The secondary metabolite sulforaphane is produced during the hypersensitive response and is involved in protection against infections.
Abstract
Plants defend themselves against microbial pathogens through a range of highly sophisticated and integrated molecular systems. Recognition of pathogen-secreted effector proteins often triggers the hypersensitive response (HR), a complex multicellular defense reaction where programmed cell death of cells surrounding the primary site of infection is a prominent feature. Even though the HR was described almost a century ago, cell-to-cell factors acting at the local level generating the full defense reaction have remained obscure. In this study, we sought to identify diffusible molecules produced during the HR that could induce cell death in naive tissue. We found that 4-methylsulfinylbutyl isothiocyanate (sulforaphane) is released by Arabidopsis (Arabidopsis thaliana) leaf tissue undergoing the HR and that this compound induces cell death as well as primes defense in naive tissue. Two different mutants impaired in the pathogen-induced accumulation of sulforaphane displayed attenuated programmed cell death upon bacterial and oomycete effector recognition as well as decreased resistance to several isolates of the plant pathogen Hyaloperonospora arabidopsidis. Treatment with sulforaphane provided protection against a virulent H. arabidopsidis isolate. Glucosinolate breakdown products are recognized as antifeeding compounds toward insects and recently also as intracellular signaling and bacteriostatic molecules in Arabidopsis. The data presented here indicate that these compounds also trigger local defense responses in Arabidopsis tissue.
Plants are constantly challenged by pathogenic microorganisms and have developed several detection and defense systems to protect themselves against the invaders. Preformed defenses include the waxy cuticle, thick cell walls, and antimicrobial compounds. After recognition of microbe-associated patterns, defense responses are induced, which include the fortification of cell walls and the production of phytoalexins (Monaghan and Zipfel, 2012). Overcoming the preformed and induced defenses of the plant hosts requires adaptation by the pathogen. Pathogenic bacteria use type III secretion to inject proteins (so-called effectors) into the host cytosol in order to overcome plant defense responses (Bent and Mackey, 2007). In turn, plants have developed systems to recognize the pathogenic effectors and mount defense. Recognition of type III effectors by plant resistance (R) proteins induces robust defense responses that frequently include the hypersensitive response (HR).
The HR is a complex defense reaction characterized by the induction of programmed cell death (PCD) in the local host tissue as well as the activation of other defense responses in both local and systemic tissue (Mur et al., 2008; Shah, 2009). Oomycetes and true fungi also secrete proteinaceous effectors that can be recognized by host R proteins (Coates and Beynon, 2010; Hückelhoven and Panstruga, 2011; Feng and Zhou, 2012). The lesions formed during the HR vary in size between different host-pathogen pairs; however, a lesion induced at one or a few cells can spread to surrounding cells (Mur et al., 2008). Since pathogens inducing HR typically fail to proliferate, the first infected cell likely releases a compound that promotes PCD in surrounding cells. This is especially clear in models with oomycete and fungal pathogens, where the localization of the pathogen and the spread of cell death around the infection site can be clearly visualized (Mur et al., 2008; Coates and Beynon, 2010). Trailing necrosis is an incomplete resistance phenotype characterized by cell death that trails, but fails to contain, the filamentous growth of the pathogen. One explanation for trailing necrosis is a failure of infected cells to produce a putative mobile defense signal required to enhance defense in neighboring cells. Farther from the site of PCD, other defense pathways are activated and systemic tissue is primed for defense.
The hunt for systemically acting compounds has been intense, and several candidates for this signal have been presented (Dempsey and Klessig, 2012). In contrast, even though the phenomenon of HR as a defense reaction was described almost a century ago (Stakman, 1915; Mur et al., 2008), compounds acting on the local tissue scale of the HR have attracted little attention. We set out to find substances released from cells undergoing the HR that could induce cell death in naive tissue. We report that leaf tissue of the model plant Arabidopsis (Arabidopsis thaliana) releases the reactive electrophilic compound sulforaphane after bacterial effector recognition. Mutants affected in sulforaphane production as well as other glucosinolate breakdown products showed delayed or reduced cell death after the recognition of pathogenic effectors and decreased resistance to an oomycete pathogen. Moreover, pretreatment of plants with sulforaphane enhanced resistance against a virulent oomycete isolate. Thus, we interpret this as that sulforaphane and likely similar compounds might both possess direct antimicrobial properties and, through a cytotoxic mechanism, act directly on plant cells to trigger defense responses.
RESULTS
Isolation and Identification of a Cell Death-Inducing Compound Released from Arabidopsis Tissue Undergoing the HR
To investigate if cells undergoing the HR release chemical signals to induce or promote cell death in noninfected tissue, a transgenic system to scale up the HR was used. The transgenic system consists of Arabidopsis harboring a dexamethasone (DEX)-inducible copy of the Pseudomonas syringae type III effector Avirulent Resistance to Pseudomonas Maculicola1 protein (AvrRpm1; Mackey et al., 2002, 2003; Andersson et al., 2006). Leaf tissue expressing the bacterial effector AvrRpm1 (DEX:AvrRpm1/Columbia-0 [Col-0]) was incubated in water with DEX. An isogenic line in the rpm1-3 background (a protein null for RPM1 and thus unable to recognize AvrRpm1) and the untransformed wild type (Col-0) were used as controls. The bathing solution was filtered, and the filtrate was run through a C18 solid-phase extraction cartridge to capture small molecules (Supplemental Fig. S1). The obtained fraction was dissolved in water and infiltrated into nontransgenic Arabidopsis leaves. The fraction obtained from DEX:AvrRpm1/Col-0 caused cell death when infiltrated into Arabidopsis wild-type leaves (Fig. 1A). In contrast, the fraction from DEX:AvrRpm1/rpm1-3 and from untransformed wild-type material had no apparent effect on plant tissue (Fig. 1B). Thus, it is apparent that Arabidopsis tissue undergoing the HR releases one or several soluble compounds that can induce cell death in naive leaf tissue.
Figure 1.
Induction of cell death by an aqueous extract from Arabidopsis tissue undergoing the HR and identification of sulforaphane. A and B, Transgenic Arabidopsis plants expressing the bacterial P. syringae effector AvrRpm1 (DEX:AvrRpm1/Col-0 and DEX:AvrRpm1/rpm1-3) were incubated in water with the inducer DEX. Small molecules recovered from the bathing solution of the Col-0 (A) and rpm1-3 (B) plants were infiltrated into wild-type (nontransgenic) plants at the indicated dilutions. C, The material obtained was further analyzed by HPLC. Fractions were collected, dried, dissolved in water, and infiltrated into wild-type leaves. Visible effects and Trypan Blue staining of leaves receiving the fractions from the DEX:AvrRpm1/Col-0 extract are shown at bottom. D, The HPLC-purified fraction from the DEX:AvrRpm1/Col-0 fraction was subjected to GC-MS with electron-impact ionization. E, Mass spectrum for the major peak. F, The fraction was also dissolved in methanol, and the UV absorption spectrum was recorded. G, Structure of the identified compound, sulforaphane. The experiments depicted in A to C were performed twice with identical results.
The active material was further fractionated by HPLC (Fig. 1C), and a fraction with the ability to induce cell death evident as visual lesions and by trypan blue staining was obtained. Gas chromatography-mass spectrometry (GC-MS) analysis of this active fraction revealed a single peak (Fig. 1D). Comparison with publicly available mass spectra libraries identified the compound as 4-methylsulfinylbutyl isothiocyanate, a compound commonly known by its trivial name, sulforaphane. The mass spectrum (Fig. 1E) showed the expected molecular ion at mass-to-charge ratio (m/z) 177 and a prominent peak at m/z 160 caused by the loss of the sulfoxide oxygen plus one hydrogen. The mass and UV absorption spectra (Fig. 1F) were identical to those reported for purified sulforaphane (Tierens et al., 2001). A sulforaphane standard coeluted with the active fraction in the semipreparative HPLC system. Thus, the purified compound isolated from Arabidopsis leaf tissue undergoing the HR was unambiguously identified as the isothiocyanate sulforaphane (Fig. 1G).
Sulforaphane Released by Arabidopsis Tissue Undergoing the HR Can Be Degraded by P. syringae SURVIVAL IN ARABIDOPSIS EXTRACTS Genes
Quantification of sulforaphane released into the bathing solution from leaf discs of DEX:AvrRpm1/Col-0 plants revealed that during 12 h post induction (hpi), about 200 nmol of sulforaphane was released per 1 g of tissue (Fig. 2A). A near-peak concentration was reached after 6 h and remained high for at least 24 h. The corresponding transgenic line in the rpm1-3 background released less than 5 nmol of sulforaphane per 1 g at 12 h after induction. The release of sulforaphane from wild-type and rpm1-3 leaf tissue infiltrated with P. syringae expressing AvrRpm1 (DC3000:AvrRpm1) was similar to that of plants harboring DEX-inducible AvrRpm1, except that the concentration of released sulforaphane decreased dramatically between 12 and 24 hpi (Fig. 2B).
Figure 2.
Release of sulforaphane during effector-induced HR. A, Transgenic Arabidopsis plants expressing the bacterial P. syringae effector AvrRpm1 (DEX:AvrRpm1/Col-0 and DEX:AvrRpm1/rpm1-3) were incubated in water with DEX. B and C, Wild type Col-0 Arabidopsis leaf discs were infiltrated with wild-type P. syringae pv tomato DC3000 (B) or the Δsax mutant (C) expressing the effector AvrRpm1 (OD600 = 0.1) and incubated in water. At the indicated times, the discs were removed and the amount of sulforaphane in the bathing solution was analyzed. Average and range values for duplicate samples are shown. The experiments in A and B were performed three times with similar results, and the experiment in C was repeated twice with similar results. Fw, Fresh weight.
It has been reported that sulforaphane formation is induced in Arabidopsis in response to several nonhost bacterial pathogens (Fan et al., 2011). Furthermore, P. syringae strains adapted to Arabidopsis were shown to harbor so-called SURVIVAL IN ARABIDOPSIS EXTRACTS (SAX) genes, which enabled the bacteria to detoxify the sulforaphane produced by the host. This spurred us to test if the presence of SAX genes in the bacteria could explain the difference in the amount of sulforaphane present in the DEX-inducible and the bacteria-infiltrated material at the 24-h time point. To this end, Δsax mutant P. syringae (Fan et al., 2011) was transformed with a vector encoding the AvrRpm1 effector. The resulting Δsax:AvrRpm1 bacteria were infiltrated into wild-type (Col-0) leaf discs and incubated in water, and the amount of sulforaphane released to the bathing solution was measured at different time points (Fig. 2C). Again, DC3000:AvrRpm1 inoculation caused a rapid induction of sulforaphane, which decreased back to low levels at 24 hpi. In contrast, inoculation with the Δsax:AvrRpm1 strain resulted in the accumulation of sulforaphane, which peaked at 12 hpi and remained high at 24 hpi.
Sulforaphane and Other Isothiocyanates Induce Cell Death in Naive Leaf Tissue
Leaf infiltration experiments were conducted in order to determine the concentration of sulforaphane needed to induce cell death. As seen in Figure 3A, visible cell death occurred after infiltration of 0.5 and 1 mm sulforaphane. In some experiments, lower concentrations were sufficient, but 0.5 mm always caused large visible lesions. This concentration also produced large necrotic lesions on leaves of several other plant species; broad bean (Vicia faba) and sunflower (Helianthus annuus) are shown in Figure 3, B and C.
Figure 3.
Infiltration of pure sulforaphane causes cell death in leaf. A to C, Sulforaphane was suspended in water to the indicated concentrations (A) or 1 mm (B and C) and syringe infiltrated into wild-type Col-0 Arabidopsis (A), broad bean (B), and sunflower (C) leaves. The left side of the leaf was infiltrated with sulforaphane, and the right side was mock infiltrated with deionized water. The leaves were detached and photographed after 24 h (A). D, Leaves were syringe infiltrated with the indicated isothiocyanates (ITC); leaf discs were punched out, washed, and incubated in water for the indicated times; and the conductance of the bathing solution was measured. Average and range values for duplicate samples are shown. Asterisks indicate statistical significance compared with mock treatment at the indicated times (one-way ANOVA, P < 0.05). The experiments were performed twice with similar results.
For a more precise measurement of cell death, electrolyte leakage after infiltration with sulforaphane was monitored. As seen in Figure 3D, 100 µm sulforaphane was sufficient to cause significant electrolyte leakage, and 200 µm caused a large loss of cellular electrolytes 48 h after infiltration. Infiltration with benzyl, butyl, and isopropyl isothiocyanate at 100, 200, and 400 µm was also tested for their ability to induce cell death. The strongest effect was observed with benzyl isothiocyanate, which at 100 µm already induced robust electrolyte leakage by 6 h. An effect of butyl isothiocyanate was apparent but weaker than that of benzyl isothiocyanate and sulforaphane, whereas isopropyl isothiocyanate seemed to lack any effect over mock infiltration.
It is believed that the major mode of action of sulforaphane in mammalian cells is to decrease the cellular glutathione pool (Valgimigli and Iori, 2009). This would be consistent with its reactive electrophile properties, and it is well known that plant defense triggers oxidative stress that contributes to cell death (Overmyer et al., 2003; Torres, 2010). Total and oxidized glutathione, therefore, were measured in leaf discs 30 min after infiltration with a sulforaphane solution (Fig. 4). Treatment with sulforaphane led to a severe depletion of the total glutathione pool and, consequently, to a large calculated increase in the redox potential of the glutathione pool.
Figure 4.
Infiltration of pure sulforaphane causes oxidation of the cellular glutathione pool. Leaf discs from wild-type Arabidopsis were vacuum infiltrated with sulforaphane suspended in water at the indicated concentrations. Reduced and oxidized glutathione contents were measured 30 min after infiltration, and the redox potential was calculated. Average and range values for triplicate samples are shown. The experiment was performed twice with similar results. Fw, Fresh weight.
Mutants with a Reduced Capacity of Sulforaphane Production Display Decreased Cell Death Response
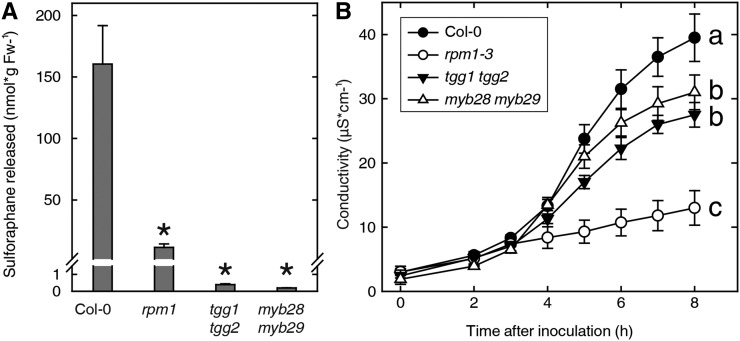
There are currently no specific biosynthesis mutants available that lack capacity to produce sulforaphane (Sønderby et al., 2010b). However, the two myeloblastosis (MYB) family transcription factors MYB28 and MYB29 have been found to control the production of aliphatic glucosinolate precursors in Arabidopsis (Beekwilder et al., 2008; Sønderby et al., 2010a). The myb28 myb29 double mutant has severely reduced levels of the aliphatic glucosinolates while maintaining wild-type levels of other glucosinolates. The myrosinase double mutant thioglucoside glucohydrolase1 (tgg1) tgg2 lacks the thioglycosidases cleaving the inactive glucosinolates and, therefore, is unable to accumulate sulforaphane and other glucosinolate breakdown products after wounding (Barth and Jander, 2006). The single mutants myb28 and myb29 and the double mutants myb28 myb29 and tgg1 tgg2 were tested for sulforaphane production after inoculation with P. syringae DC3000:AvrRpm1 (Fig. 5A; Supplemental Fig. S2). Sulforaphane release in both double mutants was reduced by more than 98% compared with wild-type Col-0.
Figure 5.
Compromised PCD in sulforaphane-deficient mutants. A, Leaf discs from Arabidopsis wild-type and mutant lines were infiltrated with P. syringae DC3000:AvrRpm1, and the amount of sulforaphane released into the bathing solution was determined at 6 hpi. Mean and range values for duplicate samples are shown. Asterisks indicate statistical significance compared with Col-0 (one-way ANOVA, P < 0.05). Fw, Fresh weight. B, Electrolyte leakage was measured at the indicated time points for the wild type (black circles), double mutant myb28 myb29 (white triangles), double mutant tgg1 tgg2 (black triangles), and rpm1-3 (white circles). Mean ± sd of six replicate samples are shown. Letters a to c indicate statistically significant groups (one-way ANOVA with Tukey’s posthoc test, P < 0.05). The experiments were performed twice with similar results.
We hypothesized that, if sulforaphane acts as a mobile signal promoting PCD in cells adjacent to the infection site, then the double mutant would show diminished death of cells neighboring those in which type III effectors are activating an R protein. In accordance with this, electrolyte leakage was markedly decreased in the double mutants infiltrated with DC3000:AvrRpm1 at low titer (Fig. 5B). We next tested if this was reflected in the ability to restrict reproduction of the pathogen in the plant tissue. Leaves of the mutants and wild-type plants were infiltrated with P. syringae DC3000:AvrRpm1, and the growth of the bacteria was measured 3 d later (Supplemental Fig. S3). The two double mutants demonstrated no change in resistance to P. syringae expressing AvrRpm1.
To further explore the role of sulforaphane in the plant’s local resistance responses, we used the oomycete Hyaloperonospora arabidopsidis (Hpa). Arabidopsis wild type Col-0 carries the Recognition of Peronospora Parasitica2 (RPP2) and RPP4 genes that mediate recognition and trigger the HR in response to the isolates Hpa Cala2 and Hpa Emwa1, respectively (van der Biezen et al., 2002; Sinapidou et al., 2004). The wild type Col-0 and the myb28 myb29 and tgg1 tgg2 double mutants were inoculated with the isolate Hpa Cala2, and the extent of cell death at the infection sites was scored after trypan blue staining (Fig. 6A). In wild-type Col-0, about 90% of the interaction sites resulted in quick localized cell death and no visible growth of the pathogen (Fig. 6B), whereas about 10% of the interaction sites demonstrated some degree of trailing necrosis and growth of hyphae (Fig. 6C) within the given time period. In the two double mutants, the situation was reversed, with 93% and 73% of the interaction sites resulting in trailing necrosis for myb28 myb29 and tgg1 tgg2, respectively. In a few instances, free hyphae, completely outgrowing the trailing necrosis, could be seen on the myb28 myb29 double mutant. At an early time point of the infection, 36 hpi, the two mutants demonstrated less cell death than the wild type (Fig. 6D), and the number of dead cells per interaction site was decreased to less than half of the wild type in the two double mutants. This may suggest that slower cell death at early stages of infection results in less restriction of pathogen growth, and consequently, the growing hyphae are followed by delayed cell death. The isolate Hpa Emwa1 also showed increased virulence on the double mutants, as evidenced by an approximately 6-fold increase in the number of sporophores produced 7 d post induction (dpi) of cotyledons (Fig. 6E).
Figure 6.
Reduced resistance to a biotrophic oomycete in sulforaphane-deficient mutants. The indicated lines were inoculated at the cotyledon stage with Hpa conidia of the isolates Cala2 (A–D) or Emwa1 (E). The cotyledons were stained with Trypan Blue 7 (A–C) or 2 (D) dpi, and the extent of cell death was determined or sporophores were counted at 7 dpi (E). Rapid cell death in the wild type is shown in B, and trailing necrosis is shown in C. Average and range values for three replicate experiments, each including 200 interaction sites, are shown in A and D. Average and sd for 15 cotyledons are shown in E. Letters a to c indicate statistically significant groups (one-way ANOVA with Tukey’s posthoc test, P < 0.05). The experiments were performed three times with similar results. Bars in B and C = 0.5 mm.
Sulforaphane Pretreatment of Arabidopsis Induces Defense against Virulent Pathogens
To test whether sulforaphane can activate defense and provide increased immunity to naive plant tissue, Arabidopsis was treated with sulforaphane before inoculation with virulent Hpa Cala2 (Fig. 7). In this case, the accession Landsberg erecta and the enhanced disease resistance1 (eds1) mutant in the Wassilewskija-0 background were used as controls, as these lines are highly susceptible to the Cala2 isolate (Parker et al., 1996; Bailey et al., 2011). Sporulation at 4 dpi was markedly decreased when plants had been pretreated with sulforaphane before inoculation with Hpa Cala2. Treatment with sulforaphane 24 hpi with Cala2 also led to decreased sporulation at 4 dpi, but the effect was significantly smaller. However, the effect of the sulforaphane was indistinguishable from that of the untreated controls at 7 dpi (Supplemental Fig. S4).
Figure 7.
Sulforaphane treatment of plants provides increased resistance against pathogens. Seedlings of wild-type Landsberg erecta (Ler) or eds1 in the Wassilewskija (Ws) background were sprayed with 200 µm sulforaphane 24 h before or after, as indicated, inoculation with Hpa isolate Cala2, and the resulting sporulation was counted at 4 dpi. Average numbers of spores and range values for three replicate samples are shown. Letters a to c indicate statistically significant groups (one-way ANOVA with Tukey’s posthoc test, P < 0.05). The experiment was repeated twice with identical results.
DISCUSSION
The HR was described over 100 years ago (Stakman, 1915; Mur et al., 2008). Eighty years later, the basic genetics of effector-triggered immunity started to be unraveled, and the past decade has seen an explosive increase in the understanding of the molecular mechanisms behind plant defense responses to pathogens. Many pathogenic effectors and their cognate plant R proteins have been identified, and several intracellular signaling components have been discovered (Desveaux et al., 2006; Torres et al., 2006; Bent and Mackey, 2007). Much effort has gone into the elucidation of phloem-mobile compounds that mediate the induction of systemic acquired resistance (Dempsey and Klessig, 2012). In contrast, comparatively little effort has been spent on compounds active at the local tissue level in plant defenses. The dead and dying cells of HR lesions might appear uniform. However, some cells directly see the bacteria and recognize the effector, whereas adjacent cells only sense the response of their immediate neighbors. Given that the HR lesion spreads to a size considerably larger than that reasonably reached by the pathogens in terms of single bacterial cells, germinated oomycetes, or fungal spores, it seems likely that signals produced by the initially responding cells contribute to propagating cell death. Importantly, cell death must also stop from spreading, and it has been described that cells adjacent to an HR site become more resistant to further pathogen attacks (Ross, 1961).
In this study, we set out to identify cell death-promoting compounds released by plant cells undergoing effector-triggered immunity. To this end, we used bioassay-guided fractionation of a diffusate obtained from transgenic Arabidopsis leaf tissue where the bacterial effector AvrRpm1 was expressed in planta. A small-molecule extract that caused necrotic lesions when infiltrated into naive wild-type tissue was obtained from a bathing solution in which the transgenic tissue had been incubated. The extract was further fractionated, and the single most active compound could be unambiguously identified as the isothiocyanate sulforaphane. We could also demonstrate that Arabidopsis wild-type tissues infiltrated with P. syringae expressing AvrRpm1 released quantities of sulforaphane sufficient to induce cell death in naive leaf tissue (Figs. 2B and 3D).
Sulforaphane belongs to a large and structurally diverse family of plant defensive compounds derived from glucosinolate precursors (Fahey et al., 2001). When these are cleaved by thioglucosidases known as myrosinases, the unstable intermediates are converted to nitriles, isothiocyanates, and thiocyanates. These compounds are found in many plant families, especially in the Brassicale order, but are far from universal in the plant kingdom (Fahey et al., 2001). In, fact, the presence of glucosinolates in plant species outside the Brassicale is a rare occurrence. The plant species that do contain glucosinolates display a large degree of variation in the composition of the side chain of their glucosinolates (Fahey et al., 2001; Agerbirk and Olsen, 2012). Sulforaphane is thus restricted to a few plant species, and the amount of both the precursor glucoraphanine and sulforaphane varies between Arabidopsis accessions (Kliebenstein et al., 2001). Thus, sulforaphane production in response to HR must be a very limited occurrence in the plant kingdom, and any conservation should be sought in function rather than in structure.
The glucosinolates were previously considered as primarily antifeeding compounds active against insect herbivores (Hopkins et al., 2009). However, recent studies on glucosinolates in Arabidopsis have firmly established roles for them in the defense against microbial pathogens as well (Bednarek et al., 2009; Clay et al., 2009; Fan et al., 2011; Bednarek, 2012a; Johansson et al., 2014). Indole glucosinolates are linked to broad-spectrum nonhost resistance against powdery mildew fungi (Bednarek et al., 2009), signaling triggered by the flagellin recognition in Arabidopsis (Clay et al., 2009), and initiation of the HR following the recognition of P. syringae effectors (Johansson et al., 2014). Sulforaphane, specifically, was previously linked to resistance against nonadapted strains of P. syringae in Arabidopsis (Fan et al., 2011), and it was reported that nonadapted bacterial strains were highly sensitive to sulforaphane, whereas strains adapted for the infection of Arabidopsis and other cruciferous plants were shown to harbor SAX genes mediating the detoxification of sulforaphane produced by the plant host. We found that when wild-type P. syringae expressing AvrRpm1 was infiltrated into Arabidopsis leaf tissue, the released sulforaphane was degraded at 24 hpi (Fig. 2B). This was not observed with the expression of transgenic AvrRpm1 (Fig. 2A), leading us to believe that the SAX genes in P. syringae are initially unable to keep up with the rapid production of sulforaphane. This was supported by the observation that the sulforaphane released was not removed by a Δsax P. syringae mutant expressing AvrRpm1 (Fig. 2C). The exact mechanism for the apparent removal of sulforaphane by P. syringae remains unclear.
Sulforaphane is but one of a family of similar compounds in Arabidopsis. The composition of the glucosinolate precursors and the cleavage products varies largely between different Arabidopsis accessions (Kliebenstein et al., 2001). The sulforaphane precursor glucoraphanine is the dominant glucosinolate in the accession Col-0 leaf tissue (Kliebenstein et al., 2001; Brown et al., 2003). We infiltrated a number of structurally dissimilar isothiocyanates into Arabidopsis Col-0 wild-type leaf tissue and measured electrolyte leakage (Fig. 3D). The results presented show that several different isothiocyanates could induce cell death. Thus, it seems likely that other glucosinolate breakdown products play a similar role in other accessions. Moreover, given the ability of sulforaphane to induce cell death in a variety of plants (Fig. 3, B and C), the activity of these compounds might be conserved across the plant kingdom. Furthermore, indole-3-acetonitrile also can induce cell death upon infiltration into leaf tissue (Johansson et al., 2014). Our admittedly very small screen of compounds did reveal very different potency depending on the nature of the side chains. This is in agreement with the dependence on the side chain for the effects of isothiocyanates in various mammalian systems as well (Wu et al., 2009). However, any conclusions about the importance of chain length, structure, or substituents would require a more thorough screen of different compounds.
To assess the in vivo function of the sulforaphane released during the HR, two different double mutant lines were used: myb28 myb29 and tgg1 tgg2. Both double mutants lack the ability to produce sulforaphane (Barth and Jander, 2006; Beekwilder et al., 2008; Sønderby et al., 2010a). Cell death induced by P. syringae expressing AvrRpm1 decreased only moderately in each of these double mutants, and there was no detectable change in the ability of the mutant plants to restrict bacterial growth (Fig. 5B; Supplemental Fig. S3). Thus, the HR pathway triggered by AvrRpm1 is able to proceed relatively unhindered despite an inability to produce sulforaphane. This is perhaps not surprising, given that the defense response induced by AvrRpm1 is particularly robust (Tsuda et al., 2009). In contrast, there was a clear cell death and resistance phenotype of the double mutants toward two different isolates of the oomycete Hpa, with clearly delayed cell death and enhanced pathogen growth and sporulation (Fig. 6, A, D, and E). The tgg and myb double mutants are arguably not ideal in their specificity toward lowered sulforaphane production, but given the very different nature of the mutated genes in the lines, it seems reasonable that the shared phenotypes are indeed caused by the lack of aliphatic glucosoinolate breakdown products rather than other indirect effects of the mutations. Since the biosynthesis pathway for the aliphatic glucosinolates is not fully understood, no specific biosynthetic mutants are available, and we deem that these two double mutants represent the best available genetic test for the involvement of sulforaphane in effector-triggered immunity. It is difficult to entirely separate the possible effects of sulforaphane on the plant cells and the possible direct effect on the pathogen in the mutant experiment. However, since sulforaphane by itself clearly affects plant tissue, at the very least both effects are likely present during the defense reaction. Future transcriptional profiling studies of the effects of sulforaphane treatment of plant tissue are likely to shed more light on the direct effects on the plant.
As sulforaphane infiltration caused a quick decrease in the cellular glutathione pool (Fig. 4), we propose the reaction of sulforaphane with glutathione and the subsequent increase in redox potential as the most straightforward explanation for the action of sulforaphane. Thus, if sulforaphane acts by simply decreasing the glutathione pool of the cell and thus making this pool more oxidative, there is no need for specific receptors. It should be noted that the redox potential and the size of the glutathione pool are well known to have profound influences on many different cellular processes, including cell death (Noctor et al., 2011). It has even been suggested that the half-cell reduction potential of cellular glutathione is a universal marker for stress and cell death (Kranner et al., 2006). The notion that lower concentrations and thus higher oxidation of the cellular glutathione pool could induce defense is supported by our observation of protection induced by sulforaphane. Treatment with sulforaphane at a concentration that did not induce visible lesions offered increased protection against a virulent Hpa isolate (Fig. 7).
It is well established that hydrogen peroxide and other reactive oxygen species (Overmyer et al., 2003; Torres, 2010) as well as other reactive electrophiles are produced during plant defense responses (Farmer and Davoine, 2007; Durand et al., 2009; Mueller and Berger, 2009; Farmer and Mueller, 2013). Furthermore, Arabidopsis tissue undergoing the HR triggered by AvrRpt2 releases salicylic acid, jasmonic acid, and 12-oxo-phytodienoic acid (Kourtchenko et al., 2007), and other studies indicate large-scale production of other electrophilic oxidation products of fatty acids (Farmer and Davoine, 2007; Tsuda et al., 2009; Farmer and Mueller, 2013). Thus, it could be envisioned that sulforaphane acts in combination with other reactive electrophilic compounds produced during plant defense responses against pathogens to execute the HR. This would provide a high degree of adaptive flexibility to plants. Several different reactive electrophiles could be utilized alone or in combination, and this would hamper the pathogen’s ability to counteract the signaling pathway. It seems clear that a cocktail of compounds, which might have both direct antimicrobial effects and signaling properties in the plant, are secreted from the tissue undergoing the HR. Some of these electrophilic compounds can also activate the expression of defense genes in neighboring tissue (Bailey et al., 2011). This specific role might be played by structurally very different compounds in different plant species. Some support for this notion comes from research on benzoxazinone glucosides, which are secondary metabolites stored in both monocot and dicot plant species (Frey et al., 2009). These compounds are also stored as inactive precursors and cleaved to unstable aglycones upon tissue disintegration or possibly other stresses. Maize (Zea mays) mutants lacking the capacity to produce these show striking similarities in pathogen defense to Arabidopsis mutants impaired in glucosinolate production (Ahmad et al., 2011; Bednarek, 2012a). Furthermore, maize benzoxazinone 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one directly reacts with glutathione and other biologically relevant thiol groups forming adducts (Dixon et al., 2012).
Our results support that sulforaphane and possibly other glucosinolate breakdown products might also function in the regulation of the expression of defense genes in neighboring tissue. It is apparent that natural products such as sulforaphane occupy a gray zone between antimicrobials and signaling compounds (Bednarek, 2012b, 2012a). However, it is important to note that, in this case, signal does not refer to a classical ligand-receptor pair. Rather, the signal is composed of multiple reactive compounds that together provide an activity that drives the cell in the direction of certain outcomes: PCD or defense priming.
To conclude, we here provide evidence that the isothiocyanate sulforaphane is released from Arabidopsis cells recognizing a bacterial effector protein. The released sulforaphane stimulates cell death and possibly other types of plant defense responses in the complex multicellular defense reaction, the HR.
MATERIALS AND METHODS
Plant Cultivation and Bacterial Strains
Arabidopsis (Arabidopsis thaliana) was cultivated under short-day conditions (8-h day and 16-h night) at 22°C daytime and 18°C nighttime and 60% relative humidity for 4 to 6 weeks. The myb28 myb29 mutant lines were confirmed by PCR using flanking and insert-specific primers (Supplemental Fig. S2). Pseudomonas syringae pv tomato DC3000 transformed with a vector carrying the avirulence gene AvrRpm1 was maintained on solid King’s B medium with appropriate antibiotics at room temperature. The vector pVSP61 containing the AvrRpm1 gene was isolated from DC3000 (Mackey et al., 2002) and transformed into the Δsax P. syringae mutant (Fan et al., 2011).
Purification of Cell Death-Inducing Substances
Ten grams of leaf tissue from 5- to 6-week-old DEX:AvrRpm1/Col-0 or DEX:AvrRpm1/rpm1-3 plants was harvested into a beaker containing 50 mL of deionized water with 20 µm DEX (Sigma-Aldrich) and incubated for 6 h with gentle agitation. The solution was filtered through filter paper and stored at −20°C under N2 as a precaution. Small molecules were captured on a preconditioned 500-mg C18 solid-phase extraction column (Discovery SPE; Supelco). The column was washed with water, and bound compounds were eluted with methanol. The eluate was dried under nitrogen, suspended in water, and used for infiltration or further purified by partitioning against chloroform:methanol:water (1:1:0.9). The chloroform phase was collected, dried, dissolved in methanol, and subjected to HPLC on a 125- × 4.5-mm C18 LiChrocart column (Merck). Flow was maintained at 1 mL min−1, and isocratic elution with methanol:water (70:30) was followed by a linear gradient to 80% (v/v) 2-propanol for 30 min. Fractions were collected manually, dried under nitrogen, dissolved in 0.5 mL of water, and used for leaf infiltration. The purified active fraction was dried in vacuo and dissolved in 100 µL of distilled ethyl acetate, and aliquots of 0.5 µL were injected into a gas chromatograph (Hewlett-Packard model 5890) equipped with a mass selective detector (Hewlett-Packard model 5970). The detector was operated in the scan mode (m/z 50–600), and a capillary column of 5% phenylmethylsiloxane (12 m, 0.33-µm film thickness) with helium as the carrier gas was used. The oven temperature was raised from 80°C to 300°C at a rate of 10°C min−1. The injector (temperature of 200°C) was operated in the split mode.
Pathogen Growth Assays, Electrolyte Leakage, and Trypan Blue Staining
Exponentially growing P. syringae pv tomato was resuspended from plates in 10 mm MgCl2 and diluted to the proper optical density at 600 nm (OD600). For bacterial growth experiments, whole leaves were pressure inoculated with the suspension (OD600 = 0.00002, corresponding to 7.1 × 103 colony-forming units [cfu]) using a needleless syringe. Plants were returned to growth chambers, and samples were collected at the indicated time points (four 7-mm leaf discs from separate plants in four replicates). The discs were homogenized in 10 mm MgCl2, serial diluted, and plated on King’s B medium agar containing rifampicin and kanamycin. The number of cfu was determined 3 d after extraction and represented as described (Morel and Dangl, 1999). Electrolyte leakage assays were performed as described (Mackey et al., 2002) with minor modifications. Bacteria suspended in 10 mm MgCl2 were diluted to OD600 = 0.01, corresponding to 3.55 × 106 cfu mL−1, and vacuum infiltrated into leaf discs (diameter, 7 mm). The discs were rinsed in deionized water and transferred to six-well microtiter plates containing 10 mL of deionized water (four discs per well in six replicates). Conductance was measured at the indicated time points. Maintenance of Hyaloperonospora arabidopsidis isolates, preparation of inoculum for experiments, and assessment of sporulation were as described (Tör et al., 2002). Arabidopsis seedlings were sprayed with 400 µm sulforaphane suspended in deionized water 24 h before or after inoculation with Hpa spores as indicated. Cell death was visualized by Trypan Blue staining as described (Koch and Slusarenko, 1990).
Synthesis of [1,1,2,2,3,3,4,4-2H8]d,l-Sulforaphane
Octadeuterio-sulforaphane was synthesized starting with [1,1,2,2,3,3,4,4-2H8]4-bromo-1-butanol (QMX Laboratories) by modification of a previously described synthetic route to unlabeled sulforaphane (Ding et al., 2006). The product was purified by reverse-phase HPLC (solvent system, acetonitrile:water, 1:3 [v/v]), affording the title compound (15 mg) as a colorless oil. The electron-impact mass spectrum showed prominent ions at m/z 185 (M+), 167 (M+-O2H), 122, 118, 90, 74 ([S=C=N-C2H2]+), and 62 ([C2H2-C2H2-C2H=C2H2]+). The identity of the material was further established by gas-liquid chromatography, reverse-phase HPLC, and UV spectrometry using a sample of authentic sulforaphane (Sigma-Aldrich) as a reference.
Quantification of the Released Sulforaphane
Two leaf discs were incubated in 1 mL of water after infiltration with P. syringae or DEX solution. At the times indicated, the discs were removed and deuterated sulforaphane was added as an internal standard to the aqueous bathing solution. The sulforaphane was extracted by partitioning against 1 mL of dichloromethane. The organic phase was dried under N2 and analyzed by GC-MS as described above or using an Agilent 7820 gas chromatograph coupled to an Agilent 5975 mass selective detector. The ions 160 and 167 m/z were used for quantification of the endogenous and deuterated internal standards, respectively.
Quantification of Total and Oxidized Glutathione
Leaf discs were vacuum infiltrated with sulforaphane suspended in water and left at room temperature for 30 min, after which 20 discs (130 mg of tissue) were ground into a fine powder in liquid nitrogen. Total and oxidized glutathione content in the tissue extracts was determined with an enzymatic method (Tietze, 1969; Queval and Noctor, 2007). Samples were prepared in triplicate and measured three times. The glutathione redox potential was calculated as described (Queval and Noctor, 2007).
Statistical Analysis
Statistical analysis was performed as described (Johansson et al., 2014) using GraphPad Prism 6 (GraphPad Software). The conductivity at the final time point (6 h) of ion leakage assays using bacterial infiltration (Fig. 5B), the electrolyte leakage at each time point for chemical infiltration compared with mock treatment (Fig. 3D), the bacterial growth at 3 dpi (Supplemental Fig. S3), the cell death progression and sporulation after Hpa inoculation (Figs. 6, A, D, and E, and 7), and the sulforaphane release following bacterial infiltration (Fig. 5; Supplemental Fig. S2) were subjected to one-way ANOVA with Tukey’s posthoc analysis, with P < 0.05 considered significant. Samples were grouped based on significant differences between them.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Experimental setup for identification of cell death-inducing compounds.
Supplemental Figure S2. Confirmation and characterization of myb mutants.
Supplemental Figure S3. In planta growth of P. syringae expressing AvrRpm1 in sulforaphane-deficient mutants.
Supplemental Figure S4. Sulforaphane pretreatment effects on Hpa infection at 7 dpi.
Supplementary Material
Acknowledgments
We thank Piero Morandini (University of Milan) for the single and double myb28 myb29 mutant lines, Georg Jander (Cornell University) for the tgg1 tgg2 double mutant, and Jun Fan (Department of Plant Pathology, China Agricultural University) for the Δsax P. syringae mutant strain.
Glossary
- HR
hypersensitive response
- PCD
programmed cell death
- DEX
dexamethasone
- Col-0
Columbia-0
- GC-MS
gas chromatography-mass spectrometry
- m/z
mass-to-charge ratio
- hpi
hours post induction
- Hpa
Hyaloperonospora arabidopsidis
- dpi
days post induction
- OD600
optical density at 600 nm
- cfu
colony-forming units
Footnotes
This work was supported by the Swedish Council for Environment, Agricultural Sciences, and Spatial Planning (grant no. 2007–1051 to M.E. and grant nos. 2007–1563 and 2009–888 to M.X.A.), the Olle Engkvist Byggmästare Foundation (to M.X.A.), the Adlerbertska Research Foundation (to M.X.A.), the Carl Tryggers Foundation (to H.A. and M.X.A.), the Swedish Research Council (to M.H. and H.A.), the Leverhulme Trust (grant no. 09 963/A to G.B. and M.T.), the U.S. Department of Agriculture (to D.M.), and the National Science Foundation (to D.M.).
References
- Agerbirk N, Olsen CE (2012) Glucosinolate structures in evolution. Phytochemistry 77: 16–45 [DOI] [PubMed] [Google Scholar]
- Ahmad S, Veyrat N, Gordon-Weeks R, Zhang Y, Martin J, Smart L, Glauser G, Erb M, Flors V, Frey M, et al. (2011) Benzoxazinoid metabolites regulate innate immunity against aphids and fungi in maize. Plant Physiol 157: 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson MX, Kourtchenko O, Dangl JL, Mackey D, Ellerström M (2006) Phospholipase-dependent signalling during the AvrRpm1- and AvrRpt2-induced disease resistance responses in Arabidopsis thaliana. Plant J 47: 947–959 [DOI] [PubMed] [Google Scholar]
- Bailey K, Cevik V, Holton N, Byrne-Richardson J, Sohn KH, Coates M, Woods-Tör A, Aksoy HM, Hughes L, Baxter L, et al. (2011) Molecular cloning of ATR5(Emoy2) from Hyaloperonospora arabidopsidis, an avirulence determinant that triggers RPP5-mediated defense in Arabidopsis. Mol Plant Microbe Interact 24: 827–838 [DOI] [PubMed] [Google Scholar]
- Barth C, Jander G (2006) Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J 46: 549–562 [DOI] [PubMed] [Google Scholar]
- Bednarek P. (2012a) Chemical warfare or modulators of defence responses: the function of secondary metabolites in plant immunity. Curr Opin Plant Biol 15: 407–414 [DOI] [PubMed] [Google Scholar]
- Bednarek P. (2012b) Sulfur-containing secondary metabolites from Arabidopsis thaliana and other Brassicaceae with function in plant immunity. ChemBioChem 13: 1846–1859 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al. (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323: 101–106 [DOI] [PubMed] [Google Scholar]
- Beekwilder J, van Leeuwen W, van Dam NM, Bertossi M, Grandi V, Mizzi L, Soloviev M, Szabados L, Molthoff JW, Schipper B, et al. (2008) The impact of the absence of aliphatic glucosinolates on insect herbivory in Arabidopsis. PLoS ONE 3: e2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Mackey D (2007) Elicitors, effectors, and R genes: the new paradigm and a lifetime supply of questions. Annu Rev Phytopathol 45: 399–436 [DOI] [PubMed] [Google Scholar]
- Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J (2003) Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62: 471–481 [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates ME, Beynon JL (2010) Hyaloperonospora arabidopsidis as a pathogen model. InVanAlfen NK, Bruening G, Leach JE, eds, Annual Review of Phytopathology, Vol 48 Annual Reviews, Palo Alto, CA, pp 329–345 [DOI] [PubMed] [Google Scholar]
- Dempsey DA, Klessig DF (2012) SOS: too many signals for systemic acquired resistance? Trends Plant Sci 17: 538–545 [DOI] [PubMed] [Google Scholar]
- Desveaux D, Singer AU, Dangl JL (2006) Type III effector proteins: doppelgangers of bacterial virulence. Curr Opin Plant Biol 9: 376–382 [DOI] [PubMed] [Google Scholar]
- Ding TJ, Zhou L, Cao XP (2006) A facile and green synthesis of sulforaphane. Chin Chem Lett 17: 1152–1154 [Google Scholar]
- Dixon DP, Sellars JD, Kenwright AM, Steel PG (2012) The maize benzoxazinone DIMBOA reacts with glutathione and other thiols to form spirocyclic adducts. Phytochemistry 77: 171–178 [DOI] [PubMed] [Google Scholar]
- Durand T, Bultel-Poncé V, Guy A, Berger S, Mueller MJ, Galano JM (2009) New bioactive oxylipins formed by non-enzymatic free-radical-catalyzed pathways: the phytoprostanes. Lipids 44: 875–888 [DOI] [PubMed] [Google Scholar]
- Fahey JW, Zalcmann AT, Talalay P (2001) The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56: 5–51 [DOI] [PubMed] [Google Scholar]
- Fan J, Crooks C, Creissen G, Hill L, Fairhurst S, Doerner P, Lamb C (2011) Pseudomonas sax genes overcome aliphatic isothiocyanate-mediated non-host resistance in Arabidopsis. Science 331: 1185–1188 [DOI] [PubMed] [Google Scholar]
- Farmer EE, Davoine C (2007) Reactive electrophile species. Curr Opin Plant Biol 10: 380–386 [DOI] [PubMed] [Google Scholar]
- Farmer EE, Mueller MJ (2013) ROS-mediated lipid peroxidation and RES-activated signaling. Annu Rev Plant Biol 64: 429–450 [DOI] [PubMed] [Google Scholar]
- Feng F, Zhou JM (2012) Plant-bacterial pathogen interactions mediated by type III effectors. Curr Opin Plant Biol 15: 469–476 [DOI] [PubMed] [Google Scholar]
- Frey M, Schullehner K, Dick R, Fiesselmann A, Gierl A (2009) Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70: 1645–1651 [DOI] [PubMed] [Google Scholar]
- Hopkins RJ, van Dam NM, van Loon JJA (2009) Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu Rev Entomol 54: 57–83 [DOI] [PubMed] [Google Scholar]
- Hückelhoven R, Panstruga R (2011) Cell biology of the plant-powdery mildew interaction. Curr Opin Plant Biol 14: 738–746 [DOI] [PubMed] [Google Scholar]
- Johansson ON, Fantozzi E, Fahlberg P, Nilsson AK, Buhot N, Tör M, Andersson MX (2014) Role of the penetration-resistance genes PEN1, PEN2 and PEN3 in the hypersensitive response and race-specific resistance in Arabidopsis thaliana. Plant J 79: 466–476 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T (2001) Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol 126: 811–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch E, Slusarenko A (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2: 437–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtchenko O, Andersson MX, Hamberg M, Brunnström A, Göbel C, McPhail KL, Gerwick WH, Feussner I, Ellerström M (2007) Oxo-phytodienoic acid-containing galactolipids in Arabidopsis: jasmonate signaling dependence. Plant Physiol 145: 1658–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranner I, Birtić S, Anderson KM, Pritchard HW (2006) Glutathione half-cell reduction potential: a universal stress marker and modulator of programmed cell death? Free Radic Biol Med 40: 2155–2165 [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL (2003) Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Mackey D, Holt BF III, Wiig A, Dangl JL (2002) RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108: 743–754 [DOI] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol 15: 349–357 [DOI] [PubMed] [Google Scholar]
- Morel JB, Dangl JL (1999) Suppressors of the Arabidopsis lsd5 cell death mutation identify genes involved in regulating disease resistance responses. Genetics 151: 305–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller MJ, Berger S (2009) Reactive electrophilic oxylipins: pattern recognition and signalling. Phytochemistry 70: 1511–1521 [DOI] [PubMed] [Google Scholar]
- Mur LAJ, Kenton P, Lloyd AJ, Ougham H, Prats E (2008) The hypersensitive response: the centenary is upon us but how much do we know? J Exp Bot 59: 501–520 [DOI] [PubMed] [Google Scholar]
- Noctor G, Queval G, Mhamdi A, Chaouch S, Foyer CH (2011) Glutathione. The Arabidopsis Book 9: e0142, doi/10.1199/tab.0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overmyer K, Brosché M, Kangasjärvi J (2003) Reactive oxygen species and hormonal control of cell death. Trends Plant Sci 8: 335–342 [DOI] [PubMed] [Google Scholar]
- Parker JE, Holub EB, Frost LN, Falk A, Gunn ND, Daniels MJ (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8: 2033–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queval G, Noctor G (2007) A plate reader method for the measurement of NAD, NADP, glutathione, and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal Biochem 363: 58–69 [DOI] [PubMed] [Google Scholar]
- Ross AF. (1961) Localized acquired resistance to plant virus infection in hypersensitive hosts. Virology 14: 329–339 [DOI] [PubMed] [Google Scholar]
- Shah J. (2009) Plants under attack: systemic signals in defence. Curr Opin Plant Biol 12: 459–464 [DOI] [PubMed] [Google Scholar]
- Sinapidou E, Williams K, Nott L, Bahkt S, Tör M, Crute I, Bittner-Eddy P, Beynon J (2004) Two TIR:NB:LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J 38: 898–909 [DOI] [PubMed] [Google Scholar]
- Sønderby IE, Burow M, Rowe HC, Kliebenstein DJ, Halkier BA (2010a) A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic glucosinolates in Arabidopsis. Plant Physiol 153: 348–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby IE, Geu-Flores F, Halkier BA (2010b) Biosynthesis of glucosinolates: gene discovery and beyond. Trends Plant Sci 15: 283–290 [DOI] [PubMed] [Google Scholar]
- Stakman EC. (1915) Relation between Puccinia graminis and plants highly resistant to its attack. J Agric Res 4: 193–200 [Google Scholar]
- Tierens KF, Thomma BPH, Brouwer M, Schmidt J, Kistner K, Porzel A, Mauch-Mani B, Cammue BPA, Broekaert WF (2001) Study of the role of antimicrobial glucosinolate-derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol 125: 1688–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietze F. (1969) Enzymic method for quantitative determination of nanogram amounts of total and oxidized glutathione: applications to mammalian blood and other tissues. Anal Biochem 27: 502–522 [DOI] [PubMed] [Google Scholar]
- Tör M, Gordon P, Cuzick A, Eulgem T, Sinapidou E, Mert-Türk F, Can C, Dangl JL, Holub EB (2002) Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA. (2010) ROS in biotic interactions. Physiol Plant 138: 414–429 [DOI] [PubMed] [Google Scholar]
- Torres MA, Jones JDG, Dangl JL (2006) Reactive oxygen species signaling in response to pathogens. Plant Physiol 141: 373–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F (2009) Network properties of robust immunity in plants. PLoS Genet 5: e1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valgimigli L, Iori R (2009) Antioxidant and pro-oxidant capacities of ITCs. Environ Mol Mutagen 50: 222–237 [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Freddie CT, Kahn K, Parker JE, Jones JDG (2002) Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J 29: 439–451 [DOI] [PubMed] [Google Scholar]
- Wu X, Zhou QH, Xu K (2009) Are isothiocyanates potential anti-cancer drugs? Acta Pharmacol Sin 30: 501–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.