (BBG) and BTSΔHHE (BBΔHG) complemented bts-1 lines by restoring root growth phenotype, rhizosphere acidification, and iron reductase activity to levels similar to the wild type (Fig. 7, B, C, and E; Supplemental Fig. S9), we propose that iron-binding HHE domains do not participate directly in these phenotypes under iron deficiency. Rather, the HHE domains are responsible for regulating the amount of active protein accumulating in the cells. Therefore, noticeable decreases in the levels of rhizosphere acidification, iron reductase activity, and AHA and IRT1 levels in the bts-1/BBΔHG 9-5 line (Fig. 7, B–E) exposed to iron deprivation could be a reflection of the higher accumulation of BBΔHG protein in the roots (Fig. 6D; Supplemental Fig. S8, E and F), which in turn could attenuate the iron deficiency response.
Accumulation of BTS is affected by binding iron through the HHE domains (Fig. 6A). This is evident from the decreased accumulation of BTS protein in the presence of both ferrous as well as ferric iron (Fig. 6B; Supplemental Fig. S8, A and B) and in vivo accumulation of truncated BBΔHG compared with BBG, irrespective of iron status in roots (Fig. 6D; Supplemental Fig. S8, E and F). These results indicate that iron binding may initiate BTS degradation, which is not necessarily in conflict with the results obtained by Kobayashi et al. (2013) showing 26S proteasome-mediated degradation of the rice orthologs OsHRZ1 and OsHRZ2. Binding iron through the BTS HHE domains could result in elevated levels of reactive oxygen species. Such accumulation could lead to the introduction of carbonyl groups into the protein and subsequently proteolytic cleavage of BTS to lower Mr products (Stohs and Bagchi, 1995; Cabiscol et al., 2000). This process could rapidly intensify in the presence of excess ferrous iron (Fig. 6, A and B; Supplemental Fig. S8B). Alternatively, but not mutually exclusive to the metal-catalyzed oxidation of protein, binding of iron could result in structural changes that expose regions of BTS that are susceptible to proteolysis by proteolytic enzymes. We propose that iron binding itself is the trigger for BTS destabilization (Supplemental Fig. S8A), which may be followed by 26S proteasome-mediated degradation. Our data also suggest that iron binding, specifically in HHE1 and HHE2 domains, is critical for BTS instability (Fig. 6C). These results are intriguing because single point mutations in either HHE1 or HHE2 domains abolish the destabilization of BTS protein, which appears to be mediated by iron. One possible explanation for this observation is that there could be cooperativity in iron binding such that the capability of one HHE domain to bind iron affects the other. However, in-depth analysis of single as well as triple point mutations of the BTS HHE domains is required to further characterize the iron-binding capabilities, E3 ligase function, and overall effect of BTS on plant iron homeostasis mechanisms.
In vitro analyses suggest that BTS interacts with (Fig. 3) and restricts the accumulation of iron-responsive bHLH transcription factors ILR3 and bHLH115 through its E3 ligase activity and can mediate proteasomal degradation of these targets even in the absence of HHE domains (Fig. 5, A–C). Notably, the mammalian protein FBXL5, unlike BTS, does not contain a RING domain and thus lacks the ability to directly ubiquitinate its targets for degradation via the 26S proteasome pathway (Salahudeen et al., 2009; Vashisht et al., 2009; Thompson et al., 2012). FBXL5 does, however, provide the specificity to the SCFFBXL5 E3 ligase complex to regulate iron homeostasis. In contrast to the mammalian iron response pathway, BTS may assume the role of the SCFFBXL5 E3 ligase complex, where both the iron-sensing and E3 ligase activities reside within a single protein (Figs. 5 and 6).
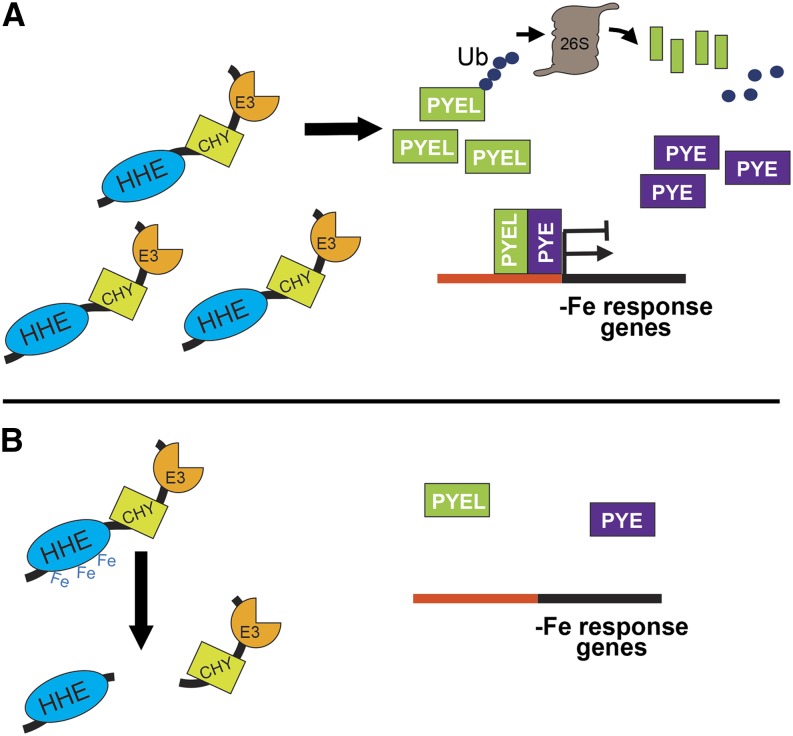
We provide evidence that BTS targets bHLH transcription factors ILR3 and bHLH115 for 26S proteasome-mediated degradation, resulting in either the disruption or inhibition of heterodimer complex formation between PYE and PYEL proteins. This could potentially perturb the downstream iron deficiency-responsive genes regulated by PYE (Long et al., 2010) or ILR3 (Rampey et al., 2006). Based on our findings, we propose a model for BTS protein stability and cellular function in Arabidopsis (Fig. 8). As plants encounter iron-deficient conditions, BTS accumulates due to both transcriptional and posttranslational control (Fig. 7A). The increase in BTS levels is reflected in an increase in iron-responsive E3 ligase activity, facilitating the degradation of PYEL proteins ILR3 and bHLH115 (Fig. 5, B and C). While the function of bHLH115 in iron homeostasis is unknown, ILR3 is thought to play a role in metal-dependent IAA-conjugate hydrolysis by controlling the expression of iron transporters (Rampey et al., 2006). By interacting with PYE, a second transcriptional regulator that induces the iron deficiency response, ILR3 and bHLH115 may form part of a functional heterodimer that accumulates under low iron and facilitates the iron deficiency response. BTS, which also accumulates under iron deprivation, could restrain the responses mediated by PYE/PYEL, thus modulating or fine-tuning the iron deficiency responses, ensuring sufficient yet not excess iron uptake, which affects overall root growth. Once sufficient iron accumulates, PYE, PYEL, and BTS are no longer transcriptionally activated, and BTS is maintained at minimal levels through iron-mediated regulation via the HHE domain. Such control would enable BTS to accumulate under low iron and consequently mediate degradation of regulatory targets, yet be rapidly decreased when iron is once again available. In conclusion, BTS is a critical iron-sensing E3 ubiquitin ligase that targets regulatory components of the iron deficiency pathway in plants. Further in-depth studies of BTS and its target bHLH transcription factor will elucidate their function in the plant iron homeostasis signaling network.
Figure 8.
Model for BTS protein stability and function. A, BTS is transcriptionally induced by iron deficiency along with bHLH transcription factors PYE and PYEL. Under low-iron conditions, BTS protein is more stable and regulates PYEL/PYE regulatory activity through its E3 ligase activity to modulate and fine-tune PYEL/PYE-mediated iron deficiency response in plants. B, Upon recovery of iron, transcriptional induction of BTS, PYE, and PYEL decreases. Elevated levels of iron are sensed through the HHE domains, altering BTS conformation and stability. This response results in the proteolysis of BTS protein, thereby further reducing the iron-responsive E3 ligase capacity of the cells.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The knockdown T-DNA insertion lines for bts-10 (SALK_087255), bts-12 (SALK_122102), and emb2454-2 (CS-16056) were reconfirmed using PCR with T-DNA left-border primer (LBb1.3) and gene-specific primers. Primer sets are listed in Supplemental Table S1. The T-DNA insertion line for bts-1 (SALK_016526) was previously confirmed (Long et al., 2010). Col-0 was used as the wild type. Seed sterilization was performed as previously described (Long et al., 2010). For plant growth on plates, iron-sufficient (+Fe) medium was standard Murashige and Skoog medium with 0.05% (w/v) MES, 1% (w/v) Suc, 1% (w/v) agar, and 0.1 mm Fe-EDTA substituted for iron sulfate. Iron-deficient (–Fe) medium was the same, with 300 μm ferrozine, an iron chelator, substituted for iron sulfate. For all experiments in which seedlings were shifted from iron-sufficient (+Fe) to either sufficient (+Fe) or deficient (–Fe) medium, 100 µm Nitex nylon mesh (Genesee Scientific) was placed on top of the solid medium and seeds were placed directly on nylon mesh. Plants were grown vertically on plates at 22°C under a 16-h-light and an 8-h-dark period in environmentally controlled plant growth chambers (Percival Scientific). Time of transfer of seedlings from +Fe to –Fe plates was dependent on individual experiments and listed for each assay. Arabidopsis (Arabidopsis thaliana) plants were grown and propagated in Metro-Mix 300 soil (SunGro Horticulture) at 22°C under a 16-h-light and an 8-h-dark period in environmentally controlled growth chambers. Nicotiana benthamiana (wild-type and RFP-Histone2B transgenic) plants were grown for 5 to 6 weeks in Metro-Mix 300 soil at 26°C under a 16-h-light and an 8-h-dark period in environmentally controlled growth chambers.
Plasmid Construction
Coding regions of bHLH transcription factors PYE (At3g47640), ILR3 (At5g54680), bHLH104 (At4g14410), and bHLH115 (At1g51070) and full-length BTS (At3g18290) were cloned into the Gateway entry vector pENTR/D-TOPO as previously described (Long et al., 2010). The full-length BTS clone was used for generating truncated derivatives of BTS (BTSΔHHE and BTSΔE3), single point mutations (E108A, E369A, and E716A), and a triple point mutation (Ex3A) and for cloning specific domains of BTS (BTS-HHE and BTS-E3 domains) into the pENTR/D-TOPO vector (Invitrogen). Primers are listed in Supplemental Table S1.
The upstream regulatory or promoter sequences of ILR3 (3,426 bp) and bHLH115 (574 bp) were amplified from genomic DNA (Arabidopsis Col-0) and cloned into the pDONRP4-P1R vector (Invitrogen). Primers are listed in Supplemental Table S1. The coding region of ILR3 (At5g54680) and bHLH115 (At1g51070) genes were cloned into the pENTR/D-TOPO vector, and GFP was cloned into the pDONRP2R-P3 vector (Invitrogen) as previously described (Long et al., 2010). The ProILR3::ILR3-GFP and Pro-115::115-GFP clones were made by Multisite Gateway cloning technology (Life Technologies). Each promoter and open reading frame was cloned into the pGREEN (Hellens et al., 2000) binary vector derivative containing a nopaline synthase terminator with a C-terminal GFP fusion and spectinomycin and BASTA resistance genes (Lee et al., 2006).
To generate the ProBTS::GUS construct, the BTS promoter sequence (3,000 bp) was subcloned from the pDONRP4P-1R ProBTS clone (Long et al., 2010) into the pENTR/D-TOPO vector to serve as an entry clone (primers are listed in Supplemental Table S1). The insert from this construct was transferred to the pMDC162 binary vector (Curtis and Grossniklaus, 2003) through Gateway cloning technology (Invitrogen) to produce ProBTS::GUS, which was transformed into wild-type Col-0 plants.
To generate constructs for transient protein expression in N. benthamiana leaves, coding regions of genes from pENTR/D-TOPO clones were transferred to pSITE-BiFC-C1cec/nec vectors (Martin et al., 2009) through Gateway cloning technology (Invitrogen) to generate vectors containing N-/C-terminal halves of EYFP fusions for BiFC assays. For protein localization studies, coding regions of genes were transferred to pSITE-2NA/CA for N-/C-terminal EGFP fusions and pSITEII-6C1 for N-terminal TaqRFP fusions proteins. These clones were introduced into Agrobacterium tumefaciens strain LBA4404 for transient expression of proteins in RFP-Histone2B transgenic N. benthamiana leaves (Martin et al., 2009).
To construct vectors for in vitro protein translation, genes from pENTR/D-TOPO clones (Long et al., 2010) were transferred to Gateway-compatible vectors pIX-3xHA, pIX-Halo (Arabidopsis Biological Resource Center), pF3K WG-BYDV, and pFN19K HaloTag T7 SP6 Flexi Vector (Promega). These vectors were used for the synthesis of epitope-tagged proteins in vitro using the TNT SP6 High-Yield Wheat Germ Protein Expression System (Promega).
Root Length Analysis
For each genotype, eight to 10 seeds were grown on +Fe medium for 4 d and then shifted to either +Fe or –Fe medium for 7 d (4 d +Fe, 7 d ±Fe). Plates were scanned when plants were 11 d old. ImageJ software (http://rsb.info.nih.gov/ij/) was used for root length measurements.
Rhizosphere Acidification
Rhizosphere acidification was performed as previously described (Yi and Guerinot, 1996). Briefly, 7-d-old (4 d +Fe and 3 d –Fe) seedlings were transferred to 1% (w/v) agar plates containing 0.006% (w/v) bromocresol purple and 0.2 mm CaSO4 (pH 6.5 adjusted with 1 m NaOH) for 24 h.
Iron Reductase Assay
Ferric reductase activity was measured as described previously (Gibbs, 1976; Yi and Guerinot, 1996). Briefly, eight roots of 10-d-old (7 d +Fe and 3 d ±Fe) seedlings were weighed and placed in assay solution containing 0.1 mm Fe(III)-EDTA and 0.3 mm ferrozine. The seedlings were incubated in the dark for 1 h. The absorbance was measured spectrophotometrically at 562 nm.
Histochemical Detection of Iron Localization
Perls staining was performed to localize iron in 7-d-old (4 d +Fe and 3 d –Fe) seedlings as previously described (Long et al., 2010). Green siliques were cleared of chlorophyll using chloral hydrate as described previously (Berleth and Jurgens, 1993). Imaging was performed with a Leica MZ FLIII stereomicroscope.
Embryo Lethality
Green siliques were cleared (Berleth and Jurgens, 1993), and the silique covers were removed with the aid of a dissecting microscope (Leica MZ12). Percentage of embryo lethality was measured by counting degraded/halted and normal embryos under a Zeiss AXIOImagerM2 microscope.
GUS Histochemical Assay
GUS staining of the wild-type/ProBTS::GUS transgenic line (T3) was performed as previously described (An et al., 1996) with the following modifications. Briefly, fresh tissue samples were fixed in ice cold 90% (v/v) acetone for 1 h and then vacuum filtrated for 30 min. Samples were vacuum infiltrated with staining buffer (50 mm PO4 buffer with 0.2% [v/v] Triton X-100, 100 mm K3Fe(CN)6, and 100 mm K4Fe(CN)6) for 10 min on ice followed by vacuum infiltration in staining buffer containing 2 mm 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid for 20 min on ice. Samples were incubated at 37°C for 30 min to 24 h followed by a 70% (v/v) ethanol wash.
Quantitative Reverse Transcription-PCR
Total RNA was isolated from roots of 7-d-old (4 d +Fe and 3 d ±Fe) seedlings using the RNeasy plant mini kit (Qiagen), and first-stand complementary DNA was synthesized using the Superscript III complementary DNA synthesis kit (Life Technologies). Quantitative reverse transcription-PCR was conducted using iTaq Universal SYBR Green Supermix (Bio-Rad) and the StepOnePlus Real-Time PCR System (Applied Biosystems). Primers are listed in Supplemental Table S1. Relative expression was calculated using the comparative cycle threshold method, normalized to β-tubulin.
Confocal Microscopy of Roots
ProILR3::ILR3-GFP and Pro115::115-GFP clones were transformed into wild-type Col-0 (BTS) and bts-1 mutant background. To examine changes in the stability of ILR3-GFP and bHLH115-GFP proteins in the wild type and bts-1 mutant background, T3 lines were grown on +Fe for 4 d and then shifted to ±Fe for 2 d. Roots were counterstained with 10 mm propidium iodide to visualize cells (Long et al., 2010) and imaged with a laser scanning microscope (Zeiss LSM 710).
Tissue Elemental Analysis
One hundred milligrams of seeds of wild-type and bts alleles was plated on Murashige and Skoog agar medium for metal ion analysis. Root and shoot tissue samples were collected from 10-d-old (7 d +Fe and 3 d ±Fe) seedlings. Shoot samples were thoroughly rinsed with deionized water, while the root samples were desorbed for 10 min with 2 mm CaSO4 and 10 mm EDTA and then rinsed for 5 min with deionized water. The tissue samples were lyophilized and analyzed for iron content by ICP-OES as previously described (Barberon et al., 2011). One hundred fifty milligrams of seeds per genotype were subjected to ICP-OES for metal content analysis.
Protein Analysis from Arabidopsis Roots
Roots from 7-d-old (4 d +Fe and 3 d ±Fe) seedlings were homogenized in buffer containing 20 mm Tris-HCl (pH 7.0), 150 mm NaCl, 1 mm EDTA, 1% (v/v) Triton X-100, 0.1% (w/v) SDS, 10 mm dithiothreitol (DTT), and 1× plant protease inhibitor cocktail (Sigma-Aldrich). Proteins were separated on 7.5% to 10% (w/v) SDS-PAGE gels. Proteins were immunodetected using anti-GFP antibody (Invitrogen), anti-H+-ATPase (Agrisera), anti-IRT1 (Agrisera), and ECL Plus chemiluminescence substrate (Pierce).
Subcellular Fractionation of Arabidopsis
Roots from 7-d-old (4 d +Fe and 3 d –Fe) seedlings were used to isolate nuclear and cytoplasmic fractions using the modified Suc-Percoll density gradient method (Luthe and Quatrano, 1980; Sikorskaite et al., 2013). For nuclei isolation, roots were homogenized in ice-cold nuclei isolation buffer (NIB; 10 mm MES-KOH, pH 5.4, 10 mm MgCl2, 10 mm NaCl, 10 mm KCl, 1 mm EDTA, 250 mm Suc, 0.5% [v/v] Triton X-100, and 1× protease inhibitor cocktail) and supplemented immediately before use with 0.1 mm spermine, 0.5 mm spermidine, and 2.5 mm DTT. The homogenate was filtered through four layers of Miracloth and centrifuged at 1,000g for 10 min at 4°C to obtain the crude nuclear pellet and supernatant. The supernatant was subjected to ultracentrifugation at 100,000g for 1 h to separate soluble cytoplasmic fraction (supernatant) from the endomembrane (membrane pellet). The crude nuclear pellet was gently resuspended in NIB buffer (without supplements) and purified by centrifugation in a discontinuous Suc-Percoll gradient (2.5 m Suc and 80%, 60%, and 40% [v/v] Percoll in NIB) at 4,080g in a swinging-bucket rotor for 30 min at 4°C. The intact nuclei banded in the 80% Percoll layer (above Suc-Percoll interface) were collected using Pasteur pipette, washed three times with five volumes of ice-cold NIB containing 0.1% (v/v) Triton X-100 to remove Percoll, and centrifuged at 1,000g for 10 min. The nuclei pellet was resuspended in 100 µL of 1× phosphate-buffered saline containing 20% (v/v) glycerol and 2.5 mm DTT. Total, cytoplasmic, and nuclear proteins were separated on a 10% (w/v) SDS-PAGE gel and immunodetected using anti-GFP antibody (Invitrogen) and ECL Plus chemiluminescence substrate (Pierce). Purity of isolated subcellular fractions was evaluated by reprobing the blots with either anti-Histone3 (for nuclear protein) or anti-ascorbate peroxidase (cytoplasmic proteins) and vice versa. The membrane pellets were washed (50 mm Tris-Cl, pH 7.5, 5 mm MgCl2, and 1× plant protease inhibitor cocktail) and centrifuged again at 100,000g for 1 h to pellet down the endomembrane. The membrane proteins were solubilized in 50 mm Tris-Cl, pH 7.5, 5 mm MgCl2, and 1% (w/v) SDS, quantified, and separated on a 10% (w/v) SDS-PAGE gel. The plasma membrane-localized AHA and IRT1 proteins were immunodetected using anti-H+-ATPase- and anti-IRT1-specific antibodies (Agrisera).
Iron-Binding Analysis of Recombinant BTS Protein
The coding regions of full-length BTS and BTSΔHHE deletion (Supplemental Fig. S5) were cloned into the expression vector pDEST-HisMBP (Nallamsetty et al., 2005) using Gateway cloning technology (Invitrogen). To generate a construct for expressing solely the HisMBP, scBTS was amplified with primers BTS_5primeSC_L_F and BTS-SC-R (Supplemental Table S1) to introduce a stop codon prior to the codon for the initiating Met. The PCR fragment was then first cloned into pENTR/D-TOPO vector and subsequently transferred into expression vector pDEST-HisMBP using Gateway cloning technology (Invitrogen). The resulting plasmids were introduced into Escherichia coli BL21(DE3)pLysS (Life Technologies) for recombinant HisMBP-BTS, HisMBP-BTSΔHHE, and HisMBP-scBTS protein expression. Cultures were grown overnight at 15°C following incubation with 0.1 mm isopropyl β-d-1-thiogalactopyranoside. Proteins were extracted in 30 mm Tris-HCl, pH 7.0, and 200 mm NaCl and purified on a Bio-Scale Mini Profinity IMAC cartridge (Bio-Rad) according to manufacture’s recommendations. Samples were dialyzed against 20 mm ammonium acetate and lyophilized. The purified proteins (1.66–3.39 mg) were subjected to ICP-MS analysis for determination of iron and zinc content.
BTS Protein Stability Assay
3xHA-BTS, 3xHA-BTSΔHHE, BTS single (E108A, E369A, and E716A) and triple (Ex3A) point mutants, and Myc-tagged bHLH transcription factors (ILR3, bHLH104, bHLH115, and PYE) were in vitro translated using the TNT SP6 High-Yield Wheat Germ Protein Expression System (Promega). The vectors (100 ng) containing respective coding regions of individual genes (as described above) were mixed with 10 µL of wheat (Triticum aestivum) germ extracts for in vitro transcription-coupled translation of proteins. The reactions were supplemented with either iron chelators (100–400 μm DFO or 400 μm ferrozine) or ferrous iron [0–12 µm Fe(II)-ascorbate] and ferric iron (400 μm FAC). Total protein from the expression reactions were separated on 9% (w/v) SDS-PAGE gels and immunodetected using tag-specific antibodies or BTS-specific antibodies directed against N-terminal (BTS amino acids 32–46) and C-terminal (BTS amino acids 1,238–1,250) peptides produced in rabbit.
BiFC Assay
BiFC assays were carried out as previously described (Selote and Kachroo, 2010). Briefly, proteins were transiently coexpressed into RFP-Histone2B-tagged N. benthamiana plants (transgenic plants expressing nuclear-localized RFP-tagged Histone2B protein). For the assay, A. tumefaciens mixtures were infiltrated using a 1-mL syringe without a needle into the abaxial side of 5- to 6-week-old N. benthamiana leaves. After 48 h of transfection, water-mounted sections of leaf tissue were examined with a laser scanning microscope (Zeiss LSM 710). Proteins were expressed as N-EYFP and C-EYFP fusions, and all interactions were tested using both combinations of N-/C-EYFP-fused reciprocal proteins.
Co-IP Assay
HA-, Myc-, and HaloTag fusion proteins were in vitro translated using the TNT SP6 High-Yield Wheat Germ Protein Expression System (Promega). The vectors (1 µg) containing respective coding regions of individual genes (as described above) were mixed with 50 µL of wheat germ extracts for in vitro transcription-coupled translation of proteins. For Co-IP assays, HA- or Halo-tagged fusion proteins (50 µL of wheat germ extract reaction) were mixed with either Myc- or HA-tagged fusion proteins (50 µL of wheat germ extract reaction), respectively, and incubated overnight at 4°C with either anti-HA agarose (Sigma-Aldrich) or HaloLink affinity resins (Promega). After Co-IP, the anti-HA or HaloLink affinity resins were extensively washed with phosphate-buffered saline containing 0.1% (v/v) Tween 20. Co-IP proteins were eluted using tag-specific peptides and separated on 9% (w/v) SDS/PAGE gel. Proteins were immunodetected using anti-HA-, anti-c-Myc- (Bethyl Laboratories), and anti-HaloTag-specific (Promega) primary antibodies and horseradish peroxidase-conjugated secondary antibody. Blots were developed using ECL Plus chemiluminescence substrate (Pierce).
In Vitro Ubiquitination Assay
Ubiquitination assays were carried out using a ubiquitinylation kit (ENZO Life Sciences). Briefly, in vitro-translated and purified 3xHA-BTS and its derivatives (0.1 μg) were incubated with 1× ubiquitinylation buffer, 1× Mg-ATP solution, 2.5 mm DTT, 100 nm human E1, 2.5 μm human E2 (Ubiquitin-conjugating enzyme H5c [UbcH5c]), 1 μg FLAG-ubiquitin, and 1 unit of inorganic pyrophosphatase (Sigma-Aldrich) at 37°C for 8 h. Proteins were separated on a 4% to 15% (w/v) gradient SDS-PAGE gel (Bio-Rad) under nonreducing conditions. Immunodetection was carried out using anti-FLAG-specific antibody (Sigma-Aldrich) as described above.
Cell-Free Degradation Assay
Cell-free protein extracts were prepared from 7-d-old wild-type, bts-1, and bts-1/ProBTS::BTSΔHHE-GFP (bts-1/BBΔHG) seedlings as previously described (Wang et al., 2009). Myc-tagged bHLH transcription factors (ILR3, bHLH104, bHLH115, and PYE) were in vitro translated and purified using anti-Myc affinity resin (Sigma-Aldrich). For the 26S proteasome-mediated degradation assay, Myc-tagged ILR3 (0.1 µg per lane), bHLH104 (0.18 µg per lane), bHLH115 (0.25 µg per lane), and PYE (0.21 µg per lane) protein were incubated with cell-free protein extracts (100 µg per lane). Reactions were carried out with 160 µm MG132 (in dimethyl sulfoxide) or without MG132 (dimethyl sulfoxide only) at 22°C for 8 h followed by separation on 10% (w/v) SDS-PAGE gel. Immunodetection was carried out using anti-Myc antibody as described above.
Statistical Analysis
All statistical significance was determined using one-way ANOVA (P < 0.001) followed by a Tukey-Kramer test (P < 0.05).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers PYE (At3g47640), ILR3 (At5g54680), bHLH104 (At4g14410), bHLH115 (At1g51070), and BTS (At3g18290).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. GUS activity in wild-type (Col-0) plant.
Supplemental Figure S2. Location of BTS T-DNA knockdown alleles.
Supplemental Figure S3. Root growth, rhizosphere acidification, and shoot and root iron content in bts alleles.
Supplemental Figure S4. Localization of BTS, PYE, and PYEL proteins in nucleus.
Supplemental Figure S5. Schematic diagram representing BTS protein.
Supplemental Figure S6. Lack of PYE and PYEL homodimer formation.
Supplemental Figure S7. Relative expression of ILR3-GFP and bHLH115-GFP in roots.
Supplemental Figure S8. In vivo and in vitro stability of BTS protein in the presence of iron.
Supplemental Figure S9. Rhizosphere acidification of bts-1 mutant lines complemented with BBG and BBΔHG.
Supplemental Table S1. List of primers used in this study.
Supplementary Material
Acknowledgments
We thank Michael Goodin (University of Kentucky) for the pSITE vectors and RFP-Histone2B-expressing transgenic N. benthamiana seeds, Eva Johannes (Cellular and Molecular Imaging Facility, North Carolina State University) for imaging assistance, Robert Franks (North Carolina State University) and Judy Callis (University of California, Davis) for technical assistance, and Kim Hutchinson (Environmental and Agricultural Testing Service, North Carolina State University) for ICP-MS/OES analysis.
Glossary
- H+-ATPase
proton-translocating adenosine triphosphatase
- T-DNA
transfer DNA
- OES
optical emission spectrometry
- ICP
inductively coupled plasma
- Y2H
yeast two-hybrid
- IAA
indole-3-acetic acid
- BiFC
bimolecular fluorescence complementation
- Co-IP
coimmunoprecipitation
- MS
mass spectrometry
- FAC
ferric ammonium citrate
- DFO
deferoxamine-B
- Col-0
ecotype Columbia
- DTT
dithiothreitol
- NIB
nuclei isolation buffer
Footnotes
This work was supported by the U.S. National Science Foundation (grant no. MCB1120937), the U.S. Department of Agriculture (grant no. NC02380), and the North Carolina Agricultural Research Service.
Articles can be viewed without a subscription.
References
- An YQ, Huang S, McDowell JM, McKinney EC, Meagher RB (1996) Conserved expression of the Arabidopsis ACT1 and ACT 3 actin subclass in organ primordia and mature pollen. Plant Cell 8: 15–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M, Zelazny E, Robert S, Conéjéro G, Curie C, Friml J, Vert G (2011) Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc Natl Acad Sci USA 108: E450–E458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth T, Jurgens G (1993) The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development 118: 575–587 [Google Scholar]
- Besold AN, Lee SJ, Michel SL, Sue NL, Cymet HJ (2010) Functional characterization of iron-substituted neural zinc finger factor 1: metal and DNA binding. J Biol Inorg Chem 15: 583–590 [DOI] [PubMed] [Google Scholar]
- Cabiscol E, Piulats E, Echave P, Herrero E, Ros J (2000) Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J Biol Chem 275: 27393–27398 [DOI] [PubMed] [Google Scholar]
- Cheng L, Wang F, Shou H, Huang F, Zheng L, He F, Li J, Zhao FJ, Ueno D, Ma JF, et al. (2007) Mutation in nicotianamine aminotransferase stimulated the Fe(II) acquisition system and led to iron accumulation in rice. Plant Physiol 145: 1647–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu HH, Chiecko J, Punshon T, Lanzirotti A, Lahner B, Salt DE, Walker EL (2010) Successful reproduction requires the function of Arabidopsis YELLOW STRIPE-LIKE1 and YELLOW STRIPE-LIKE3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol 154: 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Cassin G, Couch D, Divol F, Higuchi K, Le Jean M, Misson J, Schikora A, Czernic P, Mari S (2009) Metal movement within the plant: contribution of nicotianamine and yellow stripe 1-like transporters. Ann Bot (Lond) 103: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Panaviene Z, Loulergue C, Dellaporta SL, Briat JF, Walker EL (2001) Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409: 346–349 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Durrett TP, Gassmann W, Rogers EE (2007) The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol 144: 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs CR. (1976) Characterization and application of ferrozine iron reagent as a ferrous iron indicator. Anal Chem 48: 1197–1201 [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42: 819–832 [DOI] [PubMed] [Google Scholar]
- Holmes MA, Le Trong I, Turley S, Sieker LC, Stenkamp RE (1991) Structures of deoxy and oxy hemerythrin at 2.0 A resolution. J Mol Biol 218: 583–593 [DOI] [PubMed] [Google Scholar]
- Holmes MA, Stenkamp RE (1991) Structures of met and azidomet hemerythrin at 1.66 A resolution. J Mol Biol 220: 723–737 [DOI] [PubMed] [Google Scholar]
- Inoue H, Kobayashi T, Nozoye T, Takahashi M, Kakei Y, Suzuki K, Nakazono M, Nakanishi H, Mori S, Nishizawa NK (2009) Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J Biol Chem 284: 3470–3479 [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Suzuki M, Tsukamoto T, Suzuki K, Nakazono M, Kobayashi T, Wada Y, Watanabe S, Matsuhashi S, Takahashi M, et al. (2006) Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J 45: 335–346 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Nagasaka S, Senoura T, Itai RN, Nakanishi H, Nishizawa NK (2013) Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat Commun 4: 2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Jean M, Schikora A, Mari S, Briat JF, Curie C (2005) A loss-of-function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. Plant J 44: 769–782 [DOI] [PubMed] [Google Scholar]
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingam S, Mohrbacher J, Brumbarova T, Potuschak T, Fink-Straube C, Blondet E, Genschik P, Bauer P (2011) Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 23: 1815–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long TA, Tsukagoshi H, Busch W, Lahner B, Salt DE, Benfey PN (2010) The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22: 2219–2236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luthe DS, Quatrano RS (1980) Transcription in isolated wheat nuclei: I. Isolation of nuclei and elimination of endogenous ribonuclease activity. Plant Physiol 65: 305–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. (1995) Mineral Nutrition of Higher Plants, Ed 2.Academic Press, London [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM (2009) Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. Plant J 59: 150–162 [DOI] [PubMed] [Google Scholar]
- McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, Smith K, Thomas C, Schetter A, Zhou Q, Cushman MA, et al. (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiser J, Lingam S, Bauer P (2011) Posttranslational regulation of the iron deficiency basic helix-loop-helix transcription factor FIT is affected by iron and nitric oxide. Plant Physiol 157: 2154–2166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Cózatl DG, Xie Q, Akmakjian GZ, Jobe TO, Patel A, Stacey MG, Song L, Demoin DW, Jurisson SS, Stacey G, et al. (2014) OPT3 is a component of the iron-signaling network between leaves and roots and misregulation of OPT3 leads to an over-accumulation of cadmium in seeds. Mol Plant 7: 1455–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallamsetty S, Austin BP, Penrose KJ, Waugh DS (2005) Gateway vectors for the production of combinatorially-tagged His6-MBP fusion proteins in the cytoplasm and periplasm of Escherichia coli. Protein Sci 14: 2964–2971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampey RA, Woodward AW, Hobbs BN, Tierney MP, Lahner B, Salt DE, Bartel B (2006) An Arabidopsis basic helix-loop-helix leucine zipper protein modulates metal homeostasis and auxin conjugate responsiveness. Genetics 174: 1841–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F (2009) Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J 57: 400–412 [DOI] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Celma J, Lin WD, Fu GM, Abadía J, López-Millán AF, Schmidt W (2013) Mutually exclusive alterations in secondary metabolism are critical for the uptake of insoluble iron compounds by Arabidopsis and Medicago truncatula. Plant Physiol 162: 1473–1485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld V, Müller C, Marschner H (1984) Localization and capacity of proton pumps in roots of intact sunflower plants. Plant Physiol 76: 603–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H, Grillet L, Isaure MP, Conéjéro G, Ortega R, Curie C, Mari S (2011a) Plant cell nucleolus as a hot spot for iron. J Biol Chem 286: 27863–27866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschzttardtz H, Séguéla-Arnaud M, Briat JF, Vert G, Curie C (2011b) The FRD3 citrate effluxer promotes iron nutrition between symplastically disconnected tissues throughout Arabidopsis development. Plant Cell 23: 2725–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JC, Walker SD, Anderson SA, Eisenstein RS, Bruick RK (2013) F-box and leucine-rich repeat protein 5 (FBXL5) is required for maintenance of cellular and systemic iron homeostasis. J Biol Chem 288: 552–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salahudeen AA, Thompson JW, Ruiz JC, Ma HW, Kinch LN, Li Q, Grishin NV, Bruick RK (2009) An E3 ligase possessing an iron-responsive hemerythrin domain is a regulator of iron homeostasis. Science 326: 722–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi S, Schmidt W (2009) Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol 183: 1072–1084 [DOI] [PubMed] [Google Scholar]
- Selote D, Kachroo A (2010) RPG1-B-derived resistance to AvrB-expressing Pseudomonas syringae requires RIN4-like proteins in soybean. Plant Physiol 153: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimomura K, Nomura M, Tajima S, Kouchi H (2006) LjnsRING, a novel RING finger protein, is required for symbiotic interactions between Mesorhizobium loti and Lotus japonicus. Plant Cell Physiol 47: 1572–1581 [DOI] [PubMed] [Google Scholar]
- Shu C, Sung MW, Stewart MD, Igumenova TI, Tan X, Li P (2012) The structural basis of iron sensing by the human F-box protein FBXL5. ChemBioChem 13: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorskaite S, Rajamäki ML, Baniulis D, Stanys V, Valkonen JP (2013) Protocol: optimised methodology for isolation of nuclei from leaves of species in the Solanaceae and Rosaceae families. Plant Methods 9: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivitz A, Grinvalds C, Barberon M, Curie C, Vert G (2011) Proteasome-mediated turnover of the transcriptional activator FIT is required for plant iron-deficiency responses. Plant J 66: 1044–1052 [DOI] [PubMed] [Google Scholar]
- Sivitz AB, Hermand V, Curie C, Vert G (2012) Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS ONE 7: e44843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Patel A, McClain WE, Mathieu M, Remley M, Rogers EE, Gassmann W, Blevins DG, Stacey G (2008) The Arabidopsis AtOPT3 protein functions in metal homeostasis and movement of iron to developing seeds. Plant Physiol 146: 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohs SJ, Bagchi D (1995) Oxidative mechanisms in the toxicity of metal ions. Free Radic Biol Med 18: 321–336 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Nozawa A, Seki M, Shinozaki K, Endo Y, Sawasaki T (2009) A simple and high-sensitivity method for analysis of ubiquitination and polyubiquitination based on wheat cell-free protein synthesis. BMC Plant Biol 9: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JW, Salahudeen AA, Chollangi S, Ruiz JC, Brautigam CA, Makris TM, Lipscomb JD, Tomchick DR, Bruick RK (2012) Structural and molecular characterization of iron-sensing hemerythrin-like domain within F-box and leucine-rich repeat protein 5 (FBXL5). J Biol Chem 287: 7357–7365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzafrir I, Pena-Muralla R, Dickerman A, Berg M, Rogers R, Hutchens S, Sweeney TC, McElver J, Aux G, Patton D, et al. (2004) Identification of genes required for embryo development in Arabidopsis. Plant Physiol 135: 1206–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashisht AA, Zumbrennen KB, Huang X, Powers DN, Durazo A, Sun D, Bhaskaran N, Persson A, Uhlen M, Sangfelt O, et al. (2009) Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science 326: 718–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dédaldéchamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhu D, Huang X, Li S, Gong Y, Yao Q, Fu X, Fan LM, Deng XW (2009) Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21: 2378–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji Y, Hamada K, Yoshinuma T, Sakurai K, Yoshii A, Shimizu T, Hashimoto M, Suzuki M, Namba S, Hibi T (2010) Inhibitory effect on the tobacco mosaic virus infection by a plant RING finger protein. Virus Res 153: 50–57 [DOI] [PubMed] [Google Scholar]
- Yi Y, Guerinot ML (1996) Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J 10: 835–844 [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wu H, Wang N, Li J, Zhao W, Du J, Wang D, Ling HQ (2008) FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res 18: 385–397 [DOI] [PubMed] [Google Scholar]
- Zhai Z, Gayomba SR, Jung HI, Vimalakumari NK, Piñeros M, Craft E, Rutzke MA, Danku J, Lahner B, Punshon T, et al. (2014) OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell 26: 2249–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.