Abstract
The nonsense-mediated mRNA decay (NMD) pathway was originally discovered by virtue of its ability to rapidly degrade aberrant mRNAs with premature termination codons. More recently, it was shown that NMD also directly regulates subsets of normal transcripts, suggesting that NMD has roles in normal biological processes. Indeed, several NMD factors have been shown to regulate neurological events (for example, neurogenesis and synaptic plasticity) in numerous vertebrate species. In man, mutations in the NMD factor gene UPF3B, which disrupts a branch of the NMD pathway, cause various forms of intellectual disability (ID). Using Epstein Barr virus—immortalized B cells, also known as lymphoblastoid cell lines (LCLs), from ID patients that have loss-of-function mutations in UPF3B, we investigated the genome-wide consequences of compromised NMD and the role of NMD in neuronal development and function. We found that ~5% of the human transcriptome is impacted in UPF3B patients. The UPF3B paralog, UPF3A, is stabilized in all UPF3B patients, and partially compensates for the loss of UPF3B function. Interestingly, UPF3A protein, but not mRNA, was stabilised in a quantitative manner that inversely correlated with the severity of patients’ phenotype. This suggested that the ability to stabilize the UPF3A protein is a crucial modifier of the neurological symptoms due to loss of UPF3B. We also identified ARHGAP24, which encodes a GTPase-activating protein, as a canonical target of NMD, and we provide evidence that deregulation of this gene inhibits axon and dendrite outgrowth and branching. Our results demonstrate that the UPF3B-dependent NMD pathway is a major regulator of the transcriptome and that its targets have important roles in neuronal cells.
Keywords: intellectual disability, Nonsense-mediated mRNA decay, RNA-SEQ, UPF3A, UPF3B
Introduction
Nonsense-mediated mRNA decay (NMD) is a conserved pathway in eukaryotes ranging from Sacchromyces cerevisiae to mammals.1 NMD recognizes and degrades transcripts harboring mutations that introduce premature termination codons (PTCs), preventing the truncated proteins with possible dominant negative effects to be made. How NMD carries out its function is taxonomically dependent. In metazoan, the conserved UPF1, UPF2 and UPF3 proteins constitute the core components of the classical NMD pathway.1 UPF3 is associated with the exon-junction complex that marks the exon–exon junction during pre-mRNA splicing.2,3 UPF2 interacts with UPF3 to bridge the exon-junction complex to UPF1 and other NMD factors when the ribosome stalls at the PTC during the pioneer round of translation.1,4 UPF1 is an ATP-ase RNA helicase whose role is to trigger recruitment of downstream NMD factors to degrade transcripts bearing PTC.1,5–7 In addition to this classical pathway, it has been shown that NMD can function in alternative cascades independent of UPF2 or UPF3.8,9 The cascade studied in this paper involves UPF3 proteins, UPF3B and its ortholog UPF3A. UPF3B and UPF3A share high sequence similarity and both compete for interaction with UPF2 to activate NMD.10,11 This is part of a regulatory switch that maintains proper NMD function in different tissues where varying level of UPF3B is observed.10
NMD also regulates normal transcript levels. Microarray studies on NMD-deficient eukaryotic models and human cell lines suggested that NMD regulates 3–10% of the transcriptome.9,10,12–16 Transcripts regulated by NMD have important roles in cell survival and cell function.9,10,12,13 In fact, NMD is crucial for higher eukaryotic development as deletion of Upf1 or Upf2 in the mouse led to embryonic lethality.14,17 In man, we showed that mutations in UPF3B, an X-chromosome linked gene, cause various forms of syndromic and non-syndromic intellectual disability (ID) (Mendelian Inheritance in Man, MIM, no. 300676),18,19 thus, NMD also has a central role in regulating normal brain development and function. To date, mutations in UPF3B have been found in nine families,18–20 including one unpublished family (LSN & JG, unpublished data). These are frame-shift and missense mutations that introduce PTCs and completely abolish UPF3B function (Patient 1–4 and patient 6–9, Supplementary Table S1), or missense change in a conserved amino acid near the UPF2-binding motif, which could affect UPF3B ability to interact with UPF2 (patient 5, Supplementary Table S1). UPF3B patients present with a highly heterogeneous phenotype, which include attention-deficit hyperactivity disorder, schizophrenia, autism and ID (Supplementary Table S1). There is considerable intra- and inter-familial variability in clinical presentations in patients with UPF3B mutations. As such we propose to use the term ‘UPF3B spectrum’ to describe this. Having access to patients’ cell lines provided us with a unique opportunity to study natural consequences of compromised NMD on the human transcriptome without the need of manipulating UPF3B or NMD in vitro.
Here we identify a set of tentative NMD targets using a combination of approaches. We also show that UPF3A protein is stabilized in UPF3B patients and functionally compensates for the loss of UPF3B in a dose-dependent manner. Our data provide evidence that UPF3A and UPF3B proteins likely act on the same substrates in a redundant manner and suggest that UPF3A might be an important modifier of the UPF3B loss-of-function phenotype. We further explore the UPF3B-NMD’s role in the brain by studying the consequences of deregulation of at least one canonical NMD target, ARHGAP24, in cell models of neuronal development.
Materials and methods
RNA extraction
RNA was extracted from frozen lymphoblastoid cell pellet using Trizol (Invitrogen, Grand Island, NY, USA) and RNeasy Mini Kit (QIAGEN, Hilden, Germany), according to the manufacturer’s instruction.
RNA-SEQ, exon array and SNP (single nucleotide variants)-chip sample processing
Twenty µg of RNA (Supplementary Table S2) was sent to Geneworks (Adelaide, Australia) for library preparation and sequencing on the Illumina (San Diego, CA, USA) GAII platform. Seven µg of RNA (Supplementary Table S3) was sent to the Australian Genome Research Facility (Melbourne, Australia) for labeling and hybridizing to Affymetrix Human Exon 1.0 ST arrays (Affymetrix, Santa Clara, CA, USA). Three mg of DNA was sent to the Australian Genome Research Facility for labeling and hybridizing to the Illumina Human Omniexpress SNP chip. All patients described in Supplementary Table S1 and two controls were included in the SNP-chip analysis.
Mapping of RNA-SEQ reads
RNA-SEQ reads were mapped to the reference sequences, which includes the latest build of the human genome (HG19), exon-junction database (see below) and all known ribosomal DNA using Efficient Alignment of Nucleotide Database (ELAND, Illumina) allowing up to two mismatches in the first 32 bp seed of the reads. Reads were mapped at the original length, 65 bp, and trimmed, 50 bp (first base and the last 14 bases excluded) for comparison (Supplementary Figure S1a and S1b). Mapped reads, that have more than two mismatches along the entire reads or are mapped to ribosomal or mitochondrial DNA, were removed from downstream analysis (Supplementary Figure S1b and S1c).
Generation of exon-junction database
A comprehensive list of all known exon junctions, including alternatively spliced exons was compiled from all genes annotated in the UCSC database using script included in ELAND. Each exon junction sequence was comprised of the flanking sequence 5 bp shorter than the read length used for mapping on both sides of the exon–exon junction, that is, if read length was 65 bp or 50 bp, the flanking sequence would be 60 bp or 45 bp and the total length of the exon junction sequence would be 120 bp or 90 bp, respectively. This limit ensured that mapped reads matched at least 5 bp on either side of the exon–exon junction. For exon length less than the flanking sequence specified for the read length, the entire exon was incorporated into the exon junction sequence. A total of ~245 000 junctions were used for alignment.
Estimation of the mis-mapping rate
In order to assess the false positive rate of RNA-SEQ due to mis-mapping, we created a test set of ~75000 modified exon junctions where 5 bp was removed from the 3′ end of the leading exon and from the 5′ end of the following exon (Supplementary Figure S2a). All reads from patient 2 and control 1 were mapped to either the authentic exon junctions or the modified exon junctions. Junctions with more than two mapped reads were considered to be present (Supplementary Figure S2b). Rate of mis-mapping was calculated as ratio between the number of mapped modified junctions and authentic junctions.
Calculation of gene expression by RNA-SEQ
Gene expression was calculated using the RPKM (reads per kilobase exon model per million mapped reads) formula as previously described.21 Reads mapped to exons that are shared between two or more genes in either direction were excluded from counting. In order to correct for variation between lanes, the RPKM values were normalized using quintile normalization method.22
Analysis of affymetrix human exon 1.0. ST array
The exon array contains ~1.4 million probes targeting all known and predicted exons. We utilized the core probe sets, which target the most confidently annotated exons, for all analysis. In short, intensity values were normalized using log-scaled robust multi-array analysis23 and adjusted for batch effect (Partek, St Louis, MO, USA). Gene-expression value was taken from median of all probe sets comprised in that gene. To filter out false positives in the final analysis, we restricted the analysis of the exon array using only core probe sets (~160 000) that target the genes (~11000) that we could confidently detect by RNA-SEQ (expression threshold: RPKM⩾3). Data were transformed and analyzed using Partek Genomic Suite V6.5.
Analysis of copy-number variation
Raw data were first analyzed in Genome Bead Studio by the Australian Genomic Research Facility then exported into Partek compatible formats. Copy-number intensity of all six patients were created from allele-intensity values against one single reference control (Control 1), and adjusted for GC noise.24 Copynumber variant was detected by Genomic Segmentation algorithm25 with the following threshold: minimum markers = 10, P value threshold <0.001, signal to noise > 0.5 and expected range =0.3. All analysis was performed using Partek Genomics Suite V6.5.
Calling and validation of sequence variants
Variants were called using CASSAVA v1.6 (Illumina) with the minimum coverage threshold of six reads, and the variant called must present in at least 85% of all reads. Known SNPs (UCSC dbSNP130), which were also included on the Illumina Human Omni Express SNP chip, were considered for SNP validation. Over 95% of variants identified by RNA-SEQ have the same heterozygous/homozygous calls by the SNP chip. We estimated the false-positive rate of SNP calling by CASSAVA to be ~5%. Next, variants’ effect was predicted using SNP Effect Predictor (Ensembl).26 Non-synonymous coding SNPs were furthered examined using SIFT27 and PolyPhen28 for possible deleterious effects on protein function (Supplementary Table S7).
Analysis of transcriptome correlation between lymphoblastoid cell line (LCL) and brain
In order to assess the similarity between the transcriptome of LCL and different parts of the brain, we extracted publicly available microarray data (HU133A platform Gene Expression Omnibus no. GDS596)29 and analyzed using Partek Genomic Suite V6.5.
Statistical calculation
Pearson correlation coefficiency was used to determine similarity between two groups. Student’s t-test was used to determine significant difference between groups.
Cell culture
The Epstein-Barr virus immortalized B-cell lines, also known as LCLs, used in this study were established from peripheral blood lymphocytes of patients and controls, as previously described.19 Once established, the LCL were cultured in RPMI 1640 (Sigma) supplemented with 10% fetal calf sera, 2mM L-Glutamine, 0.017 mg ml−1 benzylpenicillin and grown at 37 °C with 5% CO2.
Primary hippocampal neurons were prepared as previously described30 with modifications. E18.5 embryos were isolated from time-mated Swiss females, and brains placed into ice-cold Hanks Balanced Salt Solution containing 10mM HEPES, pH 7.3 (Invitrogen). Hippocampi were removed under sterile conditions and placed into ice-cold Neurobasal media (Invitrogen). Trypsin (Sigma-Aldrich, St Louis, MO, USA) was added to a final concentration of 0.05% and incubated for 15 min at 37 °C. Tissue was dissociated by trituration and cells counted. Immediately following dissociation, 1 × 106 cells were transfected with 1 µg of pmaxGFP plasmid (Lonza) alone or in addition to either 2 or 5 µg (high expression) of pcDNA-Myc or pcDNA-ARH-GAP24-Myc isoform 1 using the Amaxa Mouse Neuron Nucleofector Kit (Lonza, Basel, Switzerland). Viable neurons were plated into 12-well plates on poly-D-lysine (Sigma-Aldrich)-coated coverslips at a density of 1.25 × 105 cells per well in Neurobasal media containing 2% (vol/vol) B27 (Invitrogen) and 10% fetal calf serum (Thermo Scientific, Waltham, MA, USA). Cells were maintained in a humidified incubator at 37 °C with 5% CO2. Following cell attachment (~4h), the media was replaced with Neurobasal with 2% B27. Half media was changed after 4 days.
PC12 cell line was maintained in Complete Growth Media: Dulbecco’s modified Eagle’s medium (Sigma) supplemented with 10% horse sera, 5% fetal calf sera, 2mM L-Glutamine and 0.017 mg ml−1 benzypenicillin (CLS). Cells were transfected using Lipofectamine 2000 (Invitrogen) and allowed to grow for 72 h, then collected using Trypsine (Thermo Scientific) and sub-cultured onto collagen-coated plates. Twelve hours later, differentiation was initiated by adding 50 ng ml−1 nerve growth factor to the culture.
Human HeLa cells were cultured in Dulbecco’s modified Eagle’s medium supplied with 10% fetal bovine serum, 1% glutamine and 1% penicillin/ streptomycin (Gibco, Grand Island, NY, USA).
Knocking down NMD factors in HeLa cells using RNAi
RNAi of UPF1, UPF2 and SMG1 were performed by transfecting HeLa cells with UPF1 (5′-GAUGCAGU UCCGCUCCAUU-3′),12 UPF2 (5′-CAACAGCCCUUC CAGAAUC-3′)2 and SMG1 (5′-GUGUAUGUGCGCCA AAGUA-3′).31 siRNA was purchased from Ambion (Grand Island, NY, USA). Luciferase-specific siRNA (5′-GUGCGCUGCUGGUCGCAAC-3′)32 was used as control. Cells were seated at 1.5 × 105 per well in six-well plates the day before transfection. siRNA oligonucleotides (100 nM) were mixed with lipofectamin 2000 (Invitrogen) in Opti-MEM (Gibco) and applied to the cells in culture media without penicillin/stripe. Cells were harvested 44–48 h after transfection.
Cycloheximide treatment
Control LCLs (n = 6) were treated with Cycloheximide as previously described.19 Samples were collected 6 h post treatment.
Immunofluorescence
Cells were fixed using 4% paraformaldehyde for 15 min at room temperature. Cells were blocked/ permeabilized with PBST (a solution of PBS containing 1% Tween 20) and 10% normal horse serum, Sigma-Aldrich). Primary and fluorescently tagged secondary antibodies were diluted in PBST containing 3% normal horse serum. Primary antibodies were incubated overnight at 4 °C. Secondary antibodies were incubated for 1 h at room temperature. All antibody dilutions can be found in Supplementary Table S9. Nuclei were stained using 300 nM 4,6-diamidino-2-phenylindole as per manufacturer protocols (Invitrogen). Coverslips were mounted using Slowfade anti-fade mounting media (Invitrogen).
Microscopy and analysis
Fluorescence was viewed using the Axioplan2 microscope (Carl Zeiss, Jena, Germany) fitted with an HBO 100 lamp (Carl Zeiss). Images were captured using Axiocam Mrm camera and Axio Vs40 v4.5.0.0 software (Axiovision, Carl Zeiss). Images of GFP-positive (GFP + ve) neurons were captured at 20 × magnification and collated using Adobe Photoshop (Adobe, San Jose, CA, USA). Each experiment was done in triplicate, with at least 25 representative neurons scored for in each experiment. For measurement of axonal length, axons were identified using Tau1 immuno-reactivity and then measured using ImageJ software package (National Institute of Health). For the quantitation of neurite-terminal ends, dendritic and axonal neurites were identified using MAP2 and Tau1 immmunoreactivity, respectively. Only neurites of 10µm length or longer were included, and flagged for counting using ImageJ.
In situ hybridization
A 458-bp fragment of ARHGAP24 isoform 1 was PCR amplified using primers listed in Supplementary Table S8. PCR product was ligated into pGemTEasy (Promega, Madison, WI, USA) using TA cloning and sequence verified. Probe transcription and in situ hybridization was done under contract by the MRC-Wellcome-trust Human Developmental Biology Resource In-house Gene Expression Service. The human embryonic and fetal material was provided by the Joint MRC (grant no. G0700089)/ Wellcome Trust (grant no. GR082557).
Polymerase chain reaction
cDNA was made using methods previously described.19 PCR was performed using 1 µl of Taq DNA Polymerase (ROCHE), 1 × PCR buffer with MgCl2 (stock at 10 × ), specific single-stranded DNA primers and H2O to 50 µl volume. The PCR-cycle condition was as follows: initial denaturation at 94 °C for 5 min, then 35 cycles of denaturation at 94 °C for 30 s, annealing for 30 s at specific annealing Tm of each primer pair, extension at 72 °C for 30 s, followed by final extension at 72 °C for 10 min. PCR products were visualized on the Microchip Electrophoresis System (MultiNa, Shimadzu, Tokyo, Japan). Refer to Supplementary Table S8 for the list of all primers used in this study.
Reverse-transcriptase quantitative PCR analysis
Relative standard-curve method was employed to determine gene expression as previously described.19 Reactions were carried out using a Step One Plus Real-Time PCR system (Applied Biosystems, Mulgrave, VIC, Australia). Expression values were taken from mean of triplicates.
Western blotting
Protein was extracted from frozen LCL pellet and prepared for immune-blot according to protocol described previously19 Supplementary Table S9 describes all primary, secondary antibodies and the conditions at which they were used in this study.
Results
Genome-wide identification of deregulated genes in UPF3B patients
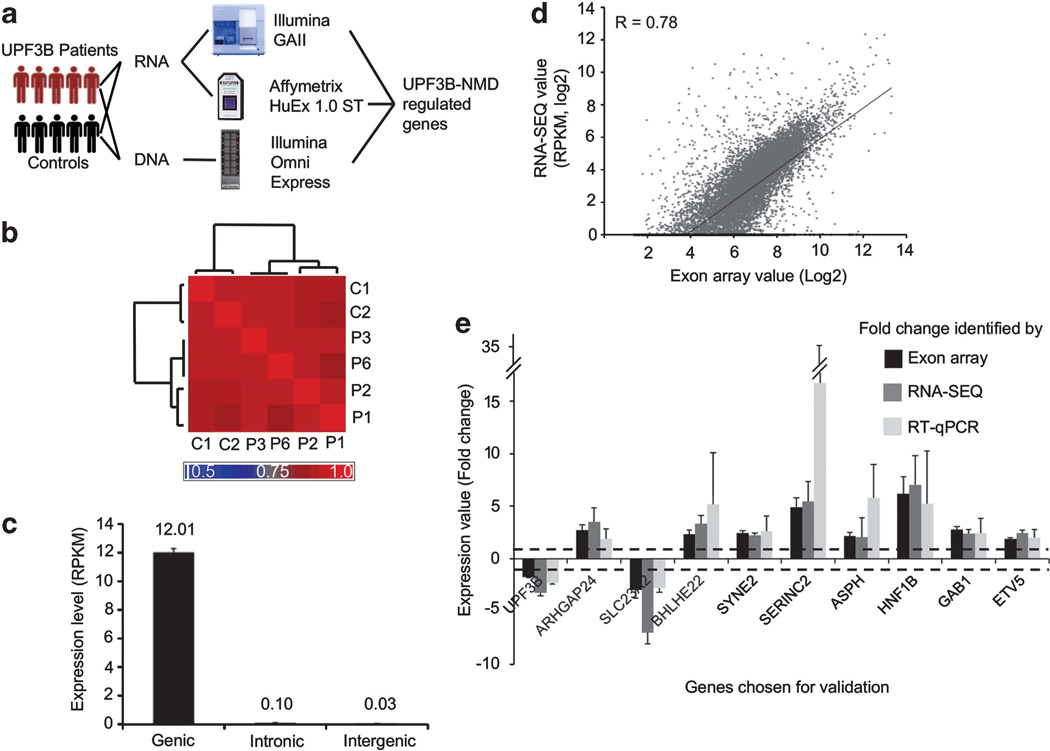
To identify the impact of complete loss of UPF3B function on the human transcriptome, we used RNA-SEQ33 to analyze polyA+ RNA from four UPF3B patients’ and two controls’ LCLs (using Illumina GAII). We obtained between 10–20 million reads per sample (Supplementary Table S2). To reduce the mismapping frequency, we trimmed the 1st and the last 14 bases of all reads before mapping (Supplementary Figure S1a). This increased the total number of mapped reads by 3% when compared with the distribution of full-length (65 bp) mapped reads (Supplementary Figure S1b). Despite the marked difference in the total number of reads in different samples (Supplementary Table S2), about ~50% of trimmed-length (50 bp) reads mapped uniquely to the genome with high accuracy (false discovery rate<0.2%, Supplementary Figure S2). About 10% of the trimmed-length reads mapped to the exon junctions in all samples (Supplementary Figure S1c). The levels of gene expression identified in all samples exhibited high correlation with each other (R > 0.9) (Figure 1b). As we have only sequenced polyA + RNA, the majority of reads were mapped to the genic regions (Figure 1c). To complement the RNA-SEQ results and to maximize the depth of whole-transcriptome analysis, we performed microarray analysis (Affymetrix Human Exon 1.0 ST array) using total RNA from five UPF3B patients (three of which were analyzed by RNA-SEQ) and five control individuals (Supplementary Table S3). We found strong concordance in absolute expression among all samples (R>0.95) (Supplementary Figure S3a) and between the two platforms we used (Figure 1d). To reduce false positives, we restricted our analysis to the core microarray probe sets targeting genes that were confidently detected by RNA-SEQ (expression threshold: RPKM >3) (Supplementary Figure S3b and S3c). About 85% of genes were found to have the same trend of either up or downregulation of expression in both platforms (Supplementary Figure S3d). Expression differences were validated by reverse-transcriptase quantitative PCR on 10 of 10 selected genes, including UPF3B (Figure 1e).
Figure 1.
Genome-wide impact of UPF3B-NMD deficiency on the transcriptome. (a) Overall design of the study. Patients and controls tested are described in detail in Supplementary Tables S1, S2 and S3. (b) RNA-SEQ samples inter correlate well at the expression level calculated by RPKM (reads per kilobase exon model per million mapped reads). Heat map illustrates the Pearson correlation coefficient among RNA-SEQ samples. Unsupervised hierarchical clustering separates samples into two distinct groups, those of patients and controls. (c) Genome-wide transcriptional activity at different genomic regions calculated as RPKM. (d) RNA-SEQ and exon array correlate well at gene transcription level, R = 0.78 (Pearson’s correlation). Exon array was analyzed using only the core probe sets. (e) Validation of RNA expression differences of 10 selected genes. Mean expression (±s.d.) of UPF3B, ARHGAP24 isoform 1, SLC23A2, BHLHE22, SYNE2, SERINC2, ASPH, HNF1B, GAB1 and ETV5 in patients carrying PTC mutations in UPF3B (n = 3), as determined by exon array (Black bar), RNA-SEQ (dark gray bar) and reverse-transcriptase quantitative PCR (RT-qPCR) (light gray bar), compared with the controls (n = 2, dotted line). Values were normalized against the ACTB level in the same sample using relative standard-curve method. Only samples that were subjected to both RNA-SEQ and exon-array analysis (patients 1,2,3 and control 1,2) were used in this analysis.
Together, we identified 526 differently expressed genes (DEGs) exhibiting ≥ two-fold change in expression level in UPF3B patients compared with controls, as determined by either RNA-SEQ or exon array, and displaying the same trend of deregulation on both platforms (Supplementary Table S4). These 526 DEGs represent ~5% of all genes significantly expressed in the LCL transcriptome, with 40% downregulated and ~60% upregulated in UPF3B patients. Using the Illumina Human Omni Express Chip (Figure 1a), we found that 8 of these 526 DEGs (<2%) had genomic copy-number variants in the patients (Supplementary Table S5), which could alternatively explain their differential expression. Interestingly, only 15 of the DEGs were the same as those found to exhibit altered expression in HeLa cells depleted of individual NMD factor: UPF1, UPF2, UPF3B, UPF3A or both UPF3B and UPF3A.9,12,13 Thus, it would appear that there is little overlap in the transcripts regulated by NMD in HeLa cells and LCLs. There was no overlap with NMD targets identified in other species (Table 1).14–16 Moreover, we did not detect any expression change in genes carrying naturally occurring SNPs that introduce PTCs. On average, there are 5 such SNPs per person detected using RNA-SEQ in our study at 95% confidence level (Supplementary Tables S6 and S7). This is consistent with our previous findings that the UPF3B-dependent branch of the NMD pathway only acts on a select subset of NMD substrate mRNAs.9
Table 1.
Overlapping DEGs identified in UPF3B patients and previous microarray studies of different NMD-deficient cell lines
| Reference | NMD-deficient models | Reported probesets |
Expressed in LCL |
DEGs (This study) |
Gene symbol |
Gene assignment | Fold change (RNA-SEQ) |
Fold change (exon array) |
|---|---|---|---|---|---|---|---|---|
| Chan et al.10 | UPF3B and UPF3A KD in HeLa cell by shRNA |
44 | 25 | 2 | ARHGAP24 | Rho GTPase-activating protein 24 | 2.4 | 3.0 |
| MCTP2 | Multiple C2 domains, transmembrane 2 |
3.6 | 4.3 | |||||
| Chan et al.9 | UPF3B KD in Hela cell by shRNA |
94 | 42 | 2 | GBP1 | Guanylate-binding protein 1 | 5.4 | 1.6 |
| MAN1A1 | Mannosidase alpha class 1A | 2.3 | 1.7 | |||||
| UPF1 KD in Hela cell by shRNA |
66 | 32 | 2 | MACROD2 | MACRO domain-containing 2 | 4.4 | 1.0 | |
| ETV5 | ETS variant gene 5 | 2.6 | 1.7 | |||||
| Mendell et al.12 | UPFl KD in HeLa cell by siRNA |
197 | 168 | 5 | TSC22D3 | TSC22 domain family, member 3 | 2.6 | 1.4 |
| GABARAPL1 | GABA(A) receptor-associated protein- like 1 |
3.0 | 1.1 | |||||
| KLHL21 | Kelch-like 21 (Drosophila) | 2.0 | 1.1 | |||||
| SAT1 | Spermidine N1-acetytransferase | 2.0 | 1.5 | |||||
| PHGDH | Phosphoglycerate dehydrogenase | 4.5 | 1.8 | |||||
| Wittmann et al.13 | UPF2 KD in HeLa cell by siRNA |
41 | 32 | 5 | TSC22D3 | TSC22 domain family, member 3 | 2.6 | 1.4 |
| MUC13 | Mucin 13, epithelial transmembrane | 2.6 | 3.0 | |||||
| TSPAN12 | Tetraspanin 12 | 2.0 | 2.0 | |||||
| PDE4B | Phosphodiesterase 4B | 2.4 | 1.3 | |||||
| SCL12A7 | Solute carrier family 12 (potassium/ chloride transporters), member 7 |
2.4 | 2.5 | |||||
| Weischenfeldt J et al.14 | Upf2 KO in mouse bone marrow-derived macrophage |
235 | 138 | 0 | ||||
| Upf2 KO in mouse thymocytes |
44 | 22 | 0 | |||||
| Rehwinkel et al.15 | upf1, upf2 and upf3 KD in fly by siRNA |
26 | 16 | 0 | ||||
| He et al.16 | upf1, nmd2 and upf3 KO in yeast |
61 | 15 | 0 |
Abbreviations: DEGs, differently expressed genes; KD, knockdown; KO, knockout; NMD, nonsense-mediated mRNA decay; shRNA, small hairpin RNA; siRNA, small-interfering RNA.
DEGs or orthologs (if found) were used for comparison. All of these studies employed the use of early microarray platforms namely Affymetrix HU133A Plus 2.0 ( × 2), HU133A, U95, U133, Mouse Genome 430 2.0, DrosGenome 1 and YG985, respectively.
UPF3A protein compensates for loss of UPF3B
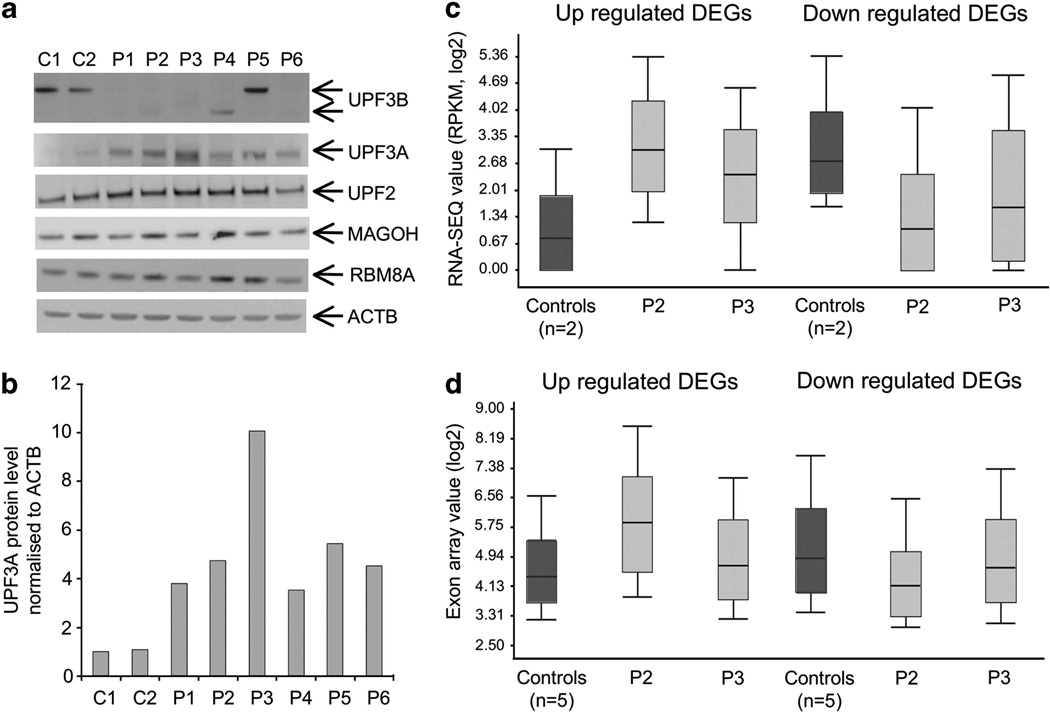
NMD is likely to elicit its effect on the transcriptome through different branches.9 The classical pathway is activated by interaction between UPF2 and either UPF3B or UPF3A, the latter of which is a paralog of UPF3B and is less efficient at activating NMD.10,11 Under normal circumstances, only UPF3B protein is present, whereas UPF3A protein is promptly degraded.10 Previously, we reported that UPF3A protein was dramatically stabilized upon UPF3B depletion.10 In the current study, we show that UPF3A protein is stabilized in all UPF3B patients (Figures 2a and b, Supplementary Figure S4a and S4b), despite unchanged UPF3A mRNA levels.19 This is true even for patient 5 with the missense change, who has apparent full-length UPF3B protein (Figure 2a). This suggests that binding between UPF2 and UPF3B is compromised in this patient’s LCL, allowing UPF3A to interact with UPF2 and become stabilized. Intriguingly, we found that the level of UPF3A protein upregulation varied between patients (Figure 2b) and the magnitude of this change was inversely correlated with the extent of transcriptome deregulation as well as the severity of the patients’ phenotype, when assessed in those who have similar genetic background (that is, patients 2 and 3 in Supplementary Table S1, Figures 2c and d). Although other NMD proteins known to directly interact with UPF3B or UPF3A (that is, UPF2, MAGOH and RBM8A), also showed some fluctuation in expression among patients (Figure 2a), this variation in protein expression did not correlate with gene expression (data not shown). We sequenced UPF3A, as well as genes encoding related or interacting proteins (UPF3B, UPF2, MAGOH and RBM8A) in our patients, and did not observe any sequence variation that could potentially explain variable UPF3A protein stabilization (data not shown). The inverse correlation between UPF3A upregulation and transcriptome deregulation suggests that UPF3A, in part, compensates for UPF3B function in the patients and that the level of UPF3A stabilization in different patients is a major modifier of their phenotype.
Figure 2.
Upregulation of UPF3A protein rescues loss of UPF3B function. (a) Western-blot analysis of UPF3B, UPF3A, UPF2, MAGOH and RBM8A. Protein lysates from two controls and six patients are shown. Full-length UPF3B protein can only be detected in controls and patient 5 (carrying a missense mutation) (top panel). UPF3A is expressed at very low level in controls and upregulated in all patients (second panel). Lower panels show full-length UPF2, MAGOH and RBM8A at comparable levels between controls and patients. ACTB (bottom panel) was used as loading control. These blots are representative of two independent experiments. (b) UPF3A protein is stabilized at different levels in different UPF3B patients. Densitometry was performed directly on image captured with low exposure time and normalized against the level of the ACTB protein in the same sample. Value was averaged from two images, and is representative of two independent experiments. (c) Box plot showing expression profiles of all upregulated differently expressed genes (DEGs) (320 genes) and downregulated DEGs (206 genes) as determined by RNA-SEQ in controls (n = 2, dark-gray bar) and patient 2 and 3 (light-gray bar). These two patients are siblings with the same mutation in UPF3B but have markedly different phenotypes (Supplementary Table S1). Notably, the extent of overall deregulation in each patient appears to be inversely proportional to the level of UPF3A stabilization as shown in panels a and b. (d) Similar trend in expression deregulation, as described in panel c, in patient 2 and 3 (light-gray bar) was also seen by exon array for all upregulated DEGs (279 genes) and downregulated DEGs (184 genes) when compared with the controls (n = 5, dark gray bar).
ARHGAP24, a natural target of NMD, regulates neuronal growth
Next we sought to identify UPF3B-regulated genes that could explain our patients’ neurological phenotypes. By reanalyzing published human tissue atlas microarray data,29 we saw a significant correlation in gene expression between LCLs and different parts of the brain (R>0.64) (Supplementary Figure S5). This correlation encouraged us to use the LCL-expression data from UPF3B patients as a surrogate for their gene expression in the brain. Using the gene ontology classifier program from Partek, we shortlisted 16 genes highly expressed in the brain with functions related to neuronal processes (Table 2). Most of these genes (9 out of 16) contain NMD-inducing features such as an upstream open-reading frame, a long 3′ untranslated region (UTR) (>1.5kb) or a PTC-containing isoform generated by alternative splicing.1,12,16,34 From this analysis, one gene, ARHGAP24, stood out as a strong candidate for further analysis for the following reasons: (i) it was the most consistently upregulated transcript in UPF3B patients (Figure 1e, Table 2 and Supplementary Table S4), (ii) it was one of only two transcripts identified as being regulated by NMD in response to depletion of both UPF3B and UPF3A factors in HeLa cells (Table 1) and (iii) it is a member of the Rho family of GTPase-activating proteins, which are known contributors to ID35,36 through their well-established roles in actin cytoskeleton remodeling.37
Table 2.
DEGs highly expressed in brain with function related to neuronal processes
| Gene ontology category |
Gene symbol |
Fold change |
Gene function | NMD features | |
|---|---|---|---|---|---|
| Exon array | RNA-SEQ | ||||
| Nervous system development |
CRMP1 | −1.2 | −4.8 | Reduction leads to loss of spinal cord neurons |
No |
| TMOD2 | −1.5 | −2.2 | Neuronal-specific regulator of actin- associated proteins |
No | |
| GPR98 | −1.2 | −2.2 | Mutation causes Usher syndrome | No | |
| RAPGEF5 | −1.0 | −2.1 | Involved in telecenphalic neuron development |
No | |
| DLG4 | 1.2 | 2.0 | Key synaptic protein | uORF | |
| EPHB1 | 1.1 | 2.1 | Mediates numerous developmental process |
AS | |
| ROBO1 | 1.0 | 3.2 | Involved in axon guidance | uORF | |
| ITM2B | 1.6 | 3.4 | Mutation causes several forms of dementia and neurodegenerative disorders |
No | |
| Central nervous system development |
ADAM23 | −2.4 | −5.9 | Membrane protein involved in neurogenesis |
Long 3′UTR |
| NRCAM | −1.7 | −2.8 | Cell–cell communication, promotes directional signaling during axonal growth |
Long 3′UTR | |
| NCKAP1 | 1.6 | 2.2 | Possible role in actin skeleton remodeling | AS | |
| Brain Development |
MDGA1 | − 1.4 | −2.2 | Schizophrenia susceptibility gene | No |
| SIX3 | 1.4 | 2.4 | Master regulator of forebrain and visual- cortex development |
uORF | |
| PHGDH | 1.8 | 4.5 | Schizophrenia susceptibility gene | uORF | |
| Not classified/ Others |
ARHGAP24 | 2.8 | 2.4 | Actin skeleton remodeling | uORF, long 3′UTR |
| SNURF-SNRPN | na | 2.2 | SNORNA host gene, in locus of PWS-AS, 15q dup syndrome, autism and schizophrenia. |
No | |
Abbreviations: AS, alternative splicing; NMD, nonsense-mediated mRNA decay; uORF, upstream open-reading frame; UTR, untranslated region.
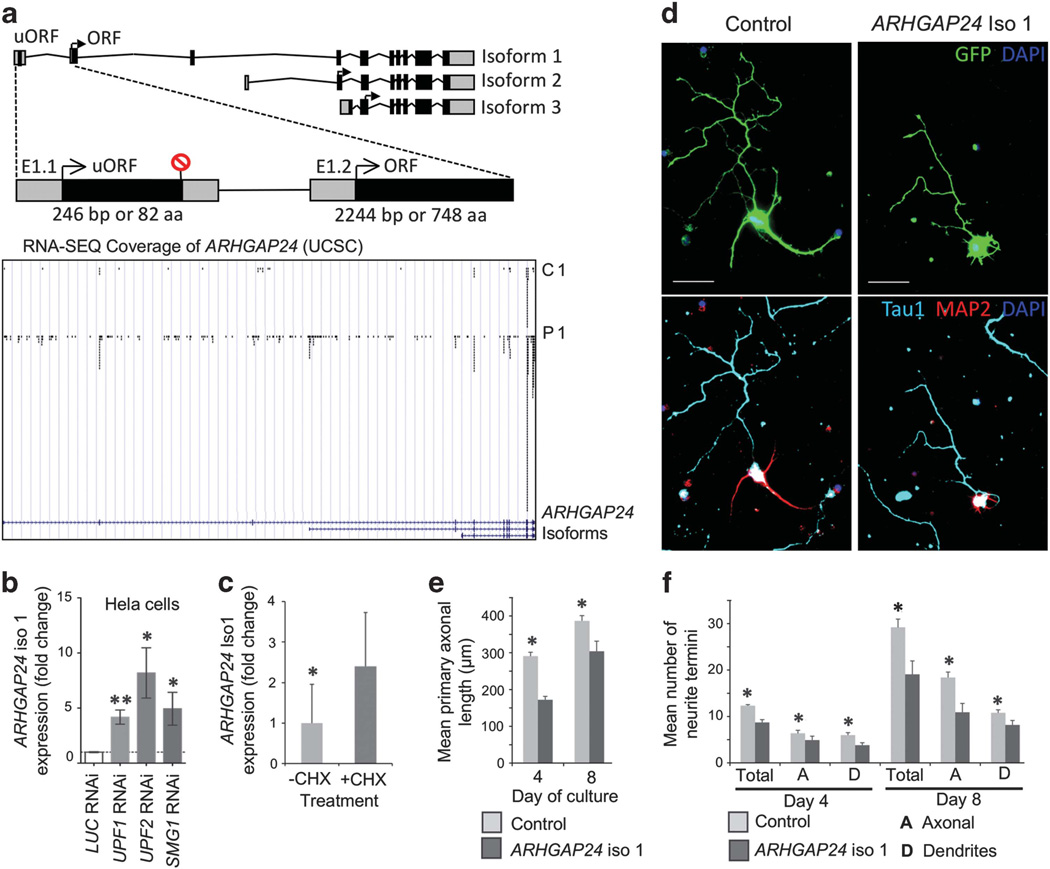
The ARHGAP24 gene gives rise to three mRNA isoforms. Among these, only the largest, isoform 1, was upregulated in UPF3B-NMD-deficient cells (Figure 3a). This isoform is also the isoform most highly expressed in the human and mouse brain (Supplementary Figure S6a and S6b).38 The level of expression in both LCL and brain of isoforms 2 and 3 was too low to be accurately measured by reverse-transcriptase quantitative PCR (data not shown). To test whether ARHGAP24 isoform 1 is a NMD substrate, we examined whether it was upregulated in response to depletion of the NMD factors UPF1, UPF2 and SMG1. We found that such manipulation of crucial NMD factors led to ARHGAP24 isoform 1 being upregulated by at least four-fold in HeLa cells (Figure 3b). We also observed a significant increase in the level of isoform 1 by at least two-fold in LCLs treated with the protein synthesis inhibitor cycloheximide (Figure 3c), which blocks NMD by preventing stop-codon recognition.39 Together, these data suggest that ARHGAP24 isoform 1 is a NMD target, possibly due to the presence of upstream open-reading frame in exon 1 (Figure 3a). Consistent with it being targeted by the UPF3B-dependent branch of NMD, ARHGAP24 isoform 1 displayed an inverse temporal-expression profile to that of UPF3B during human embryonic brain development (Supplementary Figure S6c).
Figure 3.
ARHGAP24 isoform 1 is an important regulator of neuronal morphology. (a) Schematic diagrams of ARHGAP24 isoforms (upper panels). Magnified area indicates the position of uORF (upstream open-reading frame) in isoform 1. Lower panel displays aligned RNA-SEQ reads (from Control 1 and Patient 1 only, shown as representative) to ARHGAP24 (snap shot from UCSC Genome Browser). (b) ARHGAP24 isoform 1 is a target of NMD. ARHGAP24 isoform 1 was significantly upregulated in HeLa cells depleted of UPF1, UPF2 and SMG1, relative to HeLa cells depleted of Luciferase (negative control). Expression value was measured by reverse-transcriptase quantitative PCR (RT-qPCR) using relative standard-curve method, normalized against ACTB in the same sample, and converted to fold change. *P < 0.05, **P < 0.01 by Student’s two-tailed t-test. (c) ARHGAP24 isoform 1 was significantly upregulated in normal control LCLs (n = 6) after treatment with Cycloheximide (CHX) for 6h. Expression value was measured by RT-qPCR using relative standard-curve method, normalized to ACTB level in the same sample, and converted into fold-change. Result is shown as representative of two independent experiments. *P < 0.05 by Student’s one-tailed t-test. (d) Representative images of day 4 cultures of primary hippocampal neurons exogenously expressing green fluorescent protein (GFP) only (Control) or together with ARHGAP24 isoform1. Upper panels show GFP expression within transfected cells. Bottom panels show same cells stained with MAP2 (red) and Tau1 (cyan) to reveal dendritic and axonal domains, respectively. (e) Ectopic expression of ARHGAP24 isoform 1 inhibits axonal outgrowth. Mean primary axonal length compared at day 4 and 8 of culture. (f) Ectopical expression of ARHGAP24 isoform 1 inhibits neuronal arborization. Total, axonal and dendritic neurite termini compared at day 4 and 8 of culture. Experiments in panels e and f were done in triplicate with at least 25 neurons scored per repeat. *P < 0.05 by Student’s paired two-tailed t-test.
Whereas several members of the Rho family of GTPase-activating proteins have been shown to have neuronal functions, including regulating dendrite morphology and synapse plasticity,35 the neuronal role of ARHGAP24 has not been investigated. To address this issue, we ectopically expressed ARHGAP24 isoform 1 in cultured primary mouse hippocampal neurons and found that it reduced axonal outgrowth, and hindered neuronal arborization as reflected by a reduction in the number of dendritic and axonal termini (Figures 3d–f). It also elicited neuronal cell death when transfected at higher concentrations (Supplementary Figure S7). Similar outcomes were observed upon ectopic expression of ARHGAP24 isoform 1 in PC12 cells induced to differentiate; we found that isoform 1 inhibited the outgrowth of neurites upon nerve growth factor stimulation and triggered formation of a rounded morphology in 50–60% of the cells (Supplementary Figure S8). Together, these data suggest that ARHGAP24 isoform 1 has a role in modulating neurite outgrowth and that its deregulation in patients with UPF3B mutations is a contributor to their neurological phenotype.
Discussion
Increasing evidence suggests that the NMD pathway is involved in the nervous system development and function. Studies with knockout mice have shown that the NMD-promoting exon-junction complex factor, Magoh, controls mouse cerebral cortical size by regulating neural stem-cell division.40 Another NMD-promoting exon-junction complex factor, eIF4AIII, is concentrated in neuronal mRNA granules and dendrites, and is necessary for normal synaptic strength in rat cortical neurons.41 In Drosophila melanogaster, smg1 mutants exhibit significant impairment of the neuromuscular junction synapse structure, and mutation of smg1, upf2 or smg6 greatly reduce neurotransmission and synaptic-vesicle cycling.42 More recently, we demonstrated that the miR-128 regulates NMD by acting directly on Upf1 and Mnl51, ensuring proper differentiation of the neural stem cells in mouse and that such action elevated the expression level of many NMD target genes with known neuronal functions, such as Syne1, Robo2, Nrcam and so on.43 In man, mutations in UPF3B cause ID, along with a spectrum of other neurological defects, which includes autism, attention-deficit hyperactivity disorder and schizophrenia.18–20
Here, we show that the loss of the NMD factor UPF3B impacted upon 5% of the transcriptome (Supplementary Table S4). This is in general agreement with previous studies albeit small overlap among the deregulated genes. To evaluate potential involvement of NMD in brain function, we shortlisted 16 DEGs in UPF3B patients with known neuronal functions (Table 2). We performed functional studies on one of these UPF3B-regulated transcripts, ARH-GAP24, which encodes a member of the Rho-GTPase-activating proteins that act on Rac, a Rho-GTP protein, to regulate actin remodeling37 and cell polarity.44 We showed that ARHGAP24 isoform 1 is a NMD target, which is upregulated in UPF3B patients (Figures 1e and 3b, c), and provided evidence that it is important for normal axon and dendrite growth (Figures 3e–f, Supplementary Figures S7 and S8). In man, tight regulation of the actin–cytoskeleton remodeling pathway appears to be critical for normal cognitive function. Mutations in FLNA, encoding an ubiquitously expressed actin cross-linked protein that interacts with ARHGAP24, cause periventricular nodular heterotopia, an X-linked neuronal migration disorder, which mainly affects females and is embryonically lethal in males.36,45 Mutations in other proteins that act on Rac (including FGD1, ARHGEF6, PAK3 or OPHN1) cause non-syndromic X-linked ID.35 Moreover, disruption of the formation of dendrites and synapses in the brain often leads to the clinical presentations of ID and autism, as seen in patients with mutations in SHANK3, a scaffolding protein connecting glutamate receptors to the actin–cytoskeleton,46 and CYLN2, a protein associated with the end of growing microtubules.47 Together, these data emphasize the importance of proper regulation of the actin–cytoskeleton during brain development.
In addition to ARHGAP24, there are other deregulated genes that might contribute to the neurological problems in the UPF3B patients. SIX3 is believed to be the master regulator of forebrain and visual-cortex development.48,49 Whereas mutations in SIX3 in man cause holoprosencephaly,50 a severe brain malformation phenotype, upregulation of SIX3 in the mouse developing telecephalon accelerates the cells’ clonal division and inhibits their maturation process.51 As such, upregulation of SIX3 in UPF3B patients (Table 2) could result in the enlargement of the forebrain (as seen in patient 1 and 2 in Supplementary Table S1) and visual impairment (as seen in patient 6 in Supplementary Table S1 and affected patients from the same family).18 SNURF-SNRPN, a host gene for SNORD116 and SNORD115 clusters, with known roles in pre-RNA editing and alternative splicing,52,53 is also upregulated in UPF3B patients (Table 2). Deletion of SNORD116 cluster causes Prader–Willi Syndrome (MIM no. 176270),54 and depletion of Snord115 expression in the mouse in vivo leads to increased pre-RNA editing of the 5-hydroxytryptamine (serotonin) receptor 2C gene, 5htr2c, and altered 5htr2c-mediated behaviors.52 SNURF-SNRPN lies within the frequently duplicated region 15q11-q13, which is also associated with Angelman syndrome (MIM no. 105830) and 15q duplication syndromes (MIM no. 608636).55 Whereas SNURF-SNRPN expression is also upregulated in these patients similarly to UPF3B patients,55,56 little is known about its downstream consequences. It is possible that the upregulation of the SNURF-SNRPN gene contributes to the neurological features shared among patients with these disorders and those of the UPF3B patients, which include ID, seizures and autistic behaviors.55 Nevertheless, the precise role of any of these transcripts or their relevant proteins in the phenotype of UPF3B patients remains to be determined.
The brain is the most enigmatic organ and its function requires a fine-tuned homeostatic balance.57 Such balance is maintained by several integral mechanisms that, if perturbed, cause the same defects as seen in UPF3B patients, including autism, schizophrenia, attention-deficit hyperactivity disorder and ID.57 Our findings, together with the findings of others, support a major role for NMD in normal brain development and function.40,41,43
The DEGs identified by our study differ significantly when compared with those identified using HeLa cell lines depleted of other NMD factors, including UPF3B9,10,12,13 (Table 1). These differences can be attributed to (i) different cell lines, NMD inhibition techniques, microarray platforms and analysis methods employed. This type of cross-platform and cross-laboratory errors is quite common in microarray-based studies.58; (ii) UPF1 and UPF2 also functioning in cellular pathways other than NMD,1. As such depletion of these factors likely results in non-NMD-related effects on the transcriptome; and (iii) the fact that NMD acts through multiple different branches with a broader range and different targets.8,9 In Sacchromyces cerevisiae, NMD function in a linear model and as such, loss of NMD factors upf1, upf2 or upf3 yields almost identical gene-expression changes.16,59 In higher eukaryotes, the NMD pathways are more structured with multiple alternative branches and not all dependent on UPF2 or UPF3B.8,9 This would also suggest that there might be NMD branch-specific features on specific genes; however, such features are yet to be determined. Overall, our results support major role of NMD, and UPF3B-NMD branch in particular, in regulating the transcriptome.
The UPF3B paralogous protein, UPF3A, which is normally degraded in the presence of the UPF3B protein, is stabilized in patients with different UPF3B loss-of-function mutations (Figure 2).10 The function of UPF3A and UPF3B is likely redundant also based on the repertoire of their downstream targets. The stabilization of the UPF3A protein in the absence of UPF3B offers compensatory NMD function in UPF3B patients. Compensation through UPF3A–NMD also most likely explains why the loss of UPF3B function does not lead to embryonic lethality,18–20 unlike the effect of a complete loss of Upf1 or Upf2 factors in mouse.14,17 Interestingly, we also found that the magnitude of the UPF3A protein stabilization is inversely correlated with the patients’ phenotype (Figure 2), demonstrating that varying level of stabilized UPF3A offer varying level of NMD compensation. This ‘variable’ compensation may also provide a plausible explanation for the variable phenotype associated with the loss of UPF3B function. Patients 2 and 3 (brothers), who carry the same mutation, but clinically present with markedly different phenotype (Supplementary Table S1), offer a good example. Variable NMD efficiency has previously been reported to modulate disease outcomes60 including neuropsychiatric disease.61 It is possible that a fluctuation in expression levels of different NMD factors, similar to that of UPF3A as in our patients, provide the explanation. Indeed, published data point towards the existence of expression quantitative trait loci, at least in the NMD genes UPF2 (marker rs3781646) and SMG7 (markers rs1044879, rs10911353, rs2702180 and rs6662844).62
In summary, our findings further emphasize the essential role of NMD pathways for normal brain function. More studies are needed to understand the mechanism(s) that regulate NMD efficiency and also its variability at a level of an individual. This will potentially impact significantly upon the therapeutic strategies for many genetic diseases where NMD is involved.58,59
Supplementary Material
Acknowledgments
We thank the members of the families studied for participation and Brigitte Jauffrion and Lucianne Vandeleur for help with cell culture. This work was supported by grants from the Australian NH&MRC Project Grant 453457 to JoG, and the Women’s and Children’s Hospital Foundation Grant to LJ and JoG. JoG is supported by NH&MRC Principal Research Fellowship. LSN’s PhD scholarship was supported by the MS McLeod Foundation.
Footnotes
Accession numbers
Raw and processed data have been deposited to the Gene Expression Omnibus repository under accession number GSE27433. For individual dataset, please refer to: GSE27199 (RNA-SEQ), GSE27125 (Microarray profiling) and GSE27414 (copy-number variant).
Conflict of interest
The authors declare no conflict of interest.
Author contributions: LSN performed most of the data analysis; LJ performed the overexpression of ARHGAP24 in mouse primary hippocampal neurons; CS performed the overexpression of ARHGAP24 in mouse PC12 cell line; WC, LH and MFW contributed the microarray and reverse-transcriptase quantitative PCR data of NMD knockdown in HeLa cells; LH performed the RNAi knockdown of UPF1, UPF2 and SMG1 in HeLa cells; SS performed the in situ hybridization of ARHGAP24 in human brain sample; CH and JeG generated the antibody against ARHGAP24 isoform 1 and the ARHGAP24 isoform 1 clone; FL, MR, AH, MF, JR, AKS, LY, AAM and JLR contributed with patient material or LCL lines for this work; MC provided advice on the data analysis; This project was designed and supervised by JoG; The paper was written by LSN, LJ and JoG, with advice from MFW.
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
References
- 1.Nicholson P, Yepiskoposyan H, Metze S, Zamudio Orozco R, Kleinschmidt N, Muhlemann O. Nonsense-mediated mRNA decay in human cells: mechanistic insights, functions beyond quality control and the double-life of NMD factors. Cell Mol Life Sci. 2010;67:677–700. doi: 10.1007/s00018-009-0177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE. Y14 and hUpf3b form an NMD-activating complex. Mol Cell. 2003;11:939–949. doi: 10.1016/s1097-2765(03)00142-4. [DOI] [PubMed] [Google Scholar]
- 3.Kim VN, Kataoka N, Dreyfuss G. Role of the nonsense-mediated decay factor hUpf3 in the splicing-dependent exon-exon junction complex. Science. 2001;293:1832–1836. doi: 10.1126/science.1062829. [DOI] [PubMed] [Google Scholar]
- 4.Chamieh H, Ballut L, Bonneau F, Le Hir H. NMD factors UPF2 and UPF3 bridge UPF1 to the exon junction complex and stimulate its RNA helicase activity. Nat Struct Mol Biol. 2008;15:85–93. doi: 10.1038/nsmb1330. [DOI] [PubMed] [Google Scholar]
- 5.Kashima I, Yamashita A, Izumi N, Kataoka N, Morishita R, Hoshino S, et al. Binding of a novel SMG-1-Upf1-eRF1-eRF3 complex (SURF) to the exon junction complex triggers Upf1 phosphorylation and nonsense-mediated mRNA decay. Genes Dev. 2006;20:355–367. doi: 10.1101/gad.1389006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivanov PV, Gehring NH, Kunz JB, Hentze MW, Kulozik AE. Interactions between UPF1, eRFs, PABP and the exon junction complex suggest an integrated model for mammalian NMD pathways. EMBO J. 2008;27:736–747. doi: 10.1038/emboj.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gehring NH, Kunz JB, Neu-Yilik G, Breit S, Viegas MH, Hentze MW, et al. Exon-junction complex components specify distinct routes of nonsense-mediated mRNA decay with differential cofactor requirements. Mol Cell. 2005;20:65–75. doi: 10.1016/j.molcel.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Chan WK, Huang L, Gudikote JP, Chang YF, Imam JS, MacLean JA, II, et al. An alternative branch of the nonsense-mediated decay pathway. EMBO J. 2007;26:1820–1830. doi: 10.1038/sj.emboj.7601628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan WK, Bhalla AD, Le Hir H, Nguyen LS, Huang L, Gecz J, et al. A UPF3-mediated regulatory switch that maintains RNA surveillance. Nat Struct Mol Biol. 2009;16:747–753. doi: 10.1038/nsmb.1612. [DOI] [PubMed] [Google Scholar]
- 11.Kunz JB, Neu-Yilik G, Hentze MW, Kulozik AE, Gehring NH. Functions of hUpf3a and hUpf3b in nonsense-mediated mRNA decay and translation. RNA. 2006;12:1015–1022. doi: 10.1261/rna.12506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendell JT, Sharifi NA, Meyers JL, Martinez-Murillo F, Dietz HC. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1073–1078. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 13.Wittmann J, Hol EM, Jack HM. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol Cell Biol. 2006;26:1272–1287. doi: 10.1128/MCB.26.4.1272-1287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weischenfeldt J, Damgaard I, Bryder D, Theilgaard-Monch K, Thoren LA, Nielsen FC, et al. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008;22:1381–1396. doi: 10.1101/gad.468808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA. 2005;11:1530–1544. doi: 10.1261/rna.2160905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He F, Li X, Spatrick P, Casillo R, Dong S, Jacobson A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol Cell. 2003;12:1439–1452. doi: 10.1016/s1097-2765(03)00446-5. [DOI] [PubMed] [Google Scholar]
- 17.Medghalchi SM, Frischmeyer PA, Mendell JT, Kelly AG, Lawler AM, Dietz HC. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum Mol Genet. 2001;10:99–105. doi: 10.1093/hmg/10.2.99. [DOI] [PubMed] [Google Scholar]
- 18.Laumonnier F, Shoubridge C, Antar C, Nguyen LS, Van Esch H, Kleefstra T, et al. Mutations of the UPF3B gene, which encodes a protein widely expressed in neurons, are associated with nonspecific mental retardation with or without autism. Mol Psychiatry. 2010;15:767–776. doi: 10.1038/mp.2009.14. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen LS, Tarpey PS, Raymond FL, Rodriguez J, Hackett A, Vandeleur L, et al. Mutations in UPF3B, a member of the nonsense-mediated mRNA decay complex, cause syndromic and nonsyndromic mental retardation. Nat Genet. 2007;39:1127–1133. doi: 10.1038/ng2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Addington AM, Gauthier J, Piton A, Hamdan FF, Raymond A, Gogtay N, et al. A novel frameshift mutation in UPF3B identified in brothers affected with childhood onset schizophrenia and autism spectrum disorders. Mol Psychiatry. 2011;16:238–239. doi: 10.1038/mp.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 22.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 23.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 24.Diskin SJ, Li M, Hou C, Yang S, Glessner J, Hakonarson H, et al. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36:e126. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 26.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 28.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaech S, Banker G. Culturing hippocampal neurons. Nat Protoc. 2006;1:2406–2415. doi: 10.1038/nprot.2006.356. [DOI] [PubMed] [Google Scholar]
- 31.Usuki F, Yamashita A, Kashima I, Higuchi I, Osame M, Ohno S. Specific inhibition of nonsense-mediated mRNA decay components, SMG-1 or Upf1, rescues the phenotype of Ullrich disease fibroblasts. Mol Ther. 2006;14:351–360. doi: 10.1016/j.ymthe.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Miyagishi M, Taira K. U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- 33.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rehwinkel J, Raes J, Izaurralde E. Nonsense-mediated mRNA decay: Target genes and functional diversification of effectors. Trends Biochem Sci. 2006;31:639–646. doi: 10.1016/j.tibs.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Ramakers GJ. Rho proteins, mental retardation and the cellular basis of cognition. Trends Neurosci. 2002;25:191–199. doi: 10.1016/s0166-2236(00)02118-4. [DOI] [PubMed] [Google Scholar]
- 36.Fox JW, Lamperti ED, Eksioglu YZ, Hong SE, Feng Y, Graham DA, et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron. 1998;21:1315–1325. doi: 10.1016/s0896-6273(00)80651-0. [DOI] [PubMed] [Google Scholar]
- 37.Ohta Y, Hartwig JH, Stossel TP. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat Cell Biol. 2006;8:803–814. doi: 10.1038/ncb1437. [DOI] [PubMed] [Google Scholar]
- 38.Lavelin I, Geiger B. Characterization of a novel GTPase-activating protein associated with focal adhesions and the actin cytoskeleton. J Biol Chem. 2005;280:7178–7185. doi: 10.1074/jbc.M411990200. [DOI] [PubMed] [Google Scholar]
- 39.Carter MS, Doskow J, Morris P, Li S, Nhim RP, Sandstedt S, et al. A regulatory mechanism that detects premature nonsense codons in T-cell receptor transcripts in vivo is reversed by protein synthesis inhibitors in vitro. J Biol Chem. 1995;270:28995–29003. doi: 10.1074/jbc.270.48.28995. [DOI] [PubMed] [Google Scholar]
- 40.Silver DL, Watkins-Chow DE, Schreck KC, Pierfelice TJ, Larson DM, Burnetti AJ, et al. The exon junction complex component Magoh controls brain size by regulating neural stem cell division. Nat Neurosci. 2010;13:551–558. doi: 10.1038/nn.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, et al. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 42.Long AA, Mahapatra CT, Woodruff EA, III, Rohrbough J, Leung HT, Shino S, et al. The nonsense-mediated decay pathway maintains synapse architecture and synaptic vesicle cycle efficacy. J Cell Sci. 2010;123(Part 19):3303–3315. doi: 10.1242/jcs.069468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bruno IG, Karam R, Huang L, Bhardwaj A, Lou CH, Shum EY, et al. Identification of a MicroRNA that Activates Gene Expression by Repressing Nonsense-Mediated RNA Decay. Mol Cell. 2011;42:500–510. doi: 10.1016/j.molcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura F, Heikkinen O, Pentikainen OT, Osborn TM, Kasza KE, Weitz DA, et al. Molecular basis of filamin A-FilGAP interaction and its impairment in congenital disorders associated with filamin A mutations. PLoS One. 2009;4:e4928. doi: 10.1371/journal.pone.0004928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, et al. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry. 2011 doi: 10.1038/mp.2011.57. (e-pub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoogenraad CC, Koekkoek B, Akhmanova A, Krugers H, Dortland B, Miedema M, et al. Targeted mutation of Cyln2 in the Williams syndrome critical region links CLIP-115 haploinsufficiency to neurodevelopmental abnormalities in mice. Nat Genet. 2002;32:116–127. doi: 10.1038/ng954. [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi M, Toyama R, Takeda H, Dawid IB, Kawakami K. Overexpression of the forebrain-specific homeobox gene six3 induces rostral forebrain enlargement in zebrafish. Development. 1998;125:2973–2982. doi: 10.1242/dev.125.15.2973. [DOI] [PubMed] [Google Scholar]
- 49.Lagutin OV, Zhu CC, Kobayashi D, Topczewski J, Shimamura K, Puelles L, et al. Six3 repression of Wnt signaling in the anterior neuroectoderm is essential for vertebrate forebrain development. Genes Dev. 2003;17:368–379. doi: 10.1101/gad.1059403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallis DE, Roessler E, Hehr U, Nanni L, Wiltshire T, Richieri-Costa A, et al. Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat Genet. 1999;22:196–198. doi: 10.1038/9718. [DOI] [PubMed] [Google Scholar]
- 51.Appolloni I, Calzolari F, Corte G, Perris R, Malatesta P. Six3 controls the neural progenitor status in the murine CNS. Cereb Cortex. 2008;18:553–562. doi: 10.1093/cercor/bhm092. [DOI] [PubMed] [Google Scholar]
- 52.Doe CM, Relkovic D, Garfield AS, Dalley JW, Theobald DE, Humby T, et al. Loss of the imprinted snoRNA mbii-52 leads to increased 5htr2c pre-RNA editing and altered 5HT2CR-mediated behaviour. Hum Mol Genet. 2009;18:2140–2148. doi: 10.1093/hmg/ddp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kishore S, Stamm S. Regulation of alternative splicing by snoRNAs. Cold Spring Harb Symp Quant Biol. 2006;71:329–334. doi: 10.1101/sqb.2006.71.024. [DOI] [PubMed] [Google Scholar]
- 54.Sahoo T, del Gaudio D, German JR, Shinawi M, Peters SU, Person RE, et al. Prader-Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chamberlain SJ, Lalande M. Neurodevelopmental disorders involving genomic imprinting at human chromosome 15q11-q13. Neurobiol Dis. 2010;39:13–20. doi: 10.1016/j.nbd.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 56.Runte M, Kroisel PM, Gillessen-Kaesbach G, Varon R, Horn D, Cohen MY, et al. SNURF-SNRPN and UBE3A transcript levels in patients with Angelman syndrome. Hum Genet. 2004;114:553–561. doi: 10.1007/s00439-004-1104-z. [DOI] [PubMed] [Google Scholar]
- 57.Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455:912–918. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi L, Jones WD, Jensen RV, Harris SC, Perkins RG, Goodsaid FM, et al. The balance of reproducibility, sensitivity, and specificity of lists of differentially expressed genes in microarray studies. BMC Bioinformatics. 2008;9(Suppl 9):S10. doi: 10.1186/1471-2105-9-S9-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lelivelt MJ, Culbertson MR. Yeast Upf proteins required for RNA surveillance affect global expression of the yeast transcriptome. Mol Cell Biol. 1999;19:6710–6719. doi: 10.1128/mcb.19.10.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Khajavi M, Inoue K, Lupski JR. Nonsense-mediated mRNA decay modulates clinical outcome of genetic disease. Eur J Hum Genet. 2006;14:1074–1081. doi: 10.1038/sj.ejhg.5201649. [DOI] [PubMed] [Google Scholar]
- 61.Bevilacqua L, Doly S, Kaprio J, Yuan Q, Tikkanen R, Paunio T, et al. A population-specific HTR2B stop codon predisposes to severe impulsivity. Nature. 2010;468:1061–1066. doi: 10.1038/nature09629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.