The superior manganese tolerance of a tropical legume is achieved by the coordination of internal and external manganese detoxification via enhanced malate synthesis.
Abstract
Manganese (Mn) toxicity is a major constraint limiting plant growth on acidic soils. Superior Mn tolerance in Stylosanthes spp. has been well documented, but its molecular mechanisms remain largely unknown. In this study, superior Mn tolerance in Stylosanthes guianensis was confirmed, as reflected by a high Mn toxicity threshold. Furthermore, genetic variation of Mn tolerance was evaluated using two S. guianensis genotypes, which revealed that the Fine-stem genotype had higher Mn tolerance than the TPRC2001-1 genotype, as exhibited through less reduction in dry weight under excess Mn, and accompanied by lower internal Mn concentrations. Interestingly, Mn-stimulated increases in malate concentrations and exudation rates were observed only in the Fine-stem genotype. Proteomic analysis of Fine-stem roots revealed that S. guianensis Malate Dehydrogenase1 (SgMDH1) accumulated in response to Mn toxicity. Western-blot and quantitative PCR analyses showed that Mn toxicity resulted in increased SgMDH1 accumulation only in Fine-stem roots, but not in TPRC2001-1. The function of SgMDH1-mediated malate synthesis was verified through in vitro biochemical analysis of SgMDH1 activities against oxaloacetate, as well as in vivo increased malate concentrations in yeast (Saccharomyces cerevisiae), soybean (Glycine max) hairy roots, and Arabidopsis (Arabidopsis thaliana) with SgMDH1 overexpression. Furthermore, SgMDH1 overexpression conferred Mn tolerance in Arabidopsis, which was accompanied by increased malate exudation and reduced plant Mn concentrations, suggesting that secreted malate could alleviate Mn toxicity in plants. Taken together, we conclude that the superior Mn tolerance of S. guianensis is achieved by coordination of internal and external Mn detoxification through malate synthesis and exudation, which is regulated by SgMDH1 at both transcription and protein levels.
Manganese (Mn) is an essential micronutrient for plant growth, and is a constitutive element in the Mn cluster of the oxygen-evolving complex in PSII, which is involved in the water-splitting process to oxidize water and then produce oxygen and to further provide electrons for the electron transport chain in PSII (Goussias et al., 2002; Nickelsen and Rengstl, 2013). Mn is also involved in a series of metabolic steps in plants, such as the biosynthesis of acyl lipids, flavonoids, and lignin (Lidon et al., 2004). To meet Mn requirements for growth, plants have developed a group of strategies for Mn acquisition and translocation (Sasaki et al., 2012; Yamaji et al., 2013). As the second most abundant transition metal in Earth’s crust, soil Mn can exist in several oxidation states with varying solubility that is strongly dependent on soil redox conditions and pH (Hernandez-Soriano et al., 2012). Oxidized Mn is easily reduced to Mn (II) with high solubility under the reducing conditions in soils, which could be caused by poor drainage or waterlogged conditions (Geszvain et al., 2012; Sparrow and Uren, 2014). Meanwhile, soil pH also affects Mn solubility. With a decrease of soil pH below 5.0, Mn (III) and Mn (IV) are readily reduced to Mn (II), which is the most soluble and available form for plant acquisition (Watmough et al., 2007). Therefore, excess Mn (II) accumulation is generally observed, especially on acid soils, where the occurrence of Mn toxicity trails only aluminum (Al) toxicity in limiting crop growth (Kochian et al., 2004).
Although plants exhibit a variety of responses to Mn toxicity, symptoms of Mn toxicity in plant leaves are generally observed as the formation of brown spots, chlorosis, and necrosis (Millaleo et al., 2010). Therefore, it has been suggested that leaf chloroplasts might be primary targets of Mn toxicity, which could result in impairment of thylakoid structure and disruption of the electron transport chain in photosynthesis of plants (González et al., 1998; Lidon et al., 2004). In addition, Mn toxicity might also cause reactive oxygen species accumulation and brown spot formation in leaves, and may thereby inhibit plant growth more severely (Fecht-Christoffers et al., 2006). Although soil amelioration (e.g. lime application) is conventionally used to reduce excess Mn availability, and thus alleviate Mn toxicity, it requires inordinate amounts of time and energy, and responses are strongly dependent on soil types and plant species (Hue and Mai, 2002). Therefore, developing varieties with superior Mn tolerance becomes an important complementary approach for sustainable agriculture development, which in turn requires deeper understanding of Mn tolerance mechanisms in plants.
Cumulative results recently suggested that a major Mn tolerance mechanism in plants involves detoxification through Mn sequestration into inactive subcellular sites (e.g. apoplasts, cell walls, and vacuoles) and/or inhibited reactive oxygen species generation (Millaleo et al., 2010). In cowpea (Vigna unguiculata), compartmentalization of Mn in leaf apoplasts is considered as a primary response to Mn toxicity (Horst et al., 1999). Furthermore, accumulation of peroxidases in leaf apoplasts can modulate Mn oxidation, and thereby affect Mn tolerance in cowpea (Fecht-Christoffers et al., 2006). In addition, increased cell wall binding capacity of Mn and decreased hydroxyl radical accumulation in leaf apoplasts have been suggested to alleviate Mn toxicity with silicon (Si) application (Dragišić Maksimović et al., 2012). With the aid of forward and reverse genetic analysis, another adaptive response to Mn toxicity has been observed as sequestration of excess Mn into vacuoles through the activities of several Mn transporters such as cation diffusion facilitator family member, metal tolerance protein (MTP; Pittman, 2005). In rice (Oryza sativa), OsMTP8.1 is a Mn-specific transporter, and belongs to the same cation diffusion facilitator family as ShMTP1, AtMTP11, and CsMTP8, each of which transports Mn into vacuoles (Delhaize et al., 2007; Chen et al., 2013; Migocka et al., 2014). Furthermore, disruption of OsMTP8.1 increases inhibition of plant growth under Mn toxic conditions, which suggests an important role for Mn compartmentalization in vacuoles for plant Mn tolerance (Chen et al., 2013). Although Mn species in plant vacuoles remain unknown, important roles for organic acids (e.g. malate, citrate, and oxalate) in internal Mn detoxification are suggested through analysis of foliar Mn species in Mn hyperaccumulator plants (Xu et al., 2009; Fernando et al., 2010). Furthermore, increased organic acid exudation from roots was observed in ryegrass (Lolium perenne) and white clover (Trifolium repens) subjected to different Mn levels (Rosas et al., 2007; de la Luz Mora et al., 2009), suggesting that secreted organic acids might be responsive to Mn toxicity. However, molecular mechanisms of organic acid synthesis and exudation involved in plant Mn detoxification remain largely unknown.
Stylosanthes spp. is a dominant tropical legume that is typically grown in tropical and subtropical acid soils, where Mn and Al toxicity coexist (Noble et al., 2000). By screening yeast (Saccharomyces cerevisiae) cells through expression of a Stylosanthes hamata complementary DNA (cDNA) library, a vacuole-localized Mn transporter ShMTP1 was cloned from S. guianensis, and overexpression of ShMTP1 enhanced Mn toxicity tolerance in Arabidopsis (Arabidopsis thaliana; Delhaize et al., 2003). Using two S. guianensis genotypes contrasting in phosphorus acquisition efficiency and Al toxicity tolerance, an internal Al tolerance mechanism acting through mediation of malate synthesis has been suggested (Sun et al., 2014). In this study, one S. guianensis genotype (Fine-stem) was found to have superior Mn toxicity tolerance, which was accompanied by higher internal malate concentrations and exudation rates than the contrasting genotype (TPRC2001-1). Through proteomics analysis, increased accumulation of S. guianensis Malate Dehydrogenase1 (SgMDH1) was observed in Mn toxicity-treated roots of Fine-stem. Subsequent investigation demonstrated functions of SgMDH1 in controlling malate synthesis and Mn tolerance.
RESULTS
Characterization of Mn Tolerance in S. guianensis
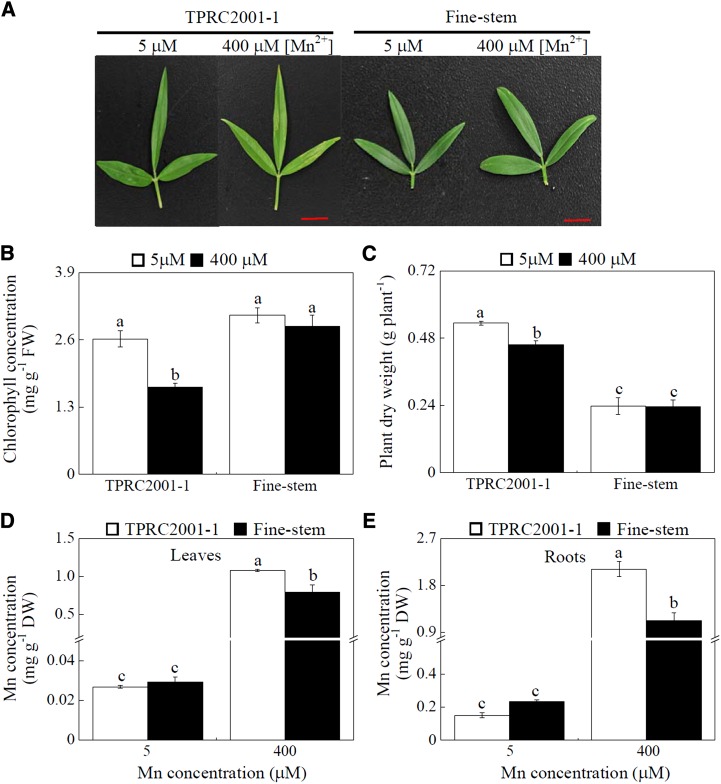
Two S. guianensis genotypes (TPRC2001-1 and Fine-stem) were hydroponically grown in nutrient solution at two Mn levels. After 10 d of treatment, leaf chlorosis was observed in TPRC2001-1 when subjected to 400 μm MnSO4, but not in Fine-stem (Fig. 1A). Leaf chlorophyll concentrations in TPRC2001-1 were 35% less with excess Mn exposure compared with those grown in the Mn control treatment, whereas Mn treatment did not significantly affect leaf chlorophyll concentrations in Fine-stem (Fig. 1B). Consistent with this result, excess Mn application resulted in inhibited growth only in TPRC2001-1, as reflected by a 14% decrease in plant dry weight (Fig. 1C). Although excess Mn application resulted in significant increases in Mn concentrations in both leaves and roots of both genotypes, Mn concentrations in TPRC2001-1 were greater than in Fine-stem by 36% and 111% for leaves and roots, respectively (Fig. 1, D and E). Two S. guianensis genotypes were further grown in the nutrient solution supplied with different Mn levels ranging from 5 to 800 µm (Supplemental Fig. S1). Different responses to Mn toxicity between the two genotypes were also observed at 200 µm Mn or above, as reflected by significant changes in both plant dry weight and chlorophyll concentrations (Supplemental Fig. S1). Taken together, these results demonstrate that Fine-stem is more Mn tolerant than TPRC2001-1 regardless of Mn toxicity levels.
Figure 1.
Growth and Mn concentrations of two S. guianensis genotypes at two Mn treatments. A, Leaves of S. guianensis after 10 d of Mn treatment. B, Leaf chlorophyll concentrations. C, Plant dry weight. D, Leaf Mn concentrations. E, Root Mn concentrations. S. guianensis seedlings were grown under normal conditions for 1 month and then treated with 5 or 400 µm MnSO4 for 10 d. F values of ANOVA analysis are as follows: B, 31.7 for genotype (P < 0.001), 15.6 for Mn treatment (P < 0.05), and 6.1 for interaction between genotype and Mn treatment (P < 0.05); C, 164.7 for genotype (P < 0.001), 3.86 for Mn treatment (no significant difference at the P = 0.05 level) and 3.38 for interaction between genotype and Mn treatment (no significant difference at the P = 0.05 level); D, 8.6 for genotype (P < 0.05), 356.4 for Mn treatment (P < 0.001), and 8.9 for interaction between genotype and Mn treatment (P < 0.05); and E, 19.3 for genotype (P < 0.001), 368.5 for Mn treatment (P < 0.001), and 29 for interaction between genotype and Mn treatment (P < 0.05). Each bar represents the mean of four independent replicates with se. Different letters indicate a significant difference at the P = 0.05 level. DW, Dry weight; FW, fresh weight. Bars = 1 cm.
Changes in Organic Acid Concentrations and Exudation in Response to Excess Mn
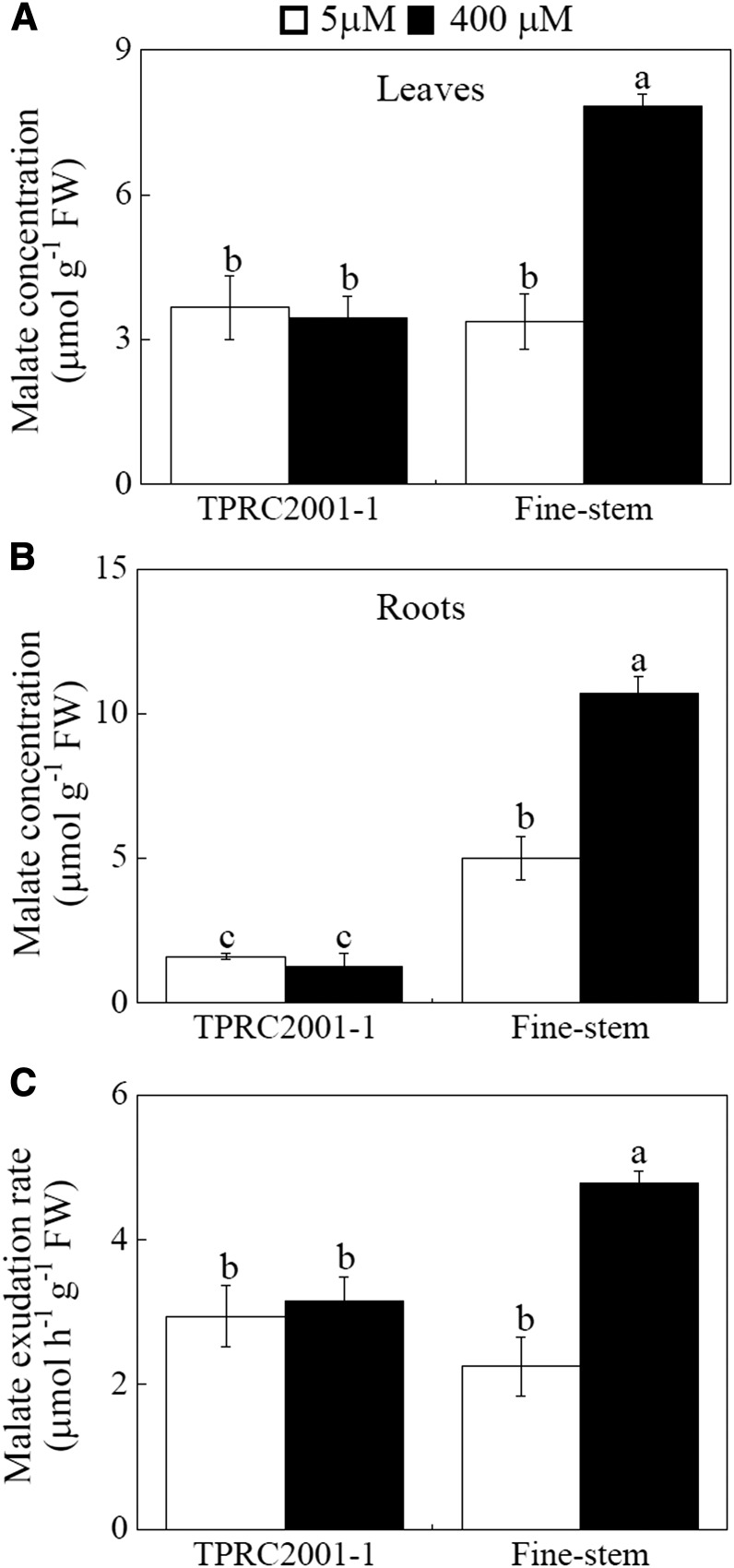
The concentrations and exudation of three organic acids (malate, citrate, and oxalate) were examined in the two S. guianensis genotypes identified as contrasting in Mn tolerance. The results showed that Mn toxicity affects internal concentrations and exudation of organic acids in a genotype-dependent matter (Fig. 2; Supplemental Fig. S2). Two-fold increases in internal malate concentrations in response to excess Mn were observed in both leaves and roots of Fine-stem (Fig. 2, A and B). However, excess Mn did not affect internal malate concentrations in either leaves or roots of TPRC2001-1, which were also significantly less than those in Fine-stem (Fig. 2, A and B). Furthermore, malate exudation increased only in Fine-stem in response to Mn toxicity, and this exudation was 66% higher in Fine-stem than in TPRC2001-1 (Fig. 2C). For the other two observed organic acids, excess Mn did not influence internal concentrations or exudation of either citrate or oxalate, except for a significant Mn-stimulated decrease in root oxalate concentrations in both genotypes (Supplemental Fig. S2). These results indicate that of the tested organic acids, excess Mn only regulates malate concentrations and exudation in the Mn-tolerant Fine-stem genotype.
Figure 2.
Malate concentrations and exudation in S. guianensis at two Mn levels. A, Malate concentrations in leaves. B, Malate concentrations in roots. C, Malate exudation from roots. S. guianensis seedlings were separately treated with 5 or 400 µm MnSO4. After 10 d, seedlings were transplanted into 40 mL of fresh nutrient solution to collect root exudates for 6 h. F values of ANOVA analysis are as follows: A, 16 for genotype (P < 0.05), 16.9 for Mn treatment (P < 0.05), and 20.9 for interaction between genotype and Mn treatment (P < 0.05); B, 148.5 for genotype (P < 0.001), 25.7 for Mn treatment (P < 0.05), and 33.1 for interaction between genotypes and Mn treatment (P < 0.001); and C, 2.2 for genotype (no significant difference at the P = 0.05 level), 18.8 for Mn treatment (P < 0.05), and 13.5 for interaction between genotype and Mn treatment (P < 0.001). Each bar represents the mean of four independent replicates with se. Different letters indicate a significant difference at the P = 0.05 level. FW, Fresh weight.
Identification of SgMDH1 through Analysis of Protein Profiles in Fine-stem Roots
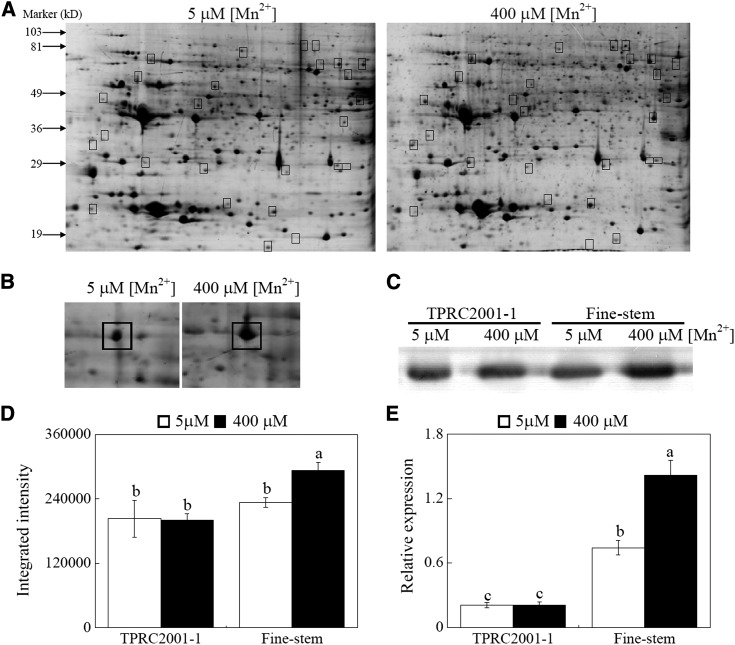
To identify differentially expressed proteins regulated by excess Mn in the roots of Fine-stem, protein profiles of Fine-stem were analyzed through two-dimensional electrophoresis (2-DE) combined with matrix-assisted laser-desorption ionization (MALDI)-time of flight (TOF)/TOF mass spectrometry (MS) analysis. In total, more than 600 protein spots were separated in the 2-DE gels, and 27 protein spots exhibited differential accumulation between the two Mn levels (Fig. 3A). Subsequently, 14 proteins were identified by MALDI-TOF/TOF MS analysis. Among them, intensity of one protein increased by more than 2-fold in response to Mn toxicity. This protein was further identified as a malate dehydrogenase (MDH) and was named SgMDH1 (Fig. 3B).
Figure 3.
Identification and expression levels of SgMDH1 in S. guianensis. A, Protein profiles of Fine-stem roots grown at two Mn levels. B, Magnified pictures of SgMDH1 accumulation in gels. C, Western-blot analysis of SgMDH1 in roots of two S. guianensis genotypes. D, The intensity of the bands in the western blot (C) was quantified by ImageJ software (http://developer.imagej.net/development) based on three independent replicates. E, Transcription levels of SgMDH1 in roots of two S. guianensis genotypes grown at two Mn levels. Boxes indicate differentially expressed proteins between the two Mn levels. F values of ANOVA analysis are as follows: D, 14.0 for genotype (P < 0.05), 2.9 for Mn treatment (no significant difference at the P = 0.05 level), and 3.6 for interaction between genotype and Mn treatment (no significant difference at the P = 0.05 level); and E, 115.8 for genotype (P < 0.001), 17.3 for Mn treatment (P < 0.05), and 17.1 for interaction between genotype and Mn treatment (P < 0.05). Each bar represents the mean of three independent replicates with se. Different letters indicate a significant difference at the P = 0.05 level.
Subsequently, the EST of SgMDH1 was amplified according to the conserved motifs of MDH in other plants. Based on the EST sequence of SgMDH1, 5′-RACE and 3′-RACE were performed to clone full-length SgMDH1 cDNA, which is 1,038 bp in length and encodes a protein with a predicated molecular mass of 36 kD. Phylogenetic analysis showed that MDH proteins in plants can be divided into two groups, and SgMDH1 belongs to group I. This S. guianensis protein exhibits high similarity to the MDH (GmAAD56659) in soybean (Glycine max; Supplemental Fig. S3). Using a SgMDH1-specific antibody, western-blot analysis was conducted to examine SgMDH1 accumulation in Fine-stem. The results showed that with excess Mn application, significant accumulation of SgMDH1 protein was observed only in the roots of Fine-stem, but not in the roots of TPRC2001-1 (Fig. 3, C and D).
Consistent to the protein accumulation patterns, transcription levels of SgMDH1 were increased by 2-fold in Fine-stem roots in response to 400 μm Mn, which was 6.8-fold higher than in TPRC2001-1 roots (Fig. 3E). These results suggest that the expression of SgMDH1 in Fine-stem roots is up-regulated by excess Mn at both transcription and protein levels. We also found that SgMDH1 expression was enhanced by excess Mn treatments ranging from 50 to 400 μm in the roots of Fine-stem, but was not responsive to 800 μm Mn treatment (Supplemental Fig. S4A). Furthermore, transcripts of SgMDH1 in roots were decreased by Al and Cu treatments, but not by the other metal stresses, including Cd, La, Fe, and Zn (Supplemental Fig. S4, B and C), suggesting that SgMDH1 expression is specifically increased by Mn toxicity.
SgMDH1 Catalyzes Malate Synthesis in Vitro, Which Is Enhanced by Mn2+ Addition
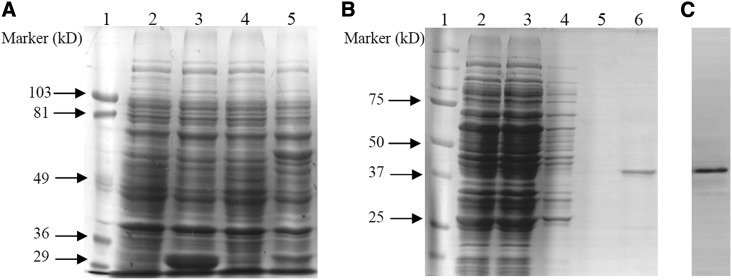
To investigate the biochemical properties of SgMDH1, the SgMDH1 encoding region was cloned and fused with a GST tag at the N terminus. Subsequently, SgMDH1 was expressed and purified from Escherichia coli lysates. SDS electrophoresis and western-blot analysis showed that the molecular mass of the purified SgMDH1 protein was approximately 36 kD after PreScission Protease digestion (Fig. 4).
Figure 4.
Purification of the recombinant SgMDH1 protein. A, SDS-PAGE analysis of SgMDH1 at different expression steps in E. coli. Lane 1, Molecular mass marker. Lane 2, Cells transformed with the empty vector pGEX-6P-3 without induction by 0.2 mm IPTG. Lane 3, Cells transformed with the empty vector pGEX-6P-3 induced by 0.2 mm IPTG. Lane 4, Cells transformed with the recombinant SgMDH1 protein without induction by 0.2 mm IPTG. Lane 5, Cells transformed with the recombinant SgMDH1 protein induced by 0.2 mm IPTG. B, SDS-PAGE analysis of SgMDH1 across purification steps in E. coli. Lane 1, Molecular mass marker. Lane 2, Total E. coli protein with the recombinant SgMDH1 protein induced by 0.2 mm IPTG. Lanes 3 to 5, Contaminate proteins at each of three steps of elution by binding buffer. Lane 6, Purified recombinant SgMDH1 protein after PreScission Protease incubation to cleave the GST tag. C, Western-blot analysis of purified recombinant SgMDH1.
Biochemical properties of SgMDH1 were further analyzed in vitro, including kinetic constants and activities affected by several cations. The SgMDH1 Km values were 0.158 mm for oxaloacetate (OAA) and 1.23 mm for malate (Table I). Furthermore, the specificity constant (catalytic constant [Kcat]/Km) of SgMDH1 was 475,000 for OAA and 126,017 for NADH, which was significantly higher than that for malate and NAD+ (Table I). These results suggest that SgMDH1 might primarily catalyze reduction of OAA with NADH to produce malate in vitro. Effects of various metal ions on SgMDH1 activity against OAA were then investigated. SgMDH1 activities were significantly increased by Mn2+ and inhibited by Fe2+, whereas other tested ions, including Cu2+, Mg2+, Zn2+, and Al3+, had no effect (Table II).
Table I. Kinetic constants and calculated kinetic parameters of the recombinant SgMDH1 protein.
Values represent the mean of three replicates with se.
| Substrate | Km | Vmax | Kcat/Km |
|---|---|---|---|
| mm | nmol−1 min−1 mg protein−1 | mm−1 min−1 | |
| OAA | 0.158 ± 0.00 | 790.6 ± 21.4 | 475,000 |
| NADH | 0.302 ± 0.05 | 400.6 ± 44.4 | 126,017 |
| Malate | 1.230 ± 0.13 | 134.1 ± 10.0 | 10,357 |
| NAD+ | 0.240 ± 0.03 | 211.7 ± 6.44 | 83,798 |
Table II. Effects of metal ions on the activity of the recombinant SgMDH1 protein.
SgMDH1 activity against 1 mm OAA was determined in reaction buffer supplied with or without the listed metal ions. Relative activity was expressed as the percentage of SgMDH1 activity without application of the metal ions. The relative activity value represents the mean of four replicates with se. Different letters in the same column indicate significant differences at the P = 0.05 level.
| Metal Ion | Concentration | Relative Activity |
|---|---|---|
| mm | % | |
| Mn2+ | 1.0 | 129.48 ± 7.72a |
| Cu2+ | 1.0 | 105.66 ± 10.00b |
| Mg2+ | 1.0 | 103.81 ± 7.71b |
| Zn2+ | 1.0 | 88.17 ± 3.10b |
| Al3+ | 1.0 | 87.13 ± 2.52b |
| Fe2+ | 1.0 | 46.75 ± 2.27c |
SgMDH1 Mediated Malate Synthesis in Yeast Cells and Plants
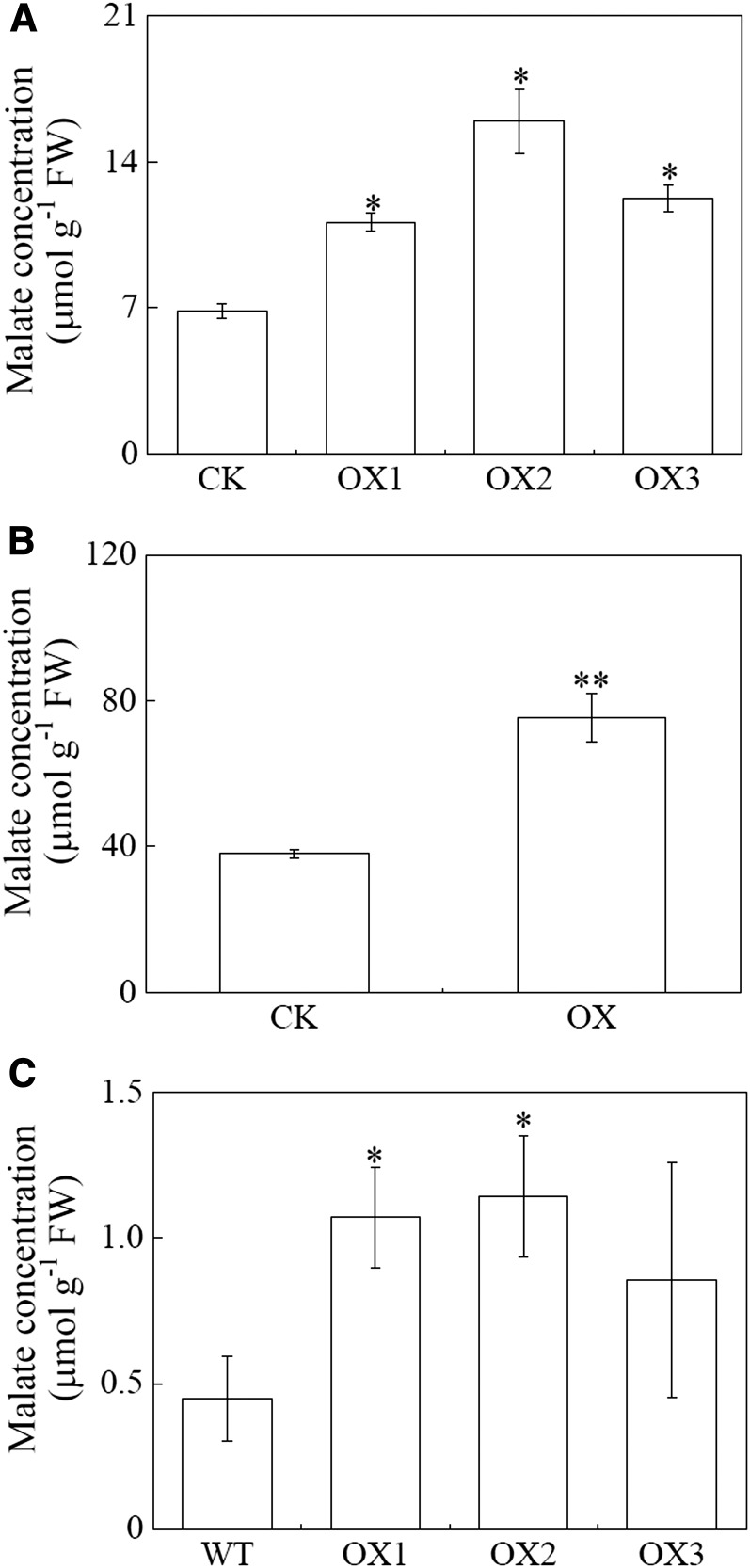
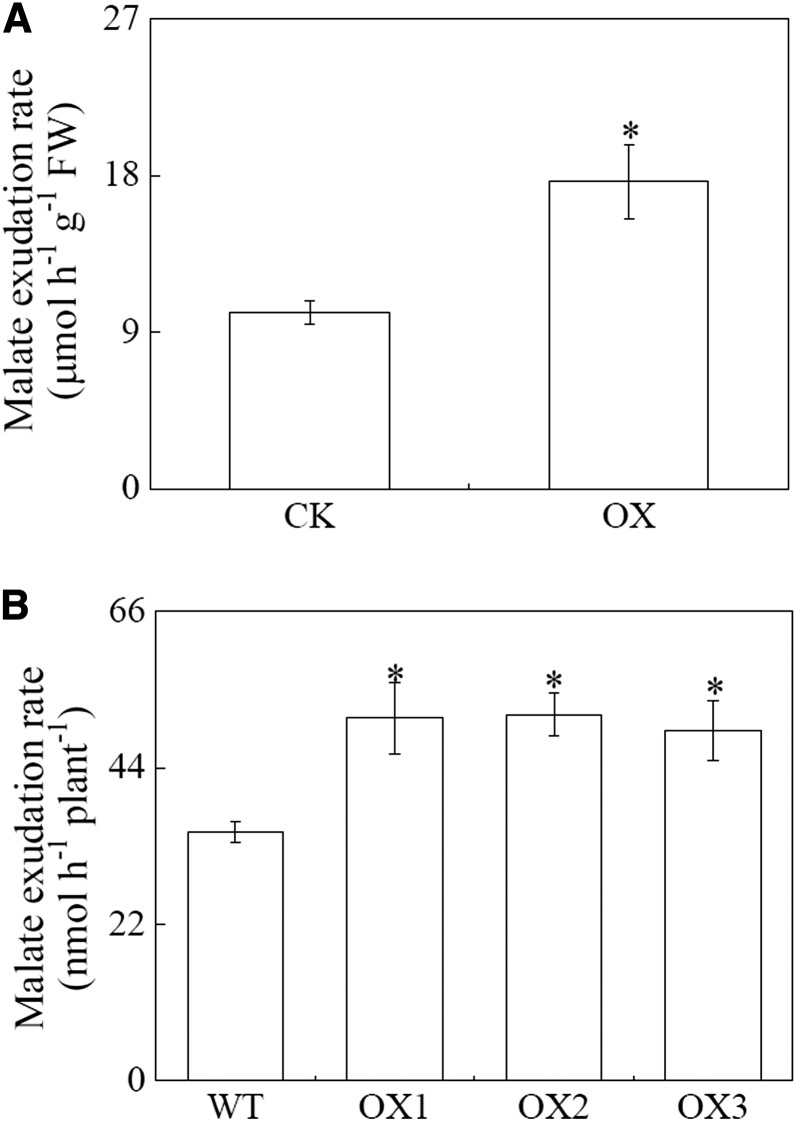
To test whether SgMDH1 functions in malate synthesis in vivo, SgMDH1 was heterologously overexpressed in yeast cells, soybean hairy roots, and Arabidopsis (Supplemental Fig. S5). Compared with controls, SgMDH1 overexpression led to increased internal malate concentrations by more than 50% in yeast cells, and more than 80% in soybean hairy roots and Arabidopsis (Fig. 5). Furthermore, increased malate concentrations associated with SgMDH1 overexpression were accompanied by increases in the rate of malate exudation over controls by 75% in soybean hairy roots and more than 41% in Arabidopsis (Fig. 6). These results demonstrate that SgMDH1 can mediate malate synthesis in yeast cells and plants.
Figure 5.
Malate concentrations in yeast cells, soybean hairy roots, and Arabidopsis with SgMDH1 overexpression. A, Malate concentrations in yeast cells. CK indicates yeast cells transformed with the empty vector pYES2. OX1, OX2, and OX3 are three clones with SgMDH1 overexpression. B, Malate concentrations in soybean hairy roots. CK indicates soybean hairy roots transformed with the empty vector. OX indicates soybean hairy roots overexpressing SgMDH1. C, Malate concentrations in Arabidopsis. WT indicates the wild-type Columbia ecotype of Arabidopsis. OX1, OX2, and OX3 are three transgenic lines overexpressing SgMDH1. Each bar presents the mean of four independent replicates with se. Asterisks indicate significant differences between the control and the SgMDH1 overexpression lines at the P = 0.05 level. A single asterisk indicates 0.01 < P < 0.05 and two asterisks indicate 0.001 < P < 0.01. FW, Fresh weight.
Figure 6.
Malate exudation rate in soybean hairy roots and Arabidopsis overexpressing SgMDH1. A, Malate exudation rate in soybean hairy roots. CK indicates soybean hairy roots transformed with the empty vector. OX indicates soybean hairy roots overexpressing SgMDH1. B, Malate exudation rate in Arabidopsis. WT indicates the wild-type Columbia ecotype of Arabidopsis. OX1, OX2, and OX3 are three transgenic lines overexpressing SgMDH1. Each bar represents the mean of four independent replicates with se. Asterisks indicate significant differences between the control and the SgMDH1 overexpression lines at the P = 0.05 level. A single asterisk indicates 0.01 < P < 0.05. FW, Fresh weight.
SgMDH1 Overexpression Confers Mn Tolerance in Yeast and Arabidopsis
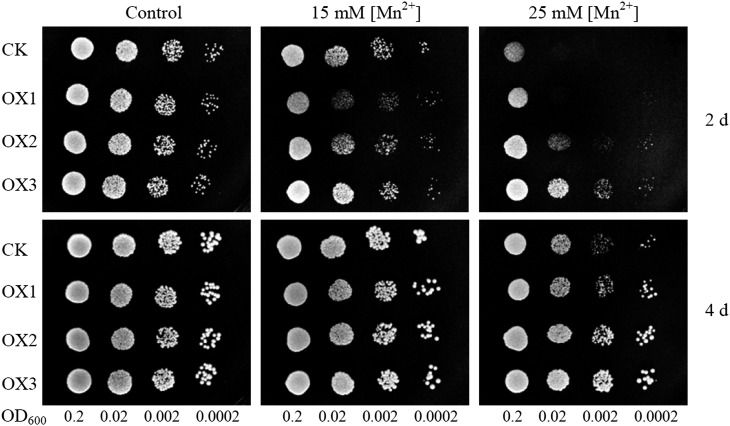
To further test whether SgMDH1 functions in tolerating Mn toxicity, SgMDH1-overexpressing yeast cells and Arabidopsis plants were separately subjected to excess Mn. As shown in Figure 7, the yeast cells harboring SgMDH1 attenuated sensitivity to excess Mn, compared with those harboring the pYES2 empty vector control, especially at the 25 mm MnSO4 level (Fig. 7).
Figure 7.
Growth performance of yeast cells overexpressing SgMDH1. Yeast cells with different optical density at 600 nm (OD600) of 0.2 and four gradient 1:10 dilutions were spotted on synthetic complete medium containing 0 (control), 15 mm, and 25 mm MnSO4 for 2 and 4 d at 30°C. CK indicates yeast cells transformed with the empty vector pYES2. OX1, OX2, and OX3 are three clones with SgMDH1 overexpression.
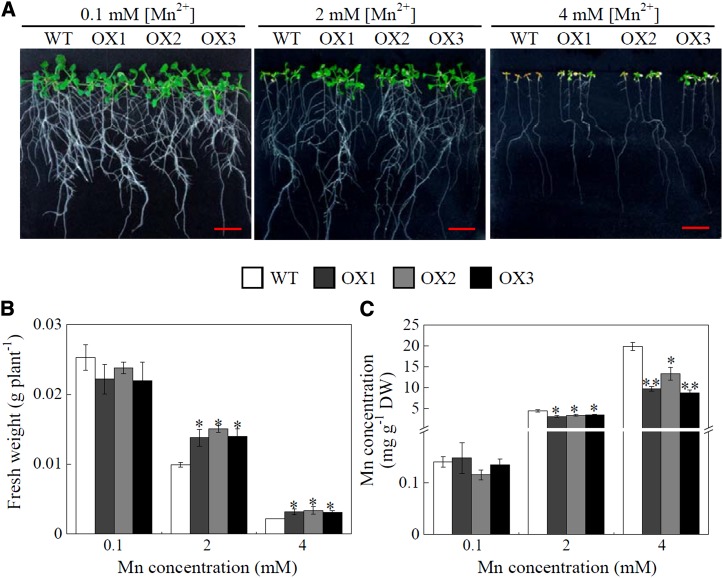
For Arabidopsis growth, no difference was observed between SgMDH1 overexpression lines and the wild type under normal conditions containing 0.1 mm MnSO4 (Fig. 8A). However, with increased Mn application, fresh weight decreased more severely in the wild type than in SgMDH1 overexpression plants, with final weight being 30% and 31% less for the wild type than that for SgMDH1 overexpression lines grown at 2 and 4 mm MnSO4 levels, respectively (Fig. 8, A and B). Furthermore, Mn concentrations in SgMDH1 overexpression lines were 20% and 30% less than those in the wild type grown at 2 and 4 mm MnSO4 levels, respectively (Fig. 8C). These results suggest that SgMDH1 overexpression confers Mn tolerance in Arabidopsis plants partially through reduced Mn accumulation in plants.
Figure 8.
Mn tolerance in Arabidopsis overexpressing SgMDH1. A, Growth performance of Arabidopsis at different Mn treatments for 7 d. B, Fresh weight of Arabidopsis plants. C, Mn concentrations in Arabidopsis. Seven-day-old seedlings were subjected to Mn treatments for 7 d before fresh weight and Mn concentrations were determined. WT indicates the wild-type Columbia ecotype of Arabidopsis. OX1, OX2, and OX3 are three transgenic lines overexpressing SgMDH1. Each bar represents the mean of four independent replicates with se. Asterisks mean significant differences between the wild type and the SgMDH1 overexpression lines. *, 0.01 < P < 0.05; **, 0.001 < P < 0.01. DW, Dry weight. Bars = 1 cm.
Exogenous Malate Alleviated Mn Toxicity in S. guianensis Plants and Yeast Cells
To investigate effects of exogenous malate on Mn tolerance in TPRC2001-1, malate was applied in Murashige and Skoog solid culture medium containing 20 mm MnSO4. Under excess Mn conditions, relative root length of S. guianensis plants with 10 μm and 40 μm exogenous malate application was more than 12% longer compared with S. guianensis plants grown in the medium without malate application (Supplemental Fig. S6, A and B). Furthermore, chlorophyll concentrations and plant fresh weight of S. guianensis plants with exogenous malate application were significantly higher than those without malate addition (Supplemental Fig. S6, C and D). Consistently, exogenous 0.1 µm malate application also helped yeast cells to grow better than those without malate addition in the medium under excess Mn conditions (Supplemental Fig. S7).
DISCUSSION
Fine-stem Has Superior Mn Tolerance
Manganese toxicity is one of the major constraints limiting crop growth on acid soils. It is generally assumed that plants well adapted to acid soils might develop an array of unique strategies to attain this adaptation, and thus exhibit a superior ability to tolerate Mn toxicity (Noble et al., 2000; Delhaize et al., 2003). As a dominant pasture crop on acid soils, superior Mn tolerance in S. guianensis has been suggested through analysis of Mn toxicity thresholds in 12 different pasture legumes. Among pasture legumes, the Mn toxicity threshold value for Stylosanthes humilis, one Stylosanthes spp., was 1.14 mg g−1 dry weight for shoots, which was much higher than that for eight other pasture legumes (Andrew and Hegarty, 1969). Similarly, Mn concentrations in TPRC2001-1 were 1.1 mg g−1 for leaves and 2.1 mg g−1 for roots (Fig. 1, D and E), which were also higher than those reported in most pasture legumes (Andrew and Hegarty, 1969). Furthermore, it has been demonstrated that treatment with 50 μm MnSO4 may result in formation of brown spots in leaves and significant growth reduction in several crops such as cowpea, barley (Hordeum vulgare), and soybean (Fecht-Christoffers et al., 2003; Führs et al., 2010; Yan et al., 2010). In this study, no symptom of Mn toxicity was observed in either of the tested S. guianensis genotypes when grown in Mn concentrations less than 200 μm (Supplemental Fig. S1A). These results suggest that S. guianensis indeed exhibits higher Mn tolerance than most other plants. Within S. guianensis, although TPRC2001-1 grows about twice as Fine-stem under normal conditions, genotypic differences in responses to Mn toxicity were observed at various Mn levels ranging from 200 to 800 μm (Fig. 1, B and C; Supplemental Fig. S1). Fine-stem has superior Mn tolerance, as reflected by its ability to avoid the significant reductions in leaf chlorophyll concentrations and plant dry weight observed in TPRC2001-1 grown at Mn levels of 200 μm or more (Supplemental Fig. S1). Therefore, shedding light on mechanisms underlying Mn tolerance in Fine-stem can not only clarify enigmatic mechanisms of S. guianensis adaptation to acid soils, but can also extend this understanding to Mn tolerance strategies throughout the plant kingdom.
Mn Tolerance of Fine-stem Is Partially Attributable to Internal Mn Detoxification through Malate Synthesis
With Mn applied in excess, internal Mn concentrations increased in both S. guianensis genotypes (Fig. 1, D and E), suggesting that detoxification of internal Mn might be required for S. guianensis to tolerate Mn toxicity. It has been well documented that internal Mn can be complexed by internal organic acids (e.g. malate, oxalate, and citrate) in plants, including cowpea and Phytolacca americana and other (hyper)accumulator plant species (Dou et al., 2009; Fernando et al., 2010; Kopittke et al., 2013). For example, with the help of x-ray absorption near-edge spectroscopy and extended x-ray absorption fine-structure spectroscopy techniques, malate has been demonstrated to chelate internal Mn within the leaf tissues in Gossia bidwillii (Fernando et al., 2010). Furthermore, in the roots of cowpea and leaves of Phytolacca acinosa, it has been estimated that 80% and 90% of internal Mn can be bound in complexes with citrate and oxalate, respectively (Xu et al., 2009; Kopittke et al., 2013). Therefore, in this study, changes in the internal concentrations of malate, oxalate, and citrate in response to Mn toxicity were examined in both S. guianensis genotypes. Interestingly, excess Mn-stimulated increases of internal malate concentrations in both leaves and roots were observed only in Fine-stem, with the internal malate concentrations increasing 2-fold to the concentrations that were significantly greater than was observed in TPRC2001-1 (Fig. 2, A and B). At the same time, no increase in internal concentrations of oxalate and citrate was observed in either S. guianensis genotype (Supplemental Fig. S2). These results suggest that malate synthesis could be partially controlled by excess Mn, and may thus be related to Mn tolerance. Furthermore, it has been documented that Mn could be complexed with oxalate, citrate, or malate at a ratio of approximately 1:1 in plants (Dou et al., 2009; Xu et al., 2009; Fernando et al., 2010). However, to further clarify the functions of malate in internal Mn detoxification, it will be valuable and necessary to investigate whether Mn-malate complexes are present in S. guianensis leaves and roots using x-ray absorption near-edge spectroscopy or extended x-ray absorption fine-structure spectroscopy techniques.
In addition, it is generally believed that compartmentalization of excess internal Mn into vacuoles is an internal Mn tolerance mechanism in plants (Delhaize et al., 2007). In this study, the gene SgMTP1 was cloned. This gene exhibits high homology to ShMTP1, which functions in sequestration of Mn into vacuoles in S. hamata (Delhaize et al., 2003). The expression of SgMTP1 in leaves and roots was significantly higher in TPRC2001-1 than in Fine-stem, and responded to excessive Mn treatments only in TPRC2001-1 roots (Supplemental Fig. S8). Therefore, variations of excess Mn tolerance between Fine-stem and TPRC2001-1 cannot be explained by SgMTP1 expression and any potentially associated vacuole sequestration.
Mn Tolerance in Fine-stem Conferred through Exclusion Occurs Primarily via Malate Exudation
Compared with TPRC2001-1, lower Mn concentrations were found in both leaves and roots of Fine-stem under excess Mn conditions (Fig. 1), suggesting that Fine-stem might have the capability to exclude Mn and prevent excess acquisition. However, few studies have tried to clarify Mn exclusion mechanisms in plants, and the results are not conclusive. For example, de la Luz Mora et al. (2009) found that Mn-tolerant ryegrass cultivars accumulate less Mn than Mn-sensitive cultivars, and this is accompanied by the release of more citrate and oxalate released from tolerant cultivars. Yet direct evidence to support Mn exclusion in plants through organic acid secretion is still lacking. Here, the Mn-tolerant genotype (Fine-stem) exhibited higher malate exudation rates than were observed in TPRC2001-1 in response to Mn toxicity (Fig. 2). Considering that secreted organic acids (e.g. malate) can chelate Al, and thus alleviate Al toxicity via the exclusion of Al from plant tissues, such as through the function of the Aluminum-Activated Malate Transporter1 (ALMT1; Sasaki et al., 2004; Liang et al., 2013), it is reasonable to assume that secreted malate might have similar functions for Mn detoxification in plants. Interestingly, we found that expression of SgALMT1 was enhanced by relatively high Mn levels (i.e. from 50 to 400 µm MnSO4) in Fine-stem, but not in TPRC2001-1 (Supplemental Fig. S9), suggesting that coordinate regulation of SgMDH1 and SgALMT1 might facilitate increased malate secretion in Fine-stem subjected to excess Mn. To test whether exogenous malate could alleviate Mn toxicity, we applied malate into the growth medium for yeast and S. guianensis under excess Mn conditions. The results showed that application of exogenous 0.1 µm malate could alleviate Mn toxicity in yeast cells, despite no effect being observed with 10 µm malate application, which might be because high malate application could affect yeast cell growth (Supplemental Fig. S7). Consistently, exogenous malate actually helps to alleviate Mn toxicity in S. guianensis (Supplemental Fig. S6). Furthermore, reduced Mn accumulation was also observed through enhanced malate exudation in transgenic Arabidopsis lines overexpressing SgMDH1 (Figs. 6 and 8). Taken together, these results provide direct evidence that secreted malate can detoxify rhizosphere Mn, and Mn exclusion through the action of organic acids might be an important strategy in plants to alleviate Mn toxicity.
SgMDH1 Plays Critical Roles in Mn Tolerance of Fine-stem through Regulation of Malate Synthesis and Exudation
To elucidate molecular mechanisms of Fine-stem tolerance to Mn toxicity, proteomics analysis was conducted with Fine-stem roots. A Mn toxicity up-regulated protein (SgMDH1) was identified (Fig. 3, A and B), and was found to exhibit higher expression levels in Fine-stem than in TPRC2001-1 under excess Mn conditions (Fig. 3, C and D). Furthermore, overexpressing SgMDH1 significantly improved Mn tolerance in yeast and Arabidopsis (Figs. 7 and 8), suggesting that SgMDH1 plays vital roles in Mn tolerance.
In plants, malate synthesis is catalyzed by several enzymes, including phosphoenolpyruvate carboxylase, MDH, malate enzyme, and malate synthase (Etienne et al., 2013). Among these, MDH (EC.1.1.1.37) is generally considered to catalyze the reversible conversion of malate to OAA (Gietl, 1992). Cumulative results generated through in vitro biochemical analysis of purified recombinant or native MDH have shown that plant MDHs can catalyze the reduction of OAA to malate (Trípodi and Podesta, 2003; Yao et al., 2011). In this study, in vitro conversion of OAA to malate was the favored direction of the reaction catalyzed by SgMDH1, as reflected by higher Kcat and Kcat/Km values for OAA reduction compared with malate oxidation (Table I). Similarly, other MDHs have exhibited higher Kcat and Kcat/Km values against OAA as substrate, and thus are suggested to drive OAA reduction to form malate in plants (Ding and Ma, 2004; Yao et al., 2011). Furthermore, SgMDH1 overexpression resulted in increased internal malate concentrations by more than 50% in yeast and by 80% in soybean hairy roots and Arabidopsis (Fig. 5). Consistent with this, it has been demonstrated that malate synthesis has been increased through overexpression of cytoplasm-localized MDH in alfalfa (Medicago sativa) and tobacco (Nicotiana tabacum; Tesfaye et al., 2001; Wang et al., 2010). We found that SgMDH1 was also localized in the cytoplasm through the transient expression of SgMDH1 in Arabidopsis mesophyll protoplasts (Supplemental Fig. S10). These results strongly indicate that SgMDH1 does indeed catalyze OAA reduction to malate, and thus mediates malate synthesis in S. guianensis. Moreover, SgMDH1 activity against OAA was enhanced by Mn application in vitro (Table II), which perfectly matches the increased malate concentrations and exudation observed in the Mn-tolerant genotype, Fine-stem. In total, these results imply critical roles for SgMDH1 in Mn tolerance of S. guianensis.
Given the results outlined above, it is reasonable to conclude that the superior Mn tolerance of Fine-stem is achieved by coordination of internal and external Mn detoxification through malate synthesis and exudation, and that these activities are regulated by SgMDH1 accumulation at both the transcription and protein levels.
MATERIALS AND METHODS
Plant Materials and Treatments
Two Stylosanthes guianensis Aubl. genotypes, TPRC2001-1 and Fine-stem, were used. Seeds of both S. guianensis genotypes were soaked in hot water (80°C) for 3 min, and germinated for 3 d as previously described (Du et al., 2009). Seedlings were transferred to a modified Hoagland nutrient solution (Sun et al., 2014), and the solution pH was adjusted to 5.8 by adding H2SO4 or KOH. After 30 d, seedlings were transplanted into nutrient solution, pH 5.0, supplied with 5 to 800 μm MnSO4. After 10 d, roots and shoots were separately harvested and stored at −80°C for further analysis. All experiments had four biological replicates.
For other metal treatments, S. guianensis seedlings were grown under normal conditions for 30 d as described above, and then the seedlings were separately transplanted into nutrient solution, pH 5.0, supplied with 10 µm CuSO4, 800 µm FeNa-EDTA and 20 µm ZnSO4 for 5 d growth, or exposed to a 0.5 mm CaCl2 solution containing 0, 100 μm AlCl3, 40 μm CdCl2, or 20 μm LaCl3 at pH 4.5 for 1 d. Roots were harvested and stored at −80°C for further analysis. All experiments had three biological replicates.
Determination of Chlorophyll Concentrations
To determine chlorophyll concentrations, 0.05 g of fresh leaf tissue was extracted with 10 mL of 80% (v/v) acetone for 2 h. Absorbance was measured at wavelengths of 440, 645, and 663 nm. The chlorophyll concentrations were calculated according to published methods (Lichtenthaler and Wellburn, 1983). All experiments had four biological replicates.
Analysis of Mn Concentrations in Plant Tissues
Shoots and roots were oven-dried and ground into powder. Approximately 0.07-g samples were subsequently burned to ash in a muffle furnace at 600°C for 10 h. After cooling to room temperature, sample ash was dissolved in 100 mm HCl, and the resulting solution was used to measure Mn concentrations through atomic absorption spectroscopy as previously described (Murphy and Riley, 1962). All experiments had four biological replicates.
Organic Acid Determination
After S. guianensis seed germination, seedlings were grown in nutrient solution for 30 d and then subjected to Mn treatments as described above for 10 d. To collect root exudates from plants, seedlings were transferred to fresh nutrient solution. After 6 h, collected solution was stored at −80°C, and then concentrated to 1 mL using a freeze-drying vacuum system (Labconco). To determine internal organic acid concentrations, approximately 0.2 g of plant tissues was ground with 1.2 mL of 0.25 m HCl, and the homogenate was then heated to 80°C for 20 min with intermittent shaking. After centrifugation at 12,000g for 15 min, the supernatant was collected for analysis. All samples were passed through 0.2-μm filters and analyzed by HPLC (1260 Infinity LC; Agilent) according to a modified previously described method (Xu et al., 2006). All experiments had four biological replicates.
Identification of Mn-Responsive Proteins through 2-DE and MALDI-TOF/TOF MS Analysis
To detect Mn-responsive proteins in Fine-stem roots, 2-DE combined with MALDI-TOF/TOF MS analysis was conducted as previously described (Chen et al., 2011). Briefly, about 3.0 g of fresh Fine-stem roots was homogenized with 8.0 mL of extraction buffer consisting of 50% (v/v) phenol, 0.45 m Suc, 25 mm EDTA, 2% (v/v) β-mercaptoethanol, 250 mm Tris-HCl, pH 8.8, 2 mm phenylmethylsulfonyl fluoride, 2% (w/v) polyvinylpyrrolidone, 0.7 m NaCl, and 20% (v/v) ethanol. Protein concentrations were determined using a two-dimensional Quant Kit (GE Healthcare,), and 800 μg of protein was loaded onto immobilized pH gradient strips (pH 4.0–7.0 linear gradient, 24 cm) for isoelectric focusing using an Etten IPGphor3 system (GE Healthcare). The second dimension of electrophoresis was performed through an Etten DALTsix electrophoresis unit (GE Healthcare). Three biological replicates were included in this experiment. PDQuest 7.0 software (Bio-Rad) was used to analyze and count differentially expressed protein spots. Selected protein spots in the gels were excised and digested with trypsin. MALDI-TOF/TOF MS analysis was used to identify these proteins using an ABI 4700 TOF-TOF system (Applied Biosystems). Data were analyzed in GPS Explorer (version 3.6) with the MASCOT (2.1) search engine run against the National Center for Biotechnology Information database (NBCInr 20100724). Only proteins with scores greater than 71 or ion scores (tandem MS) above 30 were considered to be successfully identified. Functions of identified proteins were predicted at the KEGG Web site (http://www.genome.jp/kegg/).
Isolation and Characterization of SgMDH1
Total RNA was extracted from Fine-stem roots subjected to excess Mn (400 μm MnSO4) using the Trizol reagent (Invitrogen) according to manufacturer protocols. First-strand cDNA was synthesized from 2 μg DNase I-treated RNA using Moloney-Murine Leukemia Virus reverse transcriptase (Promega). To clone SgMDH1, primers were designed according to the conserved motif sequences of four MDH genes from Lotus japonicus (BT146072), Glycine max (XM_003527163), Medicago truncatula (AF020271), and Ricinus communis (XM_002524216). A 354-bp fragment was amplified using the forward (F) and reverse (R) primers SgMDH1-EST-F and SgMDH1-EST-R (Supplemental Table S1). The PCR product was then cloned into a pMD18-T vector (Takara) and sequenced. Subsequently, the full-length cDNA of SgMDH1 was cloned through RACE according to manufacturer protocols (Clontech). Briefly, based on EST sequences, two primers were designed for PCR amplification of SgMDH1 using SgMDH1-5′-RACE-F and SgMDH1-3′-RACE-R (Supplemental Table S1). Pairs with a universal primer A mix (Supplemental Table S1) were separately used to amplify 5′ and 3′ terminals of SgMDH1 from the respective 5′-RACE and 3′-RACE constructs obtained from Fine-stem roots subjected to excess Mn. Sequences of the three fragments were analyzed and combined together in the MEGA 4.1 program to generate a full-length cDNA of SgMDH1, which was deposited in the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/, accession no. KJ123727).
Western-Blot Analysis
The specific peptide (EKEGLEKLKPEL) of SgMDH1 was synthesized and used to produce a polyclonal antibody (Abmart). Western-blot analysis was carried out to detect the expression of SgMDH1 in roots of two S. guianensis genotypes as previously described (Liang et al., 2010). Briefly, protein from roots of two S. guianensis genotypes exposed to 5 or 400 μm MnSO4 treatments was separately extracted as described above, and then electrophoretically transferred to a polyvinylidene fluoride membrane (Bio-Rad). The membrane was consecutively hybridized with the SgMDH1 antibody (1:200 dilution, v/v) and an alkaline phosphatase-tagged secondary antibody, and was then assayed as described in the reference above. The intensity of bands in western-blot analysis with three biological replicates was further quantified using ImageJ software (http://developer.imagej.net/development).
Quantitative Real-Time PCR Analysis
Quantitative real-time PCR analysis was conducted using SYBR Premix Ex Taq II (Takara) and analyzed using a Rotor-Gene 3000 quantitative real-time PCR system (Corbett Research). The primers SgMDH1-RT-F and SgMDH1-RT-R, SgMTP1-RT-F and SgMTP1-RT-R, or SgALMT1-RT-F and SgALMT1-RT-R (Supplemental Table S1) were used to assay SgMDH1, SgMTP1, and SgALMT1 expression in the two tested S. guianensis genotypes grown in control and high-Mn treatments, respectively. The housekeeping gene SgEF-1a (accession no. JX164254) was used as the internal control. Relative expression levels were calculated as the ratio of expression of the tested gene to the housekeeping gene. Gene expression levels from three biological replicates were analyzed.
Expression and Purification of SgMDH1 in Escherichia coli
The open reading frame (ORF) of SgMDH1 was amplified using SgMDH1-GST-F and SgMDH1-GST-R (Supplemental Table S1). The amplified product was digested with BamHI and EcoRI, and then cloned into the pGEX6P-3 vector (GE Healthcare). The constructs were transformed into E. coli BL21 and expressed as N-terminal fusion proteins with a GST tag. Glutathione Sepharose 4B (GE Healthcare) was used to purify SgMDH1 fusion protein according to the manual (GE Healthcare). Briefly, transformants were grown overnight in Luria-Bertani medium (10 g L−1 tryptone, 5 g L−1 yeast [Saccharomyces cerevisiae] extract, and 10 g L−1 NaCl) supplied with 0.2 mm isopropylthio-d-galactoside (IPTG) until its absorbance reached 0.4 at 600 nm. The bacteria were harvested and resuspended in binding buffer containing 140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, and 1.8 mm KH2PO4, pH 7.3. After ultrasonication, bacteria cell lysates were mixed with Glutathione Sepharose 4B. After incubation for 3 h, the mix was washed with the binding buffer three times. Subsequently, 1 mg of fused protein was treated with 20 U of PreScission Protease (GE Healthcare) in the PreScission cleavage buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, and 1 mm dithiothreitol, pH 7.5) for 4 h at 4°C. Finally, SgMDH1 protein without a GST tag was eluted using the PreScission cleavage buffer. Expression and purity of SgMDH1 was analyzed by Coomassie Blue R-250 staining, SDS-PAGE, and western-blot analyses.
SgMDH1 Activity and Biochemical Assay
SgMDH1 activity was spectrophotometrically measured at 340 nm for 2 min according to the method of López-Calcagno et al. (2009) with some modifications. To detect OAA reduction catalyzed by SgMDH1, 0.11 μg of protein was incubated in reaction buffer containing 100 mm Tris-HCl, pH 8.0, 5 mm EDTA, 0.2 mm NADH, and 1 mm OAA. For determination of malate oxidation catalyzed by SgMDH1, 0.38 μg of protein was incubated in the reaction buffer containing 100 mm Tris-HCl, pH 9.5, 5 mm EDTA, 1 mm NAD+, and 50 mm l-malate. The Km and Vmax values of the SgMDH1 catalyzed reactions were determined by extrapolation from Lineweaver-Burke plots. The effects of metal ions on the activity of SgMDH1 were separately studied upon the addition of 1 mm MnCl2, FeSO4, MgCl2, CuSO4, ZnSO4, or AlCl3 to the reaction buffer. SgMDH1 activity was presented as nanomoles of the monitored reactant NADH per minute per milligram of protein.
Yeast Transformation and Mn Tolerance Analysis
The ORF of SgMDH1 was amplified by PCR using the primers SgMDH1-pYES2-F and SgMDH1-pYES2-R (Supplemental Table S1). Amplified product was digested with BamHI and EcoRI, and inserted into the pYES2 vector (Invitrogen). The vector overexpressing SgMDH1 and the empty vector were separately introduced into the yeast strain INVSC1 according to manufacturer protocols (Invitrogen). Three clones with overexpressing SgMDH1 and one clone with the empty vector were selected for further analysis. For Mn tolerance experiments, yeast transformants were first precultured in liquid synthetic complete-uracil medium until the OD600 value reached 0.6, and then precultured yeast cells were adjusted to an OD600 value 0.2 in the same medium. Dilutions (optical densities at OD600 of 0.2, 0.02, 0.002, and 0.0002) of yeast cells were spotted on induction plates containing 2% (w/v) Gal, 1% (w/v) raffinose, 0.67% (w/v) yeast nitrogen base without amino acids, 0.1% (w/v) uracil dropout mix, and 2% (w/v) agar, which was surface plated with 0, 15, and 25 mm MnSO4, pH 5.0, for 2 and 4 d at 30°C.
To determine internal malate concentrations in yeast cells, the yeast cells were grown in 100 mL of liquid induction medium for 1 d. After centrifugation at 6,000g for 10 min at 4°C, pellets were each washed twice with 1 mL of Millipore water. Yeast samples were then ground in liquid nitrogen and dissolved in 1.2 mL of 0.25 m HCl and heated to 80°C for 20 min with intermittent shaking. Samples were collected by centrifugation at 12,000g for 10 min. Finally, malate concentrations in the supernatant were analyzed as described above.
Overexpressing SgMDH1 in Arabidopsis and Functional Analysis
To construct SgMDH1 overexpression vector for Arabidopsis transformation, the ORF of SgMDH1 was amplified with the primers SgMDH1-OX-F and SgMDH1-OX-R (Supplemental Table S1). The amplified fragment was digested with BamHI and MluI, and cloned into the pYLRNAi binary vector. The construct was transformed into Agrobacterium tumefaciens strain Gv3101 using the freeze-thaw method, and subsequently introduced into ecotype Columbia of Arabidopsis (Arabidopsis thaliana) plants as described by Clough and Bent (1998). Through semiquantitative PCR analysis, three homozygous transgenic lines (i.e. OX1, OX2, and OX3) were selected for further studies. Ecotype Columbia 0 of Arabidopsis (wild type) and transgenic seeds were surface sterilized, and then germinated on petri dishes containing modified Murashige and Skoog solid medium.
After 7 d, 50 uniform seedlings were transplanted into Murashige and Skoog nutrient solution with continuous shaking. After 7 d, seedlings were transferred into collection buffer containing 0.5 mm CaCl2, pH 5.0, for 1 d to collect root exudates for analysis of malate concentrations and exudation. Malate concentrations were measured as described above. To determine Mn concentrations in Arabidopsis, wild-type and transgenic seeds were surface sterilized and germinated on modified Murashige and Skoog solid medium. After 7 d, uniform seedlings were selected and transplanted onto Murashige and Skoog solid medium supplied with 0.1, 2, or 4 mm MnSO4. Another 7 d later, seedlings were harvested to determine fresh weight and Mn concentrations as described above. Each treatment had four biological replicates.
Generation of Transgenic Soybean Hairy Roots with SgMDH1 Overexpression
The ORF of SgMDH1 was amplified and inserted into the pYLRNAi vector as described above. Constructs were introduced into Agrobacterium rhizogenes strain K599, which then induced the formation of transgenic soybean (Glycine max) hairy roots as described by Guo et al. (2011).
Effects of Exogenous Malate on the Growth of S. guianensis under Excess Mn
S. guianensis genotype TPRC2001-1 seeds were surface sterilized, and then germinated on petri dishes containing modified Murashige and Skoog solid medium. After germination for 2 d, uniform TPRC2001-1 seedlings were exposed to 0.1 or 20 mm MnSO4 treatments on the Murashige and Skoog solid culture medium containing 0, 10, or 40 μm exogenous malate at pH 5.0 for 4 d. Seedlings were harvested to determine fresh weight and chlorophyll concentrations. Each treatment had three biological replicates.
Subcellular Localization of SgMDH1
To investigate the subcellular localization of SgMDH1, the encoding regions of SgMDH1 were amplified using SgMDH1-GFP-F and SgMDH1-GFP-R primers (Supplemental Table S1), which were subsequently digested with XbaI and BamHI, and cloned into the binary pEGFP vector. Arabidopsis mesophyll protoplasts were prepared from 3- to 4-week-old wild-type plants, and then the 35S:SgMDH1-GFP constructs and empty vector were separately transiently expressed in protoplasts according to Yoo et al. (2007). The GFP signal in Arabidopsis mesophyll protoplasts was monitored using a confocal microscope (Zeiss LSM 710 NLO) 12 h after polyethylene glycol calcium transfection.
Statistical Analysis
All statistical analysis was performed by one-way ANOVA and Student’s t tests using SPSS software (version 13.0; SPSS Institute).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Growth and chlorophyll concentrations of two S. guianensis genotypes at different Mn levels.
Supplemental Figure S2. Internal and secreted citrate and oxalate in two S. guianensis genotypes grown at two Mn treatments.
Supplemental Figure S3. Phylogenetic analysis of SgMDH1 with other plant MDHs.
Supplemental Figure S4. Expression pattern of SgMDH1 in S. guianensis subjected to toxicity of Mn and other metal ions.
Supplemental Figure S5. Expression analysis of SgMDH1 in transgenic Arabidopsis.
Supplemental Figure S6. Effects of exogenous malate on the growth of TPRC2001-1 under excess Mn conditions.
Supplemental Figure S7. Growth of yeast cells under excess Mn conditions supplied with malate.
Supplemental Figure S8. Expression analysis of SgMTP1 in two S. guianensis genotypes at two Mn levels.
Supplemental Figure S9. Expression of SgALMT1 responsive to different Mn concentrations.
Supplemental Figure S10. Subcellular localization of SgMDH1 in Arabidopsis mesophyll protoplasts.
Supplemental Table S1. List of primers used in the study for functional analysis of SgMDH1, and expression analysis of SgMTP1 and SgALMT1.
Supplementary Material
Acknowledgments
We thank Dr. Thomas Walk for critical reading and Dr. Liang Cuiyue for guiding organic analysis and yeast transformation.
Glossary
- cDNA
complementary DNA
- 2-DE
two-dimensional electrophoresis
- MALDI
matrix-assisted laser-desorption ionization
- TOF
time of flight
- MS
mass spectrometry
- MDH
malate dehydrogenase
- OAA
oxaloacetate
- ORF
open reading frame
- IPTG
isopropylthio-d-galactoside
Footnotes
This work was supported by the National Key Basic Research Special Funds of China (grant no. 2011CB100301).
References
- Andrew CS, Hegarty MP (1969) Comparative responses to manganese excess of eight tropical and four temperate pasture legume species. Aust J Agric Res 20: 687–696 [Google Scholar]
- Chen Z, Cui Q, Liang C, Sun L, Tian J, Liao H (2011) Identification of differentially expressed proteins in soybean nodules under phosphorus deficiency through proteomic analysis. Proteomics 11: 4648–4659 [DOI] [PubMed] [Google Scholar]
- Chen Z, Fujii Y, Yamaji N, Masuda S, Takemoto Y, Kamiya T, Yusuyin Y, Iwasaki K, Kato S, Maeshima M, et al. (2013) Mn tolerance in rice is mediated by MTP8.1, a member of the cation diffusion facilitator family. J Exp Bot 64: 4375–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- de la Luz Mora M, Rosas A, Ribera A, Rengel Z (2009) Differential tolerance to Mn toxicity in perennial ryegrass genotypes: involvement of antioxidative enzymes and root exudation of carboxylates. Plant Soil 320: 79–89 [Google Scholar]
- Delhaize E, Gruber BD, Pittman JK, White RG, Leung H, Miao Y, Jiang L, Ryan PR, Richardson AE (2007) A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. Plant J 51: 198–210 [DOI] [PubMed] [Google Scholar]
- Delhaize E, Kataoka T, Hebb DM, White RG, Ryan PR (2003) Genes encoding proteins of the cation diffusion facilitator family that confer manganese tolerance. Plant Cell 15: 1131–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Ma QH (2004) Characterization of a cytosolic malate dehydrogenase cDNA which encodes an isozyme toward oxaloacetate reduction in wheat. Biochimie 86: 509–518 [DOI] [PubMed] [Google Scholar]
- Dou CM, Fu XP, Chen XC, Shi JY, Chen YX (2009) Accumulation and detoxification of manganese in hyperaccumulator Phytolacca americana. Plant Biol (Stuttg) 11: 664–670 [DOI] [PubMed] [Google Scholar]
- Dragišić Maksimović J, Mojović M, Maksimović V, Römheld V, Nikolic M (2012) Silicon ameliorates manganese toxicity in cucumber by decreasing hydroxyl radical accumulation in the leaf apoplast. J Exp Bot 63: 2411–2420 [DOI] [PubMed] [Google Scholar]
- Du YM, Tian J, Liao H, Bai CJ, Yan XL, Liu GD (2009) Aluminium tolerance and high phosphorus efficiency helps Stylosanthes better adapt to low-P acid soils. Ann Bot (Lond) 103: 1239–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne A, Génard M, Lobit P, Mbeguié-A-Mbéguié D, Bugaud C (2013) What controls fleshy fruit acidity? A review of malate and citrate accumulation in fruit cells. J Exp Bot 64: 1451–1469 [DOI] [PubMed] [Google Scholar]
- Fecht-Christoffers MM, Führs H, Braun HP, Horst WJ (2006) The role of hydrogen peroxide-producing and hydrogen peroxide-consuming peroxidases in the leaf apoplast of cowpea in manganese tolerance. Plant Physiol 140: 1451–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fecht-Christoffers MM, Maier P, Horst WJ (2003) Apoplastic peroxidase and ascorbate are involved in manganese toxicity and tolerance of Vigna unguiculata. Physiol Plant 117: 237–244 [Google Scholar]
- Fernando DR, Mizuno T, Woodrow IE, Baker AJ, Collins RN (2010) Characterization of foliar manganese (Mn) in Mn (hyper)accumulators using X-ray absorption spectroscopy. New Phytol 188: 1014–1027 [DOI] [PubMed] [Google Scholar]
- Führs H, Behrens C, Gallien S, Heintz D, Van Dorsselaer A, Braun HP, Horst WJ (2010) Physiological and proteomic characterization of manganese sensitivity and tolerance in rice (Oryza sativa) in comparison with barley (Hordeum vulgare). Ann Bot (Lond) 105: 1129–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geszvain K, Butterfield C, Davis RE, Madison AS, Lee SW, Parker DL, Soldatova A, Spiro TG, Luther GW, Tebo BM (2012) The molecular biogeochemistry of manganese(II) oxidation. Biochem Soc Trans 40: 1244–1248 [DOI] [PubMed] [Google Scholar]
- Gietl C. (1992) Malate dehydrogenase isoenzymes: cellular locations and role in the flow of metabolites between the cytoplasm and cell organelles. Biochim Biophys Acta 1100: 217–234 [DOI] [PubMed] [Google Scholar]
- González A, Steffen KL, Lynch JP (1998) Light and excess manganese: implications for oxidative stress in common bean. Plant Physiol 118: 493–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goussias C, Boussac A, Rutherford AW (2002) Photosystem II and photosynthetic oxidation of water: an overview. Philos Trans R Soc Lond B Biol Sci 357: 1369–1381, discussion 1419–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Zhao J, Li X, Qin L, Yan X, Liao H (2011) A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J 66: 541–552 [DOI] [PubMed] [Google Scholar]
- Hernandez-Soriano MC, Degryse F, Lombi E, Smolders E (2012) Manganese toxicity in barley is controlled by solution manganese and soil manganese speciation. Soil Sci Soc Am J 76: 399–407 [Google Scholar]
- Horst WJ, Fecht M, Naumann A, Wissemeier AH, Maier P (1999) Physiology of manganese toxicity and tolerance in Vigna unguiculata (L.) Walp. J Plant Nutr Soil Sci 162: 263–274 [Google Scholar]
- Hue NV, Mai Y (2002) Manganese toxicity in watermelon as affected by lime and compost amended to a Hawaiian acid Oxisol. HortScience 37: 656–661 [Google Scholar]
- Kochian LV, Hoekenga OA, Piñeros MA (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 55: 459–493 [DOI] [PubMed] [Google Scholar]
- Kopittke PM, Lombi E, McKenna BA, Wang P, Donner E, Webb RI, Blamey FP, de Jonge MD, Paterson D, Howard DL, et al. (2013) Distribution and speciation of Mn in hydrated roots of cowpea at levels inhibiting root growth. Physiol Plant 147: 453–464 [DOI] [PubMed] [Google Scholar]
- Liang C, Piñeros MA, Tian J, Yao Z, Sun L, Liu J, Shaff J, Coluccio A, Kochian LV, Liao H (2013) Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiol 161: 1347–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Tian J, Lam HM, Lim BL, Yan X, Liao H (2010) Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiol 152: 854–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler HK, Wellburn AR (1983) Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem Soc Trans 11: 591–592 [Google Scholar]
- Lidon FC, Barreiro MG, Ramalho JC (2004) Manganese accumulation in rice: implications for photosynthetic functioning. J Plant Physiol 161: 1235–1244 [DOI] [PubMed] [Google Scholar]
- López-Calcagno PE, Moreno J, Cedeño L, Labrador L, Concepción JL, Avilán L (2009) Cloning, expression and biochemical characterization of mitochondrial and cytosolic malate dehydrogenase from Phytophthora infestans. Mycol Res 113: 771–781 [DOI] [PubMed] [Google Scholar]
- Migocka M, Papierniak A, Maciaszczyk-Dziubińska E, Poździk P, Posyniak E, Garbiec A, Filleur S (2014) Cucumber metal transport protein MTP8 confers increased tolerance to manganese when expressed in yeast and Arabidopsis thaliana. J Exp Bot 65: 5367–5384 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Millaleo R, Reyes-Díaz M, Ivanov AG, Mora ML, Alberdi M (2010) Manganese as essential and toxic element for plants: transport, accumulation and resistance mechanism. J Soil Sci Plant Nutr 10: 470–481 [Google Scholar]
- Murphy J, Riley JP (1962) A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta 27: 31–36 [Google Scholar]
- Nickelsen J, Rengstl B (2013) Photosystem II assembly: from cyanobacteria to plants. Annu Rev Plant Biol 64: 609–635 [DOI] [PubMed] [Google Scholar]
- Noble AD, Orr DM, Middleton CH, Rogers LG (2000) Legumes in native pasture—asset or liability? A case history with stylo. Trop Grassl 34: 199–206 [Google Scholar]
- Pittman JK. (2005) Managing the manganese: molecular mechanisms of manganese transport and homeostasis. New Phytol 167: 733–742 [DOI] [PubMed] [Google Scholar]
- Rosas A, Rengel Z, de la Luz Mora M (2007) Manganese supply and pH influence growth, carboxylate exudation and peroxidase activity of ryegrass and white clover. J Plant Nutr 30: 253–270 [Google Scholar]
- Sasaki A, Yamaji N, Yokosho K, Ma JF (2012) Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24: 2155–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Delhaize E, Matsumoto H (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J 37: 645–653 [DOI] [PubMed] [Google Scholar]
- Sparrow LA, Uren NC (2014) Manganese oxidation and reduction in soils: effects of temperature, water potential, pH and their interactions. Soil Res 52: 483–494 [Google Scholar]
- Sun L, Liang C, Chen Z, Liu P, Tian J, Liu G, Liao H (2014) Superior aluminium (Al) tolerance of Stylosanthes is achieved mainly by malate synthesis through an Al-enhanced malic enzyme, SgME1. New Phytol 202: 209–219 [DOI] [PubMed] [Google Scholar]
- Tesfaye M, Temple SJ, Allan DL, Vance CP, Samac DA (2001) Overexpression of malate dehydrogenase in transgenic alfalfa enhances organic acid synthesis and confers tolerance to aluminum. Plant Physiol 127: 1836–1844 [PMC free article] [PubMed] [Google Scholar]
- Trípodi KEJ, Podesta FE (2003) Purification and characterization of an NAD-dependent malate dehydrogenase from leaves of the crassulacean acid metabolism plant Aptenia cordifolia. Plant Physiol Biochem 41: 97–105 [Google Scholar]
- Wang QF, Zhao Y, Yi Q, Li KZ, Yu YX, Chen LM (2010) Overexpression of malate dehydrogenase in transgenic tobacco leaves: enhanced malate synthesis and augmented Al-resistance. Acta Physiol Plant 32: 1209–1220 [Google Scholar]
- Watmough SA, Eimers MC, Dillon PJ (2007) Manganese cycling in central Ontario forests: response to soil acidification. Appl Geochem 22: 1241–1247 [Google Scholar]
- Xu HW, Ji XM, He ZH, Shi WP, Zhu GH, Niu JK, Li BS, Peng XX (2006) Oxalate accumulation and regulation is independent of glycolate oxidase in rice leaves. J Exp Bot 57: 1899–1908 [DOI] [PubMed] [Google Scholar]
- Xu X, Shi J, Chen X, Chen Y, Hu T (2009) Chemical forms of manganese in the leaves of manganese hyperaccumulator Phytolacca acinosa Roxb. (Phytolaccaceae). Plant Soil 318: 197–204 [Google Scholar]
- Yamaji N, Sasaki A, Xia JX, Yokosho K, Ma JF (2013) A node-based switch for preferential distribution of manganese in rice. Nat Commun 4: 2442. [DOI] [PubMed] [Google Scholar]
- Yan W, Liu GD, Tian J (2010) Effects of manganese availability on acquisition and distribution of manganese, iron and phosphorus in soybean [Glycine max (L.) Merr.]. Plant Physiol J 46: 923–927 [Google Scholar]
- Yao YX, Li M, Zhai H, You CX, Hao YJ (2011) Isolation and characterization of an apple cytosolic malate dehydrogenase gene reveal its function in malate synthesis. J Plant Physiol 168: 474–480 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.