Abstract
During animal cell division, the cleavage furrow is positioned by microtubules that signal to the actin cortex at the cell midplane. We developed a cell-free system to recapitulate cytokinesis signaling using cytoplasmic extract from Xenopus eggs. Microtubules grew out as asters from artificial centrosomes and met to organize antiparallel overlap zones. These zones blocked interpenetration of neighboring asters and recruited cytokinesis midzone proteins including the Chromosomal Passenger Complex (CPC) and Centralspindlin. The CPC was transported to overlap zones, which required two motor proteins, Kif4A and a Kif20A paralog. Using supported lipid bilayers to mimic the plasma membrane, we observed recruitment of cleavage furrow markers, including a RhoA-GTP reporter, at microtubule overlaps. This system opens further approaches to understanding the biophysics of cytokinesis signaling.
Actomyosin-based cleavage furrows in animal cells are positioned by signals emanating from microtubule assemblies formed shortly after anaphase onset (1). In typical somatic cells, the signaling complexes Centralspindlin and CPC (Chromosomal Passenger Complex) accumulate at the center of the midzone (or central spindle), which forms in the space previously occupied by the mitotic spindle (2). It is unclear how the microtubules that position furrows are organized in much larger egg cells, and how they signal to the cortex. We addressed these questions by developing a cell-free system to reconstitute the spatial signaling characteristic of cytokinesis in a large egg cell.
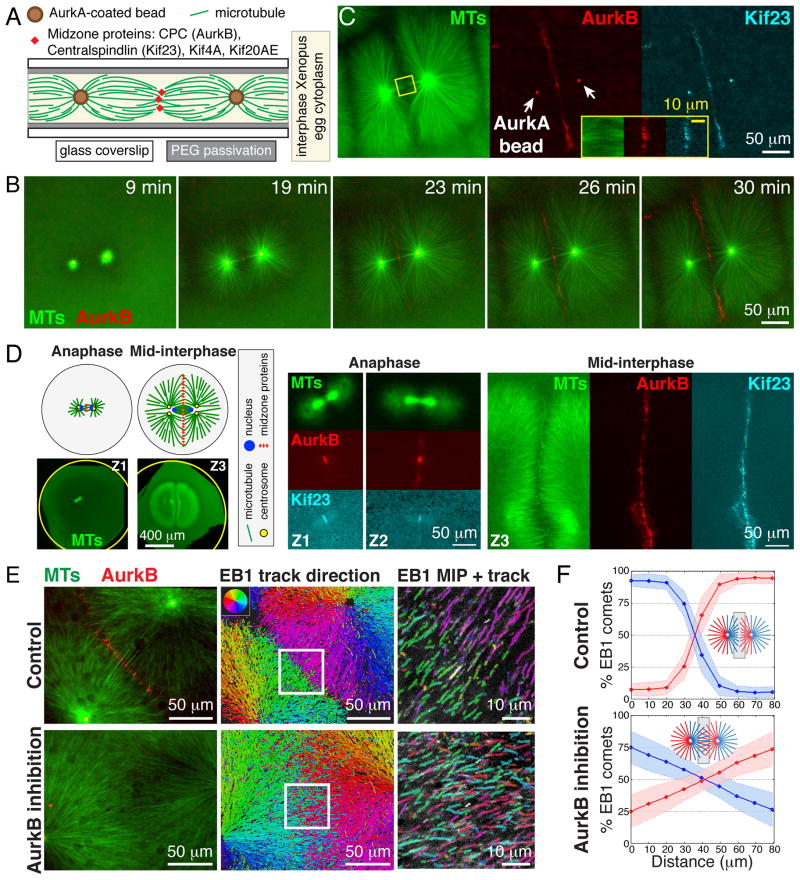
To reconstitute cytokinesis events, undiluted egg cytoplasm with intact actin (3), containing fluorescent probes and AurkA-based artificial centrosome beads (4), was treated with Ca2+ to mimic fertilization and immediately spread between two coverslips for imaging (fig. S1A). As the cell cycle progressed from metaphase to interphase (5), large microtubule asters grew out rapidly from each AurkA bead. Where the expanding edges of two neighboring asters met, antiparallel microtubule bundles formed in a boundary zone that we term the aster-aster interaction zone (AAIZ) (Fig. 1, A to C, fig. S1, and movie S1). In somatic cells, the CPC and Centralspindlin complexes are recruited to the midplane in anaphase where they specify the division plane by activating the small GTPase RhoA (2). We imaged endogenous complexes by adding labeled antibodies, and for the CPC confirmed localization with a GFP tagged DasraA subunit (5). CPC and the Kif23 subunit of Centralspindlin were recruited to the AAIZ in a 5–15 μm wide line bisecting the line between two AurkA beads (Fig. 1, B and C, fig. S1). The AAIZ was wider than a somatic cell midzone, and hundreds of microns long. To evaluate its physiological relevance, we imaged the same proteins in Xenopus zygotes fixed between mitosis and cytokinesis, which takes place at interphase in early embryonic cells (Fig. 1D) (6). The morphology of the midplane in zygotes, as defined by microtubule morphology and CPC/Centralspindlin localization, was strikingly similar to the AAIZ in extracts (Fig. 1, A to C).
Fig. 1. Aster outgrowth and interaction in Xenopus egg cytoplasm.
(A) Experimental setup. See (5) for methods and probes. (B) Widefield time-lapse showing recruitment of AurkB (red) to an AAIZ. (C) AAIZ at 30 min (widefield) showing recruitment of AurkB (CPC subunit, red) and Kif23 (Centralspindlin subunit, cyan). (D) Localization of MTs (green), AurkB (red) and Kif23 (cyan) in Xenopus zygotes fixed at consecutive stages of the first cell division (Z1–Z3) imaged by laser scanning confocal microscopy (5). (E) Quantification of microtubule orientation using EB1 tracking. Left: Spinning disc confocal images of control (top) and AurkB inhibited (bottom) AAIZs. Middle: Map of EB1-GFP trajectories. Right: 4X zoom-up of square in middle panel with maximum intensity projection (MIP) of EB1 tracks (over 15 sec). (F) Spatial distribution of EB1 tracks at AAIZs classified by direction (shading indicates standard deviations determined from N≥5 zones). (See fig. S2, S3, and (5) for data analysis.)
To measure microtubule orientation at the AAIZ we tracked GFP-tagged EB1, which binds to growing microtubule plus ends (Fig. 1E, fig. S2, movie S2) (7). Microtubules grew outward radially within each aster. At the AAIZ, EB1 comets from both directions entered antiparallel bundles, where they usually disappeared (Fig. 1E). We quantified the degree of interpenetration by categorizing EB1 comets based on their direction (fig. S3) (5). The AAIZ was characterized by a sharp change in directionality over ~20 μm, indicating a localized block to interpenetration between the asters (Fig. 1F).
Kinase activity of the Aurora B (AurkB) subunit of the CPC is required to establish midzone morphology and for furrow ingression (8). We confirmed this in Xenopus eggs (fig. S4) (5). AurkB inhibition blocked recruitment of the CPC in our cell free system (Fig. 1E), and caused much deeper interpenetration of microtubules (Fig. 1, E and F, and movie S3). Thus, AurkB activity was required to create a sharp boundary between asters.
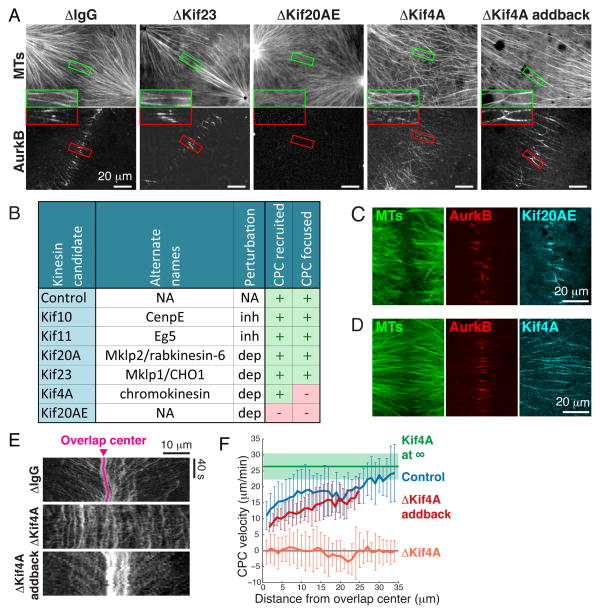
CPC is proposed to be transported to the center of midzones along microtubules by a kinesin molecular motor (9), but transport has not been observed directly. Five plus-end directed kinesins involved in cell division are candidates for CPC transport (10): Kif4A, Kif10 (also called CenpE), Kif11 (Eg5), Kif20A (Mklp2/Rabkinesin-6) and Kif23 (Mklp1, a subunit of Centralspindlin). Xenopus eggs also contain an unusual kinesin 41% identical to Kif20A by sequence (11). This is likely to be an embryo-specific paralog, and we propose the name KIF20Aembryonic, abbreviated KIF20AE. Small molecule inhibition of Kif10 and Kif11, and depletion of somatic Kif20A and Kif23, had no effect on antiparallel microtubule bundling in the AAIZ or on CPC localization (Fig. 2, A and B, figs. S5 and S6). Depletion of Kif4A did not block CPC accumulation or assembly of antiparallel bundles, but increased the width of the CPC-positive zone; adding back recombinant Kif4A rescued this phenotype (Fig. 2A, fig. S6). Depletion of Kif20AE completely blocked CPC accumulation and disorganized the AAIZ (Fig. 2A). Thus Kif20AE is absolutely required for CPC recruitment in eggs, presumably mirroring the Kif20A requirement in somatic cells (9), and Kif4A plays some role in focusing it.
Fig 2. Kinesin-dependent recruitment and motility of the CPC.
(A) Representative spinning disc confocal images of MTs and AurkB at AAIZs following kinesin immunodepletions as labeled above. (B) Summary of kinesin perturbations using drug inhibition (inh) or immunodepletion (dep) (5). (C,D) Localization of Kif20AE and Kif4A at AAIZs (5). (E) Kymographs of CPC dynamics on individual microtubule bundles in AAIZs. (F) CPC velocity as a function of distance from the center of the AAIZ (error bars are standard deviations determined from N≥3 microtubule bundles (5)).
Microtubule bundles in the AAIZ localized Kif20AE (Fig. 2C), Kif4A (Fig. 2D), and an embryonic paralog of PRC1 (PRC1E, an antiparallel microtubule crosslinker; fig. S1D), all of which have been shown to bind the CPC in pull-down experiments (9, 12–14). Time-lapse imaging showed that CPC clusters moved to the center of the overlap zone at a rate of 10–25 μm/min, slowing as they reached the center (Fig. 2, E and F, fig. S7, movie S4). Tubulin speckle imaging showed sliding of microtubules away from the overlap center at <5 μm/min (fig. S8, movies S6 to S8). Thus the observed CPC movement represents transport towards plus ends. The maximal transport rate was close to that of Kif4A in an isolated aster not part of an AAIZ (Fig. 2F, green line, fig. S9) (5). Kif4A depletion blocked CPC movement, and addback of recombinant Kif4A rescued it (Fig. 2, E and F, figs. S10, movie S5). Moving CPC and Kif4A partially co-localized by live imaging (fig. S11). Thus, the CPC is targeted to the AAIZ and transported to a narrow band by the combined action of Kif20AE and Kif4A. Transport probably slows as CPC engages binding sites in the AAIZ (5).
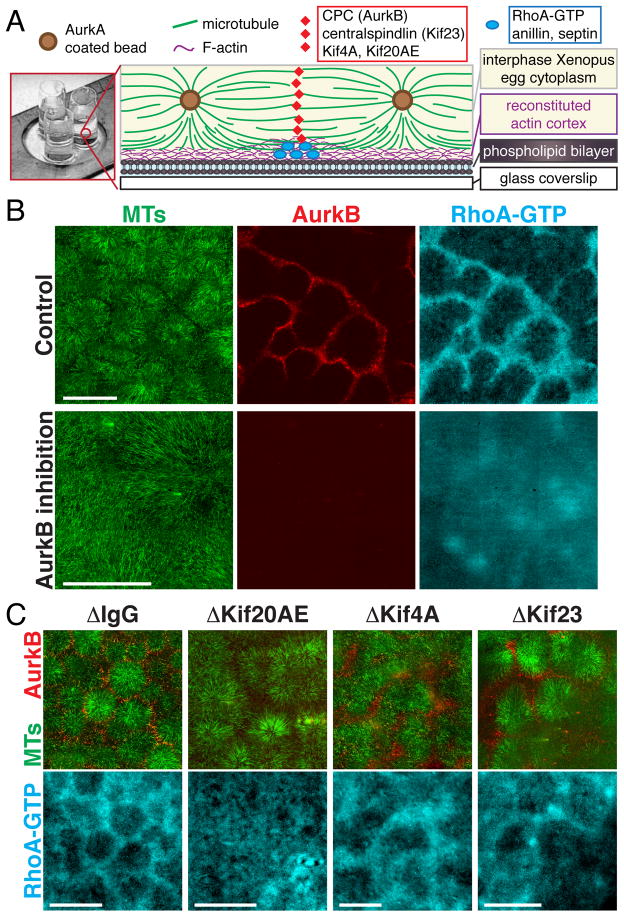
To test if AAIZs in the cell-free system can signal to the cortex, we prepared supported lipid bilayers on glass coverslips using lipids characteristic of the inner leaflet of animal cell plasma membranes, including phosphatidylinositol (PI) and phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] (5, 15). Protein recruitment was imaged using total internal reflection fluorescence (TIRF) microscopy (Fig. 3A). The small GTPase RhoA is thought to be the master organizer of the furrow (16). To localize active, GTP-bound RhoA we added an mCherry-tagged RhoA binding fragment of Rhotekin (mCherry-rGBD) (16). Using PEG-passivated coverslips, we saw no recruitment of RhoA-GTP. Using lipid bilayers, the RhoA-GTP reporter was enriched at the bilayer under AAIZs that recruited CPC and Centralspindlin (Fig. 3B, fig. S12). Inhibition of AurkB or depletion of Kif20AE completely blocked AAIZ assembly and CPC localization as expected (8, 9), and also blocked localized RhoA-GTP enrichment (Fig. 3, B and C). Kif4A depletion resulted in broadening of both CPC and RhoA-GTP zones (Fig. 3C). Depletion of Kif23, a subunit of the Centralspindlin complex, did not block AAIZ assembly or RhoA-GTP recruitment (Fig. 3C). Centralspindlin plays a central role in midzone assembly and signaling in many biological systems (2). Its dispensability in our system likely reflects functional redundancy among midzone factors and is consistent with its dispensability in Xenopus blastomere cytokinesis (17). We conclude that a CPC-positive AAIZ can locally activate RhoA in our system, although we cannot distinguish whether localized AurkB phosphorylation of cytokinetic factors or some specific organization of microtubules at the AAIZ is required for furrow signaling.
Fig. 3. Spatially localized recruitment of GTP-bound RhoA on model plasma membranes.
(A) Drawing of experimental setup. See (5) for methods and probes. (B,C) Localization of a probe for RhoA-GTP (mCherry-rGBD) (16) at lipid bilayers proximal to AAIZs. TIRF images from 3×3 to 5×5 adjacent fields were corrected for uneven illumination and stitched (5). (B) Representative control and AurkB inhibition. (C) IgG control, Kif20AE, Kif4A, and Kif23 depletions. Bar, 200 μm.
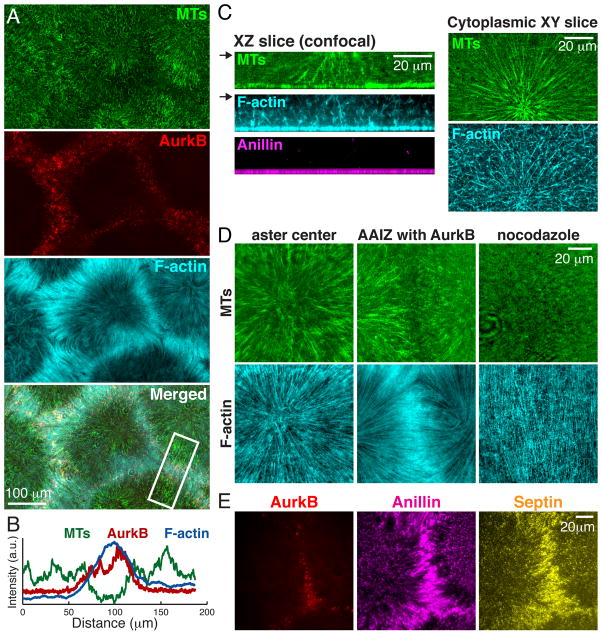
To probe cortical organization in our system we visualized F-actin using Lifeact-GFP (Fig. 4) (18). F-actin was enriched at the lipid bilayer under AAIZs (Fig. 4, A and B). Above the cortical layer, F-actin organized into branched networks that showed only partial alignment with the microtubules (Fig. 4C). At the cortical layer, F-actin organized into long, uniformly spaced cables that aligned radially with microtubules (Fig. 4D, left and middle, movie S9). The spacing between actin cables was 1.3±0.5 μm (SD; N=140), comparable to actin-containing contractile bands on the cortex of live frog eggs (19). This organization was microtubule dependent (Fig. 4D, right); we suspect it represents a configuration of actin bundles that normally precedes furrow ingression, since it has been noted in early anaphase in several systems (5). The furrow-selective actin bundling protein Anillin (20) co-localized with F-actin cables throughout the cortex, and was enriched under AAIZs along with its binding partner septin complex (Fig. 4E, movie S10).
Fig. 4. Recruitment and organization of cortical cleavage furrow proteins.
Experimental setup as per Fig. 3A. (A) F-actin (TIRF). Note enrichment proximal to CPC at AAIZs. (B) Intensity line-scans across AAIZ (boxed in A) shows enrichment of AurkB and F-actin. (C) XZ slices of 3D reconstructions using spinning disc confocal microscopy showing organization of MTs, F-actin, and Anillin (left). XY slices of MTs and F-actin at plane marked by arrow (right). (D) TIRF images of microtubules and F-actin at aster center (left) and an AAIZ (middle). Right shows a nocodazole treated sample. (E) Anillin and septin on bilayer (TIRF) co-localize with CPC at AAIZ.
Summarizing, we have developed a cell-free system that recapitulates the hallmarks of spatially organized signaling characteristic of egg cytokinesis. Midzone proteins were recruited to the overlap zone between microtubule asters in egg extract, where they signaled to a nearby lipid bilayer to locally activate the RhoA pathway. Our system confirmed known mechanisms of spatially organized signaling, for example the Aurora B kinase activity requirement for cleavage furrow induction, and revealed new mechanistic aspects that were difficult to observe in living cells, notably transport of the CPC along microtubules towards the midzone in a process requiring two kinesins. Our data favor a model in which Kif4A is a transport motor for the CPC, and Kif20AE is required for it to bind microtubules, though not necessarily as a transporter. Future pure protein reconstitution experiments are required to conclusively determine the mechanism of CPC transport and the exact roles of these two kinesins.
Two factors facilitated imaging approaches in the cell-free system: the large spatial scale of the AAIZ (~20μm) and its long time duration (>20 minutes). Both reflect the pre-cytokinesis organization of the large egg cell. By comparison, the microtubule overlap in somatic cell midzones is ~2–3μm wide and lasts only ~3 minutes before being compressed by the furrow (21). Future studies will take advantage of these features and the experimental flexibility of an extract system to probe biophysical mechanisms involved in cytokinesis.
Supplementary Material
Acknowledgments
We thank R. Ohi, D. Burgess, D. Miyamoto, E. Tan for reagents, H. Basu for preliminary work, the Nikon Imaging Center at HMS and Nikon at MBL for microscopy support, and the NXR at MBL for Xenopus animals and care. This work was supported by NIH GM39565 (TJM); MBL fellowships from the Evans Foundation, MBL Associates and the Colwin Fund (TJM, CMF); NIH GM103785 (MW).
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Supplementary content includes Materials and Methods, Supplementary Text, Figs. S1 to S13, Tables S1 to S2, Movies S1 to S10, References (22–29).
References and Notes
- 1.Rappaport R. Cytokinesis in Animal Cells. Cambridge University Press; 1996. [Google Scholar]
- 2.Glotzer M. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 3.Field CM, Nguyen PA, Ishihara K, Groen AC, Mitchison TJ. Methods Enzymol. 2014;540:399–415. doi: 10.1016/B978-0-12-397924-7.00022-4. [DOI] [PubMed] [Google Scholar]
- 4.Tsai MY, Zheng Y. Curr Biol. 2005;15:2156–2163. doi: 10.1016/j.cub.2005.10.054. [DOI] [PubMed] [Google Scholar]
- 5.Supplementary materials are available on Science Online.
- 6.Wühr M, Tan ES, Parker SK, Detrich HW, 3rd, Mitchison TJ. Curr Biol. 2010;20:2040–2045. doi: 10.1016/j.cub.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petry S, Groen AC, Ishihara K, Mitchison TJ, RD Cell. 2013;152:768–777. doi: 10.1016/j.cell.2012.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Argiros H, Henson L, Holguin C, Foe V, Shuster CB. Cytoskelet Hoboken NJ. 2012;69:840–853. doi: 10.1002/cm.21061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruneberg U, Neef R, Honda R, Nigg EA, Barr FA. J Cell Biol. 2004;166:167–172. doi: 10.1083/jcb.200403084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cross RA, McAinsh A. Nat Rev Mol Cell Biol. 2014;15:257–271. doi: 10.1038/nrm3768. [DOI] [PubMed] [Google Scholar]
- 11.Wühr M, et al. Curr Biol. 2014;24:1467–1475. doi: 10.1016/j.cub.2014.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ozlu N, et al. Mol Cell Proteomics. 2010;9:336–350. doi: 10.1074/mcp.M900308-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bieling P, Telley IA, Surrey T. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- 14.Mitchison TJ, Nguyen P, Coughlin M, Groen AC. Mol Biol Cell. 2013;24:1559–1573. doi: 10.1091/mbc.E12-12-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loose M, Fischer-Friedrich E, Ries J, Kruse K, Schwille P. Science. 2008;320:789–792. doi: 10.1126/science.1154413. [DOI] [PubMed] [Google Scholar]
- 16.Bement WM, Benink HA, von Dassow G. J Cell Biol. 2005;170:91–101. doi: 10.1083/jcb.200501131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller AL, Bement WM. Nat Cell Biol. 2009;11:71–77. doi: 10.1038/ncb1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riedl J, et al. Nat Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danilchik MV, Brown EE, Riegert K. Dev Camb Engl. 2006;133:4517–4526. doi: 10.1242/dev.02642. [DOI] [PubMed] [Google Scholar]
- 20.Straight AF, Field CM, Mitchison TJ. Mol Biol Cell. 2005;16:193–201. doi: 10.1091/mbc.E04-08-0758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mastronarde DN, McDonald KL, Ding R, McIntosh JR. J Cell Biol. 1993;123:1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oegema K, Savoian MS, Mitchison TJ, Field CM. J Cell Biol. 2000;150:539–552. doi: 10.1083/jcb.150.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groen AC, Nguyen PA, Field CM, Ishihara K, Mitchison TJ. Methods Enzymol. 2014;540:417–433. doi: 10.1016/B978-0-12-397924-7.00023-6. [DOI] [PubMed] [Google Scholar]
- 24.Lee K, Gallop JL, Rambani K, Kirschner MW. Science. 2010;329:1341–1345. doi: 10.1126/science.1191710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Applegate KT, et al. J Struct Biol. 2011;176:168–184. doi: 10.1016/j.jsb.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neef R, et al. J Cell Biol. 2003;162:863–876. doi: 10.1083/jcb.200306009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fishkind DJ, Wang YL. J Cell Biol. 1993;123:837–848. doi: 10.1083/jcb.123.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gudimchuk N, et al. Nat Cell Biol. 2013;15:1079–1088. doi: 10.1038/ncb2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Groen AC, et al. J Cell Sci. 2008;121:2293–2300. doi: 10.1242/jcs.024018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.