Abstract
Capped and polyadenylated long noncoding RNAs (lncRNAs) are shown to be degraded by a DCP2-mediated turnover mechanism by Geisler et al. (2012); this provides a new level of regulatory control for inducible genes by lncRNAs.
RNAs lead a perilous existence. From their first emergence at the site of transcription to their translation in the cytoplasm, RNAs encounter a whole host of enzymes that can degrade them. It is often underappreciated that the decay rate of an RNA has the same potential to dictate its steady-state level as its rate of synthesis (i.e., transcription). By regulating the rate of mRNA decay through, for example, small RNAs or RNA-binding proteins, the level of RNA can be dramatically altered. This will elicit a corresponding change in protein output. In yeast, the major pathway that degrades protein-coding RNAs (or mRNAs) consists of three steps: shortening of the polyA tail at their 3′ end by enzymes called deadenylases; removal of their 5′ cap by a holoenzyme composed of a catalytic subunit, DCP2, and its activator, DCP1; and decay of their “body” by the exonucleases RAT1 and XRN1, which degrade mRNAs in a 5′ → 3′ manner in the nucleus and cytoplasm, respectively (Garneau et al., 2007). The rate-limiting step for the decay of most mRNAs in eukaryotes is the first step—deadenylation—which several lines of evidence suggest leads to linearization of the mRNA circle, thereby exposing its 5′ end for decapping. While much is known about how this canonical pathway degrades mRNAs, we know little about its role in degrading capped and polyadenylated RNAs that do not encode protein. In this issue of Molecular Cell, Geisler et al. (2012) provide evidence that a significant subset of such noncoding RNAs are degraded by an alternative version of this pathway. They go on to demonstrate that this molecular circuit is vital for the rapid and robust transcriptional activation of inducible genes.
There has been an explosion of interest in noncoding RNAs in the past 10 years. Modern sensitive techniques have revealed that noncoding RNAs are incredibly pervasive; they are transcribed from a far greater proportion of many eukaryotic genomes than the genes that encode proteins (Wang and Chang, 2011). These discoveries have propelled a huge effort to determine the functional role of noncoding RNAs, which has led to the discovery of several classes of short noncoding RNAs—microRNAs, short interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs)—most of which were found to act by repressing the expression of specific target mRNAs or genes. Largely left behind in the initial flurry of research activity have been the long noncoding RNAs (lncRNAs), which are defined as being greater than 200 nt long. In fact, there has been much debate about whether these lncRNAs—which number in the hundreds in many eukaryotic genomes—are merely transcriptional noise with no function. While this may be the case for some lncRNAs, recent studies have shown that some specific lncRNAs influence key biological functions, including pluripotency, cell cycle, and innate immunity (Wang and Chang, 2011). LncRNAs have also been shown to modulate a wide range of molecular processes, including both nuclear events (retrotransposon silencing, gene dosage compensation, gene imprinting, and telomere length) and cytoplasmic events (regulation of mRNA decay and microRNA function) (Figure 1).
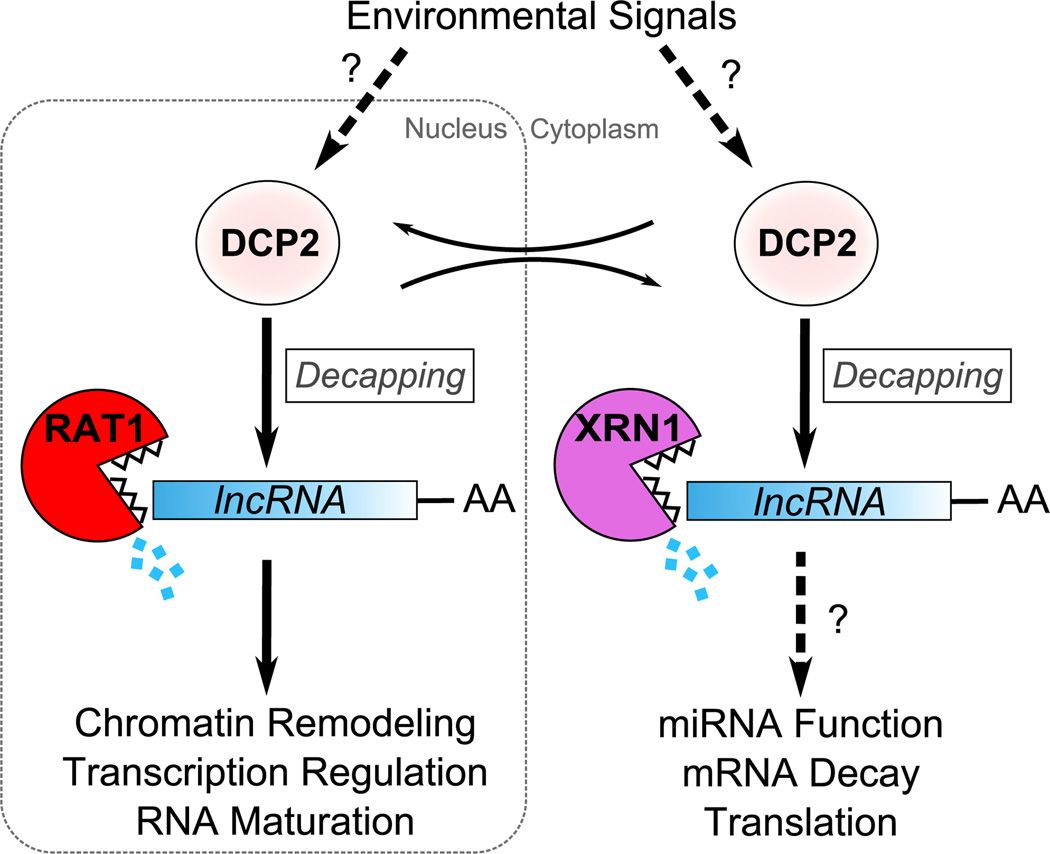
Figure 1. DCP2-Dependent LncRNA Turnover.
The decapping enzyme DCP2 exposes the body of lncRNAs to rapid decay by the nuclear and cytoplasmic 5′ → 3′ exonucleases, RAT1 and XRN1, respectively. The rapid turnover of these lncRNAs, coupled with their transcriptional downregulation in response to specific environmental cues (not shown), elicits rapid and robust transcriptional depression of their target genes and other events (Wang and Chang, 2011). Such inducible responses would be further amplified if environmental stimuli also increased DCP2 activity on lncRNAs, but there is currently no evidence for this.
Given that lncRNAs are functional, what regulates their level in different contexts? Geisler et al. set out to answer this question. Since most known lncRNAs are capped, these authors hypothesized that their decay would depend on the decapping enzyme DCP2. Indeed, using RNA-seq, they identified several lncRNAs, including ~100 previously unidentified ones, which are upregulated in DCP2-deficient yeast. Many of these lncRNAs mapped to intergenic and telomeric regions. Intriguingly, several of these DCP2-sensitive lncRNAs mapped in an antisense orientation proximal to inducible genes, many of which are tightly regulated by environmental cues and encode proteins with inducible functions such as iron sensing, inorganic phosphate uptake and metabolism, and sugar metabolism. Among these inducible genes were the GAL genes, which dictate galactose utilization. The authors chose to focus their further efforts on this set of genes for two reasons: (1) galactose utilization in yeast is a classical paradigm for understanding gene regulation, and (2) previous studies identified a functional role for a lncRNA associated with one of the GAL genes: GAL10 (Houseley et al., 2008; Pinskaya et al., 2009). Geisler et al. identified lncRNAs overlapping not only with GAL10, but also the GAL1 and GAL2 structural genes, as well as the GAL4 master regulatory gene. They then found that these GAL lncRNAs are expressed in a manner that is inversely correlated with their cognate protein-coding genes, suggesting the possibility that these lncRNAs repressed their associated GAL genes. To directly test this, they mutated the promoters of the GAL4 and GAL10 lncRNAs and observed increased steady-state levels of their respective cognate mRNAs, suggesting a direct causal role for these lncRNAs in transcriptional repression. To examine whether lncRNA stabilized by loss of DCP2 engenders enhanced transcriptional repression of its cognate target, they took advantage of previously published chromatin immunoprecipitation experiments that showed that transcriptional repression of the GAL10 lncRNA target, GAL1, is associated with repressive chromatin marks at the GAL1 promoter (Houseley et al., 2008; Pinskaya et al., 2009). Geisler et al. found that the GAL1 promoter is hypoacetylated in DCP2-deficient cells, which is consistent with the fact that these cells have higher levels of GAL10 lncRNA. The addition of galactose led to increased histone acetylation at this locus in a temporal manner that correlated with increased GAL1 mRNA and decreased GAL10 lncRNA levels. Further evidence for the role of histone acetylation in this response was that the addition of trichostatin A, a histone deacetylase inhibitor, rescued the induction of GAL1 mRNA in DCP2-deficient cells. Together, these data, along with previously published results (Houseley et al., 2008), provide support for the idea that destabilization of GAL10 lncRNA in a DCP2-dependent manner, coupled with its transcriptional downregulation, is crucial for the rapid and robust transcriptional induction of its cognate protein-coding gene, GAL1, in response to galactose.
A surprising discovery made by Geisler et al. is that the turnover of GAL lncRNAs depends on a noncanonical RNA decay pathway. Using yeast with mutations in different RNA decay pathway components, they showed that GAL lncRNAs are degraded by a pathway independent of the decapping activators DHH1 and LSM1. This was unanticipated, as the decay of mRNAs and even another lncRNA—SRG1—was previously shown to exhibit accelerated decay in response to decapping activators (Thompson and Parker, 2007; Meyer et al., 2004). Also, the decay of GAL lncRNAs was not significantly affected by loss of CCR4, a key component of the CCR4-NOT deadenylase complex. This suggests the possibility that GAL lncRNAs are degraded in a deadenylation-independent manner, perhaps by a mechanism in which decapping, not deadenylation, is the rate-limiting step. However, further experiments will be required to pin this down, including analysis of the effect of loss of other yeast deadenylase components. A perplexing observation by the authors was that GAL10 lncRNA was destabilized in response to loss of TRAMP and exosome components that mediate 3′ → 5′ exonuclease decay. This is presumably due to an indirect effect, and curiously, it is the opposite result to that obtained in a previous investigation (Houseley et al., 2008). Finally, Geisler et al. observed that lncRNAs differ with regard to the specific exonucleases that ultimately finish them off (Figure 1). In agreement with previous work (van Dijk et al., 2011), the cytoplasmic 5′ → 3′ exonuclease, XRN1, degrades many of the lncRNAs that Geisler et al. identified by RNA-seq. However, the authors found that many lncRNAs are also degraded by the nuclear 5′ → 3′ exonuclease, RAT1. The importance of RAT1 was reinforced by the finding that GAL10 lncRNA was more functionally compromised by loss of RAT1 than XRN1. Together, these observations suggest the existence of two separate, yet partially redundant, non-canonical decay pathways that function in different sub-cellular compartments to degrade lncRNAs.
Where do we go from here? One question is whether DCP2 itself is regulated in response to environmental cues. If, for example, DCP2 were more heavily recruited to GAL lncRNAs in response to galactose, this might enhance the decay of these lncRNAs and therefore elicit a more pronounced transcriptional derepression of GAL genes (Figure 1). Posttranslational modifications of DCP2 could also be regulated by environmental cues; in this regard, it has been shown that glucose deprivation triggers DCP2 hyperphosphorylation (Yoon et al., 2010). A surprising finding by Geisler et al. is that GAL lncRNAs are degraded by a unique RNA decay pathway independent of several factors crucial for the decay of most mRNAs. What underlies this? Could it be because they are not translated? Do the principles uncovered by Geisler et al. for the DCP2-mediated lncRNA turnover pathway apply only to the GAL inducible genes, or can they be extrapolated to other lncRNAs? Finally, does lncRNA stability influence the fate of other RNAs? Some microRNAs are processed from a subset of lncRNAs (Eis et al., 2005), raising the possibility that regulation of lncRNA turnover can dictate the level of some microRNAs. Regardless of the answers to these questions, the study herein adds significantly to the growing evidence that transcription and posttranscriptional events cooperate to achieve biological goals.
REFERENCES
- Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Proc. Natl. Acad. Sci. USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ. Nat. Rev. Mol. Cell Biol. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- Geisler S, Lojek L, Khalil AM, Baker KE, Coller J. Mol. Cell. 2012;45(this issue):279–291. doi: 10.1016/j.molcel.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. Mol. Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Temme C, Wahle E. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- Pinskaya M, Gourvennec S, Morillon A. EMBO J. 2009;28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson DM, Parker R. Mol. Cell. Biol. 2007;27:92–101. doi: 10.1128/MCB.01023-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk EL, Chen CL, d’Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Né P, Loeillet S, et al. Nature. 2011;475:114–117. doi: 10.1038/nature10118. [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Choi EJ, Parker R. J. Cell Biol. 2010;189:813–827. doi: 10.1083/jcb.200912019. [DOI] [PMC free article] [PubMed] [Google Scholar]